Abstract
The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on a global scale urges prompt and effective countermeasures. Recently, a study has reported that coronavirus disease-19 (COVID-19), the disease caused by SARS-CoV-2 infection, is associated with a decrease in albumin level, an increase in NETosis, blood coagulation, and cytokine level. Here, we present drug-loaded albumin nanoparticles as a therapeutic agent to resolve the clinical outcomes observed in severe SARS-CoV-2 patients. PEGylated nanoparticle albumin-bound (PNAB) was used to promote prolonged bioactivity of steroidal ginsenoside saponins, PNAB-Rg6 and PNAB-Rgx365. Our data indicate that the application of PNAB-steroidal ginsenoside can effectively reduce histone H4 and NETosis-related factors in the plasma, and alleviate SREBP2-mediated systemic inflammation in the PBMCs of SARS-CoV-2 ICU patients. The engineered blood vessel model confirmed that these drugs are effective in suppressing blood clot formation and vascular inflammation. Moreover, the animal model experiment showed that these drugs are effective in promoting the survival rate by alleviating tissue damage and cytokine storm. Altogether, our findings suggest that these PNAB-steroidal ginsenoside drugs have potential applications in the treatment of symptoms associated with severe SARS-CoV-2 patients, such as coagulation and cytokine storm.
Keywords: Albumin, Ginsenoside, COVID-19, NETosis, Cytokine storm
1. Introduction
The coronavirus disease-2019 (COVID-19) pandemic, caused by a novel severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), has brought about a major global health crisis since its emergence in December 2019 [1,2]. The COVID-19 prompted multinational pharmaceutical companies and numerous research teams to take part in a race to develop therapeutic agents [3]. Although an unprecedented scale of the global effort is put on its development, still no drugs that directly target and efficiently eliminate the SARS-CoV-2 are available. Currently, the only available option for COVID-19 patients is supportive treatments [4,5]. It is predicted that the potential vaccine for emergency-use could be available by early 2021 [6].
In order to establish an effective treatment strategy, we first explored the clinical manifestations and risk factors on severe COVID-19 patients admitted to the intensive care unit (ICU) of Yeungnam University Hospital in Korea [7,8]. According to our patient data and a recently published report [9], severe COVID-19 patients display an increase in neutrophil, a decrease in albumin, and an increase in D-Dimers in association with abnormal blood clotting and cytokine release syndrome (CRS). Moreover, it is known that the hyper-inflammatory response caused by SARS-CoV-2 infection leads to acute respiratory distress syndrome (ARDS), sepsis, uncontrollable blood pressure, and eventual death [10]. Thus, the prognosis is very poor for infected patients under clinical conditions like hypertension, diabetes, coronary heart disease, cerebrovascular illness, chronic obstructive pulmonary disease, and kidney dysfunction [1,[11], [12], [13]].
For the treatment of severe COVID-19 ICU patients, corticosteroid, such as dexamethasone [14], as well as antiviral drugs, such as hydroxychloroquine [15] and lopinavir/ritonavir (Kaletra®) [16,17], vitamins [18], and albumin [19], have been used. However, according to the interim guidance on clinical management of COVID-19 patients [20], the World Health Organization (WHO) advises against the use of steroids unless indicated for other reasons. As steroids suppress the immune response and pathogen clearance [14], their routine administration in the setting of viral pneumonia leads to increased mortality [21,22]. Nonetheless, the usage of steroids is inevitable when those patients develop sepsis and subsequent cytokine storm. Although many clinicians are actively using combinations of antiviral and anti-inflammatory therapies to control cytokine storm and promote viral clearance concomitantly [23,24], no remarkable changes have been reported in the high mortality rate of severe COVID-19 patients with ARDS or septic shock. Recently, the WHO welcomed the initial clinical trial results from the United Kingdom (UK) that showed dexamethasone, a corticosteroid, can be lifesaving for critically ill COVID-19 patients. For patients on ventilators, the treatment demonstrated a reduction in mortality by about 30%. However, the positive effect was only seen in seriously ill patients and was not observed in patients with milder disease. Similar to the principle that steroid-based drugs improve the mortality rate of COVID-19 severe patients, regulation of uncontrolled inflammatory responses is a promising treatment strategy.
Meanwhile, the convalescent plasma has been suggested as the first option for managing COVID-19 since it has been successfully used in other coronavirus outbreaks, such as SARS-CoV and MERS-CoV [25]. Recently, there have been reports on full recovery of severe COVID-19 patients infused with an immune serum that is rich in antibodies, albumin, and plasma proteins [[26], [27], [28]], of which mechanism of action is assumed to be similar with that of convalescent plasma. It is worth noting that the efficacy of the convalescent plasma and immune serum can be attributed in part to the activity of albumin that inhibits the platelet aggregation by binding to histone [29]. Indeed, in many patients under septic conditions, histone H4 was found to be released into the blood and captured by toll-like receptor-4 (TLR-4), thereby inducing excessive inflammatory responses [30,31].
In this study, we report nanoparticle albumin-bound (NAB)-steroidal ginsenoside derivatives for modulation of hyper-inflammatory responses and sepsis in severe COVID-19 patients. A rare saponin, ginsenoside Rg6, and a mixture of protopanaxatriol-type ginsenosides, Rgx365, were separated and purified from black ginseng (i.e., processed Panax ginseng, prepared by steaming and drying several times). Rg6, possessing a steroid-like structure, is well-known for its ability to suppress TLR-4-mediated systemic inflammatory responses [32]. Rgx365 also exhibits various biological activities, including anti-septic, anti-diabetic, wound-healing, immune-stimulatory, and anti-antioxidant effects [33]. The NAB technology provides a clinically-translatable nanoscale delivery system for lipophilic compounds, as in the case of Abraxane® (NAB-paclitaxel) [34,35]. We hypothesized that lipophilic compounds, including steroidal ginsenosides, can be adsorbed to the hydrophobic pockets on the surface of albumin to form injectable nanostructures via self-assembly [[36], [37], [38]]. In addition, we performed a site-specific modification of albumin via the introduction of maleimide-functionalized polyethylene glycol (PEG) moiety, to promote a prolonged circulation in the bloodstream.
The PEGylated NAB-Rg6 and Rgx365 (PNAB-Rg6 and PNAB-Rgx365, respectively) hold promises in alleviating the SARS-CoV-2-mediated hyper-inflammatory responses in the following perspectives: the long-acting ginsenosides can avoid the acute immunosuppressive side effects of steroids; can control organ damage and blood clotting accompanying excessive inflammatory reactions [39,40]; and can help to normalize the fluctuating blood pressure caused by viral infections [41]. Along with the above features, herein, we also demonstrated that the developed formulations could suppress the elevation of histone H4 and subsequent cytokine storm via down-regulation of the nuclear factor-κB (NF-κB) pathway in blood samples of patients with COVID-19. We evaluate the efficacy of these drugs in the suppression of blood clot formation and vascular inflammation by using the engineered 3D blood vessel model. The efficacy of the drugs is confirmed by using infection-induced septic mice models, encouraging further investigation of PNAB-ginsenosides as potential tools to treat SARS-CoV-2-mediated illnesses.
2. Materials and methods
2.1. Materials
BSA, phosphate monobasic, taurocholic acid (TC), deuterium oxide, dimethyl sulfoxide-d6, and albumin-fluorescein isothiocyanate conjugate were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA). mPEG-MAL (2 kDa) was purchased form Biochempeg (Watertown, NY, USA).
2.2. Plasma sample
Whole blood was collected from patients admitted at Yeungnam University Medical Center after they were diagnosed with the SARS-CoV-2 infection at a public health center in Daegu, Republic of Korea. Among the SARS-CoV-2-infected patients, those with advanced septic shock or acute respiratory syndrome disease (ARDS) were admitted to the intensive care unit (ICU), and those with mild illness were admitted to the general ward. Patients with COVID-19 sepsis were defined using criteria provided by the Sepsis Consensus Conference Committee [42]. Clinical data were collected for all the patients. Plasma samples were prepared by centrifugation at 2000×g for 5 min within 12 h after whole blood collection. The human study protocol was approved by the Institutional Review Board of Yeungnam University Hospital at Daegu in Korea (YUH-2020-03-057, 2020-05-031-001).
2.3. Blood cell count
Complete blood counts were obtained from the patients’ venous blood samples. The laboratory findings were analyzed within 24 h after admission. Blood cell count was performed using a Sysmex XE-2100 Automated Hematology System (Sysmex Corp. Kobe, Japan).
2.4. Histone H4 enzyme-linked immunosorbent assay (ELISA)
Histone H4 concentrations in plasma were determined using a human histone H4 ELISA Kit (MBS101164, MyBioSource Inc., San Diego, CA, USA).
2.5. Quantification of plasma cfDNA
For isolation of cfDNA from the plasma of SARS-CoV-2-infected patients, a Qiagen QIAamp DNA Blood Mini Kit was used according to the manufacturer's instructions (Qiagen, Valencia, CA, USA) after centrifugation of the plasma at 16,800×g for 10 min. Purified cfDNA was quantified using a NanoDrop spectrophotometry (Thermo Scientific, Loughborough, UK).
2.6. NET ELISA
Freshly isolated neutrophils (1 × 105 cells) were stimulated to generate NETs by incubating with 25 nM phorbol-myristate acetate (PMA, Sigma-Aldrich, MO, USA) or control media (RPMI 1640 supplemented with glutamine, penicillin, and streptomycin) and evaluated using a fluorometric technique as previously described [43]. NET production was measured as arbitrary fluorescent units (AFUs).
2.7. MPO and NE ELISA
To quantify the release of granule matrix proteins upon degranulation in peripheral blood mononuclear cells (PBMCs) of SARS-CoV-2-infected patients, plasma were analyzed using human MPO ELISA kit (BMS2038INST, Invitrogen, Carlsbad, CA, USA) and human neutrophil elastase (NE) ELISA Kit (MBS725923, MyBioSource Inc., San Diego, CA, USA).
2.8. Protein profiling
Plasma pools of normal individuals or SARS-CoV-2 patients were processed as indicated in the Human XL Cytokine Array Kit (R&D Systems, Minneapolis, MN, USA). Developed films were scanned, the obtained images were analyzed using ImageJ version 1.43 (NIH, Bethesda, MD, USA).
2.9. NF-kB activity
Preparation of nuclear extracts and TransAM assays were performed as previously described [44]. The activity of individual NF-κB subunits was determined using an ELISA-based NF-κB Family Transcription Factor Assay Kit (43,296; Active Motif, Carlsbad, CA, USA). Briefly, nuclear extracts (2 μg) were incubated in a 96-well plate, which was coated with NF-κB consensus oligonucleotides. The captured complexes were incubated with specific NF-κB primary antibodies and subsequently detected using HRP-conjugated secondary antibodies included in the kit. Finally, the optical density (OD) at 450 nm was measured using a Tecan Spark microplate reader (Tecan, Austria GmbH, Austria).
2.10. Cytokine ELISAs
Serum levels of inflammatory cytokines IL-1β, IL-4, IL-6, IL-8, IFN-γ, and TNF-α were determined using human ELISA kits (Quantikine ELISA, R&D Systems, Minneapolis, MN, USA) according to the manufacturer's instructions. The results were expressed as pg/mL.
2.11. WST-1 cell proliferation assay
Following exposure of PBMCs to each nanotherapeutic product, 10 μL of WST-1 reagent was added per well and incubated at 37 °C with 5% CO2. At indicated time points, measurements of absorbance were taken at 480 nm and 600 nm (background) on Tecan Spark microplate reader (Tecan, Austria GmbH, Austria).
2.12. Real-time PCR
To generate cDNA from PBMCs incubation with/without each nanotherapeutic product, 1 μg of total RNA was reverse transcribed with random hexamers using expand reverse transcription polymerase (Roche, Basel, Switzerland). Real-time PCR was performed using the LightCycler FastStart DNA Master SYBR Green I from Roche Diagnostics GmbH according to the manufacturer's protocol. The following LightCycler conditions were used: Initial denaturation at 95 °C for 10 min, followed by 45 cycles with denaturation at 95 °C for 10 min, annealing at 60 °C for 5 min, and elongation at 72 °C for 15 min. Quantities of specific mRNA in the sample were measured according to the corresponding gene-specific standard curves.
Real-time PCR primer sequences
-
1.
hSREBF2:
F-CCTTCCTGTGCCTCTCCTTTA, R-AGGCATCATCCAGTCAAACCA.
-
2.
hNOX2:
F-AACGAATTGTACGTGGGCAGA, R-GAGGGTTTCCAGCAAACTGAG.
-
3.
hNLRP3:
F-TGCCGGGGCCTCTTTTCAGT, R-CCACAGCGCCCCAACCACAA.
-
4.
hMCP-1:
F-CGCTCAGCCAGATGCAATCAATGC, R-GGTTTGCTTGTCCAGGTGGTCCA.
-
5.
hVCAM1:
F-TGTCAATGTTGCCCCCAGAGATACA, R-GGCTGTAGCTCCCCGTTAGGGA.
-
6.
hICAM1:
F-GTGTCCTGTATGGCCCCCGACT, R-ACCTTGCGGGTGACCTCCCC.
2.13. Caspases detection assay
Caspase-Glo® 1 inflammasome assay kit (G9951, Promega, Madison, WI, USA) and Caspase-Glo® 3/7 Assay System (G8090, Promega, Madison, WI, USA) were used to measure the activation of the caspases in PBMCs.
2.14. Synthesis of BSA-mPEG
mPEG-MAL (20 mg) was slowly added to BSA (100 mg) solution, and the mixture was stirred for 18 h at room temperature. The resulting solution was dialyzed against double-deionized water (DDW) using a dialysis membrane with a molecular weight cut-off of 6–8 kDa (Cellu-Sep T2; Membrane Filtration Products, Seguin, TX, USA). The dialysis product (i.e., BSA-mPEG) was lyophilized and stored at −70 °C until use.
2.15. Characterization of BSA and BSA-mPEG
BSA and BSA-mPEG were characterized by matrix-assisted laser desorption and ionization time of flight (MALDI-TOF; Voyager DE-STR, Applied Biosystems, Foster City, CA, USA). SDS-PAGE was performed using 10% polyacrylamide gel. The samples were electrophoresed at 110 V for 120 min. Coomassie blue staining of protein gels was achieved by washing the gel for 5 min three times in DDW to remove SDS, incubating the gel with Coomassie blue stain for 60 min, and rinsing the gel with distilled water for 20 min three times. To stain the PEG moieties, the gel was soaked in 5% barium chloride solution for 15 min, washed with DDW for 30 min, and stained with a 0.05 M iodide solution for 15 min.
2.16. Preparation of NAB- and PNAB-ginsenosides
Rg6 or Rgx365 (3 mg) dissolved in dimethyl sulfoxide (DMSO; 180 μL) and BSA or BSA-mPEG (5 mg) dissolved in DDW (45 μL) were vortex-mixed for 30 s. DDW (775 μL) was then added dropwise to the mixture with ultrasonication (VC-750; Sonics & Materials, Inc., Newtown, CT, USA) at an amplitude of 20% and a pulse cycle of 2 s on and 3 s off. The resulting solution was immediately lyophilized to remove the remaining DMSO. The lyophilized product was resuspended in DDW (1 mL) and filtered through a syringe filter with 0.45 μm pore size to remove unloaded ginsenosides.
2.17. Characterization of NAB- and PNAB-ginsenosides
The hydrodynamic diameter, PDI, and zeta potential of the NAB- and PNAB-ginsenosides were measured using dynamic light scattering method (Zetasizer Ultra; Malvern Panalytical, Malvern, UK). The morphology of the formulations was observed using TEM (JEM-F200 [TFEG]; JEOL Ltd., Tokyo, Japan). The formulations were placed onto the surface of a 200-mesh carbon-coated copper grid and negatively stained with uranyl acetate (UA). For the measurement of EE, an aliquot (10 μL) of NAB- or PNAB-ginsenosides was vortex-mixed with acetonitrile (ACN; 990 μL) containing 0.1% (v/v) formic acid for 5 min. The mixture was sonicated for 2 min and centrifuged at 13,000×g for 5 min. The supernatant (50 μL) was then mixed with ACN (950 μL) containing 0.1% (v/v) formic acid and internal standard (TC, 50 ng/mL), which was the analytical sample for high-performance liquid chromatography (HPLC) analysis. Chromatographic separation was performed using an Agilent 1260 Infinity HPLC system (Agilent Technologies, Palo Alto, CA, USA). An aliquot (20 μL) of the analytical sample was injected into a Kinetex 2.6 μm C18 100 Å column (100 × 4.6 mm; Phenomenex, CA, USA) with a C18 guard column (4 × 2.0 mm; Phenomenex, CA, USA) at 25 °C. The elution was carried out under an isocratic condition at a flow rate of 0.35 mL/min, and the total run time was 5 min. The mobile phase consisted of ACN and DDW with 0.1% (v/v) formic acid (80:20, v/v). Mass spectrometric detection was achieved using API 3200 LC-MS/MS (SCIEX, Framingham, MA, USA). The optimized ionization source settings of ion spray source temperature, curtain gas pressure, gas 1, and gas 2, were 250 °C, 25 psi, 50 psi, and 50 psi, respectively. The ion spray voltage, declustering potential, entrance potential, collision energy, and collision gas were set at 5500 V, 150 V, 1 V, 52 eV, and 26 V for Rg6 and -4500 V, 145 V, 5 V, 130 eV, and 28 V for TC, respectively. The monitored ion transitions (m/z) were 789.5 → 349.1 for Rg6 and 514.1 → 79.9 for TC.
2.18. Drug release test
The drug-releasing property of PNAB-Rg6 and PNAB-Rgx365 was evaluated by using a dialysis method. The formulations (PNAB-Rg6 or PNAB-Rgx365 containing approximately 450 μg of Rg6 and Rgx365) were loaded into mini-GeBAflex tubes (molecular weight cut-off: 6–8 kDa; Gene Bio-Application Ltd., Yavne, Israel), immersed into release media (1.8 mL; phosphate-buffered saline [PBS] of pH 6.6 or 7.4), and incubated in a shaking water bath at 37 °C (50 rpm). The tube was moved to another container filled with fresh media at 2, 4, 8, 24, 48, and 72 h of incubation. The collected release media (10 μL) was diluted with PBS (90 μL) and vortex-mixed with ACN (900 μL) containing 0.1% (v/v) formic acid and internal standard (TC, 20 ng/mL) for 3 min. The resulting mixture was centrifuged at 16,000×g for 5 min, and the Rg6 concentration of the supernatant was determined using the same LC-MS/MS method mentioned in Characterization of NAB- and PNAB-ginsenosides. The cumulative release (F; %) versus time (t) was plotted and fitted using the zero-order, first-order, Higuchi, and Baker-Lonsdale models, of which equations are as follows:
where k 0, k 1, k H, and k BL are release constants of each model.
2.19. Engineered blood vessel experiment
The fabrication of engineered blood vessels and the permeability assay was performed as previously described [45]. In brief, the engineered blood vessel was fabricated by using microneedles as templates. The collagen microchannels were fabricated by inserting in the sol-state type I rat tail collagen (Corning, Bedford, MA, USA) and subsequently removing them after the gelation. The human umbilical vein endothelial cells were seeded in the luminal surface of the collagen channel and cultured for five days to make a tight vascular structure. The permeability assay was performed by introducing 40 kDa FITC-Dextran solution (10 μM, Sigma-Aldrich, Saint Louis, MO, USA) into the perfusable blood vessel and monitoring the molecular transport using a confocal microscope (LSM700, Zeiss, Jena, Germany). The transendothelial permeability was quantified by using a custom-written MATLAB code (The Mathworks Inc., Natick, USA). To induce SARS-CoV-2-mediated inflammation in the engineered blood vessel, the SARS-CoV-2 patients’ plasma was introduced in the blood vessel and incubated for 2 h. Since the plasma of SARS-CoV-2 patients is coagulated, it was diluted in 1:2 ratio (plasma: media) before the injection to ensure sufficient fluidity. For the immunofluorescence imaging, phalloidin tetramethylrhodamine B isothiocyanate (for F-actin, P1951, Sigma-Aldrich, Saint Louis, MO, USA), Anti-ICAM-1 antibody (for ICAM-1, ab2213, abcam, Bristol, United Kingdom), and Anti-Fibrinogen antibody (for fibrinogen, ab118488, Bristol, United Kingdom) were utilized.
2.20. Animals and husbandry
Male C57BL/6 mice (6–7-weeks-old, weighing 18–20 g) and male ICR mice (4-weeks-old, weighing 21–24 g) were purchased from Orient Bio Co. (Sungnam, Korea) and Nara Biotech Co. (Seoul, Korea), respectively, and used after a 12-day acclimatization period. The animals were housed 5 per polycarbonate cage under controlled temperature (20–25 °C) and humidity (40%–45%) under a 12:12 h light/dark cycle, fed a normal rodent pellet diet, and supplied with water ad libitum. All animals were treated in accordance with the Guidelines for the Care and Use of Laboratory Animals issued by Chungnam National University (IRB No.; CNU-01050).
2.21. Cecal ligation and puncture
The CLP-induced septic mouse model was prepared as previously described [46]. Briefly, a 2-cm midline incision was made to expose the cecum and adjoining intestine. The cecum was then tightly ligated using a 3.0-silk suture 5.0 mm from the cecal tip, punctured with a 22-gauge needle, and then gently squeezed to extrude feces from the perforation site. The cecum was then returned to the peritoneal cavity and the laparotomy site sutured using 4.0-silk. For sham operations, the cecum of animals was surgically exposed but not ligated or punctured and then returned to the abdominal cavity.
2.22. Hematoxylin and eosin staining.
Male C57BL/6 mice underwent CLP and were administered ginsenoside Rg6, Rgx365, PNAB-Rg6, or PNAB-Rgx365 (5 mg/kg) intravenously at 24 h after CLP (n = 5). Mice were euthanized 72 h after CLP. To analyze the phenotypic change of lung in mice, lung samples were removed from each mouse, washed three times in PBS (pH 7.4) to remove remaining blood, fixed in 4% formaldehyde solution (Junsei, Tokyo, Japan) in PBS, pH 7.4 for 20 h at 4 °C. After fixation, the samples were dehydrated through ethanol series, embedded in paraffin, sectioned into 4-μm sections, and placed on a slide. The slides were deparaffinized in a 60 °C oven, rehydrated, and stained with hematoxylin (Sigma, Saint Louis, MO, USA). To remove over-staining, the slides were quick dipped three times in 0.3% acid alcohol, and counterstained with eosin (Sigma, Saint Louis, MO, USA). They are then washed in ethanol series and xylene, and then coverslipped. Light microscopic analysis of lung specimens was performed by blinded observation to evaluate pulmonary architecture, tissue edema, and infiltration of the inflammatory cells.
2.22. Clinical chemistry and cytokine level in septic mice plasma
Fresh serum was used for assaying aspartate transaminase (AST), alanine transaminase (ALT), blood urea nitrogen (BUN), creatinine, and LDH using biochemical kits (MyBioSource Inc., San Diego, CA, USA). To determine the concentrations of IL-1β, IL-6, IL-10, monocyte chemoattractant protein-1 (MCP-1), and TNF-α commercially available ELISA kits were used according to the manufacturer's protocol (R&D Systems, Minneapolis, MN, USA). Values were measured using an ELISA plate reader (Tecan, Austria GmbH, Austria).
2.23. In vivo permeability and leukocyte/Neutrophil migration assays
CLP-operated mice were injected with ginsenoside Rg6, Rgx365, PNAB-Rg6, or PNAB-Rgx365 (5 mg/kg) intravenously. After 24 h, 1% Evans blue dye solution in normal saline was injected intravenously into each mouse. 48 h later, the mice were killed, and the peritoneal exudates were collected after being washed with normal saline (5 mL) and centrifuged at 200×g for 10 min. The absorbance of the supernatant was read at 650 nm. The vascular permeability was expressed in terms of dye (μg/mouse), which leaked into the peritoneal cavity according to a standard curve of Evans blue dye. For assessment of leukocyte/neutrophil migration, CLP operated mice were treated with each molecule after CLP surgery 24 h. The mice were then sacrificed and the bronchoalveolar lavage (BAL) was washed with 0.8 mL of normal saline [47]. BAL fluid (200 μL) was counted by the auto hematology analyzer (Mindray, BC 5000 Vet, Shenzhen, China). The results were expressed as leukocyte/neutrophil × 105 per BAL fluid.
2.24. Expression of ICAMs
The expression of intercellular adhesion molecule-1 (ICAM-1) on lung tissues were determined by a direct ELISA. The lysed lung tissues were coated onto Nunc-Immuno™ MicroWell™ 96 well plates and incubated overnight at 4 °C. After washing, anti-mouse monoclonal ICAM-1 antibodies (Millipore Corporation, Billerica, MA, USA, 1:50 each) were added. After 1 h (37 °C, 5% CO2), the cells were washed three times and then 1:2000 peroxidase-conjugated anti-mouse IgG antibody (100 μL; Sigma, Saint Louis, MO, USA) was added for 1 h. The cells were washed again three times and developed using the o-phenylenediamine substrate (Sigma, Saint Louis, MO, USA). Colorimetric analysis was performed by measuring absorbance at 490 nm. All measurements were performed in triplicate wells.
2.25. Cytokine levels in the plasma of septic mice
Fresh serum was used for the analysis of AST, ALT, BUN, creatinine, and LDH levels using biochemical kits (MyBioSource Inc., San Diego, CA, USA). Values were measured using an ELISA plate reader (Tecan, Austria GmbH, Austria).
2.26. Measurement of intracellular ROS levels
Intracellular ROS level was measured in lung endothelial cells from CLP-treated mice by Fluorometric Intracellular Ros Kit (Sigma, Saint Louis, MO, USA) [48]. In brief, after treatment with CLP and each molecules for 72 h, mouse lung endothelial cells (approximately 106) were washed and resuspended in 500 μL of medium 199 and were then loaded with a cell-permeable sensor for 1 h. ROS reacts with the cell permeable sensor, resulting in a fluorometric product (λ ex = 540/λ em = 570) proportional to the amount of ROS present. Fluorescence intensity was measured by a fluorescence microplate reader (Tecan, Austria GmbH, Austria).
This is a nonpolar compound that is hydrolyzed within the cell to form a nonfluorescent derivative, which in presence of a proper oxidant converted to a fluorescent product. Fluorescence was measured through spectrofluorometer using 507 nm as excitation and 530 nm as emission wavelengths.
2.27. Statistical analysis
All experiments were performed independently at least three times. Statistically significant differences were determined using the unpaired t-test. Graphprism software 7.0 was used for statistical analyses. Data are reported as mean ± SEM with significance set at P < 0.05. p-values for each experiment are provided in the figure legends.
3. Results
3.1. Clinical background for designing PNAB-ginsenosides targeting SARS-CoV-2-mediated hyper-inflammation
For infectious diseases such as SARS-CoV-2 pneumonia, apparent clinical signatures are commonly observed in the patient blood, including blood coagulation [49,50], NETosis [51], cytokine storm [1], and inflammation [24,52]. Those clinical signatures become severe in ICU or deceased cases. Considering that there is no approved therapeutics to treat SARS-CoV-2 infection, timely regulation of the clinical signatures is the primary strategy to improve patient care. During the analysis of SARS-CoV-2 patient blood samples, we found that histone H4 level was highly upregulated in the plasma of ICU patients. Histone H4, one of the five main histone proteins composing chromatin structures in eukaryotic cells [53], has been implicated as an important risk factor in sepsis [31]. It is released into the blood circulation, causing abnormal blood coagulation and NETosis [54,55]. In a recent study, abnormal coagulation (confirmed by D-dimer concentration) [56,57] and NETosis (i.e., cfDNA, NET, MPO activity, and NE) [[58], [59], [60], [61]] were also confirmed in the SARS-CoV-2 ICU patient plasma, and the correlation between elevated histone H4 level and blood coagulation [54,62]/NETosis [63,64] was validated. According to previous studies, albumin exhibits the potential to inhibit the histone H4-induced platelet aggregation [29], suggesting albumin as a therapeutic candidate for managing blood coagulation.
We analyzed the plasma of SARS-CoV-2 patients using enzyme-linked immunosorbent assay (ELISA) and found that the level of histone H4 was highly upregulated in the SARS-CoV-2 patient plasma (Figure S1, Supporting Information). From the clinical outcome data, the level of histone H4 was higher in ICU patients than non-ICU cases (Figure S1a, Supporting Information) and in deceased cases than survival cases (Figure S1b, Supporting Information). The computed tomography (CT) images supported such ELISA analysis results, showing a significantly higher lung inflammation in the high-histone H4 cases than low-histone H4 ones (Figure S1c, Supporting Information).
3.2. Preparation and characterization of the PNAB-ginsenosides
Through the analysis of COVID-19 patient plasma samples, we found that (1) serum albumin level was decreased, (2) blood coagulation and NETosis were increased in association with the histone H4 increase, and (3) severe cytokine storm and inflammation existed in blood and lung tissue. Thus, albumin was selected as a carrier for the ginsenoside delivery to address the blood coagulation, NETosis, and the cytokine storm, simultaneously. Based on our previous findings, we designed PNAB-ginsenosides that can provide the following therapeutic benefits. First, the administration of PNAB-ginsenosides can supplement the serum albumin pool of COVID-19 patients [39], which is a widely accepted clinical intervention for treating critically ill patients with hypotension [65,66]. Unlike the conventional preparation methods for albumin nanoparticles that involve irreversible denaturing and cross-linking procedures, PNAB-ginsenosides were fabricated based on reversible hydrophobic interactions, maintaining the single protein structure [67]. Second, the delivery of ginsenosides, steroid glycosides found in ginseng plants (genus Panax), can suppress severe inflammatory responses [32]. Third, PNAB-ginsenosides were functionalized with PEG moiety for improved pharmacokinetic behaviors, such as increased exposure (due to reduced clearance) and prolonged blood circulation time, which may further enhance the efficacy of the developed formulation.
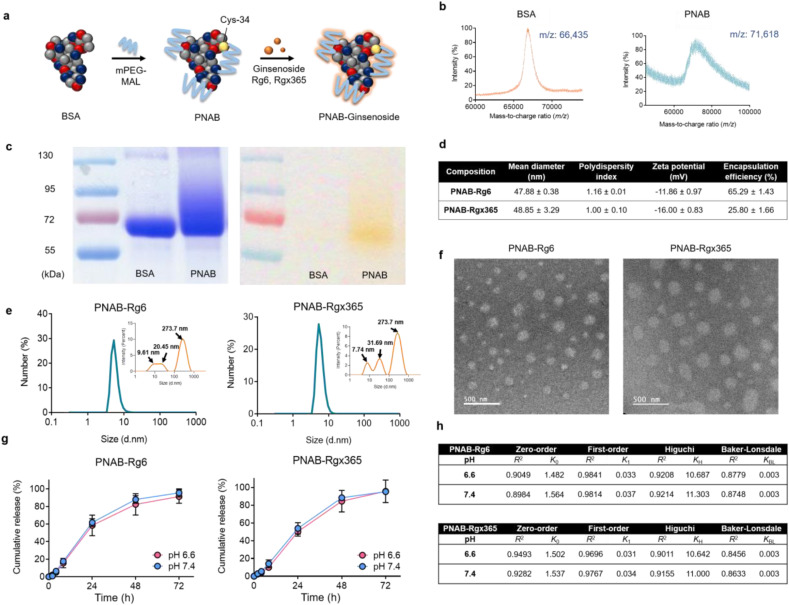
To prepare PNAB-ginsenosides, bovine serum albumin (BSA) was adopted as a core material and modified using monomethoxyPEG maleimide (mPEG-MAL), which can selectively react with thiol groups on BSA surface (Fig. 1 a) [68]. The successful conjugation of PEG was evaluated by using MALDI-TOF, where the mass-to-charge ratio (m/z) values of BSA and PEGylated BSA (BSA-mPEG) were observed to be 66,435 and 71,618, respectively (Fig. 1b). Considering the molecular weight of mPEG-MAL, the molar substitution of PEG-to-BSA of the conjugate is calculated to be 2.5. The introduction of PEG was further confirmed using barium iodide complex (BaI2) staining-assisted SDS-PAGE (Fig. 1c). The broadened band of BSA-mPEG observed in the Coomassie blue staining was colocalized with that in BaI2 staining, indicating the co-existence of protein and PEG in the band. The band broadening could mainly result from the molecular weight distribution of mPEG-MAL.
Fig. 1.
Characterizations of PNAB-Ginsenosides. (a) Scheme illustration of PNAB-ginsenosides. (b) MALDI-TOF mass spectra for BSA (left) and PEGylated Nanoparticle Albumin-Bound (PNAB) (right) at feed ratios of BSA:PEG of 1:2.5. (c) Acrylamide gels stained with Coomassie blue stain (left) and barium iodide stains for PEG (right). (d) Characteristics of PNAB-Rg6 and PNAB-Rgx365 including intensity-weighted mean hydrodynamic diameter, polydispersity index, zeta potentials, and encapsulation efficiency (EE) of PNAB-Rg6 and PNAB-Rgx365. (e) The number-weighted size distribution (inset: intensity-weighted size distribution with modal values of each peak) and (f) transmission electron microscopy (TEM) images of PNAB-Rg6 and PNAB-Rgx365 (scale bar: 500 nm). (g) The controlled-release pattern of PNAB-Rg6 and PNAB-Rgx365 under pathophysiological pH conditions (pH 6.6 for acidic inflammatory interstitial fluid and pH 7.4 for plasma) and (h) mathematical model fitting of the release profiles (zero-order, first-order, Higuchi, and Baker-Lonsdale models). The release constants (k0, k1, kH, and kBL) were estimated using the least-squares regression analysis. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Finally, Rg6 or Rgx365 was adsorbed to BSA-mPEG using ultrasonication, which then self-assembled into nanoparticles, namely, PNAB-Rg6 or PNAB-Rgx365, respectively (Fig. 1a). The intensity-weighted hydrodynamic diameter (IHD) and corresponding polydispersity index values of PNAB-ginsenosides are presented in Fig. 1d. Although the IHDs of the developed formulations were around 50 nm, the number-weighted HDs (NHDs) were below 10 nm (Fig. 1e), which supports the self-assembly phenomenon of albumin in the presence of hydrophobic cargo molecules [38,69,70]. As can be seen in the inset of Fig. 1e, the monomeric and self-assembled nanoparticle albumins are co-existing in the formulation, which explains the high polydispersity index (PDI) values of approximately 1.0 (Fig. 1d). The self-assembled morphology of PNAB-ginsenosides can also be observed in the transmission electron microscopy (TEM) images (Fig. 1f). In our preliminary study, the surface charge of NAB-ginsenosides was measured to be in a range of −18.05 to −20.7 mV. However, PNAB-ginsenosides displayed increased values from −11.86 to −16.00 mV (Fig. 1d), which is due to the shielding effect of the conjugated PEG molecules [71]. The encapsulation efficiency (EE) values of PNAB-Rg6 and PNAB-Rgx365 were 65.29% and 25.80%, respectively (Fig. 1d). The Rg6 release pattern of PNAB-ginsenosides was investigated under pathophysiologically-relevant pH conditions representing the acidity of inflammatory interstitial fluid (pH 6.6) and plasma (pH 7.4) (Fig. 1g). Notably, both formulations displayed not only sustained (up to 72 h) but also complete Rg6 release patterns. As expected from the isoelectric point of BSA (approximately 4.7), the pH dependency was barely observed, implying these formulations can efficiently release Rg6 in the inflammatory tissues, as well as blood vessels. Meanwhile, each release profile was fitted using mathematical models, including zero-order, first-order, Higuchi, and Baker-Lonsdale models, and corresponding release constants (k 0, k 1, k H, and k BL, respectively) were estimated by the least-squares regression (Fig. 1h). Among them, the first-order model exhibited the highest correlation coefficients (R 2) compared to the others, which is in good accordance with the fact that the drug-protein dissociation model is putatively expressed as first-order kinetics [72].
3.3. Increased tissue damage, inflammation, and blood coagulation in severe COVID-19 patients
Increased level of lactate dehydrogenase (LDH), which is an enzymatic indicator reflecting the tissue damage, in ICU patients’ plasma support the clinical data of increased histone H4 level and lung inflammation confirmed by CT images (Table 1 ). The level of D-dimer was highly upregulated in SARS-CoV-2 ICU patients, showing a 10-fold increase compared to the non-ICU case (Table 1). The high level of D-dimer indicates the existence of blood coagulation in response to SARS-CoV-2 viral infection. Furthermore, histone H4 is also known to promote neutrophil extracellular traps (NETs) formation [73].
Table 1.
Baseline characteristics and clinical outcomes of COVID-19 patients admitted to Yeungnam University Hospital. Data are median (IQR), n (%), or n/N (%). p values were calculated by Mann-Whitney U test.
| Total (n = 80) | Non-ICU (n = 60) | ICU (n = 20) | P value | |
|---|---|---|---|---|
| Characteristics | ||||
| Age, y | 55.6 (21–84) | 49.6 (42–58) | 77.5 (65–92) | <0·0001 |
| Sex | ||||
| Men | 35 (43.8) | 29 (48.3) | 6 (30) | |
| Women | 45 (56.2) | 31 (51.7) | 14 (70) | |
| Laboratory finding | ||||
| White blood cell count, × 10⁹/L | 6.5 (4.7–9.3) | 6.1 (4.3–7.8) | 10.2 (7.2–14.1) | <0·0001 |
| Lymphocyte count, × 10⁹/L | 1.2 (0.7–1.5) | 0.6 (0.5–0.8) | 1.1 (0.8–1.4) | <0·0001 |
| Albumin, g/L | 31.8 (28.4–35.6) | 33.4 (29.8–35.8) | 27.4 (24.5–30.2) | <0·0001 |
| Lactate dehydrogenase, U/L | 312 (221.5–421.2) | 255.7 (211.9–322.4) | 607 (401.6–823.7) | <0·0001 |
| D-dimer, μg/mL | 0.9 (0.5–3.5) | 0.6 (0.2–1.1) | 6.1 (2.1–24.2) | <0·0001 |
| Clinical outcomes | ||||
| Remained in hospital | 14 (17.5) | 11 (18.3) | 3 (15) | |
| Discharged | 49 (61.3) | 49 (81.7) | 0 (0) | |
| Died | 17 (21.2) | 0 (0) | 17 (85) | |
In addition, we also found that the blood albumin level was reduced as the severity of symptoms increases (Table 1). The decreased level of serum albumin in deceased individuals was also recently reported in Wuhan, China [74], supporting the patients’ outcomes in Daegu, South Korea case as shown in this study. The dramatically enhanced serum cytokine level, which is associated with cytokine storm, was also observed in the peripheral blood mononuclear cells (PBMCs) of SARS-CoV-2 patients [75].
3.4. PNAB-Rg6 and Rgx365 suppressed histone H4-induced NET by binding to histone H4
We aimed to resolve the three issues, (1) blood coagulation and NETosis, (2) NF-κB-mediated cytokine storm, and (3) sterol regulatory element-binding protein 2 (SREBP2)-mediated inflammasome formation using PNAB-steroidal ginsenoside nanotherapeutics. First, we validated the suppressive effect of PNAB-Rg6 and PNAB-Rgx365 on histone H4 level. For this purpose, we introduced PNAB-Rg6 and PNAB-Rgx365 in the SARS-CoV-2 ICU patients’ plasma and pull-down the compounds. The concentration of histone H4 in the supernatant and PNAB-Rg6 or PNAB-Rgx365-bound portion was measured using ELISA. As shown in Fig. 2 a, the portion of BSA-Rg6- and RSA-Rgx365-bound histone H4 was more than two-fold higher than unbound (suspended in the supernatant) portion. This result shows that the PNAB-Rg6 and PNAB-Rgx365 can capture serum histone H4.
Fig. 2.
PNAB-Rg6 and PNAB-Rgx365 suppressed histone H4-mediated neutrophil extracellular trap (NET) by binding to histone H4. (a) Binding assay of histone H4 to PNAB-ginsenoside complex. PNAB-Rg6 and PNAB-Rgx365 (50 μg/mL) were introduced in SARS-CoV-2 patients' plasma and subsequently precipitated. The histone H4 level in the precipitate (bound to BSA) and supernatant (unbound) was quantified. (b–e) Effect of PNAB-Rg6 and PNAB-Rgx365 (50 μg/mL) in the suppression of NETosis. The suppression of NETosis was validated via changes of (b) cfDNA, (c) NET, (d) MPO activity, and (e) NE (**p < 0.01).
The effect of albumin-ginsenoside in the suppression of NETosis was validated by treating PNAB-Rg6 and PNAB-Rgx365 to SARS-CoV-2 ICU patient-derived neutrophils. Those drugs reduced the levels of cell-free DNA (cfDNA) (Fig. 2b), implying the potential capability of those drugs in the treatment of sepsis. Furthermore, the protein levels related to NETosis were also recovered to normal range, including NET, myeloperoxidase (MPO) activity, and neutrophil elastase (NE) (Fig. 2c–e). These results demonstrate that PNAB-steroidal ginsenosides can effectively suppress the histone H4-mediated NET.
3.5. PNAB-Rg6 and PNAB-Rgx365 ameliorates SARS-CoV-2-mediated cytokine storm
Excessive level of serum cytokine, termed cytokine storm, is a symptom that is commonly observed in SARS-CoV-2 patients [76]. Through the cytokine array analysis, we also confirmed the substantial increase of inflammatory cytokines in SARS-CoV-2 patients' plasma (Fig. 3 a). However, upon the treatment of PNAB-Rg6 and PNAB-Rgx365 in the SARS-CoV-2 patients’ plasma, the level of inflammatory cytokines was significantly reduced (Fig. 3a).
Fig. 3.
PNAB-Rg6 and PNAB-Rgx365 ameliorate SARS-CoV-2-mediated cytokine storm via down-regulation of NF-κB activation. (a) Inhibition of cytokine storm in SARS-CoV-2 patients' plasma after the treatment of PNAB-Rg6 and PNAB-Rgx365 (50 μg/mL, 6 h). (b) Decreased NF-κB activation in SARS-CoV-2 ICU patients' PBMCs upon the PNAB-Rg6 and PNAB-Rgx365 treatment (50 μg/mL, 6 h) (*p < 0.05 and **p < 0.01). (c–h) Decreased cytokine levels in SARS-CoV-2 ICU patients' PBMCs upon the PNAB-Rg6 and PNAB-Rgx365 (50 μg/mL, 6 h) treatment including (c) IL-1β, (d) IL-4, (e) IL-6, (f) IL-8, (g) interferon (IFN)-γ, and (h) tumor necrosis factor (TNF)-α (*p < 0.05 and **p < 0.01). (i) Schematic illustration on the effect of Rgx365 in the suppression of NETosis and cytokine storm.
The PNAB-steroidal ginsenoside nanotherapeutics were also effective in the PBMCs derived from SARS-CoV-2 ICU patients, including acute respiratory distress syndrome (ARDS) and sepsis cases. For example, the treatment of PNAB-Rg6 and PNAB-Rgx365 suppressed the activation of the nuclear factor-κB (NF-κB) signaling pathway, and such suppression was more effective in PNAB-Rgx365 (Fig. 3b). The PNAB-steroidal ginsenoside drugs also reduced the level of inflammatory cytokines produced by PBMCs, such as interleukin (IL)-1β, IL-4, IL-6, IL-8, interferon (IFN)-γ, tumor necrosis factor (TNF)-α (Fig. 3c–h). Upon the treatment of PNAB-Rg6 and PNAB-Rgx365, we could modulate the cytokine storm in SARS-CoV-2 patients’ PBMCs. Overall, the steroidal ginsenosides (i.e. Rg6, Rgx365) regulate SARS-CoV-2-mediated cytokine storm in three ways as follows: (1) The Rgx365 suppresses the NETosis of SARS-CoV-2-infected neutrophils, (2) prohibits the activation of NF-κB signaling pathway which can be directly activated by SARS-CoV-2 virus, and (3) ameliorates overproduction of inflammatory cytokines (Fig. 3i).
3.6. PNAB-Rg6 and PNAB-Rgx365 inhibit SREBP-2-mediated inflammasome formation in PBMCs of SARS-CoV-2 ICU patients
Another critical signature in SARS-CoV-2 patients is the cytokine storm. In this study, we evaluated the cytokine storm issue through two signaling pathways, NF-κB and SREBP2. In the PBMCs derived from SARS-CoV-2 ICU patients, the NF-κB signaling was upregulated, in which NF-κB signaling is known to strongly correlate with cytokine production [77]. We also confirmed that the activated NF-κB signaling correlated with cytokine storm (Fig. 3). SREBP2, which belongs to the downstream signaling of NF-κB, regulates the inflammasome formation upon the infection [78].
The viability of SARS-CoV-2 ICU patients’ PBMCs decreased as a function of time due to the over-activation of inflammatory signaling pathways (Fig. 4 a). We evaluated the effect of PNAB-steroidal ginsenoside in the rescue of PBMC viability. The PNAB-Rg6 and PNAB-Rgx365 could rescue the viability of PBMCs derived from ICU patients (Fig. 4a). According to the analysis, the level of SREBP2 and caspase activities were reduced when PNAB-Rg6 was treated and further reduced when PNAB-Rgx365 was treated (Fig. 4b–d). The SREBP2 is known to be related to inflammasome formation, which is subsequently known to activate caspase-1 activity and promote the production of IL-1β [79]. In addition, the activities of caspase 1 and 3/7 were rescued, which is known to be activated by SREBP2. The real-time polymerase chain reaction (real-time PCR) supports the efficacy of PNAB-Rg6 and PNAB-Rgx365 in terms of the modulation of inflammation-related mRNA expression including SREBP2, NOX2, NRLP3, MCP1, VCAM1, ICAM1, and ELAM1 (Fig. 4e). NOX2 encodes NADPH oxidase 2 (NOX2), which forms reactive oxygen species (ROS). The protein encoded by the NLR family pyrin domain containing 3 (NRLP3) functions as an upstream activator of the NF-κB signaling pathway and regulates inflammation. Monocyte chemoattractant protein 1 (MCP1) is a chemokine regulating monocyte migration and infiltration and is also known to be correlated with the amount of atherosclerosis [80]. VCAM1, ICAM1, and ELAM1 that encode vascular cell adhesion molecule 1 (VCAM-1), intercellular adhesion molecule 1 (ICAM1), and endothelial-leukocyte adhesion molecule 1 (ELAM-1), respectively, and those proteins are related to inflammation-related cell-adhesion. The nanotherapeutics PNAB-Rg6 and PNAB-Rgx365 reduced the expression of those genes.
Fig. 4.
PNAB-Rg6 and PNAB-Rgx365 inhibit SREBP2-mediated inflammasome in PBMCs of SARS-CoV-2 ICU patients. (a) Time-course monitoring of SARS-CoV-2 ICU patients' PBMCs upon the treatment of PNAB-Rg6 and PNAB-Rgx365 (50 μg/mL, 6 h) (***p < 0.001). (b–d) Effect of PNAB-Rg6 and PNAB-Rgx365 (50 μg/mL, 6 h) in the activation level of (b) SREBP2, (c) caspase-1, and (d) caspase-3/7 in SARS-CoV-2 ICU patients' PBMC. (e) Real-time PCR analysis of NETosis- and inflammation-related mRNAs including SREBP2, NOX2, NRLP3, MCP1, VCAM1, ICAM1, and ELAM1 (*p < 0.05 and **<0.01). (f) Potential mechanism of Rg6 and Rgx365 in the suppression of SREBP2-mediated inflammasome.
3.7. PNAB-Rg6 and PNAB-Rgx365 alleviate the blood clot formation and vascular inflammation
In SARS-CoV-2 patients, vascular damage is a critical issue since the vascular disruption can induce tissue damage. The effect of PNAB-Rg6 and PNAB-Rgx365 in the recovery of vascular function was assessed by using an engineered 3D perfusable blood vessel model. To induce vascular inflammation similar to that of SARS-CoV-2 patients, SARS-CoV-2 patients’ plasma was introduced in the engineered blood vessel and incubated for 2 h (Fig. 5 a). Since the plasma of SARS-CoV-2 patients is significantly coagulated, the plasma was diluted 1:2 ratio (plasma: media) before the injection to ensure sufficient fluidity. When the PNAB-Rg6 and PNAB-Rgx365 were treated for 24 h, the vascular permeability was significantly reduced compared to the control group (PBS treated), showing the restoration of barrier function (Fig. 5b and c). Interestingly, the PNAB-Rg6 and PNAB-Rgx365 reduced the blood clot formation (stained with fibrinogen, blue) and vascular inflammation (stained with ICAM-1, green) (Fig. 5d). The efficacy of PNAB-Rg6 and PNAB-Rgx365 in the recovery of vascular function was also confirmed in the cytokine (TNF-α)-mediated vascular disruption model (Figure S3, Supporting Information). Considering the severe blood coagulation and blood vessel damage issues in SARS-CoV-2 patients, the PNAB-Rg6 and PNAB-Rgx365 exhibited a promising ability in the restoration of vascular functions.
Fig. 5.
PNAB-Rg6 and PNAB-Rgx365 alleviate blood clot formation and vascular inflammation. (a) Schematic illustration of the efficacy test of PNAB-Rg6 and PNAB-Rgx365 for the recovery of the vascular function using a 3D engineered blood vessel model. SARS-CoV-2 patients' plasma was introduced for vascular disruption. After 2 h of incubation with the SARS-CoV-2 patients' plasma, PNAB-Rg6 and PNAB-Rgx365 were treated for 24 h. (b) Molecular transport of 40 kDa FITC-Dextran out of the engineered blood vessel. (c) Quantified transendothelial permeability after the treatment of PNAB-Rg6 and PNAB-Rgx365 (**p < 0.01). (d) Immunofluorescence images of an engineered blood vessel after the treatment of PNAB-Rg6 and PNAB-Rgx365. Red: F-actin, green: ICAM-1, and blue: fibrinogen. (e) The effect of PNAB-Rg6 and PNAB-Rgx365 in the alleviation of blood clot formation and vascular inflammation. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.8. PNAB-Rg6 and PNAB-Rgx365 rescue mortality of septic mice models via inhibition of NF-κB and SREBP2 signaling pathways
The severe SARS-CoV-2 patients experience sepsis since the extraordinarily high concentration of cytokines causes vascular damage and subsequent tissue failure. To address such an issue, the efficacy of the PNAB-Rg6 and PNAB-Rgx365 was confirmed by using septic mice models. The sepsis of mice model was induced by cecal ligation and puncture (CLP) technique, which is the most widely used method for preparing septic animal models [46]. The intravenously injected PNAB-Rg6 and PNAB-Rgx365 rescued the survival rate of CLP-operated mice to 40% and 60%, respectively (Fig. 6 a and Figure S4, Supporting Information). (in Fig. 6a, the survival rate after the PNAB-Rg6 administration was noted as 40%) The histological analysis of mice lung tissue showed reduced blood leakage and inflammation after the administration of PNAB-Rg6 and PNAB-Rgx365 (Fig. 6b) and the efficacy was higher in the case of PNAB-Rgx365 administration. The treatment of those two drugs restored the transendothelial permeability, lung ICAM-1 expression (Fig. 6c), implying the reduced vascular inflammation. In terms of immune activity, the leukocyte and neutrophil migration in bronchoalveolar lavage (BAL) was reduced (Fig. 6c), suggesting the suppression of hyperactivation of the immune system. The NF-κB and SREBP2 signalings, which are known to regulate inflammation process, were recovered to normal levels after the administration of PNAB-Rg6 and PNAB-Rgx365 (Fig. 6c). As a result, the markers for tissue damage including C-reactive protein (CRP), lactate dehydrogenase (LDH), alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatinine were decreased after the administration of PNAB-Rg6 and PNAB-Rgx365 (Fig. 6d). The expression level of inflammatory cytokines including IL-1β, IL-6, IL-8, and IFN-γ, suppressing the cytokine storm (Fig. 6e and Figure S5, Supporting Information). We also evaluated the reactive oxygen species (ROS) level after the administration of PNAB-Rg6 and PNAB-Rgx365 in the CLP-operated mouse model. We found that the administration of PNAB-Rg6 and PNAB-Rgx365 could suppress the ROS level. The previous studies also suggested that the Rgx365 can prevent pulmonary injury upon particulate matter exposure by inhibiting the ROS generation [33]. Collectively, ROS assay in this study in conjunction with the previous studies suggests that the PNAB-ginsenoside can prevent tissue damage in the septic mouse model by suppressing ROS generation. The efficacy validation of PNAB-Rg6 and PNAB-Rgx365 using animal models clearly displayed the capability of those drugs for the prevention of infection-mediated sepsis.
Fig. 6.
PNAB-Rg6 and PNAB-Rgx365 rescue the survival rate of septic mice models and prevent the cytokine storm and tissue damage. (a) The time-course survival rate of CLP-operated septic mice models after the treatment of PNAB-Rg6 and PNAB-Rgx365. PNAB-Rg6 and PNAB-Rgx365 (5 mg/kg) were delivered intravenously 24 h after the CLP-operation. (b) Histological analysis of mice lung tissue after the treatment of PNAB-Rg6 and PNAB-Rgx365. (White scale bar = 75 μm). (c) Pathology-related signatures including vascular permeability, lung ICAM-1 expression, and leukocyte and neutrophil migration in bronchoalveolar lavage (BAL), and NF-κB and SREBP2 activities in lung tissue. (*p < 0.05). (d) Expression of tissue damage markers including C-reactive protein (CRP), lactate dehydrogenase (LDH), alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatinine (*p < 0.05). (e) Expression of cytokines such as IL-1β, IL-6, IL-8, and INF-γ (*p < 0.05). (f) Quantification of ROS level in mouse lung endothelial cells (**p < 0.01).
4. Discussion
It has been reported that in response to the decreased serum albumin level, the insulin-induced gene-1 protein (INSIG1) dissociates from the SREBP2-SCAP complex, and the SREBP2-SCAP complex translocates to Golgi apparatus [81]. Then the sphingosine-1-phosphate (S1P) and S2P dissociates C-term and N-term of SREBP2, and the SREBP2 N-term translocates to the nucleus and activates the inflammasome [81]. It is supposed that steroidal ginsenosides, Rg6 and Rgx365, interrupt such cleavage of SREBP2 into C- and N-term and ultimately inhibit inflammasome and sepsis (Fig. 4f). Structurally, Rg6 has a similar molecular structure with betulin, which is known as an SREBP2 suppressor (Figure S2, Supporting Information) [82]. Rgx365 is composed of Rg4 and Rg6, of which Rg4 is the main constituent, therefore we can consider that the Rgx365 component has a similar molecular structure with botulin (Figure S2, Supporting Information) [33,82].
Previously, it was shown that convalescent plasma direct neutralization of the virus, control the overactive immune systems, such as cytokine storm, Th1/Th17 ratio, and complement activation, and immunomodulate hypercoagulable state [25]. In particular, the efficacy of treatment in COVID-19 patients injected with albumin and convalescent plasma was demonstrated, in which albumin was shown to inhibit platelet aggregation by binding to histone [29]. It is supposed that another role of albumin is inhibition of platelet aggregation, pulmonary hemorrhage, and endothelial necrosis by binding with extracellular histones secreted by a viral infection in severe COVID-19 patients [40]. Overall, we could modulate the apparent clinical signatures of SARS-CoV-2 patients using PNAB-steroidal ginsenoside nanotherapeutics.
In this study, we fabricated steroidal ginsenoside saponin-bound albumin nanoparticles for the following reasons: 1) To make a long-acting steroidal ginsenoside for greater immunosuppressive effect, 2) To control organ damage and blood clotting mechanism due to excessive inflammatory reaction by removing endotoxins such as histone and inflammatory cytokines released in infectious diseases, 3) To prevent severe patients from dying of hypotension as a result of septic shock. The development of albumin formulated nanotherapeutics is of great value as a symptomatic treatment and is essential for the improvement of morbidity and mortality prior to the antiviral agent development in the current SARS-CoV-2 pandemic situation.
Numerous studies highlight the importance of materials science and nanotechnology in the field of antiviral research, proposing insightful approaches for the treatment of COVID-19 by means of innovative methodologies [[83], [84], [85]]. In this study, we presented a novel ginsenoside delivery system via NAB technology. The PNAB-Rg6 and PNAB-Rgx365 showed marked efficacy in both in vitro and in vivo models. First, the PBMCs isolated from severe SARS-CoV-2 patients demonstrated the capability for suppression of histone H4-induced NET, cytokine storm, and inflammasome. Second, the engineered blood vessel model demonstrated the effect of those drugs for the suppression of blood clot formation and vascular inflammation. The reduced blood clot formation is presumably due to the reduced fibronectin expression after the drug treatment (Fig. 5). Third, the CLP-operated septic mice model confirmed the prescribed in vitro model-based results, showing rescued survival rate, prevention of tissue damage, and suppressed cytokine storm. The confirmation through the in vitro and in vivo models supports the promising capability of these drugs as therapeutics for the severe SARS-CoV-2 patients.
5. Conclusion
In this study, we demonstrated the suppression of NETosis and cytokine storm in SARS-CoV-2 patients’ plasma and patients-derived PBMCs by using PNAB-steroidal ginsenoside nanotherapeutics. The drugs could suppress NETosis via the modulation of serum histone H4 level and cytokine storm through downregulation of NF-κB and SREBP2 signaling pathways. Furthermore, the drugs could suppress blood clot formation and vascular inflammation. The animal model-based validation showed promising efficacy in terms of the rescued survival rate, inhibited tissue damage, and suppressed cytokine storm. Considering the absence of the approved therapeutic drugs for SARS-CoV-2 pneumonia, it is anticipated that the albumin-ginsenoside-based drugs can alleviate the symptoms such as blood coagulation and cytokine storm in severe COVID-19 patients.
Credit author statement
Hee Ho Park, contributed equally to this study. Hyelim Kim, contributed equally to this study. Han Sol Lee, contributed equally to this study. Eun U Seo, contributed equally to this study. Ji-Eun Kim contributed equally to this study. Hee Ho Park: Conceptualization, Investigation, Writing – review & editing. Hyelim Kim: Methodology, Investigation. Han Sol Lee: Methodology, Investigation. Eun U Seo: Methodology, Investigation. Ji-Eun Kim: Methodology, Investigation. Jee-Hyun Lee: Resources. Yong-Hyeon Mun: Methodology, Investigation. So-Yeol Yoo: Methodology, Investigation. Jiseon An: Methodology, Investigation. Mi-Young Yun: Methodology. Nae-Won Kang: Investigation. Dea-Duk Kim: Supervision. Dong Hee Na: Supervision. Kyung Soo Hong: Resources. Jong Geol Jang: Resources. June Hong Ahn: Resources. Jong-Sup Bae: Supervision, Funding acquisition. Gyu Yong Song: Supervision, Methodology, Funding acquisition. Jae-Young Lee: Conceptualization, Supervision, Methodology, Funding acquisition, Writing – original draft. Hong Nam Kim: Conceptualization, Methodology, Supervision, Funding acquisition, Writing – original draft, Writing – review & editing. Wonhwa Lee: Conceptualization, Methodology, Validation, Supervision, Funding acquisition, Writing – original draft, review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIT) (grant no. 2018R1A2A3075013, 2021R1C1C1009320, 2018M3A7B4071204, 2017R1A5A2015385, 2020R1A4A3078645, 2020R1A2C1004131, 2021R1C1C2006896, 2020R1A4A4079817). This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI15C0001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2021.120827.
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to legal or ethical reasons.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vincent J.-L., Taccone F.S. Understanding pathways to death in patients with COVID-19. Lancet Respir Med. 2020;8(5):430–432. doi: 10.1016/S2213-2600(20)30165-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danese S., Cecconi M., Spinelli A. Management of IBD during the COVID-19 outbreak: resetting clinical priorities. Nat. Rev. Gastroenterol. Hepatol. 2020;17(5):253–255. doi: 10.1038/s41575-020-0294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Y.-H., Cai L., Cheng Z.-S., Cheng H., Deng T., Fan Y.-P., Fang C., Huang D., Huang L.-Q., Huang Q. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil Med Res. 2020;7(1):4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.She J., Jiang J., Ye L., Hu L., Bai C., Song Y. 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin. Transl. Med. 2020;9(1):1–7. doi: 10.1186/s40169-020-00271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le T.T., Andreadakis Z., Kumar A., Roman R.G., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(5):305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 7.Hong K.S., Lee K.H., Chung J.H., Shin K.-C., Choi E.Y., Jin H.J., Jang J.G., Lee W., Ahn J.H. Clinical features and outcomes of 98 patients hospitalized with SARS-CoV-2 infection in Daegu, South Korea: a brief descriptive study. Yonsei Med. J. 2020;61(5):431. doi: 10.3349/ymj.2020.61.5.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang J.G., Hur J., Choi E.Y., Hong K.S., Lee W., Ahn J.H. Prognostic factors for severe coronavirus disease 2019 in Daegu, Korea. J. Kor. Med. Sci. 2020;35(23) doi: 10.3346/jkms.2020.35.e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B., Zhou X., Qiu Y., Feng F., Feng J., Jia Y., Zhu H., Hu K., Liu J., Liu Z. MedRxiv; 2020. Clinical Characteristics of 82 Death Cases with COVID-19. [Google Scholar]
- 10.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and multiorgan response. Curr. Probl. Cardiol. 2020:100618. doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee L.Y., Cazier J.B., Starkey T., Turnbull C., Kerr R., Middleton G., U.C.C.M.P. Team COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: a prospective cohort study. Lancet. 2020:1919–1926. doi: 10.1016/S0140-6736(20)31173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehra M.R., Desai S.S., Kuy S., Henry T.D., Patel A.N. N Engl J Med; 2020. Cardiovascular disease, drug therapy, and mortality in COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020:802–810. doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell C.D., Millar J.E., Baillie J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geleris J., Sun Y., Platt J., Zucker J., Baldwin M., Hripcsak G., Labella A., Manson D., Kubin C., Barr R.G. N Engl J Med; 2020. Observational study of hydroxychloroquine in hospitalized patients with Covid-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahase E. BMJ; 2020. Covid-19: what Treatments Are Being Investigated? [DOI] [PubMed] [Google Scholar]
- 17.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant W.B., Lahore H., McDonnell S.L., Baggerly C.A., French C.B., Aliano J.L., Bhattoa H.P. Evidence that vitamin D supplementation could reduce risk of influenza and COVID-19 infections and deaths. Nutrients. 2020;12(4):988. doi: 10.3390/nu12040988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395(10237):1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Organization W.H. World Health Organization; 2020. Clinical Management of Severe Acute Respiratory Infection when Novel Coronavirus ( nCoV) Infection Is Suspected: Interim Guidance, 25 January 2020. [Google Scholar]
- 21.Delaney J.W., Pinto R., Long J., Lamontagne F., Adhikari N.K., Kumar A., Marshall J.C., Cook D.J., Jouvet P., Ferguson N.D. The influence of corticosteroid treatment on the outcome of influenza A (H1N1pdm09)-related critical illness. Crit. Care. 2016;20(1):75. doi: 10.1186/s13054-016-1230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodrigo C., Leonardi‐Bee J., Nguyen‐Van‐Tam J., Lim W.S. Corticosteroids as adjunctive therapy in the treatment of influenza. Cochrane Database Syst. Rev. 2016;3 doi: 10.1002/14651858.CD010406.pub2. [DOI] [PubMed] [Google Scholar]
- 23.Soy M., Keser G., Atagündüz P., Tabak F., Atagündüz I., Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020:1. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20(4):400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rojas M., Rodríguez Y., Monsalve D.M., Acosta-Ampudia Y., Camacho B., Gallo J.E., Rojas-Villarraga A., Ramírez-Santana C., Díaz-Coronado J.C., Manrique R. Convalescent plasma in Covid-19: possible mechanisms of action. Autoimmun. Rev. 2020:102554. doi: 10.1016/j.autrev.2020.102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson J., Schauer J., Bryant S., Graves C.R. Case Rep Women's Health; 2020. The Use of Convalescent Plasma Therapy and Remdesivir in the Successful Management of a Critically Ill Obstetric Patient with Novel Coronavirus 2019 Infection: A Case Report. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. J. Am. Med. Assoc. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valk S.J., Piechotta V., Chai K.L., Doree C., Monsef I., Wood E.M., Lamikanra A., Kimber C., McQuilten Z., So-Osman C. Convalescent plasma or hyperimmune immunoglobulin for people with COVID‐19: a rapid review. Cochrane Database Syst. Rev. 2020;5 doi: 10.1002/14651858.CD013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam F.W., Cruz M.A., Leung H.-C.E., Parikh K.S., Smith C.W., Rumbaut R.E. Histone induced platelet aggregation is inhibited by normal albumin. Thromb. Res. 2013;132(1):69–76. doi: 10.1016/j.thromres.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 30.Ekaney M.L., Otto G.P., Sossdorf M., Sponholz C., Boehringer M., Loesche W., Rittirsch D., Wilharm A., Kurzai O., Bauer M. Impact of plasma histones in human sepsis and their contribution to cellular injury and inflammation. Crit. Care. 2014;18(5):543. doi: 10.1186/s13054-014-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J., Zhang X., Pelayo R., Monestier M., Ammollo C.T., Semeraro F., Taylor F.B., Esmon N.L., Lupu F., Esmon C.T. Extracellular histones are major mediators of death in sepsis. Nat. Med. 2009;15(11):1318–1321. doi: 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paik S., Choe J.H., Choi G.-E., Kim J.-E., Kim J.-M., Song G.Y., Jo E.-K. Rg6, a rare ginsenoside, inhibits systemic inflammation through the induction of interleukin-10 and microRNA-146a. Sci. Rep. 2019;9(1):1–15. doi: 10.1038/s41598-019-40690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee W., Ku S.-K., Kim J.-E., Cho S.-H., Song G.-Y., Bae J.-S. Inhibitory effects of protopanaxatriol type ginsenoside fraction (Rgx365) on particulate matter-induced pulmonary injury. J. Toxicol. Environ. Health. 2019;82(5):338–350. doi: 10.1080/15287394.2019.1596183. [DOI] [PubMed] [Google Scholar]
- 34.Choi Y.H., Han H.-K. Nanomedicines: current status and future perspectives in aspect of drug delivery and pharmacokinetics. J Pharm Investig. 2018;48(1):43–60. doi: 10.1007/s40005-017-0370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawkins M.J., Soon-Shiong P., Desai N. Protein nanoparticles as drug carriers in clinical medicine. Adv. Drug Deliv. Rev. 2008;60(8):876–885. doi: 10.1016/j.addr.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 36.Chen Q., Liu Z. Albumin carriers for cancer theranostics: a conventional platform with new promise. Adv. Mater. 2016;28(47):10557–10566. doi: 10.1002/adma.201600038. [DOI] [PubMed] [Google Scholar]
- 37.Chen Q., Wang X., Wang C., Feng L., Li Y., Liu Z. Drug-induced self-assembly of modified albumins as nano-theranostics for tumor-targeted combination therapy. ACS Nano. 2015;9(5):5223–5233. doi: 10.1021/acsnano.5b00640. [DOI] [PubMed] [Google Scholar]
- 38.Cortes J., Saura C. Nanoparticle albumin-bound (nab™)-paclitaxel: improving efficacy and tolerability by targeted drug delivery in metastatic breast cancer. Eur. J. Canc. 2010;8(1):1–10. [Google Scholar]
- 39.Chen T.-A., Tsao Y.-C., Chen A., Lo G.-H., Lin C.-K., Yu H.-C., Cheng L.-C., Hsu P.-I., Tsai W.-L. Effect of intravenous albumin on endotoxin removal, cytokines, and nitric oxide production in patients with cirrhosis and spontaneous bacterial peritonitis. Scand. J. Gastroenterol. 2009;44(5):619–625. doi: 10.1080/00365520902719273. [DOI] [PubMed] [Google Scholar]
- 40.Ashar H.K., Mueller N.C., Rudd J.M., Snider T.A., Achanta M., Prasanthi M., Pulavendran S., Thomas P.G., Ramachandran A., Malayer J.R. The role of extracellular histones in influenza virus pathogenesis. Am. J. Pathol. 2018;188(1):135–148. doi: 10.1016/j.ajpath.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Høstmark A.T., Tomten S.E., Berg J.E. Serum albumin and blood pressure: a population-based, cross-sectional study. J. Hypertens. 2005;23(4):725–730. doi: 10.1097/01.hjh.0000163139.44094.1d. [DOI] [PubMed] [Google Scholar]
- 42.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.-D., Coopersmith C.M. The third international consensus definitions for sepsis and septic shock (Sepsis-3) J. Am. Med. Assoc. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hazeldine J., Harris P., Chapple I.L., Grant M., Greenwood H., Livesey A., Sapey E., Lord J.M. Impaired neutrophil extracellular trap formation: a novel defect in the innate immune system of aged individuals. Aging Cell. 2014;13(4):690–698. doi: 10.1111/acel.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J., Zhou P., Wang W., Sun A., Guo F. RelB, together with RelA, sustains cell survival and confers proteasome inhibitor sensitivity of chronic lymphocytic leukemia cells from bone marrow. J. Mol. Med. 2014;92(1):77–92. doi: 10.1007/s00109-013-1081-6. [DOI] [PubMed] [Google Scholar]
- 45.Kim J.A., Kim H.N., Im S.K., Chung S., Kang J.Y., Choi N. Collagen-based brain microvasculature model in vitro using three-dimensional printed template. Biomicrofluidics. 2015;9(2) doi: 10.1063/1.4917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rittirsch D., Huber-Lang M.S., Flierl M.A., Ward P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009;4(1):31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun F., Xiao G., Qu Z. Murine bronchoalveolar lavage. Bio Protoc. 2017;7(10) doi: 10.21769/BioProtoc.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee W., Seo J., Kwak S., Park E.J., Na D.H., Kim S., Lee Y.M., Kim I.S., Bae J.S. A double-chambered protein nanocage loaded with thrombin receptor agonist peptide (TRAP) and gamma-carboxyglutamic acid of protein C (PC-Gla) for sepsis treatment. Adv. Mater. 2015;27(42):6637–6643. doi: 10.1002/adma.201503093. [DOI] [PubMed] [Google Scholar]
- 49.Levi M., Thachil J., Iba T., Levy J.H. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020;7(6):e438. doi: 10.1016/S2352-3026(20)30145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnes B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Daßler-Plenker J., Guerci P., Huynh C., Knight J.S. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J. Exp. Med. 2020;217(6) doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020:1–12. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fyodorov D.V., Zhou B.-R., Skoultchi A.I., Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol. 2018;19(3):192. doi: 10.1038/nrm.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barranco-Medina S., Pozzi N., Vogt A.D., Di Cera E. Histone H4 promotes prothrombin autoactivation. J. Biol. Chem. 2013;288(50):35749–35757. doi: 10.1074/jbc.M113.509786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu N.-F., Jiang L., Zhu B., Yang D.-G., Zheng R.-Q., Shao J., Yuan J., Xi X.-M. Elevated plasma histone H4 levels are an important risk factor in the development of septic cardiomyopathy. Balkan Med. J. 2020;37(2):72. doi: 10.4274/balkanmedj.galenos.2019.2019.8.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vidali S., Morosetti D., Cossu E., Luisi M.L.E., Pancani S., Semeraro V., Consales G. D-dimer as an indicator of prognosis in SARS-CoV-2 infection: a systematic review. ERJ Open Res. 2020;6(2) doi: 10.1183/23120541.00260-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao Y., Cao J., Wang Q., Shi Q., Liu K., Luo Z., Chen X., Chen S., Yu K., Huang Z. D-dimer as a biomarker for disease severity and mortality in COVID-19 patients: a case control study. J Intensive Care. 2020;8(1):1–11. doi: 10.1186/s40560-020-00466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veras F.P., Pontelli M.C., Silva C.M., Toller-Kawahisa J.E., de Lima M., Nascimento D.C., Schneider A.H., Caetité D., Tavares L.A., Paiva I.M. SARS-CoV-2–triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217(12) doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skendros P., Mitsios A., Chrysanthopoulou A., Mastellos D.C., Metallidis S., Rafailidis P., Ntinopoulou M., Sertaridou E., Tsironidou V., Tsigalou C. Complement and tissue factor–enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020;130(11):6151–6157. doi: 10.1172/JCI141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee Y.Y., Park H.H., Park W., Kim H., Jang J.G., Hong K.S., Lee J.-Y., Seo H.S., Na D.H., Kim T.-H. Long-acting nanoparticulate DNase-1 for effective suppression of SARS-CoV-2-mediated neutrophil activities and cytokine storm. Biomaterials. 2021;267:120389. doi: 10.1016/j.biomaterials.2020.120389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arcanjo A., Logullo J., Menezes C.C.B., Giangiarulo T.C.d.S.C., Dos Reis M.C., de Castro G.M.M., da Silva Fontes Y., Todeschini A.R., Freire-de-Lima L., Decoté-Ricardo D. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19) Sci. Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-76781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vulliamy P., Gillespie S., Armstrong P.C., Allan H.E., Warner T.D., Brohi K. Histone H4 induces platelet ballooning and microparticle release during trauma hemorrhage. Proc. Natl. Acad. Sci. U.S.A. 2019;116(35):17444–17449. doi: 10.1073/pnas.1904978116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsieh I.-N., White M., Hoeksema M., Deluna X., Hartshorn K. Histone H4 potentiates neutrophil inflammatory responses to influenza A virus: down-modulation by H4 binding to C-reactive protein and Surfactant protein D. PloS One. 2021;16(2) doi: 10.1371/journal.pone.0247605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbu E.A., Dominical V.M., Mendelsohn L., Thein S.L. Detection and quantification of histone H4 citrullination in early NETosis with image flow cytometry version 4. Front. Immunol. 2020;11:1335. doi: 10.3389/fimmu.2020.01335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vasques F., Duscio E., Romitti F., Pasticci I., Caironi P., Meessen J., Latini R., Cressoni M., Camporota L., Pesenti A. Septic shock-3 vs 2: an analysis of the ALBIOS study. Crit. Care. 2018;22(1):237. doi: 10.1186/s13054-018-2169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin L., Dubovetsky D., Louzon-Lynch P. Implementation of an algorithm utilizing saline versus albumin for the treatment of intradialytic hypotension. Ann. Pharmacother. 2019;53(2):159–164. doi: 10.1177/1060028018801024. [DOI] [PubMed] [Google Scholar]
- 67.Kim D.H., Lee H.S., Kwon T.-W., Han Y.-M., Kang N.-W., Lee M.Y., Kim D.-D., Kim M.G., Lee J.-Y. Single enzyme nanoparticle, an effective tool for enzyme replacement therapy. Arch Pharm. Res. (Seoul) 2020:1–21. doi: 10.1007/s12272-020-01216-3. [DOI] [PubMed] [Google Scholar]
- 68.Mehtala J.G., Kulczar C., Lavan M., Knipp G., Wei A. Cys34-PEGylated human serum albumin for drug binding and delivery. Bioconjugate Chem. 2015;26(5):941–949. doi: 10.1021/acs.bioconjchem.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butterfield J.T., Kim H., Knauer D.J., Nevala W.K., Markovic S.N. Identification of a peptide-peptide binding motif in the coating of nab-paclitaxel nanoparticles with clinical antibodies: bevacizumab, rituximab, and trastuzumab. Sci. Rep. 2017;7(1):1–9. doi: 10.1038/s41598-017-15251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Q., Chen J., Liang C., Feng L., Dong Z., Song X., Song G., Liu Z. Drug-induced co-assembly of albumin/catalase as smart nano-theranostics for deep intra-tumoral penetration, hypoxia relieve, and synergistic combination therapy. J. Contr. Release. 2017;263:79–89. doi: 10.1016/j.jconrel.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 71.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schiel J.E., Ohnmacht C.M., Hage D.S. Measurement of drug-protein dissociation rates by high-performance affinity chromatography and peak profiling. Anal. Chem. 2009;81(11):4320–4333. doi: 10.1021/ac9000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daigo K., Takamatsu Y., Hamakubo T. The protective effect against extracellular histones afforded by long-pentraxin PTX3 as a regulator of NETs. Front. Immunol. 2016;7:344. doi: 10.3389/fimmu.2016.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park H.H., Kim H.N., Kim H., Yoo Y., Choi E.Y., Bae J.-S., Lee W. Acetylated K676 TGFBIp as a severity diagnostic blood biomarker for SARS-CoV-2 pneumonia. Sci Adv. 2020:eabc1564. doi: 10.1126/sciadv.abc1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Q., Zhou Y.-h., Yang Z.-q. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 2016;13(1):3–10. doi: 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiao H., Lu M., Lin T.Y., Chen Z., Chen G., Wang W.-C., Marin T., Shentu T.-p., Wen L., Gongol B. Sterol regulatory element binding protein 2 activation of NLRP3 inflammasome in endothelium mediates hemodynamic-induced atherosclerosis susceptibility. Circulation. 2013;128(6):632–642. doi: 10.1161/CIRCULATIONAHA.113.002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 2009;10(3):241. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Namiki M., Kawashima S., Yamashita T., Ozaki M., Hirase T., Ishida T., Inoue N., Hirata K.-i., Matsukawa A., Morishita R. Local overexpression of monocyte chemoattractant protein-1 at vessel wall induces infiltration of macrophages and formation of atherosclerotic lesion: synergism with hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2002;22(1):115–120. doi: 10.1161/hq0102.102278. [DOI] [PubMed] [Google Scholar]
- 81.Kusnadi A., Park S.H., Yuan R., Pannellini T., Giannopoulou E., Oliver D., Lu T., Park-Min K.H., Ivashkiv L.B. The cytokine TNF promotes transcription factor SREBP activity and binding to inflammatory genes to activate macrophages and limit tissue repair. Immunity. 2019;51(2):241–257 e9. doi: 10.1016/j.immuni.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang J.J., Li J.G., Qi W., Qiu W.W., Li P.S., Li B.L., Song B.L. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metabol. 2011;13(1):44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Z., Tang Z., Farokhzad N., Chen T., Tao W. Sensitive, rapid, low-cost, and multiplexed COVID-19 monitoring by the wireless telemedicine platform. Matter. 2020;3(6):1818–1820. doi: 10.1016/j.matt.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang Z., Zhang X., Shu Y., Guo M., Zhang H., Tao W. Insights from nanotechnology in COVID-19 treatment. Nano Today. 2021;36:101019. doi: 10.1016/j.nantod.2020.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tang Z., Kong N., Zhang X., Liu Y., Hu P., Mou S., Liljeström P., Shi J., Tan W., Kim J.S. A materials-science perspective on tackling COVID-19. Nat Rev Mater. 2020;5(11):847–860. doi: 10.1038/s41578-020-00247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings cannot be shared at this time due to legal or ethical reasons.