Abstract
Image labeling using convolutional neural networks (CNNs) are a template‐free alternative to traditional morphometric techniques. We trained a 3D deep CNN to label the hippocampus and amygdala on whole brain 700 μm isotropic 3D MP2RAGE MRI acquired at 7T. Manual labels of the hippocampus and amygdala were used to (i) train the predictive model and (ii) evaluate performance of the model when applied to new scans. Healthy controls and individuals with epilepsy were included in our analyses. Twenty‐one healthy controls and sixteen individuals with epilepsy were included in the study. We utilized the recently developed DeepMedic software to train a CNN to label the hippocampus and amygdala based on manual labels. Performance was evaluated by measuring the dice similarity coefficient (DSC) between CNN‐based and manual labels. A leave‐one‐out cross validation scheme was used. CNN‐based and manual volume estimates were compared for the left and right hippocampus and amygdala in healthy controls and epilepsy cases. The CNN‐based technique successfully labeled the hippocampus and amygdala in all cases. Mean DSC = 0.88 ± 0.03 for the hippocampus and 0.8 ± 0.06 for the amygdala. CNN‐based labeling was independent of epilepsy diagnosis in our sample (p = .91). CNN‐based volume estimates were highly correlated with manual volume estimates in epilepsy cases and controls. CNNs can label the hippocampus and amygdala on native sub‐mm resolution MP2RAGE 7T MRI. Our findings suggest deep learning techniques can advance development of morphometric analysis techniques for high field strength, high spatial resolution brain MRI.
Keywords: artificial intelligence, deep learning, high field MRI, segmentation
We trained a 3D convolutional neural network to label the hippocampus and amygdala on whole brain 700 μm isotropic 3D MP2RAGE MRI acquired at 7T. Our findings suggest deep learning techniques can advance development of morphometric analysis techniques for high field strength, high spatial resolution brain MRI.
1. INTRODUCTION
Improvements in spatial resolution and contrast in structural MRI acquired at 7T or higher field strength holds promise for detecting disease‐related neuroanatomical abnormalities that may not be visible on clinical imaging obtained at 3T or less. Evidence for the clinical benefit of high field neuroimaging typically relies on radiological assessment of acquired images. A limitation of radiological assessment is that it is subjective and inter‐rater variability is a significant source of error. Computational morphometric image analysis techniques are alternatives to radiological assessment used extensively in lower field strength studies to model neuroanatomical structures and statistically identify tissue changes. Although there are a variety of publicly available computational tools available for analysis of neuroanatomical MRI acquired at lower strengths, these are generally (although not always) developed for analysis of T1‐weighted MRI acquired at 1 mm isotropic resolution. There are few software tools to analyze 7T or higher field strength imaging data acquired with sub‐mm isotropic resolution (Bazin et al., 2014; Seiger et al., 2015; Zaretskaya, Fischl, Reuter, Renvall, & Polimeni, 2018).
In this study, we used a recently developed 3D convolutional neural network (CNN) architecture “DeepMedic” (Kamnitsas et al., 2017), originally developed to label brain lesions, to train a model to label hippocampi and amygdala on whole brain 700 μm isotropic MP2RAGE MRI acquired at 7T in healthy controls and individuals with epilepsy. Successful application of this technique would support the utility of CNN‐based techniques for modeling subcortical neuroanatomical structures at high spatial resolution, and allow for analysis of images with novel contrast properties that differ from standard image acquisitions such as T1‐weighted MPRAGE.
While the increased field strength 7T allows imaging at higher spatial resolution and image contrast by virtue of the increased SNR and T1 values, the MP2RAGE acquisition is also of significant advantage. As discussed by Marques et al. (2010), the multiple GRE blocks in the acquisition allows for correction of proton density variation, T2* effects, and correction of B1 receive and transmit inhomogeneity. This calculated image approach which has comparatively low B1 requirements and provides nearly pure T1 weighting, has seen widespread use at 7T and may also be useful for quantitative 3T imaging in epilepsy (Kotikalapudi et al., 2019). With these properties and the large T1 differences in white, gray matter and CSF at 7T (~1.2, 2, and 4.3 s, [Rooney et al., 2007]), MP2RAGE is therefore an attractive candidate acquisition for tissue segmentation and may be useful for deep learning‐based computational image analyses.
Existing techniques to label neuroanatomical structures on brain MRIs differ from our CNN‐based approach in that CNNs and related “deep learning” techniques do not utilize predefined atlases or templates, and do not require co‐registration of these templates to an individual MRI. Instead, CNNs encode or “learn” imaging features that discriminate between the structure of interest and surrounding tissue. Several recent studies have utilized deep learning‐based approaches to segment hippocampus and other structures, but to the best of our knowledge none have been used to label high resolution 7T MP2RAGE (Chen, Dou, Yu, Qin, & Heng, 2018; Dolz, Desrosiers, & Ben Ayed, 2018; Fedorov et al., 2017; Goubran et al., 2020; Guha Roy, Conjeti, Navab, Wachinger, & Alzheimer's Disease Neuroimaging Initiative, 2019; Henschel et al., 2020; Ito, Nakae, Hata, Okano, & Ishii, 2019; Li, Yu, Gu, Liu, & Li, 2019; Mehta, Majumdar, & Sivaswamy, 2017; Milletari et al., 2017; Nogovitsyn et al., 2019; Sun et al., 2020; Thyreau, Sato, Fukuda, & Taki, 2018).
Our primary aim was to investigate whether CNNs can segment the hippocampus and amygdala. We evaluated the performance of the technique using the DSC metric to summarize the overlap between manual labels and CNN‐derived labels. We further investigated the clinical utility of the technique by applying the trained model to epilepsy patients imaged using the same MP2RAGE acquisition protocol.
The development of computational analysis techniques that can be applied to high resolution imaging is particularly important in epilepsy since one of the most effective potential treatments for individuals who do obtain adequate seizure control on medication is surgical resection of the seizure focus. In many patients the seizure focus is coincident with subtle neuroanatomical changes identified on postsurgical histopathology; however, many of these individuals have radiologically “normal” clinical imaging at 3T or lower. The clinical benefit of 7T imaging relative to 3T or lower field strength imaging to detect epilepsy‐related neuroanatomical changes is an active field of research in epilepsy neuroimaging (Canjels et al., 2020; Colon et al., 2018; De Ciantis et al., 2016; Feldman et al., 2019; Henry et al., 2011; Pittau et al., 2018; Santyr et al., 2017; Shah et al., 2019); we believe that the computational methods described in this study will be useful for addressing this question.
To facilitate further methodological developments by other research groups, we have made imaging data, manual and CNN‐based labels and scripts available at https://sites.google.com/site/hpardoe/hacl (Pardoe et al., 2020). All software used in this study was freely available to the public at the time of publication.
2. METHODS
2.1. Participants
We imaged 21 healthy controls (9 female, mean age 39 ± 15 years) and 16 epilepsy patients (8 female, mean age 36 ± 10 years) using the same acquisition protocol. Epilepsy participants were recruited from the University of Pittsburgh Medical Center Comprehensive Epilepsy Center. The primary inclusion criteria were a diagnosis of medically refractory focal epilepsy, epilepsy surgery candidates and tolerant of 7T MR imaging (nonclaustrophobic and ferromagnetic implant‐free). In the epilepsy participants, clinical imaging was radiologically classified as MRI negative in nine patients; for the MRI positive patients a single case of unilateral hippocampal sclerosis was identified based on clinical MRI, and potential hippocampal sclerosis was observed in another participant. For the remaining five MRI positive cases abnormalities included (i) left anterior temporal focal cortical dysplasia, (ii) right frontal focal cortical dysplasia, (iii) a left anterior lateral temporal lesion, (iv) bilateral nodular heterotopia, and (v) a right posterior fossa arachnoid cyst. The potential hippocampal sclerosis case had laser ablation surgery prior to 7T imaging. We excluded this subject from quantitative analyses, however, we applied our CNN‐based hippocampus & amygdala labeling model to imaging from this participant to qualitatively determine if postsurgical remnant hippocampal tissue could be labeled using our model, since morphometric analysis of postsurgical brain changes is a potential further application of the methods presented in this study. This study was approved by the University of Pittsburgh Human Research Protection Office and conforms to standards defined in the Declaration of Helsinki. All study participants gave their informed consent prior to inclusion in the study.
2.2. MRI acquisition
All studies were performed on a Siemens Magnetom whole body 7T 8 channel pTx system with an 8 × 2 transceiver and very high order shim insert. RF shimming and 1st–4th order shimming over the whole brain were performed as previously described (Hetherington, Avdievich, Kuznetsov, & Pan, 2010; Pan, Lo, & Hetherington, 2012). Images were acquired with acquisition parameters: 0.7 mm3 isotropic, TR/TI1/TI2 6s/800/2700, acquisition time 9 min.
2.3. Hippocampus and amygdala manual labeling
Hippocampi and amygdala structures were labeled using the ITK‐SNAP software package (http://www.itksnap.org/, [Yushkevich et al., 2006]). Structures were delineated in the coronal plane on consecutive slices in a posterior‐to‐anterior direction.
2.4. CNN training and testing
Facial features were removed from MRI scans using the “pydeface” software tool (https://github.com/poldracklab/pydeface). For each subject the 1 mm isotropic MNI152 template provided as part of the FSL software package was linearly coregistered to the target MP2RAGE scan using the FLIRT software tool (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FLIRT, Jenkinson & Smith, 2001). The estimated coregistration matrix was then applied to the MNI152 brain mask to generate an approximate binary mask in the space of each individual's scan. Voxel‐wise CNN sampling and prediction was limited to voxels within this brain mask.
For these analyses, the MP2RAGE acquisition was the single input channel used for model training. Five voxel‐level output classes were predicted, corresponding to left and right hippocampi, left and right amygdala and the image background. The DeepMedic approach utilizes a dual‐pathway approach, with one path designed to identify local discriminating features and an additional pathway that operates on down‐sampled input data to identify contextual features over a larger spatial scale. The default down‐sampling factor of 3 was used for our analyses. A final fully connected Conditional Random Field was used to derive “hard” binary voxel‐level labels from the probabilistic soft segmentations output by the preceding dual pathways. Image processing (CNN training and testing) was performed on a high performance compute cluster using NVIDIA Tesla V100 GPUs. We utilized a leave‐one‐out approach for evaluation of model performance. A model was trained for every subject in the study, with all subjects except the subject of interest used to train the model, and hippocampus and amygdala labels estimated in the subject of interest using this model.
2.5. Assessing performance of CNN‐based hippocampal and amygdala labeling
Performance of the CNN was assessed by calculating the DSC between manual and CNN‐based labels in the testing groups. The DSC assesses performance of image labeling tasks. If two labels perfectly overlap, the DSC = 1; if the two labels do not have any common voxels the DSC = 0. A multiple linear regression model was used to determine if (i) diagnosis, (ii) brain structure (hippocampus or amygdala), and (iii) left/right laterality were significantly correlated with the measured DSC; in this model the DSC was the dependent variable and the three variables listed above were explanatory variables.
Volume estimates were calculated from CNN‐based and manual labels by summing the number of labeled voxels for each structure (left and right hippocampus and amygdala) for each subject. The agreement between CNN‐based and manual volume estimates was assessed using a simple linear regression model for each structure, with the CNN‐based estimate as the dependent variable and the manual volume estimate as the explanatory variable. Healthy controls and epilepsy patients were assessed separately. The linear model was constrained to pass through the origin that is, the y‐axis intercept term was set at zero. For this analysis the p‐value for statistical significance was corrected for multiple comparisons using the Bonferroni method; an adjusted threshold 6.25 × 10−3 (0.05/8) was used.
For volumetric analysis of the epilepsy group we conducted both individual and group‐level comparisons. Individual z‐scores for each individual brain structure volume estimate were calculated from fractional structure volume estimates, with each hippocampal and amygdala volume divided by the head volume estimate for all cases. Group level comparisons of hippocampal and amygdala volume changes were assessed using multiple linear regression with hippocampus or amygdala volume as the dependent variable and diagnosis, age, sex, and head size as predictor variables. For these analyses the epilepsy patients were subclassified into cases with temporal lobe involvement based on all available clinical data (N = 6) and those with primarily extratemporal seizures (N = 10).
3. RESULTS
The trained CNN‐based predictive model successfully labeled all hippocampi and amygdala in healthy controls and epilepsy cases. Figure 1 shows an example of the automated hippocampal and amygdala labeling in a healthy control. The average DSC in our testing cohort = 0.88 ± 0.03 for the hippocampus and 0.8 ± 0.06 for the amygdala (Figure 2). There were some outliers with reduced DSC observed in the analyses; in the healthy control group these corresponded to subjects with reduced image quality presumably due to in‐scanner head motion and resulted in reduced DSCs across left and right hippocampal labels and the left amygdala (see Supporting Information). The outlier in the right hippocampus epilepsy group corresponded to the case with hippocampal sclerosis. The DSC for hippocampal labels was higher than for amygdala labeling (p <2 × 10−16). There was no evidence that DSC depended on diagnosis (p = .91). There was marginal evidence that right sided structures had greater DSC than left sided structures (p = .051). Summary volume statistics are provided in Table 1. The CNN‐based volume estimates were highly correlated with manual volume estimates for both left and right hippocampi and amygdala in epilepsy cases and controls (Figure 3, p <2 × 10−16 for all linear models, slope estimate range = [0.94, 0.99]), supporting the utility of the technique in clinical populations. In the single epilepsy case with clinical MRI indicating right hippocampal sclerosis, the CNN‐based method identified reduced volume in the affected hippocampus (Figure 4). In this case, the z‐score when comparing right hippocampal volume in the individual with the healthy controls = −3.64, which indicates significantly reduced volume. Applying our hippocampus and amygdala labeling model to the epilepsy case who had undergone laser ablation surgery in the left hippocampus shows that our predictive model successfully labeled remnant posterior hippocampus (Figure 5). Group level analyses did not identify any significant hippocampal volume changes in the epilepsy patients (p = .98) however amygdala volumes were increased in the cases with temporal lobe seizures (152 mm3 or 13% increase relative to control amygdala volumes, p = .017).
FIGURE 1.
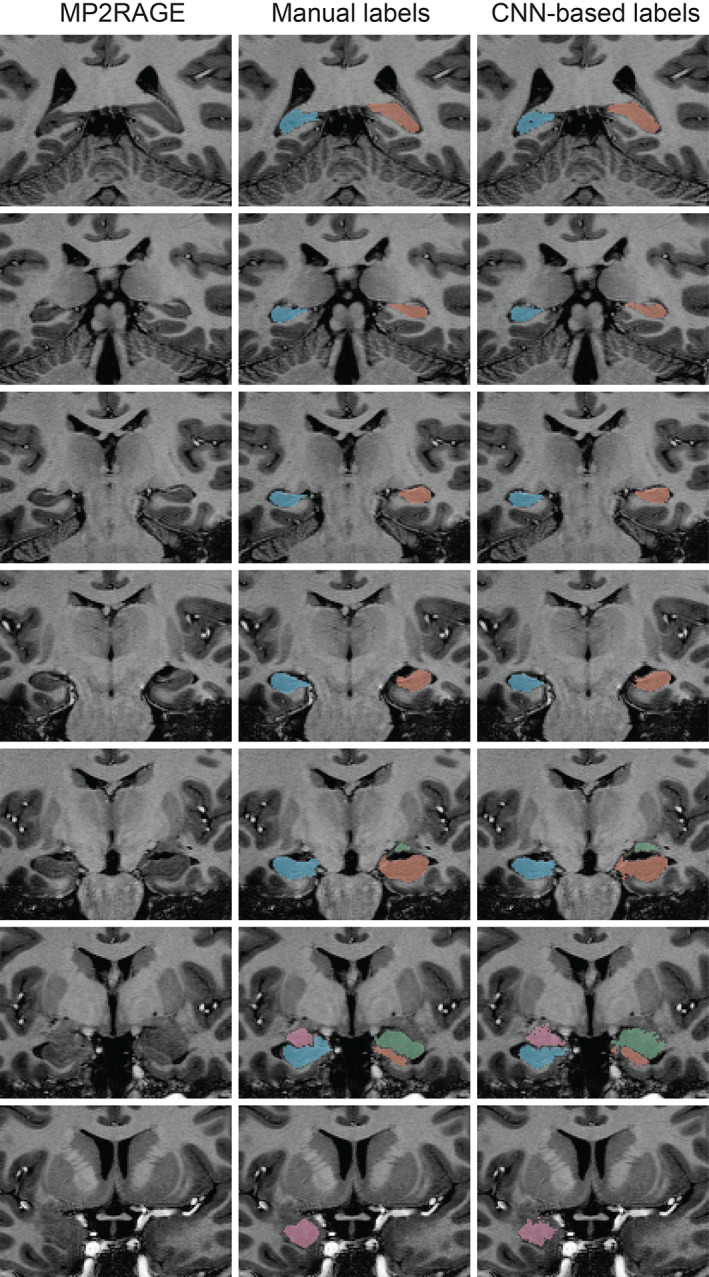
An example of automated CNN‐based labeling of the hippocampus and amygdala on whole brain 700 μm MP2RAGE acquired in a healthy control imaged at 7T. CNN‐based labels are shown on the right column, with manual labels shown in the middle column
FIGURE 2.
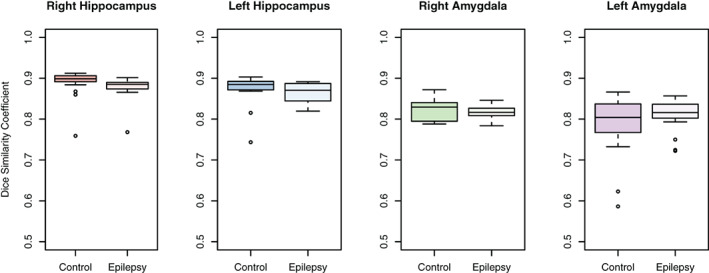
Overlap between manual and automated CNN‐based labels for hippocampi and amygdala. A DSC = 1 indicates perfect overlap of the manual and automated labels. Overlap between manual and automated labels is greater for the hippocampus than the amygdala, which is likely due to the larger volume of the hippocampus and better definition of neuroanatomical boundaries between the hippocampus and surrounding brain structures compared with the amygdala
TABLE 1.
Summary volume estimates for the hippocampus and amygdala derived using a convolutional neural network‐based labeling technique
| Right hippocampus volume (mm3, mean ± SD) | Left hippocampus volume (mm3, mean ± SD) | Right amygdala volume (mm3, mean ± SD) | Left amygdala volume (mm3, mean ± SD) | |
|---|---|---|---|---|
| Healthy control | 3,157 ± 294 | 3,046 ± 300 | 1,145 ± 217 | 1,170 ± 170 |
| Epilepsy | 2,926 ± 431 | 2,786 ± 735 | 1,215 ± 199 | 1,181 ± 259 |
FIGURE 3.
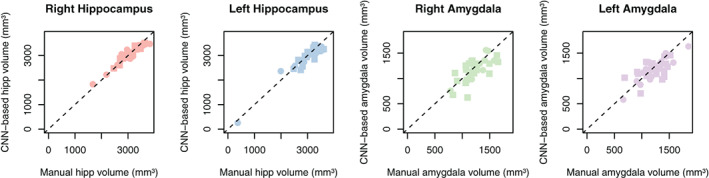
Comparison of CNN‐based volume estimates (y‐axis) with manual volume estimates (x‐axis). The plots indicate high agreement between CNN‐based volume estimates and manual volume estimates in both epilepsy patients and healthy controls
FIGURE 4.
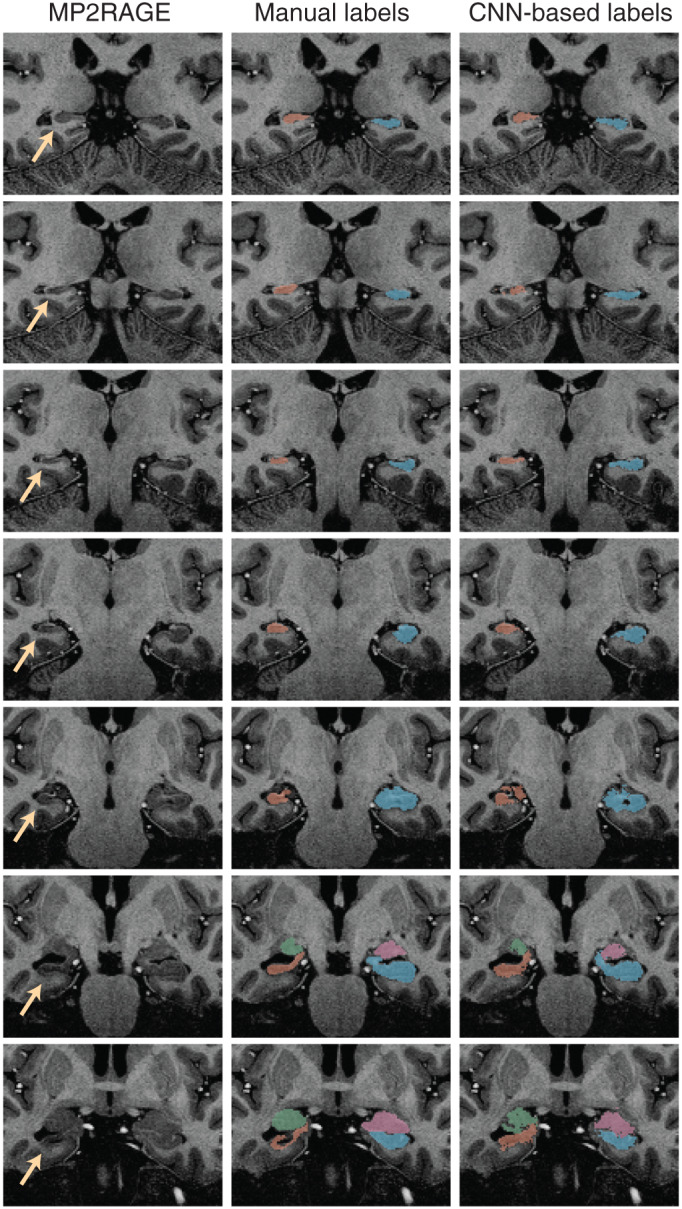
CNN‐based labeling of the hippocampus and amygdala in an individual with hippocampal sclerosis in the right hippocampus (yellow arrows). The right column shows the CNN‐based labels and the middle column shows manual labels for comparison. The figure demonstrates that the CNN‐based technique is successfully able to label the atrophic hippocampus, supporting the clinical utility of the technique for mapping hippocampal changes in temporal lobe epilepsy
FIGURE 5.
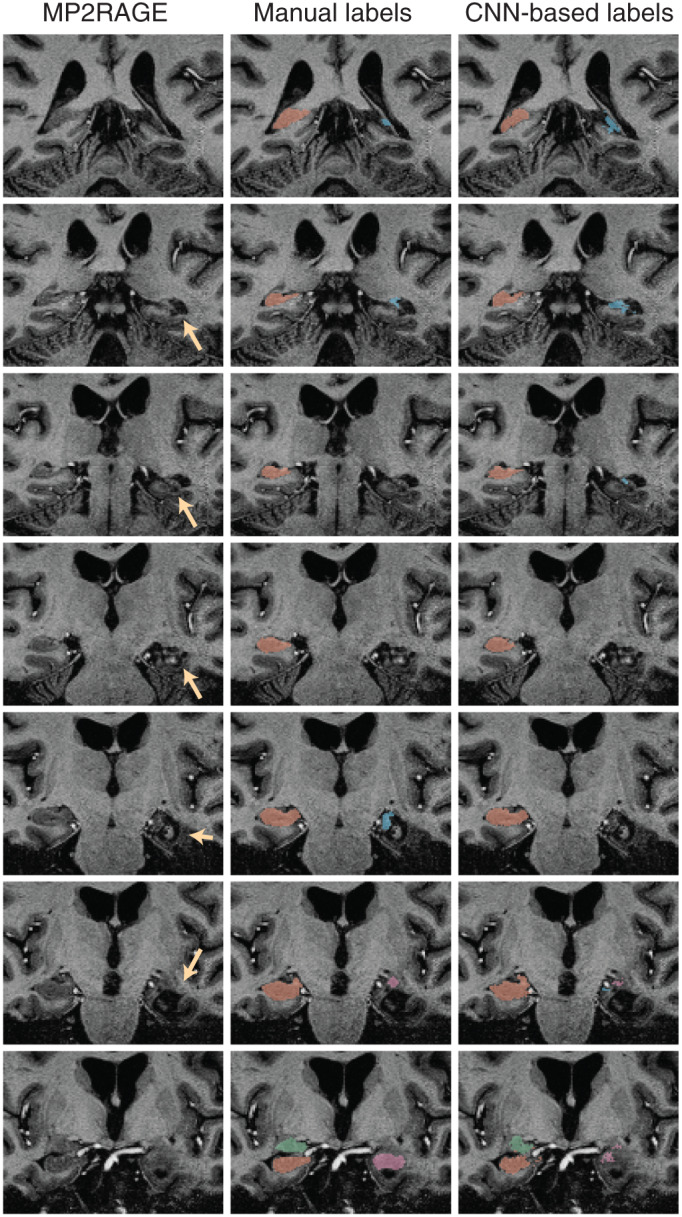
CNN‐based labeling in an epilepsy patient who has undergone laser ablation surgery. The ablated tissue region is indicated in the left column (yellow arrows). The right column indicates that the CNN‐based labeling technique is able to label remnant hippocampal tissue despite the presence of severe postsurgical neuroanatomical changes in the hippocampus and surrounding brain regions
4. DISCUSSION
Our study demonstrates that CNN‐based methods may be useful approach for labeling neuroanatomical structures on sub‐mm resolution 7T imaging. The DSC values estimated in our study are consistent with previous work; for example, Morey et al. (2009) compared automated hippocampal and amygdala volume estimates with manual segmentations using Freesurfer and FSL‐FIRST and found DSCs in the range [0.77, 0.82] for the hippocampus and [0.70, 0.75] for the amygdala, which are slightly lower than our observed DSCs of 0.88 for the hippocampus and 0.8 for the amygdala respectively. The reduced DSCs in the amygdala versus the hippocampus are likely due to the lower volume of the amygdala and relatively ambiguous neuroanatomical boundaries in the anterior end of the amygdala. Our analyses did not demonstrate that performance was reduced in epilepsy cases. We note however that our sample size is limited and medically refractory focal epilepsy is a broad category that may include a variety of distinct tissue pathologies that can vary in terms of tissue signal, contrast, and morphology. Application of the techniques described in this study to larger patient groups stratified by pathology or etiological factors will allow for more accurate assessment of CNN performance in the presence of lesional or dysmorphic tissue. Although the successful detection of reduced hippocampal volume in the patient with likely hippocampal sclerosis provides empirical support for the utility of this technique in lesional cases, our approach should be applied to a larger cohort of hippocampal sclerosis cases for a more rigorous evaluation. The inclusion of more cases with specific tissue pathology for CNN training will also likely improve performance. The finding of increased amygdala volume along with normal overall hippocampal volumes in cases with temporal lobe seizures was unanticipated, however, increased amygdala volume in temporal lobe epilepsy has been reported previously (Capizzano et al., 2019; Lv et al., 2014; Reyes et al., 2017).
Because of known problems with transmission homogeneity, a key aspect of structural imaging at 7T is the RF coil performance. In this study, the use of the 8 × 2 transceiver with RF shimming yielded sufficient B1+ homogeneity both experimentally and theoretically, so that additional methods such as the use of high permittivity pads was not necessary (Aussenhofer & Webb, 2013; Vaidya, Deniz, Collins, Sodickson, & Lattanzi, 2018). It should be noted however that it is unknown if our CNN model can be successfully applied to imaging data acquired with alternative hardware & acquisition protocols; this may require additional demonstration. Nonetheless, these datasets were acquired with routine performance, and provide target performance metrics which should be investigated in future studies. Performance is also likely to improve if a larger training dataset was used.
One of the more immediate potential applications of this technique is to segment hippocampal subfields. Arguably the canonical work in this field to date has been carried out by Wisse and colleagues using a 700 μm isotropic T2‐weighted acquisition at 7T (Wisse et al., 2012, 2014). Inspection of images provided in these publications suggests that the boundaries between hippocampal subfields are more radiologically evident than the imaging used in our study. It is possible that imaging that is predominantly T2‐weighted provides contrast that may be better suited to the task of hippocampal subfield segmentation. However it should be noted that with the minimal difference in T2 relaxation values between white and gray matter (Bartha et al., 2002; Michaeli et al., 2002), T1 lengthening that occurs at 7T, segmentation based on clarity of white‐gray matter interfaces is optimally acquired as a function of T1 weighting that is either directly included in the analysis pipeline or controlled for optimal effect (Visser, Zwanenburg, Hoogduin, & Luijten, 2010; Wisse et al., 2012).
In summary, we have demonstrated that CNNs can be trained to label the hippocampus and amygdala using high resolution MP2RAGE imaging acquired at 7T. We have provided preliminary evidence supporting the potential clinical utility of the technique in individuals with focal epilepsy. These findings suggest that CNN‐based methods may be a valuable tool for the next generation of morphometric analysis tools for high spatial resolution high‐field MRI neuroimaging.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available at https://sites.google.com/site/hpardoe/hacl, doi:10.25790/bml0cm.72.
Supporting information
Figure S1 CNN‐based hippocampal and amygdala labels estimated using scans shown in the top row had DSCs that were outliers (see Figure 2 main manuscript). Image quality is clearly reduced in these scans, presumably due to in‐scanner head motion. Images in the bottom row represent typical image quality scans with nonoutlier DSCs for hippocampal and amygdala labels.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grants NS090417, EB011639, EB024408, NS081772, and NS117990.
Pardoe HR, Antony AR, Hetherington H, et al. High resolution automated labeling of the hippocampus and amygdala using a 3D convolutional neural network trained on whole brain 700 μm isotropic 7T MP2RAGE MRI. Hum Brain Mapp. 2021;42:2089–2098. 10.1002/hbm.25348
Funding information National Institutes of Health, Grant/Award Numbers: NS117990, NS081772, EB024408, EB011639, NS090417
REFERENCES
- Aussenhofer, S. A. , & Webb, A. G. (2013). High‐permittivity solid ceramic resonators for high‐field human MRI. NMR in Biomedicine, 26(11), 1555–1561. 10.1002/nbm.2990 [DOI] [PubMed] [Google Scholar]
- Bartha, R. , Michaeli, S. , Merkle, H. , Adriany, G. , Andersen, P. , Chen, W. , … Garwood, M. (2002). In vivo 1H2O T2+ measurement in the human occipital lobe at 4T and 7T by Carr‐Purcell MRI: detection of microscopic susceptibility contrast. Magnetic Resonance in Medicine, 47(4), 742–750. 10.1002/mrm.10112 [DOI] [PubMed] [Google Scholar]
- Bazin, P. L. , Weiss, M. , Dinse, J. , Schafer, A. , Trampel, R. , & Turner, R. (2014). A computational framework for ultra‐high resolution cortical segmentation at 7Tesla. NeuroImage, 93(Pt 2), 201–209. 10.1016/j.neuroimage.2013.03.077 [DOI] [PubMed] [Google Scholar]
- Canjels, L. P. W. , Backes, W. H. , van Veenendaal, T. M. , Vlooswijk, M. C. G. , Hofman, P. A. M. , Aldenkamp, A. P. , … Jansen, J. F. A. (2020). Volumetric and functional activity lateralization in healthy subjects and patients with focal epilepsy: Initial findings in a 7T MRI Study. Journal of Neuroimaging, 30, 666–673. 10.1111/jon.12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capizzano, A. A. , Kawasaki, H. , Sainju, R. K. , Kirby, P. , Kim, J. , & Moritani, T. (2019). Amygdala enlargement in mesial temporal lobe epilepsy: an alternative imaging presentation of limbic epilepsy. Neuroradiology, 61(2), 119–127. 10.1007/s00234-018-2109-y [DOI] [PubMed] [Google Scholar]
- Chen, H. , Dou, Q. , Yu, L. , Qin, J. , & Heng, P. A. (2018). VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage, 170, 446–455. 10.1016/j.neuroimage.2017.04.041 [DOI] [PubMed] [Google Scholar]
- Colon, A. J. , Osch, M. , Buijs, M. , Grond, J. V. D. , Hillebrand, A. , Schijns, O. , … Boon, P. (2018). MEG‐guided analysis of 7T‐MRI in patients with epilepsy. Seizure, 60, 29–38. 10.1016/j.seizure.2018.05.019 [DOI] [PubMed] [Google Scholar]
- De Ciantis, A. , Barba, C. , Tassi, L. , Cosottini, M. , Tosetti, M. , Costagli, M. , … Guerrini, R. (2016). 7T MRI in focal epilepsy with unrevealing conventional field strength imaging. Epilepsia, 57(3), 445–454. 10.1111/epi.13313 [DOI] [PubMed] [Google Scholar]
- Dolz, J. , Desrosiers, C. , & Ben Ayed, I. (2018). 3D fully convolutional networks for subcortical segmentation in MRI: A large‐scale study. NeuroImage, 170, 456–470. 10.1016/j.neuroimage.2017.04.039 [DOI] [PubMed] [Google Scholar]
- Fedorov, A. , Johnson, J. , Damaraju, E. , Ozerin, A. , Calhoun, V. , & Plis, S. (2017). End‐to‐end Learning of Brain Tissue Segmentation from Imperfect Labeling. 2017 International Joint Conference on Neural Networks (IJCNN), 3785–3792.Retrieved from <Go to ISI>://WOS:000426968704006
- Feldman, R. E. , Delman, B. N. , Pawha, P. S. , Dyvorne, H. , Rutland, J. W. , Yoo, J. , … Balchandani, P. (2019). 7T MRI in epilepsy patients with previously normal clinical MRI exams compared against healthy controls. PLoS One, 14(3), e0213642. 10.1371/journal.pone.0213642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubran, M. , Ntiri, E. E. , Akhavein, H. , Holmes, M. , Nestor, S. , Ramirez, J. , … Black, S. E. (2020). Hippocampal segmentation for brains with extensive atrophy using three‐dimensional convolutional neural networks. Human Brain Mapping, 41(2), 291–308. 10.1002/hbm.24811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha Roy, A. , Conjeti, S. , Navab, N. , Wachinger, C. , & Alzheimer's Disease Neuroimaging Initiative . (2019). QuickNAT: A fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage, 186, 713–727. 10.1016/j.neuroimage.2018.11.042 [DOI] [PubMed] [Google Scholar]
- Henry, T. R. , Chupin, M. , Lehericy, S. , Strupp, J. P. , Sikora, M. A. , Sha, Z. Y. , … Van de Moortele, P. F. (2011). Hippocampal sclerosis in temporal lobe epilepsy: findings at 7 T(1). Radiology, 261(1), 199–209. 10.1148/radiol.11101651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschel, L. , Conjeti, S. , Estrada, S. , Diers, K. , Fischl, B. , & Reuter, M. (2020). FastSurfer ‐ A fast and accurate deep learning based neuroimaging pipeline. NeuroImage, 219, 117012. 10.1016/j.neuroimage.2020.117012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington, H. P. , Avdievich, N. I. , Kuznetsov, A. M. , & Pan, J. W. (2010). RF shimming for spectroscopic localization in the human brain at 7 T. Magnetic Resonance in Medicine, 63(1), 9–19. 10.1002/mrm.22182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, R. , Nakae, K. , Hata, J. , Okano, H. , & Ishii, S. (2019). Semi‐supervised deep learning of brain tissue segmentation. Neural Networks, 116, 25–34. 10.1016/j.neunet.2019.03.014 [DOI] [PubMed] [Google Scholar]
- Jenkinson, M. , & Smith, S. (2001). A global optimisation method for robust affine registration of brain images. Medical Image Analysis, 5(2), 143–156. 10.1016/s1361-8415(01)00036-6 [DOI] [PubMed] [Google Scholar]
- Kamnitsas, K. , Ledig, C. , Newcombe, V. F. J. , Simpson, J. P. , Kane, A. D. , Menon, D. K. , … Glocker, B. (2017). Efficient multi‐scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical Image Analysis, 36, 61–78. 10.1016/j.media.2016.10.004 [DOI] [PubMed] [Google Scholar]
- Kotikalapudi, R. , Martin, P. , Erb, M. , Scheffler, K. , Marquetand, J. , Bender, B. , & Focke, N. K. (2019). MP2RAGE multispectral voxel‐based morphometry in focal epilepsy. Human Brain Mapping, 40(17), 5042–5055. 10.1002/hbm.24756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. C. , Yu, Z. L. , Gu, Z. H. , Liu, H. , & Li, Y. Q. (2019). MMAN: Multi‐modality aggregation network for brain segmentation from MR images. Neurocomputing, 358, 10–19. 10.1016/j.neucom.2019.05.025 [DOI] [Google Scholar]
- Lv, R.‐J. , Sun, Z.‐R. , Cui, T. , Guan, H.‐Z. , Ren, H.‐T. , & Shao, X.‐Q. (2014). Temporal lobe epilepsy with amygdala enlargement: a subtype of temporal lobe epilepsy. BMC Neurology, 14(1), 194. 10.1186/s12883-014-0194-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, J. P. , Kober, T. , Krueger, G. , van der Zwaag, W. , Van de Moortele, P. F. , & Gruetter, R. (2010). MP2RAGE, a self bias‐field corrected sequence for improved segmentation and T1‐mapping at high field. NeuroImage, 49(2), 1271–1281. 10.1016/j.neuroimage.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Mehta, R. , Majumdar, A. , & Sivaswamy, J. (2017). BrainSegNet: a convolutional neural network architecture for automated segmentation of human brain structures. Journal of Medical Imaging (Bellingham), 4(2), 024003. 10.1117/1.JMI.4.2.024003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli, S. , Garwood, M. , Zhu, X. H. , DelaBarre, L. , Andersen, P. , Adriany, G. , … Chen, W. (2002). Proton T2 relaxation study of water, N‐acetylaspartate, and creatine in human brain using Hahn and Carr‐Purcell spin echoes at 4T and 7T. Magnetic Resonance in Medicine, 47(4), 629–633. 10.1002/mrm.10135 [DOI] [PubMed] [Google Scholar]
- Milletari, F. , Ahmadi, S. A. , Kroll, C. , Plate, A. , Rozanski, V. , Maiostre, J. , … Navab, N. (2017). Hough‐CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Computer Vision and Image Understanding, 164, 92–102. 10.1016/j.cviu.2017.04.002 [DOI] [Google Scholar]
- Morey, R. A. , Petty, C. M. , Xu, Y. , Hayes, J. P. , Wagner, H. R., 2nd , Lewis, D. V. , … McCarthy, G. (2009). A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. NeuroImage, 45(3), 855–866. 10.1016/j.neuroimage.2008.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogovitsyn, N. , Souza, R. , Muller, M. , Srajer, A. , Hassel, S. , Arnott, S. R. , … MacQueen, G. M. (2019). Testing a deep convolutional neural network for automated hippocampus segmentation in a longitudinal sample of healthy participants. NeuroImage, 197, 589–597. 10.1016/j.neuroimage.2019.05.017 [DOI] [PubMed] [Google Scholar]
- Pan, J. W. , Lo, K. M. , & Hetherington, H. P. (2012). Role of very high order and degree B0 shimming for spectroscopic imaging of the human brain at 7 tesla. Magnetic Resonance in Medicine, 68(4), 1007–1017. 10.1002/mrm.24122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoe, H. , Antony, A. , Hetherington, H. P. , Bagic, A. , Shepherd, T. , Friedman, D. , … Pan, J. W. (2020). Dataset for high resolution automated labelling of the hippocampus and amygdala using a 3D convolutional neural network trained on whole brain 700 μm isotropic 7T MP2RAGE MRI. Retrieved from https://sites.google.com/site/hpardoe/hacl [DOI] [PMC free article] [PubMed]
- Pittau, F. , Baud, M. O. , Jorge, J. , Xin, L. , Grouiller, F. , Iannotti, G. R. , … Vargas, M. I. (2018). MP2RAGE and susceptibility‐weighted imaging in lesional epilepsy at 7T. Journal of Neuroimaging, 28(4), 365–369. 10.1111/jon.12523 [DOI] [PubMed] [Google Scholar]
- Reyes, A. , Thesen, T. , Kuzniecky, R. , Devinsky, O. , McDonald, C. R. , Jackson, G. D. , … Blackmon, K. (2017). Amygdala enlargement: Temporal lobe epilepsy subtype or nonspecific finding? Epilepsy Research, 132, 34–40. 10.1016/j.eplepsyres.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney, W. D. , Johnson, G. , Li, X. , Cohen, E. R. , Kim, S. G. , Ugurbil, K. , & Springer, C. S., Jr. (2007). Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magnetic Resonance in Medicine, 57(2), 308–318. 10.1002/mrm.21122 [DOI] [PubMed] [Google Scholar]
- Santyr, B. G. , Goubran, M. , Lau, J. C. , Kwan, B. Y. M. , Salehi, F. , Lee, D. H. , … Khan, A. R. (2017). Investigation of hippocampal substructures in focal temporal lobe epilepsy with and without hippocampal sclerosis at 7T. Journal of Magnetic Resonance Imaging, 45(5), 1359–1370. 10.1002/jmri.25447 [DOI] [PubMed] [Google Scholar]
- Seiger, R. , Hahn, A. , Hummer, A. , Kranz, G. S. , Ganger, S. , Kublbock, M. , … Lanzenberger, R. (2015). Voxel‐based morphometry at ultra‐high fields. a comparison of 7T and 3T MRI data. NeuroImage, 113, 207–216. 10.1016/j.neuroimage.2015.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, P. , Bassett, D. S. , Wisse, L. E. M. , Detre, J. A. , Stein, J. M. , Yushkevich, P. A. , … Davis, K. A. (2019). Structural and functional asymmetry of medial temporal subregions in unilateral temporal lobe epilepsy: A 7T MRI study. Human Brain Mapping, 40(8), 2390–2398. 10.1002/hbm.24530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Ma, W. , Ding, X. , Huang, Y. , Liang, D. , & Paisley, J. (2020). A 3D spatially weighted network for segmentation of brain tissue from MRI. IEEE Transactions on Medical Imaging, 39(4), 898–909. 10.1109/TMI.2019.2937271 [DOI] [PubMed] [Google Scholar]
- Thyreau, B. , Sato, K. , Fukuda, H. , & Taki, Y. (2018). Segmentation of the hippocampus by transferring algorithmic knowledge for large cohort processing. Medical Image Analysis, 43, 214–228. 10.1016/j.media.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Vaidya, M. V. , Deniz, C. M. , Collins, C. M. , Sodickson, D. K. , & Lattanzi, R. (2018). Manipulating transmit and receive sensitivities of radiofrequency surface coils using shielded and unshielded high‐permittivity materials. Magma, 31(3), 355–366. 10.1007/s10334-017-0657-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, F. , Zwanenburg, J. J. , Hoogduin, J. M. , & Luijten, P. R. (2010). High‐resolution magnetization‐prepared 3D‐FLAIR imaging at 7.0 Tesla. Magnetic Resonance in Medicine, 64(1), 194–202. 10.1002/mrm.22397 [DOI] [PubMed] [Google Scholar]
- Wisse, L. E. , Biessels, G. J. , Heringa, S. M. , Kuijf, H. J. , Koek, D. H. , Luijten, P. R. , … Utrecht Vascular Cognitive Impairment Study Group . (2014). Hippocampal subfield volumes at 7T in early Alzheimer's disease and normal aging. Neurobiology of Aging, 35(9), 2039–2045. 10.1016/j.neurobiolaging.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Wisse, L. E. , Gerritsen, L. , Zwanenburg, J. J. , Kuijf, H. J. , Luijten, P. R. , Biessels, G. J. , & Geerlings, M. I. (2012). Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. NeuroImage, 61(4), 1043–1049. 10.1016/j.neuroimage.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Piven, J. , Hazlett, H. C. , Smith, R. G. , Ho, S. , Gee, J. C. , & Gerig, G. (2006). User‐guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage, 31(3), 1116–1128. 10.1016/j.neuroimage.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Zaretskaya, N. , Fischl, B. , Reuter, M. , Renvall, V. , & Polimeni, J. R. (2018). Advantages of cortical surface reconstruction using submillimeter 7 T MEMPRAGE. NeuroImage, 165, 11–26. 10.1016/j.neuroimage.2017.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 CNN‐based hippocampal and amygdala labels estimated using scans shown in the top row had DSCs that were outliers (see Figure 2 main manuscript). Image quality is clearly reduced in these scans, presumably due to in‐scanner head motion. Images in the bottom row represent typical image quality scans with nonoutlier DSCs for hippocampal and amygdala labels.
Data Availability Statement
The data that support the findings of this study are openly available at https://sites.google.com/site/hpardoe/hacl, doi:10.25790/bml0cm.72.


