Abstract
Cell therapy is revolutionizing modern medicine. To promote this emerging therapy, the ability to image and track therapeutic cells is critical to monitor the progress of the treatment. Ultrasound imaging is promising in tracking therapeutic cells but suffers from poor contrast against local tissues. Therefore, it is critical to increase the ultrasound contrast of therapeutic cells over local tissue at the injection site. Here, we describe a method to increase the ultrasound intensity of therapeutic cells with nanoparticles to make the injected therapeutic cells more visible.
Keywords: Cell tracking, Ultrasound imaging, Contrast agents, Nanoparticles, Mesenchymal stem cells
1. Introduction
Cell therapy including immune and stem cell therapies has great potential in changing modern medicine and treating various diseases [1, 2]. However, the safety, biodistribution, cell fate, and treatment efficacy of cell therapy are still big concerns [1]. Critical diagnostic information about these concerns can be obtained via conventional and emerging medical imaging tools such as MRI and PET [3]. However, the therapeutic cells usually have poor contrast to the cells in treating the disease. Therefore, it is essential to increase the contrast between therapeutic cells and local cells [4].
Ultrasound imaging plays an important role in medical diagnosis because it is safe, easily accessible, and affordable. Moreover, ultrasound imaging has a large penetration depth, good spatial resolution, and excellent temporal resolution [5–7]. Ultrasound imaging can guide cell injections into deep tissues in real-time [8]. However, ultrasound imaging in cell tracking has also been limited due to the poor contrast between transplanted cells and local soft tissues. Hence, it is important to increase the ultrasound intensity of transplanted cells [4, 9].
Nanoparticles and nanobubbles are promising ultrasound contrast agents for cell tracking because of their small size, high cellular uptake, and high impedance (product of density and velocity of sound) mismatch with cells [10–12]. Ultrasound imaging combined with nanoparticles or nanobubbles can track the location of injected stem cells in vivo [10, 11, 13, 14]. Additionally, ultrasound-based photoacoustic imaging combines the multiplexity and good contrast of optical imaging and the high penetration depth of ultrasound imaging [15–17]. Nanoparticles with photoacoustic contrast can label stem cells and then track the labeled cells in vivo via photoacoustic imaging [11, 18, 19].
This chapter will focus on stem cell tracking with ultrasound imaging and nanoparticle-based ultrasound contrast agents. We will describe the methods of labeling mesenchymal stem cells with Stöber silica nanoparticles as exemplified ultrasound contrast agents. The labeling procedure involves no transfection agents at all. Then, we will provide details on how to test the ultrasound signal of cells in vitro and how to track the nanoparticle-labeled cells in vivo via ultrasound imaging.
2. Materials
Human mesenchymal stem cells (PT-2501).
Mesenchymal stem cell growth media (PT-3001).
Trypsin/ethylenediaminetetraacetic acid (EDTA).
Ultrapure agarose powder.
EDTA titrant (0.020 M).
Sterile phosphate-buffered solution (PBS).
Ultrasound gel.
Stöber silica nanoparticles (NanoComposix, San Diego, CA, USA).
Millipore grade water with a resistivity larger than 18.2 MΩ cm at room temperature.
Contrast agent suspension: 4 mg silica nanoparticles (see Notes 1 and 2) in 2 mL PBS to make a concentrated 2 mg/mL (see Note 3) contrast agent suspension. Disperse the nanoparticles by sonicating the suspension in a sonication bath for 10 min until there is no pellet at the bottom. Use freshly made suspensions.
Agarose solution: Make this solution right before cell collection (before step 6 of Subheading 3.1). Dissolve 1 g ultrapure agarose powder into 100 mL water by heating the solution to 90 °C with stirring. Then, cool down this solution to 60 °C, seal, and maintain the temperature at 60 °C (see Note 4).
3. Methods
All procedures were carried out at room temperature unless otherwise specified. Cell culture and cell labeling should be performed in a biosafety cabinet. Cells were incubated under standard conditions: 5% carbon dioxide and a relative humidity higher than 93% at 37 °C.
3.1. Cell Labeling
Human mesenchymal stem cells were seeded into a T75 flask at a cell density of 5000 cells/cm2. The cells were incubated until the confluency is approximately 90%.
Before labeling the cells, dilute the contrast agent suspension ten times with fresh media. This results in a final concentration of 200 μg/mL.
Remove the floating dead cells by aspirating the old cell culture media from the flask. Rinse the flask with PBS if necessary (see Note 5).
Use a pipette to mix the suspension from step 2 in the original container to make sure the nanoparticles are well dispersed in the media and there are no chunks of nanoparticles. Then add 15 mL of the suspension into the T75 flask gently (see Note 6).
Put the flask back into the incubator. Allow the cells to be co-incubated with the contrast agents for 4 h (see Note 7).
Remove the media in the flask by aspirating.
Add 15 mL of PBS into the flask. Sway the flask for 10 s, place the flask vertically on the bench for 10 s, and immediately remove the PBS by aspirating.
Repeat step 7 twice or several more times until no observable removal of the nanoparticles from the growing surface of the flask (see Note 8).
Wash the cells with PBS without calcium or magnesium for the last washing step. Remove the PBS (see Note 9).
Add 2 mL of trypsin/ethylenediaminetetraacetic acid (EDTA) to cover the cells. Put the flask back into the incubator and incubate the cells for 5 min. Verify cell detachment with a microscope.
Add 8 mL of fresh media into the flask to quench the trypsin when more than 90% of cells are detached. Otherwise, continue incubating the cells and check every 3 min. The overall incubation time should be no more than 15 min.
Collect the cells into a 50-mL centrifuge tube and centrifuge the cells at 180 × g for 5 min.
Remove the supernatant with aspirating and/or pipetting (see Note 10). Resuspend the cells in 510 μL fresh media (see Note 11).
Take 10 μL of the cell suspension and then count the cell number with a hemocytometer.
3.2. In Vitro Ultrasound Signal Detection
Centrifuge the remaining cell suspension from step 12 in Subheading 3.1 at 180 × g for 5 min. Remove the supernatant and resuspend the cells with 100 μL PBS (see Note 12) to make a concentrated cell suspension (see Note 13).
Use 50 μL of the concentrated cell suspension to make 50 μL of various diluted cell suspensions (see Note 14).
Mix 50 μL of the cell suspension with 50 μL of 1% agarose solution at 60 °C. Immediately mix the cell suspension and agarose solution by pipetting and then quickly load the 80 μL mixture into a well of the 384-well plate (see Note 15). Quickly remove air bubbles by sucking the air out from the gel via pipetting if there are any (see Note 16). Repeat above steps for all the other cell suspension including nanoparticle-labeled and unlabeled cells at different concentrations. Load the cells into wells in order with the cell concentrations and mark down the sample information for each well.
Wait until the cell–agarose mixture has solidified. Fill the wells with pure agarose solution to protect the contents and avoid any air bubbles.
Tape the entire phantom on a crystallizing dish horizontally (Fig. 1). Cover the entire phantom with pure water and scan the phantom using a 40-MHz centered linear transducer (MS550) on a Vevo 2100 system (VisualSonics, Fujifilm) (see Note 17). Use the B-mode for the ultrasound signal and PA-mode for the photoacoustic signal. The gain should be optimized to produce a strong ultrasound signal without any saturation. The transducer is fixed on a motor, which moves the transducer horizontally and can image multiple cross-sections (see Note 18).
After scanning, save the frames to the ultrasound scanner (see Note 19). Export the frames using DCM format.
Open the DCM file with ImageJ. Find a good field of view, draw a region of interest (ROI) within a sample, and measure the mean gray value (Fig. 2, see Note 20). Repeat this measurement four times with different fields of view within each sample.
Calculate the average of the mean gray values for different fields of views. This value represents the ultrasound intensity of each sample. Bright images have large mean gray values.
Determine the limit of detection of cells. First, plot a curve with the ultrasound intensity (y-axis) and cell concentrations (x-axis). Then, calculate the standard deviation of the response (Sy) of the curve and slope of the calibration curve (S). The theoretical limit of detection is calculated as 3(Sy/S).
If necessary, optimize the ultrasound signal of nanoparticle-labeled stem cells by changing the nanoparticle labeling concentration (should be within the biocompatible range; see Note 3), the incubation time (less effective than the nanoparticle labeling concentration; see Note 7), and nanoparticle surface potential [10]. For the optimization, the cells should be in separate flasks, labeled with different conditions, and then scanned with the same cell numbers in the agarose phantom.
The half-life of the nanoparticles inside the cells can also be checked with the steps described above. For the half-life measurement, label cells with the exact same labeling condition. Detach these nanoparticle-labeled stem cells, seed into separate containers, and then collect the cells from each flask on different days (see Note 21). Make the in vitro cell agarose phantom right after cell collection on different days, seal the phantoms, store at 4 °C, and scan all the phantoms together after the last collection. The same cell numbers in the agarose phantoms should be the same.
Fig. 1.
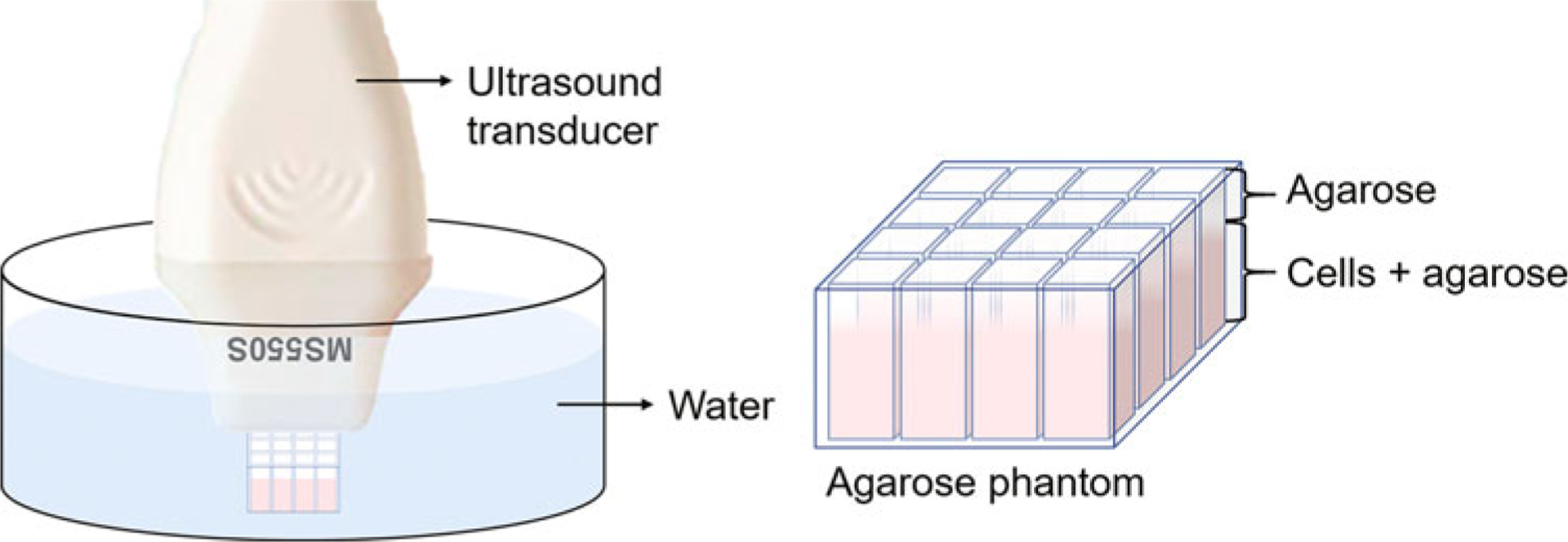
Scheme of the sample holder and sample setup for ultrasound imaging. The ultrasound transducer is connected to a 3D motor and will be moved along the direction perpendicular to the viewing plane
Fig. 2.
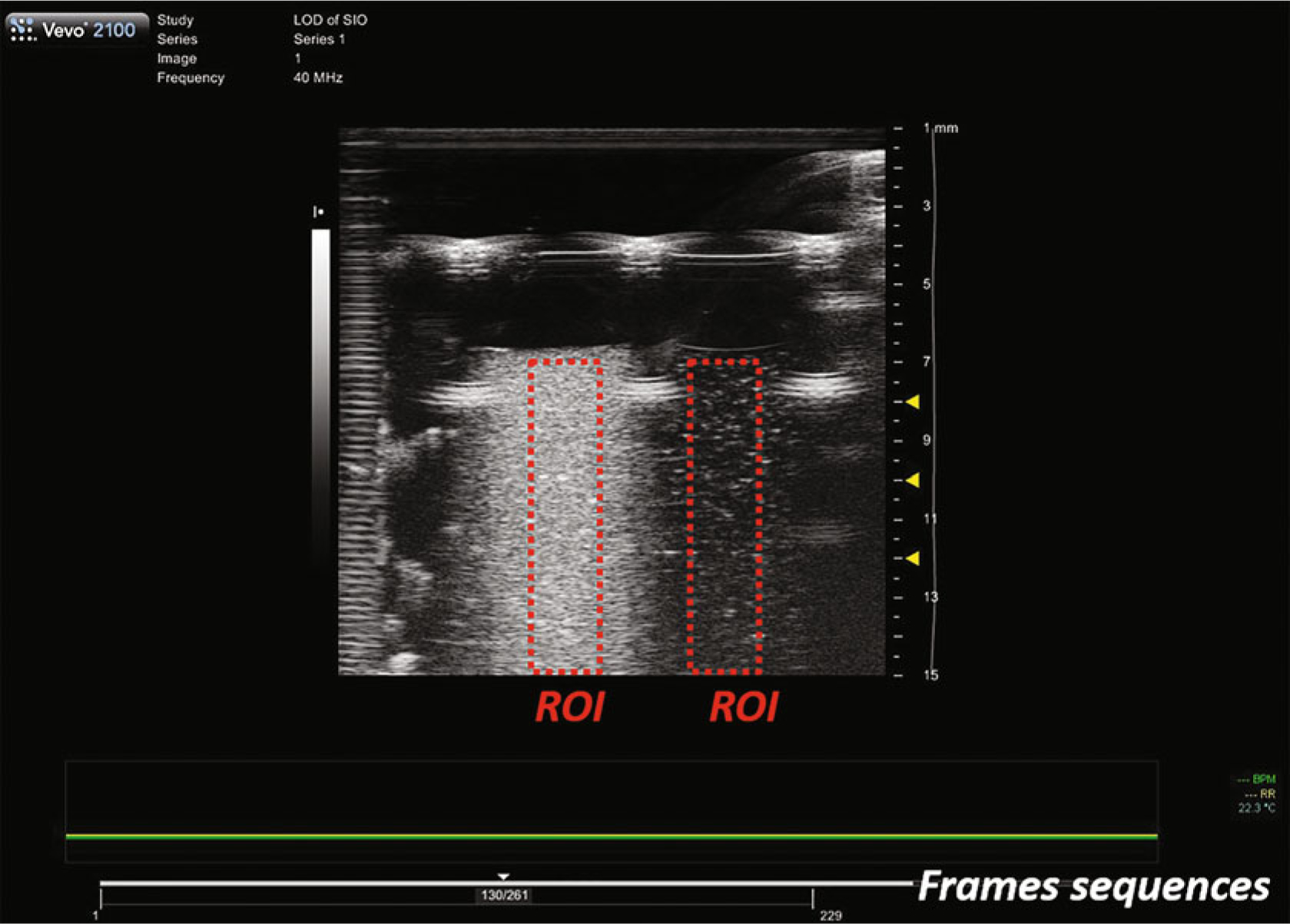
A typical ultrasound image frame of samples loaded in a sample holder based on a 384-well plate. The left and right ROIs measure the same nanoparticles but with different concentrations
3.3. In Vivo Stem Cell Tracking
Estimate the total cell numbers that are needed for in vivo injection according to the applications and disease model. Given the dead volume for injections, collect twice as many cells needed (see Note 22) from a large flask or multiple T75 flasks (see Note 23) by following the steps in Subheading 3.1.
Concentrate the collected cells by centrifugation and resuspend in growth media. The concentrated cell mixture should be twofold of the final injected cell concentration because the cells will be mixed with Matrigel. The use of Matrigel can increase cell retention.
Set up the animal for ultrasound imaging. Anesthetize the mouse with 1–2% isoflurane and then mount it on a warm animal bed with the nose inside a nozzle supplied with isoflurane.
Adjust the position of the animal bed, the animal, and the ultrasound transducer. The ultrasound transducer should be positioned above the desired scanning area.
Apply the ultrasound gel on the skin of the mouse to cover the entire desired scanning area. Remove any air bubbles with a cotton swab or apply more ultrasound gel.
Perform a 3D scan to get a baseline (or ultrasound image before cell injection). Tune the ultrasound gain and repeat the scan multiple times. It is highly recommended to include at least one landmark, e.g., the bladder per scan.
Raise the ultrasound transducer and remove the ultrasound gel on the animal carefully. The animal should still be under anesthesia. The position of the animal bed, the animal, and the transducer should be kept the same (see Note 24).
Quickly mix concentrated cell suspension with Matrigel at a volume ratio of 1:1 by pipetting. Load the cell mixture into a syringe quickly and get rid of any air bubbles (see Note 25).
Inject the cell mixture into the desired area that has been scanned for a baseline (see Note 26).
Apply ultrasound gel on the skin of the desired area. Perform a 3D scan. Use the same gain and include the same landmark from step 6 in Subheading 3.3; scan multiple times until clear and comparable images are obtained.
Raise the ultrasound transducer. Remove the ultrasound gel and clean the animal. Turn off the anesthesia system. Put the animal back into the cage when it is awake.
Repeat steps 3–5, 10, and 11 in Subheading 3.3 for later time points.
Data analysis. After the scans at each time point, save the frames to the ultrasound scanner and then export them as DCM files. Open the DCM files with ImageJ. Find a good field of view, draw a region of interest (ROI) along the outline of cell injections, and then measure the mean gray value (Fig. 3). Repeat this measurement four times with different fields of views for each injection.
Fig. 3.
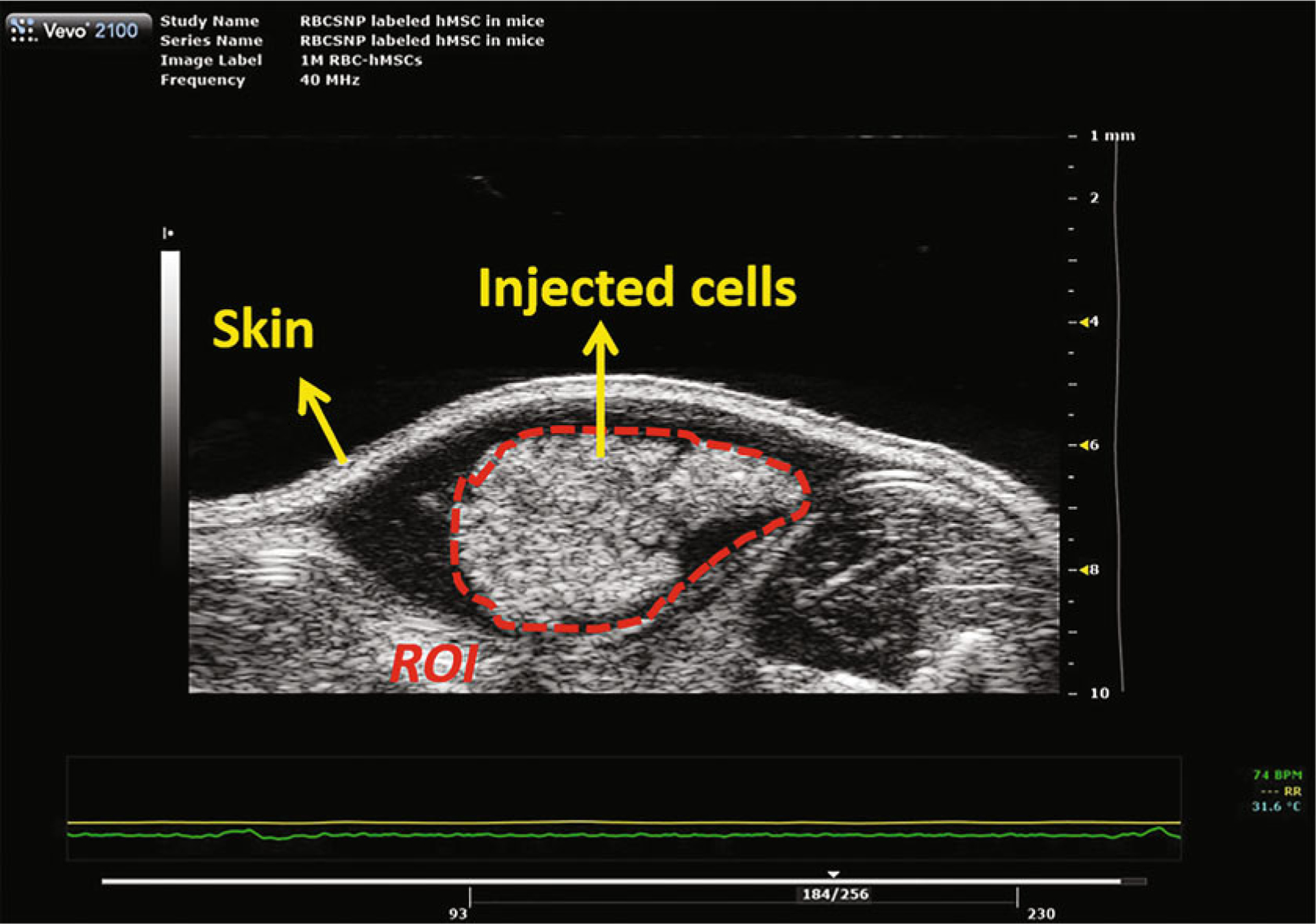
A typical ultrasound image of subcutaneously injected nanoparticle-labeled mesenchymal stem cells. The cells were co-injected with Matrigel at a volume ratio of 1:1. Cells were labeled with exosome-like silica nanoparticles reported previously [10]
4. Notes
Here we used Stöber silica nanoparticles as an example ultrasound contrast agent for stem cell tracking. Other inorganic nanoparticles can also be used to label stem cells. However, for each nanoparticle, the toxicity needs to be carefully examined. Toxicity assays include but are not limited to: metabolism, viability, live/dead, proliferation, adhesion, phenotypes, function, multipotency, migration ability, cytokine secretion, and gene expression.
The contrast agents made in the lab should be sterilized either by high temperature or UV irradiation. The sterilization method should be chosen according to the materials. For example, the bare Stöber silica nanoparticles can be calcinated at 600 °C to remove the exogenous toxins. Amine-modified Stöber silica nanoparticles should not be treated with this method because the high temperature will remove the amine groups.
The labeling concentration and incubation time of ultrasound contrast agents can affect the cellular uptake of the nanoparticles and therefore influence the ultrasound signal of the labeled cells [10]. Thus, it is necessary to optimize the labeling concentration (refer to step 10 in Subheading 3.2) of each contrast agent because a higher labeling concentration may increase the ultrasound signal of labeled cells but an excessively high concentration can also lead to toxic effects on the cells. The labeling concentration of Stöber silica nanoparticles at 200 μg/mL is biocompatible.
The 1% agarose solution will solidify when it is at room temperature. The surface of the agarose solution easily forms a heterogeneous layer. Thus, it is important to keep the temperature high and homogeneous in the solution. It is recommended to keep the agarose solution warm in a water bath while stirring.
The cells are recommended to be rinsed with PBS to remove dead cells and cell debris if there are too many dead cells in the flask under certain circumstances. For example, cells freshly thawed often have a large portion of dead cells.
For nanoparticle suspensions, the volume of suspension is as important as the nanoparticle concentration because nanoparticles are likely to settle down and therefore the total mass of nanoparticles added into the flask is important.
Similar to the cells’ labeling concentration, the labeling duration should also be optimized to achieve a high ultrasound signal (refer to step 10 in Subheading 3.2).
Some nanoparticles could be just on the surface of the cells. Thorough washing can wash away free nanoparticles and prohibit ultrasound signal that is not related to the cells. Buffer solutions with 2 mM EDTA can be used to remove weakly bound nanoparticles on the surfaces. More importantly, the entire washing process should be completed within 5 min to keep the cell healthy. Otherwise, nutrients such as FBS should be added to the washing buffer as well.
For unlabeled cells or control cells, skip steps 2–8. Unlabeled cells are necessary to elucidate the contribution of nanoparticles on the ultrasound signal of cells.
To avoid cell loss, leave about 100 μL of supernatant during aspiration and use a pipet gun to remove the rest of the supernatant.
To obtain a more accurate cell counting result, cells from a T75 flask with approximately 90% confluence should be suspended with approximately 0.4–1 mL of solution. There are roughly 0.6 million human mesenchymal stem cells when a T75 flask is 90% confluent.
For immediate in vitro cell ultrasound signal testing, cells should be suspended in PBS because cell viability is less important for this test and fewer bubbles will form while making the agarose phantom.
The exact cell concentrations can be varied based on the total cell numbers. The highest cell concentration should be no more than half of the total cell number/100 μL. Considering the dead volume of cell mixture, 100 μL of the cell and agarose mixture is needed to make a phantom in the well of 384-well plate.
It is recommended to make 5 cell concentrations between 0 and the highest cell concentration. The dilution factors between adjacent concentrations are recommended to be 2 except the blank (0 million/mL). For example, if the total cell number is 0.56 million, then the cell concentrations are recommended to be 5.6, 2.8, 1.4, 0.7, and 0 million/mL in 50 μL. Each of these concentrations will be additionally diluted 2× after mixing with the agarose.
It is very important to finish mixing and adding the cell–agarose mixture into the wells as soon as possible because the mixture will solidify quickly at room temperature. Preheating the pipet tips and containers or maintaining the temperature of the cell–agarose mixture at 37 °C can slow down the solidification.
Air bubbles will greatly increase the ultrasound signal of the samples and therefore should be removed before the mixture solidifies.
This ultrasound scanner has multiple transducers with different frequencies. Higher frequencies result in higher resolution but poorer penetration depth. For the in vitro study and small animal studies, transducers with high frequencies are recommended.
The ultrasound transducer should be parallel to the phantom. The distance between the top of the phantom and the transducer should be the same throughout the entire scan because the distance between the sample and transducer will affect the ultrasound signal intensity.
The DCM file provides multiple continuous frames through a transverse view. The ultrasound transducer moves to a position and stops, scans that field of view, and then moves to the next adjacent position. Therefore, each frame presents the ultrasound image of the phantom at a different field of view. Load these frames into a 3D rendering and use the maximum intensity projection (MIP, Max) to exhibit the images with the software Vevo LAB. The Vevo LAB can show 3D, sagittal, coronary, and transverse views in a quadrant, as shown in Fig. 4. The ultrasound signal of cells can be analyzed through each view. We exemplified the data analysis with sagittal views in this chapter.
The regions of interest should be at the same height when using sagittal and coronary views because the distance between the transducer and the sample affects the ultrasound signal intensity.
The time points should be based on the cell growth rate. For human mesenchymal stem cells, cells were collected on days 0, 1, 4, 7, and 14 to determine the half-life of nanoparticles in these cells.
The cell numbers needed can be calculated based on the injection dose and animal numbers. For example, given that 0.1 million mesenchymal stem cells in 20 μL vehicles are to be injected intramyocardially in a mouse to treat myocardial infarction and 6 mice will be injected, the collected cell number should be 1.2 million (0.1 million/mouse × 6 mice × 2).
It is ideal to use the stem cells from the same flask. If impossible, the cells should be treated with the same procedures and used at the same passage number to eliminate any variations brought by cell culturing and labeling.
Do not change the position of the animal bed, animal, and transducer after baseline scan. The 3D motor will automatically move back to the original position. Eliminating the position changes between baseline and postinjection imaging results in better visualization of the injected cells. To maintain exactly the same field of view before and after cell injection, a needle vice can be used to hold the syringe. It is important to align the needle and the transducer before the baseline scan.
Matrigel freezes at temperatures below −20 °C and solidifies at temperatures above 10 °C. Therefore, it is important to mix the Matrigel and cell suspension and load it into the syringe as fast as possible. Both the Matrigel and the cell mixture should be kept in ice to avoid early solidification. During this procedure, avoid introducing any air bubbles. Examine the loaded cell mixture in the syringe and remove air bubbles if there are any. Practice with the control injections (no cells) is recommended.
With a mechanic assistant injection arm, it is possible to image the same field of view before and after cell injection. Otherwise, the injection may move the imaging area. Moreover, for long-term cell tracking, the animal cannot be mounted all the time between adjacent time points and it is unlikely to image the same field of view after the animal is moved and remounted.
Fig. 4.
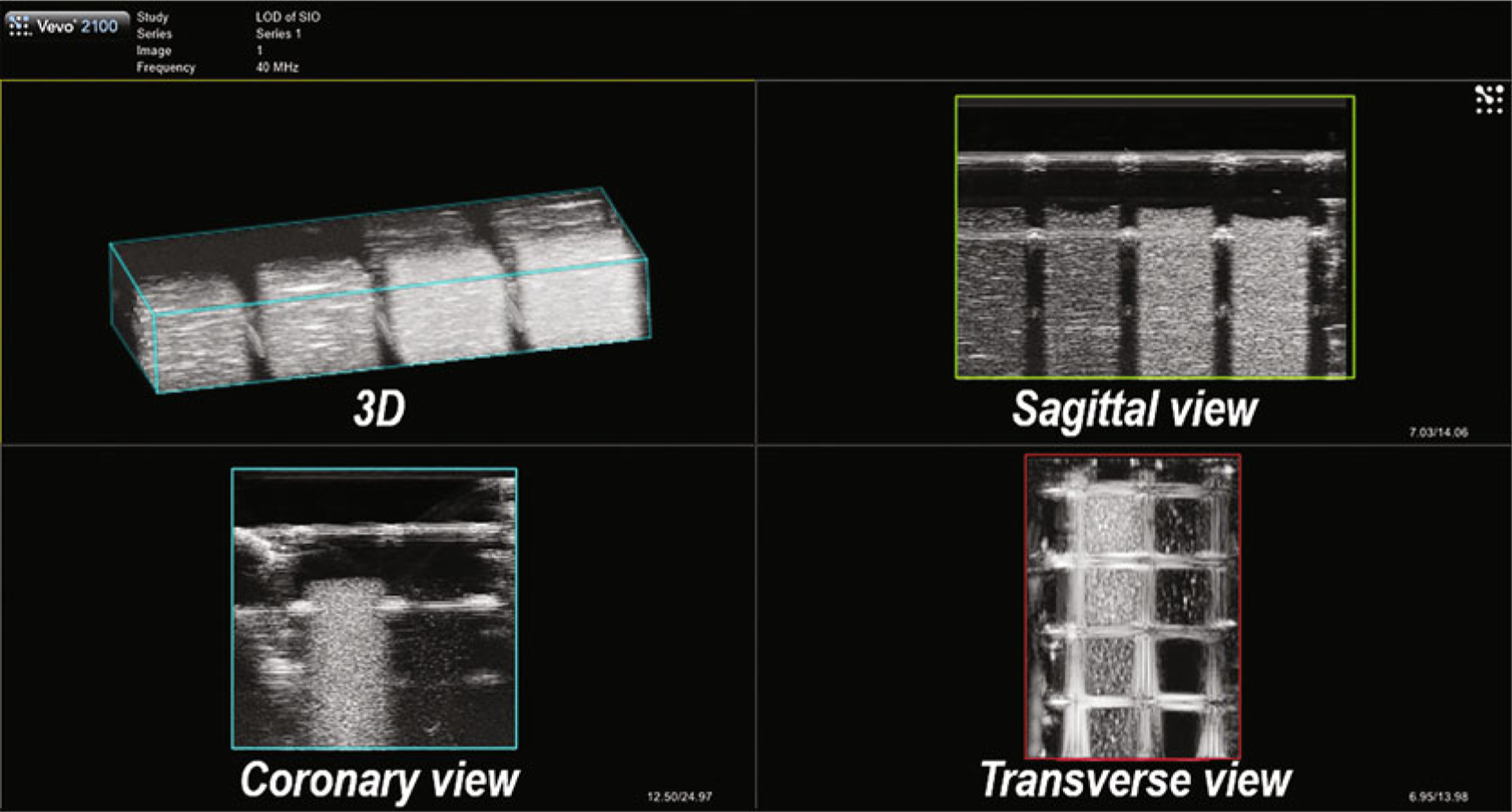
Typical views of an in vitro agarose phantom obtained with 3D B-mode ultrasound imaging shown by Vevo Lab
5. Conclusion
This chapter described a detailed method for labeling mesenchymal stem cells with inorganic nanoparticles to improve the ultrasound signal of stem cells. We explained steps for stem cell labeling, in vitro ultrasound signal detection, and in vivo stem cell tracking.
For cell labeling, we used Stöber silica nanoparticles as an example ultrasound contrast agent. While many other inorganic nanoparticles have great potential in tracking stem cells via ultrasound or photoacoustic imaging, the cytotoxicity should always be checked even if they are well known for excellent biocompatibility. This is because the toxicity of nanomaterials can be very elusive. For example, silicon carbide nanowires are biocompatible to epithelial cells but prohibited the differentiation of mesenchymal stem cells [20].
While we only introduced the in vitro ultrasound signal detection here, most updated ultrasound scanners such as the Vevo 2100 system (VisualSonics, Fujifilm) we used can also image photoacoustic signals. The 384-well plate-based agarose phantoms can also be used for the detection of photoacoustic signals of cells or nanoparticles. However, for small molecules, an agarose phantom in an unsealed container is not recommended because the small molecules can diffuse in the agarose gel. Instead, tube phantoms are recommended for small molecules [21].
The procedures for in vivo stem cell tracking are highly dependent on the injection site, animal model, and disease models. Here, we briefly introduced steps for in vivo stem cell tracking in mice. Nanoparticles can significantly increase the ultrasound signal of stem cells in vivo; however, in vivo stem cell tracking is still a relatively new area and needs many efforts in tracking not only the cell location but also the cell number, cell fate, and cell function.
Acknowledgement
This work was supported by NIH (R00 HL117048 and DP2 HL137187) and the American Cancer Society Institutional Research (grant number 14-250-42). F. Chen also acknowledges Eric Zhao for proofreading the chapter.
References
- 1.Bulte JWM, Daldrup-Link HE (2018) Clinical tracking of cell transfer and cell transplantation: trials and tribulations. Radiology 289:604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.June CH, O’connor RS, Kawalekar OU et al. (2018) CAR T cell immunotherapy for human cancer. Science 359:1361–1365 [DOI] [PubMed] [Google Scholar]
- 3.Nguyen PK, Riegler J, Wu JC (2014) Stem cell imaging: from bench to bedside. Cell Stem Cell 14:431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Jokerst JV (2016) Stem cell imaging: tools to improve cell delivery and viability. Stem Cells Int 2016:9240652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakobsen J (2001) Ultrasound contrast agents: clinical applications. Eur Radiol 11:1329–1337 [DOI] [PubMed] [Google Scholar]
- 6.Ophir J, Parker KJ (1989) Contrast agents in diagnostic ultrasound. Ultrasound Med Biol 15:319–333 [DOI] [PubMed] [Google Scholar]
- 7.Willmann JK, Van Bruggen N, Dinkelborg LM et al. (2008) Molecular imaging in drug development. Nat Rev Drug Discov 7:591–607 [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Porcel M, Gheysens O, Chen IY et al. (2005) Image-guided cardiac cell delivery using high-resolution small-animal ultrasound. Mol Ther 12:1142–1147 [DOI] [PubMed] [Google Scholar]
- 9.Ma M, Chen H, Shi J (2015) Construction of smart inorganic nanoparticle-based ultrasound contrast agents and their biomedical applications. Sci Bull 60:1170–1183 [Google Scholar]
- 10.Chen F, Ma M, Wang J et al. (2017) Exosome-like silica nanoparticles: a novel ultrasound contrast agent for stem cell imaging. Nanoscale 9:402–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemaster JE, Chen F, Kim T et al. (2018) Development of a trimodal contrast agent for acoustic and magnetic particle imaging of stem cells. ACS Appl Nano Mater 1:1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang K, Chen H, Guo X et al. (2015) Double-scattering/reflection in a single nanoparticle for intensified ultrasound imaging. Sci Rep 5:8766–8776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F, Hableel G, Zhao ER et al. (2018) Multifunctional nanomedicine with silica: role of silica in nanoparticles for theranostic, imaging, and drug monitoring. J Colloid Interface Sci 521:261–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jokerst JV, Khademi C, Gambhir SS (2013) Intracellular aggregation of multimodal silica nanoparticles for ultrasound-guided stem cell implantation. Sci Transl Med 5:177ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jokerst JV, Thangaraj M, Kempen PJ et al. (2012) Photoacoustic imaging of mesenchymal stem cells in living mice via silica-coated gold nanorods. ACS Nano 6:5920–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin CY, Chen F, Hariri A et al. (2018) Photoacoustic imaging for noninvasive periodontal probing depth measurements. J Dent Res 97:23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu MH, Wang LHV (2006) Photoacoustic imaging in biomedicine. Rev Sci Instrum 77:041101 [Google Scholar]
- 18.Kim T, Lemaster JE, Chen F et al. (2017) Photoacoustic imaging of human mesenchymal stem cells labeled with prussian blue-poly(l-lysine) nanocomplexes. ACS Nano 11:9022–9032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaster JE, Wang Z, Hariri A et al. (2019) Gadolinium doping enhances the photoacoustic signal of synthetic melanin nanoparticles: a dual modality contrast agent for stem cell imaging. Chem Mater 31:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Li G, Zhao ER et al. (2018) Cellular toxicity of silicon carbide nanomaterials as a function of morphology. Biomaterials 179:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arconada-Alvarez SJ, Lemaster JE, Wang JX et al. (2017) The development and characterization of a novel yet simple 3D printed tool to facilitate phantom imaging of photoacoustic contrast agents. Photo-Dermatology 5:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]


