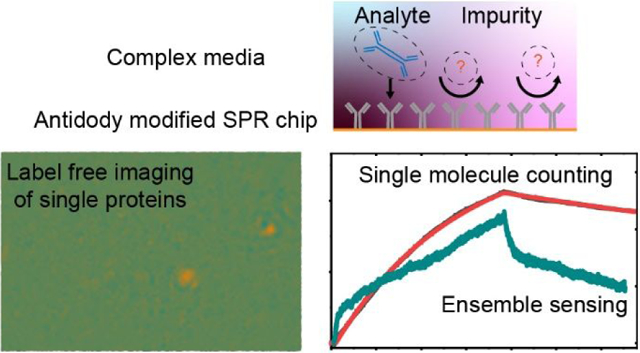
Abstract
Measuring molecular binding is critical for understanding molecule-scale biological processes and screening drugs. Label free detection technologies, such as surface plasmon resonance (SPR), has been developed for analyzing the analytes in their natural forms. However, the specificity of these methods is solely relying on surface chemistry, and often have the nonspecific binding issues when working with samples in complex media. Herein, we show that single molecule based measurement can distinct specific and nonspecific binding processes by quantifying the mass and binding dynamics of individual bound analyte molecules, thus allowing the binding kinetic analysis in complex media such as serum. In addition, this single molecule imaging is realized in a commonly used Kretschmann prism coupled SPR system, thus providing a convenient solution to realize high resolution imaging on widely used prism coupled SPR systems.
Keywords: Surface plasmon resonance, Plasmonic scattering imaging, Single proteins, Molecule interaction, Binding kinetics
Graphical Abstract
Measuring molecular binding is a central task in understanding molecular-scale biological processes1, 2, and screening drugs3, 4. To avoid the effect of labeling on intrinsic properties of molecules, label-free detection technology, such as surface plasmon resonance (SPR), has been developed for binding kinetic analysis5–7. However, label free sensor is sensitive to every objects within the detection range8, resulting in nonspecific binding signals, which is a long-standing issue for SPR measurement9–11. A reference channel is normally needed in ensemble SPR sensor to subtract out the nonspecific binding signal and interference of volatile experimental conditions, such as temperature12.
Recently, a single molecule counting approach using gold nanoparticles as labels is developed for the detection of specific protein binding, making the measurement immune to thermal or mechanical drifts13–16, a common issue in the ensemble label free sensor. Moreover, it has been demonstrated that oil-immersion objective based SPR microscopy (SPRM) combined with antibody conjugated gold nanoparticles can be used to directly differentiate nonspecific binding from specific binding by analyzing the free energy profiles of single molecules17. However, the introduce of gold nanoparticles sacrifices the main advantage of label free detection, measuring the targets in their natural forms. Besides, the ~100 nm gold nanoparticles occupy large imaging volume, limiting the system dynamic range, and may alter the analyte diffusion rate thus making this approach unsuitable for binding kinetic analysis.
Here we show that single molecule imaging without gold nanoparticle labeling can not only directly distinct specific and nonspecific binding processes based on the different binding behaviors, but also maintain the wide dynamic range of ensemble SPR and distinct strong nonspecific binding using molecular weight of analytes as barcode, which is beyond the capability of labelling approach. We demonstrate that the single molecule imaging can be realized in a Kretchmann prism coupled SPR sensor with recently developed plasmonic scattering microscopy (PSM) scheme18. The system can differentiate the individual bound proteins with different molecular weights, and also reveal different binding behaviors of specific and nonspecific binding. We then show that this approach allows the label free binding kinetic analysis in complex media such as serum. In addition, we also demonstrate that this prism coupled PSM shows better measurement reproducibility than objective based SPR that that this prism coupled PSM shows better measurement reproducibility than objective based SPR imaging system owing to the rigid mechanical structure of prism configuration, which improves the system stability and simplifies the instrument operation.
RESULTS
Principles of prism coupled plasmonic scattering imaging:
We excite surface plasmonic waves with a typical Kretschmann prism configuration, and image plasmonic waves scattered by the gold surface roughness and analytes with an objective and a camera placed on top of the sample, additional to recording the ensemble SPR signals from the bottom via another camera (Figure 1a; Figure S1 for details). PSM does not measure the strong reflection light, thus allowing incident light intensity up to 5 kW/cm2 for imaging single unlabeled proteins. The PSM image is given by |Eb + Es|2 = |Eb|2 + 2|Eb||Es|cos(θ) + |Es|2, where Eb and Es are light scattered by surface roughness and analyte, respectively, and θ is the phase difference. For thermally vacuum-evaporated gold film, PSM image intensity usually scales according to |Es|2 ~ d6 for analytes with diameter larger than 100 nm, and 2|Eb||Es|cos(θ) ~ d3, or the mass, for analytes with smaller diameter due to the condition of |Eb|2 >>|Es|2, where d is the analyte diameter. Protein molecules are typically smaller than 30 nm in diameter, so the cubic power law can be considered as a good description of the PSM image intensity for single proteins.
Figure 1.
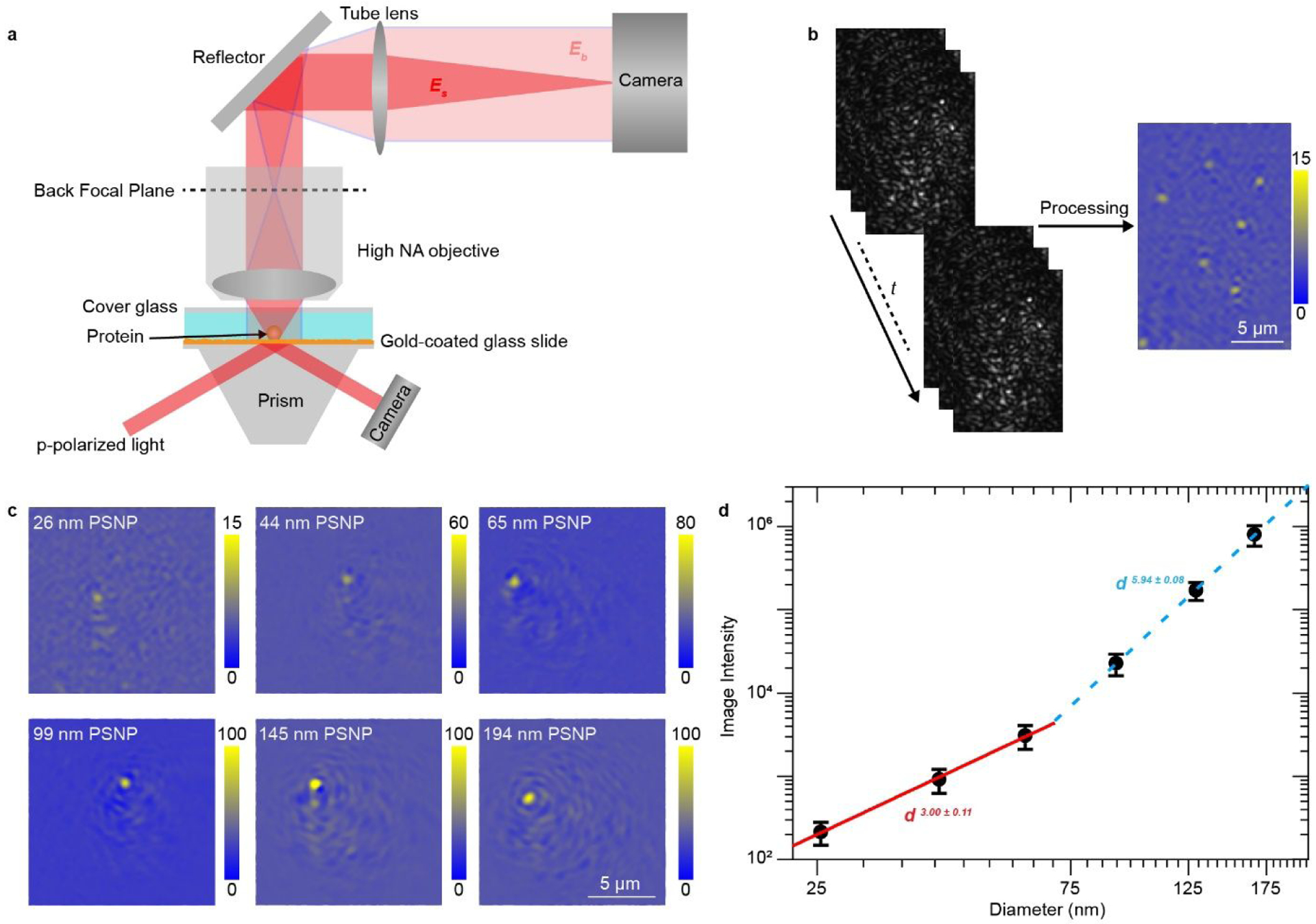
Setup and principle of prism coupled plasmonic scattering microscopy (PSM). (a) Simplified sketch of the optical setup, where surface plasmonic waves are excited by p-polarized light from the bottom of a gold-coated glass slide coupled to a prism and scattering of the plasmonic waves by a particle or protein (Es) and by the gold surface (Eb) is collected from the top to form a PSM image. A detailed description of the setup can be found in Figure S1. (b) Raw time sequence of PSM images of 26 nm polystyrene nanoparticles (left), and background and drift corrected PSM image (right). (c) PSM images of individual 26 nm, 44 nm, 65 nm, 99 nm, 145 nm, and 194 nm polystyrene nanoparticles. (d) PSM image intensity versus particle diameter. The mean image intensity for each diameter is obtained from the histograms in Figure S5. The error bars indicate the full width at half maximum of the Gaussian fitting for the corresponding histograms.
Incident light intensity and camera exposure time are 2.5 kW/cm2 and 2 ms for 26 nm, 2.5 kW/cm2 and 1 ms for 44 nm, 2.5 kW/cm2 and 0.5 ms for 65 nm, 300 W/cm2 and 2 ms for 99 nm, 300 W/cm2 and 0.3 ms for 145 nm, 300 W/cm2 and 0.03 ms for 194 nm. The image intensity was normalized with incident light intensity of 2.5 kW/cm2 and camera exposure time of 2 ms in (g).
One operation difficulty with SPRM is that it requires to accurately control the distance between the gold-coated glass slide and the objective for conditioning the incident light and maintaining the focus of images. In contrary, the prism coupled SPR imaging system does not need to maintain distance between the gold-coated glass slide and the prism, and thus the gold-coated glass slide can be firmly fixed to the prism by the thin layer of immersion oil. This mechanically stable structure permits not only shot noise limited measurement with average period up to 50 ms (Figure S2), but also only changing the gold-coated glass slides to perform different experiments without readjusting focus and incident angle. In addition, it eliminates the focusing mechanism for adjusting the slide position, which is needed for objective coupled PSM18, and provides sufficient space above the gold-coated glass slide for a high numerical aperture (NA) top objective, which usually has a short working distance. In this paper, we use an objective with NA of 0.7 and a working distance of ~2 mm to replace the long working distance objective with NA of 0.42 used in our previous objective based PSM setup. Higher NA improves the light collection efficiency and enhances the signal to noise ratio of the image (Figure S3).
After averaging the raw image sequence over 25 ms and removing the |Eb|2 with differential processing, high contrast PSM image of individual objects bound to the surface can be achieved (Figure 1b). Owing to high spatial resolution provided by the high NA top objective, the prism coupled PSM shows the effect of delocalization feature of surface plasmonic wave on the image features more clearly than objective based PSM18 (Figure 1c and Video S1). The patterns are mainly located in the same direction of surface plasmonic wave within a few microns, which is the range of the propagation length of surface plasmonic wave for gold film, as demonstrated as the features of the scattering of surface plasmonic waves19, 20. Unlike the SPRM image, where the patterns are strictly distributed as the parabolic tail along the propagation direction of surface plasmonic wave21, the patterns at different locations in PSM images reveal that they are roughly distributed along the propagation direction with spatial heterogeneity (Figure S4). However, the analysis between the analyte-induced intensity changes and random |Eb|2 reveals a missing correlation between them (Figure S5 and S6). This suggests that the delocalized scattered surface plasmonic waves by high density random nanometer-scale gold film surface roughness may create a comparable homogenous reference field with spatially heterogeneous distribution, which still requires further theoretical modeling. Considering that most energy of analyte-induced signal is concentrated within the diffraction limited spots (Figures S4) and the surrounding patterns become unobvious for the particles with diameter of 26 nm (Figure 1c), which has been larger than most proteins, we determine the PSM image intensity by integrating the intensities of all pixels within the Airy disk for current applications.
To validate the feasibility of this approach for determining image intensity, polystyrene nanoparticles with diameters (d) varying from 26 nm to 194 nm were imaged on a gold-coated glass side. For each size, the nanoparticles dissolved in PBS buffer were flowed over the bare gold surface, and the binding events were recorded over time. The incident intensity was optimized considering both signal to noise ratio and heating effect, and the camera exposure time was optimized for good image contrast while avoiding saturation for each measurement. The binding events in PSM images of individual nanoparticles are shown in Video S1. After tracking these binding events and calculating their image intensities, the intensity histograms are constructed and fitted with Gaussian distributions (Figures S7). Figure 1d shows the plot of the mean image intensity vs. nanoparticle diameter in logarithmic scale, where the effective nanoparticle diameter is employed considering that surface plasmonic wave decays exponentially from the surface. The plot clearly shows the transition from scattering to interference for PSM image intensity variation against nanoparticle diameter, agreeing with previously reported data on thermally vacuum-evaporated gold film18.
Detection of single proteins:
To obtain the calibration curve of PSM image intensity against protein molecular mass, human immunoglobulin A (IgA), human thyroglobulin (Tg), human immunoglobulin M (IgM), and low density lipoprotein (LDL) with molecular mass of 385, 660, 950, and 2300 kDa were dissolved in PBS buffer and flowed over the bare gold surface. The proteins bound to the surface via non-specific interactions. The binding events for each protein were recorded over time. Figures 2a–d show cumulative IgA, Tg, IgM, and LDL binding events at different locations, respectively. A PSM image sequence containing the binding events of individual proteins is shown in Video S2. After tracking these binding events and calculating their image intensities, the intensity histograms are constructed as shown in Figures 2a–d. The histogram width results from the intensity fluctuations of binding events, which may be caused by the protein mass, conformation and orientation heterogeneities22, 23. Gaussian distribution was employed to fit these histograms. The small secondary peaks in the histograms may be created by the formation of dimers or two proteins binding to adjacent locations within the diffraction limit simultaneously. Plotting the mean image intensity against molecular mass reveals a linear relationship (Figure 2e), confirming that the PSM image intensity is a measure of protein molecular mass.
Figure 2.
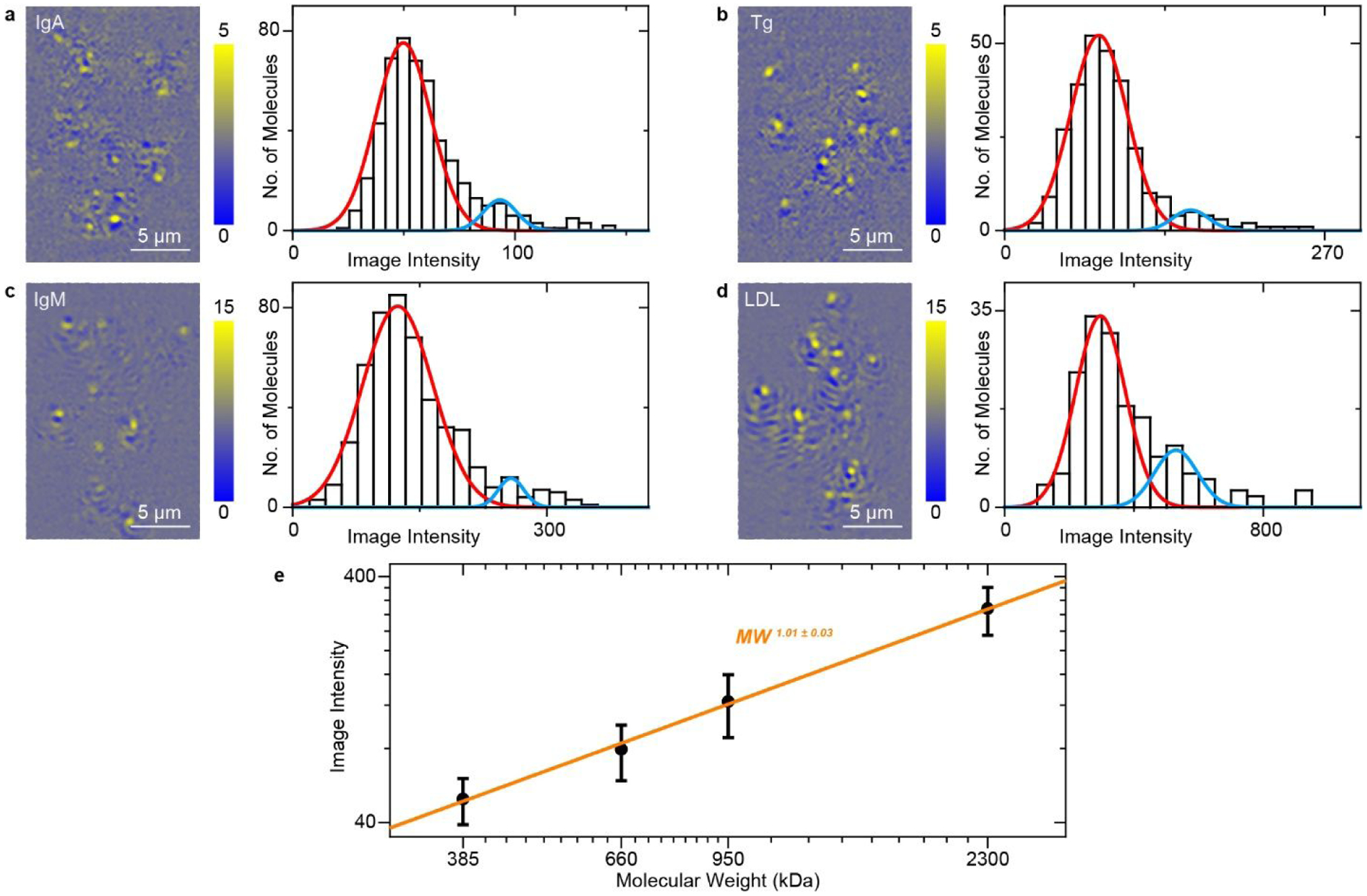
Detection of single unlabeled proteins on bare gold. PSM images of IgA, Tg, IgM and LDL proteins and corresponding image intensity histograms, where the solid lines are Gaussian fittings. A small secondary peak (blue) in each histogram is attributed to formation of dimers or two particles binding to the nearby surface simultaneously within the diffraction limit. Incident light intensity and camera exposure time are 5 kW/cm2 and 1 ms for IgA and Tg, 2.5 kW/cm2 and 2 ms for IgM and LDL. The image intensity was normalized with incident light intensity of 2.5 kW/cm2 and camera exposure time of 2 ms. (g) PSM image intensity versus molecular mass. The mean image intensity for each diameter is obtained from the mean value of the corresponding histogram in a–d. The error bars indicate the full width at half maximum of the Gaussian fitting for the corresponding histograms.
Differentiating specific and nonspecific binding at the single-molecule level:
The specificity of traditional SPR sensor relies on the target-specific molecular probes and optimized surface chemistry that prevent non-specific interactions, because any molecule binding to the surface will contribute to the signal. In contrary, PSM can differentiate specifically and non-specifically bound molecules at individual molecule level via analyzing the behaviors of individual binding events and mass discrepancy between target and non-target molecules. To demonstrate this capability, we first studied the specific binding of IgM and nonspecific binding of LDL to an anti-IgM antibody coated surface (Figure 3a and 3c). High antibody coverage of ~10000/μm2 was used throughout all experiments to ensure uniform receptor surface coverage and sufficient capture probability to detect the binding events. Bovine serum albumin (BSA) was employed to block possible non-specific binding sites. Upon flowing the IgM solution onto the sensor surface, the binding of single IgM molecules to anti-IgM took place immediately, with most proteins retaining on the surface and the number of bound proteins kept increasing over time. In contrast, when we flowed LDL solution over the anti-IgM modified surface, the LDL molecules showed transient binding and unbinding behaviors. This is because IgM molecules have high affinity to anti-IgM on the surface, but LDL molecules have very low affinity to the sensor surface due to surface blocking. Video S3 reveals the dynamic binding process, and Figures 3a and 3c show a few differential snapshots. We counted the specific binding events of individual IgM proteins and constructed a molecular mass histogram using the calibration curve in Figure 2e, showing a major peak due to single IgM molecules (Figure 3b). As additional examples for PSM revealing specific and nonspecific binding processes at single molecule level, we measured the specific binding of IgA and nonspecific binding of Tg to an anti-IgA antibody modified surface(Figures 3d and 3f), as well as specific binding of anti-BSA antibody (IgG, MW = 150 kDa) and nonspecific binding of IgA to a BSA modified surface (Figures 3g and 3i). They also show the same trend as the anti-IgM/IgM/LDL experiment, where the specific binding can be notably differentiated from non-specific binding. The full videos are presented in Video S3. We constructed molecular mass histograms using the calibration curve in Figure 2e from the specific binding events of IgG and IgA, also showing major peaks due to single IgA and IgG molecules (Figures 3e and 3h).
Figure 3.
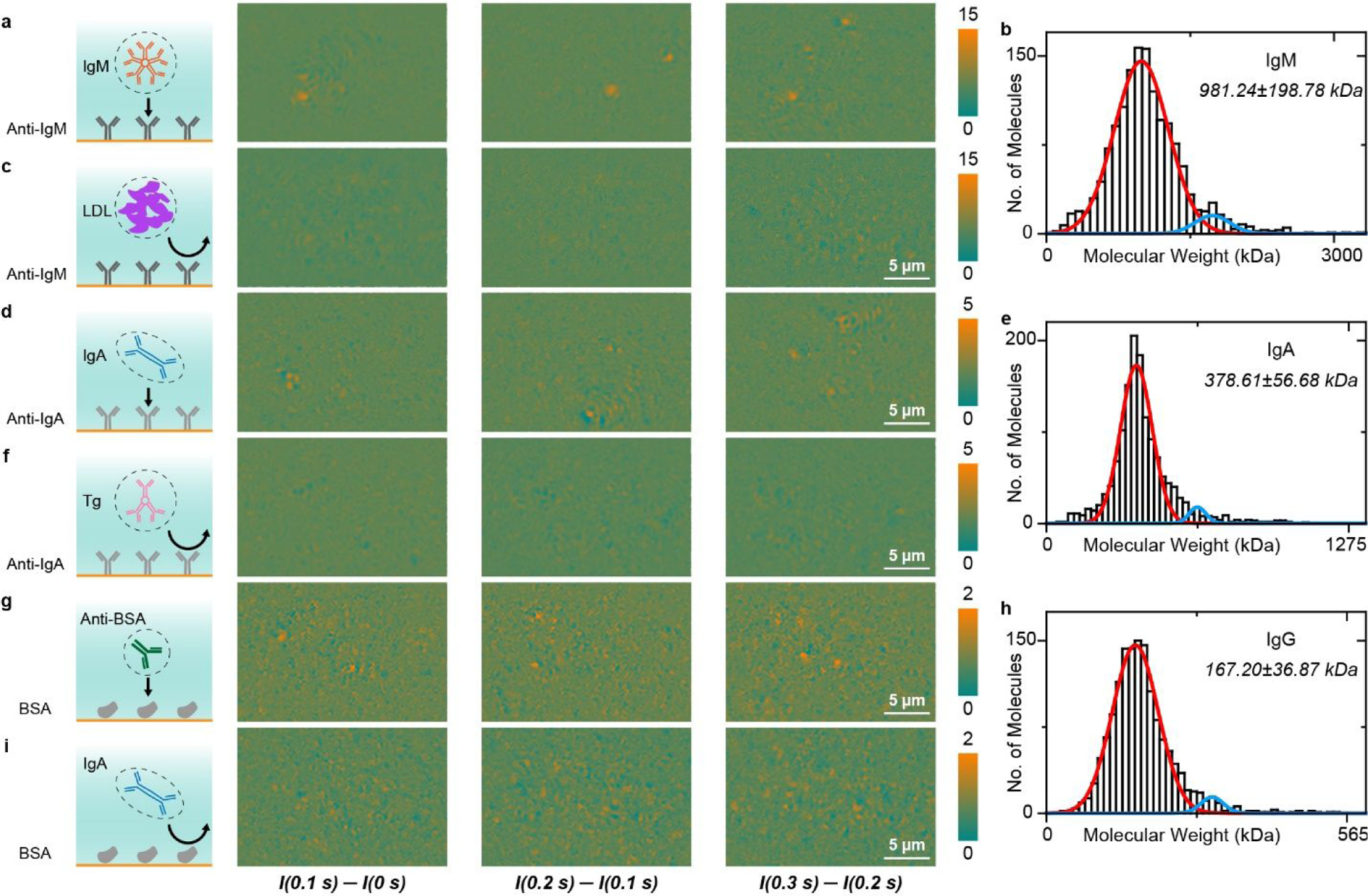
Difference between specific and nonspecific binding. (a) PSM snapshots showing specific binding of IgM to anti-IgM immobilized on the surface. (b) Molecular mass histogram of IgM molecules, where the solid lines are Gaussian fitting. (c) Negative control experiment: Introducing LDL to anti-IgM surface. (d) PSM snapshots showing specific binding of IgA to anti-IgA immobilized on the surface. (e) Molecular mass of IgA molecules, where the solid lines are Gaussian fitting. (f) Negative control experiment: Introducing Tg to anti-IgA coated surface. (g) PSM snapshots showing specific binding of anti-BSA to BSA immobilized on the surface. (h) Molecular mass histogram of IgG molecules, where the solid lines are Gaussian fitting. (i) Negative control experiment: Introducing IgA to BSA coated surface. Incident light intensity and camera exposure time are 2.5 kW/cm2 and 2 ms for (a-c), 5 kW/cm2 and 1 ms for (d-i). The molecular mass histograms were constructed with the calibration curve in Figure 2e.
For strong non-specific binding events that cannot be identified by single molecule binding behaviors, PSM can also differentiate them using molecular mass as barcode. Figure 4 shows the PSM result of binding kinetics of 5 nM IgA, which is mixed with 10 nM IgM as impurities, to anti-IgA coated surface. The molecular mass histogram (Figure 4b and 4c) constructed with the calibration curve in Figure 2e shows that the binding events with larger molecular mass notably increase compared with the measurement of purified sample (Figure 3e), indicating some IgM molecules bound to surface. After defining the specific and nonspecific binding events based on the molecular mass, the association and dissociation curves are plotted based on digital counting for IgA and IgM binding, respectively (Figure 4d). Digital counting of binding and leaving events was performed on the differential image sequence (Figure S8). The specific binding curve can be easily fitted with the first-order binding kinetics model. The association (kon) and dissociation (koff) rate constants, and equilibrium dissociation constant (KD = koff/kon) are determined to be 3.9 × 105 M−1 s−1, 1.9 × 10−4 s−1, and 487 pM, respectively. These values fall within the error range determined by measuring the kinetic of pure IgA binding onto anti-IgA in triple replicates (Figure S9), where the kon, koff and KD were determined to be 3.8 ± 0.5 × 105 M−1 s−1, 1.8 ± 0.2 × 10−4 s−1, and 484 ± 39 pM, respectively, confirming that the digital counting approach with molecular mass as barcode can differentiate specific binding from nonspecific binding at single molecule level. In contrast, the nonspecific binding curve of IgM cannot be fitted with first-order binding kinetics. The association rate of nonspecific binding is firstly linear and then slowing down, which may be saturating the nonspecific binding sites, while the dissociation curve shows a quick dip followed by very flat decay, indicating that the nonspecific binding includes both weak and strong binding sites.
Figure 4.
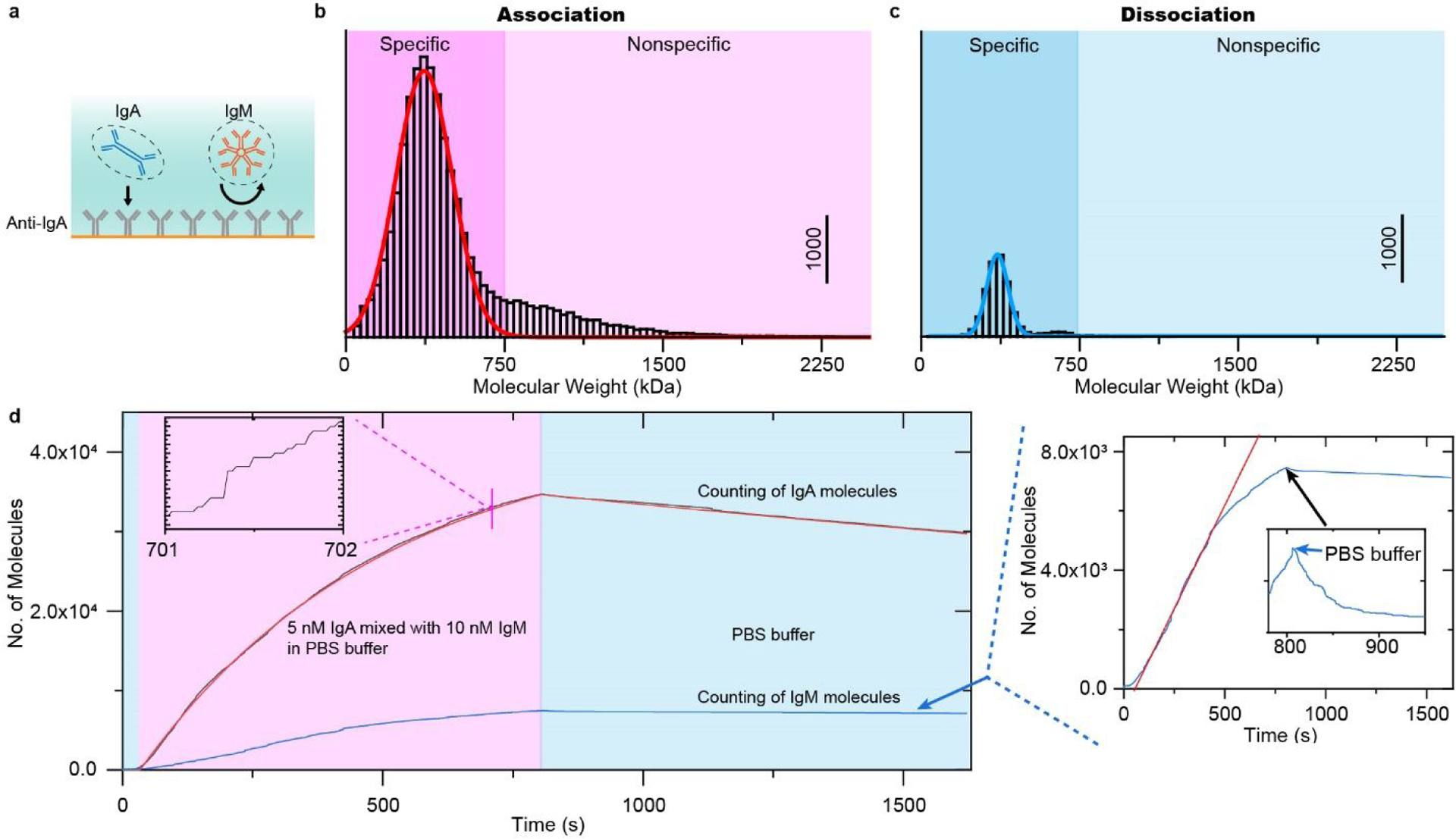
Differentiate specific binding of target protein from nonspecific binding with molecular mass as barcode. (a) 5 nM IgA and 10 nM IgM are injected onto the anti-IgA modified surface simultaneously. (b), and (c) Molecular mass histogram obtained from association, and dissociation processes, where the solid lines are Gaussian fitting. Incident light intensity and camera exposure time are 2.5 kW/cm2 and 2 ms. (d) Kinetics of IgA binding to anti-IgA and nonspecific binding of IgM to anti-IgA and BSA determined by digitally counting of the binding/unbinding of single molecules. Red colored line in the inset indicates the linear fitting of the nonspecific binding curve. The molecular mass histograms were constructed with the calibration curve in Figure 2e.
Measurement of specific binding of single unlabeled proteins in fetal bovine serum:
Figure 4 shows that PSM can differentiate the specific binding of target proteins from both weak and strong nonspecific binding, making it possible to analyze binding kinetics in complex media. Figure 5 shows the PSM result of binding kinetics of 5 nM IgA in 10% fetal bovine serum (FBS) diluted with PBS buffer to anti-IgA coated surface. Considering the weak nonspecific binding events have transient binding and unbinding behaviors, the processed differential frames were further subtracted by following frames to remove the background created by transient nonspecific binding events (Figure 5b). The molecular mass histogram (Figure 5c and 5d) constructed with the calibration curve in Figure 2e shows that there are much more binding events with larger molecular weight in serum than in pure buffer (Figure 3e, 4b and 4c), which are strong nonspecific bindings that can be differentiated based on the molecular mass. Therefore, after removing these non-specific binding events, the association and dissociation curves for specific bindings are plotted from the digital counting of IgA binding events (Figure 5e), and are well fitted with the first-order binding kinetics model. The association (kon) and dissociation (koff) rate constants, and the equilibrium dissociation constant (KD = koff/kon) are determined to be 1.3 × 105 M−1 s−1, 9.2 × 10−5 s−1, and 720 pM, respectively. The KD has ~40% difference from the result measured in PBS buffer (Figure S9). While we cannot rule out the possibility of 10% serum can actually change the IgA to anti-IgA binding kinetics, both the digital counting (Figure 5c) and ensemble SPR measurement (Figure S10) results show strong nonspecific binding from other molecules in FBS. The binding events with similar mass to IgA could be counted toward the construct the binding curve and introduce some measurement error. However, since most of the nonspecific binding events are identified and removed by digital counting method, the variation in kinetics is acceptable, because ~10 times difference is usually considered as a standard to define a different binding kinetic24, 25. In contrary, high non-specific bindings from the abundant contents of proteins in serum produced continued baseline drift and dramatically distorted the binding curve, and seriously affect the kinetic constant measurement with ensemble SPR (Figure 5e and Figure S10), which has been reported as a longstanding issue in SPR measurement for applications in diagnosis and biomarker discovery10, 26. Therefore, PSM based digital counting provided a solution with acceptable error to this longstanding issue applying SPR technology in clinical settings.
Figure 5.

Measurement of specific binding of single unlabeled proteins in fetal bovine serum. (a) schematic showing 5 nM IgA in 10% fetal bovine serum diluted with PBS buffer injected onto the anti-IgA modified surface. (b) Double differential processing to remove the weak nonspecific binding events. (c), and (d) Molecular mass histogram obtained from association, and dissociation processes, where the solid lines are Gaussian fitting. Incident light intensity and camera exposure time are 2.5 kW/cm2 and 2 ms. (e) Kinetics of IgA binding to anti-IgA determined by digital counting of the binding/unbinding of single IgA molecules (black curve and solid red fitted curve), and simultaneously recorded ensemble SPR signals (Green curve and dash red fitted line). The molecular mass histograms were constructed with the calibration curve in Figure 2e.
PSM is insensitive to bulk refractive index changes:
Another advantage of digital counting based binding kinetic measurement is the tolerance to bulk refractive index changes, which is a noise source for ensemble SPR. The running buffer and sample buffer usually need to be carefully matched in content and concentration for ensemble SPR, to avoid the bulk refractive index change induced response curve shift upon switching between buffer and sample. To demonstrate PSM is insensitive to bulk refractive index change, in the serum experiment described above, we used pure PBS as running buffer, while the sample is prepared in 10% serum. As shown in Figure 5e, switching from buffer to sample or from sample to buffer caused a huge bulk refractive index change induced shifts in ensemble SPR response curve. However, this large bulk refractive index change does not affect the response curve generated from PSM counting.
Measurement reproducibility
A major advantage of prism coupled SPR devices is system stability and high reproducibility, making it leads the commercial label-free optical detection products. Here we show that prism coupled PSM also maintains this advantage. To test sample to sample variation, we aliquoted 26 nm polystyrene nanoparticle solution into three identical samples. Then we sequentially flowed these three samples over the gold surface with an injection valve, recorded the binding events, and calculated the image intensities of the binding events. Figure 6a shows the image intensity distribution of binding events against binding time, revealing the binding frequency is consistent for all the three measurements. Figure 6b shows the histograms constructed by image intensities of binding events in three measurements, revealing the mean image intensities are also consistent for all three measurements. These indicate that the measurement on prism coupled PSM has little drift effect and maintains high measurement consistency over time.
Figure 6.
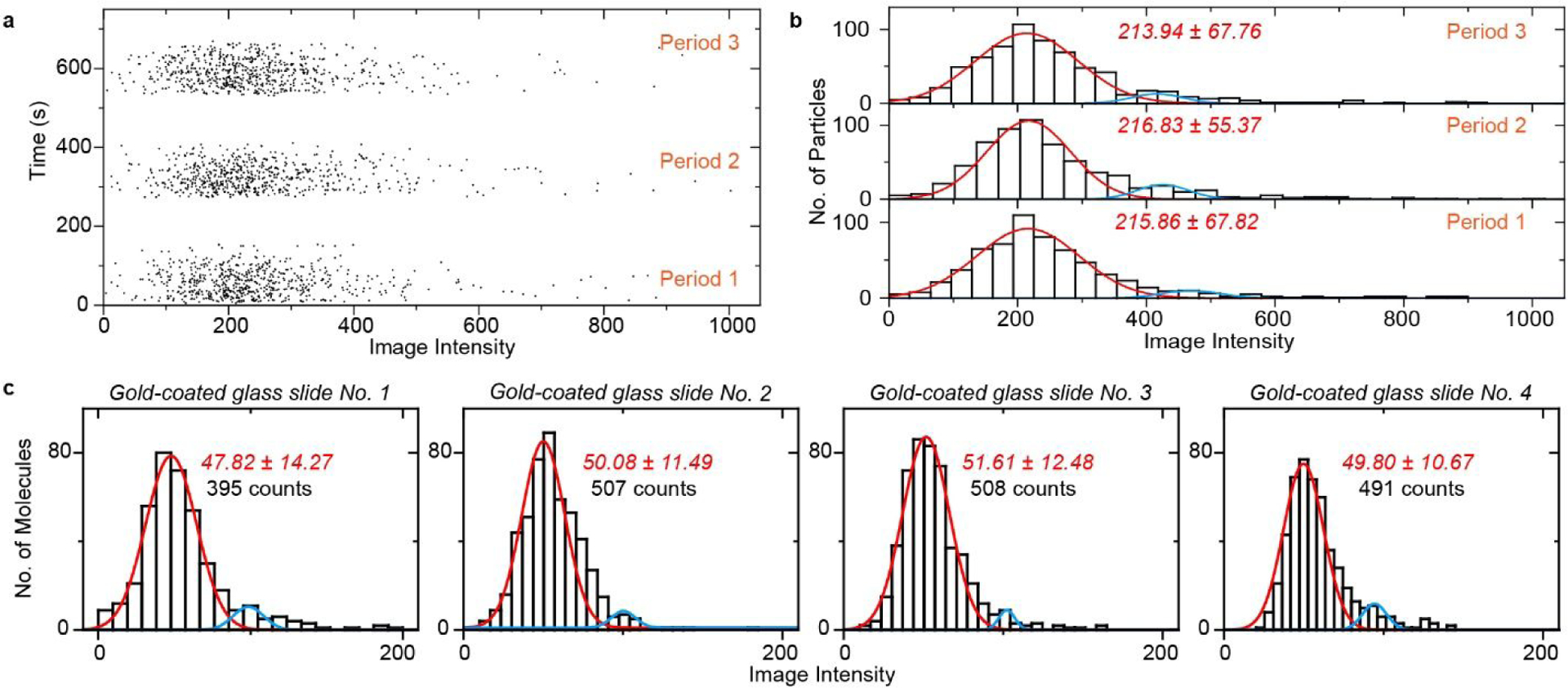
Measurement reproducibility. (a) Scatter plot of individual binding events of 26 nm polystyrene nanoparticles against binding time. (b) Resulting image intensity histograms of a. Incident light intensity and camera exposure time are 2.5 kW/cm2 and 2 ms. (c) Image intensity histograms of IgA proteins measured on 4 pieces of gold-coated glass slides, where the solid lines are Gaussian fitting. Incident light intensity and camera exposure time are 5 kW/cm2 and 1 ms. The image intensity was normalized with incident light intensity of 2.5 kW/cm2 and camera exposure time of 2 ms.
To show the chip-to-chip reproducibility, 5 nM IgA proteins were measured on 4 pieces of gold-coated glass slides, and the proteins bound to the surface via non-specific interactions within 5 minutes. The intensity histograms are constructed as shown in Figure 6c along with the mean and standard deviation values determined by Gaussian fitting. The digitally counted binding events were also presented in each figure. It shows that the PSM image intensity has only ~3% variation among different gold-coated glass slides, which is 4 times smaller than the objective configured PSM18. The number of digitally counted binding events among the glass slides has mean value of ~ 475 and standard deviation of ~ 54, and this ~10% fluctuation between different gold coated glass slides may be caused by different surface states and statistical variation of random binding events. This improvement is mainly because the incident angle can be easily fixed to resonance angle in prism coupled PSM setup, but harder to maintain the incident angle with the same stability in objective based PSM due to the small sizes of image plane and back focal plane.
DISCUSSION
The image contrast of PSM results from the interference of surface plasmonic waves, which are localized near the metal film surface with 20–30x intensity enhancement, scattered from an object and a reference, which is analogous to SPRM. However, the mechanism is different between PSM and SPRM. SPRM collects the reflection to detect the interference between planar surface plasmonic wave and scattered surface plasmonic wave by analyte27, while PSM detects the interference between surface plasmonic waves scattered by gold surface roughness and analyte. PSM avoids the strong reflection that reduces the camera signal collection efficiency, a common issue for high sensitivity imaging28, 29, allowing high contrast imaging of single unlabeled protein detection.
Nanoscopic objects invariably absorb some light, resulting in photodamage to the biological samples at high field intensities30, 31. Photodamage is also the ultimate limit when pushing the detection limit of PSM by increasing incident light intensity to suppress shot noise, a common approach to improve the signal noise ratio of single molecule imaging23. For field of view of 300 μm2, PSM usually can take the incident intensity up to ~6 kW/cm2, and higher intensity will damage the protein molecules within 5 minutes (Figure S11). An alternative approach to improve the detection limit demonstrated in this paper is increasing the collection objective NA, which is almost linearly related to signal to noise ratio (Figure S3). Owing to this, prism coupled PSM pushes the practical detection limit, which usually requires the signal to noise ratio of ~10, from IgA (385 kDa) in objective coupled PSM down to IgG (150 kDa)18. With this detection limit, one potential impactful application for PSM is for quantification of protein aggregations, a common issue in monoclonal antibody therapy32, which is becoming increasingly important for treatment of cancer and new infectious diseases, such as COVID-19.
Prism coupled PSM can recognize binding events of individual analytes, offering at least two distinct advantages over the ensemble prism coupled SPR system. First, PSM counted the binding events based on digital counting, rather than an analog signal, making it not sensitive to thermal or mechanical drift, a common issue in the label free detection. Second, PSM can distinct the specific and nonspecific binding of individual events via differences in single molecule mass or binding kinetics, which enhances the specificity of measurement and relaxes the requirement of sample purity and experimental conditions. In contrary, ensemble SPR measures changes in reflection intensity or resonance angle, which can be induced by both specific and non-specific binding, as well as solution refractive index changes. This advantage allows the PSM to analyze the specific binding kinetics in complex media, which opens the door for applications in diagnostic and biomarker discovery in liquid biopsy that requires the use of body fluid with complex background, such as blood and urine.
Compared with the oil immersion objective based PSM, prism coupled PSM permits easy fixation of each component for a stable experimental condition, thus providing better chip to chip reproducibility. This makes it unnecessary to normalize the measurement results with standard samples in each experiment, which is needed in objective based PSM. In addition, operators only need to change the gold-coated glass slides to perform different experiments on prism coupled PSM without readjusting focus and incident angle, and this will notably lower the requirement of operational skills.
We also note that PSM approach also can help the performance of commercial SPR imaging system by simply adding one dry objective on the top of sample (Figure S12). In a commercial prism coupled SPR imaging device (SPRm 200, Biosensing Instruments), we can obtain high contrast imaging of 194 nm polystyrene nanoparticles with a top mounted 10x objective with working distance of 39 mm and NA of 0.23 (Figure S12a), and obtain high contrast imaging of 145 nm polystyrene nanoparticles with a 40 × NA 0.75 objective (Figure S12b). As a comparison, SPRM constructed based on high cost inverted optical microscopy usually allows stable imaging of ~100 nm gold nanoparticles in protein interaction analysis17. It has been pointed out that small gold particles or dielectric nanoparticles create notably lower heating effect than large gold nanoparticles to reduce the damage on biological analytes and have smaller negative effect on the mode of lipid diffusion in cell imaging applications30, 33.
CONCLUSION
In summary, we have demonstrated plasmonic imaging of single unlabeled proteins by measuring scattering of plasmonic waves on top of a prism coupled SPR system. The prism coupled PSM can measure the mass of single unlabeled proteins and identify specific and nonspecific binding at single molecule level. Compared with traditional ensemble SPR, prism coupled PSM provides statistical distribution of individual objects instead of ensemble-averaging signal, thus allowing the binding kinetic analysis in complex media. Compared with objective configured PSM, prism coupled PSM provides simpler operation and better measurement reproducibility. We anticipate that prism coupled PSM approach may provide a convenient solution for realizing high resolution imaging on widely used prism configured SPR devices, allowing us to understand the biological processes at a level of detail.
METHODS
Materials.
Polystyrene nanoparticles were purchased from Bangs Laboratories. Phosphate-buffered saline (PBS) was purchased from Corning and filtered with 0.22 μm filters (Millex). Human plasma low density lipoproteins (LDL), human plasma IgM and human colostrum IgA were purchased from Athens Research and Technology. Anti-IgM, human thyroglobulin (Tg), bovine serum albumin (BSA) were purchased from Sigma-Aldrich. Anti-IgA was purchased from Bio-Rad. Fetal bovine serum, N-ethyl-N’-(dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysulfosuccinimide (Sulfo-NHS) were purchased from Thermo Fisher Scientific. Dithiolalkanearomatic PEG6-COOH was purchased from Sensopath Technologies. DI water with resistivity of 18.2 MΩ/cm was filtrated with 0.22 μm filter and used in all experiments.
Experimental setup.
An 80 mW diode laser (OBIS 660–75FP, Coherent) with central wavelength at 660 nm is used as light source. Light from the laser is conditioned by a pair of lenses with focal length of 19 mm (AC127-019-A, Thorlabs), and then focused to the back focal plane of a 100× objective by a tube lens with focal length of 300 mm (AC508-300-A, Thorlabs). Then the Gaussian beam focused on the working point of the objective was projected to the prism surface with a lens group (two of AC254-030-A and one of AC254-150-A, Thorlabs). The incident angle was adjusted by a manual translation stage (PT3, Thorlabs, Newton, NJ) to reach surface plasmon resonance. Light reflected from the gold-coated glass slide is also collected by a camera (Point Grey CM3-U3-13Y3M-CS) for helping to find the resonance angle. Scattered light from the protein and gold surface is collected by a top mounted 60× objective (NA = 0.7) equipped with 300 mm tube lens for a final 100× magnification to form a PSM image on a second camera (MQ013MG-ON, XIMEA). More detailed schematic representation of the optics can be found in Figure S1.
Surface functionalization.
Gold-coated glass slides were fabricated by coating a BK7 cover glass with 1 nm Cr followed by 47 nm gold via thermal evaporation (PVD75 E-beam/Thermal Evaporator, Kurt J. Lesker Company). Prior to coating, the gold surface was rinsed by ethanol and deionized water twice, and then annealed with hydrogen flame to remove surface contaminants. The gold surface of each glass slide was modified with carboxyl groups by incubation with 1 mM dithiolalkanearomatic PEG6-COOH for 1 hour. Then the surface was incubated in 0.05 M NHS/0.2 M EDC for 30 min to activate the carboxyl groups. After rinsing with PBS, 20 nM anti-IgA, anti-IgM, or BSA was applied to the surface and incubated for 30 min to allow immobilization. At last, the surface was incubated in 1 mg/ml BSA for 10 min to block non-specific binding sites.
Supplementary Material
ACKNOWLEDGMENT
We thank financial support from National Institutes of Health (R01GM107165 and R33CA235294). We acknowledge the use of facilities within the ASU NanoFab supported in part by NSF program NNCI-ECCS-1542160.
Footnotes
Supporting Information
The following files are available free of charge.
Details of optical design, measurements on commercial surface plasmon resonance device, and some more results (PDF)
Dynamic binding process of polystyrene nanoparticles on bare gold (MOV)
Dynamic binding process of unlabeled proteins on bare gold (MOV)
Dynamic binding process of unlabeled proteins on anti-body modified gold (MOV)
The authors declare no competing financial interest.
REFERENCES
- 1.Wang W; Yang Y; Wang S; Nagaraj V; Liu Q; Wu J; Tao N, Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nature Chemistry 2012, 4 (10), 846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos R; Ursu O; Gaulton A; Bento A; Donadi R; Bologa C; Karlsson A; Al-Lazikani B; Hersey A; Oprea T; Overington J, A comprehensive map of molecular drug targets. Nature Reviews Drug Discovery 2017, 16 (1), 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauser A; Chavali S; Masuho I; Jahn L; Martemyanov K; Gloriam D; Babu M, Pharmacogenomics of GPCR Drug Targets. Cell 2018, 172 (1–2), 41–54.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma G; Syu G; Shan X; Henson B; Wang S; Desai P; Zhu H; Tao N, Measuring Ligand Binding Kinetics to Membrane Proteins Using Virion Nano-oscillators. Journal of the American Chemical Society 2018, 140 (36), 11495–11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homola J, Surface plasmon resonance sensors for detection of chemical and biological species. Chemical Reviews 2008, 108 (2), 462–493. [DOI] [PubMed] [Google Scholar]
- 6.Yin L; Wang W; Wang S; Zhang F; Zhang S; Tao N, How does fluorescent labeling affect the binding kinetics of proteins with intact cells? Biosensors & Bioelectronics 2015, 66, 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arroyo J; Kukura P, Non-fluorescent schemes for single-molecule detection, imaging and spectroscopy. Nature Photonics 2016, 10 (1), 11–17. [Google Scholar]
- 8.Taylor R; Mahmoodabadi R; Rauschenberger V; Giessl A; Schambony A; Sandoghdar V, Interferometric scattering microscopy reveals microsecond nanoscopic protein motion on a live cell membrane. Nature Photonics 2019, 13 (7), 480–487. [Google Scholar]
- 9.Wang S; Forzani E; Tao N, Detection of heavy metal ions in water by high-resolution surface plasmon resonance spectroscopy combined with anodic stripping voltammetry. Analytical Chemistry 2007, 79 (12), 4427–4432. [DOI] [PubMed] [Google Scholar]
- 10.Mani R; Dye R; Snider T; Wang S; Clinkenbeard K, Bi-cell surface plasmon resonance detection of aptamer mediated thrombin capture in serum. Biosensors & Bioelectronics 2011, 26 (12), 4832–4836. [DOI] [PubMed] [Google Scholar]
- 11.Zhang P; Liu L; He Y; Chen X; Ma K; Wei D; Wang H; Shao Q, Composite layer based plasmon waveguide resonance for label-free biosensing with high figure of merit. Sensors and Actuators B-Chemical 2018, 272, 69–78. [Google Scholar]
- 12.Zhang P; Liu L; He Y; Xu Z; Ji Y; Ma H, One-dimensional angular surface plasmon resonance imaging based array thermometer. Sensors and Actuators B-Chemical 2015, 207, 254–261. [Google Scholar]
- 13.Jing W; Wang Y; Chen C; Zhang F; Yang Y; Ma G; Yang EH; Snozek CLN; Tao N, Gradient-Based Rapid Digital Immunoassay for High-Sensitivity Cardiac Troponin T (hs-cTnT) Detection in 1 μL Plasma. ACS Sensors 2020, DOI: 10.1021/acssensors.0c01681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jing W; Wang Y; Yang Y; Ma G; Wang S; Tao N, Time-Resolved Digital Immunoassay for Rapid and Sensitive Quantitation of Procalcitonin with Plasmonic Imaging. Acs Nano 2019, 13 (8), 8609–8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y; Jing W; Tao N; Wang H, Probing Single-Molecule Binding Event by the Dynamic Counting and Mapping of Individual Nanoparticles. ACS Sensors 2020, DOI: 10.1021/acssensors.0c02184. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y; Yang Y; Chen C; Wang S; Wang H; Jing W; Tao N, One-Step Digital Immunoassay for Rapid and Sensitive Detection of Cardiac Troponin I. Acs Sensors 2020, 5 (4), 1126–1131. [DOI] [PubMed] [Google Scholar]
- 17.Wang H; Tang Z; Wang Y; Ma G; Tao N, Probing Single Molecule Binding and Free Energy Profile with Plasmonic Imaging of Nanoparticles. Journal of the American Chemical Society 2019, 141 (40), 16071–16078. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P; Ma G; Dong W; Wan Z; Wang S; Tao N, Plasmonic scattering imaging of single proteins and binding kinetics. Nature Methods 2020, 17, 1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shchegrov A; Novikov I; Maradudin A, Scattering of surface plasmon polaritons by a circularly symmetric surface defect. Physical Review Letters 1997, 78 (22), 4269–4272. [Google Scholar]
- 20.Evlyukhin A; Bozhevolnyi S, Resonant unidirectional and elastic scattering of surface plasmon polaritons by high refractive index dielectric nanoparticles. Physical Review B 2015, 92 (24), 245419. [Google Scholar]
- 21.Yu H; Shan X; Wang S; Tao N, Achieving High Spatial Resolution Surface Plasmon Resonance Microscopy with Image Reconstruction. Analytical Chemistry 2017, 89 (5), 2704–2707. [DOI] [PubMed] [Google Scholar]
- 22.Simpson WD III; Heinrich V, Modulation of the Drag Force Exerted by Microfluidic Flow on Laser-Trapped Particles: A New Method to Assess Surface-Binding Kinetics, Analyte Size, and Solution Viscosity. Biophysical Journal 2018, 114 (3), 692a. [Google Scholar]
- 23.Piliarik M; Sandoghdar V, Direct optical sensing of single unlabelled proteins and super-resolution imaging of their binding sites. Nature Communications 2014, 5, 4495. [DOI] [PubMed] [Google Scholar]
- 24.Al Balushi AA; Gordon R, A Label-Free Untethered Approach to Single-Molecule Protein Binding Kinetics. Nano Letters 2014, 14 (10), 5787–5791. [DOI] [PubMed] [Google Scholar]
- 25.Wienken CJ; Baaske P; Rothbauer U; Braun D; Duhr S, Protein-binding assays in biological liquids using microscale thermophoresis. Nature Communications 2010, 1 (1), 100. [DOI] [PubMed] [Google Scholar]
- 26.Masson J, Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics. Acs Sensors 2017, 2 (1), 16–30. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y; Shen G; Wang H; Li H; Zhang T; Tao N; Ding X; Yu H, Interferometric plasmonic imaging and detection of single exosomes. Proceedings of the National Academy of Sciences of the United States of America 2018, 115 (41), 10275–10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole D; Young G; Weigel A; Sebesta A; Kukura P, Label-Free Single-Molecule Imaging with Numerical-Aperture Shaped Interferometric Scattering Microscopy. Acs Photonics 2017, 4 (2), 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young G; Hundt N; Cole D; Fineberg A; Andrecka J; Tyler A; Olerinyova A; Ansari A; Marklund E; Collier M; Chandler S; Tkachenko O; Allen J; Crispin M; Billington N; Takagi Y; Sellers J; Eichmann C; Selenko P; Frey L; Riek R; Galpin M; Struwe W; Benesch J; Kukura P, Quantitative mass imaging of single biological macromolecules. Science 2018, 360 (6387), 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortega-Arroyo J; Kukura P, Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Physical Chemistry Chemical Physics 2012, 14 (45), 15625–15636. [DOI] [PubMed] [Google Scholar]
- 31.Mauranyapin N; Madsen L; Taylor M; Waleed M; Bowen W, Evanescent single-molecule biosensing with quantum-limited precision. Nature Photonics 2017, 11 (8), 477–481. [Google Scholar]
- 32.van der Kant R; Karow-Zwick A; Van Durme J; Blech M; Gallardo R; Seeliger D; Assfalg K; Baatsen P; Compernolle G; Gils A; Studts J; Schulz P; Garidel P; Schymkowitz J; Rousseau F, Prediction and Reduction of the Aggregation of Monoclonal Antibodies. Journal of Molecular Biology 2017, 429 (8), 1244–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etoc F; Balloul E; Vicario C; Normanno D; Lisse D; Sittner A; Piehler J; Dahan M; Coppey M, Non-specific interactions govern cytosolic diffusion of nanosized objects in mammalian cells. Nature Materials 2018, 17 (8), 740–746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


