Abstract
The ID NOW COVID-19 assay is a promising tool for the rapid identification of COVID-19 patients. However, its performances were questioned. We evaluate the ID NOW COVID-19 in comparison to a reference RT-PCR using a collection of 48 fresh nasopharyngeal swabs sampled on universal transport media (UTM). Only 2 false negatives of the ID NOW COVID-19 were identified. They display PCR cycle threshold values of 37.5 and 39.2. The positive percent agreement and the negative percent agreement were 94.9% and 100%, respectively. The Kappa value was 0.88. The ID NOW COVID-19 combines high-speed and accurate processing. Using UTM, the ID NOW COVID-19 could be repeated in the case of invalid result. Further analyses, such as screening of genetic variants or genome sequencing, could also be performed with the same sample. As for all tests, the results should be interpreted according to clinical and epidemiological context.
Keywords: SARS-CoV-2, COVID-19, Isothermal amplification, LAMP, NEAR, RT-PCR
Introduction
In the context of the spread of the SARS-CoV-2, rapid and accurate identification of infected subjects is required. Conventional polymerase chain reaction (PCR) displays high performances [1, 2]. However, the method can be time-consuming and needed dedicated devices and specific training of operators. Therefore, it could require up to 24 h to obtain a result. This long time-to-result is not suitable for the management of patients in several situations such as the emergency departments. Antigenic tests were suggested as an alternative, but they lack sensitivity, and the results should be interpreted with caution [3, 4]. In contrast, isothermal nucleic acid amplification combines both high performances and does not require extensive expertise. It could be performed in settings with limited resources or where PCR is not available. The ID NOW (Abbott Rapid Diagnostic, Scarborough, ME, USA) is an easy to use device that provides robust and accurate results within 15 min for the biological diagnosis of flu [5, 6]. The ID NOW COVID-19 assay was developed at the beginning of the pandemic, but its performances were questioned. On May 14, the U.S. FDA issued a public warning about potential inaccurate results from using the ID NOW COVID-19 assay [7].
According to the manufacturer, the ID NOW COVID-19 should be performed on dry nasopharyngeal swabs (NPS) sampled at most in the 2 previous hours. However, when performed on a dry NPS, the swab is entirely consumed by the ID NOW COVID-19 assay. A second swab is required in case of invalid results or if further analyses are required such as genetic variant screening or viral genome sequencing.
In the present study, we aim to assess the analytical performances of the ID NOW COVID-19 using clinical NPS sampled on universal transport media (UTM).
Materials and methods
We selected a collection of 48 nasopharyngeal swabs (NPS) of which 39 SARS-CoV-2-positive NPS displaying various cycle thresholds (CT) of positivity (range 16.9–39.2) and 9 SARS-CoV-2-negative NPS. All the NPS had been sampled on universal transport media (UTM) from symptomatic patients suspected of SARS-CoV-2 infection [8]. All the patients were sampled between October 26, 2020, and November 15, 2020, in the emergency department of the Foch hospital (Suresnes, France) or the Franco-Britannique hospital (Levallois-Perret, France).
The results of the ID NOW COVID-19 were compared to those provided by the Alinity M SARS-CoV-2 RT-PCR assay (Abbott Molecular, Des Plaines, IL, USA). The ID NOW COVID-19 was performed using a volume of 200 μL of UTM. All NPS were first processed with the Alinity M SARS-CoV-2 assay and then stored at +5 °C for at most 72 h before testing by the ID NOW COVID-19. Statistical analyses were performed using Excel software (Microsoft Corporation, Albuquerque, NM, USA). The results of the ID NOW COVID-19 were compared with those of the Alinity M SARS-CoV-2 assay using the overall percentage of agreement and the Kappa test.
Results
Overall, 37 (94.9%) of the 39 positive NPS were correctly detected using the ID NOW COVID-19 assay. All the NPS displaying a CT below 35.0 were found positive with the ID NOW COVID-19 while only 6 (75.0%) over the 8 NPS displaying a CT greater than 35.0 were found positive with the ID NOW COVID-19 (Fig. 1). The 2 false negatives display a CT value of 37.5 and 39.2. Both were tested positive a second time using the Alinity M SARS-CoV-2 assay with similar CT values. None of the 6 negative SARS-CoV-2 samples provides a false-positive result with the ID NOW COVID-19 assay. No invalid results were found with all samples. The overall positive and negative percent agreements were 95.8%, 94.9%, and 100%, respectively, and the Kappa value was 0.88.
Fig. 1.
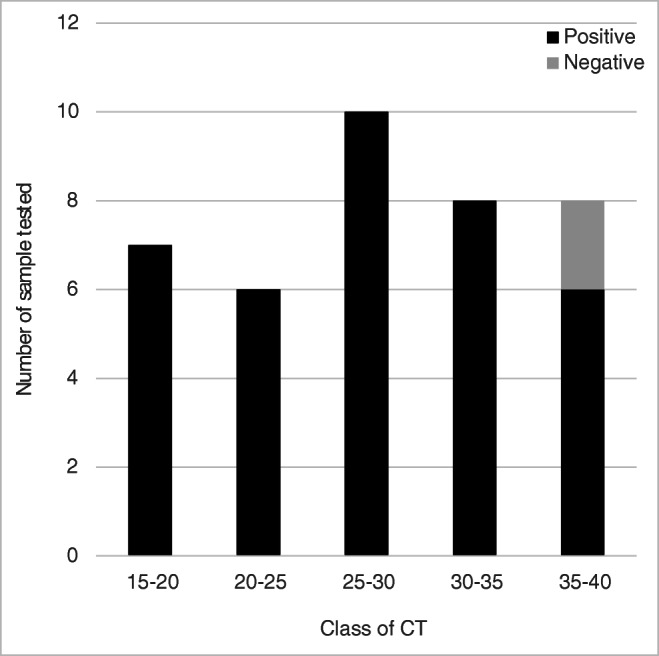
Results of the ID NOW COVID-19 according to the class of CT value of the reference RT-PCR
Discussion
Our results demonstrate high performances of the ID NOW COVID-19 assay for the detection of SARS-CoV-2 strains circulating in France comparatively to Alinity M SARS-CoV-2 RT-PCR assay. The performances of the ID NOW COVID-19 appeared related to the viral load of the samples estimated by the CT value. However, a strong heterogeneity of performances was previously reported (Table 1). A single study found low performances for samples displaying a CT value below 30.0. Harrington et al. reported a positive agreement rate with a reference RT-PCR of 74.7% (Abbott M2000) [9]. The false negatives of the ID NOW COVID-19 display a median CT value of 21.1 (CT range 6.8–30.3). But most reports showed lower performances of the ID NOW COVID-19 for samples displaying a CT value higher than 30.0. Considering these samples, Smithgall et al. reported a positive percent agreement rate of 34.3% with the Cobas SARS-CoV-2 assay [10]. Zhen et al. also found that all false negatives of the ID NOW COVID-19 were in this range of CT [11]. In another study, no discrepant result with the reference RT-PCR was reported for the samples displaying a CT value below 35.0, but the positive percent agreement was 27.7% for those displaying a CT value greater than 35.0 [12]. Basu et al. found an overall positive percent agreement of 54.5% with the Xpert Xpres SARS-CoV-2 assay, but all the discrepant results display a CT value greater than 36.0 [13]. Furthermore, in a French multicentric study showing an overall positive percent agreement of 83.5%, the lowest performances concern the samples displaying a CT value greater than 36.0 [14].
Table 1.
Reported performances of the ID NOW COVID-19
| Study | RT-PCR assay used as reference test | Positive percent agreement (PPA) | ||
|---|---|---|---|---|
| Reagent | Instrument | Overall | According to CT value | |
| This study | Alinity SARS-CoV-2 Amp kit (Abbott) | Alinity M (Abbott) | 94.6% |
CT value < 35: 100% CT value > 35: 75% |
| Harrington et al. [9] | RealTime SARS-CoV-2 Amp kit (Abbott) | M2000 (Abbott) | 74.7% | Not available |
| Smithgall et al. [10] | Cobas SARS-CoV-2 assay (Roche) | Cobas 6800 (Roche) | 73.9% |
CT value < 30: 100% CT value > 30: 34.3% |
| Zhen et al. [11] | Fusion SARS-CoV-2 (Hologic) | Panther (Hologic) | 87.7% | Not available |
| Mitchell et al. [12] |
CDC EUA* New York EUA* |
Not available | 71.7% |
CT value < 34: 100% CT value > 35: 38% |
| Basu et al. [13] | Xpert Xpress SARS-CoV-2 (Cepheid) | GeneXpert (Cepheid) | 54.8% | Not available |
| Plantamura et al. [14] | Multiple | Multiple | 82.8% |
CT value < 26: 100% CT value < 31: 98.8% CT value < 33: 98.1% CT value < 36: 94.7% CT value < 37: 94.1% CT value > 37: 25.9% |
| Serei et al. [15] | Xpert Xpress SARS-CoV-2 (Cepheid) | GeneXpert (Cepheid) | 60.0% | Not available |
| Procop et al. [16] | Multiple | Multiple | 83.3% | Not available |
| Thwe et al. [17] | Multiple | Multiple | 53.3% | Not available |
| Jin et al. [18]b | Cobas SARS-CoV-2 assay (Roche) | Cobas 6800 (Roche) | 63.7% |
CT value < 33: 95% CT value 33–34: 60% CT value 34–35: 36% CT value 35–36: 45% CT value 36–37: 15% CT value > 37: 13% |
| Cradic et al. [19] | Cobas SARS-CoV-2 assay (Roche) | Cobas 6800 (Roche) | 91% | Not available |
| Moore et al. [20] | RealTime SARS-CoV-2 Amp kit (Abbott) | M2000 (Abbott) | 75.2% | Not available |
*Emergency authorization use
Several variabilities in preanalytics and analytics processing could explain such differences in the evaluation of the ID NOW COVID-19 performances. Of them were viral inactivation by pre-heating, type of swab (dry or with transport media), type of transport media, and use of frozen or fresh samples. However, our results show high performances of the ID NOW COVID-19, false negative occurring for the samples displaying a low viral load (CT value > 37.0). They could correspond to a patient recovering from the infection, at the distance of the infection, or an inaccurate sampling. These performances are slightly lower than conventional RT-PCR but much higher than those of antigenic tests [21–23]. Therefore, when available, the ID NOW COVID-19 could likely replace rapid antigenic tests.
The manufacturer of the ID NOW COVID-19 recommends performing the assay on dry NPS. Consequently, another NPS must be sampled for comparison to the reference RT-PCR. The UTM allows performing both the ID NOW COVID-19 and the reference RT-PCR on the same sample, avoiding potential discrepant results due to sampling. The volume of 200 μL was chosen as the ID NOW influenza A&B 2 assay, which presents similar technical characteristics, could be performed using this volume of UTM. Furthermore, the ID NOW COVID-19 was previously tested using this condition [9, 10, 12, 14, 18, 24]. We confirm NPS sampled on UTM are suitable for the ID NOW COVID-19 assay with high performance. UTM allows repeating the test in case of invalid for instance. Nevertheless, no invalid was obtained in the present study, suggesting that the rate of invalid of the ID NOW COVID-19 is probably low, similar to the ID NOW Influenza A&B 2 assay. However, using 200 μL of transport media, some invalid were previously reported with the ID NOW COVID-19 [11, 14, 18]. And it was suggested that some transport media were associated with a high rate of invalid [14].
In conclusion, the ID NOW COVID-19 is a high-speed high-performance assay for the diagnosis of COVID-19. Despite the supplier recommends to perform the test on dry NPS sampled within 2 h, NPS sampled on UTM are suitable for the test. Using this sampling device, the test could be repeated in the case of invalids, and further analyses such as screening of genetic variants or genome sequencing could be performed with the same sample. However, as for all biological tests, the results of the ID NOW COVID-19 should be interpreted according to clinical and epidemiological contexts [25].
Acknowledgements
Members of the SARS-CoV-2 Foch Hospital study group: Emilie Catherinot, Colas Tcherakian, Antoine Magnan, Simon Chauveau, Sylvie Colin de Verdière, Hélène Salvatore, and Antoine Roux (service de pneumologie et transplantation pulmonaire); Benjamin Zuber, Charles Cerf, Richard Galliot, and Mathilde Neuville (service de réanimation); Morgan Le Guen and Camille Cornet (service d’anesthésie); Mathilde Roumier, Antoine Bizard, David Zucman, Ibrahim Marroun, Mathieu Groh, Erwan Fourn, Yoland Schoindre, David Khau, and Romain Paule (Service de médecine interne); Philippe Grenier, Anne-Laure Brun, and François Mellot (service d’imagerie médicale); Tiffany Pascreau, Sara Zia-Chahabi, Laurence Mazaux, and Emilie Jolly (Service de biologie clinique); Brigitte Bonan, Eve Camps and Antoine Faucheron (Service de la Pharmacie); Anne Verrat, Etienne Imhaus, Marie-Christine Ballester, Marie Desroziers, Mohamed Mouaci, and Charlotte Rachline (service des urgences); and Carline Amiel (service santé au travail).
Availability of data and material
Not applicable
Code availability
Not applicable
Declarations
Ethical approval
All the procedures were in accordance with the 1964 Helsinki Declaration and its later amendments.
Informed consent
According to French Health Public Law (CSPArticle L1121-1), this type of study did not require specific informed consent or ethics committee approval.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eric Farfour, Email: e.farfour@hopital-foch.com, Email: ericf6598@yahoo.fr.
on behalf of the SARS-CoV-2 Foch Hospital study group:
Emilie Catherinot, Colas Tcherakian, Antoine Magnan, Simon Chauveau, Sylvie Colin de Verdière, Hélène Salvatore, Antoine Roux, Benjamin Zuber, Charles Cerf, Richard Galliot, Mathilde Neuville, Morgan Le Guen, Camille Cornet, Mathilde Roumier, Antoine Bizard, David Zucman, Ibrahim Marroun, Mathieu Groh, Erwan Fourn, Yoland Schoindre, David Khau, Romain Paule, Philippe Grenier, Anne-Laure Brun, François Mellot, Tiffany Pascreau, Sara Zia-Chahabi, Laurence Mazaux, Emilie Jolly, Brigitte Bonan, Eve Camps, Antoine Faucheron, Anne Verrat, Etienne Imhaus, Marie-Christine Ballester, Marie Desroziers, Mohamed Mouaci, Charlotte Rachline, and Carline Amiel
References
- 1.Farfour E, Lesprit P, Visseaux B, Pascreau T, Jolly E, Houhou N, et al. The Allplex 2019-nCoV (Seegene) assay: which performances are for SARS-CoV-2 infection diagnosis? Eur J Clin Microbiol Infect Dis. 2020;39(10):1997–2000. doi: 10.1007/s10096-020-03930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farfour E, Jolly E, Pascreau T, Asso-Bonnet M, Mazaux L, Vasse M (2020) SARS-CoV-2 RT-PCR: at least 2 viral targets are needed. Infect Dis (Lond) 52(8):583–584. 10.1080/23744235.2020.1769178 [DOI] [PubMed]
- 3.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S et al. (2021) Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 3:CD013705. 10.1002/14651858.CD013705 [DOI] [PMC free article] [PubMed]
- 4.Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farfour E, Roux A, Ballester M, Gagneur L, Renaux C, Jolly E, et al. Improved performances of the second generation of the ID NOW Influenza A&B 2® and comparison with the GeneXpert®. Eur J Clin Microbiol Infect Dis. 2020;39(9):1681–1686. doi: 10.1007/s10096-020-03905-9. [DOI] [PubMed] [Google Scholar]
- 6.Van Nguyen JC, Caméléna F, Dahoun M, Pilmis B, Mizrahi A, Lourtet J, et al. Prospective evaluation of the Alere i Influenza A&B nucleic acid amplification versus Xpert Flu/RSV. Diagn Microbiol Infect Dis. 2016;85(1):19–22. doi: 10.1016/j.diagmicrobio.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 7.U.S. Food Drug Administration (2020) Coronavirus (COVID-19) Update: FDA informs public about possible accuracy concerns with Abbott ID NOW Point-of-Care Test | FDA [Internet]. [cited 2020 25]. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-informs-public-about-possible-accuracy-concerns-abbott-id-now-point. Accessed 14 May 2021
- 8.Santé Publique France (2020) Définition de cas d ’ infection au SARS-CoV-2 ( COVID-19 ) Cas possible Cas confirmé [Internet]. https://www.has-sante.fr/jcms/p_3165982/fr/coronavirus-covid-19. Accessed 14 May 2021
- 9.Harrington A, Cox B, Snowdon J, Bakst J, Ley E, Grajales P, et al. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020;58(8):e00798–20. doi: 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smithgall MC, Scherberkova I, Whittier S, Green DA. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the rapid detection of SARS-CoV-2. J Clin Viro. 2020;128:104428. doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhen W, Smith E, Manji R, Schron D, Berry GJ. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8):e00783–20. doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell SL, George KS. Evaluation of the COVID19 ID NOW EUA assay. J Clin Virol. 2020;128:104429. doi: 10.1016/j.jcv.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu A, Zinger T, Inglima K, Woo KM, Atie O, Yurasits L, et al. Performance of abbott id now covid-19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York city academic institution. J Clin Microbiol. 2020;58(8):e01136–20. doi: 10.1128/JCM.01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plantamura J, Bousquet A, Foissaud V, Garnotel E, Janvier F, Merens A (2020) Evaluation rapide multicentrique du test moléculaire rapide ID-NOW pour détection de l ’ ARN du SARS-CoV2, technologie de LAMP Période d’ évaluation: septembre 2020 Centres : laboratoire de biologie de 4 HIA
- 15.Serei VD, Cristelli R, Joho K, Salaru G, Kirn T, Carayannopoulous MO et al (2021) Comparison of Abbott ID NOW COVID-19 rapid molecular assay to Cepheid Xpert Xpress SARS-CoV-2 assay in dry nasal swabs [Internet]. Diagn Microbiol Infect Dis 1 [cited 2021 28];99. https://pubmed.ncbi.nlm.nih.gov/33545455/. 10.1016/j.diagmicrobio.2020.115208 [DOI] [PMC free article] [PubMed]
- 16.Procop GW, Brock JE, Reineks EZ, Shrestha NK, Demkowicz R, Cook E et al (2021) A comparison of five SARS-CoV-2 molecular assays with clinical correlations. Am J Clin Pathol 1 [cited 2021 28];155:69–78. /pmc/articles/PMC7665304/. 10.1093/ajcp/aqaa181 [DOI] [PMC free article] [PubMed]
- 17.Thwe PM, Ren P. How many are we missing with ID NOW COVID-19 assay using direct nasopharyngeal swabs? Findings from a mid-sized academic hospital clinical microbiology laboratory. Diagn Microbiol Infect Dis. 2020;98(2):115123. doi: 10.1016/j.diagmicrobio.2020.115123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin R, Pettengill MA, Hartnett NL, Auerbach HE, Peiper SC, Wang Z. Commercial SARS-CoV-2 molecular assays: superior analytical sensitivity of cobas SARS-CoV-2 relative to NxTAG Cov extended panel and ID NOW COVID-19 Test. Arch Pathol Lab Med. 2020;144:1303–1310. doi: 10.5858/arpa.2020-0283-sa. [DOI] [PubMed] [Google Scholar]
- 19.Cradic K, Lockhart M, Ozbolt P, Fatica L, Landon L, Lieber M, et al. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am J Clin Pathol. 2020;154(2):201–207. doi: 10.1093/ajcp/aqaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore NM, Li H, Schejbal D, Lindsley J. Hayden MK (2020) Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV reverse transcriptase PCR assay for the detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8):e00938–20. doi: 10.1128/JCM.00938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farfour E, Amiel C, Jolly E, Zia-Chahabi S, Mazaux L, Simonin P et al. The Panbio COVID-19 Ag Rapid Test: which performances are for COVID-19 diagnosis? Clin Lab. In press. Accessed May 14th, 2021 [DOI] [PubMed]
- 22.Gremmels H, Winkel BMF, Schuurman R, Rosingh A, Rigter NAM, Rodriguez O, et al. Real-life validation of the PanbioTM COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine. 2021;31:100677. doi: 10.1016/j.eclinm.2020.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, Gómez-Herruz P, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133:104659. doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Z, Zhang Y, Wang Y, Huang Z, Chest Song B, CT manifestations of new coronavirus disease, (COVID-19): a pictorial review. Eur Radiol. 2019;202030(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farfour E, Mellot F, Lesprit P, Vasse M. Foch TS (2020) SARS-CoV-2 RT-PCR and chest CT, two complementary approaches for COVID-19 diagnosis. Jpn J Radiol. 2020;38(12):1209–1210. doi: 10.1007/s11604-020-01016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable
Not applicable


