Abstract
Objectives
To develop and validate a machine learning model for the prediction of adverse outcomes in hospitalized patients with COVID-19.
Methods
We included 424 patients with non-severe COVID-19 on admission from January 17, 2020, to February 17, 2020, in the primary cohort of this retrospective multicenter study. The extent of lung involvement was quantified on chest CT images by a deep learning–based framework. The composite endpoint was the occurrence of severe or critical COVID-19 or death during hospitalization. The optimal machine learning classifier and feature subset were selected for model construction. The performance was further tested in an external validation cohort consisting of 98 patients.
Results
There was no significant difference in the prevalence of adverse outcomes (8.7% vs. 8.2%, p = 0.858) between the primary and validation cohorts. The machine learning method extreme gradient boosting (XGBoost) and optimal feature subset including lactic dehydrogenase (LDH), presence of comorbidity, CT lesion ratio (lesion%), and hypersensitive cardiac troponin I (hs-cTnI) were selected for model construction. The XGBoost classifier based on the optimal feature subset performed well for the prediction of developing adverse outcomes in the primary and validation cohorts, with AUCs of 0.959 (95% confidence interval [CI]: 0.936–0.976) and 0.953 (95% CI: 0.891–0.986), respectively. Furthermore, the XGBoost classifier also showed clinical usefulness.
Conclusions
We presented a machine learning model that could be effectively used as a predictor of adverse outcomes in hospitalized patients with COVID-19, opening up the possibility for patient stratification and treatment allocation.
Key Points
• Developing an individually prognostic model for COVID-19 has the potential to allow efficient allocation of medical resources.
• We proposed a deep learning–based framework for accurate lung involvement quantification on chest CT images.
• Machine learning based on clinical and CT variables can facilitate the prediction of adverse outcomes of COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00330-021-07957-z.
Keywords: COVID-19; Tomography, X-ray computed; Artificial intelligence; Prognosis
Introduction
The coronavirus disease 2019 (COVID-19), with its outbreak and rapid escalation, which range from the common cold to severe or even fatal respiratory infections caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a worldwide pandemic involving 188 countries or regions and more than 50 million individuals. About 10–20% of COVID-19 patients deteriorate to severe or critical illnesses within 7–14 days after symptom onset, characterized by acute respiratory distress syndrome (ARDS) and/or even multiorgan dysfunction syndrome (MODS), who require more intensive medical resource utilization, tend to develop nosocomial complications, and have worse prognosis with a case fatality rate about 20 times higher than that of non-severe patients [1–3]. There is no specific anti-coronavirus treatment for severe patients at present, and whether remdesivir is associated with significant clinical benefits for severe COVID-19 still requires further confirmation [4, 5]. Nevertheless, early antiviral therapy has been reported to be helpful in alleviating symptoms and shortening the duration of viral shedding in patients with mild to moderate COVID-19 [6, 7]. Thus, the key step in reducing the mortality from COVID-19 should be the prevention of progression from non-severe to severe disease stage and the subsequent development of critical illness. Early identification of patients at risk of adverse outcomes has the potential to enable more individualized treatment plans, but it is difficult for physicians solely based on their clinical experience [8, 9].
There have been several prognostic models in predicting adverse outcomes for COVID-19; however, most were established based on clinical biochemical parameters and few incorporated chest CT imaging features [10–12]. Chest CT is an exclusive tool to assess lung injury, which is the major hallmark of COVID-19 [13]. To accurately quantify the extent of lung injury using CT images, deep learning (DL)–based artificial intelligence (AI) technique may be an optimal solution, which has the advantages of good reproducibility, less time-consuming, and relieving the health systems overloads. Zhang et al have developed a clinically applicable AI system that can distinguish COVID-19 pneumonia from other common pneumonia and provide clinical prognosis for predicting the progression to critical illness and survival probability [14]. However, the clinical feasibility and benefit of machine learning–based model in the early prediction of the progression from non-severe to severe or critical illnesses in COVID-19 patients remain unclear.
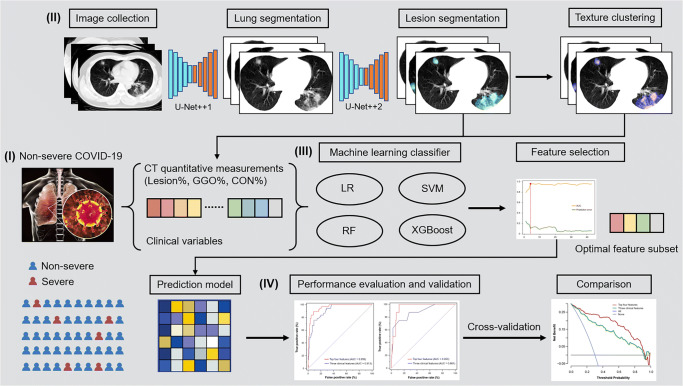
In this study, we retrospectively included patients with non-severe COVID-19 at the time of admission from multiple institutes, quantified the extent of lung injury on chest CT images using DL-based framework, constructed a machine learning model incorporating clinical characteristics and CT-derived quantitative measurement to identify the cases who developed adverse outcomes during hospitalization, determined the prediction performance and clinical use benefit, and validated these findings in an independent external cohort (Fig. 1).
Fig. 1.
Study workflow. (I) Non-severe COVID-19 patients who underwent chest CT scan on admission were included. (II) Lung and lesion segmentation were performed using DL-based framework and texture clustering was used to distinguish between GGO and CON. CT quantitative measurements including lesion%, GGO%, and CON% were calculated. (III) The optimal machine learning classifier and feature subset were selected and used for prediction model construction. (IV) The performance of the machine learning model was determined and validated in an external cohort. CON, consolidation; COVID-19, coronavirus disease 2019; CT, computed tomography; DL, deep learning; GGO, ground-glass opacification; LR, logistic regression; RF, random forest; SVM, support vector machine; XGBoost, extreme gradient boosting
Materials and methods
Study population
The Institutional Review Board of the Third Xiangya Hospital approved our study and waived the informed consent of patients for the retrospective nature of this study. The study was conducted according to the TRIPOD recommendations for prediction model development and validation [15]. Consecutive hospitalized patients with confirmed COVID-19 infection who underwent chest CT scan on admission at the Third Xiangya Hospital, First Hospital of Changsha, First Hospital of Yueyang, Second Hospital of Changde, Central Hospital of Xiangtan, Central Hospital of Shaoyang, and Central Hospital of Loudi between January 17, 2020, and February 17, 2020, were screened (n = 604). Patients who had severe or critical illnesses on admission (n = 45) and were younger than 18 years old (n = 37) were excluded. A total of 522 patients were ultimately included in this multicentre study and divided into the primary and validation cohorts according to their origin of hospital (Supplementary Figure 1). The criteria for the diagnosis and severity classification of COVID-19 infection are provided in the Supplementary Material.
Data collection
The clinical and laboratory data were obtained with data collection forms from electronic medical records. To accurately quantify the extent of lung involvement on the non-contrast chest CT images, we adopted a U-Net++ DL network developed by our team for the three-dimensional segmentation of lung and lesions (Supplementary Figure 2) [16]. Furthermore, we proposed an unsupervised multi-scale texture feature clustering method to distinguish between ground-glass opacification (GGO) and consolidation (CON) [17]. The CT lesion ratio (lesion%), GGO ratio (GGO%), and CON ratio (CON%) were then calculated, respectively. The details of data collection and CT image analysis are provided in the Supplementary Material.
Machine learning classifier and feature selection
The composite endpoint was the occurrence of severe or critical illnesses or death. The candidate feature set included 43 clinical characteristics or CT quantitative measurements, and Pearson’s correlations between features were calculated. To establish an optimal prognostic model to predict the occurrence of the composite endpoint, five supervised machine learning classifiers, namely logistic regression (LR), support vector machine with a linear kernel (SVM-Linear), SVM with a radial basis function (SVM-RBF), random forest (RF), and extreme gradient boosting (XGBoost), were employed to determine a classifier with the best performance [18]. Fivefold cross-validation was performed in the primary cohort and grid search was used for parameter tuning or hyperparameter optimization. Class weight was set at 10 to reduce the influence of inter-group unbalanced distribution. Furthermore, the average feature importance rank that indicated how valuable each feature was in the optimal classifier overall folds of cross-validation in the primary cohort was provided. With the ranked features, different feature subsets could be obtained by selecting top-n features from the ordered sequence (n = 1~43). The optimal feature subset with the highest prediction performance and minimum feature numbers was finally selected.
Model establishment and performance evaluation
The optimal machine learning classifier and feature subset were used to establish the final model. The performance to identify the patients who developed the composite endpoint in the primary and validation cohorts was assessed by the receiver operating characteristic (ROC) curve analysis. Fivefold cross-validation was performed for the machine learning classifier. The model establishment and performance evaluation of machine learning models was performed using the Python 3.7 software. Decision curve analysis was conducted to determine the clinical usefulness by quantifying the net benefits. Other statistical analyses are provided in the Supplementary Material.
Results
Patient characteristics
The main clinical characteristics of patients in the primary and validation cohorts are given in Table 1. The primary cohort that was used to train the DL-based segmentation network and construct the machine learning model consisted of 424 patients recruited from 5 hospitals, and the validation cohort that was used to externally validate the performance of the machine learning model in predicting the development of severe or critical illnesses included 98 patients recruited from 2 hospitals. There was no significant difference between the two cohorts in the prevalence of composite endpoint (8.7% vs. 8.2%, p = 0.858). The median duration from symptom onset to CT scan in all patients was 5 (range, 0–23) days.
Table 1.
Clinical characteristics of patients in the primary and validation cohorts
| Variables | Primary (n = 424) | Validation (n= 98) | p value |
|---|---|---|---|
| Age (years) | 46 (36–58) | 46 (31–53) | 0.201 |
| Male gender | 210 (49.5%) | 51 (52.0%) | 0.654 |
| Comorbidities | |||
| Any | 107 (25.2%) | 21 (21.4%) | 0.430 |
| Hypertension | 59 (13.9%) | 13 (13.3%) | 0.866 |
| Diabetes | 35 (8.3%) | 9 (9.2%) | 0.765 |
| Cardiovascular or cerebrovascular disease | 19 (4.5%) | 6 (6.1%) | 0.493 |
| COPD | 13 (3.1%) | 3 (3.1%) | 0.998 |
| Clinical outcomes | |||
| Severe or critical illnesses | 37 (8.7%) | 8 (8.2%) | 0.858 |
| Requiring mechanical ventilation | 8 (1.9%) | 3 (3.1%) | 0.466 |
| ICU admission | 14 (3.3%) | 4 (4.1%) | 0.703 |
| Death | 1 (0.2%) | 1 (1.0%) | 0.341 |
COPD, chronic obstructive pulmonary disease; ICU, intensive care unit; IQR, interquartile range
Data are presented as median (IQR) or n (percentage)
Lung lesion segmentation and quantification
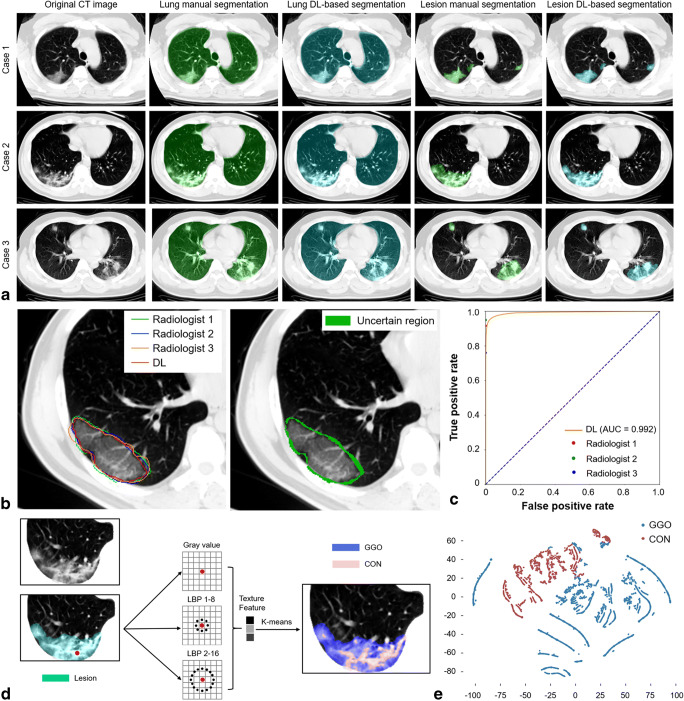
The original CT images, lung manual and DL-based segmentation, and lesion manual and DL-based segmentation of 3 example cases are illustrated in Fig. 2a, which suggested that the DL-based segmentation framework produced comparable identification of lung and lesion to manual segmentation. ROC curve analysis showed that the DL-based segmentation achieved high accuracy in identifying lesions at the pixel-level, with an AUC of 0.992, which exceeded one of three radiologists and was almost equivalent to another radiologist (Fig. 2b, c). The Dice similarity coefficient of DL-based lesion segmentation was 84.27%, while the Dice similarity coefficients of the three radiologists were 88.51%, 83.73%, and 80.92%, respectively. Furthermore, the lesion region was further subdivided into two different types (GGO and CON) using an unsupervised texture feature clustering approach based on the differences of attenuation and texture (Fig. 2d). The three lesion indicators, namely lesion%, GGO%, and CON%, of each patient in the primary and validation cohorts were yielded (Supplementary Figure 3).
Fig. 2.
DL-based lung and lesion segmentation and CT quantitative measurements. a The original CT images, lung segmentation, and lesion segmentation of 3 example cases. b The contours of 3 radiologists and lesion DL-based segmentation (left) and the uncertain region (right). c ROC curve of the pixel-level performance of DL-based segmentation to identify the lesion. d Unsupervised multi-scale texture feature clustering to distinguish between GGO and CON based on grey-level attenuation and LBP features. e t-SNE plot showing the pixel-level GGO or CON distribution. CON, consolidation; CT, computed tomography; DL, deep learning; GGO, ground-glass opacification; LBP, local binary pattern; ROC, receiver operating characteristic; t-SNE, t-distributed stochastic neighbour embedding
Machine learning classifier and feature selection
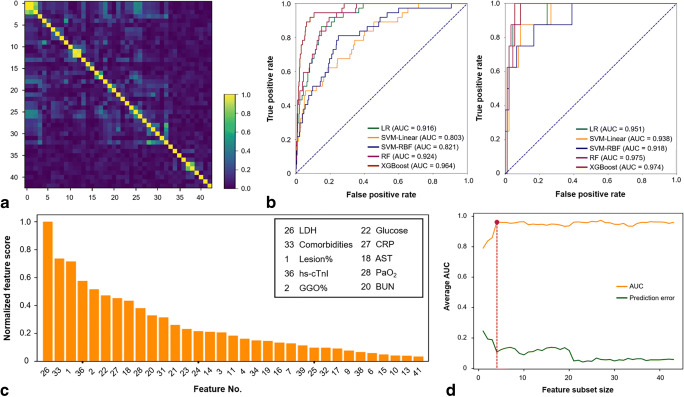
Clinical characteristics and CT quantitative measurements among patients according to whether to develop composite endpoint in the primary cohort are shown in Table 2. The correlation matrix heatmap of all 43 features is shown in Fig. 3a. The lesion% and GGO% were significantly and positively correlated with age, alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), creatine kinase, lactic dehydrogenase (LDH), and C-reactive protein (CRP) and negatively correlated with lymphocyte count (all p < 0.01), while CON% was significantly and positively correlated with AST and LDH (both p < 0.01). Considering the unobvious multicollinearity between features and specific clinical significance of each feature, we included all the features as a candidate feature set.
Table 2.
Clinical characteristics and CT quantitative measurements among patients according to whether to develop composite endpoint in the primary cohort
| Variables | Yes (n = 37) | No (n = 387) | p value |
|---|---|---|---|
| Age (years) | 58 (51–67) | 45 (35–56) | < 0.001 |
| Male gender | 20 (54.1%) | 190 (49.1%) | 0.564 |
| Smoking history | 7 (18.9%) | 33 (8.5%) | 0.068 |
| Comorbidities | |||
| Any | 25 (67.6%) | 82 (21.2%) | < 0.001 |
| Hypertension | 11 (29.7%) | 48 (12.4%) | 0.004 |
| Diabetes | 8 (21.6%) | 27 (7.0%) | 0.006 |
| Cardiovascular or cerebrovascular diseases | 7 (18.9%) | 12 (3.1%) | 0.001 |
| COPD | 7 (18.9%) | 6 (1.6%) | < 0.001 |
| Symptoms and signs | |||
| Fever | 28 (75.7%) | 220 (56.8%) | 0.026 |
| Cough | 24 (64.9%) | 199 (51.4%) | 0.118 |
| Fatigue or myalgia | 8 (21.6%) | 84 (21.7%) | 0.991 |
| Dyspnea | 4 (10.8%) | 17 (4.4%) | 0.100 |
| Temperature (°C) | 37.3 (36.8–38.0) | 36.9 (36.5–37.3) | 0.001 |
| Heart rate (/min) | 90 (80–105) | 86 (78–96) | 0.092 |
| Respiratory rate (/min) | 21 (20–22) | 20 (19–20) | 0.053 |
| Laboratory findings | |||
| Hemoglobin (g/L) | 126.5 (119.3–136.0) | 131.0 (120.0–143.0) | 0.300 |
| Platelet count (×109/L) | 148.0 (119.5–208.0) | 174.0 (139.0–228.0) | 0.067 |
| White blood cell count (×109/L) | 4.5 (3.6–6.0) | 4.6 (3.6–5.7) | 0.812 |
| Neutrophil count (×109/L) | 3.0 (2.4–4.5) | 2.9 (2.1–3.7) | 0.090 |
| Lymphocyte count (×109/L) | 0.9 (0.7–1.3) | 1.2 (0.9–1.6) | < 0.001 |
| Monocyte count (×109/L) | 0.4 (0.2–0.5) | 0.4 (0.3–0.5) | 0.618 |
| Total bilirubin (μmol/L) | 10.5 (7.1–14.6) | 11.9 (8.8–17.3) | 0.031 |
| ALT (U/L) | 23.0 (16.6–31.2) | 19.7 (14.5–28.4) | 0.124 |
| AST (U/L) | 33.2 (25.8–44.6) | 23.0 (18.3–28.3) | < 0.001 |
| Albumin (g/L) | 36.8 (34.2–39.8) | 39.3 (36.5–42.6) | 0.001 |
| BUN (mg/dL) | 4.7 (3.8–5.8) | 3.9 (3.1–4.8) | 0.002 |
| Creatinine (μmol/L) | 66.1 (53.8–86.0) | 56.4 (44.8–70.0) | 0.002 |
| Glucose (mmol/L) | 7.2 (5.8–9.2) | 5.7 (3.6–4.3) | < 0.001 |
| K+ (mmol/L) | 3.7 (3.5–4.0) | 4.0 (3.6–4.3) | 0.051 |
| Na+ (mmol/L) | 135.3 (133.0–137.6) | 137.5 (135.5–139.9) | < 0.001 |
| INR | 1.22 (0.99–1.33) | 1.10 (0.90–1.19) | 0.043 |
| D-dimer ≥ 0.5 mg/L | 16 (43.2%) | 47 (12.1%) | < 0.001 |
| Procalcitonin ≥ 0.05 ng/mL | 21 (56.8%) | 124 (32.0%) | 0.002 |
| Hs-cTnI ≥ 28 pg/mL | 5 (13.5%) | 11 (2.8%) | 0.008 |
| Creatine kinase (U/L) | 94.0 (40.0–213.5) | 72.0 (49.1–109.0) | 0.139 |
| LDH (U/L) | 265.0 (184.6–342.8) | 174.0 (141.3–214.1) | < 0.001 |
| CRP (mg/L) | 40.9 (22.9–61.0) | 10.4 (2.4–24.5) | < 0.001 |
| PaO2 (mmHg) | 71.1 (54.6–106.7) | 90.9 (76.0–115.8) | 0.009 |
| Radiological findings | |||
| Number of segments involved | 16 (12–18) | 9 (5–13) | < 0.001 |
| CT severity score | 12 (7–17) | 6 (3–9) | < 0.001 |
| CT quantitative measurements | |||
| Lesion% | 9.5 (3.5–26.6) | 3.1 (0.6–7.5) | < 0.001 |
| GGO% | 8.2 (3.3–18.9) | 2.8 (0.6–6.7) | < 0.001 |
| CON% | 1.3 (0.2–2.9) | 0.3 (0.0–0.7) | < 0.001 |
ALT, alanine aminotransferase; AST, aspartate aminotransferase; BUN, blood urea nitrogen; CON, consolidation; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; CT, computed tomography; GGO, ground-glass opacification; Hs-cTnI, hypersensitive cardiac troponin I; INR, international normalized ratio; K+, potassium; LDH, lactic dehydrogenase; Na+, sodium; PaO2, partial pressure of oxygen
Fig. 3.
Optimal machine learning classifier and feature subset selection. a The heatmap illustrating the correlations between features in the candidate feature set. b The performance of five machine learning classifiers, including LR, SVM-Linear, SVM-RBF, RF, and XGBoost, based on the candidate feature set in the primary cohort (left) and validation cohort (right). c The feature importance rank in the XGBoost classifier using fivefold cross-validation in the primary cohort. d The relationship between the feature subset size and model performance. The optimal size (red dot) was determined with the highest average AUC and a minimal number of features. The optimal feature subset contained the top 4 features, i.e. LDH, presence of comorbidity, lesion%, and hs-cTnI. AST, aspartate aminotransferase; AUC, area under the receiver operating characteristic curve; BUN, blood urea nitrogen; CRP, C-reactive protein; GGO, ground-glass opacification; hs-cTnI, hypersensitive cardiac troponin I; LDH, lactic dehydrogenase; LR, logistic regression; PaO2, partial pressure of oxygen; RF, random forest; SVM-Linear, support vector machine with a linear kernel; SVM-RBF, support vector machine with a radial basis function; XGBoost, extreme gradient boosting
We compared the performance of five machine learning classifiers based on the candidate feature set in identifying the patients who developed adverse outcomes in the primary cohort and then tested in the validation cohort. Figure 3b depicts the ROC curves of all the classifiers and the mean AUC of fivefold cross-validation, sensitivity, specificity, and accuracy are given in Table 3. The XGBoost achieved the highest performance (AUC = 0.964) in the primary cohort, followed by RF (AUC = 0.924), LR (AUC = 0.916), SVM-RBF (AUC = 0.821), and SVM-Linear (AUC = 0.803). Then, the XGBoost classifier was selected as the optimal machine learning classifier. Furthermore, the XGBoost classifier achieved comparable performance (AUC = 0.974) in the validation cohort.
Table 3.
Performance of each classifier based on the candidate feature set in the primary and validation cohorts
| Classifier | AUC | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| Primary cohort | ||||
| LR | 0.916 (0.885–0.938) | 67.6% (25/37) | 90.4% (350/387) | 0.884 (0.851–0.911) |
| SVM-Linear | 0.803 (0.760–0.838) | 51.4% (19/37) | 86.0% (333/387) | 0.830 (0.790–0.864) |
| SVM-RBF | 0.821 (0.780–0.856) | 75.7% (28/37) | 84.0% (325/387) | 0.833 (0.793–0.866) |
| RF | 0.924 (0.894–0.947) | 59.5% (22/37) | 93.0% (360/387) | 0.901 (0.867–0.927) |
| XGBoost | 0.964 (0.941–0.979) | 75.7% (28/37) | 96.4% (373/387) | 0.946 (0.919–0.965) |
| Validation cohort | ||||
| XGBoost | 0.974 (0.910–0.996) | 100% (8/8) | 85.6% (77/90) | 0.867 (0.780–0.925) |
AUC, area under the receiver operating characteristic curve; LR, logistic regression; RF, random forest; SVM-Linear, support vector machine with a linear kernel; SVM-RBF, support vector machine with a radial basis function; XGBoost, extreme gradient boosting
The feature importance rank of each feature in the XGBoost classifier is presented in Fig. 3c and Supplementary Table 2. Then, feature selection was performed in the candidate feature set, as depicted in Fig. 3d. The optimal feature subset containing the top four features, i.e. LDH, presence of comorbidity, lesion%, and hypersensitive cardiac troponin I (hs-cTnI), achieved the highest average AUC, with the minimal number of features.
Performance evaluation of machine learning model
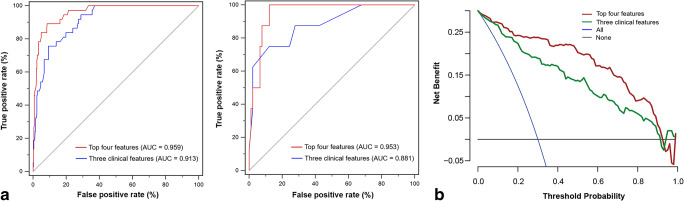
The XGBoost classifiers based on the optimal feature subset or only three clinical features in the optimal feature subset (i.e. LDH, presence of comorbidity, and hs-cTnI) were then constructed, respectively. The XGBoost classifier based on the top 4 features achieved satisfactory performance in the primary cohort, which was significantly superior to that based on only three clinical features (AUCs = 0.959 and 0.913, respectively; p = 0.007). However, no significant difference was found between the two classifiers in the validation cohort (AUCs = 0.953 and 0.881, respectively; p = 0.216). The illustration of the ROC curves in the primary and validation cohorts is shown in Fig. 4a, and the detailed model performance is listed in Table 4. The decision curve analysis for the two XGBoost classifiers in the whole cohort is presented in Fig. 4c. Our XGBoost classifier based on the top 4 features had the optimal overall net benefit, the treat-all-patients scheme, and the treat-none scheme across the majority of the range of reasonable threshold probabilities.
Fig. 4.
Performance of the XGBoost classifiers based on the top four features or only three clinical features. a ROC curves of the XGBoost classifiers in the primary cohort (left) and validation cohort (right). b Comparison of decision curves of the XGBoost classifiers in the whole cohort. AUC, area under the receiver operating characteristic curve; ROC, receiver operating characteristic; XGBoost, extreme gradient boosting
Table 4.
Performance of the XGBoost classifiers in the primary and validation cohorts
| Cohort | AUC | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|
| Primary cohort | ||||
| Top four features | 0.959 (0.936–0.976) | 89.2% (33/37) | 91.5% (354/387) | 0.913 (0.882–0.936) |
| Three clinical features | 0.913 (0.882–0.938) | 75.7% (28/37) | 90.7% (351/387) | 0.894 (0.861–0.920) |
| Validation cohort | ||||
| Top four features | 0.953 (0.891–0.986) | 100% (8/8) | 87.8% (79/90) | 0.888 (0.810–0.936) |
| Three clinical features | 0.881 (0.800–0.938) | 75.0% (6/8) | 87.8% (79/90) | 0.867 (0.786–0.921) |
AUC, area under the receiver operating characteristic curve; XGBoost, extreme gradient boosting
Discussion
Our results suggested that DL-based chest CT quantitative measurement could be combined with significant clinical variables to early identify the patients who developed adverse outcomes during hospitalization for patients with COVID-19 using machine learning algorithm. We established an XGBoost classifier incorporating LDH, presence of comorbidity, lesion%, and hs-cTnI which achieved perfectly prediction performance both in the primary and validation cohorts. These findings were derived from DL-based CT quantitative lung injury measurements with sufficient accuracy, stepwise optimal machine learning classifier and feature selection, implemented internal cross-validation and independent external validation, and heterogeneous image data from multiple hospitals; thus, we expect our results to be well generalizable. Hence, when utilized as a supportive decision tool in clinical practice, the proposed prediction of adverse outcomes for COVID-19 could accelerate the early identification of the patients with a high risk of progression enabling faster intervention and likelihood of better outcomes.
Some patients with COVID-19 develop dyspnea and hypoxemia shortly after illness onset and may further progress to ARDS or MODS even death [9]. To early identify the patients who were likely to develop adverse outcomes, our study presented a machine learning model incorporating four clinical or imaging variables, with perfect performance in the primary and validation cohorts, respectively. Zhang et al developed a clinically applicable AI-assisted model to predict the progression to critical illness with AUC, sensitivity, and specificity of 0.909, 86.71%, and 80.00%, respectively, which identified the quantitative lesion features as the most significant contributor in the clinical prognosis estimation as well as some clinical parameters relating to multiple tissues/organs function and systemic homeostasis [14]. Compared with their work, we built a model incorporating fewer significant features for clinical use, slightly improved the prediction performance, and validated these findings in an independent external cohort. As for the difference in the most important features of the machine learning model between our study and theirs, this may be explained by the differences in the machine learning algorithm adopted and study endpoint.
Previous studies reported some feasible prognostic model for the prediction of developing severe COVID-19, particularly the CALL score [11, 19]. Similar to our results, the CALL score also included four high-risk factors associated with COVID-19 progression, i.e. underlying comorbidity, age, LDH, and lymphocyte count. In our XGBoost classifier, CT-derived lesion% and hs-cTnI were also included apart from LDH and presence of comorbidity. In general, the top four features in our model were associated with multiple tissues/organs dysfunction, lung injury, and declined organ reserve function, respectively. LDH is an intracellular cytoplasmic enzyme that is widely expressed in multiple tissues and has been reported as a predictor of disease severity in several clinical conditions [20, 21]. COVID-19 involves multiple organs or systems, including the gastrointestinal tract, liver, kidney, cardiovascular system, and nervous system [22–24]. Damage to the liver, kidney, or lung in severe attacks may contribute to the cellular death and LDH leakage with consequently raised serum LDH levels in COVID-19. Meanwhile, hs-cTnI is the best laboratory parameter inflecting cardiac involvement with COVID-19, which could prompt early initiation of measures to improve tissue oxygenation. Elevated hs-cTnI concentration may be due to non-ischemic causes of myocardial injury or type 2 myocardial infarction, of which the prevalence is likely to increase in patients affected by COVID-19 [25]. Besides, it is the sensitivity of hs-cTnI testing that ensures it is one of the earliest and most precise indicators of organ dysfunction [26]. The significance of LDH and hs-cTnI as risk factors in predicting the development of ARDS or mortality has also been proposed in previous reports [9, 27]. CT-derived lesion% is a quantitative indicator directly obtained on DL-based lesion segmentation, which is associated with the extent of pulmonary infection by SARS-CoV-2. Lung involvement in COVID-19 reflects the most serious degree of damage caused by the coronavirus on various organs or systems. Furthermore, chronic comorbidity has been shown to be an independent prognostic factor associated with unfavourable outcomes in many reports [27, 28]. As expected, our analysis revealed that underlying comorbidity played an important role in the clinical progression in COVID-19 patients, which may be explained by the overactivation of the renin-angiotensin system (RAS) and enhanced susceptibility to pulmonary edema by the exhaustion of angiotensin-converting enzyme 2 (ACE2), which is the functional receptor for the SARS- CoV-2 spike protein [29, 30]. Recently, Liang et al proposed a clinical risk score incorporating 10 clinical variables to predict the occurrence of critical illness in hospitalized patients with COVID-19 [19]. By contrast, we adopted DL-derived CT quantitative measurements to accurately assess the degree of lung injury and aimed to early predict the adverse outcomes in patients with non-severe COVID-19 pneumonia on admission, and our findings further suggested that CT-derived lesion% played an important role in our XGBoost machine learning model.
To analyze the composition proportions of lung lesions, we innovatively proposed an unsupervised multi-scale texture feature clustering to distinguish GGO and CON without the need of prior annotated data for training for further quantification. Shi et al found that COVID-19 pneumonia manifested with dynamic CT abnormalities during disease evolution, with focal unilateral to diffuse bilateral GGOs that progressed to or co-exist with CONs [13]. Thus, we speculated that the extent or proportion of GGO and CON may contribute to early predicting the disease evolution. According to our results, GGO% ranked the fifth important features in identifying patients who were likely to develop severe or critical illnesses. However, to simplify the machine learning classifier with sufficient accuracy, we only included the top 4 features in our final model. Another study showed that the average infection attenuation of lung abnormalities computed automatically by a deep learning–based AI system could distinguish between the severe and non-severe COVID-19 stages [31]. However, we did not use the average attenuation of lesion to discriminate between GGO and CON in our study since there is no recognised reference threshold value. Besides, CT severity score, a semi-quantitative index associated with the lung involvement, also has been subjectively estimated and included in the candidate feature set. However, the feature importance rank indicated that the radiologist-derived CT severity score was inferior to these DL-derived CT quantitative measurements, which provides more accurate, objective, and reproducible quantification of lung involvement.
There were some limitations in our study. First, the study was retrospectively conducted and the laboratory tests were clinically driven and not systematic, which resulted in incomplete laboratory tests results in some cases. Second, the cytokine storm is the hallmark of severe ill COVID-19, which is characterized by increased amounts of serum proinflammatory cytokines [32]. The detection of cytokines may have added a further dimension to this study. Third, the utility of our model is limited by unavailable open-source segmentation software and lack of easy-to-use online tool. Also, the selection of the optimal machine learning classifier was subjective. Finally, the proportions of patients who reached the composite endpoint in the primary or validation cohorts were about 8%. Although we employed class weight adjustment to reduce the impact of imbalanced samples on the prediction performance of the machine learning classifier, our established model may be limited by the potential overfitting risk and specific cohort characteristics. The possibility to extrapolate our model to other patient populations needs to be confirmed by a larger sample.
In summary, our study presented a machine learning model incorporating four clinical or imaging variables at the time of admission with high accuracy to identify the patients who developed adverse outcomes during hospitalization, which could be used to facilitate the prediction of adverse outcomes in patients with COVID-19. Our findings may allow efficient utilization of medical resources and individualized treatment plans for COVID-19 patients.
Supplementary information
(DOCX 858 kb)
Abbreviations
- COVID-19
Coronavirus disease 2019
- CT
Computed tomography
- DL
Deep learning
- GGO
Ground-glass opacification
- hs-cTnI
Hypersensitive cardiac troponin I
- LDH
Lactic dehydrogenase
- lr
Logistic regression
- RF
Random forest
- SVM-Linear
Support vector machine with a linear kernel
- SVM-RBF
Support vector machine with a radial basis function
- XGBoost
Extreme gradient boosting
Funding
This study was supported by the National Natural Science Foundation of China (81771827, 81471715 to Rong), the Wisdom Accumulation and Talent Cultivation Project of the Third Xiangya Hospital of Central South University (2020; to Rong), and the Key Research and Development Program of Hunan Province (2020SK2097 to Shen).
Declarations
Guarantor
The scientific guarantor of this publication is Zhichao Feng, M.D.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Hongzhuan Tan kindly provided statistical advice for this manuscript.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval from the Ethics Committee of The Third Xiangya Hospital of Central South University (Changsha, China) was obtained.
Methodology
• retrospective
• case-control study/diagnostic or prognostic study
• multicentre study
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhichao Feng and Hui Shen contributed equally to this work.
Contributor Information
Dewen Hu, Email: dwhu@nudt.edu.cn.
Wei Wang, Email: cjr.wangwei@vip.163.com.
References
- 1.Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, Ling Y, Bai T, et al. COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201:1380–1388. doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung IF, Lung KC, Tso EY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Z, Li J, Yao S, et al. Clinical factors associated with progression and prolonged viral shedding in COVID-19 patients: a multicenter study. Aging Dis. 2020;11:1069–1081. doi: 10.14336/AD.2020.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2019.20153. [DOI] [PubMed] [Google Scholar]
- 9.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji D, Zhang D, Xu J, et al. Prediction for progression risk in patients with COVID-19 pneumonia: the CALL score. Clin Infect Dis. 2020;71:1393–1399. doi: 10.1093/cid/ciaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Z, Yu Q, Yao S, et al. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nat Commun. 2020;11:4968. doi: 10.1038/s41467-020-18786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang K, Liu X, Shen J, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433 e1411. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med. 2015;162:55–63. doi: 10.7326/M14-0697. [DOI] [PubMed] [Google Scholar]
- 16.Gao K, Su J, Jiang Z, et al. Dual-branch combination network (DCN): towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med Image Anal. 2021;67:101836. doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie C, Yang P, Zhang X, et al. Sub-region based radiomics analysis for survival prediction in oesophageal tumours treated by definitive concurrent chemoradiotherapy. EBioMedicine. 2019;44:289–297. doi: 10.1016/j.ebiom.2019.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angraal S, Mortazavi BJ, Gupta A, et al. Machine learning prediction of mortality and hospitalization in heart failure with preserved ejection fraction. JACC Heart Fail. 2020;8:12–21. doi: 10.1016/j.jchf.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Liang W, Liang H, Ou L, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Dong L, Zhang Y, et al. Prediction of severe acute pancreatitis using a decision tree model based on the revised atlanta classification of acute pancreatitis. PLoS One. 2015;10:e0143486. doi: 10.1371/journal.pone.0143486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muchtar E, Dispenzieri A, Lacy MQ, et al. Elevation of serum lactate dehydrogenase in AL amyloidosis reflects tissue damage and is an adverse prognostic marker in patients not eligible for stem cell transplantation. Br J Haematol. 2017;178:888–895. doi: 10.1111/bjh.14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung KS, Hung IFN, Chan PPY, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei F, Liu YM, Zhou F, et al. Longitudinal association between markers of liver injury and mortality in COVID-19 in China. Hepatology. 2020;72:389–398. doi: 10.1002/hep.31301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammadah M, Kim JH, Tahhan AS, et al. Use of high-sensitivity cardiac troponin for the exclusion of inducible myocardial ischemia: a cohort study. Ann Intern Med. 2018;169:751–760. doi: 10.7326/M18-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman AR, Bularga A, Mills NL. High-sensitivity cardiac troponin can be an ally in the fight against COVID-19. Circulation. 2020;141:1733–1735. doi: 10.1161/CIRCULATIONAHA.120.047008. [DOI] [PubMed] [Google Scholar]
- 27.Du RH, Liang LR, Yang CQ, et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. 2020;55:2000524. doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen R, Liang W, Jiang M, et al. Risk factors of fatal outcome in hospitalized subjects with coronavirus disease 2019 from a nationwide analysis in China. Chest. 2020;158:97–105. doi: 10.1016/j.chest.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaduganathan M, Vardeny O, Michel T, McMurray JJV, Pfeffer MA, Solomon SD. Renin-angiotensin-aldosterone system inhibitors in patients with Covid-19. N Engl J Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touyz RM, Li H, Delles C. ACE2 the Janus-faced protein - from cardiovascular protection to severe acute respiratory syndrome-coronavirus and COVID-19. Clin Sci (Lond) 2020;134:747–750. doi: 10.1042/CS20200363. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Zhong Z, Li Y, et al. From community-acquired pneumonia to COVID-19: a deep learning-based method for quantitative analysis of COVID-19 on thick-section CT scans. Eur Radiol. 2020;30:6828–6837. doi: 10.1007/s00330-020-07042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 858 kb)