Abstract
Mucopolysaccharidosis type IVA (MPS IVA) is a rare genetic disease caused by mutations in the GALNS gene and is inherited in an autosomal recessive manner. GALNS encodes N-acetylgalactosamine-6-sulfatase that breaks down certain complex carbohydrates known as glycosaminoglycans (GAGs). Deficiency in this enzyme causes accumulation of GAGs in lysosomes of body tissues. A human induced pluripotent stem cell (iPSC) line was generated from dermal fibroblasts of a MPS IVA patient that has compound heterozygous mutations (p.R61W and p.WT405del) in the GALNS gene. This iPSC line offers a useful resource to study the disease pathophysiology and a cell-based model for drug development.
Resource utility
This human induced pluripotent stem cell (iPSC) line is a useful tool for studies of disease phenotype and pathophysiology, and use as a cell-based disease model for drug development to treat MPS IVA patients.
Resource details
Mucopolysaccharidosis type IVA (MPS IVA, also called Morquio syndrome type A) is a rare autosomal recessive disorder caused by mutations in the GALNS gene, which encodes N-acetylgalactosamine-6-sulfatase (EC 3.1.6.4). It breaks down certain complex carbohydrates known as glycosaminoglycans (GAGs). Deficiency in N-acetylgalactosamine-6-sulfatase results in accumulations of GAGs in the lysosomes in body tissues and causes clinical symptoms including heart disease, skeletal abnormalities, vision and hearing loss, difficulty breathing, and early death (Khan et al., 2017; Peracha et al., 2018).
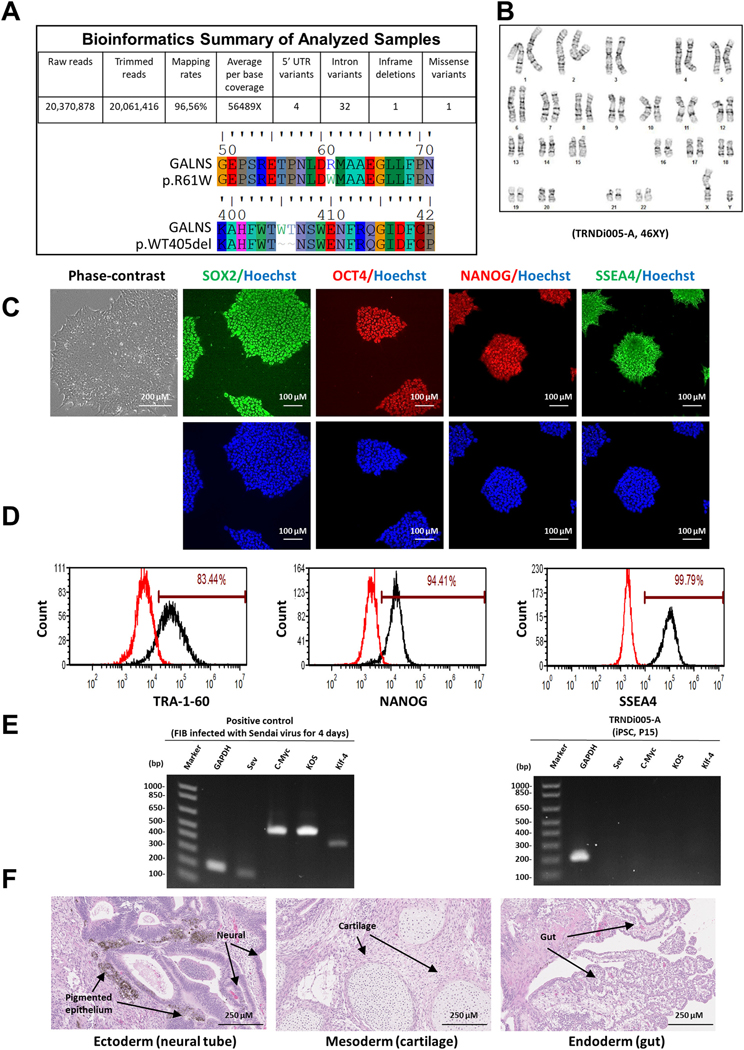
In this study, a human iPSC line was established from dermal fibroblasts of a 43-year-old male patient (GM01361, Coriell Institute) carrying compound heterozygous mutations of a p. R61W variant (c.181G > A) in exon 2 and a p.WT405del variant (c. 1213_1218del) in exon 11 of the GALNS gene (Table 1, Fig. 1A). A non-integrating CytoTune-Sendai viral vector kit (A16517, Thermo Fisher Scientific) containing OCT3/4, KLF4, SOX2 and C-MYC pluripotency transcription factors was employed to transduce the fibroblasts using the method described previously (Beers et al., 2012; Beers et al., 2015). The resulting iPSC line was named TRNDi005-A that exhibited a classical embryonic stem cell morphology (Fig. 1C), normal karyotype (46, XY), as confirmed by the G-banding karyotype at passage 11 (Fig. 1B), and expressed the major pluripotent protein markers of NANOG, SOX2, OCT4, SSEA4 and TRA-1–60 (Fig. 1C, D) evidenced by both immunofluorescence staining and flow cytometry analysis. Sendai virus vector (SeV) clearance was detected with reverse transcription polymerase chain reaction (RT-PCR) using SeV-specific primers that confirmed the vector elimination in the iPSCs by passage 15 (Fig. 1E). This iPSC line was not contaminated with mycoplasma (Supplementary Fig. S1) and were authenticated using a short tandem repeat (STR) DNA analysis, which demonstrated matching genotypes at all 16 loci examined (information available with the authors). Furthermore, the pluripotency of this iPS cell line was confirmed by a teratoma formation experiment that exhibited its ability to differentiate into cells of all three germ layers (Ectoderm, neural tube and pigmented epithelium; Mesoderm, cartilage; Endoderm, gut) in vivo (Fig. 1F).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Fig. 1 Panel C |
| Phenotype | Immunocytochemistry | SOX2, OCT4, NANOG, SSEA-4 | Fig. 1 Panel C |
| Flow cytometry | TRA-1-60 (83.44%); NANOG (94.41%) SSEA-4 (99.79%) | Fig. 1 Panel D | |
| Genotype | Karyotype (G-banding) and resolution | 46XY Resolution: 425–475 | Fig. 1 Panel B |
| Identity | Microsatellite PCR (mPCR) OR STR analysis | Not performed | N/A |
| 16 sites tested, all sites matched | Available from the authors | ||
| Mutation analysis (IF APPLICABLE) | Sequencing | Compound heterozygous mutation of GALNS p.R61W, p.WT405del | Fig. 1 Panel A |
| Southern Blot OR WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence. Negative | Supplementary Fig. S1 |
| Differentiation potential | Teratoma formation | Teratoma with three germlayers formation. Ectoderm (neural tube, pigmented epithelium); Mesoderm (cartilage); Endoderm (gut-like epithelium) | Fig. 1 Panel F |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A | |
Fig. 1. Characterization of TRNDi005-A iPSC line.
A) Detection of compound heterozygous mutations of p. R61W in exon 2 and p. WT405del in exon 11 of the GALNS gene. B) Cytogenetic analysis showing a normal karyotype (46, XY). C) Left: Phase contrast imaging of TRNDi005-A colonies grown on Matrigel at passage 10. Right: Representative immunofluorescent micrographs of iPSCs positive for stem cell markers: SOX2, OCT4, NANOG, and SSEA4. Nucleus is labelled with Hoechst (in blue). D) Flow cytometry analysis of pluripotency protein markers: TRA-1-60, NANOG and SSEA4. E) RT-PCR verification of the clearance of Sendai virus from the reprogrammed cells. Sendai virus vector transduced fibroblasts was used as positive control. F) Pathological analysis of a teratoma from TRNDi005-A iPSC, showing a normal ectodermal, endodermal and mesodermal differentiation.
Materials and methods
Cell culture
Patient skin fibroblasts were obtained from Coriell Cell Repositories (GM01361) and cultured in DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 37 °C. The iPS cells were cultured in StemFlex medium (Thermo Fisher Scientific) on Matrigel (Corning, 354,277)-coated plates at 37 °C in humidified air with 5% CO2 and 5% O2. The cells were dissociated with 0.5 mM Ethylenediaminetetraacetic acid (EDTA) and passaged when they reached 80% confluency.
Reprogramming of human skin fibroblasts
Patient fibroblasts were reprogrammed into iPS cells using the non-integrating Sendai virus technology following the method described previously (Beers et al., 2012; Beers et al., 2015).
Genome analysis
The genome analysis of variants in GALNS was conducted through ACGT, Inc. (Wheeling, IL, USA). Briefly, genomic DNA was extracted from MPS IVA patient fibroblasts using MasterPure Complete DNA and RNA Purification kit (Epicentre, Madison, WI, USA) as per manufacture instruction, followed by PCR amplification of all the exons using PrimeSTAR GXL DNA Polymerase (Takara, Mountain View, CA, USA). Equal quantities of each PCR amplicon were polled and fragmented to an average of size of 400 bp for library preparation. Sequencing of human GALNS exons by Next-Generation Sequencing (NGS) and bioinformatics data analysis was performed to identity the variants in the GALNS gene of MPS IVA patient sample. Gene variants were analyzed by using the Variant Annotation Integrator (VAI) tool from the University of California Santa Cruz Genome Browser (Casper et al., 2018). The specific primers for amplification of GALNS exons are listed in Table 2.
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-SOX2 | 1:50 | R & D systems, Cat# MAB2018, RRID: AB_358009 |
| Pluripotency Markers | Rabbit anti-NANOG | 1:400 | Cell signaling, Cat# 4903, RRID: AB_10559205 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:400 | Thermo Fisher, Cat# A13998, RRID: AB_2534182 |
| Pluripotency Markers | Mouse anti-SSEA4 | 1:1000 | Cell signaling, Cat# 4755, RRID: AB_1264259 |
| Secondary Antibodies | Donkey anti-Mouse IgG (Alexa Fluor 488) | 1:400 | Thermo Fisher, Cat# A21202, RRID: AB_141607 |
| Secondary Antibodies | Donkey anti-Rabbit IgG (Alexa Fluor 594) | 1:400 | Thermo Fisher, Cat# A21207, RRID: AB_141637 |
| Flow Cytometry Antibodies | Anti-Tra-1–60-DyLight 488 | 1:50 | Thermo Fisher, Cat# MA1–023-D488X, RRID: AB_2536700 |
| Flow Cytometry Antibodies | Anti-Nanog-Alexa Fluor 488 | 1:50 | Millipore, Cat# FCABS352A4, RRID: AB_10807973 |
| Flow Cytometry Antibodies | anti-SSEA-4-Alexa Fluor 488 | 1:50 | Thermo Fisher, Cat# 53–8843-41, RRID: AB_10597752 |
| Flow Cytometry Antibodies | Mouse-IgM-DyLight 488 | 1:50 | Thermo Fisher, Cat# MA1–194-D488, RRID: AB_2536969 |
| Flow Cytometry Antibodies | Rabbit IgG-Alexa Fluor 488 | 1:50 | Cell Signaling, Cat# 4340S, RRID: AB_10694568 |
| Flow Cytometry Antibodies | Mouse IgG3-FITC | 1:50 | Thermo Fisher, Cat# 11–4742-42, RRID: AB_2043894 |
| Primers | |||
| Target | Forward/Reverse primer (5′−3′) | ||
| Sev specific primers (RT-PCR) | Sev/181 bp | GGA TCA CTA GGT GAT ATC GAG C/ACC AGA CAA GAG TTT AAG AGA TAT GTA TC | |
| Sev specific primers (RT-PCR) | KOS/528 bp | ATG CAC CGC TAC GAC GTG AGC GC/ACC TTG ACA ATC CTG ATG TGG | |
| Sev specific primers (RT-PCR) | Klf4/410 bp | TTC CTG CAT GCC AGA GGA GCC C/AAT GTA TCG AAG GTG CTC AA | |
| Sev specific primers (RT-PCR) | C-Myc/523 bp | TAA CTG ACT AGC AGG CTT GTC G/TCC ACA TAC AGT CCT GGA TGA TGA TG | |
| House-Keeping gene (RT-PCR) | GAPDH/197 bp | GGA GCG AGA TCC CTC CAA AAT/GGC TGT TGT CAT ACT TCT CAT GG | |
| Targeted mutation analysis (PCR) | GALNS/523 bp | TCC GCG GCT CCC GTG GTT GCC AT/CCA CCT TTC CCA AGA GTA GAG | |
| Targeted mutation analysis (PCR) | GALNS/3201 bp | CCT TAG CGG AAA TGG ATT CTT G/GGT GGC TTC TTT GTG GTT TAG | |
| Targeted mutation analysis (PCR) | GALNS/6854 bp | GGT GCT CGT CTT ACC AAG AAT/CTC CAT TTA ACT CAG GAC TGC T | |
| Targeted mutation analysis (PCR) | GALNS/320 bp | GTG AGC ATG TAT GCA TAT CTG TAG/AGC TCT GGG CTT CAC TAC TT | |
| Targeted mutation analysis (PCR) | GALNS/2541 bp | AAG TAT CAA CCA AGA CCT CAC G/GGG TAT GAA TAG CAA CAG CAG A | |
| Targeted mutation analysis (PCR) | GALNS/4377 bp | CTG AGC CTT CAT TTC CTC TCT T/GAA GGC TGA CTG AAC CAA TGT A | |
Immunocytochemistry
For immunofluorescence staining, patient iPSCs were fixed in 4% paraformaldehyde for 15 mins, rinsed with Dulbecco’s Phosphate Buffered Saline (DPBS), and permeabilized with 0.3% Triton X-100 in DPBS for 15 mins. The cells were then incubated with the Image-iT™ FX signal enhancer (Thermo Fisher Scientific) for 40 mins at room temperature in a humidified environment and then followed by incubation individually with primary antibodies including SOX2, OCT4, NANOG and SSEA4, diluted in the Image-iT™ FX signal enhancer blocking buffer, overnight at 4 °C. After washing with DPBS, a corresponding secondary antibody conjugated with Alexa Fluor 488 or Alex Fluor 594 was added to the cells and incubated for 1 h at room temperature (Antibodies used are listed in Table 2). Cells were then washed and stained with Hoechst 33342 nucleic acid stain for 15 mins and imaged using an INCell Analyzer 2200 imaging system (GE Healthcare) with 20× objective lens and Texas Red, FITC and DAPI filter sets.
Flow cytometry analysis
The iPSCs were harvested using TrypLE Express enzyme (Thermo Fisher Scientific). Cells were fixed with 4% paraformaldehyde for 10 mins at room temperature and then washed with DPBS. Before fluorescence-activated cell sorting analysis, cells were permeabilized with 0.2% Tween-20 in DPBS for 10 mins at room temperature and stained with fluorophore-conjugated antibodies for 1 h at 4 °C on a shaker. Relative fluorophore-conjugated animal nonimmune Immunoglobulin were used as the negative control (Antibodies and nonimmune immunoglobulin used are listed in Table 2). Cells were then analyzed on a BD Accuri™ C6 Flow Cytometry system (BD Biosciences).
G-banding karyotype
The G-banding karyotype analysis was conducted at WiCell Research Institute (Madison, WI, USA). A total of 20 randomly selected metaphases were analyzed by G-banding for each cell line.
Short tandem repeat (STR) analysis
Patient fibroblasts and derived iPSC lines were sent to the WiCell Institute for STR analysis. Briefly, the Promega PowerPlex® 16 HS System (Promega, Madison, WI) was used in multiplex polymerase chain reaction (PCR) to amplify fifteen STR loci (D5S818, D13S317, D7S820, D16S539, vWA, TH01, TPOX, CSF1PO, D18S51, D21S11, D3S1358, D8S1179, FGA, Penta D, Penta E) plus a gender determining marker, Amelogenin (AMEL). The PCR product was capillary electrophoresed on an ABI 3500xL Genetic Analyzer (Applied Biosystems) using the Internal Lane Standard 600 (ILS 600) (Promega, Madison, WI). Data were analyzed using GeneMapper® v 4.1 software (Applied Biosystems).
Mycoplasma detection
Mycoplasma testing was performed and analyzed using the Lonza MycoAlert kit following the instructions from the company. Ratio B/A > 1.2 indicates mycoplasma positive. Ratio B/A 0.9–1.2 indicates ambiguous results and Ratio B/A < 0.9 indicates mycoplasma negative.
Testing for Sendai reprogramming vector clearance
Total RNA was extracted from TRNDi005-A iPSCs at passage 15 using RNeasy Plus Mini Kit (Qiagen). Human fibroblasts (GM05659, Coriell Institute) after transduction with Sendai virus for 4 days was used as the positive control. A total of 1 μg RNA/reaction was reverse transcribed with SuperScript™ III First-Strand Synthesis SuperMix kit and PCR was performed using Platinum II Hot-Start PCR Master Mix (Thermo Fischer Scientific). The amplifications were carried out using the following program: 94 °C, 2 mins; 30 cycles of [94 °C, 15 s, 60 °C, 15 s and 68 °C, 15 s] on Mastercycler pro S (Eppendorf) with the primers listed in Table 2. The products were then loaded to the E-Gel® 1.2% with SYBR Safe™ gel, run at 120 V electric field, and then imaged by G: Box Chemi-XX6 gel doc system (Syngene, Frederick, MD).
Teratoma formation assay
Patient iPSCs cultured in 6- well plates were dissociated with DPBS containing 0.5 mM EDTA and approximately 1 × 107 dissociated cells were resuspended in 400 μl culture medium supplied with 25 mM HEPES (pH 7.4) and stored on ice. Then, 50% volume (200 μl) of cold Matrigel (Corning, 354277) was added and mixed with the cells. The mixture was injected subcutaneously into NSG mice (JAX No. 005557) at 150 μl per injection site. Visible tumors were removed 6–8 weeks post injection, and were immediately fixed in 10% Neutral Buffered Formalin. The fixed tumors were embedded in paraffin and stained with hematoxylin and eosin.
Supplementary Material
Resource table.
| Unique stem cell line identifier | TRNDi005-A |
| Alternative name(s) of stem cell line | HT438A |
| Institution | National Institutes of Health National Center for Advancing Translational Sciences Bethesda, Maryland, USA |
| Contact information of distributor | Dr. Wei Zheng, wei.zheng@nih.gov |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Age: 43-year-old Sex: Male Ethnicity: Caucasian |
| Cell Source | Skin dermal fibroblasts |
| Clonality | Clonal |
| Method of reprogramming | Integration-free Sendai viral vectors |
| Genetic Modification | NO |
| Type of Modification | N/A |
| Associated disease | Mucopolysaccharidosis type IVA (MPS IVA) |
| Gene/locus | GALNSR61W; GALNSWT405del |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 06-29-2017 |
| Cell line repository/bank | N/A |
| Ethical approval | NIGMS Informed Consent Form was obtained from patient at time of sample submission. Confidentiality Certificate: CC-GM-15-004 |
Acknowledgement
We would like to thank Dr. Zu-xi Yu of the pathology Core of National Heart, Lung and Blood Institute, National Institutes of Health for sectioning and staining the teratoma. We also would like to thank Jacob Roth at National Center for Advancing Translational Sciences for mycoplasma testing service. This work was supported by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health, United States. CJAD was supported by Pontificia Universidad Javeriana, Colombia [Grant ID 7204 and 7520] and a scholarship from Fulbright Colombia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2019.101408.
References
- Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G, 2012. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc 7, 2029–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers J, Linask KL, Chen JA, Siniscalchi LI, Lin Y, Zheng W, Rao M, Chen G, 2015. A cost-effective and efficient reprogramming platform for large-scale production of integration-free human induced pluripotent stem cells in chemically defined culture. Sci. Rep 5, 11319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, Hinrichs AS, Haeussler M, Guruvadoo L, Navarro Gonzalez J, Gibson D, Fiddes IT, Eisenhart C, Diekhans M, Clawson H, Barber GP, Armstrong J, Haussler D, Kuhn RM, Kent WJ, 2018. The UCSC genome browser database: 2018 update. Nucleic Acids Res. 46, D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Almeciga-Diaz CJ, Sawamoto K, Mackenzie WG, Theroux MC, Pizarro C, Mason RW, Orii T, Tomatsu S, 2017. Mucopolysaccharidosis IVA and glycosaminoglycans. Mol. Genet. Metab 120, 78–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracha H, Sawamoto K, Averill L, Kecskemethy H, Theroux M, Thacker M, Nagao K, Pizarro C, Mackenzie W, Kobayashi H, Yamaguchi S, Suzuki Y, Orii K, Orii T, Fukao T, Tomatsu S, 2018. Molecular genetics and metabolism, special edition: diagnosis, diagnosis and prognosis of Mucopolysaccharidosis IVA. Mol. Genet. Metab 125, 18–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.