SUMMARY
Brain sex differences are established developmentally and generate enduring changes in circuitry and behavior. Steroid-mediated masculinization of the rat amygdala during perinatal development produces higher levels of juvenile rough-and-tumble play by males. This sex difference in social play is highly conserved across mammals, yet the mechanisms by which it is established are unknown. Here, we report that androgen-induced increases in endocannabinoid tone promote microglia phagocytosis during a critical period of amygdala development. Phagocytic microglia engulf more viable newborn cells in males; in females, less phagocytosis allows more astrocytes to survive to the juvenile age. Blocking complement-dependent phagocytosis in males increases astrocyte survival and prevents masculinization of play. Moreover, increased astrocyte density in the juvenile amygdala reduces neuronal excitation during play. These findings highlight novel mechanisms of brain development whereby endocannabinoids induce microglia phagocytosis to regulate newborn astrocyte number and shape the sexual differentiation of social circuitry and behavior.
In Brief
VanRyzin et al. demonstrate that microglia in the developing amygdala engulf and kill otherwise viable newborn astrocytes, establishing sex differences in social circuits. This process, which depends on gonadal hormones and endocannabinoid signaling, promotes juvenile play by males.
INTRODUCTION
Brain sex differences are established by steroid hormone exposure during the perinatal period. The fetal testis produces androgens as early as the second trimester in humans and the latter third of gestation in rodents, resulting in higher levels of circulating testosterone in males. Testosterone readily gains access to the brain, where it either acts directly on androgen receptors or is locally converted into estradiol and acts via estrogen receptors. Activation of steroid hormone receptors initiates the process of sexual differentiation, whereby region-specific mechanisms masculinize the brain and program lasting behavioral differences between males and females (reviewed in Zuloaga et al., 2008; McCarthy et al., 2017).
It has long been known that sexual differentiation of the amygdala mediates a male bias toward greater intensity and frequency of juvenile rough-and-tumble play (Meaney et al., 1981; Meaney and McEwen, 1986). We previously discovered that the developing amygdala of males has fewer newborn cells than that of females. The sex difference in newborn cell number and juvenile play are both driven by a parallel and inverse sex difference in endocannabinoid (eCB) tone, being higher in the male amygdala (Krebs-Kraft et al., 2010). The eCB system is active early in brain development and comprises two principal ligands, 2-arachidonoylglycerol (2-AG) and anandamide (AEA), which act on the type-1 and type-2 cannabinoid receptors (CB1R and CB2R; reviewed in Maccarrone et al., 2014). We previously demonstrated that mimicking the “male-like” eCB tone in female rat pups via administration of cannabinoid (CB) receptor agonists masculinizes both newborn cell number and later juvenile play (Krebs-Kraft et al., 2010; Argue et al., 2017). However, the mechanism by which eCBs regulate cell number and impact play circuitry and behavior was unknown. To this end, we investigated microglia as a putative link between developmental sex differences in eCB tone and later life social behavior.
Microglia are the brain’s innate immune cells and are increasingly recognized as important modulators of brain development. They both promote and prune synaptic connectivity (Paolicelli et al., 2011; Schafer et al., 2012; Ji et al., 2013; Lenz et al., 2013; Squarzoni et al., 2014; Miyamoto et al., 2016) and regulate progenitor cell populations by providing trophic support or inducing cell death (Marín-Teva et al., 2004; Sierra et al., 2010; Cunningham et al., 2013; Ueno et al., 2013; Shigemoto-Mogami et al., 2014). Microglia express both CB1Rs and CB2Rs (reviewed in Stella, 2009), making them likely candidates to influence eCB-mediated sexual differentiation.
Here, we test the hypothesis that microglia program sex differences in the developing rat amygdala by phagocytosing newborn cells. We find that testosterone-induced elevations in eCB tone drive microglia to engulf viable newborn astrocytes in a complement-dependent manner. By developmentally eliminating astrocyte precursors, microglia alter neural excitation selectively in one node of the play circuit. Together, these findings reveal a novel mechanism for establishing developmental sex differences that involves a convergence of the eCB system and the brain’s immune system to control cell number and thereby regulate social behavior.
RESULTS
More Microglia Are Phagocytic in the Developing Male Amygdala
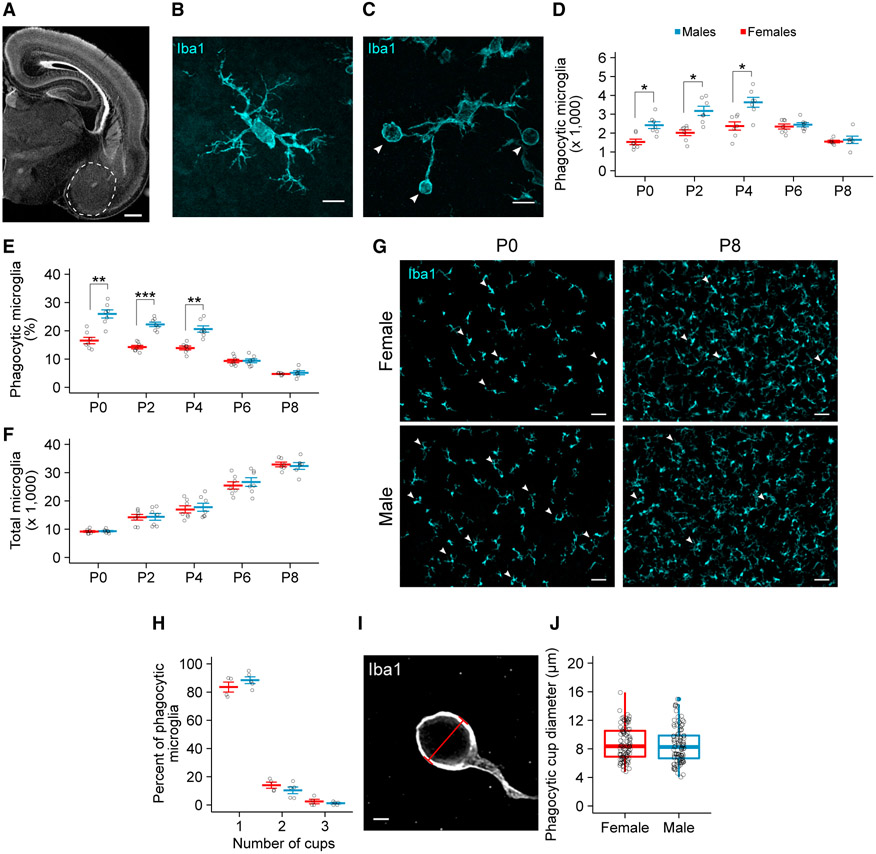
We began by characterizing the microglia population in the developing amygdala over the first postnatal week (Figure 1A), a time that encompasses the middle and end of the critical period for sexual differentiation. Using an antibody for ionized calcium binding adaptor molecule 1 (Iba1) to visualize microglia via immunohistochemistry, we found more microglia engaged in phagocytosis (defined by the presence of a phagocytic cup; Figures 1B and 1C, white arrowheads) in the amygdala of males from postnatal day 0 (P0) (birth) to P4 than in females (Figure 1D). Phagocytic microglia made up a substantial portion of the total microglia population in both males and females (25.9% ± 1.4% in males and 16.5% ± 1.1% in females on P0; Figures 1E and 1G) and gradually waned as the total microglia number increased over the first postnatal week (Figure 1F).
Figure 1. Males Have More Phagocytic Microglia in the Developing Amygdala.
(A) Nissl-stained coronal section of the P0 brain. Dashed white line indicates the boundaries of the amygdala used for analysis. Scale bar represents 500 μm.
(B and C) Maximum intensity projection of a non-phagocytic (B) and phagocytic microglia (C) immunolabeled for Iba1. White arrowheads indicate phagocytic cups in (C). Scale bars represent 10 μm.
(D–F) Quantification of the number of phagocytic microglia (D; two-way ANOVA sex × age interaction F(4, 58) = 4.763; p = 0.00216), the percentage of microglia that are phagocytic (E; two-way ANOVA sex × age interaction F(4,58) = 12.19; p = 2.96e–07), and the total number of microglia (F; two-way ANOVA main effect of age F(4, 58) = 135.419; p < 2e–16) in the developing amygdala. Holm-Bonferroni post hoc comparison between males and females at each age. p, postnatal day. n = 6 or 7 rats per sex per age.
(G) Representative 20× field of view of Iba1 labeling from the P0 and P8 amygdala in females and males. White arrowheads indicate phagocytic microglia. Scale bars represent 50 μm.
(H) Quantification of the percentage of phagocytic microglia with 1, 2, or 3 phagocytic cups. n = 4 females (140 cells) and 5 males (236 cells).
(I) Single confocal image through a phagocytic cup. Red bar demonstrates how the diameter of phagocytic cups was measured for (J). Scale bar represents 2 μm.
(J) Boxplot of the distribution of microglia phagocytic cups diameters. Open circles indicate values for individual phagocytic cups. n = 4 females (96 cups) and 4 males (97 cups).
Red and blue bars represent the mean ± SEM of females and males, respectively. Boxplot represents the median, 25%–75% range, and whiskers show max and min values. Open circles represent individual data points for each animal or values for each cell (J). *p < 0.05, **p < 0.01, and ***p < 0.001.
See also Figure S1.
To determine whether the observed sex difference in phagocytic microglia was a consequence of the cell’s activational state, we analyzed microglial morphology and CD68 immunore-activity by confocal microscopy (Figure S1A). We scored microglia along a scale ranging from 0 (highly ramified, low CD68) to 5 (amoeboid, high CD68) on P4, as previously described (Schafer et al., 2012). The distribution of microglia activation did not differ between the sexes (Figure S1B), nor were there sex differences in activation state of phagocytic microglia (Figure S1C). Microglia at the midpoint of the 0–5 scale were the most frequently phagocytic, but some microglia along the entire scale possessed phagocytic cups.
We next sought to determine whether the sex difference in phagocytic microglia number could be attributed to intrinsic differences in phagocytic activity between males and females. We analyzed the number of phagocytic cups per microglia in vivo and found no difference between males and females at P4 (Figure 1H). Moreover, most phagocytic microglia possessed just one phagocytic cup (83.6% ± 3.58% in females; 88.5% ± 2.4% in males). Because sex differences can also arise from the sex chromosomes (McCarthy and Arnold, 2011; e.g., XX versus XY), we next assessed microglia phagocytic capacity in vitro to determine whether our findings in vivo could be attributed to intrinsic differences in genetic composition. We harvested microglia from sex-specific mixed glia cultures, incubated them with fluorescent beads, and then analyzed microglial phagocytosis by flow cytometry (Figure S1D). In the absence of local cues present in the in vivo environment, microglia derived from male and female cultures engulfed similar numbers of fluorescent beads (Figure S1E), and the percentage of phagocytic microglia did not differ between the sexes (72.47% ± 3.42% in females; 79.30% ± 2.87% in males; Figure S1F). Together, these results suggest that microglia in the developing amygdala are highly phagocytic in vivo and that local cues in the newborn male amygdala make them more phagocytic than in females.
Phagocytic Microglia Engulf Newborn Cells in the Developing Amygdala
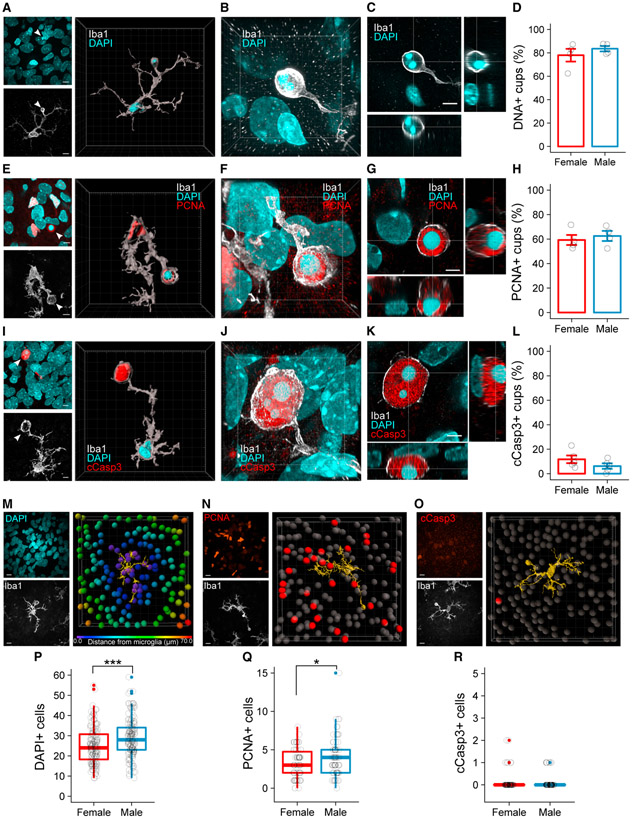
To identify the targets of microglial engulfment, we measured the diameter of phagocytic cups in the P4 amygdala using confocal microscopy (Figure 1I). We found no difference in average cup diameter between males and females (Figure 1J), and the large size (8.75 ± 0.31 μm for females; 8.55 ± 0.26 μm for males) suggested microglia were engulfing objects consistent in size with cell bodies rather than synapses or small cellular debris. To confirm that microglia were phagocytosing cell bodies, we analyzed the contents of phagocytic cups at P4 by co-immunolabeling with Iba1 and DNA binding dyes (Figures 2A-2C). Three-dimensional visualization of phagocytic cups showed clear engulfment of DAPI (Figure 2A), and as seen in Figure 2C, the DAPI signal was contained within, not apposed to, phagocytic cups. The majority of phagocytic cups contained nuclear material in both males (83.54% ± 2.29%) and females (77.94% ± 5.42%; Figure 2D). These observations led us to hypothesize that microglia engulf newborn cells. We tested this possibility by immunolabeling for proliferating cell nuclear antigen (PCNA), a marker for recently divided cells, and again analyzed the contents of phagocytic cups in males and females (Figures 2E-2G). Nearly 60% of phagocytic cups co-labeled with PCNA in both sexes (62.55% ± 4.11% in males; 59.25% ± 4.09% in females; Figure 2H), indicating microglia predominantly engulf recently born cells in the developing amygdala. To confirm that the majority of cells being engulfed were viable as opposed to undergoing apoptosis, we visualized cleaved caspase-3 (cCasp3) and again analyzed phagocytic cups (Figures 2I-2K). cCasp3 localized to only 10%–15% of phagocytic cups (Figure 2L), compared to ~60% for PCNA (Figure 2H). Overall, there were ~20× fewer cCasp3+ cells than PCNA+ cells in the P4 amygdala (Figures S2A-S2F), indicating that the differences in newborn cell number cannot be driven by apoptosis alone.
Figure 2. Phagocytic Microglia Engulf Newly Proliferated Cells.
(A) Maximum intensity projection (MIP) of DAPI (top left) and Iba1 (bottom left). White arrowhead indicates phagocytic cup. Three-dimensional rendering of a phagocytic microglia shows DAPI colocalized within the phagocytic cup (right). Scale bars represent 7 μm; grid lines represent 5 μm.
(B) Three-dimensional projection of DAPI localized within the phagocytic cup. Grid lines represent 2 μm.
(C) Orthogonal views of a phagocytic cup demonstrating colocalization of DAPI within the cup. Scale bar represents 4 μm. Voxel size = 0.065 × 0.065 × 0.1 mm.
(D) Quantification of the percentage of cups that contain DAPI. n = 4 females (95 cups) and 4 males (97 cups).
(E) MIP of PCNA and DAPI (top left) and Iba1 (bottom left). White arrowhead indicates phagocytic cup. Three-dimensional rendering of a phagocytic microglia shows PCNA and DAPI colocalized within a phagocytic cup (right). Scale bars and grid lines represent 5 μm.
(F) Three-dimensional projection of PCNA and DAPI localized within the phagocytic cup. Grid lines represent 2 μm.
(G) Orthogonal views of a phagocytic cup demonstrating colocalization of PCNA and DAPI within the cup. Scale bar represents 3 μm. Voxel size = 0.02 × 0.02 × 0.2 μm.
(H) Quantification of the percentage of cups that contain PCNA. n = 4 females (100 cups) and 4 males (98 cups).
(I) MIP of cCasp3 and DAPI (top left) and Iba1 (bottom left). White arrowhead indicates phagocytic cup. Three-dimensional rendering of a phagocytic microglia shows cCasp3 and DAPI colocalized within a phagocytic cup (right). Scale bars and grid lines represent 5 μm.
(J) Three-dimensional projection of cCasp3 and DAPI localized within the phagocytic cup. Grid lines represent 2 μm.
(K) Orthogonal views of a phagocytic cup demonstrating colocalization of cCasp3 and DAPI within the cup. Scale bar represents 3 μm. Voxel size = 0.02 × 0.02 × 0.2 μm.
(L) Quantification of the percentage of cups that contain cCasp3. n = 5 females (253 cups) and 5 males (277 cups).
(M) Representative model of a “microglia interactome.” MIP of DAPI (top left) and Iba1 (bottom left) show the raw image data for the model (right). DAPI+ cells colored by distance from the microglia surface (yellow) are shown. Only cells within 10 μm from the microglia surface were quantified in (P). Scale bars and grid lines represent 10 μm.
(N) Representative model of a microglia interactome. MIP of PCNA (top left) and Iba1 (bottom left) show the raw image data for the model (right). Only PCNA+ cells (red) within 10 μm of the microglia surface (yellow) were quantified in (Q). Scale bars and grid lines represent 10 μm.
(O) Representative model of a microglia interactome. MIP of cCasp3 (top left) and Iba1 (bottom left) show the raw image data for the model (right). Only cCasp3+ cells (red) within 10 μm of the microglia surface (yellow) were quantified in (R). Scale bars and grid lines represent 10 μm.
(P) Quantification of DAPI+ cells within 10 μm of a microglia surface. Welch’s t test t(299.52) = −3.7187; p = 0.000239. n = 150 microglia (5 females) and 152 microglia (5 males).
(Q) Quantification of PCNA+ cells within 10 μm of a microglia surface. Welch’s t test t(131.79) = −2.2802; p = 0.0242. n = 70 microglia (5 females) and 72 microglia (5 males).
(R)Quantification of cCasp3+ cells within 10 μm of a microglia surface. n = 80 microglia (5 females) and 80 microglia (5 males).
Bars represent the mean ± SEM. Boxplots represent the median, 25%–75% range, and whiskers show max and min values. Open circles represent individual data points for each animal or values for each cell (P, Q, and R). All images and quantifications were taken from P4 amygdala. *p < 0.05 and ***p < 0.001.
See also Figure S2.
To further explore the dynamics of microglia interactions with neighboring cells, we created a “microglia interactome,” in which we identified individual microglia and measured the numberof DAPI+, PCNA+, or cCasp3+ cells within 10 μm of that microglia’s processes. We found significantly more DAPI+ and PCNA+ soma in close proximity to male microglia (Figures 2M, 2N, 2P, and 2Q). In contrast, the few cCasp3+ cells detected were rarely found near microglia processes (Figures 2O and 2R). Collectively, these data indicate male microglia engage more cells overall than females and are biased toward newborn cells as opposed to dying cells. Given the higher number of phagocytic microglia in males, our data indicate that sex differences in newborn cell number are largely determined by microglia engulfment in the developing amygdala.
Testosterone Programs 2-AG Content, Newborn Cell Number, and Phagocytic Profile in the Developing Amygdala to Masculinize Juvenile Play
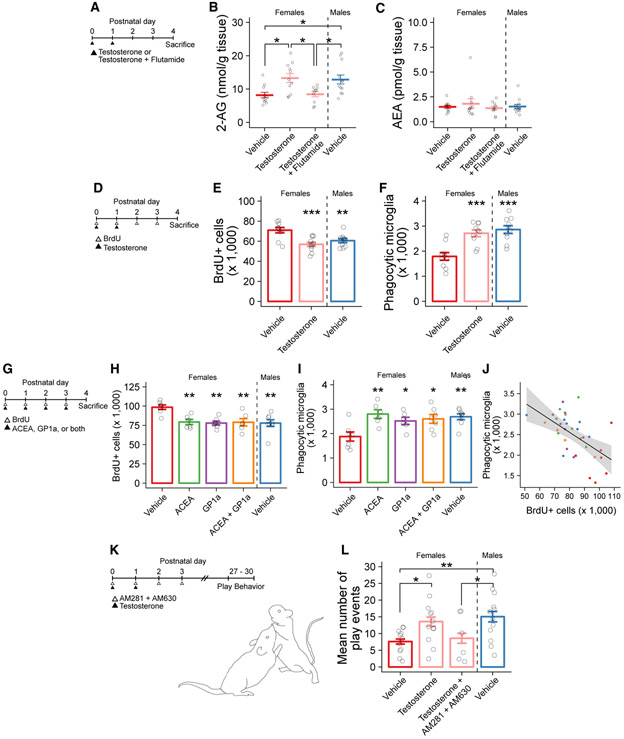
We next sought to determine whether the observed sex difference was androgen or estrogen dependent. As the critical period for sexual differentiation extends into the first few postnatal days, we treated female pups on P0 and P1 with a masculinizing dose of testosterone, or testosterone combined with the androgen receptor antagonist flutamide, and measured amygdala 2-AG content via mass spectrometry on P4 (Figure 3A). Consistent with prior results, vehicle-treated males had higher 2-AG, but not AEA, content than vehicle-treated females. Masculinizing females with testosterone increased their 2-AG, but not AEA, content, which was prevented by co-administration with flutamide (Figures 3B and 3C). In a separate experiment, administering a masculinizing dose of estradiol to female pups had no effect on eCB content (Figures S3A-S3C), confirming that hormonal programming of eCB content is androgen driven.
Figure 3. Testosterone Masculinizes Female 2-AG Content, Newborn Cell Number, and Phagocytic Microglia Number in the Developing Amygdala.
(A) Schematic showing the treatment paradigm and timeline for (B) and (C).
(B) Quantification of 2-AG levels measured by mass spectrometry in amygdala tissue. ANOVA F(3, 40) = 5.83; p = 0.00211. Holm-Bonferroni post hoc comparisons between groups are shown. n = 10–12 rats per group.
(C) Quantification of AEA levels measured by mass spectrometry in amygdala tissue. n = 10–12 rats per group.
(D) Schematic showing the treatment paradigm and timeline for (E) and (F).
(E) Quantification of the number of BrdU+ cells. ANOVA F(2, 28) = 10.38; p = 0.000423. Dunnett’s post hoc comparisons to female vehicle are shown. n = 10–11 rats per group.
(F) Quantification of the number of phagocytic microglia. ANOVA F(2, 27) = 15.75; p = 2.92e–05. Dunnett’s post hoc comparisons to female vehicle are shown. n = 10 rats per group.
(G) Schematic showing the treatment paradigm and timeline for (H)–(J).
(H) Quantification of the number of BrdU+ cells. ANOVA F(4, 29) = 5.95; p = 0.00127. Dunnett’s post hoc comparisons to female vehicle are shown. n = 6–8 rats per group.
(I) Quantification of the number of phagocytic microglia. ANOVA F(4, 29) = 4.976; p = 0.00353. Dunnett’s post hoc comparisons to female vehicle are shown. n = 6–8 rats per group.
(J) Correlation between the number of BrdU+ cells and phagocytic microglia in the same histological sections (data from H and I). Pearson’s product-moment correlation r(32) = −0.566; p = 0.004788. Filled circles indicate individual data points for each animal; colors indicate treatment group as used in (H) and (I). Black line indicates linear regression; gray shaded region indicates 95% confidence interval.
(K) Schematic showing the treatment paradigm and timeline for (L).
(L) Quantification of the mean number of play events from P27 to P30. ANOVA F(3, 37) = 3.532; p = 0.024. Post hoc Welch’s t test for specific comparisons is shown. n = 6–12 rats per group over 4 days of play testing.
Bars represent the mean ± SEM. Vertical dashed line indicates separation of male from female groups. Open circles represent individual data points for each animal. *p < 0.05, **p < 0.01, and ***p < 0.001.
See also Figure S3.
Microglia are capable of producing and secreting eCBs in vivo and in vitro (Carrier et al., 2004; Gabrielli et al., 2015; Viader et al., 2016). In order to determine whether microglia are also the source of eCBs in the developing amygdala, we depleted microglia via liposomal clodronate injection intracerebroventricularly (i.c.v.) on P0–P2 and measured eCB content in the amygdala on P4 (Figure S3D). Liposomal clodronate treatment decreased microglia by ~85% (Figure S3E) but did not affect either 2-AG or AEA content in males or females (Figures S3F and S3G).
As testosterone was sufficient to masculinize female 2-AG content in the developing amygdala, we predicted that testosterone should similarly masculinize the number of newborn cells and phagocytic microglia. We treated pups with the thymidine analog 5-bromo-2′-deoxyuridine (BrdU) on P0–P3 to mark cells born during the first few postnatal days and quantified both newborn cells and phagocytic microglia in the amygdala on P4 (Figure 3D). As before, males had fewer BrdU+ cells in the amygdala than females, and they also had more phagocytic microglia. Testosterone treatment in females reduced the number of BrdU+ cells to that of males (Figure 3E) and correspondingly increased the number of phagocytic microglia (Figure 3F). Together, these data indicate that testosterone programs the natural sex difference in eCB content, which correlates with a decrease in the number of newborn cells and an increase in the number of phagocytic microglia.
Having established that testosterone increases eCB content and increases microglial phagocytosis but reduces the number of newborn cells, we next tested the prediction that eCBs directly induce phagocytosis. Because 2-AG is an agonist at both the CB1R and CB2R, we administered the agonists ACEA (CB1R agonist) and GP1a (CB2R agonist) alone or in combination to comprehensively mimic the endogenous actions of 2-AG during this early developmental window (Figure 3G). In females, treatment with ACEA, GP1a, or a combination of ACEA + GP1a all decreased the number of newborn cells to the level normally seen in males (Figure 3H). In the same animals, CB receptor agonist treatments increased the number of phagocytic microglia in females to male levels (Figure 3I), exhibiting a significant negative correlation (r = −0.556; p < 0.001), such that animals with higher numbers of phagocytic microglia also had fewer newborn cells (Figure 3J).
To verify the effects of CB receptor agonist treatment on newborn cell number, we employed the additional approach of flow cytometry to quantify Ki-67, a protein expressed during cell proliferation (Figures S3I and S3J). We repeated the previous experiment and treated female pups with testosterone on P0 and P1 and compared the percentage of Ki-67+ (newborn) cells to vehicle-treated males and females (Figure S3K). Similar to BrdU+ cell counts, both vehicle-treated males and testosterone-treated females had fewer Ki-67+ cells than vehicle-treated females (Figure S3L). We then applied this approach to investigate endocannabinoid-induced masculinization. We administered the monoacylglycerol lipase (MAGL) inhibitor KML29 to inhibit 2-AG hydrolysis in female pups and thereby increase endogenous 2-AG levels (Figure S3M), which decreased the percentage of Ki-67+ cells compared to vehicle-treated females (Figure S3N). Thus, CB receptor activation, either by receptor agonism or endogenous increases in eCB content, is sufficient to drive sexual differentiation of the developing amygdala.
To test the hypothesis that androgen-mediated masculinization of play occurs via increasing eCBs, we treated neonatal females with a masculinizing dose of testosterone alone or combined with CB1R (AM281) and CB2R (AM630) antagonists and analyzed play behavior beginning at P27 (Figure 3K). As expected, females masculinized with testosterone exhibited male-like social play, which was not observed if combined with simultaneous CB1R and CB2R antagonism (Figure 3L), demonstrating that eCB signaling is necessary for androgen-mediated masculinization of rough-and-tumble juvenile play behaviors.
Microglia Engulf Viable Newborn Cells in a Complement-Dependent Manner to Masculinize Juvenile Play
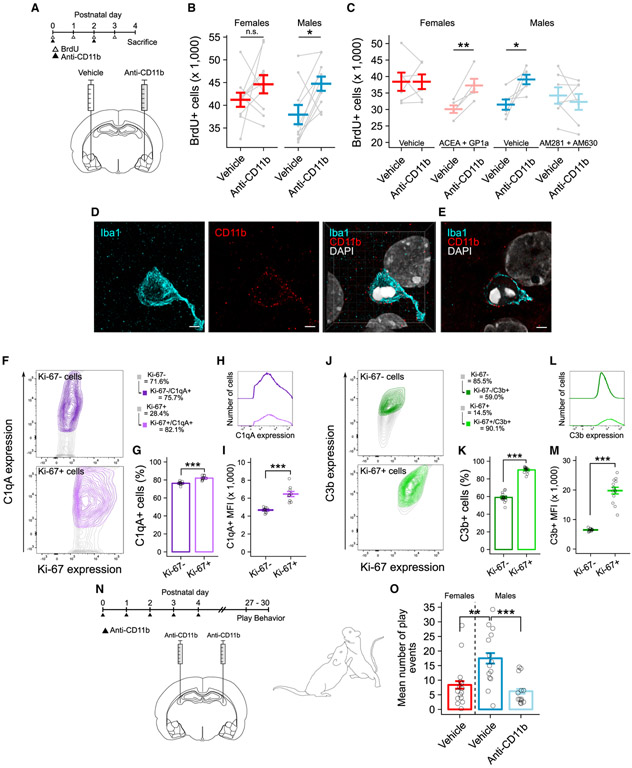
To test the hypothesis that newborn cells targeted for engulfment by microglia are otherwise capable of surviving, we used a function-blocking antibody against the CD11b component of complement receptor 3 (CR3) to prevent phagocytosis. We predicted that, if newborn cells were undergoing apoptosis, blocking phagocytosis would have no effect on total newborn cell number. However, if newborn cells were being targeted and killed by microglia, blocking phagocytosis would increase the number of newborn cells. We first confirmed the efficacy of the anti-CD11b antibody. Using a within-subject design, we administered anti-CD11b antibody directly into the amygdala of one hemisphere and vehicle solution into the amygdala of the opposite hemisphere on P2 (Figure S4A). Within 5 h post-injection, the phagocytic microglia population was reduced by ~65% compared to the vehicle-treated hemisphere (Figure S4B). To examine the effects of blocking phagocytosis on newborn cell number, we repeated the same experimental design and administered anti-CD11b antibody or vehicle on P0 and P2 (Figure 4A). We found significantly more BrdU+ cells in the anti-CD11b-treated hemisphere compared to the control hemisphere only in males, having no effect on BrdU+ cell number in females (Figure 4B).
Figure 4. Endocannabinoids Direct Male Microglia to Engulf Viable Newborn Cells in a Complement-Dependent Manner.
(A) Schematic showing the treatment paradigm and timeline for (B) and (C).
(B) Quantification of the number of BrdU+ cells between anti-CD11b treated and untreated hemispheres. Paired t test: females t(9) = −1.1924, p = 0.2636; males t(9) = −3.1369, p = 0.01199. n = 10 rats per sex.
(C) Quantification of the number of BrdU+ cells between anti-CD11b-treated and untreated hemispheres. Paired t test: female vehicle t(5) = −0.0004, p = 0.9997; female agonist t(4) = −5.199, p = 0.006521; male vehicle t(5) = −3.2157, p = 0.02358; male antagonist t(6) = 0.8718, p = 0.4168. n = 5–7 rats per group.
(D) Maximum intensity projection of Iba1 (left) and CD11b (middle) and resulting merged three-dimensional projection (right) of the phagocytic cup showing membrane localization of CD11b and DAPI colocalization within the cup. Scale bars and grid lines represent 2 μm.
(E) Single confocal plane of the merged image in (D). Scale bar and grid lines represent 2 μm.
(F) Contour plots showing the relationship between Ki-67 and C1qA expression analyzed by flow cytometry on P4. Data are quantified in (G) and (I). See Figures S4G and S4I for gating strategy.
(G) Quantification of the percent of C1qA+ cells. Paired t test t(9) = −10.282; p = 2.873e–06. n = 10 independent samples, both sexes combined.
(H) Median fluorescence intensity (MFI) plot of C1qA expression on Ki-67+ and Ki-67− cell populations. Data are quantified in (I).
(I) Quantification of C1qA MFI. Paired t test t(8) = −7.5241; p = 3.124e–08. n = 9 independent samples, both sexes combined.
(J) Contour plots showing the relationship between Ki-67 and C3b expression analyzed by flow cytometry on P4. Data are quantified in (K) and (M). See Figures S4H and S4J for gating strategy.
(K) Quantification of the percent of C3b+ cells. Paired t test t(13) = −21.086; p = 1.947e–11. n = 14 independent samples, both sexes combined.
(L) MFI plot of C3b expression on Ki-67+ and Ki-67− cell populations. Data are quantified in (M).
(M) Quantification of C3b MFI. Paired t test t(12) = −12.478; p = 3.124e−08. n = 13 independent samples, both sexes combined.
(N) Schematic showing the treatment paradigm and timeline for (O).
(O) Quantification of the mean number of play events from P27 to P30. ANOVA F(2, 40) = 8.807; p = 0.00677. Post hoc Welch’s t test for specific comparisons is shown. n = 13–16 rats per group over 4 days of play testing.
Bars represent the mean ± SEM. Open circles represent individual data points for each sample. Gray lines connect data points between hemispheres for each individual animal in (B) and (C). Contour lines in (F) and (J) represent 95% of the data at 5% intervals. *p < 0.05, **p < 0.01, and ***p < 0.001
See also Figure S4.
The unexpected male-specific effect led us to predict that the eCB tone may influence the impact of anti-CD11b antibody administration on phagocytic blockade. Given that CB receptor activation was sufficient to masculinize microglia phagocytosis and reduce newborn cell number and that the eCB tone is elevated in the developing male amygdala, we repeated the same anti-CD11b treatment (P0 and P2) and also administered CB receptor agonists (ACEA and GP1a) to females or CB receptor antagonists (AM281 and AM630) to males from P0 to P3. Reversing the eCB tone in males and females also reversed the effects of anti-CD11b treatment. We found increased BrdU+ cell number in control males and females masculinized with CB receptor agonists, but not in control females or males treated with CB receptor antagonists (Figure 4C). Thus, a higher eCB tone is necessary and sufficient to drive microglial phagocytosis of newborn cells, regardless of the sex of the pup.
We next tested the possibility that sex differences in CD11b expression may underlie the observed differences in the effectiveness of anti-CD11b treatment. Using confocal imaging, we found CD11b enriched at the microglial membrane—particularly at the phagocytic cup—in both males and females (Figures 4D and 4E). Relative quantification of CD11b levels by western blot revealed no differences between males and females on P4 (Figure S4F), indicating that differences in microglial phagocytosis are not due to CD11b. We then investigated the involvement of C1q and C3, two ligands for the phagocytic receptor CR3, which are essential to microglia-mediated synaptic pruning (Stevens et al., 2007; Veerhuis et al., 2011; Schafer et al., 2012). We acutely isolated cells from the P4 amygdala and used flow cytometry to analyze their expression on Ki-67+ and Ki-67− cells (Figures S4G-S4J). Complement proteins C1qA and C3b were both highly localized to and heavily enriched on Ki-67+ cells compared to Ki-67− cells, regardless of sex (Figures 4F, 4G, 4J, and 4K).
To determine the functional impact of developmental phagocytosis on sex differences in juvenile social play, we treated males with anti-CD11b antibody from P0 to P4 and assessed play behavior beginning at P27 (Figure 4N). As expected, vehicle-treated males played more than females, and neonatal anti-CD11b treatment reduced juvenile male play to female levels (Figure 4O). Taken together, these data demonstrate that microglia in the neonatal amygdala sense complement proteins on newborn cells via the CD11b component of CR3. Moreover, developmental phagocytosis is necessary for masculinizing juvenile play.
Neonatally Born Cells Largely Differentiate into Astrocytes in the Juvenile Posterodorsal Medial Amygdala
To determine the identity of neonatally born cells in the juvenile amygdala, we used BrdU birth dating and co-immunolabeled with cell phenotype markers. We treated pups with BrdU from P0 to P3 and euthanized them at P26 (Figure 5A). By this age, the subregions of the medial amygdala are differentiated and include the anterodorsal (MeAD), anteroventral (MeAV), posterodorsal (MePD), and posteroventral (MePV) regions, all of which process social stimuli (Meaney et al., 1981; Choi et al., 2005; Bergan et al., 2014; Hong et al., 2014; Li et al., 2017).
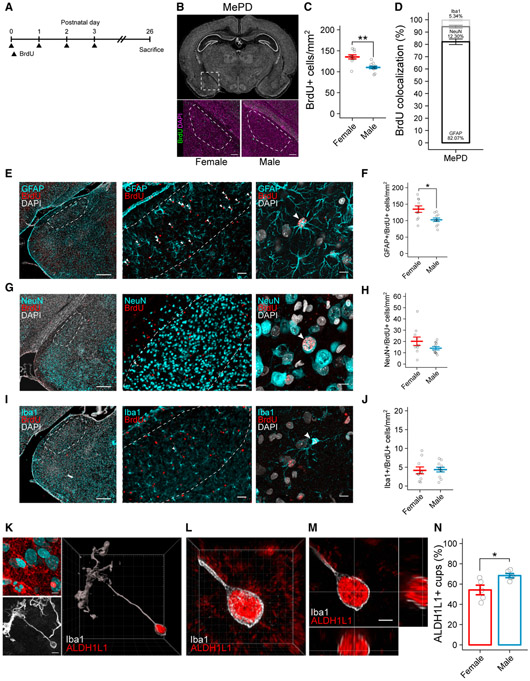
Figure 5. Neonatally Born Cells Differentiate into Astrocytes by the Juvenile Age, with More in the Female Posterodorsal Medial Amygdala.
(A) Schematic showing the treatment paradigm and timeline for (B)–(J).
(B) Nissl-stained coronal section of the P26 brain (top). Dashed box indicates location of the MePD. Representative images of female (bottom left) and male (bottom right) sections immunolabeled for BrdU and DAPI are shown. Dashed lines indicate boundaries of the MePD. Scale bars represent 150 μm.
(C) Quantification of the density of BrdU+ cells in the MePD at P26. Welch’s t test t(16.338) = 3.9763; p = 0.004184. n = 10 or 11 rats per sex.
(D) Quantification of the percent of BrdU+ cells colocalized with GFAP, NeuN, and Iba1 in the MePD. Data represent both sexes combined.
(E) Representative coronal section of the P26 amygdala immunolabeled for GFAP, BrdU, and DAPI (left). Representative 20×field of view shows GFAP and BrdU labeling in the MePD (middle). Maximum intensity projection (MIP) shows GFAP, BrdU, and DAPI colocalization (right). Dashed lines indicate boundaries of the MePD. White arrowheads indicate GFAP+/BrdU+ colocalization. Scale bars represent 400 μm (left), 50 μm (middle), and 10 μm (right).
(F) Quantification of the density of GFAP+/BrdU+ cells in the MePD. Welch’s t test t(13.904) = 2.8891; p = 0.01196. n = 10 or 11 rats per sex.
(G) Representative coronal section of the P26 amygdala immunolabeled for NeuN, BrdU, and DAPI (left). Representative 20× field of view shows NeuN and BrdU labeling in the MePD (middle). MIP shows NeuN, BrdU, and DAPI colocalization (right). Dashed lines indicate boundaries of the MePD. White arrowheads indicate NeuN+/BrdU+ colocalization. Scale bars represent 400 μm (left), 50 μm (middle), and 10 μm (right).
(H) Quantification of the density of NeuN+/BrdU+ cells in the MePD. n = 10 or 11 rats per sex.
(I) Representative coronal section of the P26 amygdala immunolabeled for Iba1, BrdU, and DAPI (left). Representative 20× field of view shows Iba1 and BrdU labeling in the MePD (middle). MIP shows Iba1, BrdU, and DAPI colocalization (right). Dashed lines indicate boundaries of the MePD. White arrowheads indicate Iba1+/BrdU+ colocalization. Scale bars represent 400 μm (left), 50 μm (middle), and 10 μm (right).
(J) Quantification of the density of Iba1+/BrdU+ cells in the MePD. n = 10 or 11 rats per sex.
(K) MIP of ALDH1L1 and DAPI (top left) and Iba1 (bottom left). White arrowhead indicates phagocytic cup. Three-dimensional rendering of a P4 phagocytic microglia shows ALDH1L1 colocalized within a phagocytic cup (right). Scale bars and grid lines represent 5 μm.
(L) Three-dimensional projection of ALDH1L1 localized within the phagocytic cup. Grid lines represent 1 μm.
(M) Orthogonal views of the phagocytic cup demonstrating colocalization of ALDH1L1 within the cup. Grid lines represent 2 μm. Voxel size = 0.0124 × 0.0124 × 0.1 μm.
(N) Quantification of the percentage of phagocytic cups that contain ALDH1L1. Welch’s t test t(5.4415) = −2.7408; p = 0.03728. n = 5 females (120 cups) and 5 males (120 cups).
Bars represent the mean ± SEM. Open circles represent individual data points for each animal. MePD, posterodorsal medial amygdala. *p <0.05and ***p < 0.001.
See also Figure S5.
We found more BrdU+ cells in females in the MePD, but not the other subregions (Figures 5B, 5C, and S5A-S5E). To determine the cellular phenotype of the neonatally born cells in the MePD, we co-immunolabeled sections with BrdU and astrocyte-specific (glial fibrillary acidic protein [GFAP]; Figure 5E), neuron-specific (NeuN; Figure 5G), or microglia-specific (Iba1; Figure 5I) antibodies. Approximately 80% of BrdU+ cells co-labeled with GFAP in both sexes (Figure 5D); however, females had a higher density of BrdU+/GFAP+ cells than males (Figure 5F) and a corresponding increase in total GFAP+ cell density (Figure S5F). Far fewer BrdU+ cells were also NeuN+ (Figure 5H) or Iba1+ (Figure 5J), with no sex difference in density of either neurons or microglia (Figures S5G and S5H).
Based on these data, we hypothesized that microglia drive the sex difference in the amygdala by phagocytosing cells that were destined to differentiate into astrocytes. To test this, we used confocal microscopy to analyze microglia in the P4 amygdala co-immunolabeled for Iba1 and the astrocyte marker ALDH1L1, as this labels astrocytes earlier in development than GFAP. Some microglia were found to have ALDH1L1 within the phagocytic cup (Figures 5K-5M), indicating the engulfment of astrocytic material. A significantly greater percentage of microglial cups co-labeled with ALDH1L1+ in males (68.35% ± 2.05%) compared to females (54.17% ± 4.75%; Figure 5N). These data support our hypothesis that microglia phagocytose more astrocytes in the developing amygdala of males, resulting in a lower astrocyte density by the juvenile age.
Neonatal Microglia Phagocytosis Programs Juvenile Play Behavior, Astrocyte Density, and Neuronal Activation in the MePD
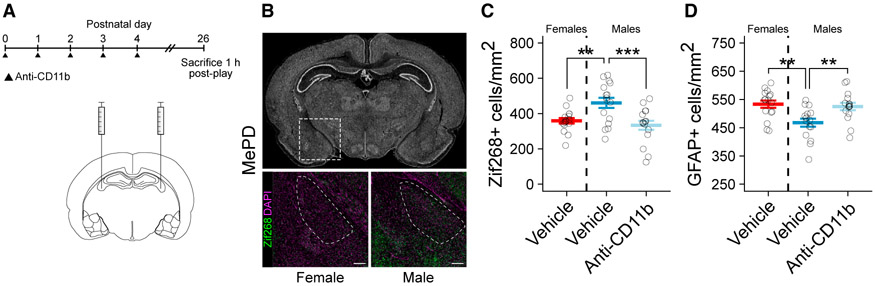
To test the dual hypothesis that developmental phagocytosis regulates astrocyte density and likewise impacts neuronal excitation during play, we administered anti-CD11b to males from P0 to P4, raised them to P26, and euthanized them 1 h after a single 10-min exposure to a novel play partner (Figure 6A). We again found greater astrocyte density in the MePD of females than males, and males neonatally treated with anti-CD11b had an astrocyte density indistinguishable from females (Figure 6D). We assessed neuronal activation by visualizing the immediate early gene zif268 (Egr1). In the MePD, males had more zif268+ neurons than females, and anti-CD11b treatment reduced the number of zif268+ cells in males to female levels (Figures 6B and 6C). These data demonstrate that preventing developmental phagocytosis feminizes neuronal activation in the MePD in response to social interactions.
Figure 6. Developmental Microglia Phagocytosis Programs Juvenile Play Behavior, Astrocyte Density, and Neuronal Activation in the MePD.
(A) Schematic showing the treatment paradigm and timeline for (B)–(D).
(B) Nissl-stained coronal section of the P26 brain (top). Dashed box indicates location of the MePD. Representative images of female (bottom left) and male (bottom right) sections immunolabeled for zif268 and DAPI are shown. Dashed lines indicate boundaries of the MePD. Scale bars represent 200 μm.
(C) Quantification of the density of zif268+ cells in the MePD. ANOVA F(2, 45) = 7.877; p = 0.00117. Dunnett’s post hoc comparisons to male vehicle are shown. n = 16 rats per group.
(D) Quantification of the density of GFAP+ cells in the MePD. ANOVA F(2, 45) = 7.266; p = 0.00184. Dunnett’s post hoc comparisons to male vehicle are shown. n = 16 rats per group.
Bars represent the mean ± SEM. Open circles represent individual data points for each animal. **p < 0.01 and ***p < 0.001.
See also Figure S6.
Our observation that developmental phagocytosis selectively regulates astrocyte density in the MePD suggests this region is the critical node for determining sex differences in play. To determine whether development of other regions in the play and social reward circuitry are also regulated by phagocytosis, we quantified microglia in the prefrontal cortex (PFC), paraventricular nucleus of the hypothalamus (PVN), and the nucleus accumbens (NAc) across the first 4 days of life. There were no sex differences in the number of total or phagocytic microglia across any of these regions (Figures S6A, S6D, and S6G). We also quantified zif268+ cell number in these regions following play and found no sex difference in the PFC (Figures S6B and S6C) and found higher numbers of zif268+ cells in females in both the PVN (Figures S6E and S6F) and NAc (Figures S6H and S6I). Thus, only in the amygdala is developmental phagocytosis greater in males and associated with increased neuronal activation during juvenile play.
DISCUSSION
Rough-and-tumble play is a unique form of social behavior with strong face validity for humans. Social play is widely expressed across the animal kingdom and in most species is restricted to the juvenile period. In virtually every species that exhibits physical play, including humans and non-human primates (Leresche, 1976; Palagi et al., 2007), males are characterized by higher frequency and intensity of interaction (Pellis et al., 1997). Social play comprises a complex sequence of coordinated behaviors that requires appropriate initiation and reciprocal response to social cues (Pellis and McKenna, 1992; Argue and McCarthy, 2015). Here, we identify the amygdala as an essential node mediating the sex difference in juvenile social play, which is established developmentally by the phagocytic action of microglia. Testosterone-induced endocannabinoids drive microglia phagocytosis of newborn astrocytes, which ultimately modulates neuronal activity in the MePD and promotes a higher frequency of play.
Androgens Elevate Endocannabinoid Tone in the Amygdala during the Critical Period for Sexual Differentiation
Sexual differentiation of the brain begins prenatally, induced by a testosterone surge produced by the fetal male testis. Testosterone circulates and reaches the brain, where it acts on androgen receptors or is locally converted into estradiol (McCarthy et al., 2017). Multiple mechanisms are invoked by steroids simultaneously in distinct brain regions in the service of differentiating a variety of functional endpoints. One such mechanism, as identified here, is androgen-dependent upregulation of eCB tone within the developing amygdala. By birth, this process is well underway with the male-specific increase in local eCB content observable at P0 and persisting for the first few days of life (Krebs-Kraft et al., 2010). However, neither the source of eCBs nor the mechanism by which androgens lead to their increase (i.e., synthesis or degradation) is clear. Microglia are capable of producing 2-AG (Carrier et al., 2004; Witting et al., 2004) and express distinct synthetic and hydrolytic enzymes (Witting et al., 2004; Viader et al., 2016). Although they would seem to be likely candidates for mediating differential eCB production, our data indicate otherwise, as neonatal microglia depletion did not alter the developmental eCB tone. This raises the question: where are neonatal amygdala eCBs coming from? There are two non-exclusive possibilities. First, mature neurons and astrocytes could be responsible for the developmental eCB tone, as both are critical to 2-AG metabolism in the adult brain (Viader et al., 2015, 2016). Second, the newborn cells themselves may be the source of the eCB tone (Aguado et al., 2005, 2006). The former hypothesis predicts that earlier born and mature cells influence the number of cells that subsequently develop, possibly as a mechanism for attaining appropriate cell numbers and density. Conversely, the latter hypothesis predicts that control of cell number in the developing amygdala is self-regulated by the pool of stem cells, perhaps related to their ultimate fate as astrocytes instead of neurons.
Endocannabinoids Provide a Local Cue that Directs Phagocytosis of Viable Newborn Cells in a Complement-Dependent Manner
Sex differences in the morphology and activational state of microglia are evident developmentally in multiple brain regions (Schwarz et al., 2012; Lenz et al., 2013; Hanamsagar et al.,2017). We found no differences in microglia number or functional profile; instead, only the propensity for phagocytosis differed, being higher in males and promoted by endocannabinoids. Quantification of CD68+ lysosomal activity has been used to quantify microglia engulfment (Schafer et al., 2012) and to infer microglia functional or metabolic state (De Biase et al., 2017). Microglia along the entire spectrum of functional states were phagocytic, but the majority were at the midpoint between fully activated and surveying, reinforcing the notion that microglia can effectively phagocytose their targets without becoming activated, as seen in the adult hippocampus (Sierra et al., 2010).
Sex differences can arise both from gonadal hormone exposure and directly from the sex chromosomes. We found that, when removed from the body and hence any circulating steroids, microglia did not differ in their intrinsic phagocytic capacity, suggesting the sex difference is the result of the microenvironment of the developing amygdala, consistent with other findings (De Biase et al., 2017). We identified eCBs as the microenvironment cue, with the sex difference in developmental phagocytosis tightly linked to transient changes in eCB tone. How eCBs induce a phagocytic state is unknown. 2-AG can act as a chemoattractant for microglia, and its role as a “find me” signal has been proposed in the context of neuroinflammation (Walter et al., 2003). The higher eCB tone may induce more microglial mobility and motility in males, increasing surveillance and driving microglia toward newborn cells, which are then engulfed due to the high localization of “eat me” signals. Our findings support this notion, based on an interactome in which the proximity of microglia to newborn cells was mapped and measured. On average, more newborn cells were in close proximity to microglia in males. In developing neurons, 2-AG is critical for appropriate pathfinding, directing axonal outgrowth to appropriate targets, and assisting in the formation of cortical circuits (Mulder et al., 2008; Vitalis et al., 2008; Wu et al., 2010). Thus, eCB signaling may provide chemoattractant signals for both neurons and microglia, albeit for different purposes, in early life. Notably, the effects of eCBs on microglia in the immature brain equally involve CB1 and CB2 receptors, with no additive effects, suggesting a convergence on a common signal transduction pathway.
Phagocytosis is usually associated with the engulfment of dead or dying cells, but phagocytosis of viable cells, termed primary phagocytosis or “phagoptosis” (reviewed in Brown and Neher, 2012, 2014), is an evolutionarily conserved mechanism during brain development that eliminates excess progenitor cells (Hoeppner et al., 2001; Reddien et al., 2001; Cunningham et al., 2013). Microglia also actively induce cell death via the production of reactive oxygen species in the developing cerebellum (Marín-Teva et al., 2004) and hippocampus, which is dependent upon CD11b/CR3 (Wakselman et al., 2008). We found tightly regulated phagoptosis of newborn cells, producing a sex difference in density of mature astrocytes. Moreover, complement proteins, recognized by microglial CR3, trigger the phagocytosis of these otherwise viable newborn cells. We found newborn cells are more likely to express eat me signals and, when they do so, at a higher level than non-newborn cells. C3b was substantially more enriched than C1qA, perhaps reflecting its role as a “tagging and/or signal amplification” factor in the complement cascade (Veerhuis et al., 2011). There may also be additional factors, such as “don’t eat me” signals (Lehrman et al., 2018), expressed by mature cells that protect them from phagocytosis.
Juvenile Social Play Is Masculinized by Microglia-Mediated Reduction in Astrocyte Density of the MePD
The neural circuitry of play is largely redundant with the canonical social behavior circuit and intersects with components of the reward circuit (see Vanderschuren et al., 2016 for review). Both males and females engage in social play but males more so. The higher levels of play by males could be a distributed property of the social and/or reward networks or it could be due to a specific node that boosts responsiveness to cogent stimuli and promotes more frequent interactions. It has long been known that the medial amygdala is important to promoting higher levels of play by males (Meaney et al., 1981, 1983; Meaney and McEwen, 1986), but why that is so and whether it is uniquely so has been unknown. We have filled that gap, implicating microglial control of astrocyte density as essential to higher play by males. The sex difference in the astrocyte population of the MePD could either impact the formation of this node, perhaps by impacting synapse formation as in other sexually dimorphic brain regions (Mong et al., 1999; Amateau and McCarthy, 2002), or MePD astrocytes may perform “real-time” synaptic gating of social information during a play bout (Martin-Fernandez et al., 2017). In support of the latter scenario, male MePD neurons were more active during play, unless the astrocyte density was increased by blocking developmental phagocytosis, in which case both neuronal activity and frequency of play were the same as seen in females. Independently manipulating microglia phagocytosis, without changing steroid hormone profile or endocannabinoid tone, causally connects the density of surviving astrocytes and the propensity for play. Furthermore, we found no evidence that developmental phagocytosis programs other critical nodes of the social and reward circuits, confirming the centrality of the MePD.
In conclusion, we present a series of novel mechanisms mediating masculinization of the medial amygdala to promote higher levels of juvenile play. An androgen-induced upregulation of the endocannabinoid tone promotes microglia phagocytosis of astrocytic precursors via a complement-dependent process. This results in a reduced density of astrocytes in the amygdala of juvenile males, which allows for greater neuronal activation that in turn promotes more intense play. Through this series of cellular events, a reliable and enduring sex difference is established for a complex social behavior.
Identifying the biological origins of sex differences provides new insights into basic mechanisms and sources of individual variability in brain development. Our data highlight a potential consequence of cannabis use by pregnant women, as delta-9-tetrahydrocannibinol (THC) and other cannabinoid molecules can cross the placenta (Grant et al., 2018) and could thereby influence brain development in a sex-dependent manner. To date, prenatal exposure to cannabis has been found to elicit subtle, yet potentially significant, effects on brain development (Berghuis et al., 2005; Tortoriello et al., 2014). As Western society enters a new era with increasing medicalization, decriminalization, and legalization of marijuana, our results compel us to understand the basic biological mechanisms by which the brain develops and can be perturbed by exogenous neuromodulatory compounds in a sex-dependent manner.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Margaret McCarthy (mmccarthy@som.umaryland.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal studies
Adult Sprague-Dawley rats, obtained from Charles River Laboratories, were maintained on a 12:12h reverse light/dark cycle with ad libitum food and water. Animals were mated in our facility, and pregnant females were allowed to deliver naturally with the day of birth being designated as postnatal day 0 (P0). On P0, pups were sexed, treated, and culled to no more than 14 pups per dam. Male and female pups were used in these studies, and treatment groups and sexes were balanced across litters. All animal procedures were performed in accordance with the Animal Care and Use Committee’s regulations at the University of Maryland School of Medicine.
Primary cell culture
To generate sex-specific mixed glia cultures, rat pups were sexed on the day of birth and rapidly decapitated, and their brains were removed and placed into ice cold HBSS (without divalent ions), stripped of meninges, and minced with a razor blade. Individual brains were incubated for 15 min in 0.5% trypsin at 37°C, then dissociated by trituration with a pipette. The resulting suspension was pelleted by centrifugation at 400g for 4 min. Excess supernatant was aspirated and the suspension was resuspended in culture media (DMEM/F12 supplemented with 0.45% D-glucose, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum). One P0 rat brain was used to seed a T75 flask, which was incubated at 37°C with 5% CO2. One day after seeding, the culture media was completely replaced to remove unattached cells. Subsequently, 50% of the media was replaced every 4 days, and cultures were allowed to grow until DIV14. On DIV14, flasks were shaken on a rotating platform at 100 rpm for 1 h to detach microglia from the astrocyte layer. Media was removed from the flask and centrifuged at 400g for 4 min to pellet microglia, and resuspended in serum-free culture media (DMEM/F12, 0.45% D-glucose, 100 U/ml penicillin, 100 μg/ml streptomycin).
METHOD DETAILS
Animal treatments
The following drugs and doses were used in these studies:
For studies involving hormonal modulation, drugs were dissolved in sesame oil and delivered subcutaneously. Testosterone propionate (100 μg; Sigma-Aldrich Cat#T1875), flutamide (100 μg; Sigma-Aldrich Cat#F9397), and estradiol benzoate (10 μg; Sigma-Al-drich Cat#E8515) were injected in a volume of 0.1 mL per pup per day.
Cannabinoid receptor agonists ACEA (1 mg/kg; Tocris Cat#1319) and GP1a (1 mg/kg; Tocris Cat#2764) and the MAGL inhibitor KML29 (1 mg/kg; Tocris Cat#4872) were dissolved in ethanol at 5 mg/ml and further diluted in saline and delivered intraperitoneally in a volume of 0.1 mL per pup per day.
Cannabinoid receptor antagonists AM281 (1 mg/kg; Tocris Cat#1115) and AM630 (1 mg/kg; Tocris Cat#1120) were dissolved in DMSO at 5 mg/ml and further diluted in saline and delivered intraperitoneally in a volume of 0.1 mL per pup per day.
BrdU (50 mg/kg; Sigma-Aldrich Cat#B5002) was dissolved in saline and delivered intraperitoneally in a volume of 0.1 mL per pup per day.
Liposomal clodronate (Encapsula Nanosciences Cat#CLD-8909) was delivered by bilateral intracerebroventricular injection (1 μL per hemisphere) at the following coordinates: −1.0 mm, caudal from bregma; ± 1.0 mm, lateral from midline; −3.0 mm, ventral from surface of the skull.
Anti-CD11b antibody (0.5 mg/ml; OX-42 clone; Bio-Rad Cat#MCA275GA, RRID:AB_566455), anti-F4/80 antibody (0.2 mg/ml; Santa Cruz Cat#sc-52664, RRID:AB_629466) or phosphate buffered saline vehicle was delivered by intra-amygdalar injection at the following coordinates: −0.8 mm, caudal from bregma; ± 3.0 mm, lateral from midline; −5.0 mm, ventral from surface of the skull).
Intracerebroventricular and intra-amygdalar injections were performed under cryoanesthesia using a 23 gauge Hamilton syringe attached to a stereotaxic instrument. The time the pups were separated from the dam was kept to a minimum, between 15 min to 1 h for all procedures.
Immunohistochemistry
Rat pups were deeply anesthetized with Fatal Plus (Vortech Pharmaceuticals) and transcardially perfused with phosphate-buffered saline (PBS; 0.1M, pH 7.4) followed by 4% paraformaldehyde (PFA; 4% in PBS, pH 6.8). Brains were removed and postfixed for 24 h in 4% PFA at 4°C, then kept in 30% sucrose at 4°C until fully submerged. Coronal sections were cut at a thickness of 45 μm (for developmental studies) or 20 μm (for juvenile studies) on a cryostat (Leica CM2050S) and directly mounted onto silane-coated slides.
For fluorescent imaging, slide-mounted sections were washed in PBS and blocked with either 5% normal goat serum (NGS) or 10% bovine serum albumin (BSA) in PBS + 0.4% Triton X-100 (PBS-T) for 1 h. Slides were incubated in primary antibody solution (either 5% NGS or 5% BSA in PBS-T) overnight. The following day, slides were incubated in secondary antibody solution (either 5% NGS or 5% BSA in PBS-T) for 2 h and coverslipped with ProLong Diamond Antifade (Thermo Fisher Scientific). For quantification of DNA material in phagocytic cups, sections were incubated in NucRed Dead 647 (Invitrogen Cat#R37113) or DAPI (Hoechst; Thermo Fisher Scientific Cat#62249) according to manufacturer’s instructions. Sections used for fluorescent Nissl imaging were stained with NeuroTrace (Thermo Fisher Scientific Cat#N21480) according to manufacturer’s instructions.
For DAB staining, slide-mounted sections were washed in Tris-buffered saline (TBS; 0.05M, pH 7.6), incubated in 0.3% hydrogen peroxide in TBS for 30 min at room temperature. Sections were blocked with 5% NGS or 10% BSA in TBS + 0.4% Triton X-100 (TBS-T). Sections were incubated in primary antibody in solution (either 5% NGS in TBS-T or 5% BSA in TBS-T) overnight. The following day, sections were incubated in biotinylated secondary antibody for 1 h, followed by incubation in ABC reagent (1:500 dilution; Vectastain Elite ABC Kit, Vector Laboratories) in TBS-T for 1 h, and visualized using DAB chromagen or nickel-enhanced DAB chromagen (0.05% 3,3′-diaminobenzidine, 0.2% nickel (II) sulfate, 0.006% hydrogen peroxide; all from Sigma-Aldrich). The DAB reaction was allowed to proceed until completion, as confirmed under a microscope. Sections were counterstained with either hematoxylin or methyl green, dehydrated in an ascending ethanol series, defatted in xylene, and coverslipped with DPX mounting medium.
Antigen retrieval (0.01M sodium citrate, pH 6.0 for 20 min at 99°C) was used prior to the blocking step to enhance immunolabeling for PCNA, ALDH1L1, and zif268. Primary antibodies used included the following: Rabbit anti-Iba1 (1:1000; Wako Cat#019-19741, RRID:AB_839504), goat anti-Iba1 (1:1000; Abcam Cat#ab5076, RRID:AB_2224402), mouse anti-BrdU (1:500; BD Biosciences Cat#347580, RRID:AB_400326), mouse anti-CD68 (1:500; Abcam Cat#ab31630, RRID:AB_1141557), mouse anti-CD11b (1:500; OX-42 clone; Abcam Cat#ab1211, RRID:AB_442947), rabbit anti-NeuN (1:1000; Abcam Cat#ab177487, RRID:AB_2532109), rabbit anti-GFAP (1:1000; Abcam Cat#7260, RRID:AB_305808), mouse anti-PCNA (1:1000; Abcam Cat#ab29, RRID:AB_303394), rabbit anti-cleaved caspase-3 (Asp175) (1:500; Cell Signaling Technology Cat#9661, RRID:AB_2341188), rabbit anti-ALDH1L1 (1:1000; Abcam Cat#ab87117, RRID:AB_10712968), rabbit anti-Egr1 (zif268) (1:1000; Cell Signaling Technology Cat#4153, RRID: AB_2097038). Secondary antibodies used included the following: biotinylated anti-rabbit (1:500; Vector Laboratories Cat#BA-1000, RRID:AB_2313606), biotinylated anti-mouse (1:500; Vector Laboratories Cat#BA-2000, RRID:AB_2313581), Alexa Fluor 488- or 594-conjugated antibodies to rabbit (488 Cat#A21206, RRID:AB_2535792; 594 Cat#A21207, RRID:AB_141637), goat (488 Cat#A11055, RRID:AB_2534102; 594 Cat#A11058, RRID:AB_2534105), or mouse (488 Cat#A21202, RRID:AB_141607; 594 Cat#A21203, RRID:AB_2535789) (1:500; Thermo Fisher Scientific, all raised in donkey).
Unbiased stereological cell counting
Stereological cell counts were performed using StereoInvestigator (MBF Bioscience) on a computer interfaced with a Nikon Eclipse E600 microscope and MBF Bioscience CX9000 camera. Every third section (45 μm thick) was used for analysis for a total of four sections, and the amygdala, prefrontal cortex, nucleus accumbens, and paraventricular nucleus of both hemispheres were quantified. The boundaries of each region were drawn with a 4x objective, referencing a neonatal rat atlas (Ashwell and Paxinos, 2008), and the optical fractionator method was used to quantify microglia and BrdU+ cells at 40x magnification, using a 100 μm x 100 μm counting grid with a 250 μm x 250 μm sampling grid for microglia and a 50 μm x 50 μm counting grid with a 250 μm x 250 μm sampling grid for BrdU+ cells. The optical dissector height was set to 12 μm with a 2 μm guard zone on the top and bottom for both quantifications. Microglia were counted based on the presence of an observable cell body within the designated counting region and determined to be phagocytic if the microglia contained an observable phagocytic cup that was distinctly identifiable from the cell body. BrdU+ cells were counted if the nuclear staining was uniformly dark and was within the designated counting region.
Image acquisition
For all experiments, confocal fluorescence images were acquired with the following: Zeiss LSM 710 microscope equipped with 488, 561, and 633 lasers, using a 20x (1.0 NA) water-immersion or 100x (1.46 NA) oil-immersion objective and Zeiss Zen software; Nikon CSU-W1 or A1 microscope equipped with 405, 488, 561, and 647 lasers, using a 20x (0.75 NA), 60x (1.49 NA) TIRF oil-immersion, or a 100x (1.45 NA) oil-immersion objective and Nikon Elements software. Widefield fluorescence and brightfield images were captured on a Keyence BZ-X700 microscope using a 4x (0.2 NA), 10x (0.45 NA) or a 20x (0.75 NA) objective and BZ-X Viewer software.
Quantification of microglial activation state
45 μm thick fixed coronal sections were immunolabeled for Iba1 and CD68 and imaged with a Zeiss 710 confocal microscope. Six fields of view (1 per hemisphere from 3 sections total) were taken with a 20x objective using 2 um z steps through the entire tissue thickness. Subsequent maximum intensity projections were used to quantify microglia morphology and CD68 expression. The activation state was categorized on a 0 (lowest activation) to 5 (highest activation) according to the following criteria: morphology was scored as 0 (5+ processes with at least secondary branches), 1 (1 - 4 processes with at least secondary branches), 2 (1+ processes with no secondary branches), and 3 (round with no clear processes). CD68 expression was scored as 0 (no clear expression), 1 (punctate expression) or 2 (aggregated expression throughout the cell). For each cell, the morphology and CD68 scores were combined to produce a final 0-5 score, as described in Schafer et al. (2012).
Quantification of phagocytic cup contents, diameter, and three-dimensional rendering of microglia
For three-dimensional rendering, confocal z stacks were taken with a 100x objective to achieve an x-y pixel size between 0.012 - 0.070 μm and a z-step size between 0.050 - 0.200 μm. Images were deconvolved using the automatic deconvolution algorithm in Nikon Elements, and were reconstructed in three dimensions in Imaris (Bitplane, RRID:SCR_007370). Individual microglial cells were reconstructed using the Surfaces module to create a volumetric boundary of the cell. The resulting microglial surface was then used as a mask to process the channels containing the material to be colocalized. After masking, the Surfaces module was again used to generate a new volume of the engulfed material, and the two surfaces (microglia and engulfed material) were merged to create the final rendering.
For phagocytic cup analyses, single confocal images were taken with a 20x objective through the middle of the phagocytic cup. Images were then assessed for colocalization or measured for diameter in ImageJ (RRID:SCR_001775).
Quantification of local cell-microglia interactions (“Microglia Interactome”)
Three-dimensional renderings of microglia were generated using the Surfaces module in Imaris as described above. The Spots module was used to mark the location of surrounding cells (DAPI+, PCNA+, cCasp3+), and the DistanceTransform function was used to calculate the distance of each spot (cell) from the microglia surface. Spots within a 10 μm distance from the microglia surface were included in analyses. Additionally, the total number of PCNA+ and cCasp3+ cells were quantified using 20x field of view images of the amygdala using ImageJ (RRID:SCR_001775).
Quantification of cell fate
Quantification of the number of BrdU+ cells across the four MeA subregions was performed using Neurolucida software (MBF Bioscience, RRID:SCR_001775) on a computer interfaced with a Nikon Eclipse E600 microscope and MBF Bioscience CX9000 camera. Contours outlining the boundaries of each subregion were drawn at 4x magnification, and the area of each subregion was recorded. Sections from three alternate series were used to quantify the total number of BrdU+ cells and the number colocalized with either GFAP, NeuN or Iba1. Cells in each subregion were counted at 20x magnification across both hemispheres of the MeA. Additionally, the total number of GFAP+ cells was quantified only in the MePD. The data were normalized to the area of the subregion to account for any volumetric differences in the subregions between the sexes and averaged across hemispheres to generate a density estimate. In all cases, BrdU+ cells were counted if the nuclear staining was uniformly dark and present within the boundaries of the designated subregion. BrdU+ cells were counted as colocalized if a well-defined BrdU+ nucleus was associated with an immunopositive (i.e., GFAP+, NeuN+, Iba1+) cell body. For a subset of samples, colocalization criteria was confirmed by confocal microscopy.
Juvenile social play testing
Animals were weaned on P21 and housed in same-sex, same-treatment sibling pairs. On P26, animals were tested for 10 min in an open field (78 × 78 cm, 40 cm high), underlaid with a grid delineating perimeter and center regions. Total locomotion and center time were analyzed to rule out confounds due to differences in activity or anxiety-like behavior on play. Once per day from P27-30, same-sex, same-treatment non-sibling pairs of animals were placed in an enclosure (49 × 37 cm, 24 cm high) with TEK-Fresh cellulose bedding (Harland Laboratories). Animals were allowed to acclimate for 2 min, then video recorded for 10 min. Behavior testing took place during the dark phase of the animal’s light/dark cycle under red light illumination. Videos were scored offline to determine the number of pounces, pins, and boxing behaviors.
Quantification of zif268 expression following a single social exposure
Juvenile animals were given access to a novel sex-, treatment-, and age-matched play partner for 10 min as described above. Immediately following, animals were returned to their original cages. After 1 h, they were euthanized and brains were collected for histological analysis. Sections were labeled for zif268 and imaged using a Keyence BZ-X700 microscope at 4x magnification. Using the Hybrid Cell Count module in BZ-X Analyzer software (Keyence), contours were drawn to outline each region of interest, images were thresholded to account for background signal, and zif268+ cells were quantified. The number of zif268+ cells was normalized to the area of each contour to account for any volumetric differences.
In vitro phagocytosis assay
Microglia harvested from sex-specific mixed glia cultures were plated into 6 well plates at a density of 50,000 cells per well and kept in serum-free culture media at 37°C with 5% CO2. After 24 h, carboxylated latex beads (FluoSpheres, 1 μm; Thermo Fisher Scientific Cat#F8816) were added to each well in a 1:100 cell:bead ratio and allowed to incubate for 1 h at 37°C with 5% CO2. Immediately following this, plates were placed on ice to halt any further phagocytosis, washed with ice-cold PBS containing 2 mM EDTA, and vigorously pipetted to detach cells from the plate. Cells were centrifuged at 400g for 4 min, resuspended in FACS buffer (1.0% BSA, 0.1% sodium azide, in HBSS), and incubated with propidium iodide for flow cytometry analysis. Flow cytometry was performed on a LSR II (BD Biosciences) with FACSDiva software, and analysis performed using FlowJo X (FlowJo). Live cells were gated based on propidium iodide staining and bead fluorescence determined using live cells that were not exposed to beads.
Flow cytometry for complement proteins
On P4, male and female pups were rapidly decapitated, and the amygdala of both hemispheres was dissected out from the brain on ice. Tissue was immediately dissociated using the Neonatal Neuronal Dissociation (P) Kit (Miltenyi Bioscience Cat#130-092-628). The resulting single cell suspension was fixed in 4% paraformaldehyde for 20 min and stored in a 1:1 ratio of glycerol:HBSS at −20°C until staining. Cells were permeabilized with saponin and stained with a combination of the following antibodies: rabbit anti-C3b (1:50; Abcam Cat#ab11887, RRID:AB_298669), rabbit anti-C1qA (1:50; Abcam Cat#ab189922), and mouse anti-Ki-67-647 (1:10; BD Biosciences Cat#558615, RRID:AB_647130). Unconjugated primary antibodies were labeled using Alexa Fluor goat anti-rabbit-FITC (Cat#11-4839-81, RRID:AB_1210845) or Alexa Fluor goat anti-mouse-PE (Cat#12-4010-82, RRID:AB_11063706) (1:100; Thermo Fisher Scientific). Flow cytometry was performed on a LSR II (BD Biosciences) with FACSDiva software, and analysis performed using FlowJo X. Debris was eliminated based on forward and side scatter gating. Appropriate gates for C3b, C1qA, and Ki-67 were determined using an “empty channel” with the appropriate FMO samples to determine the negative population and autofluorescence.
Mass spectrometry
Lipid extraction and mass spectrometry was performed as previously described (Morena et al., 2015; Qi et al., 2015). On P4, rat pups were rapidly decapitated and the amygdala dissected out and immediately flash frozen until analysis. Brain tissue was weighed and placed into borosilicate glass culture tubes containing 2 mL of acetonitrile with 5 pmol of AEA and 5 nmol of 2-AG for extraction. Tissue was homogenized with a glass rod, sonicated for 30 min in ice water, and incubated overnight at −20°C to precipitate proteins. Subsequently, samples were then centrifuged at 1500g to remove particulates, and the supernatants transferred to a new glass tube and evaporated to dryness under nitrogen gas. The samples were reconstituted in 200 μL of acetonitrile and stored at −80°C until further analysis. Analysis of AEA and 2-AG was performed by liquid chromatography tandem mass spectrometry as previously described (Qi et al., 2015).
Western blot
On P4, rat pups were rapidly decapitated and the amygdala dissected out and immediately flash frozen until analysis. Tissue was homogenized in RIPA buffer with added phosphatase (Sigma; 1:1000) and protease (Sigma; 1:1000) inhibitors. After homogenization, samples were centrifuged at 3000 rpm at 4°C for 10 min. The protein supernatant was collected and total protein concentration determined by Bradford assay using an Infinite M1000 Pro (Tecan). 25 mg protein was loaded per sample and run on a 8%–16% tris-glycine gel (Invitrogen). Protein was transferred to a polyvinyl difluoride membrane (Bio-Rad), blocked for 1 h in Odyssey Blocking Buffer (LI-COR) diluted 1:1 with TBS and incubated overnight at 4°C with rabbit anti-CD11b (1:1000; Abcam Cat#ab133357, RRID: AB_2650514), goat anti-Iba1 (1:1000; Abcam Cat#ab5076, RRID:AB_2224402), rabbit anti-β actin (1:1000; Abcam Cat#ab8229, RRID:AB_796208) and rabbit anti-GAPDH (1:20,000; Sigma-Aldrich Cat#G9545, RRID:796208) in diluted Odyssey Blocking Buffer with 0.1% Tween-20 (BioRad). Subsequently, membranes were rinsed and incubated for 1 h with IRDye 680RD donkey anti-rabbit (Cat#925-68073, RRID:AB_2716687) and IRDye 800CW donkey anti-goat (Cat#925-68074, RRID:AB_2650427) antibodies (1:20,000; LI-COR) in diluted Odyssey Blocking Buffer with 0.1% Tween-20 and 0.02% sodium dodecyl sulfate (Sigma). Membranes were imaged in both the 700 and 800 nm channels using an Odyssey CLx scanner (LI-COR) and quantified using ImageStudio software (LI-COR). The CD11b signal was normalized to Iba1, GAPDH, and β actin, and the resulting values averaged for each sample. Each sample was normalized to a standard internal control sample to allow for comparison across membranes.
QUANTIFICATION AND STATISTICAL ANALYSIS
All values are shown as the mean ± SEM, with the exception of boxplots, which depict the median, 25th and 75th percentiles, with whiskers depicting the minimum and maximum values. Statistical analyses were performed using R (R Core Team, 2018; version 3.4.4). Statistical details of experiments can be found in figure legends and in the text (tests used, exact n, p value). Comparisons between two experimental groups were performed using a two-tailed Welch’s t test (for independent samples) or paired t test (for dependent samples). Data including multiple experimental groups were analyzed using one-way or two-way analysis of variance (ANOVA) when appropriate. In experiments in which females were treated with agents predicted to induce masculinization, Dunnett’s post hoc comparisons were calculated using female vehicle as a control to test the hypothesis that each treatment masculinized the end point. In some experiments, post hoc pairwise t tests were calculated for specific comparisons to determine differences in means between the treated female groups and male and female vehicle groups. Linear correlation was calculated using Pearson’s r. A p value of < 0.05 was used as the criterion for significance.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat polyclonal anti-Iba1 | Abcam | Cat#ab5076; RRID: AB_2224402 |
| Rabbit polyclonal anti-Iba1 | Wako | Cat#019-19741; RRID: AB_839504 |
| Mouse monoclonal Anti-BrdU | BD Biosciences | Cat#347580; RRID: AB_400326 |
| Mouse monoclonal anti-CD68 (ED1) | Abcam | Cat#ab31630; RRID: AB_1141557 |
| Mouse monoclonal anti-CD11b, clone OX-42 | Bio-Rad | Cat#MCA275GA; RRID: AB_566455 |
| Mouse monoclonal anti-CD11b antibody (OX42) | Abcam | Cat#ab1211; RRID: AB_442947 |
| Rabbit monoclonal anti-NeuN | Abcam | Cat#ab177487; RRID: AB_2532109 |
| Rabbit polyclonal Anti-GFAP | Abcam | Cat#7260; RRID: AB_305808 |
| Mouse monoclonal Anti-PCNA | Abcam | Cat#ab29; RRID: AB_303394 |
| Rabbit polyclonal anti-ALDH1L1 | Abcam | Cat#ab87117; RRID: AB_10712968 |
| Rabbit monoclonal anti-Egr1 (zif268) | Cell Signaling Technology | Cat#4153; RRID: AB_2097038 |
| Rabbit anti-cleaved caspase-3 (Asp175) | Cell Signaling Technology | Cat#9661; RRID: AB_2341188 |
| Biotinylated goat anti-rabbit | Vector Laboratories | Cat#BA-1000, RRID: AB_2313606 |
| Biotinylated horse anti-mouse | Vector Laboratories | Cat#BA-2000; RRID: AB_2313581 |
| Donkey anti-rabbit, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat#A21206; RRID: AB_2535792 |
| Donkey anti-goat, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat#A11055; RRID: AB_2534102 |
| Donkey anti-mouse, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat#A21202; RRID: AB_141607 |
| Donkey anti-rabbit, Alexa Fluor 594 conjugate | Thermo Fisher Scientific | Cat#A21207; RRID: AB_141637 |
| Donkey anti-goat, Alexa Fluor 594 conjugate | Thermo Fisher Scientific | Cat#A11058; RRID: AB_2534105 |
| Donkey anti-mouse, Alexa Fluor 594 conjugate | Thermo Fisher Scientific | Cat#A21203; RRID: AB_2535789 |
| NucRed Dead 647 | Invitrogen | Cat#R37113 |
| Hoechst 33342 Solution | Thermo Fisher Scientific | Cat#62249 |
| NeuroTrace 500/525 Green Fluorescent Nissl Stain | Thermo Fisher Scientific | Cat#N21480 |
| Rat monoclonal anti-F4/80 | Santa Cruz | Cat#sc-52664; RRID: AB_629466 |
| Rabbit polyclonal Anti-C3b | Abcam | Cat#ab11887; RRID: AB_298669 |
| Rabbit monoclonal anti-C1qA | Abcam | Cat#ab189922 |
| Alexa Fluor 647 mouse anti-Ki-67 | BD Biosciences | Cat#558615; RRID: AB_647130 |
| Alexa Fluor goat anti-rabbit-FITC | Thermo Fisher Scientific | Cat#11-4839-81; RRID: AB_1210845 |
| Alexa Fluor goat anti-mouse-PE | Thermo Fisher Scientific | Cat#12-4010-82; RRID: AB_11063706 |
| Rabbit monoclonal anti-CD11b | Abcam | Cat#ab133357; RRID: AB_2650514 |
| Rabbit polyclonal anti-β actin | Abcam | Cat#ab8229; RRID: AB_306374 |
| Rabbit polyclonal anti-GAPDH | Sigma-Aldrich | Cat#G9545; RRID: AB_796208 |
| IRDye 680RD donkey anti-rabbit | LI-COR | Cat#925-68073; RRID: AB_2716687 |
| IRDye 800CW donkey anti-goat | LI-COR | Cat#925-68074; RRID: AB_2650427 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Testosterone propionate | Sigma-Aldrich | Cat#T1875 |
| Flutamide | Sigma-Aldrich | Cat#F9397 |
| Estradiol benzonate | Sigma-Aldrich | Cat#E8515 |
| Cannabinoid Receptor agonist ACEA | Tocris | Cat#1319 |
| Cannabinoid Receptor agonist GP1a | Tocris | Cat#2764 |
| MAGL inhibitor KML29 | Tocris | Cat#4872 |
| Cannabinoid Receptor antagonist AM630 | Tocris | Cat#1120 |
| Cannabinoid Receptor antagonists AM281 | Tocris | Cat#1115 |
| Liposomal coldronate | Encapsula Nanoscienes | Cat#CLD-8909 |
| FluoSpheres carboxylate-modified microspheres, 1 μm | Thermo Fisher Scientific | Cat#F8816 |
| 5-Bromo-2′-deoxyuridine (BrdU) | Sigma-Aldrich | Cat#B5002 |
| Critical Commercial Assays | ||
| Neonatal Neuronal Dissociation Kit (P) | Miltenyi Bioscience | Cat#130-092-628 |
| Experimental Models: Organisms/Strains | ||
| Sprague-Dawley rats | Charles River Laboratories | N/A |
| Software and Algorithms | ||
| Stereoinvestigator | MBF Bioscience | https://www.mbfbioscience.com/stereo-investigator |
| Imaris | Bitplane | https://www.bitplane.com/imaris; RRID: SCR_007370 |
| ImageJ | NIH | https://imagej.net/Welcome; RRID: SCR_003070 |
| Neurolucida | MBF Bioscience | https://www.mbfbioscience.com/neurolucida; RRID: SCR_001775 |
| FlowJo X | FlowJo | https://www.flowjo.com/ |
| ImageStudio | LI-COR | https://www.licor.com/bio/products/software/image_studio/ |
| R, version 3.4.4 | R Core Team | https://cran.r-project.org/ |
| BZ-X Analyzer | KEYENCE | https://www.keyence.com/ss/products/microscope/bz-x700/product/index.jsp |
Highlights.
Microglia are more phagocytic in the male amygdala during neonatal development
Androgen-induced endocannabinoids increase phagocytosis in males
Microglia engulf viable newborn astrocytes in a complement-dependent manner
Developmental phagocytosis produces a sex difference in juvenile social play
ACKNOWLEDGMENTS
This work was funded by the National Institutes of Health (R01MH52716 and R01DA039062 to M.M.M.) and an operating grant from the Natural Sciences and Engineering Council of Canada (NSERC) to M.N.H. Salary support was provided to M.N.H. in the form of a Tier II Canada Research Chair from the Canadian Institutes of Health Research (CIHR), and H.A.V. is a Vanier Scholar (CIHR) and receives studentships from Alberta Innovates and BranchOut Neurological Foundation. We thank the Confocal Microscopy Core Facility at the University of Maryland School of Medicine, including J. Mauban, for the technical assistance. Flow cytometry analyses were performed at the University of Maryland Marlene and Stewart Greenebaum Comprehensive Cancer Center Flow Cytometry Shared Service, and we thank K. Underwood and X. Fan for the technical assistance. We would also like to acknowledge the Southern Alberta Mass Spectrometry Centre, located in and supported by the Cumming School of Medicine, University of Calgary, for their services in targeted liquid chromatography tandem mass spectrometry. Finally, we thank B. Alger, J. Cheer, M.K. Lobo, D. Loane, and R. Ritzel for their helpful comments and critiques of the manuscript.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and can be found with this article online at https://doi.org/10.1016/j.neuron.2019.02.006.
REFERENCES
- Aguado T, Monory K, Palazuelos J, Stella N, Cravatt B, Lutz B, Marsicano G, Kokaia Z, Guzmän M, and Galve-Roperh I (2005). The endocannabinoid system drives neural progenitor proliferation. FASEB J. 19, 1704–1706. [DOI] [PubMed] [Google Scholar]
- Aguado T, Palazuelos J, Monory K, Stella N, Cravatt B, Lutz B, Marsicano G, Kokaia Z, Guzmän M, and Galve-Roperh I (2006). The endocannabinoid system promotes astroglial differentiation by acting on neural progenitor cells. J. Neurosci 26, 1551–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amateau SK, and McCarthy MM (2002). A novel mechanism of dendritic spine plasticity involving estradiol induction of prostaglandin-E2. J. Neurosci 22, 8586–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argue KJ, and McCarthy MM (2015). Characterization of juvenile play in rats: importance of sex of self and sex of partner. Biol. Sex Differ 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argue KJ, VanRyzin JW, Falvo DJ, Whitaker AR, Yu SJ, and McCarthy MM (2017). Activation of both CB1 and CB2 endocannabinoid receptors is critical for masculinization of the developing medial amygdala and juvenile social play behavior. eNeuro 4, ENEURO.0344–16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell KWS, and Paxinos G (2008). Atlas of the Developing Rat Nervous System, Third Edition (Academic Press; ). [Google Scholar]
- Bergan JF, Ben-Shaul Y, and Dulac C (2014). Sex-specific processing of social cues in the medial amygdala. eLife 3, e02743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghuis P, Dobszay MB, Wang X, Spano S, Ledda F, Sousa KM, Schulte G, Ernfors P, Mackie K, Paratcha G, et al. (2005). Endocannabinoids regulate interneuron migration and morphogenesis by transactivating the TrkB receptor. Proc. Natl. Acad. Sci. USA 102, 19115–19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC, and Neher JJ (2012). Eaten alive! Cell death by primary phagocytosis: ‘phagoptosis’. Trends Biochem. Sci 37, 325–332. [DOI] [PubMed] [Google Scholar]
- Brown GC, and Neher JJ (2014). Microglial phagocytosis of live neurons. Nat. Rev. Neurosci 15, 209–216. [DOI] [PubMed] [Google Scholar]
- Carrier EJ, Kearn CS, Barkmeier AJ, Breese NM, Yang W, Nithipatikom K, Pfister SL, Campbell WB, and Hillard CJ (2004). Cultured rat microglial cells synthesize the endocannabinoid 2-arachidonylglycerol, which increases proliferation via a CB2 receptor-dependent mechanism. Mol. Pharmacol 65, 999–1007. [DOI] [PubMed] [Google Scholar]
- Choi GB, Dong HW, Murphy AJ, Valenzuela DM, Yancopoulos GD, Swanson LW, and Anderson DJ (2005). Lhx6 delineates a pathway mediating innate reproductive behaviors from the amygdala to the hypothalamus. Neuron 46, 647–660. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martínez-Cerdeño V, and Noctor SC (2013). Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J. Neurosci 33, 4216–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase LM, Schuebel KE, Fusfeld ZH, Jair K, Hawes IA, Cimbro R, Zhang HY, Liu QR, Shen H, Xi ZX, et al. (2017). Local cues establish and maintain region-specific phenotypes of basal ganglia microglia. Neuron 95, 341–356.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli M, Battista N, Riganti L, Prada I, Antonucci F, Cantone L, Matteoli M, Maccarrone M, and Verderio C (2015). Active endocannabinoids are secreted on extracellular membrane vesicles. EMBO Rep. 16, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant KS, Petroff R, Isoherranen N, Stella N, and Burbacher TM (2018). Cannabis use during pregnancy: pharmacokinetics and effects on child development. Pharmacol. Ther 182, 133–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, Alter MD, Block CS, Sullivan H, Bolton JL, and Bilbo SD (2017). Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia 65, 1504–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner DJ, Hengartner MO, and Schnabel R (2001). Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412, 202–206. [DOI] [PubMed] [Google Scholar]
- Hong W, Kim DW, and Anderson DJ (2014). Antagonistic control of social versus repetitive self-grooming behaviors by separable amygdala neuronal subsets. Cell 158, 1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji K, Akgul G, Wollmuth LP, and Tsirka SE (2013). Microglia actively regulate the number of functional synapses. PLoS ONE 8, e56293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs-Kraft DL, Hill MN, Hillard CJ, and McCarthy MM (2010). Sex difference in cell proliferation in developing rat amygdala mediated by endocannabinoids has implications for social behavior. Proc. Natl. Acad. Sci. USA 107, 20535–20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, Walker AJ, Heller MD, Umemori H, Chen C, and Stevens B (2018). CD47 protects synapses from excess microglia-mediated pruning during development. Neuron 100, 120–134.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz KM, Nugent BM, Haliyur R, and McCarthy MM (2013). Microglia are essential to masculinization of brain and behavior. J. Neurosci 33, 2761–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leresche LA (1976). Dyadic play in Hamadryas baboons. Behaviour 57, 190–205. [Google Scholar]
- Li Y, Mathis A, Grewe BF, Osterhout JA, Ahanonu B, Schnitzer MJ, Murthy VN, and Dulac C (2017). Neuronal representation of social information in the medial amygdala of awake behaving mice. Cell 171, 1176–1190.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccarrone M, Guzmän M, Mackie K, Doherty P, and Harkany T (2014). Programming of neural cells by (endo)cannabinoids: from physiological rules to emerging therapies. Nat. Rev. Neurosci 15, 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Teva JL, Dusart I, Colin C, Gervais A, van Rooijen N, and Mallat M (2004). Microglia promote the death of developing Purkinje cells. Neuron 41, 535–547. [DOI] [PubMed] [Google Scholar]
- Martin-Fernandez M, Jamison S, Robin LM, Zhao Z, Martin ED, Aguilar J, Benneyworth MA, Marsicano G, and Araque A (2017). Synapse-specific astrocyte gating of amygdala-related behavior. Nat. Neurosci 20, 1540–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, and Arnold AP (2011). Reframing sexual differentiation of the brain. Nat. Neurosci 14, 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, de Vries GJ, and Forger NG (2017). Sexual differentiation of the brain: a fresh look at mode, mechanisms, and meaning. In Hormones, Brain and Behavior, Pfaff and DW Joëls M, eds. (Academic Press; ), pp. 3–32. [Google Scholar]
- Meaney MJ,and McEwen BS (1986).Testosterone implants into the amygdala during the neonatal period masculinize the social play of juvenile female rats. Brain Res. 398, 324–328. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Dodge AM, and Beatty WW (1981). Sex-dependent effects of amygdaloid lesions on the social play of prepubertal rats. Physiol. Behav 26, 467–72. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Stewart J, Poulin P, and McEwen BS (1983). Sexual differentiation of social play in rat pups is mediated by the neonatal androgen-receptor system. Neuroendocrinology 37, 85–90. [DOI] [PubMed] [Google Scholar]
- Miyamoto A, Wake H, Ishikawa AW, Eto K, Shibata K, Murakoshi H, Koizumi S, Moorhouse AJ, Yoshimura Y, and Nabekura J (2016). Microglia contact induces synapse formation in developing somatosensory cortex. Nat. Commun 7, 12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mong JA, Glaser E, and McCarthy MM (1999). Gonadal steroids promote glial differentiation and alter neuronal morphology in the developing hypothalamus in a regionally specific manner. J. Neurosci 19, 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morena M, De Castro V, Gray JM, Palmery M, Trezza V, Roozendaal B, Hill MN, and Campolongo P (2015). Training-associated emotional arousal shapes endocannabinoid modulation of spatial memory retrieval in rats. J. Neurosci 35, 13962–13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder J, Aguado T, Keimpema E, Barabás K, Ballester Rosado CJ, Nguyen L, Monory K, Marsicano G, Di Marzo V, Hurd YL, et al. (2008). Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proc. Natl. Acad. Sci. USA 105, 8760–8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palagi E, Antonacci D, and Cordoni G (2007). Fine-tuning of social play in juvenile lowland gorillas (gorilla gorilla gorilla). Dev. Psychobiol 49, 433–445. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. [DOI] [PubMed] [Google Scholar]
- Pellis SM, and McKenna MM (1992). Intrinsic and extrinsic influences on play fighting in rats: effects of dominance, partner’s playfulness, temperament and neonatal exposure to testosterone propionate. Behav. Brain Res 50, 135–145. [DOI] [PubMed] [Google Scholar]
- Pellis SM, Field EF, Smith LK, and Pellis VC (1997). Multiple differences in the play fighting of male and female rats. Implications for the causes and functions of play. Neurosci. Biobehav. Rev 21, 105–120. [DOI] [PubMed] [Google Scholar]
- Qi M, Morena M, Vecchiarelli HA, Hill MN, and Schriemer DC (2015). A robust capillary liquid chromatography/tandem mass spectrometry method for quantitation of neuromodulatory endocannabinoids. Rapid Commun. Mass Spectrom 29, 1889–1897. [DOI] [PubMed] [Google Scholar]
- R Core Team (2018). R: a language and environment for statistical computing (R Foundation for Statistical Computing; ). https://www.R-project.org/. [Google Scholar]
- Reddien PW, Cameron S, and Horvitz HR (2001). Phagocytosis promotes programmed cell death in C. elegans. Nature 412, 198–202. [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Sholar PW, and Bilbo SD (2012). Sex differences in microglial colonization of the developing rat brain. J. Neurochem 120, 948–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y, and Sato K (2014). Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J. Neurosci 34, 2231–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A, Encinas JM, Deudero JJ, Chancey JH, Enikolopov G, Overstreet-Wadiche LS, Tsirka SE, and Maletic-Savatic M (2010). Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, and Garel S (2014). Microglia modulate wiring of the embryonic forebrain. Cell Rep. 8, 1271–1279. [DOI] [PubMed] [Google Scholar]
- Stella N (2009). Endocannabinoid signaling in microglial cells. Neuropharmacology 56 (Suppl 1), 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Tortoriello G, Morris CV, Alpar A, Fuzik J, Shirran SL, Calvigioni D, Keimpema E, Botting CH, Reinecke K, Herdegen T, et al. (2014). Miswiring the brain: Δ9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. EMBO J. 33, 668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, and Yamashita T (2013). Layer V cortical neurons require microglial support for survival during postnatal development. Nat. Neurosci 16, 543–551. [DOI] [PubMed] [Google Scholar]
- Vanderschuren LJ, Achterberg EJ, and Trezza V (2016). The neurobiology of social play and its rewarding value in rats. Neurosci. Biobehav. Rev 70, 86–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerhuis R, Nielsen HM, and Tenner AJ (2011). Complement in the brain. Mol. Immunol 48, 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viader A, Blankman JL, Zhong P, Liu X, Schlosburg JE, Joslyn CM, Liu QS, Tomarchio AJ, Lichtman AH, Selley DE, et al. (2015). Metabolic interplay between astrocytes and neurons regulates endocannabinoid action. Cell Rep. 12, 798–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viader A, Ogasawara D, Joslyn CM, Sanchez-Alavez M, Mori S, Nguyen W, Conti B, and Cravatt BF (2016). A chemical proteomic atlas of brain serine hydrolases identifies cell type-specific pathways regulating neuroinflammation. eLife 5, e12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis T, Lainé J, Simon A, Roland A, Leterrier C, and Lenkei Z (2008). The type 1 cannabinoid receptor is highly expressed in embryonic cortical projection neurons and negatively regulates neurite growth in vitro. Eur. J. Neurosci 28, 1705–1718. [DOI] [PubMed] [Google Scholar]
- Wakselman S, Béchade C, Roumier A, Bernard D, Triller A, and Bessis A (2008). Developmental neuronal death in hippocampus requires the microglial CD11b integrin and DAP12 immunoreceptor. J. Neurosci 28, 8138–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter L, Franklin A, Witting A, Wade C, Xie Y, Kunos G, Mackie K, and Stella N (2003). Nonpsychotropic cannabinoid receptors regulate microglial cell migration. J. Neurosci 23, 1398–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting A, Walter L, Wacker J, Möller T, and Stella N (2004). P2X7 receptors control 2-arachidonoylglycerol production by microglial cells. Proc. Natl. Acad. Sci. USA 101, 3214–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Zhu J, Wager-Miller J, Wang S, O’Leary D, Monory K, Lutz B, Mackie K, and Lu HC (2010). Requirement of cannabinoid CB(1) receptors in cortical pyramidal neurons for appropriate development of corticothalamic and thalamocortical projections. Eur. J. Neurosci 32, 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuloaga DG, Puts DA, Jordan CL, and Breedlove SM (2008). The role of androgen receptors in the masculinization of brain and behavior: what we’ve learned from the testicular feminization mutation. Horm. Behav 53, 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.