Abstract
Herein, we introduce a flexible, biocompatible, robust and conductive electrospun fiber mat as a substrate for flexible and stretchable electronic devices for various biomedical applications. To impart the electrospun fiber mats with electrical conductivity, poly(3,4-ethylenedioxythiophene) (PEDOT), a conductive polymer, was interpenetrated into nitrile butadiene rubber (NBR) and poly(ethylene glycol) dimethacrylate (PEGDM) crosslinked electrospun fiber mats. The mats were fabricated with tunable fiber orientation, random and aligned, and displayed elastomeric mechanical properties and high conductivity. In addition, bending the mats caused a reversible change in their resistance. The cytotoxicity studies confirmed that the elastomeric and conductive electrospun fiber mats support cardiac cell growth, and thus are adaptable to a wide range of applications, including tissue engineering, implantable sensors and wearable bioelectronics.
Keywords: poly(3,4-ethylene dioxythiophene); stretchable; electrochemical polymerization; electrospinning; cardiac tissue engineering; flexible electronics
Introduction
Flexible and stretchable electronics have recently gained an enormous interest in healthcare and a myriad of advanced biomedical applications, such as epidermal sensors, soft contact lenses, artificial muscles, neuron-machine interfaces, implantable medical devices and artificial skin.[1] Current commercially available sensors are chiefly based on conventional rigid electronics with no stretchability,[1b,2] while wearable sensors provide sensing without compromising user comfort or constraining movement.[2,3] Therefore, high demand has recently emerged to fabricate flexible sensors, along with seeking new sensing elements.
Conductive polymers (CPs) are a new generation of “smart”, stimuli-responsive biomaterials that integrate the electrical properties of metals and semiconductors with the favorable characteristics of conventional polymers, namely flexibility, ease of synthesis and functionalization.[4] In addition, the CPs, which are not biodegradable or have long degradation time, have demonstrated nontoxic and excellent biocompatibility in biological applications in vitro and in vivo.[5] There are numerous publications about CP-based implantable devices that do not cause any side effect, especially in neural applications.[6] Furthermore, the biodegradability of the conductive polymers can be controlled via chemical modification and composite systems with biodegradable polymers.[7]
Therefore, CPs are gaining significant interest in various research areas of the biomedical field, owing to their unique inherent features. As such, CPs have been investigated as transducers in biosensors and bioactuators,[8] drug delivery,[9] implantable monitoring devices,[10] tissue engineering scaffolds[11] and smart wearables.[12] Despite the rapid research growth in flexible biosensors and bioelectronics, there is still a need for better integration of electrically active materials with biological systems, including the better matching of the mechanical properties of these materials with the biological entities[13] and the improvement of the robustness of the interfaces. Consequently, recent research has focused on creating CP materials and their composites with enhanced properties and functionality.[4,8b,14]
Electrospinning is a simple approach to fabricate fibrous polymeric mats and the technique has regained significant popularity in recent years.[15] This technique generates structures analogous to the native extracellular matrices (ECM),[16] as it can produce flexible, porous, large surface area substrates, with spatially interconnected fibers of suitable mechanical properties.[17] Another important advantage of this fabrication technique is the possibility to modify the fiber morphology and orientation using unique collector designs along with the ability to control the electrospinning conditions. As such, one can indirectly tune the mechanical properties of the mats by adjusting the degree of fiber alignment.[18] Electrospinning provides flexibility to fabricate fibrous mats from a broad range of precursors, including natural or synthetic polymers, semiconductors, and ceramics, individually or in combination.[17a] Furthermore, the fibers are amenable to post electrospinning modification that allows the introduction of other functionality such as embedding with PEDOT post electrospinning, introducing conductivity and electrochemical responsiveness. There are different approaches for introducing electrical conductivity to the formed fibers. One approach consists of introducing an conductive material into an electrospinning solution, such as gold nanoparticles,[19] carbon nanotubes,[20] graphene,[21] or CPs.[15b,22]
A second approach consists of incorporating conductive nanoparticles[23] or CPs into/onto the fibers post electrospinning.[24] The latter approach has been employed more commonly, owing to the wider range of materials available to it. Therefore, the versatility of fabrication approaches makes flexible conductive electrospun fiber mats excellent candidates for wearable bioelectronics.
Among the various conductive polymers, poly(3,4-ethylenedioxythiophene) (PEDOT)[25] is one of the most widely used in biomedical applications, due to its high electrical conductivity, chemical stability and biocompatibility.[4b,26] However, the stiff and brittle mechanical properties and insolubility of pristine PEDOT hinder its use for flexible electronics.[21b,27] We overcame PEDOT’s drawbacks by incorporating it into flexible and stretchable electrospun rubbery fiber mats forming a semi-interpenetrating polymer network of two polymers. In this way, the formed mats preserved the elastomeric properties of rubber and the electrical properties of PEDOT.[24]
In this work, a stretchable, biocompatible, robust and conductive electrospun fiber mat is introduced as a substrate or scaffold for flexible and stretchable devices for various biomedical applications. The electrospun fiber mat is specifically composed of a blend of nitrile butadiene rubber (NBR) and poly (ethylene glycol)dimethacrylate (PEGDM) was used for its compatibility of the formed fibers with PEDOT and its effect on ionic conductivity.[24,28] The resulting electrospun mats are flexible, conductive and contain fibers that are orientated either randomly or aligned. Herein, we show that the resistance of the mats changes with bending-relaxation cycles and stretching. Importantly, the presented electrospun fiber mats have been shown to support cardiomyocyte adhesion and proliferation, thus proving a promising platform for biocompatible and flexible substrates for bioelectronics.
Results and Discussion
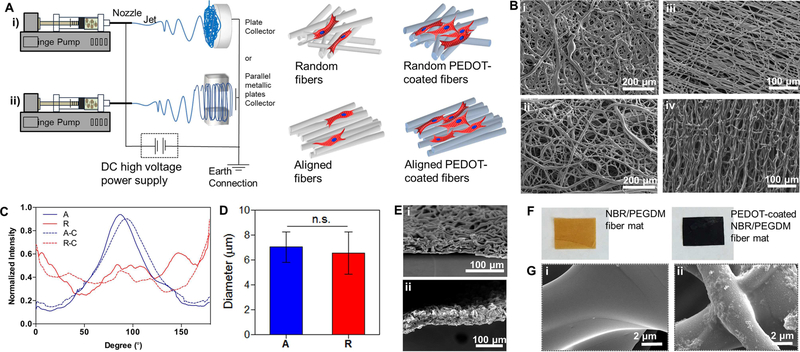
To create flexible fiber substrates, the electrospun fiber mats (shown in Figure 1) were prepared from a solution of NBR and PEGDM, with partial photo-crosslinking induced during the electrospinning process. Additional crosslinking was achieved by curing at 80°C in an oven with the use of a thermal crosslinker, as previously reported by us.[24] The random fibers were formed by using a stationary plate collector, while the aligned fibers were fabricated using a custom electrospinning apparatus (Figure 1A). During initial fabrication, the aligned fibers experienced high tensile stress leading to fracture, aggregation, and the eventual fusion of the fibers (Figure S2). As a solution, high molecular weight poly(ethylene oxide) (PEO) was introduced to the electrospinning solution of NBR/PEGDM to facilitate the electrospinning process by having excellent solubility and being compatible with the fibers (i.e., PEGDM). These properties allowed the aligned fibers to be electrospun without loss of fiber morphology, and the PEO could be leached out from the final fibers after the NBR/PEGDM had been crosslinked. Therefore, we successfully obtained random and aligned NBR/PEGDM fiber mats as shown in Figure 1B (i) and (iii). PEDOT was then polymerized within the NBR/PEGDM network to form the conductive electrospun mats via oxidative chemical polymerization of EDOT with FeCl3. Scanning electron microscope (SEM) images of the intact aligned and random fiber mats after the polymerization process of EDOT can be seen in Figure 1B (ii) and (iv). To quantify the fiber alignment after PEDOT polymerization, the SEM images were characterized by fast Fourier transform (FTT)-based image analysis method, which confirmed the conservation of the fiber alignment after embedding with PEDOT (Figure 1C). In addition, we analyzed averaged diameters of aligned and random fibrous scaffolds. As shown in Figure 1D, there is no significant difference between fiber diameters of aligned (7.0±1.2 μm) and random (6.6±1.7 μm) pristine NBR/PEGDM fiber mats. Furthermore, the random and aligned fiber mats after PEDOT polymerization showed similar density and thickness around 60 μm (Figure 1E). To confirm whether PEDOT has been polymerized, the color change of the fiber mats following the polymerization process is a good indicator, as PEDOT turns dark blue after a successful polymerization.[29] After PEDOT polymerization, we observed the dark blue/black color of the PEDOT-embedded fiber mat (Figure 1F) compared with the pristine NBR/PEGDM fiber mats. In addition, the SEM images confirmed the deposition of PEDOT into the NBR/PEGDM fiber network (Figure 1G).[24] The PEDOT-embedded fibers appeared to have a rough surface in comparison to the pristine NBR fibers. Atomic force microscope (AFM) analysis also confirmed that the surface roughness of the random and aligned fiber mats increased after PEDOT polymerization (Figure S3).
Figure 1.
(A) Schematic of the preparation steps for (i) random and (ii) aligned fibers. (B) SEM images of (i) random pristine NBR/PEGDM fiber mat, (ii) random PEDOT-embedded fiber mat, (iii) aligned pristine NBR/PEGDM fiber mat and (iv) aligned PEDOT-embedded fiber mat. (C) Degree of alignment in different types of electrospun fibers. Legend: A aligned, R random, A–C aligned-PEDOT embedded and R–C random-PEDOT embedded. (D) The averaged fiber diameters of aligned and random pristine NBR/PEGDM fiber mats. Data were expressed as mean± standard. n.s. not significant via Mann–Whitney U test. (n=13 in G) (E) SEM images showing the cross-section of (i) random and (ii) aligned random PEDOT-embedded fiber mats, (iii) aligned pristine NBR/PEGDM fiber mat and (iv) aligned PEDOT-embedded fiber mat. (F) Fabricated i) yellowish NBR/PEGDM vs. ii) dark blue PEDOT-embedded NBR/PEGDM electrospun sheets. (G) Magnified SEM images of the surface of the electrospun sheets (i) pristine NBR/PEGDM and (ii) PEDOT-embedded fibers.
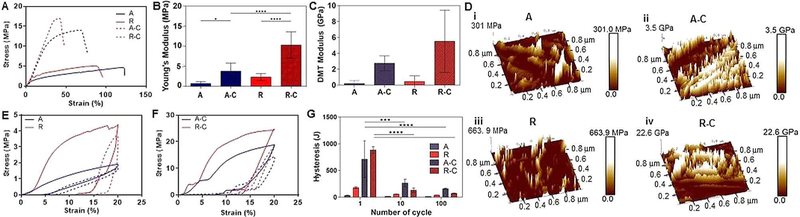
To assess the practicality of the conductive electrospun mats as flexible and stretchable substrates for bioelectronics, the mechanical properties of aligned and random fiber mats, with and without PEDOT embedding, were characterized by uniaxial tensile tests (Figure 2). For all mechanical tests, the aligned fibers were oriented parallel to the direction of the applied mechanical stress. In Figure 2A, PEDOT-embedded fiber mats exhibited a notably higher Young’s modulus and decreased elongation compared to the pristine NBR/PEGDM fiber mats. PEDOT embedding induced an increase in the Young’s modulus from 0.7±0.4 MPa to 3.8±1.8 MPa in the aligned fibers, and from 2.3±0.8 MPa to 10.3±3.0 MPa in the random fibers (Figure 2B). This macroscale mechanical behavior of a composite material is largely impacted by mechanical partitioning.[30] When a macroscopic mechanical load is applied to a multiphase material, the phase with the larger Young’s modulus will bear the majority of stress compared to the phase with the lower Young’s modulus.[31] The elastic properties are expected to differ from one phase to another in multiphase structures.[32] It is, therefore, essential to understand and determine the local elastic properties of polymerized PEDOT to achieve an accurate understanding of the macroscale measurements. In the present study, the nanomechanical characteristics of the electrospun fiber mats with and without PEDOT embedding were analyzed using the nanoindentation module of the AFM. We assess the nanoscale mechanical properties of the multiphasic PEDOT embedded electrospun fiber mats with peakforce quantitative nanomechanical property mapping (PFQNM), a method which maps the local elastic properties on the surface of the single electrospun fiber (diameter: 15±5 μm) with lateral nanometer resolution.[33] The PEDOT-embedded aligned and random fiber mats showed significantly higher Derjaguin-Muller-Toporov (DMT) modulus compared with that of the non-embedded fiber mats (Figure 2C and D). Also, we observed large error bars on the PEDOT embedded electrospun fiber mats, most likely because the aggregated PEDOT on the fiber mats as shown in Figure 1F (ii) induced a high DMT modulus. Consequently, the high DMT modulus in conductive fibers comes from the polymerized PEDOT within the NBR/PEGDM network that has formed a semi-interpenetrated polymer network (s-IPN). The DMT moduli measured at nanoscale follows the same trend as the macroscale Young’s moduli; however, the nanoscale elastic moduli were observed to be larger than macroscale moduli. Such disparity is attributed to the higher PEDOT content closer to the fiber surface, also observed through EDX mapping as reported in our previous work.[24] Therefore, it is concluded that embedded PEDOT acts as a reinforcing agent in conductive fiber mats resulting in an increase in the overall elastic modulus of the fiber mats. In addition, the pristine NBR/PEGDM fiber mats, both aligned and random, showed elongations of 122.4±12.6% and 84.9±11.1%, respectively, whereas the PEDOT-embedded aligned and random fibers exhibited only 75.1±18.1% and 48.3±6.4% maximum strain, respectively (Figure 2A). In fact, PEDOT embedding has led to threefold reduction of elongation at break. However, the PEDOT-embedded fiber mats still possessed favorable flexible and stretchable properties.
Figure 2.
(A) Representative stress-strain curves. (B) Young’s modulus of electrospun fiber mats. (C) Nanoscale measurement with AFM of DMT modulus and (D) represented image of (i) aligned, (ii) aligned-PEDOT embedded, (iii) random and (iv) random-PEDOT embedded electrospun fibers. Hysteresis curves for (E) aligned and random pristine NBR/PEGDM fiber mat and (F) aligned and random PEDOT-embedded fiber mats. (Solid line: cycle 1, Dashed line: cycle 100). (G) Summary of energy loss over 100 cycles of tensile stretching up to 20% strain. (n=5 in A–G)
Furthermore, we observed the different behavior of aligned and random fiber mats that showed a correlation between the material strength and fiber alignment. The tensile test of aligned fiber mats was performed in line with the alignment of the fibers. Directional alignment of fibers allowed for larger elongation at lower mechanical strength with the elongation parallel to the direction of tension compared with random fiber mats. With the fewer network junction points in the aligned fiber mat, the stress could not be dissipated in multiple directions in these mats, and as a result, aligned fiber mats were easier to stretch in one direction at low mechanical strength. Moreover, when stretched in a cyclic manner with 20% elongation, all electrospun fiber mats showed nonlinear restoring force and significant hysteresis differences between 1 and 10 cycles (Figures 2E–G). However, no significant reduction was observed in the hysteresis after 10 cycles. Such observation suggests that all electrospun fiber mats were stabilized, especially the PEDOT-embedded fiber mats, which showed similar behavior to elastic restoring force after 100 cycles of tensile stretching. As a result, PEDOT incorporation reduced stretchability and flexibility of the fiber mats but improved the restoring force to return their original shape after stress removal.
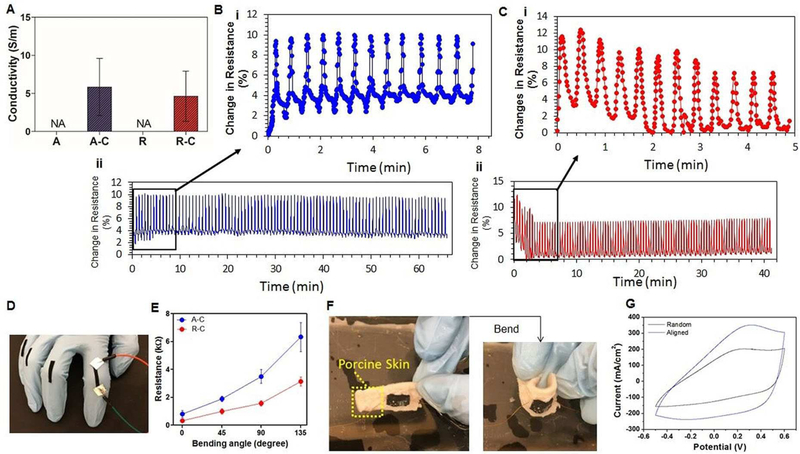
The conductivity of the PEDOT-embedded fiber mats was found to be 5.8±3.6 S/cm and 4.6±3.1 S/cm for the aligned and random fibers, respectively (Figure 3A). The results indicate that the conductivity is not impacted by fiber orientation. To evaluate mechanical and electrical stability, the change in resistance of the PEDOT-embedded fiber mats was measured over 100 extension and relaxation cycles with 20% strain rate. The change in resistance was plotted over time for the PEDOT-embedded aligned (Figure 3B) and random fibers (Figure 3C) with 100 cycles. In both samples, the peaks of resistance change are roughly in sync with the peaks of extension, illustrating a correlation between the elongation and the resistance of the fiber. The aligned PEDOT-embedded fiber mats showed stable and reversible changes in resistance compared to that of the random PEDOT-embedded fiber mats during the first 10 cycles (Figure 3B (ii) and 3C (ii)). It is hypothesized that stabilization of fiber resistance during the unloading process correlates with the elasticity and hysteresis of the fiber mats, and stable electrically conductive junction points. Therefore, the aligned fibrous network of the electrospun mats allows for anisotropic electrical connectivity, thus creating stable and reversible changes in resistance. On the other hand, the random PEDOT-embedded fiber mats showed a high fibrous network density that has electrically conductive junction points along with a high young’s modulus compared to the aligned fiber mats, thus they will experience difficulty in changing the distance of the electrically conductive junction points during cyclic elongations, a matter which leads to decreasing resistance changes, especially for the first 10 cycles compared with aligned fibers. However, after 10 cycles, both aligned and random PEDOT-embedded fiber mats showed stable and reversible resistance change along with similar magnitudes of resistance change (~6%).
Figure 3.
(A) Electrical conductivity of aligned, aligned-PEDOT embedded, random and random-PEDOT embedded electrospun fibers. Cyclic stress and resistance measurements with 20% strain rate for (B) PEDOT-embedded aligned fibers and (C) PEDOT-embedded random fibers, and their (i) the first 10 cycles and (ii) 100 cycles. (D) Highly stretchable and bendable PEDOT-embedded fiber mats attached to the finger. (E) Resistance vs. bending angle in PEDOT-embedded random and aligned fibers measured in the air. (F) Photograph of bendable PEDOT-embedded fiber mats on porcine skin in PBS. (G) Parallel and perpendicular CV curve of PEDOT-embedded random and aligned fibers in PBS. (n=3 in A–G)
As proposed earlier, the conductive electrospun mats provide an excellent platform for monitoring muscle contraction.[15a,34] It is shown that for many applications, the conductive mats can survive without significant performance degradation after many elongation and contraction cycles. Here, as a proof-of-concept, we used the electrospun mats to measure the resistance change in response to the bending angle of finger joints (Figure 3D and E). To measure the resistance of the finger joints, copper wires were attached to the ends of a small conductive electrospun fiber mat on the finger joints of a gloved hand. The fingers were bent, and the resistance of fiber mats was measured for different bending angles. Figure 3E demonstrates the change in resistance of the PEDOT-embedded aligned and random fibers when the finger is bent in the air. The results revealed that the resistance increased as the bending angle was continuously increased and remained unchanged while the finger remained bent. The resistance change of the aligned PEDOT-embedded fibers was larger than that of the random embedded fibers at all bending angles (0°, 45°, 90°, and 135°). Therefore, the aligned fiber mats will be more useful to monitor the repeated finger motions response as they undergo reversible electrical resistance changes against repeated bending behavior. Furthermore, a PEDOT-embeded nanofibrous sheet was tested on the surface of porcine skin and the resistance was likewise changed upon bending (Figure 3F).
Additionally, the PEDOT-embedded fiber mats were characterized by cyclic voltammetry in biological solutions, such as PBS, to assess their electrochemical properties (Figure 3G). The obtained CV curves for all samples are attributed to the improved rates of ionic mass transfer at the PEDOT-embedded fiber mat-electrolyte interface. This quality is achieved mainly due to the unique morphology of PEDOT-embedded fiber mats, which results in larger surface area, shorter average conducting pathway, and higher fiber density. Furthermore, the obtained CV curves in aligned vs. random conductive fibers may reflect higher electrical conductivity of samples in biological conditions, with no requirement for specific electrolytes for PEDOT with high salt concentration or low/high pH solutions,[35] all of which negatively affect the cellular viability. Conclusively, the PEDOT-embedded random and aligned fiber mats are eligible to be used as stand-alone electrode materials in biological systems, eliminating the need for any cytotoxic electrolytes.
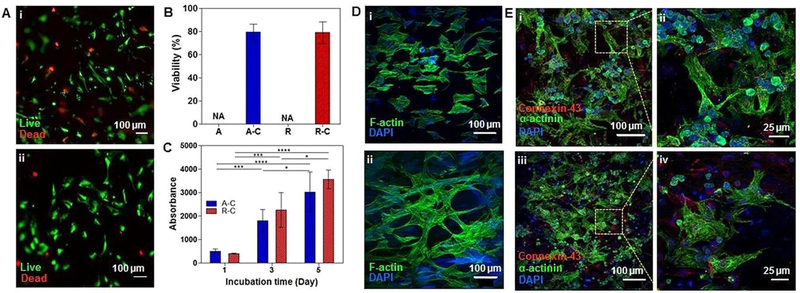
Such large changes in resistances in response to mechanical strain up to 20% makes this material a good strain sensor. Especially, in the human heart, a mechanical strain is considered to be cyclic, ranging from 2% to 20% depending on pathological states.[36] Therefore, the PEDOT-embedded fiber mats will be good strain sensor candidates for monitoring the heart’s pathological states. To evaluate the cytotoxicity of the electrospun fiber mats, cardiac fibroblasts were cultured on the fiber mats, and cell viability was examined using a Live/Dead assay after 24 hours of incubation (Figure 4A). The viability results were recorded as 79.7±7.0% and 79.1±9.5% for PEDOT-embedded aligned and random PEDOT embedded fiber mats, respectively (Figure 4B). This ~20% cell death might be inudced during cardiac fibroblasts’ and cardiomyocytes’ isolation process from neonatal rat hearts or from the seeding process that might be harsh, especially to primary cells.[37] However, no cardiac fibroblasts were observed on the pristine NBR/PEGDM fiber mats following 24 hours of culture. The chemical structure of PEDOT contains a slight positive charge on the polymer backbone (Figure S1) which allows PEDOT to act as an electrical binding site for the negatively charged cell membrane, as well as promoting protein adsorption.[38] The proliferation rate of cells was significantly increased on PEDOT-embedded scaffolds, as shown in Figure 4C. These results indicate that the PEDOT-embedded scaffolds might have accelerated adhesion and cell proliferation by either diminishing the antifouling properties of the PEGDM,[39] such that cells would adhere favourably,[40] or through an electrostatic interaction.[41] Furthermore, AFM measurements confirmed that the surface roughness of the electrospun mats was increased after PEDOT deposition (Figure S3), providing another reason for the enhanced cellular adhesion and proliferation on the PEDOT-embedded scaffolds compared to the pristine NBR/PEGDM fiber mats.[38b] The adhered cell morphology and cytoskeletal structure of the seeded cardiac fibroblasts were stained with F-actin and were imaged 5 days after incubation using a confocal laser scanning microscope (Figure 4D). The images show that the adhered cells stretched on the aligned and random PEDOT-embedded scaffolds. Nevertheless, the random PEDOT-embedded electrospun fibers exhibited more F-actin elongation, possibly as a response of the cells to the topographical cues of the electrospun fibers.[42]
Figure 4.
(A) Live/Dead staining of cardiac fibroblasts seeded on (i) PEDOT-embedded aligned fibers and (ii) PEDOT-embedded random fibers. (B) Quantification of Live/Dead images. (C) Metabolic activity of cardiac fibroblasts seeded on the PEDOT-embedded aligned and random fibers (n=3 in B, C). (D) F-actin and DAPI staining of cardiac fibroblasts seeded on PEDOT-embedded (i) aligned and (ii) random fibers. (E) Immunostaining of sarcomeric α-actinin and Cx-43 of cardiomyocytes seeded on (i and ii) PEDOT-embedded aligned fibers and (iii and iv) PEDOT-embedded random fibers.
The adhesion and spreading of cardiomyocytes were studied after seeding the cells onto the scaffolds, and cell morphology was observed after 5 days of incubation (Figure 4E). Cardiomyocytes were stained for the specific cardiac markers, namely, sarcomeric α-actinin (green), and gap junction protein, and as connexin (Cx-43, red), while the nucleus was counterstained for DAPI (blue). The staining indicated positive expression of sarcomeric α-actinin striations, which in turn indicated the presence of contractility and cell maturation.[43] Although clear alignment of cells on the aligned conductive scaffold was not noticed, both the embedded aligned and random fibers showed positive expression of Cx-43, a necessary component in promoting cell-cell communication through electrical and mechanical junctions.[42] Cell alignments has been controlled by nano- and micro-topography features such as the density and height of nano- or micro-patterns. Since these aligned or random nanofibers are dense with relatively low height, cardiac cells had not been able to sense these topography features and could not spread at following alignment direction as expected.[44] While the Young’s modulus of the embedded electrospun mats is higher than that of cardiac or skeletal muscles, with Young’s modulus ranging from 10 kPa to 50 kPa,[46] the successful expression of the cardiac biomarkers suggests that the PEDOT-embedded electrospun fiber mats would serve as an appropriate material for cardiac tissue engineering. For tissue regeneration application, the PEDOT-embedded electrospun sheets are very thin (~100 μm), making it difficult to create thick tissue constucts. Therefore, to create thick tissue contrscuts, they will be stacked along with cell layers following previously reported studies.[47]
Conclusion
In the present study, we fabricated flexible, porous, and conductive electrospun scaffolds with tunable fiber orientation which can be used in bioengineering applications. The electro-spun fiber mats were comprised of NBR and PEGDM to provide flexibility and mechanical integrity, and were embedded with PEDOT to provide electrical conductivity (~5 S/cm). The addition of PEDOT also alters the mechanical properties of the fiber mats by increasing the Young’s modulus and surface roughness. Fiber alignment introduces anisotropic effects on the mechanical and electrochemical properties of the fiber mats. The PEDOT-embedded fiber mats were stabilized and showed similar behavior to elastic restoring force after 100 cycles of tensile stretching. Consecutive bending cycles and repetitive strain cycles showed that the resistance of the fiber mat is responsive to the bending angle and the amount of applied strain up to 20% stretching. In addition, the PEDOT-embedded fiber mats showed high cellular viability along with excellent adhesion, proliferation, and cellular maturation of cardiac cells. Therefore, we demonstrated that the conductive electrospun fiber mats could serve effectively as biocompatible strain sensors for monitoring muscle contraction and body movement.
Experimental Section
Materials and reagents:
Phosphate-Buffered Saline (PBS), Fetal Bovine Serum (FBS), Dulbecco’s Modified Eagle Medium (DMEM), LIVE/DEAD® Viability/Cytotoxicity Kit, Alexa Fluor 488 actin conjugate (F-actin staining), and antibodies (sarcomeric α-actin, connexin-43 (Cx-43)) were purchased from Life Technologies. NBR with 44% acetonitrile (ACN) was kindly provided by Cédric Plesse from LPPI, Institut des Materiaux who received it from Lanxess (Mw=230 000 gmol−1, Per-bunan 4456F). Chloroform, methanol, benzoyl peroxide (BPO), poly(ethylene glycol) dimethacrylate (PEGDM, Mn=550 gmol−1), 2,2-dimethoxy-2-phenylacetophenone (Irgacure 651), acetone, poly(ethylene oxide) (PEO, 100 000 D) and phosphate buffer saline solution were purchased from Sigma Aldrich. Anhydrous iron III chloride was purchased from Acros, 3,4-ethylenedioxythiophene (EDOT) was purchased from AK Scientific. EDOT was doubly distilled under reduced pressure before use. All materials were used as received unless otherwise stated.
Preparation of random PEDOT embedded NBR/PEGDM fibers:
PEDOT embedded electrospun NBR mats were prepared through oxidative chemical polymerization of EDOT with FeCl3 as previously reported[24] (Scheme S1). Briefly, a 10%wt/v solution of NBR in chloroform was prepared to which 6%wt/v PEGDM, 1% wt/v Irgacure 651 and 0.5%wt/v BPO were added. The solution was stirred until homogeneous, then electrospun under high intensity UV light at 1 ml/h with an 18 G needle at a 10 cm working distance. The fibers were then further crosslinked in a vacuum oven at 80°C for 16 h. These fiber mats were then swollen with EDOT for 5 h in a vacuum and subsequently immersed in a 1.5 M FeCl3 solution for 20 min. The PEDOT-embedded mats were then washed and soaked in methanol for 2 days to remove any unreacted FeCl3 and monomer.
Preparation of aligned PEDOT embedded NBR/PEGDM fibers:
Aligned fibers were produced using the oriented electrospinning apparatus from Plant and Food Research Ltd, New Zealand. To improve the solution’s original mechanical properties, high molecular weight PEO (100 000 D) was added into the electrospinning solution at 2:1 NBR to PEO weight ratio. To ensure that all PEO capable of leaching out was removed, the aligned fibers were soaked in chloroform for 3 days. Lastly, PEDOT was incorporated into the fibers following the same method in random fibers.
Alignment characterization:
The alignment of the fibers was characterized by analyzing the scanning electron microscope (SEM) images in ImageJ software. The directionality function preformed a fast Fourier transform (FTT) transformation and performed a radial summation on the resulting pixel distribution image from 0 to 360° at 2° intervals. The resulting data plotted from 0 to 180°, as the FTT is symmetric around the horizontal axis.
SEM analysis:
SEM analysis of the samples was conducted using a Philips XL30S FEG with a SiLi (Lithium drifted) detector with a Super Ultra-Thin Window energy dispersive X-ray spectrometry (EDS) detector and with a LEICA LEO S430i SEM. The samples were coated with a Quorum Q150RS sputter coater prior to PEDOT incorporation to receive a double layer of platinum coating.
Mechanical and electrical properties of the electrospun fibers:
The Young’s Modulus of the fibers was measured using a uniaxial tensile loading machine (Instron 5542, USA) with 100 N load cell at a rate of 20 mm/min. Fibers were cut to a rectangular size of 15 mm×3 mm. The Young’s Modulus was calculated as the slope in the initial linear region in the stress-strain curve. The aligned fibers were stretched in the direction of fiber alignment. Cyclic tensile tests were performed to determine the hysteresis and elasticity of the fiber over time. Fibers were cut to a size of 15 mm×3 mm, and were cyclically stretched for 100 cycles at a rate of 20 mm/min. The hysteresis or energy loss was calculated as the area inside the loading and unloading curves. Again, aligned fibers were loaded in the direction of fiber alignment. To determine electrical properties over time, the change in resistance was measured simultaneously during cyclic tensile testing. Fibers were cut into a 20 mm×3 mm rectangle and the fibers were cyclically tensed to 20% strain for 10 cycles. The changes in resistance and elongation were measured by a digital multimeter and then plotted over time for the conductive random and aligned fibers. Aligned fibers were oriented in the direction of fiber alignment.
Nanoindentation-atomic force microscope (AFM):
Quantitative three-dimensional high magnification images were obtained at room temperature using a Bruker Dimension Icon AFM (Bruker Corporation, CA, USA) to determine the surface topography, as well as the nanomechanical properties. The images were obtained with quantitative nanomechanical property mapping (QNM) imaging mode in air, at a scan rate of 1.0 Hz, using a TESPA tip (Veeco, Santa Barbara, CA, USA) with a spring constant of 34 N/m, and a resonant frequency of 250 kHz (50 kHz). The tip was calibrated initially on a glass surface in order to set the deflection sensitivity before being used for peakforce QNM (PF-QNM) imaging. 250 samples per line were recorded, and for higher resolution images, 500 samples per line were recorded.
Conductivity measurements:
Conductivity was determined with a Jandel cylindrical four-point probe, with 1 mm spacing. The samples were measured five times and were averaged to account for their geometry and surface roughness.
Cyclic voltametry (CV):
CV was used to characterize the electro-spun mats. CV was performed using a CHI660E (CH instrument, Inc.) potentiostat in PBS solution under pH 7.4 at 20 mV/s and 5 cycles. For all experiments, the fiber mats operated as the working electrode with a Ag/AgCl reference and a platinum wire counter electrode.
Cell isolation and culture:
Neonatal cardiomyocytes and cardiac fibroblasts were isolated from two-day-old Sprague-Dawley rats based on a previously established approved protocol by the Institute’s committee on Animal Care.[48] Briefly, the hearts were subjected to three collagenase treatments and then pre-plated for 1 h to separate the cardiomyocytes from the cardiac fibroblasts based on time of attachment. The cardiomyocytes were removed from the flask and used immediately. The cardiac fibroblasts were cultured for later experiments in DMEM supplemented with 10% FBS, and 1% penicillin-streptomycin (P/S). Cardiac fibroblasts were used at third passage or lower. Fibers were washed with ethanol, Dulbecco’s phosphate-buffered saline (DPBS) with 1% P/S twice and DMEM, all for 30 minutes. Two hours prior to seeding, the fibers were coated with 50 μg/ml fibronectin (Sigma-Aldrich). Cardiac fibroblasts and cardiomyocytes were seeded at a concentration of 2.5×105 cells/mL.
Viability was measured using a LIVE/DEAD® Viability/Cytotoxicity Kit. Samples were imaged using an inverted fluorescent microscope (Zeiss, Axio Observer.D1, Germany). ImageJ software was used for the analysis of the images. Metabolic activity of the fibers was tested on days 1, 3, and 5 using a PrestoBlueTM kit (Life Technologies). Samples were incubated according to the manufacturer’s instructions, and absorbance was measured using spec-trophometry (Synergy HT-Reader, BioTek, Winooski, VT). On the 5th day of culture, samples were fixed with 4% paraformaldehyde at room temperature for 30 min. Following fixation, samples were permeabilized with 0.1% Triton X-100 for 30 minutes. Cardiomyocyte-seeded samples were stained for sarcomeric α-actinin, and Cx-43, 2 cardiomyoctye specific antibodies. The samples were incubated with the antibodies overnight at 4°C at a dilution of 1:200. The samples were then incubated with the secondary antibody for 40 minutes at room temperature. Finally, the samples were counterstained for 15 min with 4′,6-diamidino-2-phenylindole (DAPI) prepared at a dilution of 1:1000. Imaging was performed using an inverted laser scanning confocal microscope (Leica SP5X MP, Germany). Samples seeded with cardiac fibroblasts were fixed and permeabilized following the above steps, and were stained for f-actin with Alexa Fluor 488 phalloidin at a 1:40 dilution for 30 minutes. The nuclei were counterstained with DAPI. The samples were then imaged on an inverted laser scanning confocal microscope.
Statistical analysis:
Statistical data was analyzed using a one-way ANOVA (GraphPad Prism 5.02, GraphPad Software) when applicable. The data represented the mean, and the error bars represented standard deviation (SD) of each sample. Data was classified as significant if p<0.05, following a Tukey’s multiple comparison test.
Supplementary Material
Acknowledgements
The authors declare no conflict of interests in this work. The authors also acknowledge funding from the National Institutes of Health (EB021857, AR074234, and EB026824) and the Qatar national Research Fund (a part of Qatar Foundation, NPRP9–144-3-021). S.R.S. would like to recognize and thank Brigham and Women’s Hospital President Betsy Nabel, MD, and the Reny family, for the Stepping Strong Innovator Award through their generous funding. R.O.R. acknowledges the Ph.D. scholarship SFRH/BD/97658/2013 granted by FCT – Fundação para a Ciência e a Tecnologia, and the Fulbright Research Grant 2017 awarded from Fulbright Portugal. Thomas Kerr-Phillips would like to acknowledge the MacDiarmid Institute for Advanced Materials and Nanotechnology for his PhD scholarship. M.A.H. thanks the National Strategic Technologies and Innovation Program of King Abdulaziz City for Science and Technology (KACST), grant number 11-NAN1544–03 for their support and funding.
Footnotes
Supporting information for this article is available on the WWW under https://doi.org/10.1002/cnma.201900146
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].a) Ponce Wong RD, Posner JD, Santos VJ, Sens. Actuators A 2012, 179, 62–69; [Google Scholar]; b) Shin SR, Farzad R, Tamayol A, Manoharan V, Mostafalu P, Zhang YS, Akbari M, Jung SM, Kim D, Comotto M, Annabi N, Al-Hazmi FE, Dokmeci MR, Khademhosseini A, Adv. Mater 2016, 28, 3280–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kenry, Yeo JC, Lim CT, Microsystems & Nanoengineering 2016,2, 16043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Larimi SR, Nejad HR, Oyatsi M, O’Brien A, Hoorfar M, Najjaran HJS, A. A. Physical 2018, 271, 182–191; [Google Scholar]; b) Sadeqi A, Nejad HR, Alaimo F, Yun H, Punjiya M, Sonkusale SRJISJ, 2018, 18, 9137–9144; [Google Scholar]; c) Nejad HR, Punjiya MP, Sonkusale S, in 2017 19th International Conference on Solid-State Sensors, Actuators and Microsystems (TRANSDUCERS), IEEE, 2017, pp. 1183–1186; [Google Scholar]; d) Alaimo F, Nejad HR, Sadeqi A, Demarchi D, Sonkusale S, in 2018 IEEE SENSORS, IEEE, 2018, pp. 1–4. [Google Scholar]
- [4].a) Balint R, Cassidy NJ, Cartmell SH, Acta Biomater 2014, 10, 2341–2353; [DOI] [PubMed] [Google Scholar]; b) Kaur G, Adhikari R, Cass P, Bown M, Gunatillake P, RSC Adv 2015, 5, 37553–37567. [Google Scholar]
- [5].a) Hackett AJ, Malmström Jenny, Travas-Sejdic Jadranka, Prog. Polym. Sci 2017, 70, 6; [Google Scholar]; b) Wallace G, Spinks G, Soft Matter 2007, 3, 7; [DOI] [PubMed] [Google Scholar]; c) Irimia-Vladu MJCSR, 2014, 43, 588–610. [DOI] [PubMed] [Google Scholar]
- [6].a) Abidian MR, Corey JM, Kipke DR, Martin D. C. J. s., 2010, 6, 421–429; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Green R, Abidian MRJAM, 2015, 27, 7620–7637; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Richardson-Burns SM, Hendricks JL, Foster B, Povlich LK, Kim D-H, Martin DCJB, 2007, 28, 1539–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Llorens E, Armelin E, del Mar Pérez-Madrigal M, del Valle L, Alemán C, Puiggalí JJP, 2013, 5, 1115–1157; [Google Scholar]; b) Pal RK, Farghaly AA, Wang C, Collinson MM, Kundu SC, Yadavalli VKJB, Bioelectronics 2016, 81, 294–302. [DOI] [PubMed] [Google Scholar]
- [8].a) Peng H, Soeller C, Travas-Sejdic J, Macromolecules 2007, 40, 909–914; [Google Scholar]; b) Aydemir N, Chan E, Baek P, Barker D, Williams DE, Travas-Sejdic J, Biosens. Bioelectron 2017, 97, 128–135; [DOI] [PubMed] [Google Scholar]; c) Guimard NK, Gomez N, Schmidt CE, Prog. Polym. Sci 2007, 32, 876–921. [Google Scholar]
- [9].a) Abidian MR, Kim DH, Martin DC, Adv. Mater 2006, 18, 405–409; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li Y, Neoh KG, Kang ET, J. Biomed. Mater. Res. Part A 2005, 73, 171–181. [DOI] [PubMed] [Google Scholar]
- [10].a) Kim DH, Abidian M, Martin DC, J. Biomed. Mater. Res. Part A 2004, 71, 577–585; [DOI] [PubMed] [Google Scholar]; b) Cui X, Martin DC, Sens. Actuators B 2003, 89, 92–102. [Google Scholar]
- [11].Guex AG, Puetzer JL, Armgarth A, Littmann E, Stavrinidou E, Giannelis EP, Malliaras GG, Stevens MM, Acta Biomater 2017, 62, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu S, Liu P, Zhang Y, Zhang H, Qin X, Sens. Actuators B 2017, 252, 697–705. [Google Scholar]
- [13].Someya T, Bao Z, Malliaras GG, Nature 2016, 540, 379. [DOI] [PubMed] [Google Scholar]
- [14].a) Chan EWC, Baek P, Barker D, Travas-Sejdic J, Polym. Chem 2015, 6, 7618–7629; [Google Scholar]; b) Aznar-Cervantes S, Roca MI, Martinez JG, Meseguer-Olmo L, Cenis JL, Moraleda JM, Otero TFJB, 2012, 85, 36–43; [DOI] [PubMed] [Google Scholar]; c) Balint R, Cassidy NJ, Cartmell S. H. J. t. e., 2014, 2, 7; [Google Scholar]; d) Li M, Guo Y, Wei Y, MacDiarmid AG, Lelkes PIJB, 2006, 27, 2705–2715. [DOI] [PubMed] [Google Scholar]
- [15].a) Kerr-Phillips TE, Aydemir N, Chan EWC, Barker D, Malmström J, Plesse C, Travas-Sejdic J, Biosens. Bioelectron 2018, 100, 549–555; [DOI] [PubMed] [Google Scholar]; b) Zou Y, Qin J, Huang Z, Yin G, Pu X, He D, ACS Appl. Mater. Interfaces 2016. [DOI] [PubMed] [Google Scholar]
- [16].a) D’Amore A, Soares JS, Stella JA, Zhang W, Amoroso NJ, Mayer JE Jr, Wagner WR, Sacks MS, J. Mech. Behav. Mater 2016, 62, 619–635; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jafari M, Paknejad Z, Rad MR, Motamedian SR, Eghbal MJ, Nadjmi N, Khojasteh A, J. Biomed. Mater. Res. Part A 2017, 105, 431–459. [DOI] [PubMed] [Google Scholar]
- [17].a) Wang X, Ding B, Li B, Mater. Today 2013, 16, 229–241; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Huang Z-M, Zhang YZ, Kotaki M, Ramakrishna S, Compos. Sci. Technol 2003, 63, 2223–2253. [Google Scholar]
- [18].Arts T, Costa KD, Covell JW, New York 2001, 280, 2222–2229. [Google Scholar]
- [19].a) Marx S, Jose MV, Andersen JD, Russell AJ, Biosens. Bioelectron 2011, 26, 2981–2986; [DOI] [PubMed] [Google Scholar]; b) Shevach M, Maoz BM, Feiner R, Shapira A, Dvir T, 2013, 1, 2050–2750. [DOI] [PubMed] [Google Scholar]
- [20].a) Saeed K, Park S-Y, Lee H-J, Baek J-B, Huh W-S, 2006, 47, 8019–8025; [Google Scholar]; b) Crowder SW, Liang Y, Rath R, Park AM, Maltais S, Pintauro PN, Hofmeister W, Lim CC, Wang X, Sung H-J, Nanomedicine 2013, 8, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Pant HR, Pokharel P, Joshi MK, Adhikari S, Kim HJ, Park CH, Kim CS, Chem. Eng. J 2015, 270, 336–342; [Google Scholar]; b) Wang J, Naguib HE, Bazylak A, Proceedings of SPIE—The International Society for Optical Engineering 2012, 8342. [Google Scholar]
- [22].Reddy NR, Anandhan S, J. Mater. Sci. Mater. Electron 2016, 27, 13329–13337. [Google Scholar]
- [23].Jin L, Wu D, Kuddannaya S, Zhang Y, Wang Z, ASC Applied Materials and Interfaces 2016, 8, 5170–5177. [DOI] [PubMed] [Google Scholar]
- [24].Kerr-Phillips TE, Woehling V, Agniel R, Nguyen GT, Vidal F, Kilmartin P, Plesse C, Travas-Sejdic J, J. Mater. Chem. B 2015, 3, 4249–4258. [DOI] [PubMed] [Google Scholar]
- [25].a) Gao J, Crapo PM, Wang Y. J. T. e., 2006, 12, 917–925; [DOI] [PubMed] [Google Scholar]; b) Guex AG,Puetzer JL, Armgarth A, Littmann E, Stavrinidou E, Giannelis EP,Malliaras GG, Stevens M. M. J. A. b., 2017, 62, 91–101; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ma Z, Kotaki M, Inai R, Ramakrishna S. J. T. e., 2005, 11, 101–109. [DOI] [PubMed] [Google Scholar]
- [26].Luo SC, Mohamed Ali E, Tansil NC, Yu HH, Gao S, Kantchev EA,Ying JY, Langmuir 2008, 24, 8071–8077. [DOI] [PubMed] [Google Scholar]
- [27].Ichikawa S, Toshima N, Polym. J 2015, 47, 5. [Google Scholar]
- [28].Festin N, Maziz A, Plesse C, Teyssié D, Chevrot C, Vidal F, Smart Mater. Struct 2013, 22, 104005. [Google Scholar]
- [29].Martinez JG, Berrueco B, Otero TF, Front Bioeng Biotechnol 2015, 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Odegard GM, Clancy TC, Gates TS, Polymer 2005, 46, 553–562. [Google Scholar]
- [31].Morales-Rivas L, González-Orive A, Garcia-Mateo C, Hernández-Creus A, Caballero FG, Vázquez L, Sci. Rep 2015, 5, 17164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gniiupel-HeroldlJ T, Brand PC, Prask HJ, 1998.
- [33].a) Pittenger B, Erina N, Su C, Application Note AN128, Bruker Nano Surfaces Division 2010; [Google Scholar]; b) Adamcik J, Berquand A, Mezzenga R, Appl. Phys. Lett 2011, 98, 193701. [Google Scholar]
- [34].a) Yang G, Kampstra KL, Abidian MR, Adv. Mater 2014, 26, 4954–4960; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ouyang Z, Li J, Wang J, Li Q, Ni T, Zhang X, Wang H, Li Q, Su Z, Wei G, J. Mater. Chem. B 2013, 1, 2415–2424. [DOI] [PubMed] [Google Scholar]
- [35].a) Zhao D, Zhang Q, Chen W, Yi X, Liu S, Wang Q, Liu Y, Li J, Li X, Yu H, ACS Appl. Mater. Interfaces 2017, 9, 13213–13222; [DOI] [PubMed] [Google Scholar]; b) Casado N, Hilder M, Pozo-Gonzalo C, Forsyth M, Mecerreyes D, ChemSusChem 2017, 10, 1783–1791. [DOI] [PubMed] [Google Scholar]
- [36].a) Ugolini GS, Pavesi A, 2017, 6; [Google Scholar]; b) Balachandran K, Sucosky P, Jo H, Yoganathan AP, American journal of physiology. Heart and circulatory physiology 2009, 296, H756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].a) Rosellini E, Zhang YS, Migliori B, Barbani N, Lazzeri L, Shin SR,Dokmeci MR, Cascone M. G. J. J. o. B. M. R. P. A., 2018, 106, 769–781; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim SB, Nikkhah M, Khabiry M, Azize M, Kong J. J. A. n., 2013, 7, 2369–2380; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhu K, Shin SR, van Kempen T, Li YC, Ponraj V, Nasajpour A, Mandla S, Hu N, Liu X, Leijten J. J. A. f. m., 2017, 27, 1605352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].a) Rad AT, Ali N, Kotturi HS, Yazdimamaghani M, Smay J, Vashaee D, Tayebi L, J. Biomed. Mater. Res. Part A 2014, 102, 4169–4181; [DOI] [PubMed] [Google Scholar]; b) Maione S, Gil AM, Fabregat G, Del Valle LJ, Triguero J, Laurent A, Jacquemin D, Estrany F, Jimenez AI, Zanuy D, Cativiela C, Aleman C, Biomater. Sci 2015, 3, 1395–1405. [DOI] [PubMed] [Google Scholar]
- [39].Wagner VE, Koberstein JT, Bryers JD, Biomaterials 2004, 25, 2247–2263. [DOI] [PubMed] [Google Scholar]
- [40].del Valle LJ, Aradilla D, Oliver R, Sepulcre F, Gamez A, Armelin E, Alemán C, Estrany F, Eur. Polym. J 2007, 43, 2342–2349. [Google Scholar]
- [41].a) Horbett T. a., Waldburger JJ, Ratner BD, Hoffman a. S., J. Biomed. Mater. Res 1988, 22, 383–404; [DOI] [PubMed] [Google Scholar]; b) Ishizaki T, Saito N, Takai O, Langmuir 2010, 26, 8147–8154; [DOI] [PubMed] [Google Scholar]; c) Ren J, Xu Q, Chen X, Li W, Guo K, Zhao Y, Wang Q, Zhang Z, Peng H, Li YGJAM, 2017, 29, 1702713. [Google Scholar]
- [42].a) Nikkhah M, Edalat F, Manoucheri S, Khademhosseini A, Biomaterials 2012, 33, 5230–5246; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kharaziha M, Nikkhah M, Shin S-R, Annabi N, Masoumi N, Gaharwar AK, Camci-Unal G, Khademhosseini A, Biomaterials 2013, 34, 6355–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tandon V, Zhang B, Radisic M, Murthy SK, Biotechnol. Adv 2013, 31, 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Boengler K, Dodoni G, Rodriguez-Sinovas A, Cabestrero A, Ruiz-Meana M, Gres P, Konietzka I, Lopez-Iglesias C, Garcia-Dorado D, Di Lisa F, Heusch G, Schulz R, Cardiovasc. Res 2005, 67, 234–244. [DOI] [PubMed] [Google Scholar]
- [45].a) Song L, Wang K, Li Y, Yang YJC, Biointerfaces SB 2016, 148, 49–58; [DOI] [PubMed] [Google Scholar]; b) Ventre M, Causa F, Netti P. A. J. J. o. t. R. S. I., 2012, 9, 2017–2032; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhou Q, Kühn PT, Huisman T, Nieboer E, Van Zwol C, Van Kooten TG, Van Rijn P. J. S. r., 2015, 5, 16240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].a) Lieber RL, Skeletal Muscle Structure, Function, and Plasticity, Lippincott Williams & Wilkins, 2002; [Google Scholar]; b) Chen Q-Z, Harding SE, Ali NN, Lyon AR, Boccaccini AR, Mater. Sci. Eng. R 2008, 59, 1–37. [Google Scholar]
- [47].a) Kim MS, Lee B, Kim HN, Bang S, Yang HS, Kang SM, Suh K-Y, Park S-H, Jeon NLJB, 2017, 9, 015029; [DOI] [PubMed] [Google Scholar]; b) Lovett M, Lee K, Edwards A, Kaplan DLJTEPBR, 2009, 15, 353–370; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sofi HS, Ashraf R, Beigh MA, Sheikh FA, in Cutting-Edge Enabling Technologies for Regenerative Medicine, Springer, 2018, pp. 49–78. [Google Scholar]
- [48].Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim S. b., Nikkhah M, Khabiry M, Azize M, Kong J, Wan K-T, Palacios T, Dokmeci MR, Bae H, Tang X, Khademhosseini A, ACS Nano 2013, 7, 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.