Abstract
Detection of surrounding organisms in the environment plays a major role in the evolution of interspecies interactions, such as predator–prey relationships. Nematode-trapping fungi (NTF) are predators that develop specialized trap structures to capture, kill, and consume nematodes when food sources are limited. Despite the identification of various factors that induce trap morphogenesis, the mechanisms underlying the differentiation process have remained largely unclear. Here, we demonstrate that the highly conserved pheromone-response MAPK pathway is essential for sensing ascarosides, a conserved molecular signature of nemaotdes, and is required for the predatory lifestyle switch in the NTF Arthrobotrys oligospora. Gene deletion of STE7 (MAPKK) and FUS3 (MAPK) abolished nematode-induced trap morphogenesis and conidiation and impaired the growth of hyphae. The conserved transcription factor Ste12 acting downstream of the pheromone-response pathway also plays a vital role in the predation of A. oligospora. Transcriptional profiling of a ste12 mutant identified a small subset of genes with diverse functions that are Ste12 dependent and could trigger trap differentiation. Our work has revealed that A. oligospora perceives and interprets the ascarosides produced by nematodes via the conserved pheromone signaling pathway in fungi, providing molecular insights into the mechanisms of communication between a fungal predator and its nematode prey.
Keywords: nematode-trapping fungi, Arthrobotrys oligospora, pheromone-response pathway, MAPK, predator–prey interaction
Introduction
The ability to sense and respond to the environment is essential for survival across the tree of life. For example, predators often display a swift change in behavior upon sensing prey. At the molecular level, cells sense and respond to the environment via surface receptors and signaling pathways that initiate transcriptional responses, which regulate downstream biological processes. Sensing environmental cues and responding accordingly by activating conserved signaling pathways is a common feature among organisms. Many fundamental processes in biology, such as sex and development, are coordinately regulated via conserved signaling pathways, and such systems have been well documented in fungi (Lengeler et al. 2000). Multiple fungal lineages have evolved to become predatory under certain environmental conditions. Amongst them, the nematode-trapping fungi (NTF) are a group of specialized predators that can sense, capture, and consume nematodes under nutritional deprivation (Nordbring-Hertz 2004; Jiang et al. 2017). The vegetative hyphae of NTF can differentiate into specialized mycelial structures known as “traps” that capture nematode prey. Although NTF develop various trap types, a common requirement for trap morphogenesis is the presence of biotic and abiotic factors related to nematodes and nutrients (Su et al. 2017). Generally, the presence of nematodes and a lack of nutrients can trigger trap formation (Scholler and Rubner 1994). Previous study has demonstrated that ascarosides, a highly conserved family of nematode-secreted pheromones, are sufficient to trigger trap morphogenesis in several species of NTF (Hsueh et al. 2013). Despite the identification of ascarosides as major cues inducing trap formation, it remains unclear how NTF sense these prey-derived signals.
Mitogen-activated protein kinase (MAPK) signaling cascades are essential regulators of cell differentiation and virulence in fungal pathogens of plants and animals (Hamel et al. 2012; Tong and Feng 2019). Studies of Saccharomyces cerevisiae budding yeast have paved the way for studies of MAPK signaling pathways in other eukaryotes (Chen and Thorner 2007). Three major MAPKs and corresponding cascades that regulate cell physiology in S. cerevisiae are Fus3/Kss1 (pheromone-response and filamentous growth pathway), Slt2 (cell wall integrity pathway), and Hog1 (hyperosmolarity pathway). The Fus3/Kss1 cascades of budding yeast are critical in responses to sex pheromones and nutrient deprivation (Madhani and Fink 1997). Conserved orthologs of yeast Fus3/Kss1 MAPKs are essential for differentiation in filamentous fungi, including mating, cell fusion, and pathogenesis (Widmann et al. 1999; Chen and Thorner 2007; Hamel et al. 2012). Furthermore, the MAK-2 MAPK cascade of Neurospora crassa is required for cell fusion and formation of protoperithecia (Li et al. 2005; Fu et al. 2011). In the phytopathogenic rice blast fungus Magnaporthe oryzae, Pmk1 MAPK plays a vital role in host-sensing, formation of the appressorium, and penetration of the plant cell cuticle (Xu and Hamer 1996; Sakulkoo et al. 2018). Similarly, deletion of Fmk1 MAPK in the plant fungal pathogen Fusarium oxysporum results in strains that fail to differentiate penetration hyphae (Di Pietro et al. 2004).
Following activation, MAPKs phosphorylate downstream substrates, including transcriptional factors with regulatory outputs Ste12 is the transcription factor acting downstream of the yeast Fus3/Kss1 MAPK cascades that control cellular differentiation, and conserved Ste12 orthologs are involved in the development and pathogenicity of multiple fungal systems (Chou et al. 2006; Wong Sak Hoi and Dumas 2010). The Ste12-like ortholog in N. crassa, PP-1, acts downstream of MAK-2 MAPK and is involved in protoperithecia development and hyphal fusion (Li et al. 2005; Fu et al. 2011). Mst12 is the Ste12-like transcription factor of rice blast fungi M. grisea, and its mutation results in defective appressoria formation and reduced virulence (Park et al. 2004).
Although the roles of the Fus3/Kss1 MAPK pathway and its downstream targets remain largely unexplored in NTF, a previous study has demonstrated that disruption of GPB1, the homolog of S. cerevisiae G-protein beta subunit STE4 that lies upstream of the Fus3/Kss1 MAPK signaling cascade, caused a strong defect in Arthrobotrys oligospora trap morphogenesis (Yang et al. 2020). This result suggests that the pheromone-response MAPK signaling might play a role in sensing nematode-derived cues, including ascarosides.
To test this hypothesis, we here investigate the roles of the MAPK cascade and the downstream transcription factor Ste12 for responses to nematode-derived cues, including ascarosides. Using a recently established reference strain of A. oligospora (Yang et al. 2020), we identified MAPK FUS3, its upstream kinase (MAPKK) STE7, and the transcription factor STE12 as key components of the signaling cascade downstream of trap morphogenesis and ascaroside perception in NTF. In addition, we found that Ste7, Fus3, and Ste12 are important for growth, but only the kinases Ste7 and Fus3 are required for conidiation, implying that Fus3 likely activates other downstream targets in addition to Ste12 to regulate conidiation. Furthermore, we have characterized the Ste12-dependent regulon upon nematode contact, and our transcriptomic analyses have revealed additional candidate genes that may be important for trap morphogenesis. Our study demonstrates that the conserved pheromone-response MAPK pathway functions in cross-kingdom pheromone sensing and predator–prey communication in the NTF A. oligospora.
Materials and methods
Strains and culture conditions
Fungal strains used in this study are listed in Supplementary Table S1. Arthrobotrys oligospora cultures were maintained on potato dextrose agar (PDA; Difco). All trap induction assays were performed on low-nutrient medium (LNM: 2% agar, 1.66 mM MgSO4, 5.4 µM ZnSO4, 2.6 µM MnSO4, 18.5 µM FeCl3, 13.4 mM KCl, 0.34 µM biotin, and 0.75 µM thiamin). Caenorhabditis elegans N2 strain was maintained on standard NGM with Escherichia coli (OP50) as the food source.
Phylogenetic analysis of MAPKs from A. oligospora
Phylogenetic relationships of MAPKs from A. oligospora and model fungi were assessed by aligning full-length protein sequences using ClustalW (KEGG IDs of sequences used are listed in Supplementary Table S2). A neighbor-joining phylogenetic tree of the MAPK sequences was created using Mega 7 (Kumar et al. 2016), with 3000 bootstrap replications to evaluate clade support. A hypothetical model of MAPK signaling in A. oligospora was illustrated in BioRender (https://biorender.com).
Phylogeny of A. oligospora and other fungal species was built based on nucleotide and protein sequence databases of INSDC and NCBI taxonomy using phyloT (https://phylot.biobyte.de). The following species and corresponding NCBI taxonomy IDs were used as input: A. oligospora (13349), S. cerevisiae (4932), Schizosaccharomyces pombe (4896), N. crassa (5141), F. oxysporum (5507), M. oryzae (318829), Aspergillus nidulans (162425), Cryptococcus neoformans (5207), and Ustilago maydis (5270).
Generation of targeted gene deletion mutants and complemented strains
Targeted gene deletions in the A. oligospora TWF154 strain were carried out using a homologous recombination strategy (Yang et al. 2020). We fused 1.5–2 kb of the 5′ and 3′ sequences flanking the open reading frame of the target gene to a drug marker by means of overlap PCR. Three drug resistance markers were used in this study: the hygromycin-B resistance cassette from vector pAN7-1 (Punt et al. 1987), the clonNAT resistance cassette of pRS41N (Chee and Haase 2012), and the G418 resistance cassette of pUMa1057 (Youssar et al. 2019). The gene deletion constructs were introduced into protoplasts of either wild type or the ku70 background strain of TWF154 via PEG-mediated transformation. For generation of protoplasts in A. oligospora, 5 × 106 conidia were cultured in 100 ml of potato dextrose broth (PDB) for 48 h at 25 °C, 200 rpm. Mycelia were washed with MN buffer (0.3 M MgSO4, 0.3 M NaCl), mixed with 60 mg/ml VinoTastePro lytic enzyme in MN buffer, and incubated for 5–6 h at 25 °C, 200 rpm. Protoplasts were collected by filtering through two layers of miracloth and washed with STC buffer (1.2 M sorbitol, 50 mM CaCl2, 10 mM Tris–HCl pH 7.5). For transformation, 1 × 106 protoplasts were mixed with 5 µg of construct DNA and incubated on ice for 30 min, after which five volumes of PTC buffer (40% PEG 4000 [w/v], 10 mM Tris–HCl pH 7.5, 50 mM CaCl2) was added and placed at room temperature for 20 min. Then, molten regeneration agar (1% agar, 0.5 M sucrose, 0.3% yeast extract [w/v], 0.3% casein acid hydrolysate [w/v]) containing 100 µg/ml hygromycin B, 200 µg/ml clonNAT, or 350 µg/ml G418 was added to the protoplasts and poured into Petri dishes. Resistant transformants were then screened for gene deletion. Primers used for targeted gene deletion and genotyping are listed in Supplementary Table S3.
For gene complementation, a wild-type copy of the target gene was amplified (2 kb upstream and 1 kb downstream of the open reading frame), fused with a drug marker, and transformed into protoplasts of mutant strains. The resulting complemented strains were genotyped to confirm gene rescue.
CRISPR-Cas9-mediated gene deletion in A. oligospora
We employed in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with homologous repair template (Al Abdallah et al. 2017) to achieve targeted deletion of MAPK SLT2. Two single-guide RNAs targeting the 5′ and 3′ ends of the SLT2 (EYR41_004559) open reading frame were generated, and the repair template was built by fusing 1.5 kb of the 5′ and 3′ flanking sequences of the SLT2 ORF to the hygromycin-B resistance cassette. The Cas9-sgRNA ribonucleoproteins were assembled by mixing 10 µl of Cas9 MgCl2 buffer (20 mM HEPES, 0.15 M KCl pH7.5, 10% glucose [w/v], 10 mM MgCl2, 1 mM β-mercaptoethanol, 1 mM DTT), 5 µl of 5 µM sgRNA, and 5 µl of 5 µM Cas9 protein, and then incubating the mixture at 37 °C for 10 min. Transformations of A. oligospora protoplasts were carried out as described in “Generation of targeted gene deletion mutants and complemented strains,” by adding two Cas9-sgRNA ribonucleoproteins and repair template to protoplasts. The transformants were screened by PCR and Southern blotting to confirm STL2 deletion.
Southern blot
Digoxigenin (DIG)-based Southern blots were carried out to examine events of ectopic integrations. Genomic DNA was acquired using a CTAB-PVP extraction buffer (Barbier et al. 2019), followed by chloroform purification and column clean up (DNA Clean and Concentrator Kits, Zyom Research). Digested gDNA was separated in a TAE agarose gel and transferred to a nylon membrane. DIG-labeled probes were generated using direct PCR labeling (PCR DIG Probe Synthesis Kit, Roche). Probe hybridization was carried out in hybridization buffer (0.5 M Na-phosphate buffer pH7, 7% [w/v] SDS) followed by subsequent washes. The membrane was then hybridized with an anti-DIG antibody (Anti-Digoxigenin-AP, Fab fragments, Roche), and detection of DIG-labeled DNA was accomplished using CDP-Star solution (CDP-Star, Roche).
Evaluation of trap morphogenesis in response to C. elegans
To quantify the number of fungal traps induced by nematodes, A. oligospora was cultured on an LNM plate (2.5 cm) at 25 °C for 2 days. Then, 30 synchronized wild-type L4-stage C. elegans nematodes were added to the plates and removed 6 h later. Twenty-four hours after nematode induction, plates were observed under a dissecting microscope and three images were randomly taken within 0.5 cm from the edge of the plate at 40× magnification. Traps in those images were quantified using “Fungal Feature Tracker” (Vidal-Diez de Ulzurrun et al. 2019), and trap numbers from all three images of each plate were summed.
For traps induced by ascarosides, fungal mycelium was cultured on an LNM plate (5 cm) for 3 days. One microliter of 1 mM ascaroside ascr#18 solution (Zhang et al. 2017) was dropped on the mycelium at a marked position (∼0.5 cm from the edge of mycelium distal tips). After 24 h, traps within a 1.5-cm radius of the site of the ascaroside drop were imaged.
Total RNA isolation, RNA-seq library preparation, and data analysis
Arthrobotrys oligospora ku70 and ste12 strains were cultured on LNM agar plates (9 cm) for 5 days at 25 °C. Then, 2000 L4-stage N2-strain C. elegans were placed on each plate to induce trap morphogenesis. The nematodes were washed away after 4 h with deionized water. The fungal cultures were immediately harvested with a cell scraper and frozen in liquid nitrogen, followed by lyophilization. Total RNA was extracted using a Trizol-based protocol (Schumann et al. 2013), treated with Turbo DNase, and cleaned by ethanol precipitation. RNA quality and quantity were determined using Bioanalyzer and Qubit systems, respectively.
Library preparation for total RNA samples was carried out by the Genomics Core Facility (Institute of Molecular Biology, Academia Sinica) using an Illumina TruSeq stranded mRNA library preparation kit. Generated libraries were sequenced according to 150-bp paired-end sequencing protocols on an Illumina NovaSeq 6000 system platform. The read library was converted into FASTQ format using the Illumina package Bcl2fastq v2.20.
Paired-end raw reads were first subjected to adapter trimming and quality filtering using Fastp (Chen et al. 2018). The Salmon program (Patro et al. 2017) was then used for read mapping and quantification against a reference transcriptome of A. oligospora TWF154 (Yang et al. 2020). The transcript abundance estimates from Salmon were converted to a Sleuth-compatible format using the Wasabi package, and transcript differential expression was assessed using Sleuth (Pimentel et al. 2017). First, a Sleuth object was created using the transcript estimates from Salmon, before computing a reduced and full model. Then, likelihood ratio and Wald tests were performed to establish overall and differential expression for each transcript. For a transcript to be considered differentially expressed, the absolute |betas| value (approximately equivalent to log2 fold-change) had to be >2, and the adjusted P-value <0.05. MA plots depicting betas for genome-wide transcripts among conditions were drawn using scatterplot.online (https://scatterplot.online). Gene expression data are available in Gene Expression Ommibus (GEO) with accession number GSE159974.
Gene ontology enrichment analysis was carried out in Blast2GO 5 PRO (Conesa et al. 2005) with Fisher’s exact test using the InterPro GO IDs (filter criterion: P-value <0.05). Transcripts without annotation were excluded from enrichment analysis to enable more accurate predictions of significant GO terms.
Protein extraction and Western blot
The procedures for fungal culture, nematode induction, sample harvesting, and sample lyophilization are identical to those described above in “Total RNA isolation, RNA-seq library preparation, and data analysis.” A TCA-acetone precipitation method was employed for protein extraction. Mycelial powder was resuspended in 20% (w/v) trichloroacetic acid (TCA) in acetone containing 0.75% (v/v) β-mercaptoethanol and precipitated for 1 h at −20 °C, followed by centrifugation (15,000 × g, 4 °C, 15 min). Protein pellets were washed with acetone containing 0.75% (v/v) β-mercaptoethanol and resuspended in HU loading buffer (8 M Urea, 5% SDS; 0.2 M Tris–HCl (pH 6.5); 1 mM EDTA; 0.01% bromophenol blue) supplemented with 6% (v/v) 2 M Tris and 5% (v/v) β-mercaptoethanol for loading in SDS-PAGE (GenScript, SurePAGE).
Protein on gels was transferred to PVDF membranes and blocked in TBST with 1% (w/v) casein. Membranes were then hybridized with phospho-p44/42 MAPK antibody (Cell Signaling Technology, #9101) and tubulin antibody (Abcam, ab184970) diluted in blocking solution. After washes with TBST, membranes were then incubated with AffiniPure goat anti-rabbit IgG H+L (Jackson ImmunoResearch, AB_2337913) for 1 h. For signal detection, membranes were incubated with ECL solution (Bio-Rad, Clarity Western ECL substrate) and subjected to standard film-developing procedures.
Data availability
Strains and plasmids are available upon request. RNA-seq raw data (.fastq) and differential expression analysis outputs are available at GEO with the accession number GES159974.
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13225307.
Results
Identification and phylogenetic analysis of mitogen-activated protein kinases in Arthrobotrys oligospora
In order to establish which MAPKs are encoded in the genome of A. oligospora, we conducted a BLAST analysis of full-length Fus3, Kss1, Slt2, Hog1, and Ime2 MAPK protein sequences from S. cerevisiae and respective orthologs in N. crassa, F. oxysporum, M. oryzae, and C. neoformans against the genome of A. oligospora strain TWF154 using BLASTP in the Blast2GO software (Conesa et al. 2005). We identified A. oligospora homologs for all these MAPKs and corresponding components in their respective signaling cascades (Supplementary Figure S1A). A phylogenetic analysis of the A. oligospora MAPKs and their orthologs in model fungi revealed segregation into well-established clades (Supplementary Figure S1B). The segregation of A. oligospora MAPKs from other filamentous ascomycetes (N. crassa, F. oxysporum, and M. oryzae) is consistent with phylogeny based on NCBI genome database (Supplementary Figure S1C).
Disruption of core components of the pheromone signaling MAPK pathway leads to defects in growth, conidiation, and trap morphogenesis in A. oligospora
To explore the roles of the pheromone-signaling MAPK cascade during prey sensing in A. oligospora, we generated targeted gene deletion mutants of STE7 (EYR41_011623) and FUS3 (EYR41_011934). Gene deletion in A. oligospora is inefficient due to its low rate of homologous recombination (Yang et al. 2020). To enhance the recombination rate, we used a non-homologous end-joining-deficient ku70 strain as the recipient strain for several gene deletion mutants (Kuo, Chen, et al. 2020). Although we also attempted targeted gene deletion utilizing CRISPR-Cas9 ribonucleoproteins coupled with a homologous repair template (Supplementary Figure S2A), the deletion strain we obtained contained multiple ectopic integrations of the repair DNA (Supplementary Figure S2B). We therefore used the ku70 strain to obtain gene deletion mutants of interest in this study. Southern blots were performed to confirm targeted gene deletions and lack of ectopic integrations in the mutant strains (Supplementary Figure S2, C–F).
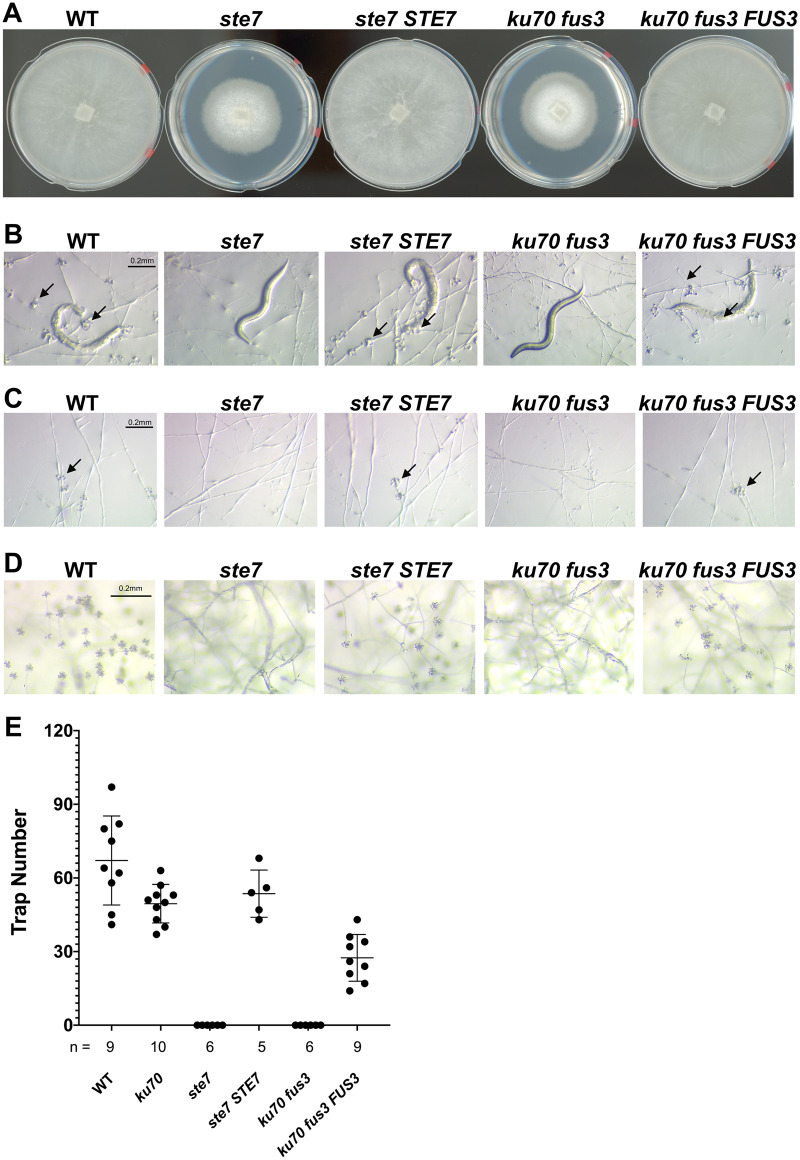
Deletion of the MAPKK STE7 and MAPK FUS3 resulted in slow growth of vegetative hyphae, indicating that the Fus3 MAPK cascade plays critical roles in physiological growth of A. oligospora (Figure 1A). Furthermore, both mutants were defective in trap development when they encountered the nematode C. elegans. In contrast to the wild-type strain, which had developed numerous traps and consumed the nematode prey within 24 h, no traps were developed in the ste7 and fus3 mutants when exposed to nematodes (Figure 1B). To examine whether the defect in trap formation is due to slow growth, we incubated the ste7 and fus3 strains in the presence of more than 1000 C. elegans for a week. No traps were developed in the ste7 and fus3 mutant plates, demonstrating that the defects in trap morphogenesis cannot be attributed to slow growth. Moreover, the ste7 and fus3 mutants did not respond to ascr#18 (Choe et al. 2012), a conserved nematode pheromone that is a potent trap-inducing molecule (Figure 1C). Together, these observations demonstrate that the Fus3 MAPK cascade is essential for prey-sensing and trap morphogenesis in A. oligospora. One additional prominent phenotypic defect shared by the ste7 and fus3 mutants was the absence of conidiation. In contrast to wild-type colonies, which produced abundant conidia when grown on nutrient-rich PDA medium, conidiation was completely abolished in the ste7 and fus3 mutants, indicating that the pheromone-response MAPK pathway is essential for asexual sporulation (Figure 1D). Complementation of the mutants with the wild-type alleles of STE7 or FUS3, respectively, rescued these phenotypic defects (Figure 1, A–E). Collectively, our results indicate that disruption of the conserved pheromone-response MAPK signaling cascade induces defects in physiological growth, trap morphogenesis, and conidiation in A. oligospora.
Figure 1.
Disruption of the pheromone-response MAPK pathway leads to defects in growth, conidiation, and trap morphogenesis in A. oligospora. (A) Colony morphologies of WT, ste7, ku70 fus3, and complemented strains after incubation for 4 days at 25 °C on PDA plates (5 cm diameter). (B) Trap morphologies of WT, mutants, and complemented strains. Trap formation was induced by adding 30 C. elegans nematodes to fungal culture on LNM plates (2.5 cm), and images were taken 24 h after induction. The arrows indicate traps produced by A. oligospora. (C) Trap morphogenesis induced by the addition of ascr#18 (1 mM, 1 µl). Images were taken 24 h after induction. The arrows indicate traps produced by A. oligospora. (D) Conidiation of mutant and complemented strains were recorded after culturing for 4 days on PDA plates. (E) Quantification of trap numbers among mutants and complemented strains that were induced by exposure to 30 C. elegans nematodes (mean ± SEM; n shown along the x-axis).
As disruption of the Fus3 MAPK cascade resulted in defective trap formation, we hypothesized that Fus3 is activated by phosphorylation upon nematode contact (Hamel et al. 2012). To explore the phosphorylation state of Fus3 in response to C. elegans, we used a phospho-p44/42 MAPK antibody that has been demonstrated to detect phosphorylation of both the Slt2 (cell wall integrity) and Fus3 (pheromone response) MAPKs in several model fungi (Lev et al. 2009; Kamei et al. 2016; Li, Gao, et al. 2017). We have previously reported that A. oligospora initiates trap formation ∼2.5 h after being exposed to nematodes and that complete adhesive networks (traps) forms ∼12 h post-induction (Yang et al. 2020). To monitor the phosphorylation dynamics of Fus3 throughout trap morphogenesis, we conducted a time-course assessment of A. oligospora mycelium that had been exposed to nematodes using the phospho-p44/42 MAPK antibody in Western blotting. Fus3 was phosphorylated during the initial stages (2 and 4 h time-points) of nematode contact, but the signal had disappeared at 24 h post-induction (Figure 2A) when all captured nematodes had been partially or completely digested by the fungal hyphae. These observations reveal that nematode signals indeed activate the Fus3 MAPK pathway in A. oligospora.
Figure 2.
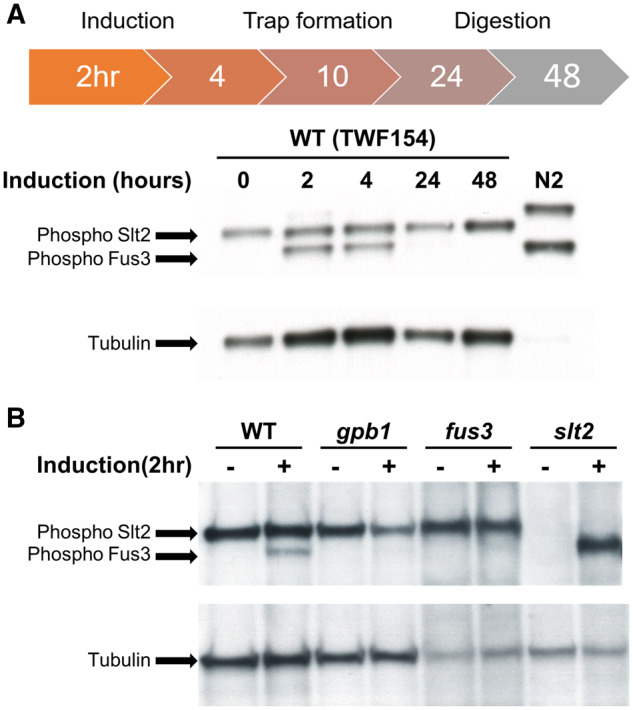
Fus3 MAPK is phosphorylated upon nematode induction in A. oligospora. (A) Time-course phosphorylation status of Slt2 and Fus3 MAPKs in A. oligospora upon nematode induction. Mycelia were cultivated on LNM agar plates for 7 days at 25 °C, and then exposed to C. elegans for 2, 4, 24, and 48 h. Protein extraction and Western blot were then performed on harvested fungal cultures using a phospho-p44/42 MAPK antibody. Loading control: anti-tubulin. N2: C. elegans control. (B) Phosphorylation status of Slt2 and Fus3 MAPKs in different mutant backgrounds of A. oligospora. Fungal mycelia were exposed to C. elegans N2 for 2 h and then collected for protein extraction. −: untreated control. +: nematode-treated.
The gpb1 mutant of A. oligospora is highly defective in trap formation (Yang et al. 2020), and the respective homologous protein in the budding yeast S. cerevisiae, Ste4, operates upstream of Fus3 in that MAPK cascade. Accordingly, we speculated that Gpb1 may also act upstream of Fus3 in A. oligospora and that Fus3 activation may be diminished or absent in the gpb1 mutant. Thus, we examined the phosphorylation states of Fus3 and Slt2 MAPKs in different mutant backgrounds and observed that both the gpb1 and fus3 mutants lacked apparent Fus3 phosphorylation signal upon nematode induction, whereas Slt2 was consistently phosphorylated in the wild type, gpb1, and fus3 mutants (Figure 2B). The slt2 deletion mutant of A. oligospora displayed severe defects in vegetative growth, conidiation, and trap morphogenesis (Supplementary Figure S3). These findings indicate that nematode presence induces phosphorylation of Fus3 in A. oligospora, whereas Slt2 MAPK appears to be constitutively active. As Fus3 was still phosphorylated with the presence of nematodes in slt2 background, we examined whether slt2 mutant was still able to develop traps after prolonged incubation with C. elegans. Indeed, in line with this evidence, we observed that slt2 mutant could develop traps after 3 days of nematode exposure, suggesting that Slt2 MAPK is not essential for trap development.
STE12 is required for proper trap development in A. oligospora
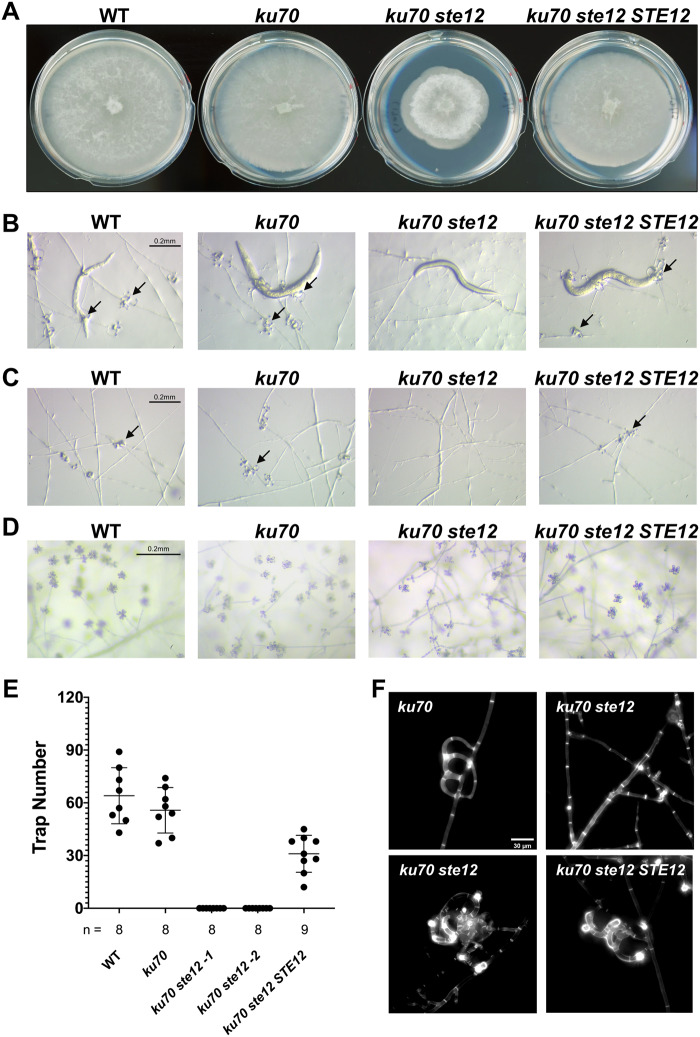
The Ste12 transcription factor and its orthologs play key roles in cellular differentiation and act downstream of the Fus3 MAPK cascade in budding yeasts and filamentous fungi (Wong Sak Hoi and Dumas 2010). Hence, we hypothesized that A. oligospora Ste12 could be a downstream target of Fus3 to orchestrate transcriptional regulation during trap morphogenesis. We characterized the roles of the A. oligospora STE12 ortholog (EYR41_007450) by generating a targeted deletion mutant. The corresponding ku70 ste12 mutant displayed reduced growth without an obvious defect in the formation of aerial hyphae (Figure 3A). In support of our hypothesis, the ste12 mutant did not form traps when exposed to nematodes or ascr#18 (Figure 3, B, C, and E). After prolonged incubation in the presence of nematodes, ste12 mutant developed a few traps with abnormal morphology (Figure 3F, lower left image), whereas majority of the hyphae remained undifferentiated (Figure 3F, top right image). We were able to rescue this defect in trap morphogenesis by introducing the wild-type STE12 allele into the ste12 mutant (Figure 3, A–F). Intriguingly, conidiation was not affected in the ste12 mutant, generating conidia when cultured on PDA medium just like the wild-type strain (Figure 3D), suggesting that Fus3 activates additional downstream targets other than Ste12 to regulate conidiation in A. oligospora (Figure 3D). Together, these results support our model in which the pheromone-response MAPK pathway activates the downstream transcription factor Ste12 to regulate expression of “nematode-responsive genes” that trigger trap morphogenesis in A. oligospora.
Figure 3.
The Ste12 transcription factor is required for proper and rapid trap formation. (A) Colony morphologies of WT, ku70, ku70 ste12, and a complemented strain (ku70 ste12 STE12) after incubation for 4 days at 25 °C on PDA plates (5 cm diameter). (B) Trap morphologies of the WT, ku70, ku70 ste12, and the complemented strain after 24 h of exposure to 30 C. elegans nematodes for fungal cultures grown on LNM plates (2.5 cm). The arrows indicate traps produced by A. oligospora. (C) Trap morphologies after 24 h of induction by ascr#18 (1 mM, 1 µl). The arrows indicate traps produced by A. oligospora. (D) Conidiation of ste12 mutant and the complemented strain after 4 days of culture on PDA plates. (E) Quantification of trap number induced by presence of 30 C. elegans nematodes for WT, ku70, two independent ku70 ste12 mutants, and the STE12 complemented strain (mean ± SEM; n shown along the x-axis). (F) Images of traps formed by the ku70, ku70 ste12, and ku70 ste12 STE12 strains after 30 h of continuous induction with 50–60 nematodes. Vegetative hyphae and traps of A. oligospora are stained with SR2200, which specifically binds to fungal cell walls.
Transcriptional profiling of A. oligospora upon nematode contact reveals a set of Ste12-dependent nematode-responsive genes with various functions
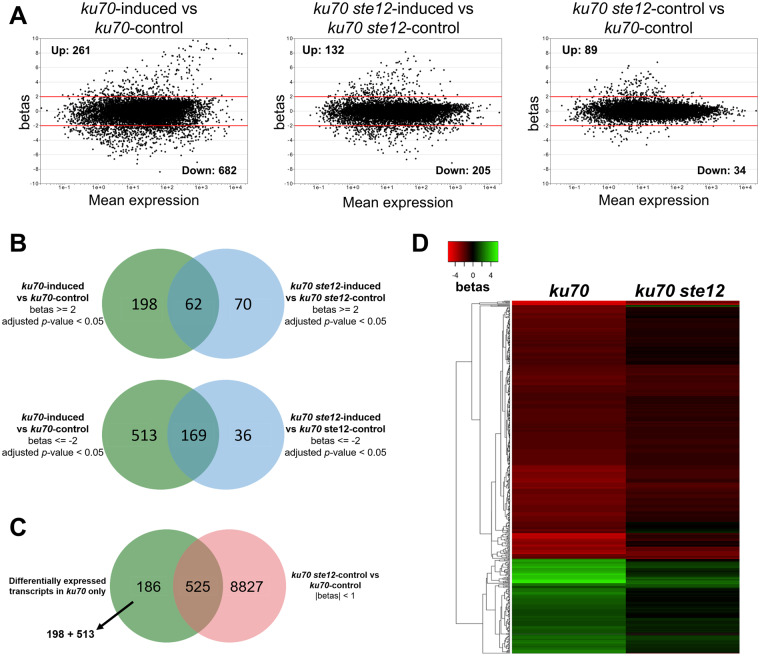
To identify the potential Ste12-dependent nematode-responsive genes, we conducted RNA-seq analyses to examine the gene expression profiles of the ku70 and the ku70 ste12 mutants in the absence or presence of C. elegans. Total RNAs were extracted from ku70 and ku70 ste12 mutants that had been cultured alone or upon exposure to C. elegans for 4 h, coinciding with a peak in trap initiation, and then subjected to RNA-seq analyses. A principal component analysis revealed that the nematode-induced transcriptomes of ku70 were well separated from the non-induced controls (Supplementary Figure S4A), whereas the transcriptomes of the nematode-exposed and non-exposed ku70 ste12 mutants were much closer to each other, implying that transcriptional response to nematodes among the mutants were less pronounced relative to the wild type. To identify the Ste12-dependent nematode-responsive genes, we performed the following analyses. First, we examined differential gene expression upon nematode induction in the ku70 line that had a |betas| value (approximately equal to fold-change) ≥2 and identified a total of 261 upregulated and 682 downregulated transcripts (Figure 4A: left panel). Next, we used the same criteria to identify the differentially expressed transcripts in the ste12 mutant in response to the presence of C. elegans and found that only 132 and 205 transcripts were up- or downregulated, respectively (Figure 4A: middle panel). In addition, we compared the ku70 and ste12 mutants under non-induced conditions (Figure 4A: right panel) and observed that only 0.93% of all transcripts (123 out of 13,142; 89 upregulated and 34 downregulated) were differentially expressed. Next, we compared the differentially expressed transcripts between the ku70 and ku70 ste12 mutants and identified common genes that were differentially expressed in both strain backgrounds. In total, 62 and 169 transcripts were commonly up- or downregulated, respectively, in both the ku70 and the ku70 ste12 mutants, indicating that these genes are regulated independently of Ste12 (Figure 4B). Therefore, we considered the 711 transcripts that were only differentially expressed in the ku70 line but not in the ku70 ste12 mutant to be candidate Ste12-dependent nematode-responsive genes (Figure 4B). From the 711 transcripts, we identified those exhibiting similar expression between the ku70 and ku70 ste12 lines in the absence of nematodes and defined this set of 525 transcripts (accounting for ∼4% of the A. oligospora genome) as “Ste12-dependent nematode-responsive genes” or the “Ste12-dependent regulon” (Figure 4C, Supplementary Table S4).
Figure 4.
Transcriptional profiling to identify Ste12-dependent genes differentially regulated upon nematode induction. (A) MA plots of genome-wide RNA-seq data for different comparisons. The y-axis represents the |betas| value (approximately equivalent to log2 fold-change) between the indicated strains and corresponding conditions. The x-axis represents the level of expression (transcripts per million, tpm) for each gene averaged between the two conditions. Threshold of differential expression: |betas| ≥ 2, adjusted P-value < 0.05. (B) Venn diagrams displaying the overlap of genes that were differentially expressed in both the ku70 and ku70 ste12 lines in response to nematodes. Upper diagram: overlapping upregulated genes. Lower diagram: overlapping downregulated genes. (C) Venn-diagram illustrating the Ste12-dependent regulon upon nematode induction. The green circle includes transcripts significantly up- and downregulated (|betas| ≥2, adjusted P-value < 0.05) only in the ku70 line upon nematode induction (from Figure 4B). The red circle represents transcripts not differentially expressed (|betas| < 1) for the ku70 ste12-control vs ku70-control comparison. (D) Heatmap displaying the Ste12-dependent regulon in terms of expression |betas| values.
We found that the 141 upregulated and 384 downregulated transcripts of the Ste12-dependent regulon remained unchanged in the ste12 mutant background (Figure 4D, left panel). GO enrichment analysis of the Ste12-dependent regulon revealed “integral component of membrane” and “oxidation–reduction process” as two of the most significant terms (Supplementary Figure S4B). The “integral component of membrane” group consists of transporters for ions, sugars, and amino acids, among others. The “oxidation–reduction process” group comprises genes involved in various biological processes, including polyketide synthase (EYR41_000001), lipoxygenase (EYR41_002416), alcohol dehydrogenases (EYR41_000524, EYR41_008901, EYR41_010593), and NOX enzyme (EYR41_011461).
A notable feature of the Ste12-dependent regulon is the high percentage of transcripts that only seem to exist in NTF. Of the 525 transcripts of this regulon, we discovered through protein BLAST analyses that 231 (44%, 231 out of 525) of them generated significant hits only in NTF (Supplementary Figure S4C). Upon further investigation, these NTF-predominant transcripts may provide profound insights into the mechanism of trap morphogenesis and the switch to a predatory lifestyle of NTF. Many of these NTF-specific genes that our analysis identified lack annotations or well-defined functional domains. Among the 231 NTF-specific genes of the Ste12-dependent regulon, 186 lack functional descriptions. This result suggests that activation of the Ste12 transcription factor in A. oligospora by nematodes orchestrates transcription of nematode-responsive genes with diverse functions that might play critical roles in NTF’s predatory lifestyle.
Discussion
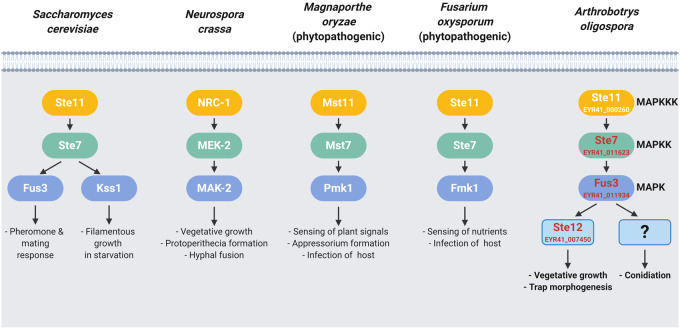
The development of specialized hyphal structures that function as traps to capture nematodes is an iconic feature of NTF. The molecular mechanisms underlying trap morphogenesis have remained largely unknown. Our data show that kinases and a transcription factor within the conserved pheromone-response MAPK pathway of A. oligospora have significant roles in vegetative growth, conidiation, and trap development. In the budding yeast S. cerevisiae, the Fus3 MAPK cascade is essential for mating morphogenesis in response to sex pheromones; whereas, the paralogous Kss1 MAPK is critical for filamentous growth in response to starvation (Bahn et al. 2007). The conserved pheromone-signaling MAPK pathway is crucial for pheromone sensing and sexual development in the Basidiomycete C. neoformans and U. maydis (Davidson et al. 2003; Müller et al. 2003), but in the filamentous Ascomycetes, this pathway play additional roles and might not directly involved in pheromone signaling (Figure 5). For instance, in N. crassa, the MAK-2 MAPK cascade controls the formation of protoperithecia (female reproductive structures), but it does not regulate pheromone sensing (Li et al. 2005). In the rice blast fungus M. oryzae, the Pmk1 MAPK stimulates appressorium formation in response to plant signals (Zhao et al. 2005; Liu et al. 2011), but its role during the sexual cycle remains unclear. For the fungal pathogen F. oxysporum, sensing of sex pheromones is mediated by the cell wall integrity Mpk1 MAPK; the Fus3/Kss1 ortholog, Fmk1, controls F. oxysporum chemotropism toward nutrients (Turra et al. 2015). We have shown that the Fus3 MAPK cascade of A. oligospora is essential for trap morphogenesis and virulence (Figure 1). However, whether the same MAPK cascade is involved in sex pheromone recognition or sexual development in NTF remains to be elucidated.
Figure 5.
The pheromone-response MAPK pathway is essential for trap morphogenesis in the nematode-trapping fungus A. oligospora. Different roles of Fus3/Kss1 MAPK signaling cascades in A. oligospora and other Ascomycetes. In A. oligospora, nematode-derived cues activate the Fus3 MAPK signaling cascade to turn on the trap morphogenesis via the conserved downstream transcription factor Ste12. The Fus3 MAPK signaling also regulates vegetative growth and conidiation in A. oligospora.
Trap formation is a complex morphogenesis process, which likely requires the coordination of multiple signaling pathways. Previous finding has shown that Slt2 MAPK of the cell wall integrity pathway is required for trap morphogenesis of A. oligospora (Zhen et al. 2018). However, we demonstrated that Slt2 is not essential for trap development. Our group recently reported that the conserved high osmolarity pathway mediated by Hog1 MAPK plays a minor role in predation of A. oligospora; the hog1 mutant has reduced trap number and predation efficiency, but this MAPK is dispensable for trap morphogenesis (Kuo, Chen, et al. 2020).
Ascarosides, a group of conserved nematode pheromones, can trigger trap differentiation in several species of NTF (Choe et al. 2012; Hsueh et al. 2013). Trap induction by ascarosides thus represents an example for a cross-kingdom interaction based on recognition of a conserved molecular signature. In another cross-kingdom interaction, ascarosides have been shown to activate MAPK pathways and to trigger an immune response in Arabidopsis thaliana (Manosalva et al. 2015). Our results demonstrate that ascaroside responses in both plants and fungi depend on MAPK signaling.
One attractive hypothesis is that GPCRs and G-protein signaling act upstream of the Fus3 MAPK cascade in NTF. In support of this possibility, recent studies have reported that GPCRs are critical for the pathogenesis of multiple phytopathogenic fungi, such as F. graminearum (Dilks et al. 2019) and M. oryzae (Kou et al. 2017). Here, we have revealed that a gpb1 mutant strain is unable to induce Fus3 phosphorylation (Figure 2B) and that both gpb1 and fus3 mutant lines are defective in trap formation. However, the gpb1 mutant did not exhibit slow growth or impaired conidiation, suggesting that Fus3 plays a broader role in the physiology of the cell, so that when disrupted, resulted in pleiotropic phenotypes (Yang et al. 2020).
Apart from the Fus3 MAPK cascade, we also explored the functions of the transcription factor Ste12 in A. oligospora. Ste12-like orthologs in plant-pathogenic fungi display conserved roles in virulence and mostly act downstream of Fus3/Kss1-type MAPKs (Wong Sak Hoi and Dumas 2010). In several fungal pathogens, Ste12 is involved in only some MAPK-regulated functions. For example, MST12 is the STE12 ortholog of M. oryzae, and the mst12 mutant generates appressoria that cannot penetrate plant hosts, whereas the upstream kss1 MAPK mutant completely lacks appressoria (Park et al. 2004). Similarly, we found that the fus3 mutant of A. oligospora cannot form traps, whereas the ste12 mutant could still form a few abnormal traps after prolonged exposure to C. elegans (Figure 3F). Moreover, we have shown that conidiation is not regulated by Ste12 but by other putative components downstream of Fus3. Importantly, our ste12 mutant exhibits impaired trap morphogenesis, and the Ste12-dependent regulon provides clues to additional molecular players in the differentiation process. Intriguingly, we identified two transcripts encoding NOX enzymes as being Ste12-dependent (Supplementary Table S4; NoxR, EYR41_003117 and Nox2, EYR41_011461), with expression of both being highly induced in ku70 but not in the ste12 mutant. NOX enzymes are essential for the production of reactive oxygen species (ROS), and they have been linked to growth, cell fusion, asexual development, and pathogenicity in fungi (Cano-Domínguez et al. 2008; Roca et al. 2012; Dirschnabel et al. 2014; Rossi et al. 2017; Zhou et al. 2017). Magnaporthe oryzae mutant lines of NOX enzymes can form appressoria but fail to penetrate host plant surfaces (Egan et al. 2007). In N. crassa, the NOX enzymes NOX-1 and NOR-1 are regulated by the Ste12 homolog PP-1, and they are essential for sexual differentiation and hyphal fusion (Cano-Domínguez et al. 2008; Leeder et al. 2013). It has been demonstrated previously that NoxA is required for ROS production and trap formation in A. oligospora (Li, Kang, et al. 2017), although we did not find NoxA being upregulated during prey sensing in our transcriptomic analysis. It is likely that the regulatory output of Fus3 and Ste12 leads to activation of NOX enzymes and consequent ROS production, which act as secondary messengers to induce trap development and hyphal fusion. Other genes identified from our Ste12-dependent regulon include a putative polyketide synthase (Supplementary Table S4; EYR41_000001). Polyketide secondary metabolites have long been hypothesized as nematotoxins in NTF (Olthof and Estey 1963). Despite recent metabolomics studies of A. oligospora (Wang et al. 2018; Kuo, Yang, et al. 2020), little is known about the functions of its secreted compounds. Upregulation of polyketide synthases may contribute to the production of virulence factors that are crucial to trap development or nematicidal activity. Other targets of Ste12 could be coupled to cell cycle progression. The yeast autophagy regulatory protein Atg2 is required for vesicle formation during autophagy under starvation (Wang et al. 2001), and we observed the corresponding A. oligospora homolog EYR41_001564 being downregulated in response to nematode presence, indicating that it may play a role in exiting starvation and entering active cell cycle.
In conclusion, our results demonstrate that the pheromone-response MAPK pathway acts as a central regulator for physiological growth, conidiation, and the cross-kingdom prey sensing that induces trap morphogenesis in A. oligospora (Figure 5). Genes regulated by the Ste12 transcription factor provide insights into the molecular switches and downstream players controlling trap formation in NTF. Elucidating the molecular mechanisms that control trap differentiation will not only develop our understanding of how fungi and nematodes have coevolved but may also inform strategies for genetically engineering these predatory fungi to tackle parasitic nematodes and enable development of novel biological control agents.
Acknowledgments
We are grateful to Steve Lin from Institute of Biological Chemistry, Academia Sinica, and Tsui-Fen Chou from Caltech for providing the Cas9 proteins. We thank the IMB Genomics core for conducting Illumina sequencing and the Transgenic core facility for gRNAs synthesis. We thank Meng-Chao Yao, Pedro Gonçalves, and John O’Brien for their comments on this article and Paul Sternberg for his supports and encouragements.
Funding
This work was supported by Taiwan Ministry of Science and Technology (Grant Nos. 106-2311-B-001-039-MY3 and 109-2311-B-001-023) to Y.P.H.
Conflicts of interest
None declared.
Literature cited
- Al Abdallah Q, Ge W, Fortwendel JR. 2017. A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere. 2:e00446-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J, et al. 2007. Sensing the environment: lessons from fungi. Nat Rev Microbiol. 5:57–69. [DOI] [PubMed] [Google Scholar]
- Barbier FF, Chabikwa TG, Ahsan MU, Cook SE, Powell R, et al. 2019. A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods. 15:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Domínguez N, Álvarez-Delfín K, Hansberg W, Aguirre J, 2008. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell. 7:1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee MK, Haase SB. 2012. New and redesigned pRS plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3 (Bethesda). 2:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RE, Thorner J. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1773:1311–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe A, von Reuss SH, Kogan D, Gasser RB, Platzer EG, et al. 2012. Ascaroside signaling is widely conserved among nematodes. Curr Biol. 22:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Lane S, Liu H. 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 26:4794–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 49:469–485. [DOI] [PubMed] [Google Scholar]
- Di Pietro A, García-MacEira FI, Méglecz E, Roncero MI. 2004. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol Microbiol. 39:1140–1152. [PubMed] [Google Scholar]
- Dilks T, Halsey K, De Vos RP, Hammond-Kosack KE, Brown NA. 2019. Non-canonical fungal G-protein coupled receptors promote Fusarium head blight on wheat. PLoS Pathog. 15:e1007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirschnabel DE, Nowrousian M, Cano-Domínguez N, Aguirre J, Teichert I, et al. 2014. New insights into the roles of NADPH oxidases in sexual development and ascospore germination in Sordaria macrospora. Genetics. 196:729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Wang ZY, Jones MA, Smirnoff N, Talbot NJ. 2007. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci U S A. 104:11772–11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Iyer P, Herkal A, Abdullah J, Stout A, et al. 2011. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot Cell. 10:1100–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Duplessis S, Ellis BE. 2012. Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell. 24:1327–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Mahanti P, Schroeder FC, Sternberg PW. 2013. Nematode-trapping fungi eavesdrop on nematode pheromones. Curr Biol. 23:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Xiang M, Liu X. 2017. Nematode-trapping fungi. Microbiol Spectr. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei M, Yamashita K, Takahashi M, Fukumori F, Ichiishi A, et al. 2016. Involvement of MAK-1 and MAK-2 MAP kinases in cell wall integrity in Neurospora crassa. Biosci Biotechnol Biochem. 80:1843–1852. [DOI] [PubMed] [Google Scholar]
- Kou Y, Tan YH, Ramanujam R, Naqvi NI. 2017. Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 214:330–342. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CY, Chen SA, Hsueh YP. 2020. The high osmolarity glycerol (HOG) pathway functions in osmosensing, trap morphogenesis and conidiation of the nematode-trapping fungus Arthrobotrys oligospora. J Fungi (Basel). 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo TH, Yang CT, Chang HY, Hsueh YP, Hsu CC. 2020. Nematode-trapping fungi produce diverse metabolites during predator-prey interaction. Metabolites. 10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeder AC, Jonkers W, Li J, Glass NL. 2013. Early colony establishment in Neurospora crassa requires a MAP kinase regulatory network. Genetics. 195:883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D’Souza C, Harashima T, Shen WC, et al. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 64:746–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S, Tal H, Rose MS, Horwitz BA. 2009. Signaling by the pathogenicity-related MAP kinase of Cochliobolus heterostrophus correlates with its local accumulation rather than phosphorylation. Mol Plant Microbe Interact. 22:1093–1103. [DOI] [PubMed] [Google Scholar]
- Li D, Bobrowicz P, Wilkinson HH, Ebbole DJ. 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics. 170:1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao C, Li L, Liu M, Yin Z, et al. 2017. MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae. PLoS Pathog. 13:e1006449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kang YQ, Luo YL, Zhang KQ, Zou CG, et al. 2017. The NADPH oxidase AoNoxA in Arthrobotrys oligospora functions as an initial factor in the infection of Caenorhabditis elegans. J Microbiol. 55:885–891. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou X, Li G, Li L, Kong L, et al. 2011. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7:e1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 275:1314–1317. [DOI] [PubMed] [Google Scholar]
- Manosalva P, Manohar M, von Reuss SH, Chen S, Koch A, et al. 2015. Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat Commun. 6:7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Weinzierl G, Brachmann A, FeldbrüGge M, Kahmann R, 2003. Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot Cell. 2:1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordbring-Hertz B. 2004. Morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora—an extensive plasticity of infection structures. Mycologist. 18:125–133. [Google Scholar]
- Olthof T, Estey R. 1963. A nematotoxin produced by the nematophagous fungus Arthrobotrys oligospora. Nature. 197:514–515. [Google Scholar]
- Park G, Bruno KS, Staiger CJ, Talbot NJ, Xu JR. 2004. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol Microbiol. 53:1695–1707. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 14:687–690. [DOI] [PubMed] [Google Scholar]
- Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 56:117–124. [DOI] [PubMed] [Google Scholar]
- Roca MG, Weichert M, Siegmund U, Tudzynski P, Fleißner A. 2012. Germling fusion via conidial anastomosis tubes in the grey mould Botrytis cinerea requires NADPH oxidase activity. Fungal Biol. 116:379–387. [DOI] [PubMed] [Google Scholar]
- Rossi DCP, Gleason JE, Sanchez H, Schatzman SS, Culbertson EM, et al. 2017. Candida albicans FRE8 encodes a member of the NADPH oxidase family that produces a burst of ROS during fungal morphogenesis. PLoS Pathog. 13:e1006763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakulkoo W, Osés-Ruiz M, Oliveira Garcia E, Soanes DM, Littlejohn GR, et al. 2018. A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science. 359:1399–1403. [DOI] [PubMed] [Google Scholar]
- Scholler M, Rubner A. 1994. Predacious activity of the nematode-destroying fungus Arthrobotrys oligospora in dependence of the medium composition. Microbiol Res. 149:145–149. [DOI] [PubMed] [Google Scholar]
- Schumann U, Smith NA, Wang MB. 2013. A fast and efficient method for preparation of high-quality RNA from fungal mycelia. BMC Res Notes. 6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Zhao Y, Zhou J, Feng H, Jiang D, et al. 2017. Trapping devices of nematode-trapping fungi: formation, evolution, and genomic perspectives. Biol Rev. 92:357–368. [DOI] [PubMed] [Google Scholar]
- Tong SM, Feng MG. 2019. Insights into regulatory roles of MAPK-cascaded pathways in multiple stress responses and life cycles of insect and nematode mycopathogens. Appl Microbiol Biotechnol. 103:577–587. [DOI] [PubMed] [Google Scholar]
- Turra D, Ghalid ME, Rossi F, Pietro AD. 2015. Fungal pathogen uses sex pheromone receptor for chemotropic sensing of host plant signals. Nature. 527:521–524. [DOI] [PubMed] [Google Scholar]
- Vidal-Diez de Ulzurrun G, Huang TY, Chang CW, Lin HC, Hsueh YP. 2019. Fungal feature tracker (FFT): a tool for quantitatively characterizing the morphology and growth of filamentous fungi. PLoS Comput Biol. 15:e1007428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BL, Chen YH, He JN, Xue HX, Yan N, et al. 2018. Integrated metabolomics and morphogenesis reveal volatile signaling of the nematode-trapping fungus Arthrobotrys oligospora. Appl Environ Microbiol. 84:e02749-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Kim J, Huang WP, Abeliovich H, Stromhaug PE, et al. 2001. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 276:30442–30451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 79:143–180. [DOI] [PubMed] [Google Scholar]
- Wong Sak Hoi J, Dumas B. 2010. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot Cell. 9:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JR, Hamer JE. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10:2696–2706. [DOI] [PubMed] [Google Scholar]
- Yang CT, Vidal-Diez de Ulzurrun G, Goncalves AP, Lin HC, Chang CW, et al. 2020. Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi. Proc Natl Acad Sci U S A. 117:6762–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssar L, Wernet V, Hensel N, Yu X, Hildebrand HG, et al. 2019. Intercellular communication is required for trap formation in the nematode-trapping fungus Duddingtonia flagrans. PLoS Genet. 15:e1008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YK, Sanchez-Ayala MA, Sternberg PW, Srinivasan J, Schroeder FC. 2017. Improved synthesis for modular ascarosides uncovers biological activity. Org Lett. 19:2837–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Kim Y, Park G, Xu JR. 2005. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell. 17:1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Z, Xing X, Xie M, Yang L, Yang X, et al. 2018. MAP kinase Slt2 orthologs play similar roles in conidiation, trap formation, and pathogenicity in two nematode-trapping fungi. Fungal Genet Biol. 116:42–50. [DOI] [PubMed] [Google Scholar]
- Zhou TT, Zhao YL, Guo HS. 2017. Secretory proteins are delivered to the septin-organized penetration interface during root infection by Verticillium dahliae. PLoS Pathog. 13:e1006275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. RNA-seq raw data (.fastq) and differential expression analysis outputs are available at GEO with the accession number GES159974.
Supplementary material is available at figshare DOI: https://doi.org/10.25386/genetics.13225307.