Abstract
Mutations of the Drosophila melanogaster insulin/IGF signaling system slow aging, while also affecting growth and reproduction. To understand this pleiotropy, we produced an allelic series of single codon substitutions in the Drosophila insulin receptor, InR. We generated InR substitutions using homologous recombination and related each to emerging models of receptor tyrosine kinase structure and function. Three mutations when combined as trans-heterozygotes extended lifespan while retarding growth and fecundity. These genotypes reduced insulin-stimulated Akt phosphorylation, suggesting they impede kinase catalytic domain function. Among these genotypes, longevity was negatively correlated with egg production, consistent with life-history trade-off theory. In contrast, one mutation (InR353) was located in the kinase insert domain, a poorly characterized element found in all receptor tyrosine kinases. Remarkably, wild-type heterozygotes with InR353 robustly extended lifespan without affecting growth or reproduction and retained capacity to fully phosphorylate Akt. The Drosophila insulin receptor kinase insert domain contains a previously unrecognized SH2 binding motif. We propose the kinase insert domain interacts with SH2-associated adapter proteins to affect aging through mechanisms that retain insulin sensitivity and are independent of reproduction.
Keywords: insulin receptor, longevity, aging, reproduction, Drosophila, kinase insert domain
Introduction
Insulin/insulin growth factor (insulin/IGF) signaling provides a powerful avenue to study aging. Early work extended lifespan through mutations of the insulin-like receptors daf-2 of C. elegans and InR of Drosophila (Kenyon et al. 1993; Kimura et al. 1997; Tatar et al. 2001; Altintas et al. 2016). These invertebrates have single insulin-like receptors that simultaneously regulate the metabolic and growth functions of mammalian insulin receptor (IR) and insulin growth factor receptors (IGFR) (Finch and Ruvkun 2001; Tatar et al. 2003). As in mammals, Daf-2 and InR are stimulated by insulin-like peptides to induce intracellular signals through Akt (also called Protein kinase B), Ras (Proto-oncogene protein P21), and TOR (Target of rapamycin) (Teleman 2010), which collectively mediate transcription factors (Lin et al. 1997; Hsu et al. 2003) and cellular metabolism (Jia et al. 2004; Burkewitz et al. 2014; Post et al. 2018). These pathways ultimately assure survival through mechanisms that include translation rate, autophagy, mitochondrial uncoupling, REDOX, genomic stability, and lipogenesis (Honda and Honda 1999; Murphy et al. 2003; Mostoslavsky 2008; Perez and Van Gilst 2008; Zarse et al. 2012; Bai et al. 2013). Altered insulin/IGF function also affects aging in mammals. With some variation in outcome, knockdown of IR, IGF1R, and insulin receptor substrates (IRS1, IRS2) is associated with retarded aging in mice (Blüher et al. 2003; Taguchi et al. 2007; Selman et al. 2008; Xu et al. 2014; Merry et al. 2017). In humans, polymorphisms of IGF1R and the transcription factor FOXO3A are associated with exceptional lifespan (Suh et al. 2008; Bao et al. 2014). Overall, insulin/IGF signaling integrates cells, tissue and physiology to control adult survival and lifespan.
The breadth of insulin/IGF function challenges our ability to understand how it affects aging. Insulin/IGF signaling simultaneously regulates many traits including growth, metabolism, cell proliferation, differentiation, Dauer/diapause, and reproduction (Kido et al. 2001; Bartke et al. 2013; Ashpole et al. 2017; Ewald et al. 2018). Furthermore, these traits are mediated by a single insulin-like receptor in C elegans and Drosophila while these invertebrates produce many insulin-like ligands (Brogiolo et al. 2001; Pierce et al. 2001; Gronke et al. 2010). The insulin-like receptor is a molecular switch-board taking many incoming calls and routing each to distinct signaling destinations. Broadly, understanding such pleiotropy is a longstanding problem in receptor tyrosine kinase biology (Marshall 1995; Zinkle and Mohammadi 2018). In the biology of aging, a key aspect of pleiotropy involves trade-offs between lifespan and reproduction where alleles of a gene are favored that optimize the conflicting functions supporting these life-history traits (Tabatabaie et al. 2011; Maklakov et al. 2017). In this context, the mechanistic basis for how a protein such as the insulin-like receptor jointly controls lifespan and reproduction is unknown.
One approach to understand insulin/IGF receptor pleiotropy is to study alleles in cultured cells (Tazearslan et al. 2011; Hall et al. 2020), among polymorphic animals (Paaby et al. 2014), and even in humans with inherited insulin and IGF resistance (Klammt et al. 2011; Ardon et al. 2014). Insight is developed by relating the observed amino acid substitutions to receptor structure and function. In aging biology, this strategy was pioneered by work with C. elegans daf-2 (Gems et al. 1998; Patel et al. 2008). About two dozen daf-2 alleles extend lifespan, and variously also affect Dauer, fertility, and growth. Classes of these traits were defined based on where substitutions fell in the ectodomain relative to the kinase domain, but understanding how the substitutions jointly control traits was limited by the structural data of the time.
Structural analysis of receptor tyrosine kinases has advanced in recent years and we can now apply this to Drosophila. The Drosophila insulin-like receptor was initially identified in the lab of Rosen (Petruzzelli et al. 1986). EMS mutagenesis was subsequently used to generate a series of InR alleles that were viable as trans-heterozygotes yet otherwise recessive lethal (Azpiazu and Frasch 1993; Fernandez et al. 1995; Chen et al. 1996). Sequencing of several alleles identified substitutions in the kinase domain (Brogiolo et al. 2001). These genotypes variously reduced cell growth, fecundity, and receptor kinase activity (Chen et al. 1996; Brogiolo et al. 2001). In early work with the Drosophila insulin receptor, aging was slowed by one EMS-generated mutant (InRE19) when heterozygous over a P-element insertion allele (Tatar et al. 2001). Here, we characterize how other InR mutations from the original EMS collection affect aging using the method of quantitative complementation testing (QCT) (Flatt 2004; Geiger-Thornsberry and Mackay 2004). We then validate and extend these results by independently regenerating the putative substitutions through homologous recombination gene replacement in a separate genetic background and evaluate lifespan, growth, development rate, fecundity, and insulin sensitivity. Finally, recent advances provide new insights into the structure of activated human insulin receptors (Uchikawa et al. 2019; Chen et al. 2020) that we apply to understand how the Drosophila insulin-like receptor modulates aging.
We propose three conclusions. (1) Drosophila InR mediates aging in independent ways. One affects reproduction and thereby extends lifespan through reduced survival costs, the other affects mechanisms to assure longevity independent of reproduction. (2) The biochemical impact of substitutions within InR may be understood in a context of asymmetric transphosphorylation (Zinkle and Mohammadi 2018; Chen et al. 2020). Aging is slowed when one activated protomer contains a mutation that alters the quality or quantity of signaling. (3) Mutations slow aging by altering the receptor’s kinase activity or protein domain interactions. We identify a dominant substitution in the kinase insert domain (Locascio and Donoghue 2013) that robustly increases lifespan yet produces normal growth, high fecundity, and wild-type insulin sensitivity. This kinase insert domain contains a SH2 binding motif. We propose this site interacts with as yet to be identified adapter proteins to regulate longevity.
Materials and methods
EMS-induced mutations of InR
The Drosophila insulin/IGF-receptor InR is on the third chromosome. Lines with EMS-induced mutations in InR, balanced by the third chromosome inversion InR+TM3, were provided by M. Frasch (The Mount Sinai School of Medicine) (Azpiazu and Frasch 1993; Fernandez et al. 1995). We tested the ability of 17 EMS mutations to produce viable adults when complemented to the EMS-induced mutation InRE19 (provided by J. Jack, University of Connecticut) (Chen et al. 1996; Tatar et al. 2001). Nine Frasch alleles produced viable adults when complemented to InRE19: InR74, InR76, InR211, InR246, InR262, InR327, InR351, InR353, and InR373. Subsequently, these nine alleles and the InRE19 allele were crossed six generations into the inbred Samarkand I-236 (Sam) background using the balancer stock Sam 1; Sam 2; InR+TM3/TM6B (provided by T. Mackay, North Carolina State University).
Wild-type InRNC alleles
Iso-female lines were established from wild-type Drosophila melanogaster caught at the Raleigh Farmer’s Market (provided by T. Mackay, North Carolina State University) (De Luca et al. 2003). Single females from each line were crossed to males of Sam 1; Sam 2; InR+TM3/TM6B and backcrossed to extract third chromosomes balanced by InR+TM3. Eighteen chromosome extraction lines from this collection provided a sample of wild-type InR alleles (denoted InR NC series). In addition, the InRNC442 line was used to normalize survival data across test blocks.
Genotypes for quantitative complementation test
Quantitative complementation testing measures the phenotypic difference of one allele complemented to a standard hypomorph relative to when it is complemented to a standard wild-type allele (Long et al. 1996; Mackay and Fry 1996; Flatt 2004; Geiger-Thornsberry and Mackay 2004; Turner 2014). We made reciprocal crosses between InRE19 (using adult InRE19/InR+TM3) and the nine viable Frasch InR EMS-alleles (using InREMS/InR+TM3), and between the 18 InRNC wild-type alleles (using InRNC/InR+TM3). Offspring from reciprocal crosses were pooled to measure phenotypes. Phenotypes of every InREMS/InRE19 and InRNC/InRE19 offspring were compared to a contemporaneous cohort of the allele when complemented by wild-type InR (InREMS/InR+TM3 or InRNC/InR+TM3).
Demographic quantitative complementation
Life tables (number of deaths per 2-day census period) were generated for adult offspring of each genotype (details in Supplementary Methods). Cohorts were initiated with three replicate demography cages of ∼150 newly eclosed adults at ∼1:1 sex ratio per genotype. In total, we assessed the lifespan of 16,057 males and 17,223 females. Each 2 days, food vials were changed and dead flies were removed and counted. Life tables collected in five test blocks (j = 1–5) were made with data from replicate cages combined. Within each test block, we included the wild-derived line InRNC442 to normalized genotypes across blocks. One life table was generated for each InREMS allele and InRNC allele expect for duplicates generated in independent blocks for InR211(EMS), InR74(EMS), and InR353(EMS).
We used proportional hazard analysis (Parmer and Machin 1995) to conduct QCT with cohort life table data, executed in JMP (SAS Institute, Cary, NC). We quantified the proportional mortality difference (hazard coefficient βij) within each block j for each InR allele i (EMS or NC) when complemented to InRE19 relative to when complemented to InR+TM3. In each block, we estimated and calculated the adjusted mortality coefficient for each InR allele as . Negative values of indicate mortality rate is reduced in InRi/InRE19 relative to InRi/(InR+TM3); values near zero indicate there is no effect on mortality and values greater than zero indicate there is increased mortality. The transformation estimates the fold change in mortality risk (hazard ratio) induced by each InR allele complemented to InRE19 relative to when it is complemented to InR+TM3. Life expectancy was also standardized among blocks: for each block j we calculated the difference in life expectancy between InRNC442/InRE19 and InRNC442/(InR+TM3) denoted . The adjusted difference in life expectancy for each InR allele i is .
Body size quantitative complementation
Head capsule width was measured from 20 females from each InREMS mutant and InRNC wild-type allele when complemented to InRE19 and to InR+TM3. Adults were reared simultaneously on standard diet from vials seeded with 100 eggs, and collected across three replicate vials. The relative body size of each tested InRi allele was measured as (mean of InRi/InRE19)/(mean of InRi/InR+TM3).
Homologous recombination InR alleles
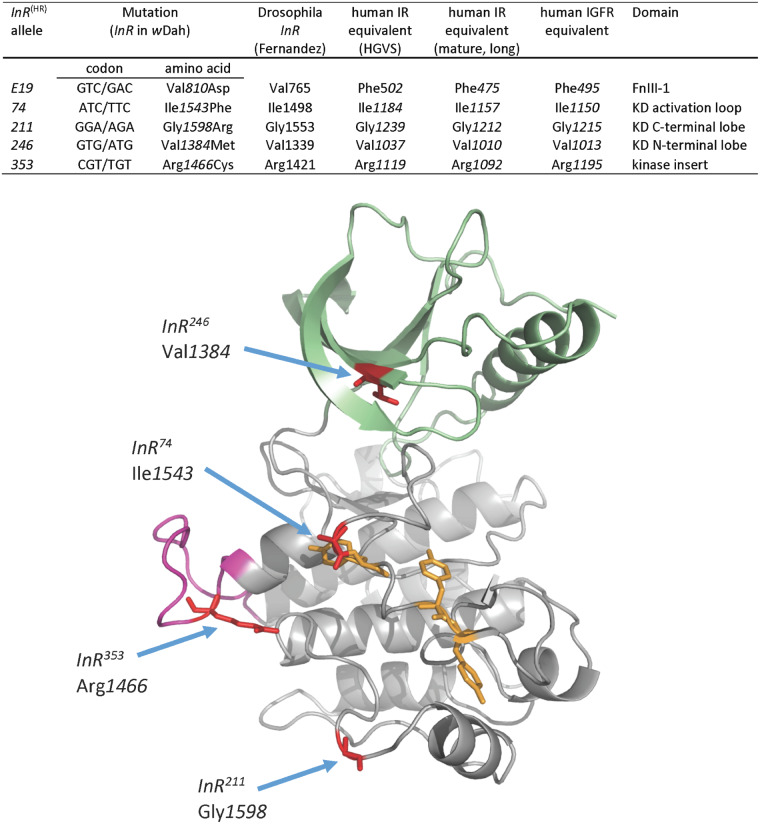
To validate and extend the results of QCT, we focused on the EMS alleles InRE19, InR211, InR74, and InR353 based on their ability to reduce mortality, and InR246 as a contrast where QCT showed effects body size but not mortality. Ron Kohanski (Johns Hopkins University, personal communication) provided cDNA sequences for the InRE19, InR74, and InR246 alleles that respectively contained codon substitutions as Val810Asp, Ile1543Phe, and Val1384Met (Figure 3). Codon substitutions for the EMS alleles InR353 and InR211 were reported by Brogiolo et al. (2001) (Figure 3). In previous work as in our QCT, the causal effect of these substitutions has not been independently verified. Accordingly, we used ends-out homologous recombination gene replacement to regenerate each codon substitution in a coisogenic wDah background. We made five single replacement lines (designated InRE19(HR), InR74(HR), InR211(HR), InR246(HR), InR353(HR)). We produced independent, replicate accessions for InRE19(HR) (denoted InRE19(HR)-2, InRE19(HR)-14, InRE19(HR)-22) and for InR353(HR) (denoted InR353(HR)-8, InR353(HR)-15, InR353(HR)-20). We replaced the native wDah sequence with itself to produce the wild-type control InR+(HR) (accession denoted “29B”). We generated a double substitution allele (InRE19; 74(HR)), and a null InR allele (InRnull(HR)).
Figure 3.
Sites, nomenclature, and structural location of InR alleles generated by homologous recombination. Amino acids numbered from the translation initiation site of the wDah stock used as the progenitor wild-type InR in homologous recombination (GenBank accession MT_563159). This TIS is 45 additional N-terminal amino acids from the TIS reported in Fernandez et al. (1995). Human insulin receptor sequence is numbered following the nomenclature of the Human Genome Variation Society (HGVS; http://www.hgvs.org/rec.html) and based on the mature, long-form type IR cDNA of Ebina et al. (1985). The human IGF1R amino acid sequence is based on NCBI Reference Sequence: NP_000866.1. KD: kinase domain. FnIII: extracellular fibronectin domain III. Ribbon model of kinase domain (based on hIR), indicating conserved sites of amino acid substitutions (red). N-terminal lobe: green; C-terminal lobe: gray; kinase insert domain: magenta; Tyrosine residues of the activation loop: yellow.
Detailed methods for the homologous recombination are provided in the Supplementary Materials. Targeting arms for InR cloned from wDah (Staber et al. 2011) were inserted into the pW25.2 vector (Drosophila Genomics Resource Center, Bloomington, IN) and injected into w1118 embryos by Genetic Services (Sudbury, MA). Targeting arms contained single nucleotide substitutions to produce the desired amino acid replacements. Flies with the white+ eye color marker mapped to the first or second chromosomes were used for homologous recombination. These lines were crossed to flies expressing flipase and the restriction enzyme I-Sce1 to execute excision and linearization of the transgenic construct. Candidate insertion lines were selected based on white+ mapped to chromosome 3. Chromosomes with replaced alleles were crossed to flies expressing cre recombinase to remove the white+ marker. The white+ marker was retained in an accession of the wild-type replacement allele to produce InRnull(HR). All replacement alleles were validated by sequencing.
Phenotypes of InR homologous recombination alleles
Viability, development time, and adult size
Reciprocal crosses were made among all InR(HR) alleles. Four females and males were housed for mating and to lay eggs over 24 h in replicate vials with yeast-supplemented standard media. We recorded stage of lethality (larvae, prepupae, or pupae) and the number of emerging adults each subsequent 24 h. Adult size (thorax length) was measured from 20 off-spring females of every viable cross.
Life table demography
Newly eclosed males and females were collected, transferred to bottles with fresh medium and kept at 25°C for 2 days (details in Supplementary Methods). Mated females were transferred to demography cages maintained at 25°C at an initial density of 125 females per cage. Three to five replicate cages were initiated for each genotype. Food was changed every two days, at which time dead flies were removed and counted. Life tables were constructed with genotype replicate cages combined for survival analysis. In total, we assessed the lifespan of 17,184 homologous recombination females.
Fecundity and ovariole number
Fifteen newly eclosed females of each genotype were placed with 15 wDah males into three replicate demography cages, modified for egg collection (details in Supplementary Methods). For the following 15 days, egg plates were supplemented with yeast paste and changed twice a day at which time eggs were counted. Dead females were removed and counted daily. Daily fecundity (mean eggs per day) is the number of eggs summed across replicate cages per surviving female. Average fecundity is the mean of daily fecundity across the sample period. Ovariole number was measured from fifteen 10-day old females of each genotype. Ovaries were dissected in PBS, fixed with 4% paraformaldehyde, and scored for total number of ovarioles across both ovaries.
Insulin stimulation of pAkt
We evaluated the ability of IRs to induce phosphorylation of Akt from larval fat bodies stimulated by human insulin, as in Musselman et al. (2011). For each biological sample, wandering L3 larvae were rinsed in PBS, bisected in Schneider’s medium (Gibco Cat#21-720-001) and the anterior half everted. This tissue was transferred to Eppendorf tubes containing 500 µl Schneider’s medium with 0, 1, 2, or 5 µg/ml human insulin (Santa Cruz Biotechnology; Cat#SC-360248) and incubated at room temperature with agitation for 20 min. Samples were transferred to 75 µl of sample buffer containing phosphatase inhibitors, homogenized, and used for Western-blot (Invitrogen NUPage system). Drosophila specific phospho-Akt antibody (Ser505) and pan-Akt were obtaining from Cell Signaling Technology (PO4-Akt Cat#4054, pan-Akt Cat#4691). Blots were imaged and quantified with ImageLab (Bio-Rad).
Data availability
Insulin receptor alignments and data access
Figure 3 and Supplementary Figure S1 provide amino acid sequence annotation of Drosophila InR, and human IR and IGF1R. We number the human IR as the mature long isoform of Ebina et al. (1985) (NCBI NP_000199.2). For cross-reference, the Human Genome Variation Society annotation includes an additional 27 a.a. localization sequence (Hall et al. 2020). The human IGF1R sequence is numbered as NCBI NP_000866.1. Throughout this manuscript we number Drosophila InR beginning at the first translation initiation site in wDah wildtype (deposited in GenBank as accession MT_563159), which is +45 amino acids relative to the TIS reported by Fernandez et al. (1995) (NCBI NM_079712.6). The wDah sequence also differs from that of Fernandez by an insertion/deletion (wDah lacks H144) and the substitution H146Q. This H144/H146 polymorphism segregates along latitudinal clines of Drosophila populations (Paaby et al. 2014). Supplementary Figures S4 and S5 provide the verified sequence alignments of the homologous recombination alleles.
Contents of Supplementary materials
Supplementary Methods contain detailed methods for demography, stocks, homologous recombination, and all PCR primers. Supplementary Materials contain figures for InR/hIR/hIGFR alignment (Supplementary Figure S1), eclosion time distribution (Supplementary Figure S2), and the verified sequence alignments of the homologous recombination alleles (Supplementary Figures S4 and S5). Supplementary Figure S3 provides conceptual diagrams of dimer complementation and a model for receptor domain function in aging. Tables in the Supplemental Materials provide survival statistics of the QCT (Supplementary Table S1), survival data for wild-type alleles in QCT (Supplementary Table S2), survival data of hemizygotes (Supplementary Table S3), and survival data of all HR alleles and accessions (Supplementary Table S4).
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13377023.
Results
Demographic quantitative complementation
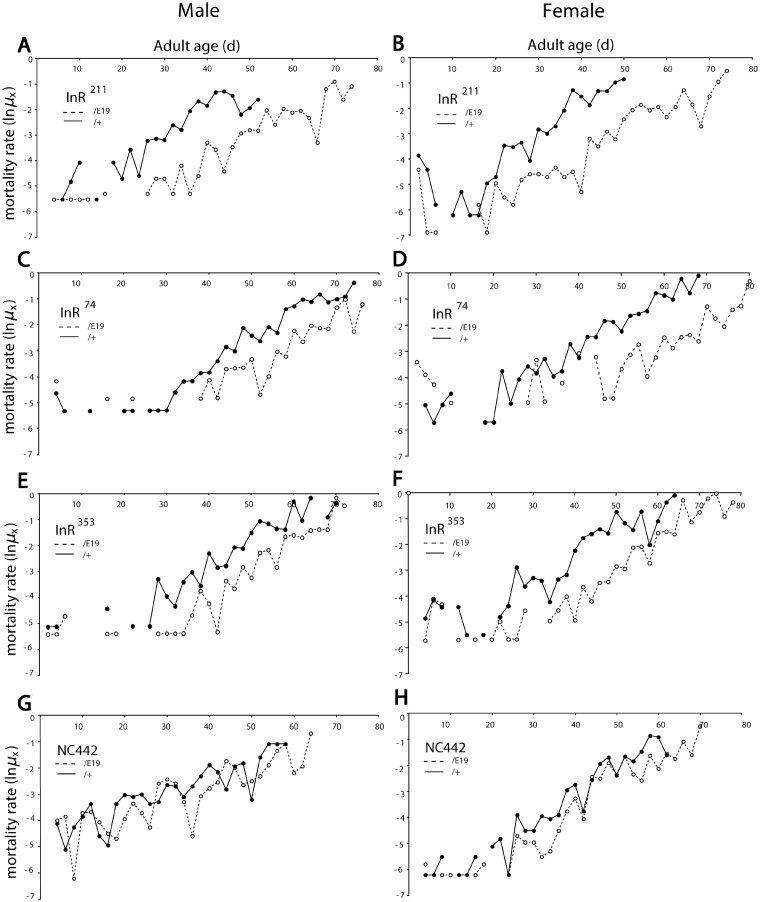
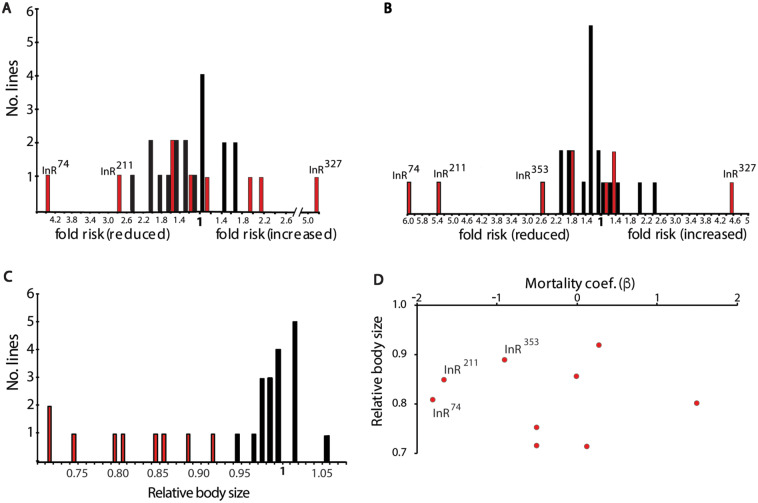
We used QCT to determine how the viable Frasch InR EMS alleles (InR74, InR76, InR211, InR246, InR262, InR327, InR351, InR353, and InR373) affect aging. Figure 1 illustrates mortality for several mutants complemented to InRE19 relative to when complemented to wild-type InR (InR+TM3) (Figure 1, Supplementary Table S1). Mortality rates in all cohorts increased exponentially once the death rate exceeded the lower limit of demographic power (Promislow et al. 1999). Life tables were generated in five blocks, with the natural wild-type allele InRNC442 measured in each to control for block effects (Supplementary Table S2). Relative to InRNC442 within each block, the EMS mutations variously reduced mortality (block adjusted βiadj = -1.80), had little effect (βiadj 0) or substantially increased mortality (βiadj = 1.64) (Supplementary Table S1). Because InRNC442 is just one instance of a wild-type allele, we applied QCT to a collection of 18 wild-type InR alleles extracted from the North Carolina Market population (NC series) and evaluated how each EMS mutant performed relative to the wild-type distribution (Supplementary Table S1). Six EMS InR mutants did not significantly reduce mortality relative to wild-type alleles (InR76, InR246, InR262, InR273, InR327, and InR351), while InR327 strongly increased mortally. InR74 and InR211 significantly reduced mortality in males and females. The InR353 allele reduced female mortality but not significantly so relative to the wild-type distribution. The transformation estimates fold change in mortality: InR74 and InR211 alleles reduced male and female mortality 2.6- to 6-fold, while InR353 reduced female mortality about 2-fold (Figure 2, A and B).
Figure 1.
Mortality of EMS- and wild-type InR alleles in quantitative complementation test. Log mortality rate plotted from observed age-specific mortality (qx), lnµx = ln(−ln(1 − qx)). Block 3 data (Supplementary Table S3): InRallele/InRE19 (dashed line), InRallele/(InR+, TM3 Sb) (solid line). N0: initial cohort size combined genotypes. Cox proportional hazard analysis estimates mortality coefficient β for each allele as InRallele/InRE19 relative to InRallele/(InR+ TM3 Sb); β < 1.0 indicates InRallele/InRE19 has reduced mortality, P < 0.0001 (survival statistics in Supplementary Table S1). (A, B) InR211: males (N0 = 484, β = −1.65, SE = 0.11); females (N0 = 712, β = −1.82, SE = 0.10). (C, D) InR74: males (N0 = 337, β = −0.98, SE = 0.12); females (N0 = 442, β = −1.68, SE = 0.13). (E, F) InR353: males (N0 = 394, β = −0.99, SE = 0.11); females (N0 = 560, β = −1.42, SE = 0.099). (G, H) NC442 third chromosome (wild-type InR): males (N0 = 443, β = −0.504, SE = 0.099); females (N0 = 571, β = −0.97, SE = 0.11).
Figure 2.
Distributions of allelic effects estimated from Quantitative Complementation Test comparing EMS- and wild-type InR alleles. (A) Males, (B) females; distribution of proportional hazard (fold risk of mortality) for nine EMS mutants (red bars) and 18 wild-type (NC series) alleles (black bars); estimated as exp(|β|). EMS InR alleles with significantly different mortality are labeled (Statistics in Supplementary Table S1). (C) Distribution of relative body size (head capsule width) among mutant (red) and wild-type (black) females. All mutants were smaller than the expected based on the distribution of wild-type alleles, z-test, P < 0.0007. (D) No significant correlation between relative female mortality (β) and relative body size among InR alleles assessed in QCT (r2 = 0.08, P = 0.85).
Body size quantitative complementation
To describe the allelic effects on female body size, we quantified the ratio of each EMS allele complemented to InRE19 relative to InR+TM3. All EMS InR alleles significantly reduced body size, with relative effects ranging from 0.9 to less than 0.75 (Figure 2C). Among the EMS InR alleles, relative body size does not correlate with a change in mortality (Figure 2D).
Homologous recombination InR alleles
Quantitative complementation testing has limitations: it only measures recessive allelic effects; it confounds epistatic interactions with potential co-segregating mutations; the tested alleles were not derived from the wild-type InR (InR+TM3) used for complementation; each tested InR may contain unidentified substitutions; allelic effects are relative rather than absolute; and the measured effect of InR is confounded by a deleterious effect of the TM3 balancer chromosome. Our newly generated series of homologous recombination InR alleles resolves these issues.
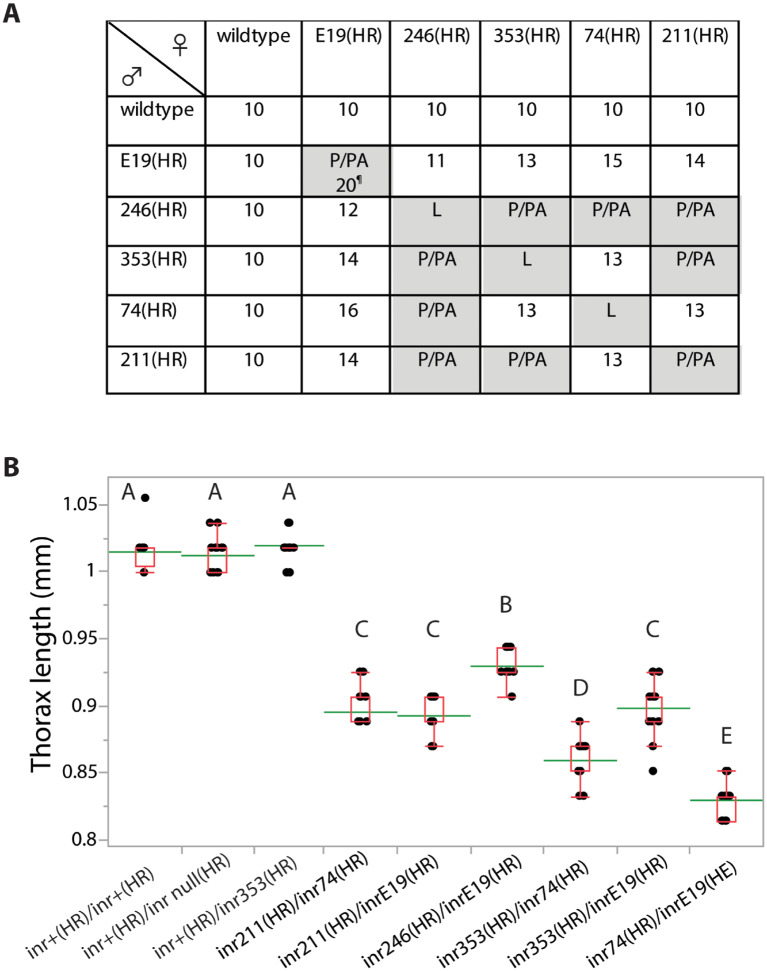
We evaluated viability, eclosion time, and adult size for all allele combinations (Figure 4, A and B, Supplementary Figure S2). Every mutation had normal size and development time when heterozygous with InR+(HR). All mutations were homozygous lethal except for rare escapes in the case of InRE19(HR)/InRE19(HR). As expected from our QCT, all kinase domain mutations (InR74(HR), InR211(HR), InR246(HR), and InR353(HR)) produced small adults with delayed development when heterozygous with InRE19(HR) (an ectodomain mutation). Several kinase domain mutations were viable as trans-heterozygotes: InR74(HR)/InR353(HR) and InR74(HR)/InR211(HR), and these adults were small and developed slowly.
Figure 4.
Growth characteristics of homologous recombination InR(HR) alleles. (A) Development time (days to first eclosion), or stage of lethality for all allele combinations with reciprocal crosses. Lethality at first or second stage larvae (L), late pupae (P) or pharate adult (PA). ¶: InRE19(HR)/InRE19(HR) produce few minute adults at 20 days, but otherwise were pupal/pharate lethal. (B) Adult size (thorax length, mm) of InR(HR) alleles across genotypes. Means, std. dev. and range shown, N = 20 females each genotype. Means not connected by same letter are significantly different (one-way ANOVA, Tukey–Kramer HSD comparison, P < 0.05).
Demography of InR hemizygotes
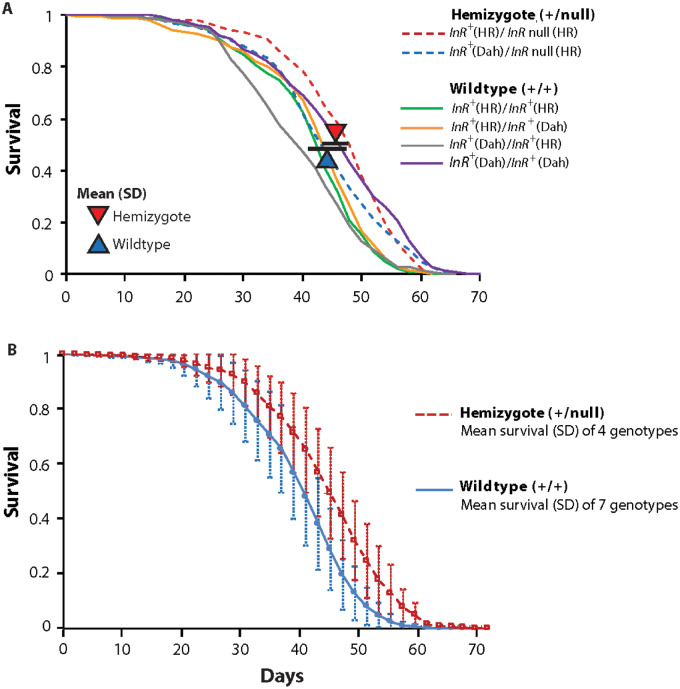
Insulin receptors are preformed dimers assembled with protomers translated from either allele (Supplementary Figure S3). Because homozygotes of mutant alleles are inviable, we infer homodimeric mutant receptors are nonfunctional. Viable mutant trans-heterozygotes must produce functional heterodimers, but they will also make fewer functional dimers because half of all potential receptors are mutant homodimers. To determine if simply having fewer functional receptors extends longevity, we evaluated the survival of hemizygote InR+(HR)/InRnull(HR) adults, which produce about 25% less InR mRNA (quantitative PCR in Supplementary Table S3). These adults increased life expectancy between 2 and 4 days when tested in replicate trials (Figure 5; significant only in Trial 2; Supplementary Table S3). This difference provides an approximate benchmark: we may infer a genotype affects receptor function when its gain in longevity is more than that caused by numerical loss of receptors modeled in InR hemizygotes.
Figure 5.
Impact of InR hemizygosity upon adult survival. (A) Trial 1. Two hemizygotes (InRnull(HR)/InR+(HR) and InR null(HR)/InR+Dah) compared to four wild-type (generated from reciprocal, pairwise combinations of InR +(HR) and InR +Dah alleles). Triangles indicate the average of median lifespan of the genotype group (bars: standard deviation). Hemizygotes are 2 days longer lived, arising from lower proportional hazard mortality (β = −0.14, χ2 = 36.4, P < 0.0001). (B) Trial 2. Four hemizygotes compared to seven wildtypes; genotype details in Supplementary Table S3. Survival plots show mean values among genotypes within the genotype class, with standard deviation (per census interval). Hemizygotes are 4 days longer lived, arising from lower proportional hazard mortality (β = −0.25, χ2 = 275.1, P < 0.0001).
Demography of InR homologous recombination adults
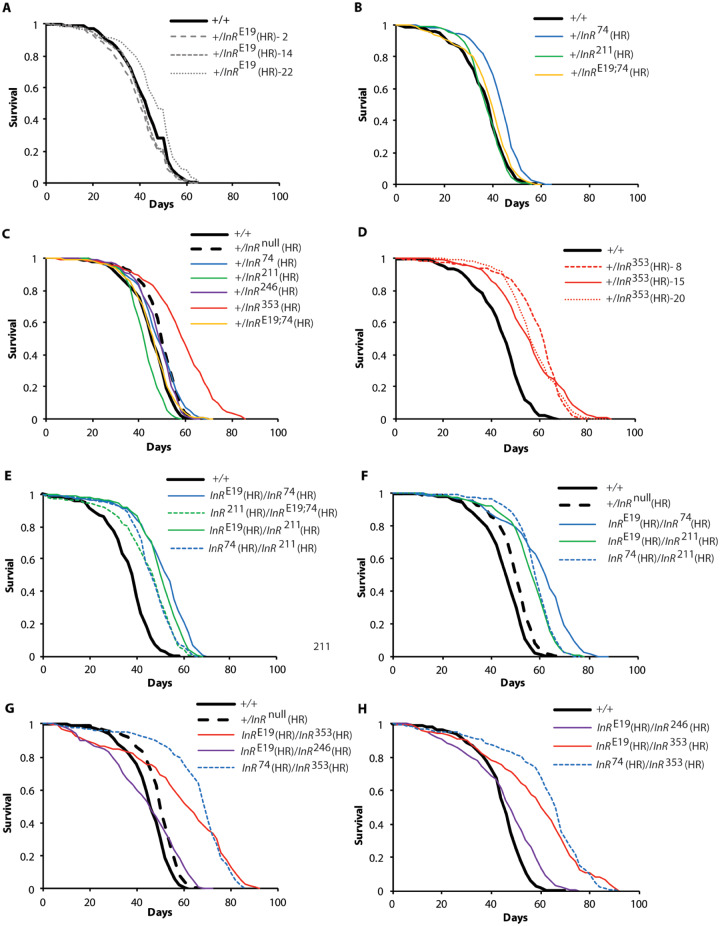
The heterozygote InRE19(HR)/InR+(HR) does not extend lifespan (Figure 6A): median life expectancy averaged among InRE19(HR) accessions was 43.3 days, compared to 44 days for wildtype (Supplementary Table S4). Likewise, the intracellular kinase domain alleles InR74(HR), InR211(HR), and InR246(HR) when heterozygous with InR+(HR) did not extend lifespan more than expected from InR hemizygotes (Figure 6, B and C; Supplementary Table S4). In contrast, in replicated trials with independent accessions, InR+(HR)/InR353(HR) increased life expectancy on average 12.5 days by decreasing mortality about fourfold (Figure 6, C and D; Supplementary Table S4).
Figure 6.
Survival of homologous recombination InR alleles. Females. Plots represent independent trials except for repeated controls shown in (B and E), and in (C, F, and G). In all trials, wild-type and null allele controls were homologous recombinant InR+(HR) and InRnull(HR) respectively. (A) Wild-type heterozygotes made with three accessions of ectodomain mutant InRE19(HR). (B) Wild-type heterozygotes made with mutants of the kinase domain A-loop (InR74(HR)) and C-terminal lobe (InR211(HR)). (C) Wild-type heterozygotes made with mutants of kinase domain N-terminal lobe (InR246(HR)), A-loop, KID (InR353(HR)), and C-terminal lobe; hemizygotes shown for contrast. (D) Wild-type heterozygotes made with three accessions of the kinase insert domain mutant InR353(HR). (E) Transheterozygotes of InRE19(HR), InR74(HR), and InR211(HR). (F) Independent, replicate trails of transheterozygotes of InRE19(HR), InR74(HR), and InR211(HR). (G) Transheterozygotes of InRE19(HR), InR74(HR), InR246(HR) and InR353(HR). (H) Independent, replicate trail of transheterozygotes of InRE19(HR), InR74(HR), InR246(HR), and InR353(HR). Life table summaries and proportional hazard statistics in Supplementary Table S4.
In QCT, kinase domain alleles reduced mortality when complemented to InRE19 relative to when complemented to InR+. From the homologous recombination alleles, we now estimate the absolute allelic effect of each mutation. Across replicate trials, the genotypes InR74(HR)/InRE19(HR) and InR211(HR)/InRE19(HR) extended lifespan 6–14 days (Figure 6, E and F). We also find the kinase domain heterozygote InR74(HR)/InR211(HR) extended lifespan (Figure 6, E and F). In every case, these longevity benefits exceeded that of the InR hemizygote (Supplementary Table S4).
Because the trans-heterozygote InRE19(HR)/InR74(HR) slowed aging, we asked if the mutations of these individual alleles extend lifespan when the substitutions were on the same protomer. They did not: InRE19,74(HR) had little effect on survival when heterozygous over a wild-type allele (Figure 6B). We likewise tested InRE19,74(HR) when heterozygous with InR211(HR). These adults extended longevity to the same extent as trans-heterozygote InR74(HR)/InR211(HR) (Figure 6E). Longevity is only extended when the substitutions of these two alleles occur on opposing protomers.
In QCT, the InR246 allele did not slow aging although it reduced body size. These observations are confirmed with InR246(HR)/InRE19(HR): adults were small (Figure 4) but not long-lived (Figure 6, G and H). However, the survival plot of InR246(HR)/InRE19(HR) intersects that of wildtype, suggesting the mutant genotype may have high age-independent mortality which obscures a beneficial effect of reduced demographic aging.
While InR353(HR) extended lifespan when heterozygous with wildtype (10–16 days), InR353(HR) also slowed aging when heterozygous with InRE19(HR) (14–15 days). When heterozygous with InR74(HR), InR353(HR) extended lifespan 21–22 days, an effect caused by decreasing age-dependent mortality sixfold (Figure 6, G and H; Supplementary Table S4).
The homologous recombination alleles validate inferences from QCT but now measure absolute effects and are tested in an independent genetic background. We describe new trans-heterozygous genotypes that extend lifespan (InR74(HR)/InR211(HR); InR74(HR)/InR353(HR); InRE19(HR)/InR353(HR)) and document a dominant longevity benefit conferred by InR353(HR) (InR+(HR)/InR353(HR)).
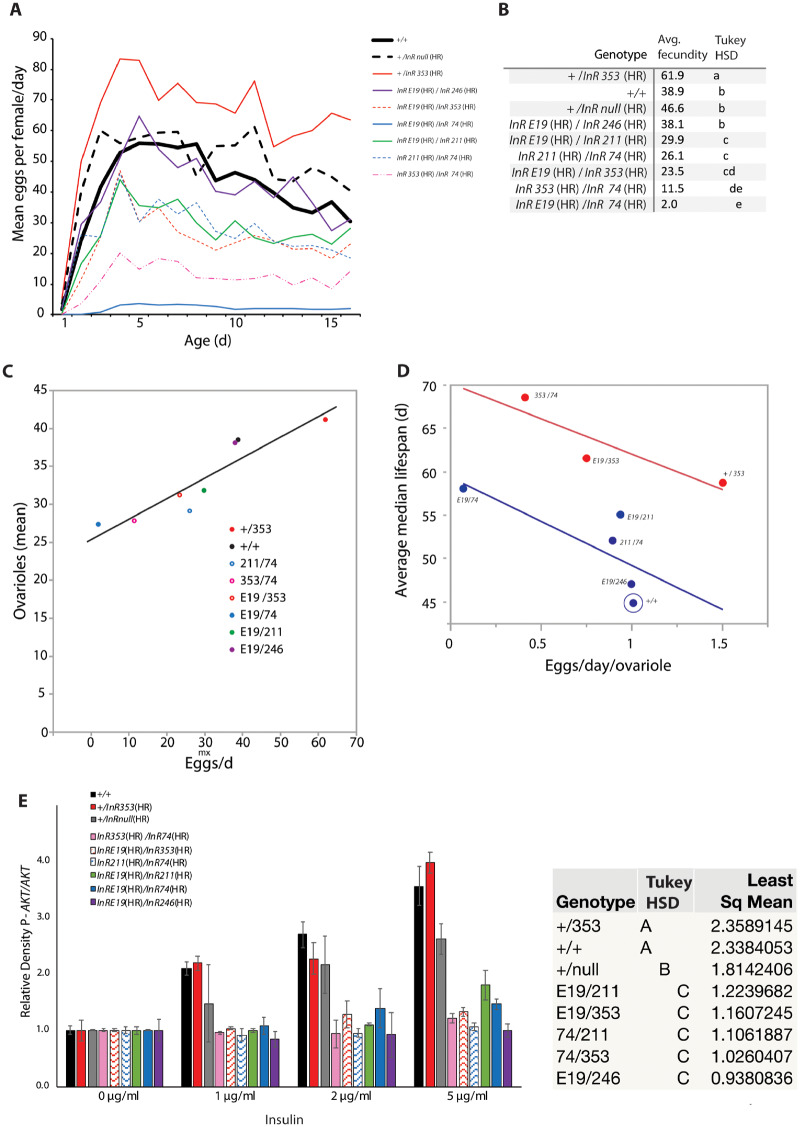
Fecundity
Drosophila insulin signaling modulates fecundity (LaFever and Drummond-Barbosa 2005; Hsu et al. 2008). Because reproduction is a common trade-off with survival (Flatt 2011), we determined how the InR alleles affect egg production (Figure 7, A and B). Fecundity was similar among wildtype, hemizygote, and InRE19(HR)/InR246(HR) females. Fecundity was reduced in all long-lived trans-heterozygotes. In contrast, long-lived InR+(HR)/InR353(HR) females produced more eggs than wildtype. The elevated fecundity of InR+(HR)/InR353(HR) could arise because these females have more ovarioles (subunits of the ovary) or produce more eggs per ovariole. We therefore measured ovariole number for all genotypes and regressed this trait against daily egg production. Fecundity positively associates with ovariole number with a slope less than one (Figure 7C): differences in daily fecundity derive in part from differences in the rate of egg production within ovarioles. We therefore regressed genotype lifespan against egg production per ovariole (eggs/day/ovariole), treating the InR353(HR) allele as a covariate (Figure 7D). A striking pattern emerges. Egg production per ovariole negatively associates with life expectancy (N = 8, P = 0.06), but the regression intercept for genotypes with one InR353(HR) allele is ∼12 days greater than genotypes without this allele (P = 0.013). The InR353(HR) allele confers longevity that is additive to and independent of how reproduction associates with survival.
Figure 7.
Fecundity and insulin-simulated pAkt. (A) Mean number of daily eggs laid from eclosion (age 0) through 15 days. (B) Average of daily fecundity. Genotypes not connected by same letter are significantly different, ANOVA with Tukey HSD post hoc analysis. (C) Regression of genotype mean ovariole number upon average fecundity; R2 = 0.85, F = 34.1, P < 0.001. (D) With InR353(HR) as a covariate, regression of genotype lifespan (average of median lifespan is calculated among independent cohorts) and egg production per ovariole (average daily fecundity/mean ovariole number). ANCOVA: β(fecundity/ovariole) = −9.15, t = −2.58, P = 0.061; β (InR353) = 6.23, t = 4.25, P = 0.013; β (fecundity/ovariole x InR353) = 1.01, t = 0.28, P = 0.79). (E) Quantity of phosphorylated Akt (pAKT) measured from fat body stimulated by human insulin; mean (SE) from densitometry of two independent immunoblots relative to total Akt of each sample, normalized to mean of unstimulated tissue. Two-way ANOVA; genotype, dose, and genotype-by-dose interaction, all effects P < 0.0001. Squares mean genotype differences not connected by same letter are significantly different (P < 0.05); Tukey HSD post hoc analysis.
Kinase activity of InR homologous recombination alleles
The InR353(HR) substitution (R1466C) lies in the kinase insert domain, a region of receptor tyrosine kinases thought to modulate substrate interaction rather than receptor kinase activity (Locascio and Donoghue 2013). We therefore assessed Akt phosphorylation of fat body stimulated with insulin. As expected, pAkt was strongly induced in tissue from wild-type flies (Figure 7E). Fat body from the hemizygote InR+(HR)/InRnull(HR) was moderately insulin resistant. Fat body from trans-heterozygotes involving InRE19(HR), InR74(HR), InR246(HR), and InR211(HR) induced little pAkt; they were insulin resistant. In contrast, fat body from InR+(HR)/InR353(HR) induced wild-type levels of pAkt. The InR353(HR) allele dominantly extends lifespan without loss of insulin-stimulated kinase activity.
Discussion
Mutations of insulin/IGF receptors are thought to slow aging and reduce growth because they diminish the total amount of insulin signaling. Alternatively, mutations of the IR may alter protein-receptor interactions in specific domains that control particular phenotypes, as proposed for receptor tyrosine kinases in general by Zinkle and Mohammadi (2018). Here, we explore these ideas to explain how Drosophila InR alleles modulate aging, growth, and reproduction.
Alleles in the quantitative complementation test
We used QCT to identify EMS InR mutations that slow aging. Three alleles reduced mortality more than expected from a sample of natural wild-type alleles. We also quantify how alleles affect growth. While all alleles significantly reduced adult size, there was no association between relative size and relative mortality among genotypes.
As noted, QCT has caveats (Flatt 2004; Turner 2014). The EMS-derived InR mutations do not share a common genetic background. Epistatic interactions with unknown second site mutations on the third chromosome could affect traits attributed to InR. QCT only estimates recessive effects of the InR mutations, and we could not evaluate complementation besides with InRE19. Unidentified polymorphisms within each InR locus might influence the complementation. Finally, QCT measures relative rather than absolute allelic effects, and these relative effects were confounded by how the balancer haplotype itself reduced lifespan.
To address these issues, we used ends-out homologous recombination to reconstitute the putatively causal InR substitutions of selected EMS alleles. These new alleles provide single, defined substitutions derived from a common wild-type progenitor in a controlled genetic background. We produced five homologous recombination (HR) alleles (InR74(HR), InR211(HR), InR246(HR), InR353(HR), InRE19(HR)), an HR-derived wild-type allele, a null allele, and a cis-allele with the E19 and 74 substitutions upon the same protomer (InRE19,74(HR)).
Receptor quantity moderately affects longevity
InR is a dimeric transmembrane receptor. Flies with two wild-type alleles generate normal protomers so that every dimer is functional. On the other hand, trans-heterozygotes produce some functional dimers with protomers of complementing alleles, as well as nonfunctional homodimers from protomers of each single mutant allele (Supplementary Figure S3). Thus, the lifespan of trans-heterozygote adults may be slowed simply because they have fewer functional receptors. To test this idea, we studied the hemizygote InR+(HR)/InRnull(HR). This genotype reduced InR mRNA about 25% and presumably this reduced the total number of receptors. Hemizygotes moderately impaired phosphorylation of Akt and only modestly affected longevity. The number of receptors in these hemizygotes is expected to be similar to that of recessive lethal alleles when heterozygous with wildtype (InR+(HR)/InRE19(HR), InR+(HR)/InR74(HR), InR+(HR)/InR211(HR)) due to their loss of mutant-mutant dimers (Supplementary Figure S3). Yet, those adults have normal longevity. Together these data suggest that genotypes with robustly extended lifespan either further reduce the quantity of receptors, or alter the quality or quantity of signaling from their functional receptors.
InRE19(HR): extracellular fibronectin domain
The InRE19(HR) substitution Val810Asp occurs in a linker sequence between the L2 and FnIII-1 ectodomains, corresponding to Phe475 of human IR and to Phe495 of human IGF-1R (Figure 3; Supplementary Figure S1). Insulin induces a hinge motion in this linker to swing the fibronectin domain inward, bringing together the intracellular kinase domains of each protomer (Gutmann et al. 2018; Scapin et al. 2018; Uchikawa et al. 2019). This induced proximity permits asymmetric kinase transphosphorylation, a non-reciprocal process whereby the intrinsic kinase activity of one protomer phosphorylates the opposing activation loop (A-loop) (Zinkle and Mohammadi 2018; Chen et al. 2020; Ferguson et al. 2020). The Val810Asp substitution of Drosophila InR will destabilize the fibronectin domain movement (Uchikawa et al. 2019) and thus prevent transphosphorylation of its associated intracellular kinase (Chen et al. 2020). The InR+(HR)/InRE19(HR) genotype has wild-type traits because the Val810Asp protomer retains its ability to transphosphorylate the complementing wild-type kinase domain, which is sufficient for full receptor kinase function.
InR74(HR): kinase domain activation loop
The activation loop (A-loop) of the Drosophila insulin/IGF-like receptor is homologous to that of human IR and IGF1R (Cabail et al. 2015). The Drosophila A-loop begins with a conserved DFG motif, followed by five amino acids (MTRDI), a seven amino acid sequence containing three tyrosine residues that participate in transphosphorylation (human IR: Tyr1158, Try1162, Tyr1163) and 13 final residues (Favelyukis et al. 2001). The A-loop blocks its adjacent kinase catalytic cleft in unstimulated receptors. Ligand binding induces A-loop transphosphorylation to alleviate this autoinhibition; this also provides a binding platform for substrate proteins (Hubbard et al. 1994; Hubbard 1997; Till et al. 2001). Drosophila InR74(HR) is an Ile1543Phe substitution within the MTRDI motif. This change may reduce the ability for Tyr1544 to be phosphorylated and thus diminish the domain catalytic activity or destabilize the loop binding platform (Cama et al. 1993; Ablooglu and Kohanski 2001; Favelyukis et al. 2001).
InR +(HR)/InR74(HR) has normal phenotypes, suggesting that the Ile1543Phe protomer can transactivate its complementing wild-type protomer to provide full receptor function. In contrast, only the Ile1543Phe protomer can be transactivated in InRE19(HR)/InR74(HR) adults, and the lone hypomorphic function of Ile1543Phe increases life span. This conclusion is consistent with how InRE19(HR)/InR74(HR) reduces insulin-stimulated kinase signaling (pAkt). To further test this explanation, we produced a double mutant where Val810Asp and Ile1543Phe were on the same protomer (InRE19,74(HR)). This allele was homozygous lethal but the genotype InR+/InRE19,74(HR) had normal growth and lifespan. The data show that aging is slowed when receptors only activate a protomer with a Ile1543Phe kinase domain.
InR246(HR): kinase domain N-terminal lobe
The InR246(HR) allele is a Val1384Met substitution at the invariant ATP binding loop of the N-terminal lobe (Supplementary Figure S1) (Hubbard 1997). This substitution will reduce the catalytic rate of the activated protomer (Chou et al. 1987; Odawara et al. 1989) and thus repress insulin-stimulated phosphorylation of Akt, as we observe from InRE19(HR)/InR246(HR) fat body. The normal traits of the heterozygote InR+(HR)/InR246(HR) suggests the InR246(HR) protomer fully transactivates wild-type protomers within heterodimers. In contrast, InRE19(HR)/InR246(HR) adults are small, as expected from their loss of kinase activity. It is unclear why this genotype does not also extend longevity although their cross-over survival pattern suggests these adults suffer high age-independent mortality that could obscure slow demographic aging.
InR211(HR): kinase domain C-terminal lobe
The Gly1598Arg substitution of InR211(HR) is in the kinase domain C-terminal lobe, a site conserved in human IR (Gly1212 Ebina) and IGF1R (Gly1215). These sites are three conserved residues from the substitution found in daf-2(e1370), the canonical IR longevity allele of C. elegans (Supplementary Figure S1). The homologous amino acid segment in human fibroblast growth factor receptor FGFR3 stabilizes the dimer interface during transphosphorylation (Chen et al. 2020). In Drosophila InR, the Gly1598Arg substitution will disrupt hydrophobic interactions among transactivating protomers. Consequently, as we observed from their limited insulin-stimulated pAkt, Gly1598Arg protomers will have reduced kinase catalytic activity. This molecular outcome extends lifespan, reduces growth, and limits reproduction.
InR353(HR): kinase insert domain
The InR353(HR) allele is exceptional. The heterozygote InR+(HR)/InR353(HR) extends lifespan yet adults have normal size and elevated fecundity. InR353(HR) combined with InRE19(HR) and InR74(HR) likewise produce long-lived adults, although these are small and have reduced fecundity.
InR 353(HR) contains the substitution Arg1466Cys. This arginine resides at a site found in human IRs (Arg1092) (Brogiolo et al. 2001) within in the conserved Arg-Pro-Glu sequence of the kinase insert domain (KID) (Locascio and Donoghue 2013) (Supplementary Figure S1) where Arg1092 stabilizes kinase domain dimerization (Chen et al. 2020). Notably, an Arg1092Glu mutation in human IR produces Donohue syndrome (Takahashi et al. 1997). Insulin receptors with one Arg1092Glu substitution localize normally to cell membranes (Hall et al. 2020) and humans with one mutant allele appear largely normal, but individuals homozygous for this substitution have severe insulin resistance, retarded growth, and low juvenile viability (Takahashi et al. 1997).
InR +(HR)/InR353(HR) has wild-type insulin sensitivity, demonstrating these receptors retain kinase catalytic activity unlike other longevity assurance mutations of InR. How then are InR+(HR)/InR353(HR) adults long-lived? The KID of Drosophila InR is 12 amino acids longer than the KID of human IR (Hubbard et al. 1994; Hanks and Hunter 1995), and contains the SH2-adaptor protein binding motif Tyr1477-Leu-Asn. This SH2 motif is found in mammalian IRS-2 where it binds Grb2 (Growth factor receptor-bound protein 2) (Patti et al. 1995) and within Drosophila Chico (homolog of IRS1-4) where it recruits Grb2/Drk (Grb2/Downstream-of-kinase) (Oldham et al. 2002). Grb2 is implicated in aging. Deletion of IRS-2 extends mouse lifespan (Taguchi et al. 2007; White 2014) as do null mutations of Drosophila chico (Clancy et al. 2001; Tu et al. 2002), and a Y243A substitution at the Chico SH2 site extends lifespan by repressing Grb-to-Ras signaling (Oldham et al. 2002; Slack et al. 2015).
We hypothesize the SH2 motif of the Drosophila InR kinase insert domain regulates lifespan through its interaction with adaptor proteins. Drosophila InR has been previously recognized to contain adaptor protein binding sites in the C-terminal tail and in the juxtamembrane domain (Li et al. 2013). Juxtamembrane domain SH2B sites recruit Lnk to modulate interactions with Chico, and mutation of lnk extends lifespan (Werz et al. 2009; Slack et al. 2010; Song et al. 2010; Almudi et al. 2013). We hypothesize the Drosophila InR kinase insert domain itself has the capacity to recruit Grb/Drk through its SH2 motif, and the Arg1466Cys substitution destabilizes this specific interaction to extend longevity without retarding receptor kinase activity.
Drosophila InR domains modulate aging through distinct mechanisms
InR+ (HR)/InR353(HR) presents a paradox: adults are highly fecund and yet long-lived. This positive association, which has been seen in other genetic manipulations of C. elegans and Drosophila (Kenyon et al. 1993; Gems et al. 1998; Jenkins et al. 2004), challenges a central idea of life-history theory where longevity is a trade-off with reproduction (Leroi et al. 2005). Our data suggest an explanation: InR regulates survival through two mechanisms (Supplementary Figure S3).
The first mechanism involves reproductive costs (Flatt 2011). Reduced germline stem cell proliferation in Drosophila limits the rate of egg production within ovarioles (Drummond-Barbosa 2008) and increases longevity (Flatt et al. 2008). Genotypes with the alleles InRE19(HR), InR74(HR), InR211(HR) may be long-lived, in part, because their reduced egg production mitigates survival costs-of-reproduction. Genotypes with these alleles are also insulin resistant and small. We propose these growth and life history phenotypes are caused by how each amino acid substitution reduces insulin-stimulated kinase catalytic activity.
The second mechanism confers longevity assurance independent of reproductive costs. We propose the kinase insert domain recruits substrate proteins through its SH2 binding motif. The Arg1466Cys substitution destabilizes this interaction to induce systems that promote somatic survival while maintaining kinase catalytic activity. Thus, InR+(HR)/InR353(HR) gains about 12 days of longevity. The genotypes InRE19(HR)/InR353(HR) and InR74(HR)/InR353(HR) also gain this reproduction-independent longevity, but now in addition to benefits conferred by reduced costs-of-reproduction.
Our explanation is consistent with how we currently understand Drosophila chico to regulate aging (Bohni et al. 1999; Clancy et al. 2001; Tu et al. 2002; Bai et al. 2015). As noted, Chico contains an SH2 phosphotyrosine binding site for Grb2/Drk. It also has sites to recruit p85/p60 of the PI3K complex, which regulates signaling through Akt (Oldham et al. 2002). Mutation of chico to block Grb2/Drk extends fly lifespan without reducing growth or fertility; these later traits are maintained because this mutated Chico still signals to Akt through p85/p60-PI3K (Oldham et al. 2002; Alic et al. 2014; Slack et al. 2015). Notably, InR353(HR) extends fly lifespan without reducing growth or fertility and maintains its ability for insulin to stimulate Akt phosphorylation. Chico mutated at the p85/p60 sites, on the other hand, extends life span but represses growth and reproduction. This suite of traits is seen in genotypes with the alleles InRE19(HR), InR74(HR), and InR211(HR), which also do not phosphorylate Akt in response to insulin.
The Drosophila IR appears to regulate aging through distinct systems modulated through structure-defined domains. This view reflects an emerging concept whereby protein tyrosine receptors control varied phenotypes through the action of specific protein interactions regulated by structural domains (Oldham and Hafen 2003; Liu et al. 2012; Li et al. 2013; Zinkle and Mohammadi 2018). Future biochemical and genetic studies will test our mechanistic hypotheses.
Acknowledgments
For patient mentoring on insulin receptor structure and function that made this work possible, we thank P. de Meyts, R. Kohanski, B. Garofalo, B. Forbes, Y. Suh, X. Bai, M. Mohammadi, S. Takahashi, and W. Peti. M. Frasch generously shared his archival collection of InR EMS mutants. T. Mackay provided the third chromosome extraction lines in the Samarkand background, derived from wild flies of the Raleigh Farmer’s Market. R. Kohanski shared unpublished data on the sites of substitutions in the Frasch alleles. W. Peti provided the structural diagram of substitutions mapped to the kinase domain. Thanks for supporting work in the Tatar lab is due to Aleksandra Norton, Miyo Malouf, Richard Martinez, Dante Ordantella, Colette Toal, and Roy Hsu.
R.Y., M.P., H.K., and N.C-J. conducted the experiments. R.Y., M.P., and M.T. designed the studies. M.T. and R.Y. analyzed the data and wrote the article.
Funding
This work was provided by the National Institutes of Health (R01AG16632 and R37AG024360).
Conflicts of interest
None declared.
Literature Cited
- Ablooglu AJ, Kohanski RA. 2001. Activation of the insulin receptor's kinase domain changes the rate-determining step of substrate phosphorylation. Biochemistry. 40:504–513. [DOI] [PubMed] [Google Scholar]
- Alic N, Giannakou ME, Papatheodorou I, Hoddinott MP, Andrews TD, et al. 2014. Interplay of dFOXO and two ETS-family transcription factors determines lifespan in Drosophila melanogaster. PLoS Genet. 10:e1004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almudi I, Poernbacher I, Hafen E, Stocker H. 2013. The Lnk/SH2B adaptor provides a fail-safe mechanism to establish the Insulin receptor-Chico interaction. Cell Commun Signal. 11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altintas O, Park S, Lee SJ. 2016. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Rep. 49:81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardon O, Procter M, Tvrdik T, Longo N, Mao R. 2014. Sequencing analysis of insulin receptor defects and detection of two novel mutations in INSR gene. Mol Genet Metab Rep. 1:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashpole NM, Logan S, Yabluchanskiy A, Mitschelen MC, Yan H, et al. 2017. IGF-1 has sexually dimorphic, pleiotropic, and time-dependent effects on healthspan, pathology, and lifespan. Geroscience. 39:129–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. 1993. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7:1325–1340. [DOI] [PubMed] [Google Scholar]
- Bai H, Kang P, Hernandez AM, Tatar M. 2013. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet. 9:e1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Post S, Kang P, Tatar M. 2015. Drosophila longevity assurance conferred by reduced insulin receptor substrate chico partially requires d4eBP. PLoS One. 10:e0134415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao JM, Song XL, Hong YQ, Zhu HL, Li C, et al. 2014. Association between FOXO3A gene polymorphisms and human longevity: a meta-analysis. Asian J Androl. 16:446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A, Sun LY, Longo V. 2013. Somatotropic signaling: trade-offs between growth, reproductive development, and longevity. Physiol Rev. 93:571–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M, Kahn BB, Kahn RC. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 299:572–574. [DOI] [PubMed] [Google Scholar]
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, et al. 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 97:865–875. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, et al. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 11:213–221. [DOI] [PubMed] [Google Scholar]
- Burkewitz K, Zhang Y, Mair WB. 2014. AMPK at the nexus of energetics and aging. Cell Metab. 20:10–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabail MZ, Li S, Lemmon E, Bowen ME, Hubbard SR, et al. 2015. The insulin and IGF1 receptor kinase domains are functional dimers in the activated state. Nat Commun. 6:6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama A, de la Luz Sierra M, Quon MJ, Ottini L, Gorden P, et al. 1993. Substitution of glutamic acid for alanine 1135 in the putative "catalytic loop" of the tyrosine kinase domain of the human insulin receptor. A mutation that impairs proteolytic processing into subunits and inhibits receptor tyrosine kinase activity. J Biol Chem. 268:8060–8069. [PubMed] [Google Scholar]
- Chen C, Jack J, Garofalo RS. 1996. The Drosophila insulin receptor is required for normal growth. Endoccrinology. 137:846–856. [DOI] [PubMed] [Google Scholar]
- Chen L, Marsiglia WM, Chen H, Katigbak J, Erdjument-Bromage H, et al. 2020. Molecular basis for receptor tyrosine kinase A-loop tyrosine transphosphorylation. Nat Chem Biol. 16:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CK, Dull TJ, Russell DS, Gherzi R, Lebwohl D, et al. 1987. Human insulin receptors mutated at the ATP-binding site lack protein tyrosine kinase activity and fail to mediate postreceptor effects of insulin. J Biol Chem. 262:1842–1847. [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, et al. 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 292:104–106. [DOI] [PubMed] [Google Scholar]
- De Luca M, Roshina NV, Geiger-Thornsberry GL, Lyman RF, Pasyukova EG, et al. 2003. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat Genet. 34:429–433. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D. 2008. Stem cells, their niches and the systemic environment: an aging network. Genetics. 180:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina Y, Ellis L, Jarnagin K, Edery M, Graf L, et al. 1985. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 40:747–758. [DOI] [PubMed] [Google Scholar]
- Ewald CY, Castillo-Quan JI, Blackwell TK. 2018. Untangling longevity, Dauer, and healthspan in Caenorhabditis elegans insulin/IGF-1-signalling. Gerontology. 64:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favelyukis S, Till JH, Hubbard SR, Miller WT. 2001. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol. 8:1058–1063. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Hu C, Lemmon MA. 2020. Insulin and epidermal growth factor receptor family members share parallel activation mechanisms. Protein Sci. 29:1331–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. 1995. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14:3373–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE, Ruvkun G. 2001. The genetics of aging. Annu Rev Genom Hum Genet. 2:435–462. [DOI] [PubMed] [Google Scholar]
- Flatt T. 2004. Assessing natural variation in genes affecting Drosophila lifespan. Mech Ageing Dev. 125:155–159. [DOI] [PubMed] [Google Scholar]
- Flatt T. 2011. Survival costs of reproduction in Drosophila. Exp Gerontol. 46:369–375. [DOI] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, et al. 2008. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci USA. 105:6368–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger-Thornsberry GL, Mackay TF. 2004. Quantitative trait loci affecting natural variation in Drosophila longevity. Mech Ageing Dev. 125:179–189. [DOI] [PubMed] [Google Scholar]
- Gems D, Sutton ML, Sundermeyer PS, Albert KV, King ML, et al. 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 150:129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6:e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann T, Kim KH, Grzybek M, Walz T, Coskun U. 2018. Visualization of ligand-induced transmembrane signaling in the full-length human insulin receptor. J Cell Biol. 217:1643–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Yu H, Choi E. 2020. Insulin receptor endocytosis in the pathophysiology of insulin resistance. Exp Mol Med. 52:911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576–596. [PubMed] [Google Scholar]
- Honda Y, Honda S. 1999. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 13:1385–1393. [PubMed] [Google Scholar]
- Hsu A-L, Murphy CT, Kenyon C. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 300:1142–1145. [DOI] [PubMed] [Google Scholar]
- Hsu HJ, LaFever L, Drummond-Barbosa D. 2008. Diet controls normal and tumorous germline stem cells via insulin-dependent and -independent mechanisms in Drosophila. Dev Biol. 313:700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR. 1997. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. Embo J. 16:5572–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Wei L, Ellis L, Hendrickson WA. 1994. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 372:746–754. [DOI] [PubMed] [Google Scholar]
- Jenkins NL, McColl G, Lithgow GJ. 2004. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc R Soc Lond B. 271:2523–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. 2004. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 131:3897–3906. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature. 366:461–464. [DOI] [PubMed] [Google Scholar]
- Kido Y, Nakae J, Accili D. 2001. Clinical review 125: The insulin receptor and its cellular targets. J Clin Endocrinol Metab. 86:972–979. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 277:942–946. [DOI] [PubMed] [Google Scholar]
- Klammt J, Kiess W, Pfaffle R. 2011. IGF1R mutations as cause of SGA. Best Pract Res Clin Endocrinol Metab. 25:191–206. [DOI] [PubMed] [Google Scholar]
- LaFever L, Drummond-Barbosa D. 2005. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science. 309:1071–1073. [DOI] [PubMed] [Google Scholar]
- Leroi AM, Bartke A, De Benedictis G, Franceschi C, Gartner A, et al. 2005. What evidence is there for the existence of individual genes with antagonistic pleiotropic effects? Mech Ageing Dev. 126:421–429. [DOI] [PubMed] [Google Scholar]
- Li CR, Guo D, Pick L. 2013. Independent signaling by Drosophila insulin receptor for axon guidance and growth. Front Physiol. 4:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 14:1319–1322. [DOI] [PubMed] [Google Scholar]
- Liu BA, Engelmann BW, Jablonowski K, Higginbotham K, Stergachis AB, et al. 2012. SRC homology 2 domain binding sites in insulin, IGF-1 and FGF receptor mediated signaling networks reveal an extensive potential interactome. Cell Commun Signal. 10:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio LE, Donoghue DJ. 2013. KIDs rule: regulatory phosphorylation of RTKs. Trends Biochem Sci. 38:75–84. [DOI] [PubMed] [Google Scholar]
- Long AD, Mullaney SL, Mackay TFC, Langley CH. 1996. Genetic interactions between naturally occurring alleles at quantitative trait loci and mutant alleles at candidate loci affecting brisle number in Drosophila melanogaster. Genetics. 144:1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Fry JD. 1996. Polygenic mutation in Drosophila melanogaster: genetic interactions between selection lines and candidate quantitative trait loci. Genetics. 144:671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklakov AA, Carlsson H, Denbaum P, Lind MI, Mautz B, et al. 2017. Antagonistically pleiotropic allele increases lifespan and late-life reproduction at the cost of early-life reproduction and individual fitness. Proc Biol Sci. 284: 20170376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 80:179–185. [DOI] [PubMed] [Google Scholar]
- Merry TL, Kuhlow D, Laube B, Pohlmann D, Pfeiffer AFH, et al. 2017. Impairment of insulin signalling in peripheral tissue fails to extend murine lifespan. Aging Cell. 16:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R. 2008. DNA repair, insulin signaling and sirtuins: at the crossroads between cancer and aging. Front Biosci. 13:6966–6990. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 424:277–284. [DOI] [PubMed] [Google Scholar]
- Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, et al. 2011. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech. 4:842–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odawara M, Kadowaki T, Yamamoto R, Shibasaki Y, Tobe K, et al. 1989. Human diabetes associated with a mutation in the tyrosine kinase domain of the insulin receptor. Science. 245:66–68. [DOI] [PubMed] [Google Scholar]
- Oldham S, Hafen E. 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13:79–85. [DOI] [PubMed] [Google Scholar]
- Oldham S, Stocker H, Laffargue M, Wittwer F, Wymann M, et al. 2002. The Drosophila insulin/IGF receptor controls growth and size by modulating PtdInsP(3) levels. Development. 129:4103–4109. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, Schmidt PS. 2014. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution. 68:3395–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmer MKB, Machin D. 1995. Survival analysis: a practical approach. Chichester: Wiley. [Google Scholar]
- Patel DS, Garza-Garcia A, Nanji M, McElwee JJ, Ackerman D, et al. 2008. Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics. 178:931–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Sun XJ, Bruening JC, Araki E, Lipes MA, et al. 1995. 4PS/insulin receptor substrate (IRS)-2 is the alternative substrate of the insulin receptor in IRS-1-deficient mice. J Biol Chem. 270:24670–24673. [DOI] [PubMed] [Google Scholar]
- Perez CL, Van Gilst MR. 2008. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 8:266–274. [DOI] [PubMed] [Google Scholar]
- Petruzzelli L, Herrera R, Arenas-Garcia R, Fernandez R, Birnbaum MJ, et al. 1986. Isolation of a Drosophila genomic sequence homologous to the kinase domain of the human insulin receptor and detection of the phosphorylated Drosophila receptor with an anti-peptide antibody. Proc Natl Acad Sci USA. 83:4710–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, et al. 2001. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans gene family. Genes Dev. 15:672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post S, Karashchuk G, Wade JD, Sajid W, De Meyts P, et al. 2018. Drosophila insulin-like peptides DILP2 and DILP5 differentially stimulate cell signaling and glycogen phosphorylase to regulate longevity. Front Endocrinol (Lausanne)). 9:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promislow DE, Tatar M, Pletcher SD, J.R C. 1999. Below-threshold mortality: implications for studies in evolution, ecology and demography. J Evol Biol. 12:314–328. [Google Scholar]
- Scapin G, Dandey VP, Zhang Z, Prosise W, Hruza A, et al. 2018. Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature. 556:122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, et al. 2008. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 22:807–818. [DOI] [PubMed] [Google Scholar]
- Slack C, Alic N, Foley A, Cabecinha M, Hoddinott MP, et al. 2015. The Ras-Erk-ETS-signaling pathway is a drug target for longevity. Cell. 162:72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack C, Werz C, Wieser D, Alic N, Foley A, et al. 2010. Regulation of lifespan, metabolism, and stress responses by the Drosophila SH2B protein, Lnk. PLoS Genet. 6:e1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Ren D, Li W, Jiang L, Cho KW, et al. 2010. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 11:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber CJ, Gell S, Jepson JE, Reenan RA. 2011. Perturbing A-to-I RNA editing using genetics and homologous recombination. Methods Mol Biol. 718:41–73. [DOI] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho M-O, Hwang D, Liu B, et al. 2008. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci. 105:3438–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabaie V, Atzmon G, Rajpathak SN, Freeman R, Barzilai N, et al. 2011. Exceptional longevity is associated with decreased reproduction. Aging (Albany NY). 3:1202–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Wartschow LM, White MF. 2007. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 317:369–372. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kadowaki H, Momomura K, Fukushima Y, Orban T, et al. 1997. A homozygous kinase-defective mutation in the insulin receptor gene in a patient with leprechaunism. Diabetologia. 40:412–420. [DOI] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. 2003. The endocrine regulation of aging by insulin-like signals. Science. 299:1346–1351. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, et al. 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 292:107–110. [DOI] [PubMed] [Google Scholar]
- Tazearslan C, Huang J, Barzilai N, Suh Y. 2011. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging Cell. 10:551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA. 2010. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem J. 425:13–26. [DOI] [PubMed] [Google Scholar]
- Till JH, Ablooglu AJ, Frankel M, Bishop SM, Kohanski RA, et al. 2001. Crystallographic and solution studies of an activation loop mutant of the insulin receptor tyrosine kinase: insights into kinase mechanism. J Biol Chem. 276:10049–10055. [DOI] [PubMed] [Google Scholar]
- Tu M-P, Epstein D, Tatar M. 2002. The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homolog chico. Aging Cell. 1:75–80. [DOI] [PubMed] [Google Scholar]
- Turner TL. 2014. Fine-mapping natural alleles: quantitative complementation to the rescue. Mol Ecol. 23:2377–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa E, Choi E, Shang G, Yu H, Bai XC. 2019. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 8:e48630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werz C, Kohler K, Hafen E, Stocker H. 2009. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 5:e1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF. 2014. IRS2 integrates insulin/IGF1 signalling with metabolism, neurodegeneration and longevity. Diabetes Obes Metab. 16:4–15. [DOI] [PubMed] [Google Scholar]
- Xu J, Gontier G, Chaker Z, Lacube P, Dupont J, et al. 2014. Longevity effect of IGF-1R(+/-) mutation depends on genetic background-specific receptor activation. Aging Cell. 13:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, et al. 2012. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 15:451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkle A, Mohammadi M. 2018. A threshold model for receptor tyrosine kinase signaling specificity and cell fate determination. F1000Res. 872:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Insulin receptor alignments and data access
Figure 3 and Supplementary Figure S1 provide amino acid sequence annotation of Drosophila InR, and human IR and IGF1R. We number the human IR as the mature long isoform of Ebina et al. (1985) (NCBI NP_000199.2). For cross-reference, the Human Genome Variation Society annotation includes an additional 27 a.a. localization sequence (Hall et al. 2020). The human IGF1R sequence is numbered as NCBI NP_000866.1. Throughout this manuscript we number Drosophila InR beginning at the first translation initiation site in wDah wildtype (deposited in GenBank as accession MT_563159), which is +45 amino acids relative to the TIS reported by Fernandez et al. (1995) (NCBI NM_079712.6). The wDah sequence also differs from that of Fernandez by an insertion/deletion (wDah lacks H144) and the substitution H146Q. This H144/H146 polymorphism segregates along latitudinal clines of Drosophila populations (Paaby et al. 2014). Supplementary Figures S4 and S5 provide the verified sequence alignments of the homologous recombination alleles.
Contents of Supplementary materials
Supplementary Methods contain detailed methods for demography, stocks, homologous recombination, and all PCR primers. Supplementary Materials contain figures for InR/hIR/hIGFR alignment (Supplementary Figure S1), eclosion time distribution (Supplementary Figure S2), and the verified sequence alignments of the homologous recombination alleles (Supplementary Figures S4 and S5). Supplementary Figure S3 provides conceptual diagrams of dimer complementation and a model for receptor domain function in aging. Tables in the Supplemental Materials provide survival statistics of the QCT (Supplementary Table S1), survival data for wild-type alleles in QCT (Supplementary Table S2), survival data of hemizygotes (Supplementary Table S3), and survival data of all HR alleles and accessions (Supplementary Table S4).
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13377023.