SUMMARY
The type 2 cytokine-high asthma endotype (T2H) is characterized by IL-13-driven mucus obstruction of the airways. To further investigate this incompletely understood pathobiology, we characterize IL-13 effects on human airway epithelial cell cultures using single-cell RNA sequencing, finding that IL-13 generates a distinctive transcriptional state for each cell type. Specifically, we discover a mucus secretory program induced by IL-13 in all cell types which converts both mucus and defense secretory cells into a metaplastic state with emergent mucin production and secretion, while leading to ER stress and cell death in ciliated cells. The IL-13-remodeled epithelium secretes a pathologic, mucin-imbalanced, and innate immunity-depleted proteome that arrests mucociliary motion. Signatures of IL-13-induced cellular remodeling are mirrored by transcriptional signatures characteristic of the nasal airway epithelium within T2H versus T2-low asthmatic children. Our results reveal the epithelium-wide scope of T2H asthma and present candidate therapeutic targets for restoring normal epithelial function.
In Brief
Using airway epithelial cell cultures, Jackson et al. show that IL-13, a driver of type 2-high asthma, induces emergent mucus secretory expression states for each cell type. This program universally diminishes innate airway defense, produces a pathologic mucus secretome that arrests mucociliary movement, and is recapitulated in type 2 inflamed children.
Graphical Abstract
INTRODUCTION
Asthma is a chronic disease of the airways marked by clinical features that include airway hyperresponsiveness and reversible airway obstruction. Although united by these clinical features, asthmatics exhibit heterogeneous pathobiology, with distinct endotypes driving development and persistence of disease (Fahy, 2015). The most common asthma endotype is characterized by airway inflammation triggered by type 2 (T2) cytokines (IL-4, IL-5, and IL-13). Referred to as T2-high (T2H), this endotype affects not only 50% of asthmatics but also a subgroup of chronic obstructive pulmonary disease (COPD) patients (Barceló et al., 2006; Christenson et al., 2015; George and Brightling, 2016). Thus, the particular features of T2H disease, including severe hyperresponsiveness, eosinophilia, and mucus overproduction, may underlie a more general form of airway pathobiology relevant to many airway and lung diseases (Poole et al., 2014; Woodruff et al., 2009).
Although T2H asthma involves multiple tissue types, dysfunction of the airway epithelium is central to development, pathology, and persistence of this disease endotype (Kuperman et al., 2002). Positioned at the interface between the inhaled environment and the lung, the airway epithelium forms a physical barrier and generates the mucociliary escalator that clears inhaled particulates and microorganisms. Moreover, the airway epithelium generates defensive secretions critical to innate immunity that contain growth factors, cytokines, and chemokines that stimulate and direct the activity of airway immune cells. These higher order epithelial functions are the cumulative products of a functionally diverse set of cell types and intermediate states that populate the healthy airway in particular proportions (Montoro et al., 2018; Plasschaert et al., 2018). Studies using bulk transcriptomics and histology make clear that both the function and relative frequency of epithelial cell types are altered in T2H asthma and chronic airway diseases (Bergeron et al., 2009; Fahy, 2015; Poole et al., 2014), yet the imprecision (bulk RNA sequencing [RNA-seq]) and limited scope (histology) of these approaches have thus far prevented comprehensive study of cell type-specific responses.
The effects of T2 inflammation on the airway epithelium have been shown to be mediated primarily by T2 cytokines, particularly IL-4, IL-5, and IL-13. For example, human airway epithelial cells (AECs) were recently shown to express a functional IL-5 receptor that, upon stimulation, exhibited differential expression (DE) of >3,000 genes (Barretto et al., 2020). Although these and other regulators of T2 inflammation can play distinctive and collaborative roles, using divergent pathways that orchestrate particular aspects of disease (Bao and Reinhardt, 2015), transgenic overexpression specifically of IL-13 in mice has been shown to be sufficient to recapitulate most of the cardinal features of T2H asthma, occurring both within the airway epithelium and in other tissues (Zhu et al., 1999). Consequently, IL-13 has successfully been used to model the effects of T2 inflammation on the airway epithelium in both in vivo mouse models (Chen et al., 2009; Tyner et al., 2006; Zhen et al., 2007) and in vitro human AEC cultures (Gerovac and Fregien, 2016; Nicodemus-Johnson et al., 2016; Zhen et al., 2007), revealing some of the key genes, regulators, and cellular shifts brought on by this cytokine. Mouse models have specifically been used to show that binding of IL-13 to its receptor induces activation of SPDEF (Park et al., 2007), leading to the overproduction of mucus and the metaplastic conversion of secretoglobin-secreting club cells into MUC5AC-producing mucous cells (Chen et al., 2009). Supporting translation of this mouse model to human disease, bronchial epithelial expression of MUC5AC is upregulated in T2H asthmatics (Lachowicz-Scroggins et al., 2016; Woodruff et al., 2009), while in mature human mucociliary AEC cultures, IL-13 increases the frequency of MUC5AC+ mucus cells at the numerical expense of ciliated cells (Gomperts et al., 2007; Turner et al., 2011). However, many questions regarding the effects of IL-13 on the human airway epithelium remain to be answered, including (1) to what extent metaplastic events or reprogramming of cell fates underlies the observed increases in MUC5AC transcription and mucus secretion, (2) how IL-13 specifically modifies gene expression programs and protein secretions of secretory cell states residing in the healthy airway, and (3) whether changes in behavior and frequency of other cell types (basal, ciliated, club secretory) contribute to IL-13-induced pathology.
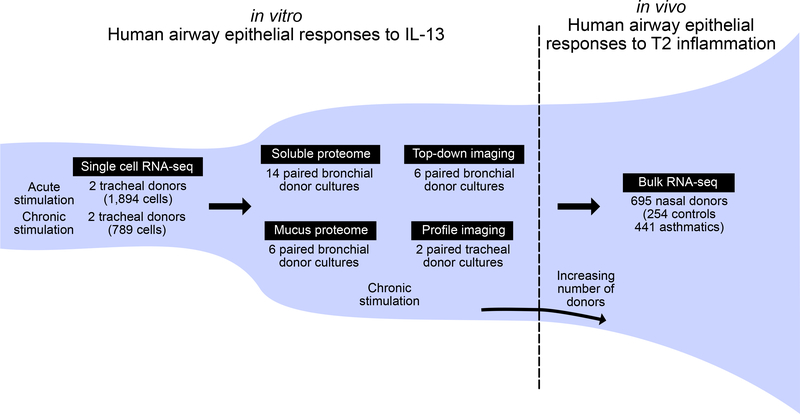
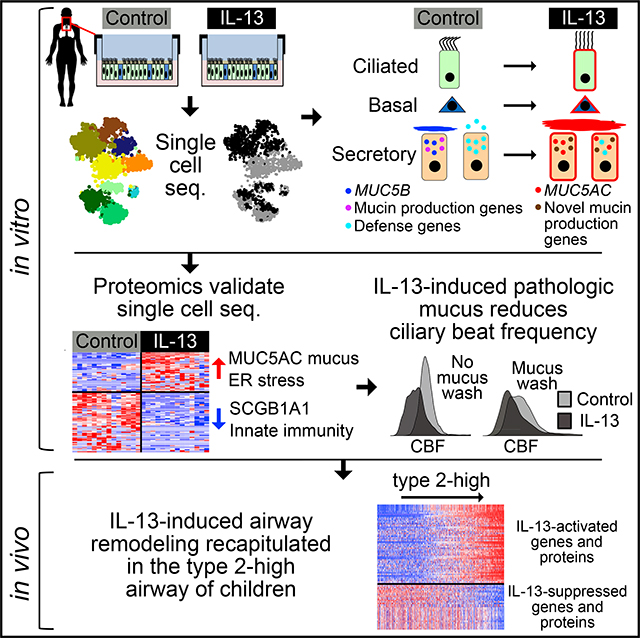
To address these gaps in our understanding of T2 airway inflammation, here, we investigate the whole-transcriptome responses to IL-13 of single cells from differentiated human AEC cultures. Using this dataset, we define the transcriptional cell states that populate the unperturbed human airway epithelium and then show how IL-13 stimulation alters these states to create new metaplastic cell populations. We relate these transcriptional effects to IL-13-induced changes to the mucus secretome of the airway epithelium and its accompanying influence on mucociliary function. Finally, we examine the extent to which cellular remodeling and secretory changes observed in response to IL-13 in vitro mimics transcriptional reprogramming in children with T2H asthma disease (Figure 1).
Figure 1. Outline of the Study Design and Generated Data.
The study proceeded in three in vitro phases with a fourth in vivo validation phase. In the in vitro phase, IL-13 transcriptomic effects on a cell type-specific level in human AECs were determined, effects were translated to the secreted proteome, and influences of this altered proteome on mucociliary motion and ciliary beat frequency were determined. In the in vivo phase, in vitro IL-13 effects were validated using in vivo transcriptome data from the nasal airway epithelial brushings of a large cohort of asthmatic and healthy children.
RESULTS
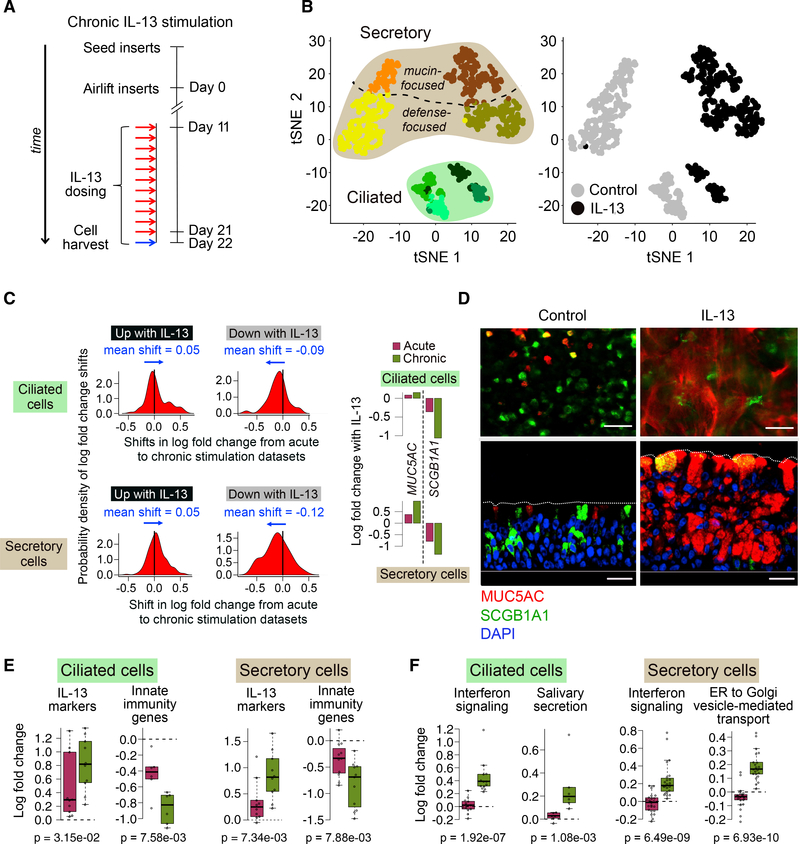
Acute IL-13 Stimulation Drives Cellular Remodeling of the Mucociliary Epithelium
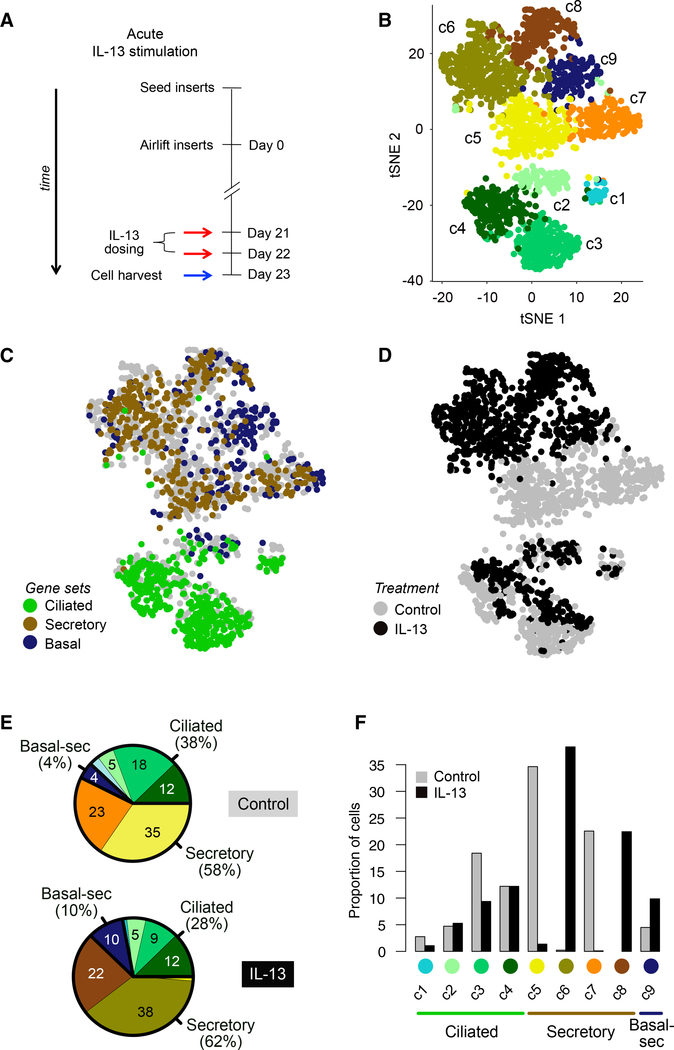
We first identified transcriptionally defined cell populations in mature mucociliary air-liquid interface (ALI) cultures of human to represent ciliated cells, on the basis of expression of genes from cilia motility-related Gene Ontology categories (Figures 2C and S1C). Four non-ciliated clusters (c5–c8) expressed genes that mark secretory cell populations (e.g., SCGB1A1, SLPI) in the airway epithelium, suggesting that these cells carry out secretory functions (Figures 2C and S1D). Although the fifth non-ciliated cluster (c9) exhibited some secretory gene expression, it was most distinguished by expression of basal cell genes (KRT5 and TP63). Moreover, genes differentially expressed in c9 cells were enriched for airway epithelial basal cell genes (hypergeometric test; control cell false discovery rate [FDR] = 1.93 × 10−14, IL-13 cell FDR = 6.07 × 10−11) (Hackett et al., 2011), indicating that this population represents basal cells and/or basal cells differentiating into secretory cells (Figures 2C and S1E).
Figure 2. Acute IL-13 Stimulation Drives Cellular Remodeling of the Mucociliary Epithelium.
(A) Schematic detailing human AEC ALI mucociliary differentiation followed by acute stimulation with IL-13; n = 2 human tracheal epithelial cell (HTEC) donors (T71, T72).
(B) t-SNE plot of 1,894 cells depicting nine unsupervised scRNA-seq clusters.
(C) t-SNE plot in (B), with cells colored by characteristic expression of ciliated, secretory, or basal cell gene signatures. See also Figures S1C–S1E.
(D) t-SNE plot in (B), with cells colored by treatment.
(E) Pie charts of differences in cell state proportions within major groups (ciliated, secretory, and basal-secretory) between control and IL-13-stimulated epithelia.
(F) Bar-plot depiction of data in (E).
We observed a dramatic secretory cell state “switch” with acute IL-13 treatment (Figure 2D). Namely, although the frequency of secretory cell clusters (c5–c8) remained constant between IL-13 (62%) and control (58%) treatments, secretory cell AECs and investigated how these populations were affected by acute (48 h) IL-13 stimulation. To do this, we analyzed RNA-seq data from 1,894 single cells (1,025 IL-13-stimulated cells, 869 controls) isolated from mature (day 21) ALI cultures derived from the basal AECs of two tracheal donors (Figures 2A, S1A, and S1B; Table S2). To identify and visualize cell populations, we performed t-distributed stochastic neighbor embedding (t-SNE), followed by the plotting of all cells using the top two t-SNE dimensions and overlaying shared nearest neighborhood (SNN) clustering results. These analyses revealed nine discrete clusters (Figure 2B). Four of the cell clusters (c1–c4) appeared population membership changed from being overwhelmingly c5 and c7 cluster dominated in the control cultures to being c6 and c8 cluster dominated with IL-13 treatment (Figures 2E and 2F). Specifically, secretory clusters c5 and c7 constituted 35% and 23% of all control culture cells, while these clusters were nearly absent from the IL-13-stimulated cultures (1.4% and 0.1%, respectively; Figures 2E and 2F). In contrast, the c6 and c8 secretory clusters encompassed 38% and 22% of IL-13-treated cells, but altogether only 0.2% of control culture cells. The basal-secretory cluster (c9) contained a mix of control and IL-13-stimulated cells, although t-SNE segregation of IL-13 and control cells within the cluster (Figure 2D) indicates reprogramming in this cluster as well (Ordovas-Montanes et al., 2018). Finally, acute IL-13 treatment significantly decreased the percentage of ciliated cells within the cultures, from 38% to 28% (p < 1 × 10−5; Figure 2E). A mix of IL-13 and control cells was observed in each of the four ciliated cell clusters, although clusters c3 and c4 exhibited clear t-SNE partitioning of IL-13 and control cells (Figure 2D). These results illustrate how acute IL-13 stimulation is sufficient to thoroughly transform cellular composition of the mature airway epithelium.
Normal Defense and Mucus Secretory Cells Are Reprogrammed by IL-13 into a Distinct Pathologic Mucus Secretory State
Studies in mice have established that IL-13 transforms the epithelium through a metaplastic rather than a hyperplastic process (Chen et al., 2009). In support of a metaplastic basis to the observed secretory cell "switch," we found that neither proliferation nor apoptosis increased with IL-13 stimulation (Figure S2A). In mice, the IL-13-induced generation of mucin 5ac (MUC5AC+) goblet cells occurs through metaplasia of secretoglobin 1a1 (SCGB1A1+) club cells (Evans et al., 2004). Investigating whether a similar process might underlie the secretory cell switch in humans, we stained control and IL-13-stimulated cultures for both MUC5AC and SCGB1A1 protein. In the control cultures, we found that among cells staining positive for one or both markers, 79% were exclusively SCGB1A1+, 15% were exclusively MUC5AC+, and only 5.8% were double-positive (Figures 3A and S2B). In contrast, after acute IL-13, only 20% of cells were exclusively SCGB1A1+, while 48% were exclusively MUC5AC+ (3.2-fold increase), and 32% were double-positive (5.6-fold increase), suggesting that SCGB1A1+ cells are a source of MUC5AC+ cells (Chen et al., 2009; Evans et al., 2004; Figures 3A and S2B). A 2.2-fold reduction of SCGB1A1 expression and a 1.5-fold increase of MUC5AC in the two IL-13 treatment-dominant secretory populations (c6 and c8) compared with the control-dominant populations (c5 and c7) further support this SCGB1A1+ to MUC5AC+ transition.
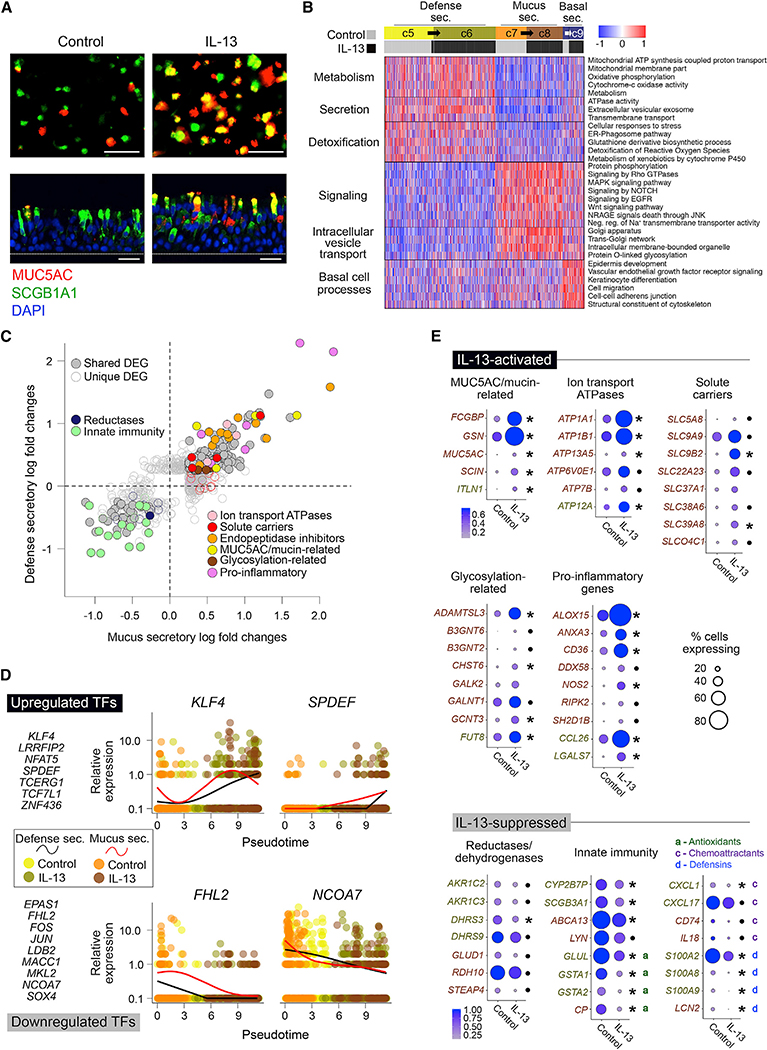
Figure 3. Normal Defense and Mucus Secretory Cells Are Reprogrammed by IL-13 into a Distinct Pathologic Mucus Secretory State.
(A) Immunofluorescence (IF) labeling of ALI-differentiated human AEC cultures in top-down (top) and side (bottom) views show control cultures (left) dominated by SCGB1A1+ (green) and MUC5AC− (red) cells transformed into cultures (right) with prominent co-staining (yellow) after acute IL-13. n = 3 HTEC donors (T73, T76, T79); scale bar, 30 mm; DAPI nuclei labeling (blue).
(B) Heatmap shows shared functions between pairs of control and IL-13-dominant secretory populations (c5/c6, c7/c8, and c9-control/c9-IL-13). Mean scaled expression of population-defining DEGs within enriched functionally annotated sets is shown.
(C) Scatterplot compares responses to IL-13 in defense (y) and mucus (x) secretory cells. Points, expression log fold changes with IL-13; filled, differentially expressed in both populations; unfilled, differentially expressed in one population; colored, belonging to specified function categories.
(D) TFs whose expression significantly changes across a pseudotime trajectory transitioning from baseline to IL-13-stimulated cell states. Left: TFs increasing (top) or decreasing (bottom) across pseudotime. Right: scaled normalized expression (points) and smoothed trends (lines) for exemplar TFs across pseudotime-ordered cells. Point color, population as in (B); line color, defense-secretory (black), mucus-secretory (red).
(E) Dot plots show shifts in level (dot color) and ubiquity (dot size) of expression for genes "activated" (top) or "suppressed" (bottom) by IL-13 in one or both secretory populations. Functional groups as in (C). Gene label font color: gold, defense; brown, mucus (population with strongest effect is shown). Same direction responses in both population pairs are indicated by asterisk (significant) or point (not significant).
See also Figure S2.
We thus aimed to investigate the genes and pathways underlying IL-13-induced metaplasia of secretory cells in our dataset. However, prior to doing this, we needed to (1) identify which baseline populations transition into which IL-13 populations and (2) characterize the role of these secretory populations in the airway epithelium. First, because we observed four secretory cell populations, two consisting of SCGB1A1+ control cells and two consisting of MUC5AC+ IL-13 cells, we reasoned that two distinct baseline secretory cell populations each transitioned into one of the corresponding IL-13 populations. To match analogous baseline and IL-13-induced populations, we compared gene signatures among them and found highly concordant signatures between populations c5 (control) and c6 (IL-13) and between populations c7 (control) and c8 (IL-13), strongly suggesting that IL-13 stimulation caused c5 to transition into c6 and c7 to transition into c8 (Figures S2C and S2D).
Having identified analogous baseline and IL-13 populations, we next characterized the shared function of these population pairs. To do this, we first performed pathway analysis on the defining genes of c5/c6 secretory cells when compared with c7/c8 cells, which resulted in enriched terms related to airway detoxification, protein secretion, and metabolism (Figure 3B). Active protein secretion of c5/c6 cells was supported by enrichments for extracellular vesicular exosome, transmembrane transport, and ATPase activity. Moreover, these populations more highly expressed defense genes, such as SCGB1A1, WFDC2, LYN, SLPI, and B2M, as well as genes related to the glutathione-derivative biosynthetic process/metabolism of xenobiotics (e.g., GSTP1, MGST1) and detoxification of reactive oxygen species, strongly suggesting that c5/c6 cells are airway defense-secretory cells, similar to the club cell in mice.
We similarly performed pathway analysis on the genes defining c7/c8 cells compared to c5/c6 cells, resulting in enrichments for gene groups involved in MAPK, NOTCH, and EGFR signaling, all of which have been implicated in mucin production and secretion, as well as in the folding (e.g., trans-Golgi network), O-linked glycosylation, and transport and storage of mucins (Figure 3B). Examination of gel-forming mucin expression (MUC5B and MUC5AC) was impeded by poor detection of these transcripts with our single-cell RNA-seq [scRNA-seq] method, despite immunochemistry revealing an abundance of MUC5B+ cells (Figure S2E) or MUC5AC+ cells (Figures 3A and S2B) in control or IL-13-induced cultures, respectively. Notably, we did find that the two most common transmembrane mucins (MUC4 and MUC16) were both more highly expressed in these clusters. Together these data suggest that c7/c8 cells are more specialized for mucin production and mucus secretion than are c5/c6 cells.
Having characterized these secretory cell populations, we next examined how gene expression, and thus function, of these defense and mucus secretory populations were altered by IL-13. We performed DE analysis between control and IL-13-treated populations for defense secretory (c5 versus c6) and mucus secretory cells (c7 versus c8). We identified 396 and 535 differentially expressed genes (DEGs) in defense and mucus secretory cells, respectively. A shared IL-13 response between the two cell types was evident from 208 overlapping DEGs, which exhibited a strong correlation in expression fold changes between cell types (R2 = 0.86; Figure 3C). This group of shared IL-13-response genes was enriched for those DEGs with the largest fold changes and included genes known to characterize both IL-13 stimulation of the airway epithelium in vitro and T2 inflammation of the airway epithelium in vivo, such as CDH26, DPP4, FCGBP, MUC5AC, and POSTN. This shared IL-13 response also included upregulation of genes involved in mucin production and secretion (e.g., ion transport ATPases, glycosylation genes, mucins and mucin-related genes; Figure 3C) and downregulation of genes involved in airway defense (Figure 3C), indicating that the mucus/defense-secretory balance in the epithelium was significantly shifted in both secretory cell types. To identify gene regulators of these shared IL-13-mediated transcriptional changes, we constructed a pseudotime trajectory of IL-13 stimulation using all control and IL-13-secretory cells (Figure S2F). In addition to observing increased expression of SPDEF across pseudotime, we identified multiple novel transcription factors (TFs) that significantly increased and/or decreased expression across pseudotime, including many known regulators of Wnt signaling (e.g., repressors [KLF4, NFAT5, TCF7L1] and activators [LRRFIP2, SOX4, FHL2, and EPAS1]; Figure 3D), suggesting that Wnt signaling may help modulate the IL-13 response in secretory cells.
Interestingly, the vast majority of IL-13 DEGs were markers neither of baseline mucus secretory cells (89%) nor of defense secretory cells (95%). "Activated" genes (those not among cell type signature genes) included a majority of the shared DEGs highlighted in Figure 3C relating to mucin production and secretion (Figure 3E), suggesting that IL-13 has significantly altered the manner in which mucus secretory cells produce and secrete mucus and also that many of these emergent promucus pathways are similarly activated in defense-focused secretory cells. For example, a set of glycosylation genes were activated, likely modifying how mucins are being glycosylated, while activation of ion transport ATPases and solute carriers signals a modification in how air surface liquid hydration is maintained (Figure 3E). Additionally, IL-13-induced expression of the mucin-interacting proteins ITLN1 and FCGBP likely indicates alterations to the physical properties of IL-13-stimulated mucus (Figure 3E). Induction of this novel mucus-secretory program coincided with suppression of the two most abundant transmembrane mucins at baseline (MUC4 and MUC16) and the enhanced expression of MUC5AC, in line with histology (Figure 3A). Acquisition of this new IL-13 mucus-secretory function was accompanied by a loss of airway defense function in both the mucus and defense-secretory populations (e.g., antioxidants, defensins, and immune cell chemoattractants; Figure 3E). Together, these results show that IL-13 induces transcriptional reprogramming of secretory cells through a metaplastic process that converts secretory populations focused on airway defense or healthy mucus production into distinct states focused on production of a MUC5AC-enriched pathological mucus.
Ciliated Cells Acquire Secretory Cell Expression Patterns with IL-13 Stimulation at the Expense of Ciliogenic and Innate Immunity Functions
As with the secretory populations, we first determined the distinct nature of four ciliated cell clusters using DE analysis. The c1 ciliated cell population was most distinguished by high expression of centriole biogenesis genes, suggesting that these cells were in an early stage of multiciliogenesis (Ruiz Garciá et al., 2019; Figure 4A). Marker genes for c2 were enriched in gene groups involved in the endomembrane system, which may contribute to development of the ciliary membrane (Ishikawa and Marshall, 2011), c3 exhibited upregulation of genes relating to ciliary motility and axoneme generation, and c4 appeared to be particularly focused on intraflagellar transport, while also exhibiting a distinct immune, metabolic, and secretory profile (Figure 4A).
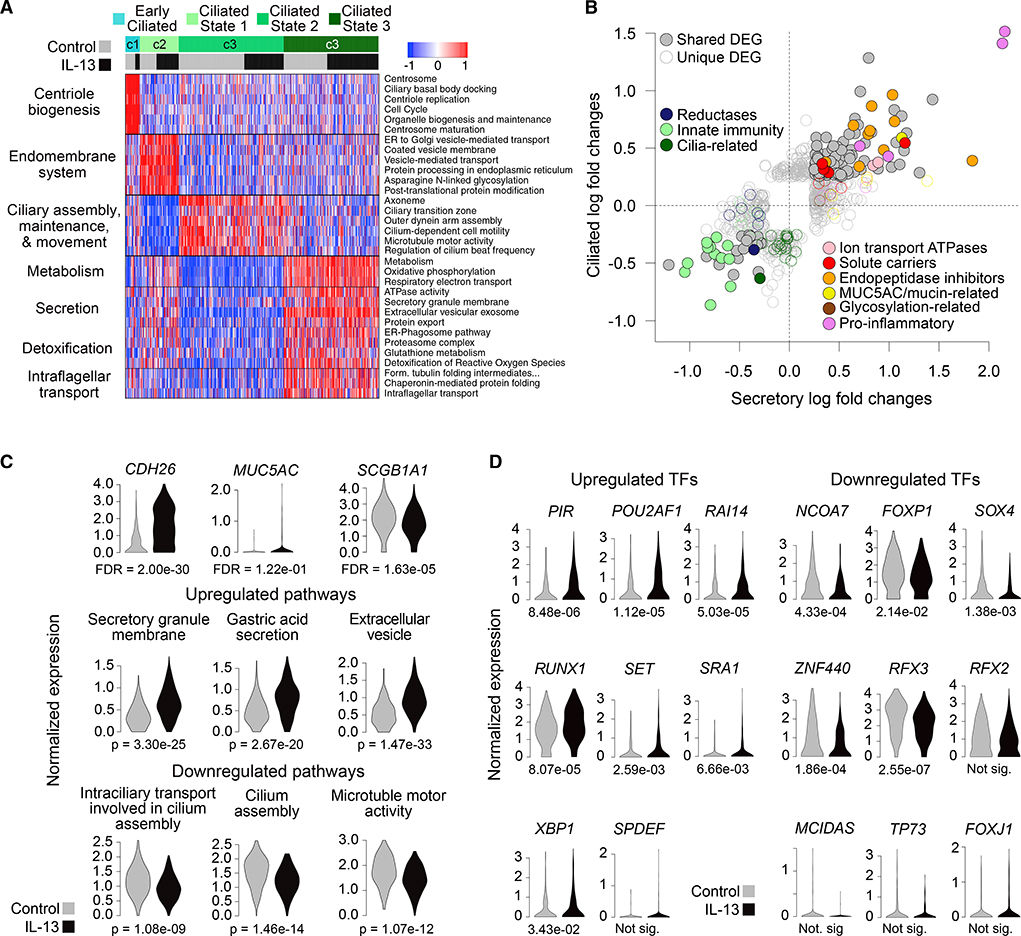
Figure 4. Ciliated Cells Acquire Secretory Cell Expression Patterns with IL-13 at the Expense of Ciliogenic and Innate Immunity Functions.
(A) Heatmap shows expression of functionally annotated gene groups distinguishing four ciliated cell populations. Mean scaled expression of population-defining DEGs within enriched functionally annotated sets is shown.
(B) Scatterplot compares ciliated and secretory (defense + mucus) cell responses to IL-13. Points, expression log fold changes with IL-13; filled, differentially expressed in both populations; unfilled, differentially expressed in one population; colored; belonging to specified function categories.
(C) Violin plots show expression in ciliated cells of key genes (top row) and mean expression of sets of genes up or downregulated with IL-13 that belong to enriched pathways (middle and bottom rows). FDRs are based on DE analysis; p values (p) are based on one-sided Wilcoxon tests.
(D) Violin plots of expression of TFs up and downregulated with IL-13 in ciliated cells. Numbers below plots give FDRs based on DE analysis (where "not sig" denotes differences that were not significant).
To characterize IL-13 effects on ciliated cells, we carried out DE analysis between control and IL-13 cells for all ciliated populations combined, yielding 201 upregulated DEGs and 136 downregulated DEGs. Interestingly, we found that 24 of the top 25 IL-13 DEGs in ciliated cells were also DEGs in secretory cells, including T2 biomarkers, ALOX15, CCL26, CDH26,and SERPINB4. Furthermore, the expression fold changes for ciliated and secretory IL-13 DEGs were highly correlated (R2 = 0.44; Figure 4B), with upregulation of many of the same ion transport ATPases (ATP1A1, ATP1B1), solute carriers (SLC9B2, SLC9A9), and endopeptidase inhibitors (CAPN14, CSTA) and downregulation of many of the same innate immunity-related genes, including SCGB1A1 (Figure 4C), ABCA13, CYP2B7P, WFDC2, antioxidants such as GLUL, CP, GSTA1/2, and defensins such as S100A2 and S100A8 (Figure 4B). Additionally, while secretion-related genes were upregulated with IL-13, many components of the expansive ciliated cell transcription program were downregulated, including ciliary assembly and motility genes (Figures 4B and 4C). Although MUC5AC expression was activated in ciliated cells, they failed to contain MUC5AC protein. Moreover, many prominent IL-13-secretory cell DEGs thought to be directly involved in mucin formation and structure (e.g., glycosylation genes, FCGBP, ITLN1, and SCIN) were not induced by IL-13 in ciliated cells. This suggests that although the normal ciliogenesis expression program is blocked and an IL-13-secretory program is transcriptionally induced in ciliated cells, these cells are not actively secreting mucus.
Despite many similarities in the IL-13 responses of ciliated and secretory cells, activated TFs in ciliated cells included several NF-kB-mediated pro-inflammatory regulators (PIR, POU2AF1, RUNX1, XBP1) that were not significantly upregulated in secretory cells (Figure 4D), suggesting that inflammatory signaling was particularly activated in ciliated cells. This may be triggered by enhanced endoplasmic reticulum (ER) stress, resulting from the induction of a secretory transcriptional program in these cells. TFs downregulated by IL-13 in ciliated cells (Figure 4D) included RFX3, an important regulator of ciliogenesis.
Collectively, these results show that ciliated cells are reprogramed by IL-13 to downregulate ciliogenesis and innate immunity functions, while gaining secretory function associated with inflammation.
Chronic IL-13 Stimulation Brings to Completion Metaplasia and Promotes ER Stress and Interferon Immune Responses
Although acute stimulation was able to model the initial transcriptional effects of IL-13 on the naive airway epithelium, we also generated a longer term stimulation model to identify changes in the T2H asthmatic airway that are dependent on more persistent IL-13. Specifically, we applied IL-13 daily for 11 days (Figure 5A) to mucociliary cultures grown from the same two tracheal donors used in the acute experiment. We then analyzed RNA-seq data generated for 789 single cells derived from these cultures.
Figure 5. Chronic IL-13 Completes Epithelium-wide Metaplasia and Promotes Both ER Stress and Interferon Responses in Ciliated Cells.
(A) Schematic detailing human AEC ALI mucociliary differentiation followed by chronic stimulation with IL-13 of cells; n = 2 HTEC donors (T71, T72).
(B) t-SNE plot overlaying cell clusters (left) or cell treatment (right) for 789 cells from experiment in (A).
(C) Density plots show probability distribution (on the basis of a kernel density function) of shifts in log fold change between acute and chronic datasets for genes up (left) or downregulated (right) with IL-13 in ciliated (top) or secretory (mucus + defense) cells (bottom). All shifts significant with t test p values < 0.05. Bar plots compare log fold changes in MUC5AC and SCGB1A1 between acute and chronic IL-13.
(D) Immunofluorescence (IF) labeling of ALI-differentiated human AEC cultures in top-down (top) and side (bottom) views show control cultures (left) dominated by SCGB1A1+ (green)/MUC5AC− (red) cells transformed by chronic IL-13 into SCGB1A1−/MUC5AC+-dominated cultures (right). Co-staining (yellow); DAPI nuclei (blue); scale bar, 30 μm; n = 3 HTEC donors (same as in Figure 3A).
(E) Boxplots show mean log fold change of IL-13 markers and innate immunity genes responding significantly more strongly with chronic than acute IL-13. p values (p) are based on one-sided Wilcoxon tests.
(F) Boxplots show mean log fold change of enriched annotated gene sets composed of genes exhibiting significantly stronger responses with chronic compared with acute IL-13. p values (p) are based on one-sided Wilcoxon tests.
See also Figure S3.
Using an approach similar to our acute dataset analysis, we inferred eight cell populations (Figure 5B), with marker expression revealing four secretory and four ciliated cell clusters (Figure S3A). Secretory cell clusters included both defense and mucus types observed in the 48 h data, one of each specific to control and IL-13 cultures (Figure 5B), suggesting that the same secretory cell "switch" observed in the 48 h dataset also occurred with chronic stimulation (Figures S3B and S3C). Supporting this, of the mucus secretory genes activated or suppressed with acute IL-13 in both defense and mucus secretory cells, 91% or 75% of these were also upregulated or downregulated, respectively, in one or both of the secretory populations after chronic stimulation. Similarly, IL-13 responses in ciliated cells were broadly concordant between experiments, with 84% or 46% of the top 50 up- or downregulated DEGs in acutely stimulated ciliated cells being also up- or downregulated, respectively, in chronically stimulated ciliated cells.
Although the chronic IL-13 response contained many of the same transcriptional shifts observed with acute stimulation, fold changes in expression of IL-13-response genes shared between the two datasets tended to be more extreme with chronic stimulation, regardless of the direction of change or cell type (Figure 5C). For example, downregulation of SCGB1A1 and upregulation of MUC5AC in both secretory and ciliated cells were stronger with chronic than with acute stimulation (Figure 5C). This more extreme chronic response extended to the protein level, with the SCGB1A1/MUC5AC double-positive cells induced with acute IL-13 (Figure 3A) becoming highly positive for MUC5AC and negative for SCGB1A1 after 11 days of stimulation (Figure 5D). Furthermore, we found the ratio of defense to mucus secretory cells to be reduced from 3:1 in control cultures to near 1:1 with chronic IL-13 (Figure S3D). This shift is consistent with defense secretory cells, which gained new mucus secretory properties with acute IL-13, eventually making a full transition to the IL-13 mucus secretory state after prolonged inflammation. Enrichment analysis on the basis of genes exhibiting a greater response with chronic, compared with acute IL-13, revealed that innate immunity genes suppressed with acute IL-13 in both ciliated and secretory cells were even further downregulated after 11 days (Figure 5E). Overall, these data show that prolonged IL-13 stimulation exacerbates many of the expression changes initiated under acute IL-13.
Additionally, some responses to IL-13 were observed only after chronic stimulation. For example, DEGs annotated by enriched terms related to secretion (e.g., salivary secretion in ciliated cells and ER to Golgi transport in secretory cells) were activated only after 11 days of IL-13 (Figure 5F), accompanied by the chronic-dependent upregulation of AGR2 and ERO1A in both secretory and ciliated cells, two known markers of ER stress induced by mucin production. Also indicative of an ER stress response being uniquely activated with chronic stimulation was the chronic-dependent induction of a suite of type I interferon-induced genes and their regulators (e.g., IFI27, IFITM2, IFITM3, IFI6, IFIH1, IRF6, and DDX58) as well as inflammatory genes (e.g., HLA-DRA, HLA-DRB1, HLA-A/B/C), most strongly in ciliated cells, but also in secretory cells (Figure 5F), suggesting that there is a unique burden of mucin and secretory-related proteins with long-term IL-13, culminating in interferon-related inflammation.
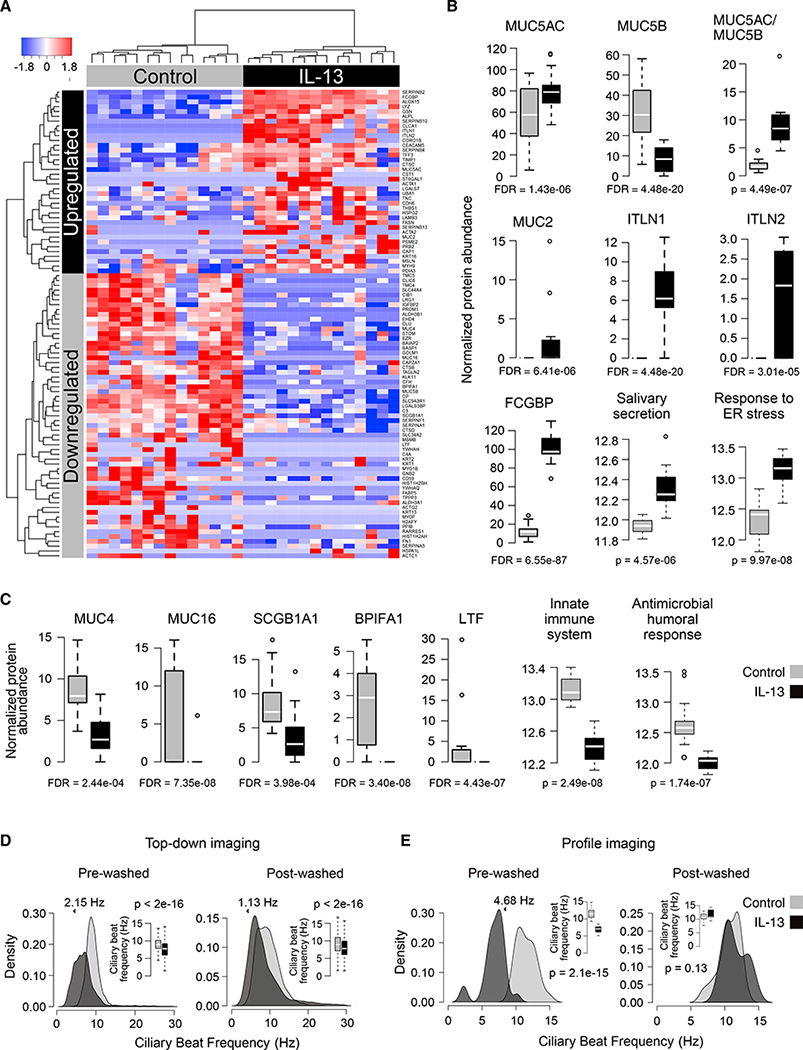
The IL-13-Induced Epithelial Secretome Exhibits a Highly Modified Mucus and Weakened Innate Immunity, Consistent with Transcriptomic Shifts
Our scRNA-seq data suggested that defensive secretory function is lost and new mucus-secretory function is acquired in airway epithelia chronically stimulated with IL-13. To investigate whether these transcriptomic changes are also observed at the level of secreted proteins, we collected apical secretions from paired ALI cultures of bronchial epithelia from 14 donors, after 10 days of mock or IL-13 stimulation, for use in mass spectrometry-based proteomic analysis. We analyzed the soluble component of these secretions in all subjects (n = 14) and the mucus solids in a subset (n = 6). In the soluble fraction, 980 unique proteins were detected across samples, with an average of 272 unique proteins per sample (results for the mucus fraction were broadly similar; see Figures S4A–S4C). We tested whether protein levels differed with IL-13 stimulation, identifying 39 upregulated and 58 downregulated proteins in the soluble fraction (Figure 6A), which exhibited log fold changes that were remarkably correlated with those from the acute (R2 = 0.5) and chronic (R2 = 0.4) transcriptome experiments (Figures S4D and S4E). Of particular interest, we found that the two most prominent gel-forming mucins in the human airway, MUC5AC and MUC5B, were strongly upregulated (1.68-fold) and downregulated (3.85-fold), respectively, leading to a strong increase in their average ratio (5.02-fold) (Figure 6B). Additionally, we found that MUC2, thought to be primarily a gut gel-forming mucin, was induced in four of the IL-13 cultures, while not present in any control cultures. At the same time, transmembrane mucins, MUC4 (2.36-fold) and MUC16 (10.22-fold) were downregulated (Figure 6C). These mucins can be liberated from the airway surface by cleavage and have previously been observed in human airway mucus. Relatedly, we observed strong upregulation of mucin domain-containing protein, FCGBP, and mucin crosslinkers, ITLN1 and ITLN2, which were absent in all controls. Upregulated proteins were also enriched for salivary secretion and ER stress response, consistent with an enhanced ER load due to both quantitative and qualitative changes in mucin secretory activity under IL-13.
Figure 6. The IL-13-Stimulated Epithelium Downregulates Secretion of Innate Defensive Proteins and Activates Secretion of Mucus-Related Proteins, while Decelerating Ciliary Beat Frequency and Mucociliary Transport.
(A) Heatmap of normalized spectral counts for soluble apical secretome proteins significantly up- or downregulated with IL-13; n = 14 paired HBEC cultures.
(B) Boxplots compare soluble apical secretome protein abundance between control and IL-13-stimulated samples for proteins or enriched terms upregulated with IL-13. FDRs are based on DE analysis; p values are based on one-sided Wilcoxon tests.
(C) Same as (B) but based on proteins downregulated with IL-13.
(D) Density plots and boxplots (insets) show distributional shift in CBF with chronic IL-13 on the basis of top-down imaging before (left) and after (right) apical washing. Specified shifts and p values from a linear mixed model; outliers excluded from boxplots; n = 6 paired HBEC cultures.
(E) Same as (D) but based on profile imaging; n = 2 paired HTEC cultures.
In contrast, IL-13 downregulated many of the most prominent and highly secreted defense proteins in the airway epithelium, including SCGB1A1 (2.31-fold), BPIFA1 (23.15-fold), and LTF (27.55-fold). Furthermore, enriched terms for downregulated proteins encompassed those related to detoxification, the innate immune system, and antimicrobial humoral response, revealing a broad loss in normal defensive protein secretion within the IL-13-stimulated airway.
IL-13-Modified Mucus Slows Ciliary Beat Frequency and Arrests Mucociliary Transport
We expected that dramatic changes in both the amount and type of mucins, mucus structural proteins, and other products secreted under IL-13 would modify properties of mucociliary movement in these cultures. Video examination of mucociliary movement through fluorescent bead tracking revealed its near complete loss in cultures stimulated chronically with IL-13 (Video S1). This loss could result from dysfunction or reduction of cilia (Laoukili et al., 2001) and/or to changes in mucus properties, the latter of which was suggested by our proteomic results. To investigate this, we used differential dynamic microscopy (DDM) and particle image velocimetry (PIV) to measure ciliary beat frequency (CBF) from top-down and profile imaging, respectively, of mock and IL-13-stimulated cultures, before and after application of apical washes. These disulfide bond-reducing washes were meant to both hydrate and remove mucus and break down its structure, increasing fluidity. High-speed profile videos of ciliary beating show that although ciliated cells remained able to beat with IL-13 stimulation, this beating was not productive in generating movement of the mucus layer in unwashed cultures (Video S2). Moreover, analysis of top-down CBF data revealed a significant decrease in mean CBF (2.15 Hz) with IL-13 in unwashed cultures, while washing resulted in partial restoration in CBF (1.08 Hz recovery; Figure 6D; Table S1) and mucociliary movement (Video S2). CBF measures based on profile imaging validated these results, showing an even more pronounced reduction in CBF with IL-13 in the unwashed cultures (4.68 Hz) and an almost complete restoration to baseline CBF after washing (Figure 6E). These results illustrate how deceleration of ciliary beating and arrest of mucociliary motion with chronic IL-13 stimulation are strongly related to modified physical properties of mucus generated by these cultures.
The Airway Epithelium of T2H Children Exhibits Expression Changes Indicative of an IL-13-Remodeled Epithelium with a Modified Mucus Secretome
Finally, we sought to determine the extent to which IL-13-induced changes to cell composition, gene expression, and protein secretome in AEC cultures accurately modeled characteristics of the airway epithelium in T2H subjects. To do this, we generated whole-transcriptome data from nasal AEC brushings of 695 children (441 asthmatics and 254 healthy controls) belonging to the Genes-Environments & Admixture in Latino Asthmatics (GALA II) cohort. We performed co-expression-based gene network analysis to identify a network of T2 inflammation to be used in the classification of subjects into T2H and T2-low (T2L) groups. This analysis yielded a network containing 47 genes highly enriched in well-established epithelial markers of T2 airway inflammation (e.g., POSTN, CLCA1, IL1RL1, DPP4). Moreover, expression of this network exhibited a highly significant correlation with levels of both blood eosinophil (Pearson’s r = 0.56, p = 4.4 × 10−59) and IgE (Pearson’s r = 0.54, p = 1.3 × 10−48), two strong biomarkers of T2 inflammation. Hierarchical clustering of subjects on the basis of expression of these T2 network genes segregated them into two groups exhibiting high (T2H) or low (T2L) expression of these genes, respectively (Figure 7A). We found remarkable concordance between in vitro IL-13 responses of the airway epithelium and the genes defining the in vivo T2H airway epithelium (Figures 7A and S5). For example, the same endopeptidase inhibitors, pro-inflammatory genes, and genes related to mucus production and secretion induced by IL-13 were upregulated in T2H subjects, whereas, the same IL-13 downregulated genes, including those related to innate immunity and ciliary function, were also strongly reduced in T2H subjects (Figure 7A).
Figure 7. IL-13-Activated Expression and Secretion Responses of the Airway Epithelium Are Mirrored by Changes in the Airway Epithelium of T2H Children.
(A) Heatmap shows functional gene groups responding to IL-13 in vitro are also differentially regulated between T2L and T2H children in vivo; n = 695. Genes and functional groups are the same as in Figures 3C, 4B, S4D, and S4E. Samples are sorted from left to right by increasing T2 inflammation signatures.
(B) Boxplots show how average expression of genes coding for proteins up- or downregulated by IL-13 in the soluble apical secretome shows similar trends in T2H individuals.
(C) Boxplots compare expression of key AEC genes between T2L and T2H donors.
(D) Schematic summarizing metaplastic and dysregulatory transformation of the T2L human airway epithelium (top) under T2 inflammation (bottom), on the basis of both our in vitro IL-13 model and transcriptomes of a large in vivo cohort.
See also Figure S5.
Importantly, this concordance extended to the response of genes in the epithelial secretome. We found mean expression of genes encoding IL-13 up- and downregulated secreted proteins to be highly upregulated (p = 2.6 × 10−109) and downregulated (p = 2.4 × 10−14), respectively, among T2H subjects (Figure 7B). Among these, the gel-forming mucins MUC5AC and MUC5B exhibited a 1.61-fold (p = 1.9 × 10−32) increase and 2.42-fold (p = 3.0 × 10−17) decrease, respectively, in T2H subjects, resulting in a 4.03-fold average shift in the MUC5AC/MUC5B ratio (p = 1.4 × 10−28)(Figure 7C). The rarer gel-forming airway mucin MUC2 was similarly upregulated in T2H subjects (2.11-fold, p = 4.9 × 10−33). In contrast, markers of healthy airway function, such as FOXJ1, a master regulator of ciliogenesis, and the secretory defense marker, SCGB1A1, were strongly downregulated in T2H subjects (Figure 7C). Taken together, these results demonstrate that the cellular remodeling and changes to both the epithelial secretome and properties of mucus identified by our IL-13 model faithfully capture transformation of the in vivo airway epithelium of T2H children (Figure 7D).
DISCUSSION
Leveraging the resolution and scope of scRNA-seq, we have presented a more nuanced view into the effects of IL-13 on the airway epithelium. We find that rather than simply increasing secretion of mucus or MUC5AC protein in mucus secretory cells, IL-13 transcriptionally and functionally transforms the very nature of all cell types in the epithelium. This metaplastic epithelium, characterized by a distinct cellular composition and transcriptional programming, produces pathologic secretions that contain new mucus constituents while being bereft of homeostatic innate defense proteins, results that are consistent with the fact that this mechanism evolved to combat complex and potentially fatal parasitic infections. Translating this work to the in vivo human airway epithelium, we find strong evidence that these mechanisms operate in the airway of children with T2H asthma. Our results not only provide insight into T2H asthma pathobiology, which can be harnessed for development of targeted disease therapies, but also illustrate the remarkable plasticity of the airway epithelium in response to stimulus.
By identifying cell types among human AECs and their responses to IL-13 with single-cell resolution, we have shown that T2-induced metaplastic events that occur in mice (Chen et al., 2009; Evans et al., 2004) have an analog in humans. However, in contrast to one SCGB1A1+ cell population reported in mice, we observed two distinct secretory populations expressing SCGB1A1 at baseline, one characteristic of a club defense cell and the other more focused on mucus production. We show that IL-13 initiated a metaplastic process in both cell populations, culminating in a new type of MUC5AC+ mucus secretory cell. These IL-13 mucus secretory cells exhibited extensive reprogramming, which includes activation of a battery of new glycosylation genes, solute carriers, and mucin-interacting proteins. Importantly, this transcriptional divergence generated a mucus layer that arrested mucociliary motion and slowed CBF in a mucus-dependent fashion. Although prior work has suggested that increases in MUC5AC protein and the MUC5AC-mediated tethering of mucus are the source of depressed mucociliary movement driven by IL-13 (Bonser and Erle, 2017), we observed 39 IL-13-induced secreted proteins that might also play a role. Indeed, proteins exclusive to the IL-13 condition included the gut gel-forming mucin (MUC2), the mucin-interacting FCGBP, and lectin proteins that may mediate mucin crosslinking (ITLN1 and ITLN2). These and other mucus-related proteins, genes, and pathways that underlie production and secretion of IL-13-modified mucus by metaplastic secretory cells may present excellent targets for the specific disruption of these cells and their accompanying mucus.
Although there is general consensus that a ciliated cell cannot transdifferentiate into a mucus secretory cell, two studies have reported that ciliated cells express MUC5AC with IL-13 (Turner et al., 2011; Tyner et al., 2006). Investigating this at a single-cell transcriptome level, we find that much of the IL-13-induced mucus secretory expression program is remarkably acquired by ciliated cells, including MUC5AC. Moreover, although we failed to observe MUC5AC protein in ciliated cells, their strong ER stress response under IL-13 suggests to us a hypothesis that the particularly strong interferon response in ciliated cells induced by chronic IL-13 results from the stress involved in these cells trying to generate functional mucin proteins that they are not fully equipped to manufacture. This interferon response adds to the T2 inflammatory environment and may eventually lead to ciliated cell death, as supported by a decrease in ciliated cell frequency with IL-13. Moreover, surviving ciliated cells downregulate ciliogenesis genes, likely compromising function, and we confirmed that this ciliated cell dysfunction occurs in the airway of T2H compared with T2L children, on the basis of transcriptional shifts. The interferon response to chronic IL-13 is particularly interesting given previous studies finding a suppression of interferons and interferon-stimulated genes when the T2 inflamed airway epithelium is subjected to virus, which is thought to enhance susceptibility to virus infections in those with asthma or COPD (Chen et al., 2014; Contoli et al., 2015; Korfhagen et al., 2012; Wark et al., 2005). It is possible that under chronic T2 inflammation, antiviral responses that are initially downregulated by T2 pathways, as a way to limit type 1 inflammation, may be reinstated if cells become sufficiently stressed because of extended mucin protein burden.
We also found an underappreciated pan-epithelial loss of innate airway defenses due to IL-13, affecting all cell types and including many secreted antimicrobial and detoxification proteins and genes. Although previous research has shown that IL-13 can limit some pathways of innate defense in the human AECs (Beisswenger et al., 2006; Chen et al., 2014;Wu et al., 2012), our study defines the transcriptome-wide genes involved and demonstrates the pervasiveness of this loss of function across all cell types. Moreover, we confirmed that MUC5B, critical for antibacterial defenses in mice (Livraghi-Butrico et al., 2017; Roy et al., 2014), was transcriptionally downregulated in T2H children, along with an assortment of other innate defense-related genes. Furthermore, we show that not only did IL-13 reduce defense expression, but also the frequency of defense-focused secretory cells in the airway epithelium, when applied chronically. Taken together, this reduction in innate airway defense and loss of ciliary function, when coupled with an airway awash with adhesive MUC5AC+ mucus, provides a mechanistic explanation for the increased vulnerability of allergic asthmatics to respiratory infections that require antibiotics and exacerbate disease (Juhn, 2014; Woehlk et al., 2018).
Although our scRNA-seq analysis has enabled us to study the cell type-specific nature of IL-13-induced metaplasia in the airway epithelium, additional sampling of both more subjects and more cells would enable us to extend this analysis further into cell types that we were unable to detect with our sampling, including different types of basal cells as well as rare epithelial cell types that are particularly relevant to disease. Furthermore, in vivo scRNA-seq data, such as from subjects in the GALA II asthma cohort, would be an incredibly powerful tool for validating that the cell type-specific transformations we observe in vitro translate into cell type-specific effects of T2 inflammation.
T2 cytokine inhibitors have recently become available, enjoying some success in reducing exacerbation rates (Bousquet et al., 2005; Castro et al., 2015; Nair et al., 2017), but their limited ability to improve lung function (Liu et al., 2013; Noonan et al., 2013; Russell et al., 2018) suggests either that not all core T2 inflammatory pathways are blocked by these drugs or that the T2 airway epithelium can be locked into a metaplastic state, despite blockade of the T2 stimulus. Thus, treating T2 airway disease may ultimately require us to "reverse-remodel" the airway epithelium. As such, improved therapeutics will likely target epithelial pathways controlling production and release of T2 cytokine-inducing alarmins such as TSLP, IL-33, and IL-25, while reversing the metaplastic events underlying mucus obstruction and loss of immunity inherent to disease. Epithelial responses to other environmental toxicants and pathogens that underlie airway disease, such as cigarette smoke, pollution, and viruses, may similarly involve widespread remodeling of the airway epithelium. Elucidation of how the epithelium is remodeled in asthma and other chronic obstruction lung diseases may hold the key to development of new therapies that provide lasting improvements for these patients.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Max A. Seibold, Ph.D. (seiboldm@njhealth.org).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Raw and processed RNA-seq data used in the study are available at the National Center for Biotechnology Information/Gene Expression Omnibus (GEO) under repository accession number GEO: GSE145013 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE145013] for the scRNA-seq data and under GEO: GSE152004 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152004] for bulk RNA-seq data from GALA II. Proteomics and CBF data used to generate Figures 6 and S4 are available in Table S6. Code used to carry out data analysis is available on GitHub [https://github.com/seiboldlab/SingleCell_IL13].
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human tracheal samples
For single cell RNA sequencing, human tracheal epithelial cells (HTEC) were isolated from two de-identified donors (T71 and T72) whose lungs were not suitable for transplantation (see Table S2). HTEC samples were isolated from several additional donors for use in cell imaging (T73, T76, and T79 were used for imaging in Figures 3A, 5D, and S2B and T147 and T148 were used in Figure S2D). All lung specimens were obtained from the International Institute for the Advancement of Medicine (Edison, NJ) and the Donor Alliance of Colorado. The National Jewish Health Institutional Review Board (IRB) approved the research under IRB protocols HS-3209 and HS-2240.
Human bronchial samples
Human Bronchial Epithelial Cells (HBEC) were acquired from the National Jewish Live Cell Core/Mucosal Immunity Program (Denver, CO) for use in proteomic and functional assays. The National Jewish Health Institutional Review Board (IRB) approved the research under IRB protocol HS-2240. Basal cells were obtained from 14 age- and sex-matched donors (4 control and 10 asthmatic) with no history of smoking (see Table S3).
GALA II nasal airway epithelial samples
Nasal AECs used in this study came from subjects recruited as part of the Genes-environments & Admixture in Latino Americans II (GALA II) childhood asthma cohort, which was approved by local institutional review boards (University of California, San Francisco, IRB number 10–00889, reference number 153543; National Jewish Health, IRB number HS-2627). All subjects and their parents provided written informed assent and consent, respectively (Neophytou et al., 2016; Nishimura et al., 2013). Demographic and clinical variables for GALA II tissue donors used in this study are listed in Table S4.
Whole transcriptome RNA-seq data were collected from 254 control subjects and 441 asthma subjects, where asthmatics were defined as those having received an asthma diagnosis by a physician and having exhibited two or more symptoms within the two years prior to enrollment. See Borrell et al. (2013) and Kumar et al. (2013) for more in depth descriptions of the study.
Air-liquid interface in vitro model
To model the transcriptional, proteomic, and functional responses to T2 inflammation in the human airway epithelium, we used cell cultures established and grown at air-liquid interface (ALI) (Everman et al., 2018). These ALI cultures were initially seeded with primary basal cells from tracheal or bronchial epithelial samples and then allowed to differentiate into functional, mucociliary airway epithelia consisting of a mix of basal, secretory, and ciliated cells that respond to environmental insults in ways expected to be similar to their responses in vivo (de Jong et al., 1994; Kesimer et al., 2009; Pezzulo et al., 2011). We stimulated T2 inflammation using interleukin-13 (IL-13), which is known to recapitulate many of the features of T2H asthma, both in vivo (Chen et al., 2009; Tyner et al., 2006; Zhen et al., 2007) and in vitro (Gerovac and Fregien, 2016; Nicodemus-Johnson et al., 2016; Zhao et al., 2009).
METHOD DETAILS
ALI culture differentiation and IL-13 stimulation - HTEC stimulation experiments for scRNA-seq
Primary basal HTECs from the two donors were isolated and expanded for 2 passages on NIH 3T3 fibroblast feeders using F-medium (67.5% DMEM-F, 25% Ham’s F-12, 7.5% FBS, 1.5 mM L-glutamine, 25 ng/mL hydrocortisone, 12 5 ng/mL EGF, 8.6 ng/mL cholera toxin, 24 μg/mL Adenine, 0.1% insulin, 75 U/mL pen/strep) with ROCK1 Inhibitor (RI, 10 μM) and antibiotics (1.25 μg/mL amphotericin B, 2 μg/mL fluconazole, 50 μg/mL gentamicin). Passage 2 cell suspensions were harvested by trypsinization, treated with DNase (12.5 μg/ml) in HBSS for 5min at 37°C, and seeded onto bovine collagen-coated 6.5 mm transwell inserts (2×104 cells/insert for acute stimulation, 1×104 cells/insert for chronic stimulation) in ALI expansion medium (50% BEBM, 50% DMEM-C, 0.5 mg/mL BSA, 80 μM ethanolamine, 10 ng/mL hEGF, 0.4 μM MgSO4, 0.3 μM MgCl2, 1 μM CaCl2, 30 ng/mL retinoic acid, 0.8X insulin*, 0.5X transferrin*, 1X hydrocortisone*, 1X epinephrine*, 1X bovine pituitary extract*, 1X gentamicin/amphotericin* (*relative to BEGM Bullet Kit aliquot) with RI. RI was removed after 24 hours and confluent monolayers were airlifted 6 (acute stimulation) or 7 (chronic stimulation) days post seed and differentiated using complete PneumaCult ALI media made as per manufacturer’s instructions. Starting at day 21 (acute stimulation) or day 11 (chronic stimulation) post-airlift, paired cultures for each donor were stimulated with IL-13 or BSA for 48 hours (acute stimulation) or 11 days (chronic stimulation), after which cells were harvested on day 23 (acute stimulation) or day 22 (chronic stimulation).
ALI culture differentiation and IL-13 stimulation - HBEC stimulation experiments for proteomic and functional assays
Primary basal HBECs were isolated and expanded for 2 passages on NIH 3T3 fibroblast feeders using F-medium with ROCK1 Inhibitor (RI, 10 μM). Cell suspensions were harvested by trypsinization, treated with DNase (12.5 mg/ml) in HBSS for 5 minutes at 37°C, and seeded onto 6.5 μm, 0.4 μm pore transmembrane inserts pre-coated with PureCol bovine collagen (2×104 cells/insert) in ALI expansion medium with RI, and RI was removed after 24 hours. Upon confluence, air-liquid interface was established and cultures were allowed to differentiate using PneumaCult ALI (PC-ALI) differentiation media. At day 11 post-airlift, paired cultures for each donor were stimulated with IL-13 (10 ng/ml) or BSA daily for 10 days until day 21. For functional mucociliary assays, cultures were used for cilia beat frequency analysis as described below. For apical proteomic analysis, following 10 day IL-13 stimulation, each insert was washed three times with warm PBS using a new plate well to remove any contaminating media proteins carried over from the cultures and plasticware. Inserts were then stimulated either with IL-13 or BSA alone in base PneumaCult ALI media in the absence of suggested additives and media components. After 24 hours, the apical chambers of each well were washed two times with warm PBS, collected, and pooled. Soluble and insoluble fractions were isolated and processed separately, as detailed below.
Harvesting of cells for scRNA-seq
Replicate mature stimulated ALI cultures were washed with warm PBS/DTT (10 mM) for 5 minutes at 37°C, followed by a warm PBS wash of both chambers. Cultures were then harvested from the filter to single cell suspensions using apical dissociation solution (Accutase with 5 mM EDTA and 5 mM EGTA) for 30 minutes at 37°C with occasional manual agitation (Everman et al., 2018). Single cell suspensions were diluted, centrifuged, and washed once with PBS/DTT and twice with PBS before sorting for live singlets. Cells from both tracheal donors and treatments were dispensed into each of two 5,184-nanowell version 1 microchips (91497 and 94576) (for the acute IL-13 experiment) or into a single version 2 microchip (97570) (for the chronic IL-13 experiment) using the ICELL8 platform (Takara Bio USA, Inc.), after which non-doublet cells were selected for library preparation and sequencing.
Single cell RNA isolation and sequencing
Library preparation was carried out using ICELL8 single-cell poly-(A)+ transcriptome amplification hands-on workflow (with Triton X-100 for cell lysis), protocol D07–000040-003 rev4. For the acute IL-13 experiment, libraries were generated for a total of 2,385 selected cells from the two tracheal donors (plus positive and negative controls), after which they were sequenced as 100 base pair single-end reads on an Illumina HiSeq 2500 (Illumina, Inc., San Diego, CA), with cells from each of the two microchips sequenced across three flow cell lanes. For the chronic IL-13 experiment, the same approach was used, except that 802 single cells were selected for sequencing and then sequenced across three flow cell lanes. See Table S5 for a summary of sequencing statistics for cells stratified by dataset, treatment, and donor.
Immunofluorescence microscopy
Intact differentiated ALI culture inserts were fixed for 15 minutes at room temperature in 3.2% paraformaldehyde. For histological sections, fixed ALIs were embedded in paraffin and sectioned onto microscope slides. Sections were stripped of paraffin using HistoChoice Clearing Agent and rehydrated using a decreasing gradient of alcohol washes (100%, 90% 70%, 50%, 30%, 0%), followed by antigen retrieval conducted using Citric Acid-based Antigen Unmasking Solution (pH 6.0). Histologic sections were blocked and permeabilized (3% BSA, 0.4% Triton X-100, 1X TBS) for 30 minutes at room temperature, and immunolabeled with primary antibodies (3% BSA, 0.1% Triton X-100, 1X TBS) overnight at 4°C with dilutions as follows: SCGB1A1 (1:1500), MUC5AC (1:500). Whole-mount samples were blocked and permeabilized for 30 minutes using 3% BSA/0.1% Triton X-100 in tris-buffered saline (TBS) followed by primary labeling using 3% BSA/0.1% Triton X-100 in TBS for 1 hour with antibodies against MUC5AC (1:500), SCGB1A1 (1:500). All samples were washed twice using TBS/0.1% Triton X-100 (TBST) and secondary labeling was conducted using DAPI (1:1000), and either donkey anti-rabbit IgG or anti-mouse IgG (1:500) conjugated to AlexaFluor488 or AlexaFluor 594 dyes for 30 minutes. Slides were washed twice and mounted with Vectashield HardSet Antifade Mounting Media. Images were acquired using an Echo Revolve R4 microscope.
Fluorescence in situ hybridization (RNAScope)
ALI cultures rinsed with PBS were fixed in 10% neutral buffered formalin for 24+/− 8 hours at room temperature, washed with 1X PBS, paraffin-embedded, and sectioned onto SuperFrost Plus slides. Sections were dried overnight, baked for 1 hour at 60°C, and stored for 1 week before proceeding with the RNAScope Multiplex Fluorescent v2 assay according to the manufacturer’s instructions with the following modifications: target retrieval was performed for 15 minutes in a boiling beaker, Protease III was used for 30 minutes pre-treatment at 40°C, and hybridized slides were left overnight in 5X SSC before proceeding with the amplification steps. Opal fluors were applied at 1:1500 dilution and before mounting. Immunofluorescence labeling (E-cadherin: 1:2500; FOXJ1: 1:500) was performed as outlined above, except with 1 hour of primary application at room temperature. Slides were imaged on an Echo Revolve R4 microscope (Discover Echo, Inc) or a Zeiss LSM700 confocal microscope.
Proteomic sample preparation
Apical washes from paired BSA and IL-13-treated ALI cultures were collected in Fisher 2 mL low binding microfuge tubes and then samples were centrifuged at 225 × g at 4°C to remove intact cells from the washes. Supernatants were collected from each sample and the visible insoluble (i.e., mucus) fraction was removed from each wash, dried in a SpeedVac at 45 C, and stored at −80°C. The remaining soluble apical fraction was mixed with 3x volume of ice cold LC/MS grade methanol followed by incubation at −80°C for 1 hour. Proteins were then pelleted by centrifugation at 18,000 × g and the supernatant was removed. Protein pellets were dried in a SpeedVac at 45°C and frozen at −80°C. For trypsin digestion, protein pellet pairs were denatured and reduced using 50% trifluoroethanol in 50 mM ammonium bicarbonate with 0.1 mM DTT at 65°C for 45 minutes. Proteins were then alkylated for 1 hour in the dark using 0.32 mM iodoacetamide, and excess IAA was then reduced using 0.08 mM DTT for 1 hour in the dark. Samples were diluted with 400 μL of 25 mM ammonium bicarbonate buffer and digested with 0.3 μg of trypsin for 5 hours at 37°C. Digests were then stopped using 2 μL of formic acid and duplicate samples were combined and dried at 45°C in a SpeedVac concentrator. Dried samples were stored at 70°C until mass spectrometry analysis.
Mass spectrometry
Mass spectrometry analysis was carried out for the soluble fraction from all 14 donors and for the mucus fraction from a subset of six donors. Samples were first resuspended with 10 μL of 3% Acetonitrile + 0.1% formic acid and the total sample was analyzed using a nanoAdvanced UPLC (Bruker) with a 15 cm × 100 μm ProntoSil C18AQ column and 2 cm trap column (nanoLCMS Solutions). Mobile phase was H2O + 0.1% FA (A) and acetonitrile + 0.1% FA (B), peptides were separated using a gradient of 2%–40% B over 30 minutes 2 at a flow rate of 800 nL/minute and with the column temperature kept constant at 40°C. The column was connected through a Captive Spray nano source to an Impact Q-tof (Bruker). Data were collected over a mass range of 150–2200 m/z using a 1Hz MS scan and a total duty cycle time of 2 s, a setting that allows the Impact to adjust the acquisition time for each fragment scan based on precursor intensity while still collecting data on as many precursors as possible without exceeding the 2 s duty cycle. Data were processed using DataAnalysis 4.2 (Bruker), compounds were searched against the SwissProt database using Mascot 2.4 (Matrix Science) with the percolator algorithm, and protein and peptide results were assessed and filtered with ProteinScape 3.0 (Bruker).
Top-down imaging and measurement of ciliary beat frequency
Using six paired HBEC ALI cultures chronically stimulated with IL-13 or BSA as described above, we measured ciliary beat frequency (CBF) based on top-down imaging in order to assess functional response to IL-13 in these epithelia. ALI cultures were first washed once in 37°C PBS for 10 minutes, after which the wash was removed and cultures were imaged by placing the plastic insert in a glass-bottom Petri dish with 1 mL of PC-ALI medium.
Digital high-speed videos were recorded on 5 different fields of view across the sample, (FOV = 1920 × 1024 px, 1 px = 0.146 μm), under bright-field illumination and at a sampling frequency of 150 fps using a Nikon Eclipse Ti-E inverted microscope (Nikon Instruments, Japan) with a 40x objective (Plan Apo λ 40 ×, N.A. 0.95, Nikon) fitted with a Grasshopper 3 GS3-U3–23S6M-C CMOS camera (FLIR Integrated Imaging Solutions GmbH, Germany). Samples of epithelial cells were imaged in a custom-made chamber, where temperature, CO2 and humidity were continuously monitored and maintained at values of 37°C, 5%, and 90%, respectively, by using custom software running on a Linux machine. Imaging was repeated as described above for these same inserts after they were subjected to a PBS + 10 mM DTT wash (10 minutes at 37°C) aimed at hydrating and denaturing the mucus layer. CBF was quantified using Differential Dynamic Microscopy, as previously described (Chioccioli et al., 2019a; Feriani et al., 2017). Briefly, each field of view was first divided into tiles of 64 × 64 px (~9 mm per side). The distribution of CBF in the field of view was then measured by running DDM on the tiles that showed motion and fitting its output with an empirical oscillating function (the Image Structure Function). The CBF distributions for each treatment were then built by pooling the values of CBF (in units of Hz) measured on the "active" tiles of all the fields of view imaged across the six sample. Total data points per treatment were as follows: control, unwashed = 11,782; IL-13, unwashed = 7,896; control, washed = 10,511; and IL-13 washed = 7,444.
Profile imaging of ciliary beating pattern
In order to capture a detailed ciliary beating pattern of the single cilium under IL-13 and control conditions, we carried out a similar experiment as described above using two paired HTEC donor ALI cultures chronically stimulated with IL-13 or BSA and then processed as previously described (Chioccioli et al., 2019b). Briefly, to perform high speed video microscopy on the cilia, the bottom membrane supporting the epithelium was excised from the plastic ALI insert. The membrane was then folded in half and placed between two coverslips, together with a few μl of medium. The profile videos were then obtained by imaging the edge of the folded membrane. High speed videos were acquired at room temperature on an Eclipse Ti-E inverted microscope (Nikon, Japan) using a 60X water immersion objective (NA = 1.2, Nikon, Japan) and an additional 1.5X magnification in the optical path, and a CMOS camera (GS3-U3–23S6M-C, FLIR Integrated Imaging Solutions GmbH, Germany). The image acquisition was run by a custom software running on a Linux machine. About 10 videos were acquired per insert, at four different locations along the membrane’s edge. Videos were at least 10 s long, recorded with a framerate in excess of 200 fps (1px = 0.065 μm).
To estimate CBF, videos were first analyzed using Particle Image Velocimetry implemented in PIVlab (Thielicke and Stamhuis, 2014), which yields a velocity field for each couple of subsequent frames. Using a custom MATLAB script, we then assessed at how this velocity field behaves as a function of the height from the cell surface (i.e., along the cilium). Specifically, we manually identified the surface of the ciliated cells, after which the script automatically found four measuring lines at distance 2.5 μm, 3.5 μm, 4.5 μm and 5.5 μm from the cell surface. The CBF was then obtained at each of the measuring lines by taking the Fourier transform of the lag time dependence of the autocorrelation of the kymograph of the component of the velocity parallel to the cell surface. Specifically, we first took the velocity measured at each frame by PIV along each measuring line and, at each point along it, we defined a frame of reference with two axes respectively parallel and perpendicular to the measuring line (which is in turn parallel to the cell surface). By projecting the velocity on the parallel axis, we obtained the component of the velocity that is parallel to the cell surface. By doing so we measured the velocity parallel to the cell surface at each point along the measuring line. We then build a kymograph of this quantity by repeating the procedure above on each of the frames analyzed by PIV. We then took the autocorrelation of the kymograph over time and space, and Fourier transform its behavior with the time lag. The frequency that corresponds to the peak in the Fourier spectrum is the average CBF along the measuring line.
Mucociliary movement analysis using particle tracking
Using the chronic stimulation model, ALI cultures were treated with IL-13 or BSA prior to particle tracking of cultures (n = 6). Inserts were placed onto a plastic Petri dish, and Cy5-labeled 5 mm beads were added to the apical chamber of each culture. For each sample, 10sec videos were captured on 4x magnification over 4 fields of view per culture. Videos were processed and particle tracking analysis was conducted using the Particle Tracker plug-in by ImageJ.
RNA extraction, library preparation, and sequencing for GALA II nasal epithelial brushings
Primary nasal AEC brushes obtained from GALA II cohort subjects were placed into RLT Plus lysis buffer for RNA and DNA extraction using the AllPrep DNA/RNA MiniPrep Kit (QIAGEN). RNA normalization, library construction using KAPA Stranded mRNA-Seq Kit with KAPA mRNA Capture Beads (KAPA Biosystems), and library pooling for a total of 695 subjects were all performed on the Beckman Coulter FXP automation system. Briefly, RNA samples were randomized over the normalization plate in alternating control and asthmatic conditions at 250 ng per well. Illumina Dual-Index adapters (Integrated DNA Technologies) were used to barcode libraries using 12 cycles of amplification. Samples were pooled using equal concentrations of six libraries/pool and pair-end sequenced with Illumina HiSeq 2500 System sequencing.
QUANTIFICATION AND STATISTICAL ANALYSIS
Pre-processing of scRNA-seq data
We trimmed and culled raw demultiplexed cDNA reads in FASTQ files using Cutadapt (Martin, 2011), trimming poly A tails and 5' and 3' ends with q < 20 and removing any reads shorter than 25 base pairs. We then aligned trimmed reads to the hg38 human genome with GSNAP (Wu and Nacu, 2010), setting "max-mismatches=0.05" and accounting for both known Ensembl splice sites and SNPs. We quantified gene expression using HTSeq (Anders et al., 2015) with "stranded=yes," "mode=intersection-nonempty," and "t=gene" and then summed the number of unique molecular identifiers (UMIs) for each gene across runs for each cell to obtain a UMI count matrix that we used for all downstream analysis.
Although samples comprising multiple cells were removed during the cell selection stage, to safeguard against undetected doublets, we removed 34 outlier cells expressing > 46,000 UMIs (chip 91497 cells) or > 28,500 UMIs (chip 94576 cells) from the acute stimulation dataset and 10 outlier cells expressing > 60,000 UMIs from the chronic stimulation dataset. For the acute dataset, we also removed 11 cells for which the percentage of reads that mapped to genes was < 50%. Prior to downstream analysis, we removed select mitochondrial and ribosomal genes (those beginning with MTAT, MT-, MTCO, MTCY, MTERF, MTND, MTRF, MTRN, MRPL, MRPS, RPL, RPS) and very lowly expressed genes (those genes not expressed in at least 0.5% of cells). Finally, we removed 446 cells expressing fewer than 2,500 UMIs from the acute dataset and three cells expressing fewer than 2,000 UMIs from the chronic dataset. The final QCed acute dataset contained 1,894 cells with a mean of 8,736 UMIs and 3,159 genes per cell and the final QCed chronic dataset contained 789 cells with a mean of 12,970 UMIs and 4,580 genes per cell.
Pre-processing of bulk RNA-seq data
Sequencing reads were trimmed with Skewer (Jiang et al., 2014) (—quality 15,—mean-quality 25,—min 30) and then aligned to human reference genome GRCh38 with GSNAP using splice site and SNP databases from iGenomes. Transcript quantification of these aligned reads on iGenomes GTF file were conducted with HTSeq ("stranded=no," "mode=intersection-nonempty," and "t=exon").
Clustering and visualization of scRNA-seq data
For the acute simulation dataset, we accounted for differences in coverage across cells by dividing each cell’s UMI count by the sum of all the cell’s UMIs, multiplying by 10,000, and then taking the natural log. For input into dimensionality reduction and clustering analyses and for plotting relative expression in heatmaps and dot plots, we also fitted normalized expression of each gene to a set of predictors that included the sum of the UMI counts per cell and the identity of the originating donor and chip, and then mean centered (subtracted from each gene count the average expression of that gene) and scaled (divided each centered gene count by that gene’s standard deviation) the residuals. We then used the top eight PC dimensions based on the top 8,070 most informative genes to visualize and cluster cells, where “informativeness” was defined by gene dispersion (i.e., the log of the ratio of expression variance to its mean) across cells, calculated after accounting for its relationship with mean expression. We visualized these top components of variation in two dimensions using the Barnes-Hut implementation of t-distributed neighborhood embedding (tSNE; perplexity = 45). We also carried out unsupervised clustering by first constructing a shared nearest neighbor (SNN) graph based on k-nearest neighbors (k = 30) calculated from PC dimensions and then determined the number and composition of clusters using a modularity function optimizer based on the Louvain algorithm (resolution = 1.1). We used the Seurat R package (Satija et al., 2015) to carry out all data normalization and scaling as well as downstream dimensionality reduction, clustering, tSNE plot overlaying, and differential expression.
Due to batch-based differences in expression between the two donors in the chronic stimulation dataset, we reduced the dimensionality of variation in that dataset using canonical correlation analysis (CCA), which identifies the strongest components of gene correlation structure that are shared across datasets (Seurat’s RunMultiCCA function). After normalizing and scaling datasets for the two donors separately, we carried out CCA based on the 3,350 genes contained within the union of the top 2,000 most informative genes from each donor. Correlated expression across donors based on the top three CCA dimensions was then projected into a common subspace using Seurat’s AlignSubspace function, allowing us to identify and visualize universal populations of cells, rather than those driven by donor-specific variation. We removed 33 cells that were not well explained by these shared components (i.e., any gene for which variance explained by CCA was < 2-fold that explained by PCA). We then visualized these subspace-aligned CCA dimensions in two tSNE dimensions (perplexity = 30) and performed SNN clustering. Initial SNN clustering yielded six clusters (resolution = 0.6), four of which corresponded to secretory cell populations and two of which corresponded to ciliated cell populations (one ciliated cluster was dominated by IL-13 cells and the other was dominated by control cells). We identified further subgroups of ciliated cells that proved functionally distinct by carrying out dataset dimensionality reduction and SNN clustering on the two ciliated clusters alone. For this, we used the top three PC dimensions based on the top 1,278 most informative genes to cluster cells (resolution = 0.7), yielding four clusters, which we overlaid, along with the four secretory cell clusters, onto the tSNE plot (Figure 5B).
Plotting expression across cells
We overlaid characteristic expression of three major cell types in the airway epithelium (ciliated, secretory, and basal cells) onto tSNE plots by measuring mean expression across cell-type specific markers in each cell and then identifying cells as being characteristic of a given cell type if mean expression for that cell type was above the 70th percentile while falling below the 70th percentile for the other two cell types. For ciliated cell markers, we used 32 genes annotated to the Gene Ontology (GO) 2017 terms, "cilium movement" and "cilium-dependent cell motility." For secretory markers, we used 18 genes known to be highly expressed in airway epithelial secretory cells (B4GALT5, WFDC2, SLPI, MECOM, ALDH1A1, SERPINB3, SERPINB4, ADH7, PIGR, BPIFB1, SCGB1A1, KRT4, SLC16A9, SLC6A14, MUC5AC, MUC5B, CD55, TFF3). For the basal cell signature, we used 574 genes shown to be expressed at least 10-fold more in basal epithelial cells than in cells of the differentiated airway epithelium (Hackett et al., 2011).
For overlaying group expression in dot plots for panels of genes, we plotted normalized expression along a continuous color scale. All heatmaps show scaled normalized gene expression (or the average of expression across a function gene panel) along a continuous color scale across cells and were produced using Heatmap3 (Zhao et al., 2014). Break scales were set as indicated in the color legends.
For plots of expression across functional gene groups in heatmaps (see Figures 3B and 4A), violin plots (see Figure 4C), and boxplots (see Figures 5E, 5F, 6B, 6C, S4B, and S4C, mean expression across overlapping genes annotated for significantly enriched terms or pathways were used (see the "Functional enrichment analysis" section below for the libraries used), except in the cases outlined below.
The functional gene groups plotted in log fold change scatterplots in Figures 3C, 4B, and S4D contained the following genes: Ion transport ATPases: ATP1A1, ATP1B1, ATP13A5, ATP6V0E1, ATP7B, ATP12A; Solute carriers: SLC5A8, SLC9A9, SLC9B2, SLC12A2, SLC22A23, SLC31A1, SLC37A1, SLC38A6, SLC39A7, SLC39A8, SLCO3A1, SLCO4C1, SLC6A14; Endopeptidase inhibitors: CAPN14, CSTB, FETUB, SERPINB2, SERPINB6, SERPINB10, SPINK5, CSTA, TIMP1, SERPINB4, SERPINB3; MUC5AC/mucin-related: FCGBP, GSN, ITLN1, MUC5AC, SCIN; Glycosylation-related: ADAMTSL3, B3GNT6, B3GNT2, CHST6, GALK2, GALNT1, GCNT3, FUT8, GALNT3; Pro-inflammatory: ALOX15, ANXA3, CCL26, CD36, DDX58, LGALS7, NOS2, RIPK2, SH2D1B; Reductases: AKR1C2, AKR1C3, DHRS3, DHRS9, GLUD1, RDH10, STEAP1B, STEAP4; Innate immunity: ABCA13, CP, CYP2B7P, GLUL, GSTA1, GSTA2, LYN, SCGB1A1, SCGB3A1, WFDC2, CD74, CXCL1, CXCL17, IL18, S100A2, S100A6, S100A8, S100A9, LCN2; Cilia related ( Figure 4B only): DNAH5, ARMC4, EFHC1, CFAP46, DNAH9, CFAP54, DRC3, CEP126, PCM1, IFT81, DZIP1L, RFX3, CC2D2A, DYNC2H1, TTC21B, DYNLRB2, DNAH3, DNAH7, KIF21A.
The "IL-13 markers" used in boxplots in Figure 5E included the following genes for ciliated cells: PRB1, CST1, CST4, DPP4, SERPINB10, SERPINB13, GALNT7, FCGBP, SH2D1B, GSN; and the following genes for secretory cells: PRB1, MUC5AC, CST1, CST4, DPP4, POSTN, CDH26, SLC7A1, GALNT7, SH2D1B. The "Innate immunity genes" used in boxplots in Figure 5E included the following genes for ciliated cells: SCGB1A1, SCGB3A1, BPIFA1, S100A2, GSTA1, CYP2B7P; and the following genes for secretory cells: GSTA3, GSTA2, AKR1C1, GSTA1, MGST1, SCGB1A1, SCGB3A1, BPIFA1, BPIFB1, AKR1C1, AKR1C2, CXCL.
Differential gene expression analysis
Differential expression (DE) for each gene between various single cell groups specified in the text was tested using a non-parametric Wilcoxon rank sum test carried out with Seurat. Except where noted, we limited each comparison to genes exhibiting both an estimated log fold change > +0.25 and detectable expression in > 10% of cells in one of the two groups being compared. We corrected for multiple hypothesis testing by calculating false discovery rate (FDR)-adjusted p values using the Benjamini-Hochberg method. Genes were considered to be differentially expressed when FDR < 0.05.
In the acute stimulation scRNA-seq analysis, when investigating which control secretory cell types (c5 or c7) were most similar to which IL-13-secretory cell types (c6 or c8), our approach was to first establish which genes were upregulated in control cells of groups c5 and c7 relative to each other as well as in IL-13 cells from groups c6 and c8 relative to each other using DE analysis. This allowed us to obtain "cell type signature" for the two pairs of secretory populations within their respective treatment milieus. We then compared overlap of these defining genes between control and IL-13 populations to identify analogous populations that share gene signatures (see Figures S2C and S2D.
When defining cell states recovered in the chronic stimulation dataset in terms of those recovered in the acute stimulation dataset, we carried out DE analysis among pairs of clusters belonging to the same cell type (ciliated and secretory) and treatment (BSA and IL-13) and then compared differentially expressed gene (DEG) lists from these comparisons with those from parallel comparisons we’d carried out previously for the chronic stimulation dataset (see Figures S3B and S3C).
When investigating the chronic IL-13 response, we compared IL-13 cells within each of the IL-13 cell-dominated secretory clusters (mucus and defense secretory) to control cells within their respective control cell-dominated secretory cluster. We did the same for ciliated cells, but lumping together the ciliated subclusters belonging to the same treatment group. To construct the density plots in Figure 5C, we obtained IL-13 DEGs for secretory cells by lumping the defense and mucus-focused subgroups together for both acute and chronic stimulation datasets. We then subtracted acute log fold changes from chronic log fold changes for each overlapping gene for secretory and ciliated cells in turn.
To obtain genes more activated with chronic stimulation compared to acute stimulation, we repeated DE analyses for both acute and chronic datasets between control and IL-13-stimulated cells, stratified by cell membership to secretory (combined) and ciliated clusters. To allow us to evaluate more subtle patterns, for these DE tests we did not exclude genes with < 0.25 log fold change. Genes were considered "significantly" more responsive with chronic than acute stimulation if either 1) chronic FDR was < 0.05 while acute FDR was < 0.05 and chronic log fold change was at least 0.1 higher than acute log fold change or 2) chronic and acute FDRs were both < 0.05, but chronic log fold change was at least 0.25 higher than acute log fold change. It is from these lists of genes with a stronger response after chronic stimulation that enriched gene sets were obtained for Figures 5E and 5F.
Functional enrichment analysis
We tested for gene overrepresentation of all target lists within a panel of annotated gene databases (Gene Ontology [GO] Biological Process [BP] 2017, GO Molecular Function [MF] 2017, GO Cellular Component [CC] 2017, Kyoto Encyclopedia of Genes and Genomes [KEGG] 2016, and Reactome 2016) using hypergeometric tests implemented with Enrichr (Chen et al., 2013), as automated using the python script, EnrichrAPI (https://github.com/russell-stewart/enrichrAPI). We report only terms and pathways that were enriched with FDR < 0.05.
Lineage trajectories
We constructed lineage trajectories for both the defense and mucus-focused secretory cell populations, in turn, to better understand the transcriptional regulators that govern the transition from normal secretory cells to T2-induced cells upon stimulation with IL-13. To produce these trajectories, we used Monocle v2.8 (Trapnell et al., 2014) to first carry out dimensionality reduction using the DDRTree algorithm, and to then order cells along a trajectory of pseudotime (using Monocle’s orderCells function) based on their expression across the 500 most differentially expressed genes (sorted by FDR) between control and IL-13-dominated clusters (inferred using Monocle’s differentialGeneTest function). For the top informative genes from each of the two groups of cells, based on the combined mean and dispersion of expression (2,116 and 2,699 genes for defense and mucus-focused cells, respectively), we tested for differential expression as a function of pseudotime. Of the pseudotime-dependent genes (FDR < 0.05), there were 16 identified as transcription regulators by Ingenuity Pathway Analysis (IPA) that were significantly associated with pseudotime in both defense and mucus-secretory cell trajectories (Figure 3D). Because pseudotime scaling was the same for the two sets of cells, we overlaid relative expression, as well as smoothed expression curves, of exemplar regulators for the two trajectories across a common pseudotime (Figure 3D).
Differential protein abundance
For all proteins observed in at least two samples, we used edgeR (Robinson et al., 2010) to investigate differential abundance in response IL-13. To do this, raw spectral counts were normalized with scaling factors inferred using the weighted trimmed mean of M-values (TMM) method, and then for each protein, tagwise negative binomial dispersions were estimated using Cox-Reid profile-adjusted likelihoods, after which proteins fitted to a general linear model were tested for differential abundance between IL-13 and control samples, accounting for donor pairing, using an empirical Bayes quasi-likelihood F-test. We used Benhamini-Hochberg-derived FDR < 0.05 as a cutoff for inferring differentially secreted proteins.
For plotting of protein abundance in boxplots (Figures 5B and S4B) we used spectral count data normalized by size factors calculated using the median ratio method in DESeq2 (Love et al., 2014).
Analyzing ciliary beat frequency
To estimate the shift in CBF induced by chronic stimulation of IL-13 for pre- and post-washed inserts separately, we used the data from each washing regime in turn to fit linear mixed-effects models (LMM) that model the effects of IL-13 treatment (fixed effect) and donor identity (random effect) on CBF. To estimate the interaction between IL-13 treatment and washing status, we used a combined pre- and post-washed dataset to fit an LMM that models the effects of IL-13 treatment (fixed effect), wash status (fixed effect), donor identity (random effect), and an interaction between IL-13 treatment and wash status (fixed effect). Model coefficients as well as degrees of freedom and p values based on Satterthwaite approximations were calculated using the lmerTest package (Kuznetsova et al., 2017). The CBF experiment was carried out using both top-down and profile imaging. Full results from these models are in Table S1.
Normalization of bulk RNA-seq data
Prior to plotting expression in boxplots, we normalized counts by size factors calculated using DESeq2. For plotting expression in the heatmap and for carrying out weighted gene co-expression network analysis (WGCNA, discussed below), we used VST counts as a way to minimize mean-dependence of variation in expression.
Assignment to type 2-high/low status
To identify T2H and T2L individuals within the in vivo dataset, we carried out weighted gene co-expression network analysis (WGCNA) to detect modules of co-expressed genes, which may share common biological functions (Zhang and Horvath, 2005). Briefly, the WGCNA method measures pairwise Pearson correlations among all expressed genes in the dataset, constructs a gene network based on these correlations, calculates topological overlap among genes in the network, and then, by carrying out hierarchical clustering of a distance metric based on the topological overlap matrix, identifies discrete clusters of genes. We ran the method using 17,473 genes and 695 samples. We set the soft-thresholding power to 9, which best maximized the number of connections in the gene network, while still meeting the scale-free network assumptions of the method, and we retained the direction of correlations among indirect connections in the network (i.e., analysis was ‘signed’). Minimum module size was set to 30 and the deepSplit parameter was set to 2.
WGCNA identified 28 modules, one of which was highly enriched for known T2H markers, including IL1RL1, CLCA1, and POSTN. We hierarchically clustered GALA II samples based on expression of this module and used the first split as the basis for assignment to T2H and T2L status groups.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-MUC5AC | Invitrogen | MA1–38223; RRID:AB_2266697 |
| rabbit anti-SCGB1A1 | BioVendor | RD I 81022220–01; RRID:AB_344578 |
| mouse anti-E-cadherin | ECM Biosciences | CM1681; RRID:AB_2076812 |
| donkey anti-rabbit IgG (H+L) | Invitrogen | A-21207, A-31573; RRID:AB_141637, RRID:AB_2536183 |
| donkey anti-mouse IgG (H+L) | Invitrogen | A-21203, A-31571; RRID:AB_141633, RRID:AB_162542 |
| Biological Samples | ||
| Human tracheal epithelial cells (HTECs) | Seibold Laboratory at National Jewish Health | N/A |
| Human bronchial epithelial cells (HBECs) | National Jewish Live Cell Core | N/A |
| Human nasal epithelial cells from Genes-environments & Admixture in Latino Americans II (GALA II) | Seibold Laboratory at National Jewish Health | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DMEM-F (high glucose 4.5g/L) | Fisher Scientific | 10–017-CV |
| Ham’s F-12 | ThermoFisher Scientific | 11765–054 |
| FBS (heat-innactivated) | LifeTechnologies | 10082–147 |
| L-glutamine | Fisher Scientific | MT25005CI |
| Penicillin/Streptomycin (P/S) | Fisher Scientific | MT30002CI |
| Hydrocortisone | Sigma | H-0888 |
| EGF (for F-media) | Fisher Scientific | 17005042 |
| Cholera Toxin | Sigma | C-8052 |
| Insulin | Sigma | I9278 |
| Adenine | Sigma | A8626 |
| Rockl Inhibitor (RI) (Y-27632 dihyrochloride) | APExBio | A3008 |
| Amphotericin B | Sigma | A2942 |
| Fluconazole | Fagron, Inc. | 803890 |
| Gentamicin Reagent | ThermoFisher Scientific | 15710064 |
| PureCol Type I Bovine Collagen | Advanced Biomatrix | 5005–100ml |
| BEBM Basal Medium | Lonza | CC-3171 |
| DMEM-C (low glucose 1g/L) | ThermoFisher Scientific | 11885084 |
| Bovine Serum Albumin (BSA) | Sigma | A2153 |
| Ethanolamine | Sigma | E0135 |
| human EGF (hEGF) (for ALI Expansion Medium) | EMD Millipore | 01–107 |
| MgS04 | Sigma | M7506 |
| MgCI2 | Sigma | M8266 |
| CaCI2 | Sigma | C4901 |
| Retinoic Acid | Sigma | R2625 |
| BEGM Brachial Epithelial SingleOuots Kit | Lonza | CC-4175 |
| PneumaCult ALI Medium Kit | StemCell Technologies | #05001 |
| Heparin Solution (2mg/ml) | StemCell Technologies | #07980 |
| Hydrocortisone Stock Solution | StemCell Technologies | #07925 |
| Interleukin 13 (IL-13) | R&D Systems | 213-ILB-005 |
| 1x PBS (without Ca/Mg) | Fisher Scientific | 21 –040-CV |
| Trypsin | Fisher Scientific | 25–053-Cl |
| 1X HBSS | Fisher Scientific | 21 –021 -CV |
| DNase | Sigma | DN25 |
| Accutase | Fisher Scientific | NC9839010 |
| EDTA (0.5M) | Fisher Scientific | BP2482–500 |
| EGTA (0.5M) | Fisher Scientific | 50–255-957 |
| Paraformaldehyde (16%), MeOH Free | Fisher Scientific | AA433689M |
| HistoChoice Clearing Agent | Sigma | H2779 |
| Molecular Grade Ethanol (100%) | Fisher Scientific | BP28184 |
| Antigen Unmasking Solution, Citrate based | Vector Labs | H-3300 |
| Triton X-100 | Sigma | T8787 |
| Tris-Buffered Saline (TBS - 20x) | Fisher Scientific | AAJ60877K3 |
| DAPI (4′,6-diamidino-2-phenylindole) | Sigma | D9542 |
| VectaShleld HardSet Antifade Mounting Media | Vector Laboratories | H-1400 |
| 10% Neutral Buffered Formalin | Sigma | HT501128 |
| LC/MS grade Methanol | Fisher Scientific | A456–500 |
| Trypsin (proteomlcs grade) | Promega | V5280 |
| Trlfluoroethanol | Fisher Scientific | AC13975–1000 |
| Ammonium Bicarbonate | Fisher Scientific | BP2413500 |
| Dithiothreitol (DTT) | Biorad | 1610611 |
| Lodoacetamide | Biorad | 1632109 |
| Formic Acid | Fisher Scientific | PI28905 |
| Acetonitrile | Fisher Scientific | A955–4 |
| Deposited Data | ||
| Single cell RNA-seq data (fastq and processed data files) | GEO | https://www.ncbi.nim.nih.gov/geo/query/acc.cgi?&acc=GSE145013 |
| Bulk RNA-seq data from GALA II (fastq and processed data files) | GEO | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152004 |
| Experimental Models: Cell Lines | ||
| NIH 3T3 Fibroblasts (feeders) | ATCC | #CRL-1658 |
| Software and Algorithms | ||
| DataAnalysis 4.2 | Bruker | https://www.bruker.com/ |
| Mascot 2.4 | Matrix Science | http://www.matrixscience.com/index.html |
| Percolator (algorithm) | Matrix Science | http://www.matrixscience.com/help/percolator_help.html |
| ProteinScape 3.0 | Bruker | https://www.bruker.com/ |
| Particle Tracker - ImageJ | ImageJ | https://imagej.nih.gov/ij/ |
| iGenomes GTF (hg38) | UCSC | https://emea.support.illumina.com/sequencing/sequencing_software/igenome.html |
| PIVlab | Thielicke and Stamhuis, 2014 | https://www.mathworks.com/matlabcentral/fileexchange/27659-pivlab-particle-image-velocimetry-piv-tool |
| Cutadapt version 2.4 | Martin, 2011 | https://cutadapt.readthedocs.io/en/stable/ |
| Skewer version 0.2.2 | Jiang et al., 2014 | https://github.com/relipmoc/skewer |
| GSNAP version 2016_05_01 | Wu and Nacu, 2010 | http://research-pub.gene.com/gmap/ |
| HTSeq-count version 0.8.0 | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.11.1/count.html |
| DESeq2 version 1.16.1 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| edgeR version 3.24.0 | Robinson et at, 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Seurat version 2.3.0 | Satija et at, 2015 | https://cran.r-project.org/web/packages/Seurat/index.html |
| heatmap3 version 1.1.1 | Zhao et at, 2014 | https://cran.r-project.org/web/packages/heatmap3/index.html |
| EnrichrAPI | Chen et at, 2013 | https://github.com/russell-stewart/enrichrAPI |
| Monocle version 2.8 | Trapnell et at, 2014 | https://bioconductor.org/packages/release/bioc/html/monocle.html |
| WGCNA version 1.61 | Zhang and Horvath, 2005 | https://cran.r-project.org/web/packages/WGCNA/index.html |
| ImerTest version 3.0 | Kuznetsova et at, 2017 | https://cran.r-project.org/web/packages/ImerTest/index.html |
| Custom Code | This study | https://github.com/seiboldlab/SingleCell_IL13 |
| Other | ||
| Echo Revolve R4 Microscope | Discover Echo, Inc. | https://discover-echo.com/revolve |
| Beckman Coulter FXp Automation System | Beckman Coulter | https://www.beckrnancoulter.com/ |
| HiSeq 2500 System | Illumina | https://www.illumina.com/ |
| Bruker Mass Spec | Bruker | https://www.bruker.com/ |
| Eclipse Ti-E inverted microscope | Nikon | https://www.nikon.com/ |
| CMOS camera | FLIR Integrated Imaging Solutions Inc. | GS3-U3–23S6M-C |
| 6.5mm transwell inserts, 0.4um PET membrane | Corning | 3470 |
| SuperFrost Plus Sides | Fisher Scientific | 12–550-15 |
| Siliconized protein low-bind eppendorf tubes (2ml) | Fisher Scientific | 02–681-332 |
| ProntoSil C18AQ Column (15cm x 100um) | nanoLCMS Solutions | 31001 |
| Capillary 3jj 200A ProntoSIL C18AQ Trap | nanoLCMS Solutions | 41002 |
| SwissProt | UniProtKB/Swiss-Prot | https://www.expasy.org/proteomics/ |
Highlights.
IL-13 alters secretory expression states in all airway epithelial cell types
Chronic IL-13 weakens innate defense but spurs interferon signaling in ciliated cells
IL-13 reprogramming leads to pathologic mucus that slows ciliary beat frequency
IL-13-induced metaplasia recapitulates reprograming in type 2 inflamed children
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 HL135156, R01 MD010443, R01 HL128439, P01 HL132821, U01 HL138626, the National Jewish Health REGEN Program, and the European Union (EU)-European Research Council (ERC) CoG HydroSync grant.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107872.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao K, and Reinhardt RL (2015). The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine 75, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló B., Pons J, Fuster A, Sauleda J, Noguera A, Ferrer JM, and Agustí AGN (2006). Intracellular cytokine profile of T lymphocytes in patients with chronic obstructive pulmonary disease. Clin. Exp. Immunol. 145, 474–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto KT, Brockman-Schneider RA, Kuipers I, Basnet S, Bochkov YA, Altman MC, Jarjour NN, Gern JE, and Esnault S (2020). Human airway epithelial cells express a functional IL-5 receptor. Allergy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisswenger C, Kandler K, Hess C, Garn H, Felgentreff K, Wegmann M, Renz H, Vogelmeier C, and Bals R (2006). Allergic airway inflammation inhibits pulmonary antibacterial host defense. J. Immunol. 177, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Bergeron C, Al-Ramli W, and Hamid Q (2009). Remodeling in asthma. Proc. Am. Thorac. Soc. 6, 301–305. [DOI] [PubMed] [Google Scholar]
- Bonser LR, and Erle DJ (2017). Airway mucus and asthma: the role of MUC5AC and MUC5B. J. Clin. Med. 6, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell LN, Nguyen EA, Roth LA, Oh SS, Tcheurekdjian H, Sen S, Davis A, Farber HJ, Avila PC, Brigino-Buenaventura E, et al. (2013). Childhood obesity and asthma control in the GALA II and SAGE II studies. Am. J. Respir. Crit. Care Med. 187, 697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet J, Cabrera P, Berkman N, Buhl R, Holgate S, Wenzel S, Fox H, Hedgecock S, Blogg M, and Cioppa GD (2005). The effect of treatment with omalizumab, an anti-IgE antibody, on asthma exacerbations and emergency medical visits in patients with severe persistent asthma. Allergy 60, 302–308. [DOI] [PubMed] [Google Scholar]
- Castro M, Zangrilli J, Wechsler ME, Bateman ED, Brusselle GG, Bardin P, Murphy K, Maspero JF, O’Brien C, and Korn S (2015). Reslizumab for inadequately controlled asthma with elevated blood eosinophil counts: results from two multicentre, parallel, double-blind, randomised, placebo-controlled, phase 3 trials. Lancet Respir. Med. 3, 355–366. [DOI] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, and Whitsett JA (2009). SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J. Clin. Invest. 119, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Karp CL, Impey S, Xu Y, Randell SH, Kitzmiller J, Maeda Y, Haitchi HM, Sridharan A, et al. (2014). Foxa3 induces goblet cell metaplasia and inhibits innate antiviral immunity. Am. J. Respir. Crit. Care Med. 189, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioccioli M, Feriani L, Kotar J, Bratcher PE, and Cicuta P (2019a). Phenotyping ciliary dynamics and coordination in response to CFTR-modulators in Cystic Fibrosis respiratory epithelial cells. Nat. Commun. 10, 1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioccioli M, Feriani L, Nguyen Q, Kotar J, Dell SD, Mennella V, Amirav I, and Cicuta P (2019b). Quantitative high-speed video profiling discriminates between DNAH11 and HYDIN variants of primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 199, 1436–1438. [DOI] [PubMed] [Google Scholar]
- Christenson SA, Steiling K, van den Berge M, Hijazi K, Hiemstra PS, Postma DS, Lenburg ME, Spira A, and Woodruff PG (2015). Asthma-COPD overlap. Clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 191, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contoli M, Ito K, Padovani A, Poletti D, Marku B, Edwards MR, Stanciu LA, Gnesini G, Pastore A, Spanevello A, et al. (2015). Th2 cytokines impair innate immune responses to rhinovirus in respiratory epithelial cells. Allergy 70, 910–920. [DOI] [PubMed] [Google Scholar]
- de Jong PM, van Sterkenburg MA, Hesseling SC, Kempenaar JA, Mulder AA, Mommaas AM, Dijkman JH, and Ponec M (1994). Ciliogenesis in human bronchial epithelial cells cultured at the air-liquid interface. Am. J. Respir. Cell Mol. Biol. 10, 271–277. [DOI] [PubMed] [Google Scholar]
- Evans CM, Williams OW, Tuvim MJ, Nigam R, Mixides GP, Blackburn MR, DeMayo FJ, Burns AR, Smith C, Reynolds SD, et al. (2004). Mucin is produced by clara cells in the proximal airways of antigen-challenged mice. Am. J. Respir. Cell Mol. Biol. 31, 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everman JE, Rios C, and Seibold MA (2018). Utilization of air-liquid interface cultures as an in vitro model to assess primary airway epithelial cell responses to the type 2 cytokine interleukin-13. Methods Mol. Biol. 1799, 419–432. [DOI] [PubMed] [Google Scholar]
- Fahy JV (2015). Type 2 inflammation in asthma—present in most, absent in many. Nat. Rev. Immunol. 15, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feriani L, Juenet M, Fowler CJ, Bruot N, Chioccioli M, Holland SM, Bryant CE, and Cicuta P (2017). Assessing the collective dynamics of motile cilia in cultures of human airway cells by multiscale DDM. Biophys. J. 113, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George L, and Brightling CE (2016). Eosinophilic airway inflammation: role in asthma and chronic obstructive pulmonary disease. Ther. Adv. Chronic Dis. 7, 34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerovac BJ, and Fregien NL (2016). IL-13 inhibits multicilin expression and ciliogenesis via Janus kinase/signal transducer and activator of transcription independently of notch cleavage. Am. J. Respir. Cell Mol. Biol. 54, 554–561. [DOI] [PubMed] [Google Scholar]
- Gomperts BN, Kim LJ, Flaherty SA, and Hackett BP (2007). IL-13 regulates cilia loss and foxj1 expression in human airway epithelium. Am. J. Respir. Cell Mol. Biol. 37, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett NR, Shaykhiev R, Walters MS, Wang R, Zwick RK, Ferris B, Witover B, Salit J, and Crystal RG (2011). The human airway epithelial basal cell transcriptome. PLoS ONE 6, e18378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, and Marshall WF (2011). Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12, 222–234. [DOI] [PubMed] [Google Scholar]
- Jiang H, Lei R, Ding SW, and Zhu S (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhn YJ (2014). Risks for infection in patients with asthma (or other atopic conditions): is asthma more than a chronic airway disease? J. Allergy Clin. Immunol. 134, 247–257, quiz 258–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesimer M, Kirkham S, Pickles RJ, Henderson AG, Alexis NE, Demaria G, Knight D, Thornton DJ, and Sheehan JK (2009). Tracheobronchial air-liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L92–L100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfhagen TR, Kitzmiller J, Chen G, Sridharan A, Haitchi HM, Hegde RS, Divanovic S, Karp CL, and Whitsett JA (2012). SAM-pointed domain ETS factor mediates epithelial cell-intrinsic innate immune signaling during airway mucous metaplasia. Proc. Natl. Acad. Sci. U S A 109, 16630–16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Nguyen EA, Roth LA, Oh SS, Gignoux CR, Huntsman S, Eng C, Moreno-Estrada A, Sandoval K, Peñ aloza-Espinosa RI, et al. (2013). Factors associated with degree of atopy in Latino children in a nationwide pediatric sample: the Genes-environments and Admixture in Latino Asthmatics (GALA II) study. J. Allergy Clin. Immunol. 132, 896–905.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, Elias JA, Sheppard D, and Erle DJ (2002). Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med. 8, 885–889. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, and Christensen RHB (2017). lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26. [Google Scholar]
- Lachowicz-Scroggins ME, Yuan S, Kerr SC, Dunican EM, Yu M, Carrington SD, and Fahy JV (2016). Abnormalities in MUC5AC and MUC5B protein in airway mucus in asthma. Am. J. Respir. Crit. Care Med. 194, 1296–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili J, Perret E, Willems T, Minty A, Parthoens E, Houcine O, Coste A, Jorissen M, Marano F, Caput D, and Tournier F (2001). IL-13 alters mucociliary differentiation and ciliary beating of human respiratory epithelial cells. J. Clin. Invest. 108, 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S, Li DW, and Jiang SJ (2013). Efficacy of anti-interleukin-5 therapy with mepolizumab in patients with asthma: a meta-analysis of randomized placebo-controlled trials. PLoS ONE 8, e59872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livraghi-Butrico A, Grubb BR, Wilkinson KJ, Volmer AS, Burns KA, Evans CM, O’Neal WK, and Boucher RC (2017). Contribution of mucus concentration and secreted mucins Muc5ac and Muc5b to the pathogenesis of muco-obstructive lung disease. Mucosal Immunol. 10, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10. [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair P, Wenzel S, Rabe KF, Bourdin A, Lugogo NL, Kuna P, Barker P, Sproule S, Ponnarambil S, and Goldman M; ZONDA Trial Investigators (2017). Oral glucocorticoid-sparing effect of Benralizumab in severe asthma. N. Engl. J. Med. 376, 2448–2458. [DOI] [PubMed] [Google Scholar]
- Neophytou AM, White MJ, Oh SS, Thakur N, Galanter JM, Nishimura KK, Pino-Yanes M, Torgerson DG, Gignoux CR, Eng C, et al. (2016). Air pollution and lung function in minority youth with asthma in the GALA II (Genes-Environments and Admixture in Latino Americans) and SAGE II (Study of African Americans, Asthma, Genes, and Environments) studies. Am. J. Respir. Crit. Care Med. 193, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemus-Johnson J, Naughton KA, Sudi J, Hogarth K, Naurekas ET, Nicolae DL, Sperling AI, Solway J, White SR, and Ober C (2016). Genome-wide methylation study identifies an IL-13-induced epigenetic signature in asthmatic airways. Am. J. Respir. Crit. Care Med. 193, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura KK, Galanter JM, Roth LA, Oh SS, Thakur N, Nguyen EA, Thyne S, Farber HJ, Serebrisky D, Kumar R, et al. (2013). Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am. J. Respir. Crit. Care Med. 188, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan M, Korenblat P, Mosesova S, Scheerens H, Arron JR, Zheng Y, Putnam WS, Parsey MV, Bohen SP, and Matthews JG (2013). Dose-ranging study of lebrikizumab in asthmatic patients not receiving inhaled steroids. J. Allergy Clin. Immunol. 132, 567–574.e12. [DOI] [PubMed] [Google Scholar]
- Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, Wadsworth MH, 2nd, Hughes TK, Kazer SW, Yoshimoto E., et al. (2018). Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 560, 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, Khurana Hershey GK, Chen G, and Whitsett JA (2007). SPDEF regulates goblet cell hyperplasia in the airway epithelium. J. Clin. Invest. 117, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo AA, Starner TD, Scheetz TE, Traver GL, Tilley AE, Harvey BG, Crystal RG, McCray PB Jr., and Zabner J (2011). The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol 300, L25–L31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, Galanter JM, Gignoux CR, Roth LA, Kumar R, et al. (2014). Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J. Allergy Clin. Immunol. 133, 670–8.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, Alexander SN, Bellinghausen LK, Song AS, Petrova YM, et al. (2014). Muc5b is required for airway defence. Nature 505, 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Garciá S, Deprez M, Lebrigand K, Cavard A, Paquet A, Arguel MJ, Magnone V, Truchi M, Caballero I., Leroy S et al. (2019). Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 146, dev177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RJ, Chachi L, FitzGerald JM, Backer V, Olivenstein R, Titlestad IL, Ulrik CS, Harrison T, Singh D, Chaudhuri R, et al. ; MESOS study investigators (2018). Effect of tralokinumab, an interleukin-13 neutralising monoclonal antibody, on eosinophilic airway inflammation in uncontrolled moderate-to-severe asthma (MESOS): a multicentre, double-blind, randomised, placebo-controlled phase 2 trial. Lancet Respir. Med 6, 499–510. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thielicke W, and Stamhuis EJ (2014). PIVlab—towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw 2, e30. [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Roger J, Fitau J, Combe D, Giddings J, Heeke GV, and Jones CE (2011). Goblet cells are derived from a FOXJ1-expressing progenitor in a human airway epithelium. Am. J. Respir. Cell Mol. Biol. 44, 276–284. [DOI] [PubMed] [Google Scholar]
- Tyner JW, Kim EY, Ide K, Pelletier MR, Roswit WT, Morton JD, Battaile JT, Patel AC, Patterson GA, Castro M, et al. (2006). Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J. Clin. Invest. 116, 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wark PAB, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, Holgate ST, and Davies DE (2005). Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J. Exp. Med. 201, 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlk C, von Bülow A, Kriegbaum M, Backer V, and Porsbjerg C (2018). Allergic asthma is associated with increased risk of infections requiring antibiotics. Ann. Allergy Asthma Immunol. 120, 169–176.e1. [DOI] [PubMed] [Google Scholar]
- Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, Koth LL, Arron JR, and Fahy JV (2009). T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit. Care Med. 180, 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, and Nacu S (2010). Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Jiang D, Smith S, Thaikoottathil J, Martin RJ, Bowler RP, and Chu HW (2012). IL-13 dampens human airway epithelial innate immunity through induction of IL-1 receptor-associated kinase M. J. Allergy Clin. Immunol. 129, 825–833.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, and Horvath S (2005). A. General Framework for Weighted Gene Co-expression Network Analysis, Volume 4 (The Berkeley Electronic Press). [DOI] [PubMed] [Google Scholar]
- Zhao H, Albino AP, Jorgensen E, Traganos F, and Darzynkiewicz Z (2009). DNA damage response induced by tobacco smoke in normal human bronchial epithelial and A549 pulmonary adenocarcinoma cells assessed by laser scanning cytometry. Cytometry A 75, 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Guo Y, Sheng Q, and Shyr Y (2014). Advanced heat map and clustering analysis using heatmap3. BioMed Res. Int. 2014, 986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen G, Park SW, Nguyenvu LT, Rodriguez MW, Barbeau R, Paquet AC, and Erle DJ (2007). IL-13 and epidermal growth factor receptor have critical but distinct roles in epithelial cell mucin production. Am. J. Respir. Cell Mol. Biol. 36, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, and Elias JA (1999). Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 103, 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed RNA-seq data used in the study are available at the National Center for Biotechnology Information/Gene Expression Omnibus (GEO) under repository accession number GEO: GSE145013 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE145013] for the scRNA-seq data and under GEO: GSE152004 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152004] for bulk RNA-seq data from GALA II. Proteomics and CBF data used to generate Figures 6 and S4 are available in Table S6. Code used to carry out data analysis is available on GitHub [https://github.com/seiboldlab/SingleCell_IL13].