Abstract
Objective:
Epigenetic modifications are gaining focus due to their indirect association with tumorigenesis. DNA methylation plays a prime role in regulation of gene expression. Any aberrations in this gene family may lead to chromosomal instability and increased magnitude of tumour progression. In line with the above fact, the present study has been designed to identify genetic alterations in the genes of the DNMT (DNA methyl-transferase) family among head and neck squamous cell carcinoma patients (HNSCC).
Methods:
The present study follows an observational design employing computational tools for analysis. The TCGA-Firehose Legacy data was assessed using the cBioportal database. The dataset comprised of 530 samples from HNSCC patients which were assessed for genetic alterations in the DNMT family. Furthermore, the protein stability analysis and pathogenicity of the mutations were assessed using I-Mutant Suite and PROVEAN tools.
Results:
Almost all genes of the DNMT family harboured gene amplification. The TRDMT1 and DNMT3L genes showed deep deletions. Apart from these several non-synonymous, truncating and splice-site mutations were also documented. Protein stability and pathogenicity analysis revealed that majority of the mutations were found to decrease the stability and impose pathogenicity. Upon probing for reported mutations using gnomAD database, around six reference single nucleotide polymorphisms were identified which were found to exhibit a minor allele frequency less than 0.01.
Conclusions:
Screening of an exhaustive collection of patient’s samples could provide immense knowledge about the disease pathogenesis and identification of therapeutic leads. The variants identified in the present study could be used as diagnostic markers. However, further experimental analysis through genotyping assay is warranted to validate the present findings.
Key Words: HNSCC, in silico, methyltransferases, mutations
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common type of cancer which is responsible for over 350,000 deaths every year (McDermott and Bowles, 2019). According to the GLOBOCAN report 2012, cancer of lip and oral cavity was found to be the 12th most common cancer in Asia (Gupta et al., 2016). Recent reports by the global cancer observatory, GLOBOCAN, 2018, revealed an increase in the mortality rate due to cancer of the lip and oral cavity especially in males, with a significant proportion of cases arising from south Asian countries (Bray et al., 2018). These two reports have projected a steep increase in the incidence of the disease. The risk for oral cancer is precipitated by several factors such as tobacco chewing, chronic alcoholism, smoking, HPV (Human Papilloma Virus) infection, sharp tooth in addition to the genetic makeup (Ram et al., 2011). The incidence of HNSCC is high in males when compared to females, especially in eastern Europe and India with over 20 males affected per 100, 000 individuals (Fitzmaurice et al., 2017). Smoking tobacco such as cigarettes and pipes along with smokeless tobacco increased the risk of HNSCC by 2-4 fold (Wyss et al., 2013, Zhou et al., 2013). The practice of betel nut chewing, a habit most commonly seen in the Asian subcontinent increased the risk of HNSCC from 2 - 15 fold (Guha et al., 2014).
Epigenetic alterations in cancer have gained a lot of attention in the recent years. Genetic alterations spanning the genes encoding the enzymes methyltransferases have been shown to be associated with numerous cancers and pre-cancerous lesions. The process of methylation inactivates crucial genes without interfering with the sequence. Hypermethylation of promoter regions of cell cycle control genes, tumor suppressor genes are known to diminish the function, thereby contributing to the malignant transformation of the cells (Rotondo et al., 2018; Casalino and Verde, 2020). An in-depth knowledge on the expression profile of genes coding for methyltransferases under normal and pathological conditions would aid in identifying the triggers associated with these epigenetic marks. In line with the above facts, DNMTs are known to play a crucial role in the process of tumorigenesis. Defects in DNMTs is accompanied by chromosomal imbalances which results in the remodelling of chromatin, instability of genome and inactivation of genes. Although global hypomethylation is common with tumor cells, hypermethylation of specific regions have resulted in silencing of crucial genes involved in the repair or cell cycle regulatory pathways (Rodriguez-Paredes and Esteller, 2011). There are five DNMTs discovered in the human genome till date viz., DNMT1, DNMT2, DNMT3A, DNMT3B and DNMT3L. The DNMT1, DNMT3A and DNMT3B are canonical enzymes which take part in the catalytic activity, whereas the other DNMTs, i.e., DNMT2 and DNMT3L are non-canonical family members as they do not possess the catalytic activity (Lyko, 2019). A few genes known to be methylated or otherwise silenced in oral premalignant lesions are p16, MGMT, RAR-beta -2, cadherin and DAP kinase (Díez-Pérez et al., 2011). Other genes have been found to be hypermethylated in oral cancer, such as ZNF582, PAX1 (Cheng et al., 2018; Cheng et al., 2016) and EYA4 (Towle et al., 2016). The DNMT inhibitors have been tested to assess their efficacy on animal models. One such experiment included the DNMT inhibitor, 5-Aza-2’-deoxycytidine in combination with a low dose of trans-retinoic acid on a murine model of oral carcinoma, induced using the carcinogen, 4-nitroquinoline 1-oxide (4-NQO). The results revealed that the combination was effective in attenuating tongue lesion severity when compared to the other treatment modalities used for the purpose. Hence, a combination of DNA demethylating drugs could serve as a promising strategy to reduce cancer of oral cavity (Tang et al., 2009).
Several studies have identified polymorphic variants in DNMT genes with both positive and negative associations with different cancer types. Promoter polymorphisms viz.,-149C>T, -283T>C, and -579G>T located in the DNMT3B gene did not produce any significant association with nasopharyngeal carcinoma in Taiwanese population (Chang et al., 2008). A meta-analysis conducted by Zhu et al., (2015) suggested that the risk of head and neck cancer was associated with heterozygous variant of DNMT3B which is -149C/T. Duan et al., demonstrated that the variants (rs2424913 C/T, rs1569686 G/T, rs6087990 T/C and rs2424908 T/C) were either negatively associated with cancer risk or confer protection against cancer in Asian population.
Materials and Methods
Sample data set
The cBioPortal for Cancer Genomics (http://cbioportal.org) integrates an exhaustive collection of molecular profiling information from cancer tissues and cell lines (Cerami et al., 2012; Gao et al., 2013). The database is user friendly and hosts genetic, epigenetic and proteomic information of the cases registered. The sample data set includes 528 HNSCC cases (530 samples) of which 504 samples harboured information about copy number variations and mutation data. The demographic details of cases in the head and neck squamous cell carcinoma (TCGA, Firehose Legacy) dataset were recorded (Table 1).
Table 1.
Demographic Details of Patients Analyzed in the Present Study (as Obtained from the cBioportal Site)
| Variable | Number |
|---|---|
| Gender | |
| Male | 386 |
| Female | 142 |
| Mutation count | 6-3181 |
| Diagnosis age | 19-90 years |
| Smoking status | |
| Smokers | 515 |
| Data not available | 12 |
| Unknown | 1 |
| Alcohol history | |
| Yes | 352 |
| No | 165 |
| Data not available | 11 |
| Neoplasm Histologic grade | |
| Grade 1 | 63 |
| Grade 2 | 311 |
| Grade 3 | 125 |
| Grade 4 | 7 |
| Grade GX | 18 |
| Data not available | 4 |
| Race category | |
| White | 452 |
| African | 48 |
| Asian | 11 |
| American Indian or Alaska native | 2 |
| Data not available | 15 |
Oncoprint data analysis
Submission of user defined query of 5 genes belonging to the family of DNA methyltransferase (Data not shown) returned a window with OncoPrint data which demonstrated the presence of alterations in 5 crucial genes of the DNMT family viz., DNMT1, TRDMT1, DNMT3A and DNMT3B, DNMT3L. The somatic mutation frequency and the site of mutation in the candidate genes were documented (Cerami et al., 2012; Gao et al., 2013) (Table 2).
Table 2.
Description on the Genes, Protein Encoded, Genetic Alterations, Loci, Frequency of Variant Allele in Tumor Sample in DNA Methyltransferase Family of Genes
| Gene | Protein | Alteration | Loci | % of alteration |
Variant allele frequency in tumor sample | gnomAD frequency data |
|---|---|---|---|---|---|---|
| DNMT1 | DNA methyltrasferase 1 | Gene amplification | 19q13.2 | 2.6 | ||
| E559Q | 0.03 | Novel | ||||
| P1330S | 0.17 | Novel | ||||
| P1325S | 0.2 | Novel | ||||
| E912Q | 0.05 | Novel | ||||
| S1352G | 0.2 | Novel | ||||
| P692S | 0.21 | Novel | ||||
| H370Y | 0.03 | Novel | ||||
| T616M | 0.42 | Novel | ||||
| R325L | 0.09 | Novel | ||||
| X891_splice | 0.05 | Novel | ||||
| P1106H | 0.04 | Novel | ||||
|
TRDMT1
(DNMT2) |
TRNA Aspartic Acid Methyltransferase 1 | Gene amplification | 10p13 | 1.8 | ||
| Deep deletions | ||||||
| A198E | 0.44 | rs200204830 | ||||
| S272* | 0.22 | Novel | ||||
| DNMT3A | DNA methyltrasferase 3 alpha | Gene amplification | 2p23.3 | 2.8 | ||
| E371* | 0.31 | Novel | ||||
| R729Q | 0.52 | rs757211277 | ||||
| E491D | 0.12 | Novel | ||||
| S689C | 0.17 | Novel | ||||
| R556M | 0.16 | Novel | ||||
| R488Q | 0.13 | rs566390868 | ||||
| Q374L | 0.16 | Novel | ||||
| P419L | 0.12 | Novel | ||||
| P106S | 0.08 | Novel | ||||
| G150V | 0.12 | Novel | ||||
| W893R | 0.45 | Novel | ||||
| E907K | 0.14 | rs769590067 | ||||
| R899C | 0.18 | Novel | ||||
| DNMT3B | DNA methyltrasferase 3 beta | Gene amplification | 20q11.21 | 5 | ||
| R576* | 0.14 | Novel | ||||
| K441Efs*11 | 0.41 | Novel | ||||
| C496= | 0.05 | Novel | ||||
| Q328E | 0.26 | Novel | ||||
| E464Q | 0.16 | Novel | ||||
| R840Q | 0.25 | rs121908946 | ||||
| R485L | 0.12 | Novel | ||||
| P845S | 0.22 | Novel | ||||
| H132R | 0.29 | rs770751820 | ||||
| Q196H | 0.42 | Novel | ||||
| DNMT3L | DNA methyltrasferase 3 like | Gene amplification | 21q22.3 | 1.6 | ||
| Deep deletions | ||||||
| R47* | 0.13 | Novel | ||||
| P310S | 0.22 | Novel | ||||
| S325N | 0.21 | Novel |
gnomAD analysis
The genome aggregation database (gnomAD) hosts a comprehensive information of data spanning 125,748 exome sequences and 15,708 whole genome sequences from unrelated individuals sequenced and deposited as part of various disease-specific or population genetic studies. A subtractive analysis was carried out to identify those variants which were not previously reported. Hence, such variants were designated as novel variants. The search also provided an insight about the minor allele frequency of the variants in the population by which the nature of the variants can be ascertained (Karczewski et al., Version 4, 2020) (Table 2).
Protein stability analysis
The I-Mutant server was used for prediction of protein stability changes upon single nucleotide mutations leading to change in the amino acid being encoded by the triplet codon. The server uses either protein sequence or structure to predict stabilisation and destabilisation of protein structure in majority of cases. The protein sequence coded by the genes were downloaded in the FASTA format from the public domain (https://www.ncbi.nlm.nih.gov/protein/). These sequences were further analysed upon substitution with the variant amino acid. The changes in the stability of the proteins were assessed using the free energy stability change (ΔΔG) value. A value greater than zero implies increase in protein stability and a value less than zero is considered to decrease the protein stability (Capriotti et al., 2005) (Table 3).
Table 3.
Consequences of Non-Synonymous Variations on Protein Stability and Pathogenicity as Assessed by IMutant and PROVEAN Tools
| Gene | Variation | I-Mutant prediction | Score | PROVEAN Score | Prediction |
|---|---|---|---|---|---|
| DNMT1 | E559Q | Decrease | -0.33 | -2.024 | Neutral |
| P1330S | Decrease | -1.09 | -6.84 | Deleterious | |
| P1325S | Decrease | -1.47 | -6.973 | Deleterious | |
| E912Q | Decrease | -0.67 | -1.683 | Neutral | |
| S1352G | Decrease | -3.01 | -2.551 | Deleterious | |
| P692S | Decrease | -0.11 | -7.006 | Deleterious | |
| H370Y | Decrease | -0.11 | -2.633 | Deleterious | |
| T616M | Decrease | -0.84 | -4.531 | Deleterious | |
| R325L | Decrease | -0.13 | -0.594 | Neutral | |
| P1106H | Decrease | -0.25 | -1.505 | Neutral | |
| TRDMT1 (DNMT2) | A198E | Decrease | -1.22 | -0.585 | Neutral |
| DNMT3A | R729Q | Decrease | -0.21 | -3.695 | Deleterious |
| E491D | Increase | 0.05 | -2.742 | Deleterious | |
| S689C | Decrease | -1.25 | -3.432 | Deleterious | |
| R556M | Increase | 0.97 | -5.288 | Deleterious | |
| R488Q | Decrease | -1.39 | -1.513 | Neutral | |
| Q374L | Increase | 0.74 | -4.303 | Deleterious | |
| P419L | Increase | 0.38 | -2.962 | Deleterious | |
| P106S | Increase | 0.03 | -1.025 | Neutral | |
| G150V | Decrease | -1.61 | -0.38 | Neutral | |
| W893R | Decrease | -1.36 | -13.22 | Deleterious | |
| E907K | Decrease | -0.7 | -3.13 | Deleterious | |
| R899C | Decrease | -1.7 | -7.55 | Deleterious | |
| DNMT3B | Q328E | Increase | 0.94 | -0.838 | Neutral |
| E464Q | Decrease | -0.95 | -2.561 | Deleterious | |
| R840Q | Decrease | -1.9 | -3.531 | Deleterious | |
| R485L | Decrease | -0.37 | -3.598 | Deleterious | |
| P845S | Decrease | -1.72 | -7.328 | Deleterious | |
| H132R | Decrease | -0.12 | -0.699 | Neutral | |
| Q196H | Decrease | -0.79 | -1.144 | Neutral | |
| DNMT3L | P310S | Decrease | -1.62 | 0.389 | Neutral |
| S325N | Decrease | -0.62 | -0.9 | Neutral |
Pathogenicity analysis
PROVEAN (Protein Variation Effect Analyzer) predicts the consequences of an amino acid substitution in a protein during a mutational event (Table 3). The present analysis employs a user defined query of missense variants identified in a specific gene entered along with the reference sequence of the protein derived from the NCBI database. The default cut-off value of -2.5 was set prior to analysis. Based on the scores the amino acid substitutions are classified as either neutral or deleterious (Choi and Chan, 2015). A score < -2.5 or > -2.5 was considered to be deleterious and neutral respectively (Table 3).
Gene expression analysis
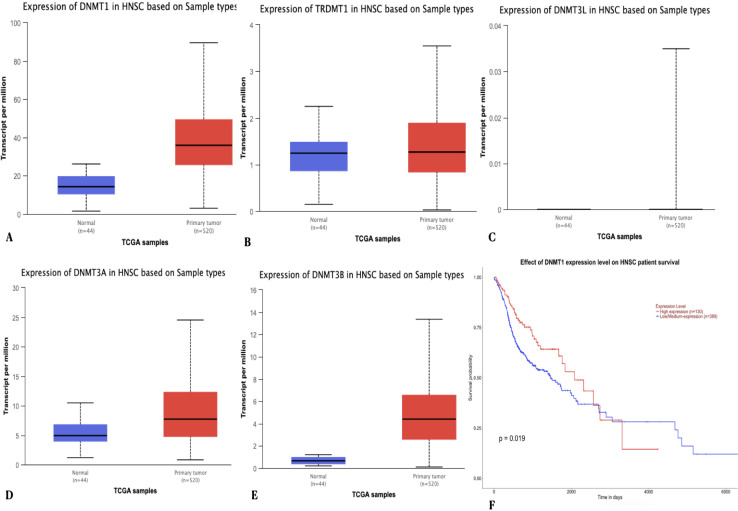
Gene expression analysis on the DNMT1, TRDMT1, DNMT3A, DNMT3B and DNMT3L genes were carried out using the UALCAN database (http://ualcan.path.uab.edu/cgi-bin/TCGA- survival1.pl?genenam) (Figure 2). Expression data was represented as transcripts per million (TPM) which is a normalization method for RNA- seq data. Further, these TPM values were used for the generation of box- whisker plots which was employed to determine the significant difference between the groups. Survival effect analysis of gene expression were assessed using multivariate Kaplan - Meier survival analysis (Chandrasekar et al., 2017).
Figure 2.
The Box Whisker Plot and Kaplan Meier Survival Analysis Demonstrating Differential Gene Expression Pattern and Survival Probability of HNSCC Patients Respectively. Upregulation of genes (A) DNMT1 (B) TRDMT1 (C) DNMT3A (D) DNMT3B and (E) DNMT3L when compared to normal samples. (F) The survival plot showed that the low/medium level expression of DNMT1 gene resulted in lower survival probability when compared to patients with high level expression
Results
Demographic data
The dataset (TCGA, Firehose Legacy) included in the present study had information on 528 HNSCC patients (530 samples) of which mutations and copy number variation data was available for 504 samples. The male:female ratio was found to be 2.7:1, with age group ranging from 19 - 90 years. The number of individuals with the history of smoking and alcohol was roughly around 98% and 67%. The dataset had samples from patients of American (85.6%), African (9.1%), Asian (2.1%) and American Indian (0.4%) decent. The distribution of patients based on the histologic grade of neoplasm is given in Table 1, of which 59% of patients had grade 2 tumor.
Oncoprint data analysis
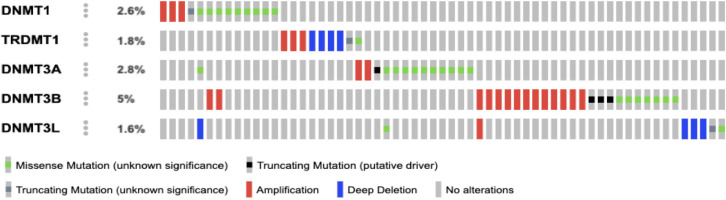
The oncoprint data analysis revealed gene amplification in all five genes viz., DNMT1, TRDMT1, DNMT3A, DNMT3B and DNMT3L, of which DNMT3B was found to harbour highest frequency of gene amplification. The genes TRDMT1 and DNMT3L demonstrated deep deletions, whereas gene amplification was observed in all the genes investigated. The DNMT3B gene possessed the highest frequency of variations/mutations (5%) from among all the genes identified with alterations (Table 2). Several truncating and mis-sense variants of unknown significance have been documented (Figure 1). Truncating mutations identified in DNMT3A gene (E371*) and DNMT3B gene (C496=, K441Efs*11, R576*) were found to be putative drivers. The clinical implication states that they are likely oncogenic with a biological effect involving loss of function. Novel variants were common among the genes DNMT1 and DNMT3L.
Figure 1.
The Oncoprint Data Depicts the Gene Alterations in DNMT Family of Genes. Each of the grey bar represents HNSC patients. The colour codes represents genetic alterations in individual patient
gnomAD analysis
A total of 6 reported variants were identified using gnomAD analysis viz., rs200204830 in TRDMT1, rs757211277, rs566390868, rs769590067 in DNMT3A and rs121908946, rs770751820 in DNMT3B genes. All the variants identified in the present study had a minor allele frequency < 0.01, implying the fact that these rare variants might be associated with risk of a particular disease (Table 2).
Protein stability and pathogenicity analysis
Stability of the protein largely affects the biological function of the protein. Hence, protein stability was assessed for all the non-synonymous variants identified in the study. Majority of mis-sense variants observed were found to decrease the stability of the protein product, thereby giving away a chance for influencing the catalysis process. A few of the non-synonymous mutations in DNMT3A gene showed an increase in stability. Although, presented with decreased stability all the variants were not found to lead to a deleterious phenotype. Majority of the variants produced deleterious effect with exception in a few gene variants exhibiting neutral outcomes viz., TRDMT1 and DNMT3L (Table 3).
Gene expression analysis
The gene expression data revealed that almost all the genes in the study panel presented with a statistically significant differential gene expression pattern (Figures 2A - 2E). The p values for each of the gene as computed is as follows: DNMT1 (<1 x 10-12), TRDMT1 (7.64 x 10-4), DNMT3A (1.62 x 10-12), DNMT3B (<1 x 10-12) and DNMT3L (2.22 x 10-3). The Kaplan-Meier survival analysis for DNMT1 gene expression returned a significant result between the patients with high level and low/medium level expression (p value = 0.019). The patients with low/medium level gene expression showed a decreased survival probability when compared to patients with high level expression of DNMT1. A p value less than 0.05 was considered to be significant (Figure 2F).
Discussion
The most important epigenetic marks which is considered to be the key players in gene silencing is DNA methylation (Zemach et al., 2010; Jeltsch and Gowher, 2019). The C5 (methylcytosine - 5mC), N6 (methyladenine - 6mA) methylations are known types of methylations identified in the eukaryotic genome. Similar catalytic mechanisms are exhibited by the family of DNA methyltransferases. The promoter methylation is usually encountered in the early stage of carcinogenesis process. A very recent study by Sánchez-Siles et al., (2019) investigated the association between T allele of the rs16906252 and O16-methylguanine-DNA methyltransferase (MGMT) with oral lichen planus. The results were convincing showing association between the genotype and the promoter region of MGMT gene. Since the process of DNA methylation is related to the silencing of genes, methylation of tumor suppressor genes may inevitably lead to uncontrolled cell division and accumulation of lesions in DNA. The present study has identified several novel variants which might have a profound effect on the clinical presentation of HNSCC. Variants previously reported viz., rs200204830 of TRDMT1, rs757211277, rs566390868, rs769590067 of DNMT3A, rs121908946, rs770751820 of DNMT3B. Protein stability and pathogenicity analysis revealed three variants R729Q, E907K (TRDMT1) and R840Q (DNMT3B) which were highly pathogenic with scores less than 3.0.
The relationship between hypermethylation of CDKN2A (p16) and OSCC was reported in a study by Yakushiji et al., (2003) The p16 gene was found to be hypermethylated in about 48% of the samples, with significant downregulation observed in both mRNA and protein expressions. Demethylation assay was also performed on OSCC derived cell lines which eventually resulted in the reactivation of p16 gene expression. The DNMTs over-expressed were DNMT1, DNMT3A and DNMT3B at 72%, 56% and 64% respectively. Although a definitive conclusion could not be drawn by the researchers about the connection between hypermethylation of p16 genes and up-regulation of DNMTs, they concluded by reporting the fact that both the processes are observed in oral carcinoma acting through different mechanisms. A similar kind of trend was observed in the present study, wherein an increase in gene expression was observed in all the genes selected. The differential gene expression observed was statistically significant for all the genes with DNMT1, DNMT3A and 3B showing a dramatic increase in the gene expression when compared to the normal group.
Gene expression and genotyping analysis performed from the samples obtained from OSCC patients revealed over-expression of DNMT1 (36.9%), DNMT3A (26%) and DNMT3B (23%). The over-expression of DNMT1 was found to influence the overall survival of patients presenting with a p value of 0.003. Also, over-expression of DNMT1 was considered to be an independent prognostic factor which showed a 2.385 times higher risk for the tumor to relapse in comparison to a lower expression level. Furthermore, the analysis of 2 polymorphisms in DNMT1 (rs2228612) and DNMT3B (rs406193) by TaqMan SNPs genotyping assays demonstrated the fact that the former variant was associated with reduced survival of OSCC patients (p = 0.036). The present study has also identified several variants which led to protein de-stabilization exhibiting a deleterious consequences (Supic et al., 2017). The survival analysis of the HNSCC patients exhibiting a differential expression profile of the DNMT1 gene exhibited a significant difference in the survival period. The patients with a high level expression of DNMT1 had an increased survival rate in comparison to those with low/medium level expression. This fact was found to contradictory to the study conducted by Supic et al. Probing more into the genetic variations and influence of genetic and epigenetic factors could provide clues on the contrasting results obtained.
Chen et al., (2014) demonstrated through in vitro and in vivo experiments, that the inhibition of DNMT3bB resulted in slower progression of tumor, attenuated tumor invasion and epithelial mesenchymal transition. The activation of inflammatory marker IL-6 could be the vital signal responsible for the induction and over-expression of DNMT3B in oral cancer. In addition, the up-regulation of DNMT3B was significantly linked to the risk of lymph node involvement, recurrence of the disease and shorter survival period in patients in advanced stage of oral cancer. Hence, the researchers suggest that DNMT3B could be used as a promising therapeutic target for oral cancer. A recent study identified an anti-aging gene Klotho to be associated with carcinogenesis. The study group analyzed the association between methylation and the expression of Klotho which returned significant results indicating that the expression of DNMT3B was increased with a simultaneous reduction in the expression of Klotho gene in OSCC patients. Therefore it was concluded that downregulation of Klotho was associated with overexpression of DNMT3B in tumor tissues. As reported by several studies, DNMT3B was found to be the most commonly studied gene in relation to oral cancer. The present study also reported highest frequency of alteration in the DNMT3B gene when compared to the other members of the family.
A very recent study by Shiah et al., (2020) demonstrated that epigenetic silencing of genes related retinoic acid signaling had a significant role to play in the pathogenesis and clinical outcome of OSCC. Genome-wide expression and methylation profile analysis revealed that genes such as ADHFE1 (alcohol dehydrogenase, iron containing 1) and ALDH1A2 (aldehyde dehydrogenase 1 family, member A2) were frequently hyper-methylated and downregulated in OSCC samples. Furthermore, the research group identified microRNAs viz., miR-30a and miR-379 which can influence the expression of DNMT3B that is involved in the process of methylation. Then it was shown that tobacco usage and betel quid chewing resulted in the dysregulation of microRNAs which ultimately resulted in the upregulation of DNMT3B. Chou et al., showed that arecoline, a betel-nut alkaloid recruited DNMT3B which in-turn regulates the miR-486-3p/DDR1 genes. The upregulation of miR-486-3p exhibited a tumor suppressor activity by inhibiting tumor growth and inducing apoptosis, by targeting the 3’-UTR of Discoidin domain receptor-1 (DDR1).
Computational approaches have been considered to be the boon for biologist. An exhaustive collection of data can be analyzed within a short time and cost-effective way. Several studies are emerging out in identifying novel genetic markers which can be further applied for population based screening (Vijayashree and Paramasivam, 2020; Anita et al., 2020). The limitations of the study were (a) the dataset selected had individuals from different ethnic groups or populations, which made data analysis and interpretation to be more generic rather than specific, (b) since both genetic and epigenetic factors act congruently to exhibit a disease phenotype, investigations on epigenetic factors such as influence of methylation, histone modification, microRNA interference could provide clues on trans-generational effects. Accumulation of knowledge on the methylation process, the identification of DNMT inhibitors have revolutionised the field of molecular medicine (Abreu et al., 2008). More details on the effect of variants especially the promoter and regulatory variants of the DNMT gene would open new avenues towards identification of prognostic and diagnostic markers. In conclusion, authors declare that advancements of computational approaches in biology have lead to the development of methods which can be used for identification of genetic abnormalities within a short span of time in a cost effective manner. The present study is one such attempt to accumulate information related to the genetic abnormalities in the DNMT and HNSCC. Findings of the study might aid in solving the puzzle underlying the epigenetic mechanisms and their role in HNSCC.
Acknowledgements
The authors thank all the consorts and groups who were involved in the compilation of data from patients for public use. Our sincere thanks to all the patients who have indirectly contributed to the scientific community by providing consent for sharing their data for research use.
Funding statement
The authors Dr.Vijayashree Priyadharsini.J and Dr. Paramasivam.A thank the Science and Engineering Research Board (SERB), Government of India for the financial support rendered through the core research grant (CRG) (No. CRG/2019/003756).
Statement of conflict of interest
None.
References
- Abreu PA, Dellamora-Ortiz G, Leão-Ferreira LR, et al. DNA methylation: a promising target for the twenty-first century. Expert Opin Ther Targets. 2008;12:1035–47. doi: 10.1517/14728222.12.8.1035. [DOI] [PubMed] [Google Scholar]
- Adhikari BR, Uehara O, Matsuoka H, et al. Immunohistochemical evaluation of Klotho and DNA methyltransferase 3a in oral squamous cell carcinomas. Med Mol Morphol. 2017;50:155–60. doi: 10.1007/s00795-017-0156-9. [DOI] [PubMed] [Google Scholar]
- Anita R, Paramasivam A, Vijayashree Priyadharsini J, et al. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res. 2020;10:2546–54. [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortalityworldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Casalino L, Verde P. 2020) Multifaceted Roles of DNA Methylation in Neoplastic Transformation, from Tumor Suppressors to EMT and Metastasis. Genes (Basel) 11:E922. doi: 10.3390/genes11080922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Casadio R. I-Mutant2. 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;2005:306–10. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KP, Hao SP, Tsang NM, et al. Gene expression and promoter polymorphisms of DNA methyltransferase 3B in nasopharyngeal carcinomas in Taiwanese people: a case-control study. Oncol Rep. 2008;19:217–22. [PubMed] [Google Scholar]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WC, Chen MF, Lin PY. Significance of DNMT3b in oral cancer. PLoS One. 2014;9:e89956. doi: 10.1371/journal.pone.0089956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SJ, Chang CF, Ko HH, et al. Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head Neck. 2018;40:355–68. doi: 10.1002/hed.24958. [DOI] [PubMed] [Google Scholar]
- Cheng SJ, Chang CF, Lee JJ, et al. Hypermethylated ZNF582 and PAX1 are effective biomarkers for detection of oral dysplasia and oral cancer. Oral Oncol. 2016;62:34–43. doi: 10.1016/j.oraloncology.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Peng HY, Mo KC, et al. MicroRNA-486-3p functions as a tumor suppressor in oral cancer by targeting DDR1. J Exp Clin Cancer Res. 2019;38:281. doi: 10.1186/s13046-019-1283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez-Pérez R, Campo-Trapero J, Cano-Sánchez J, et al. Methylation in oral cancer and pre-cancerous lesions (Review) Oncol Rep. 2011;25:1203–1209. doi: 10.3892/or.2011.1205. [DOI] [PubMed] [Google Scholar]
- Duan F, Cui S, Song C, et al. Systematic evaluation of cancer risk associated with DNMT3B polymorphisms. J Cancer Res Clin Oncol. 2015;141:1205–20. doi: 10.1007/s00432-014-1894-x. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2012;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha N, Warnakulasuriya S, Vlaanderen J, et al. Betel quid chewing and the risk of oral and oropha- ryngeal cancers: a meta-analysis with implications for cancer control. Int J Cancer. 2014;135:1433–43. doi: 10.1002/ijc.28643. [DOI] [PubMed] [Google Scholar]
- Gupta N, Gupta R, Acharya AK, et al. Changing Trends in oral cancer - a global scenario. Nepal J Epidemio. 2016;6:613–9. doi: 10.3126/nje.v6i4.17255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A, Gowher H. Editorial-Role of DNA Methyltransferases in the Epigenome. Genes (Basel) 2019;10:574. doi: 10.3390/genes10080574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in humans. bioRxiv. 2020;141:531210. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19:81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- McDermott JD, Bowles DW. Epidemiology of head and neck squamous cell carcinomas: Impact on Staging and Prevention Strategies. Curr Treat Options Oncol. 2019;20 doi: 10.1007/s11864-019-0650-5. [DOI] [PubMed] [Google Scholar]
- Ram H, Sarkar J, Kumar H, et al. Oral cancer: risk factors and molecular pathogenesis. J Maxillofac Oral Surg. 2011;10:132–7. doi: 10.1007/s12663-011-0195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotondo JC, Borghi A, Selvatici R, et al. Association of Retinoic Acid Receptor β gene with onset and progression of Lichen Sclerosus-associated vulvar squamous cell carcinoma. JAMA Dermatol. 2018;154:819–23. doi: 10.1001/jamadermatol.2018.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17 doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Sánchez-Siles M, Aliaga-Sánchez A, Medina S, et al. Genotyping of the C>T allele of rs16906252, predictor of O16-methylguanine-DNA methyltransferase (MGMT) promoter methylation status, in erosive atrophic lesions of oral lichen planus. Int J Dermatol. 2019;58:1078–82. doi: 10.1111/ijd.14473. [DOI] [PubMed] [Google Scholar]
- Shiah SG, Hsiao JR, Chang HJ, et al. MiR-30a and miR-379 modulate retinoic acid pathway by targeting DNA methyltransferase 3B in oral cancer. J Biomed Sci. 2020;27:46. doi: 10.1186/s12929-020-00644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supic G, Kozomara R, Zeljic K, et al. Prognostic value of the DNMTs mRNA expression and genetic polymorphisms on the clinical outcome in oral cancer patients. Clin Oral Investig. 2017;21:173–82. doi: 10.1007/s00784-016-1772-9. [DOI] [PubMed] [Google Scholar]
- Tang XH, Albert M, Scognamiglio T. A DNA methyltransferase inhibitor and all-trans retinoic acid reduce oral cavity carcinogenesis induced by the carcinogen 4-nitroquinoline 1-oxide. Cancer Prev Res (Phila) 2009;2:1100–10. doi: 10.1158/1940-6207.CAPR-09-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towle R, Truong D, Garnis C. Epigenetic mediated silencing of EYA4 contributes to tumorigenesis in oral dysplastic cells. Genes Chromosomes Cancer. 2016;55:568–76. doi: 10.1002/gcc.22360. [DOI] [PubMed] [Google Scholar]
- Vijayashree P, Paramasivam A. Virtual screening of mutations in antioxidant genes and its putative association with HNSCC: An in silico approach. Mutat Res Fund Res Fund Mol M. 2020;821 doi: 10.1016/j.mrfmmm.2020.111710. 111710 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Wyss A, Hashibe M, Chuang SC, et al. Cigarette, cigar, and pipe smoking and the 22 risk of head and neck cancers: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Am J Epidemiol. 2013;178:679–90. doi: 10.1093/aje/kwt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushiji T, Uzawa K, Shibahara T, et al. Over-expression of DNA methyltransferases and CDKN2A gene methylation status in squamous cell carcinoma of the oral cavity. Int J Oncol. 2003;22:1201–7. [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, et al. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zhou J, Michaud DS, Langevin SM, et al. Smokeless tobacco and risk of head and neck cancer: evidence from a case-control study in New England. Int J Cancer. 2013;132:1911–7. doi: 10.1002/ijc.27839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Du S, Zhang J, et al. Polymorphism of DNA methyltransferase 3B -149C/T and cancer risk: a meta-analysis. Med Oncol. 2015;32:399. doi: 10.1007/s12032-014-0399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]