Abstract
Severe coronavirus disease 2019 (COVID-19) caused by the Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) is characterized by an unpredictable disease course, with variable presentations of different organ systems. The clinical manifestations of COVID-19 are highly variable ranging from mild presentations to severe, life-threatening symptoms and the wide individual variability may be due to the broad heterogeneity in the underlying pathologies. There is no doubt that early management may have a major influence on the outcome. This led the scientists to search for ways to monitor disease progression or to predict outcomes in COVID-19. Although it is not yet possible to predict who will progress to the severe forms or in what time, numerous prospective and longitudinal studies represent the evidence for determining the potential immunological risk factors of COVID-19 critical disease and death. The kinetics and breadth of immune responses during COVID-19 appear to follow a trend which is consistent to the predominant pathological alterations. Recent publications have used these biomarkers to help identify patients who will develop the severe acute COVID-19. Of particular interest is the relationship between the kinetics of peripheral leukocytes and clinical progress of the disease in COVID-19. Although research is ongoing in this area, we present details about the current status of the evaluation. Understanding of the COVID-19 related alterations of the innate and adaptive immune responses may help to promote the vaccine development and immunological interventions.
Keywords: 2019 novel coronavirus disease (COVID-19), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Immune response kinetics, Disease progression, Fatal outcome, Leukocyte, Lymphocyte
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a new member of the Coronaviridae family, comprising of the alpha- and beta-coronaviruses that are considered to be mammalian pathogens and the gamma and delta-coronaviruses which mostly infect birds [3]. Beta-coronaviruses have been responsible for prior outbreaks of respiratory infections in humans with coronaviruses such as SARS and Middle East respiratory syndrome (MERS) in the years 2002 and 2012, respectively. SARS-CoV-2 belonging to the beta-coronaviruses is markedly different from other coronaviruses with 79.5% nucleotide sequence similarity with SARS and to a less extent (60% similarity) with MERS [4]. Although SARS-CoV-2 share some homology in genomic and phenotypic characteristics with the related SARS and MERS viruses, it has been noticed that the pathology of the disease due to SARS-CoV-2 exhibit some distinct features with high risk of person-to-person transmission and widespread prevalence [5].
SARS-CoV-2 infected patients show typical pneumonia and develop complications that might result in death due to massive lung damage and progressive respiratory failure [6]. Clinical manifestations of SARS-CoV-2 infection include spectrum of asymptomatic or mild illness to severe condition with pneumonia, acute respiratory distress syndrome (ARDS), and multiorgan dysfunction [7]. It has been reported that SARS-CoV-2 patients with mild symptoms tend to have a better overall outcome after one week, while the patients with severe infection have an elevated risk of progression to acute lower respiratory infection leading to respiratory failure and/or multi-organ dysfunction syndrome and death [8]. According to the reports from the Chinese Center for Disease Control and Prevention, from a total of 44,500 confirmed infections the vast majority (80%) of cases had mild disease experiencing either mild pneumonia or no complications, 14% of the patients exhibited a documented severe disease and 5% developed critical illness (defined as having respiratory failure, multi-organ failure or systemic shock) [9]. It is assumed that approximately 20–30% of hospitalized patients with COVID-19-associated pneumonia may ultimately require intensive care unit (ICU) support [10,11].
A key question for hospitalized patients with COVID-19, then, is how immune responses alter over time in the course of COVID-19. Complete and comprehensive clinical assessment focusing on the immunological characteristics is fundamental to the appropriate selection of treatment for the patient groups and for reliable analysis of experimental results [12,13]. A proper comprehension of viral immunopathogenesis may help with earlier management of the severe complications and clarify the best approach in managing this disease and better monitoring of the treatment response as well as clinical course [14,15]. With this review, we aim to contribute to the understanding of the clinical spectrum of the infection resulted from SARS-CoV-2 by summarizing the kinetic changes in immune-cell populations at systemic level during the course of illness in mild, moderate and severe conditions.
2. The kinetics of lymphocyte subsets in the peripheral blood during COVID-19
2.1. Total lymphocytes and neutrophil counts
Following infection with COVID-19, circulating neutrophils increase in numbers while total lymphocyte counts decrease. Studies show a correlation between the degree of lymphopenia and severity of the SARS-CoV-2 infection [16]. The study of 41 hospitalized cases in Wuhan revealed that the proportions of patients with elevated neutrophils, and decreased lymphocyte counts were statistically different in patients of ICU vs. non-ICU care and the severity of these adverse events positively correlated with disease severity and fatal outcome in COVID-19 patients [17]. In the fatal cases of SARS-CoV or MERS-CoV, prolonged ongoing inflammation, part of which includes increased neutrophil, is consistently observed [18,19]. Higher neutrophil counts can be useful to discriminate between those who develop ARDS and those without ARDS by the findings that the COVID-19 cases with pneumonia who had developed ARDS showed elevated levels of neutrophils in peripheral blood than did those without ARDS [20]. Increased neutrophil count followed by a further activation of neutrophils leads directly to the release of chemokines and cytokines contributing to cytokine storm. Neutrophilia is also defined as a risk factor associated with ARDS progression to death among COVID-19 patients [20,21]. Of particular interest is the evidence that though the lymphocyte count was correlated with the development of ARDS in SARS-CoV-2, it was not associated with death. Patients might protected from developing ARDS, but not from death by increased frequency of CD3+ and CD4+ T-cells [20].
Together with the disease improved condition, the lymphocyte count returns to normal, which is most contributed by the enhancement of both CD4+ and CD8+ T cells whereas NK- and B cells stably remain in reduced counts [22]. In a retrospective cohort study of patients who were hospitalized with COVID-19 in Wuhan, China, significant higher numbers of lymphocytes were found in survivors versus non-survivors, among whom severe lymphopenia was observed until death, suggesting that patients with lymphopenia had a high risk of death. Alternatively, leukopenia showed the opposite trend, being more prevalent among survivors (20%) than non-survivors (9%) [23]. It has been documented that over activation of innate immune responses can be suppressed by the T-cell responses during viral infection [24,25]. Thus, one hypothesis is that a loss of T cells in the course of SARS-CoV-2 infection may participate in worsen inflammatory responses, while T cell number restoration may resolve inflammation [26]. In line with this assumption are the results of an analysis of lymphocyte responses indicating a reverse correlation between the changes of T cell counts and the cytokine kinetic alterations in peripheral blood of severe COVID-19 patients [16]. Still, there is a lack of overlap between the associations of fatality with lymphocyte counts in different studies, possibly because of limited patient populations in most studies. Thus, larger cohort studies are warranted to further define the disease risk factors and clinical characteristics. What is clearly demonstrated by the above mentioned data is that the vast majority of patients with severe diseases have lymphopenia [27,28]. Other studies also confirm the kinetic alterations of various leukocyte subsets and demonstrate the use of aforementioned criteria as prognostic parameters for the severe course of the COVID-19 infection [16]. Recent researches have reported other prognostic factors, besides the degree of increase in neutrophils and decrease in lymphocyte counts, such as the ratio of neutrophils to different lymphocyte subsets [29]. A recent study in a 40-patient cohort identified neutrophil-to-lymphocyte ratio (NLR) and neutrophil-to-CD8+ T cell ratio (N8R) as independent factors to predict the incidence of severe disease in COVID-19 patients [16].
The mechanism underlying the observed lymphocyte reduction is still not clarified. One prevalent hypothesis has been the activation-induced cell death of lymphocytes by apoptosis induced by Fas/Fas ligand pathway as well as effects of TNF-related apoptosis-inducing ligand (TRAIL) [30]. In addition, there is some evidence that SARS-CoV-2 can directly infect lymphocytes, particularly T cells, that initiate or provoke cell death and thus would eventually result in lymphopenia [[31], [32], [33], [34]]. There may also be lymphocyte trafficking and sequestration in the infected tissue induced by cytokine storm [35] considering that the majority of the infiltrating adaptive immune cells in the lungs from severe COVID-19 patients are likely T cells [36].
2.2. CD4+ and CD8+ T cell responses
T cells are the main mediators of cell-mediated adaptive immunity which function in the killing of cells that are affected by a virus or a tumor [26]. There exist two principal populations of T cells, the CD4+ helper T (Th) cells and the cytotoxic CD8+ T cells. CD8+ cytotoxic T cells through a variety of effector mechanisms, which include perforin, granzyme, and interferons (IFNs) are principally responsible for eliminating viruses from the host body [37]. Successful virus elimination also depends on CD4+ T cells helping cytotoxic T cells and B cells [38,39]. At present, however, significant reduction in total lymphocytes is part of routine blood analysis in COVID-19, variations in the lymphocyte subset counts is not clearly known. Research into lymphocyte subsets is of great value to ensure immune system functionality [40]. Cell-mediated immunity involving T cells generates efficient antiviral responses against SARS-CoV-2 [41]. COVID-19 patients have shown deficient T-cell responses, related to both qualitative and quantitative perturbations of T-cell subsets [42]. In a cohort study of 452 patients, CD4+ T cells counts were reduced below normal levels in laboratory-confirmed COVID-19 patients, and the decrease was more evident in the severe cases. By contrast, no significant changes were observed in the numbers of CD8+ T cells of the infected patients in the cohort proposing a role for dysregulated immune responses in COVID-19 pathogenesis.
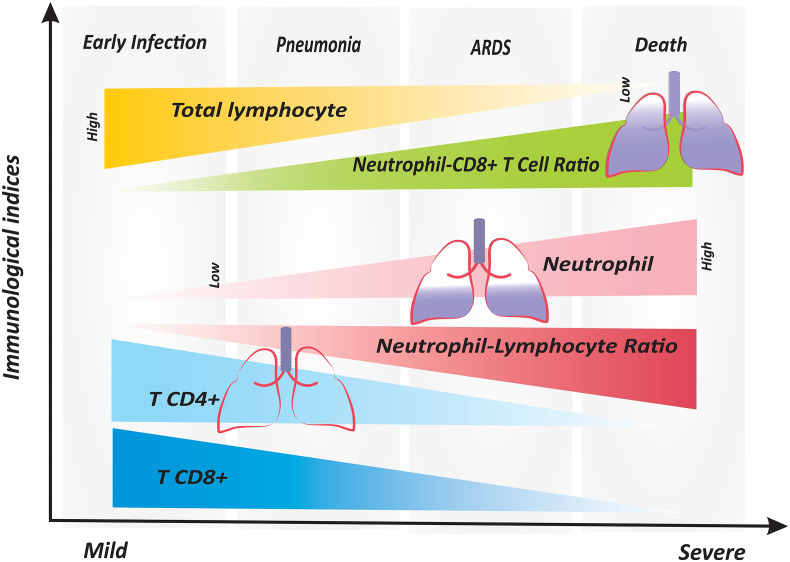
SARS-CoV-2 infection may also have adverse effects on lymphocyte homeostasis independent of the absolute numbers of major T cell subsets. Different from that reported earlier, Zheng et al. observed that the absolute numbers of major circulating leukocytes in both mild and severe/critical patients with COVID-19 remained similar to healthy group [42]. Surprisingly, in that study, IL-6 and TNF-α plasma concentrations failed to reach statistical significance among the three groups. However, the perturbed T cell response was shown in that study by focusing on molecules concerned with activation and regulation of T cells. Accordingly, TNF-α and IFN-γ levels in CD4+ T cells were found lessen in the severe group compared to those in the mild group, whereas the higher amounts of cytotoxic mediators granzyme B and perforin were detected in CD8+ T cells from the severe patients compared to those of the mild group [42], which is in consistent with a recent case report [43]. Furthermore, CD8+ T cell subset showed a higher activation state with an increased expression of activation molecules HLA-DR and TIGIT in severe cases than mild cases, whereas these molecules showed no difference in CD4+ T cells. Functional and phenotypic characterization of T cell subsets indicated that the proportion of non-functional (IFN-γ − TNF-α − IL-2−) CD4+ and exhausted (PD-1 − CTLA-4 − TIGIT−) CD8+ T cells was significantly higher in the severe group than that in the mild and healthy groups [42]. In another study, similar data were obtained for CD8 molecule of the T-lymphocyte subgroups which showed the protein expression of CD8 was increased by mean fluorescence intensity (MFI) analysis of COVID-19 infected patients; whereas, MFI of CD4 marker revealed a non-significant change in patients compared to the healthy group [41]. Collectively, these results highlight that COVID-19 in the fatal course of the disease is associated with impaired CD4+ T cell function and excessive activation of CD8+ T cells that may lead to exhaustion and thus appear similar to chronic infections [43]. More importantly, CD3+ CD8+ T-cells ≤75 cells μL−1 has been reported to be a reliable predictor for identifying high risk of death in COVID-19 pneumonia cases [44]. Parallel analyses of CD4+ and CD8+ T cell populations identified a decline in CD3+ CD8+ T-cells, but not CD3+ CD4+ T-cell, in the circulation in deceased patients compared to survivors [44]. Clearly, CD8+ T cell response holds the key for successful viral control and do protect against SARS-CoV-2 virus [45]. Thus, as suggested by another research group, identification of relatively reduced frequency of CD4+ and CD8+ T cell on admission has been linked to a worse prognosis [46]. Levels of CD4+ and CD8+ T lymphocytes, together with other markers, have potential to serve as routine laboratory biomarkers for diagnosis and severity prediction of COVID-19 in the clinic due in part to the increasing number of outcome studies that have validated the significance of lymphocyte detection [14]. As shown in Fig. 1 there exist continuing alterations in the immune response compartments over time from mild COVID-19 illness to severe ARDS causing death in some cases.
Fig. 1.
The alterations of peripheral blood immune indices in different stages of the COVID-19 disease. There is a specific trend of changes in immune system effectors which is maintained from early infection to severe lethal stages of the disease. Neutrophil counts and the ratios of neutrophils to lymphocytes and to CD8+ T cells pursue an obviously increased pattern in COVID-19 patients who progress to the advanced stages of the disease. However, total lymphocytes and the number of CD4+ and CD8+ T cells tend to decrease from the early to severe stages. ARDS; Acute respiratory distress syndrome, Neu; Neutrophil, NLR: Neutrophil to lymphocyte ratio, N8R; Neutrophil to CD8+ T ratio.
Several lines of evidence have consistently indicated that although COVID-19 illness affects the population of peripheral blood CD4+ and CD8+ T lymphocytes [48], the ratio of CD4+ T/CD8+ T may not be influenced significantly [41,48]. Based on experience with 249 cases of mild type, 45.5% of COVID-19 patients suffered from reduced CD4+ T cells, and 92.8% of the patients showed the T CD4+/T CD8+ ratio in the normal range [48]. Another related abnormality is the inconsistent count and function of T cells in COVID-19 patients [49]. A recently published study enrolling 65 COVID-19 patients demonstrated that lymphocytes were in a hyper function state with higher percentages of IFN-γ–producing CD4+ and CD8+ T cells in severe and extremely severe patients than those in mild stage. Moreover, the increased IFN-γ–producing ability of T cells were simultaneous with a substantial decrease in total numbers of CD3+ T cells, CD4+ T cells, and CD8+ T cells [49]. This discrepancy provides evidence that robust adaptive immune responses exist in the final stage of COVID-19 patients.
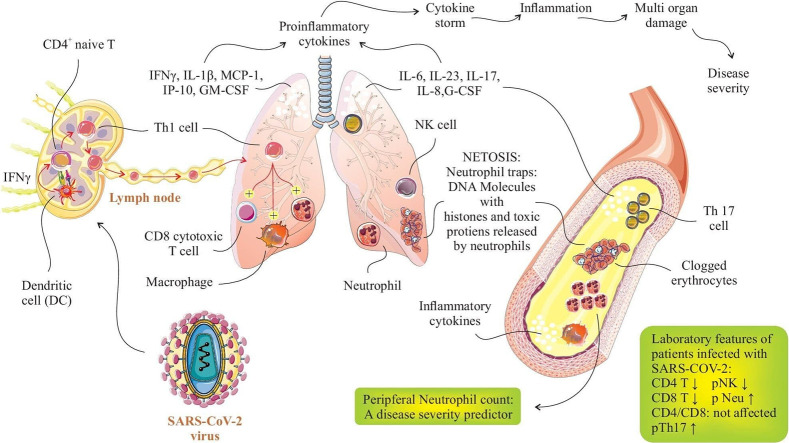
The findings also suggest that a COVID-19-related dysregulated immune mechanisms exist, that consist of an increased naive CD4+ T-cell subpopulations and smaller populations of memory T cells as well as a higher naive to memory cell ratio [50]. Bronchoalveolar lavage fluid (BALF) analysis has shown higher proportion of CD8+ T cells in mild patients than the severe patients. Increased clonal expansion of CD8+ T cells is observed in mild cases highlighting the role of CD8+ T cells recruited from blood and recovered in BALF [51]; this may reflect the importance of virus-specific CD8+ T cells in viral clearance in mild to moderate COVID-19 illness, as in influenza virus infection [45]. Assessing the functional status of T cell subsets is becoming increasingly important in identifying the disease course of COVID-19 patients. These T cells have been analyzed functionally (by gene expression analysis) regarding production of effector molecules and the results show high levels of effector molecules in CD8+ T cells [45]. As noted, patients infected with 2019-nCoV display a cytokine signature associated with activated T-helper-1 (Th1) cell responses. The infection also initiates the boosted production of Th2 type cytokines IL-4 and IL-10 anti-inflammatory cytokines that suppress inflammation [11]. This effect is in contrast to the previously documented modulatory effects of SARS-CoV infection in Th2 response which does not enhance the Th2 pathway [52].
2.3. Th1/Th2/Th17/Tfh responses
Initially, Th1 cells are important in defense against viral infection by producing cytokines and inducing the CD8+ T cell, neutrophil, and macrophage immune responses. Relying on previous studies, an early increase in CD8+ T cells and a predominance of Th1 responses have been reported in MERS-CoV infection. The proven key role of Th1 responses in SARS-CoV and MERS-CoV infections supports the important role of Th1 cells in SARS-CoV-2, as well [53,54]. In advanced or severe stage of SARS-CoV-2 infection, a low number of T helper subsets (Th1, Th2, and Th17) has been reported [50]. It is thought that Th1 response accompanied by Th17 cell immunity crucially contribute to aggravate the inflammation, disease severity, and multi-organ damage by driving cytokine storm following the production of proinflammatory cytokines [[55], [56], [57]]. As expected, the elevated proportions of the Th1 and Th17 cells, as crucial inflammatory effectors, were found in severe COVID-19 cases. In severe COVID-19 patients, a large amount of inflammatory immune cells like Th1 cells and monocytes, which infiltrate into pulmonary circulation, are substantially responsible for cytokine storm, inflammation, immune system disturbance, and severe lung pathology [43,58,59].
Accordingly, Zhou et al. showed that immediately after the onset of SARS-CoV-2 infection, CD4+ cells become active and differentiate into Th1 cells, leading to the release of inflammatory cytokines (GM-CSF and IL-16), which in turn activate monocytes and neutrophils. It has been found that the above mentioned mechanisms elicit the inflammation and progression of the disease to a severe condition [58]. In a study by Grifoni et al. the secreted cytokines were measured to determine the CD4+ T cell polarization and function against SARS-CoV-2. Based upon the findings, it seems that T cells were polarized to Th1 type due to the considerable production of IFN-γ than IL-4, IL-5, IL-13, or IL-17A cytokines [60]. As the same findings, GM-CSF+ and IL-6+ CD4+ T cells were in an increased percentage in ICU-admitted patients with severe pneumonia when compared to non-ICU cases and healthy individuals. Pathogenic Th1 cells expressing the IFN-γ and GM-CSF cytokines, were found only in ICU-admitted COVID-19 patients, but not in non-ICU and healthy subjects [58]. Confirming the important role of Th1 cells in inflammation and SARS-CoV2 pathogenesis, Neidleman et al. reported the higher expression levels of T-bet transcription factor and IFN-γ in COVID-19 patients, mostly in severe cases, which approved the differentiation and activation of Th1 cells in the course of the disease [61]. This claim has been also proven in other studies showing that the overexpression of IFN-γ is associated with the upregulated Th1 responses in COVID-19 patients [60,62]. These data propose the critical role of Th1 cells in the induction of hyper-inflammation and disease progression in severe SARS-CoV-2.
Th17 cells have been identified as inflammatory immune cells that produce proinflammatory cytokines, such as IL-17, IL-21, IL-22, and GM-CSF to defense against extracellular bacterial and fungal pathogens [63]. In the course of infections, the presence of IL-1β and IL-18, and lack of IL-12 induce the IL-17 production, which elicit the CD4+ T cell differentiation into Th17 cell [64,65]. During the SARS-CoV-2 infection, IL-1β and TNF-α provoke the responses of circulating peripheral Th17 cells, which play a role in generating cytokine storm and inflammation [55]. Additionally, overexpression of the IL-6 and IL-23 cytokines during the infection may explain the enhanced level of Th17 cells [43,58]. Th17 responses contribute to neutrophil and macrophage recruitment as well as the induction of inflammation derived from cytokine storm. It is noteworthy that Th17 may have a dominant immune response in pulmonary pathogenesis of severe COVID-19 patients with ARDS by relying on the increased levels of IL-1β, IL-8, IL-17, G-CSF, and GM-CSF cytokines as well as a neutrophil count during the infection [[66], [67], [68], [69]]. The results of another study on a COVID-19 patient who died from a severe stage of the disease showed the enriched levels of CCR4+ CCR6+ Th17 cells in the peripheral blood of patients, which supported the inflammatory role of Th17 cells in SRAS-CoV-2 infection [43]. Similarly, elevated frequency of Th17 cells and IL-17 cytokine were detected in the circulating peripheral blood of severe COVID-19 patients which resulted in cytokine storm, pulmonary edema, and ARDS [43,70].
It is found that the dominant responses of Th17 in the course of COVID-19 occur as a result of host immune system disability in recognizing the viral infection as well as its misleading by the virus. Increased proportions of Th17 cells were previously reported in MERS-CoV and SARS-CoV infections, which support the upregulation of Th17 responses in SARS-CoV-2, as well [71,72].
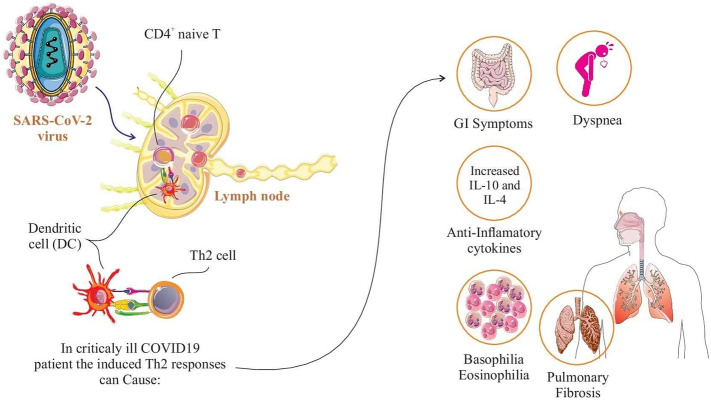
It is well established that Th2 cells as a distinct subset of CD4+ T cells direct the immune responses against extracellular pathogens [73]. Unlike the available information on Th1 and Th17 responses in COVID-19, the pathologic role of Th2 cells is not clarified. In a study on severe hospitalized COVID-19 patients receiving intensive care, the cytological signals of Th2 immune responses were observed in patients characterized by basophilia, degranulated eosinophils, eosinophilia as well as T cytotoxic lymphopenia. On the other hand, in severe COVID-19 patients requiring intensive care, it was found that the immune system tends to shift the responses toward Th2 cells to control the infection by the contribution of macrophages and T cytotoxic cells. It may be associated with antigen cross-reactivity, the type of antigen presenting cells (APCs) with which T cells interact, patient viral load, and Th1 and T cytotoxic breakdown [74]. This can also be explained by the observation of gastrointestinal symptoms and dyspnea in up to 30% of SARS-CoV-2 infected patients, which are considered to be the defense mechanisms of Th2 cells [75,76]. In addition to the production of IFN-γ, IL-1β, MCP-1, and IP-10 cytokines in SARS-CoV-2 infection that confirm the substantial immunopathological role of Th1 cells, IL-10 secretion was also detected in patients explaining the role of Th2 responses in COID-19 [52]. Interestingly, the enhanced levels of the anti-inflammatory cytokines like IL-4 and IL-10 in severe COVID-19 cases compared to those in moderate stage documented the upregulated Th2 responses and pulmonary interstitial fibrosis in the course of the disease [77]. Fig. 2 shows the spectrum of clinical manifestations and adverse events associated with Th2 immune responses in severe cases of COVID-19.
Fig. 2.
Clinical manifestations associated with Th2 immune responses in critically ill COVID-19 patients. Severe or critical COVID-19 infection do enhance Th2 immune pathways which is associated with clinical symptoms including shortness of breath, gastrointestinal symptoms (eg, diarrhea, vomiting), pulmonary interstitial fibrosis, and basophilia/eosinophilia.
Follicular helper T (Tfh) cell, a T cell helper subset, activates the B-cell responses and induces the antibody production. Greater number of circulating Tfh cells has been reported after vaccination and in viral infections. A study conducted on 32 COVID-19 patients revealed that the proportions of cytotoxic Tfh cells, as well as cytotoxic T helper cells (CD4- CTLs), were considerably enriched in the severe stage of disease when compared to the mild stage and healthy controls. Moreover, overexpression of Tfh-relevant cytotoxicity-associated transcripts, including PRF1 and GZMB were found in these cases, which justify the increased responses of Tfh cells. Moreover, it was found that the cytotoxic function and B cell destruction in severe COVID-19 patients correlate with an increased level of Tfh cells [78].
2.4. T regulatory (Treg) cell responses
Since regulatory T cells are important elements in dampening immune response, the elimination of these cells could result in enhanced innate immune activity leading to uncontrolled inflammatory response [79]. As in many viral diseases, maintaining the balance between anti-viral immune functions and inflammation derived from cytokine storm is critically needed in SARS-CoV-2 infection. Results show that although the proportion of total Treg is comparable between the severe and moderate COVID-19 patients, severe cases show a remarkably reduced proportion of CD45RA+ naive Tregs (nTregs) as well as a slightly upregulated proportion of CD45RO+ memory Tregs (mTregs) [74,75]. According to a comparison of 32 severe and mild 2019-nCoV patients using the large-scale single-cell transcriptomic analysis of viral antigen-reactive CD4+ T cells, there were a reduced number of Treg cells in severe illness [38]. It suggested that the dysregulated immune system has a defect in the production of immunosuppressive SARS-CoV-2-reactive Treg cells in the severe stage. In another study, the dysregulated immune system was investigated in a cohort of 452 COVID-19 patients. Findings revealed the reduced percentage of Tregs (CD3+ CD4+ CD25+ CD127low+) in COVID-19 patients, particularly in severe cases, which resulted in aggravated inflammation, disease severity, and lung injury [67]. In addition, Duerr et al. introduced the CD8/Treg/monocyte ratio to be used as a progression parameter for cardiac involvement in COVID-19 [80]. That ratio was found to be related to an inadequate immune response associated with myocardial inflammation. That study showed that patients who developed respiratory failure or died had a high CD8/Treg/monocyte ratio, but a low ratio was found in COVID-19 patients being admitted with pericardial effusion (PE) who survived and those without PE [80]. To date, inconsistent findings have been reported in terms of the increased or decreased levels of Tregs during the SARS-CoV-2 infection, which needs further investigation to determine their exact role [67,81,82].
3. How innate immune responses against SARS-CoV-2 change in different stages of COVID-19
3.1. NK cell response
Natural killer (NK) cells are known as vital innate immune cells in the first line of defense against viral infections. Alterations in NK cell responses have also been reported in COVID-19 cases due to the disturbance of immune system balance. Based upon studies on SARS-CoV-2, the decreased number of peripheral NK cells has been detected in the severe groups compared to the mild ones, as well as healthy individuals [56,[83], [84], [85]], which is consistent to the published results in SARS-CoV [86]. Mechanistically, increased expression level of CXCR3 ligands as well as the enriched number of CXCR3-ligand-producing monocytes in lung tissue of COVID-19 patients support the NK cell recruitment from peripheral blood into the lungs, which partly explain the decreased levels of NK cells in the peripheral blood [87,88]. In SARS-CoV-2 infected patients, NK cells mostly appear with mature CD16+ KIR+ CD56dim phenotype, displaying the cytotoxic function [89].
According to Zheng et al. study [83], a significant reduction in the absolute count of NK and CD8+ cells was found in SARS-CoV-2 patients. Functional exhaustion of NK and CD8+ cells, along with the overexpression of the NKG2A inhibitory receptor were detected in 2019-nCoV patients compared to healthy subjects. Comparatively, the percentage of NK cells was noticeably downregulated in severe disease patients when compared with those in mild illness and healthy controls. The proportions of IFN-γ + NK, TNF-α + NK, IL-2+ NK, and CD107a + NK cells were found to reduce in COVID-19 patients, as well. Altogether, these findings suggest the lower count and functional exhaustion of NK cells in the course of SARS-CoV-2 infection. Interestingly, downregulated expression of granulysin, Ksp37, CD107a, and granzyme B, and impaired secretion of IFN-ɣ and TNF-a, predict the decrease and dysfunction of circulating NK cells in severe 2019-nCoV patients [83,84].
In a similar study, Wen et al. reported the alterations of the peripheral blood mononuclear cells (PBMCs) in the early and late recovery stage (ERS and LRS, respectively) of COVID-19 patients and healthy controls [56]. Accordingly, a lower population of NK and T cells was found in both ERS and LRS patients in comparison with healthy groups. Noteworthy, a higher level of NK cells was detected in LRS patients than those in ERS. Some other studies evidence a remarkably diminished proportion of NK cells in severe cases of COVID-19 than mild patients and healthy subjects [10,50,85,90]. Contrarily, scRNA-seq analysis of BALF manifested a higher percentage of NK cells in mild and severe COVID-19 patients than controls, which supported the peripheral NK cell trafficking into the lungs [45]. Collectively, it has been found that the functional exhaustion and lower level of peripheral NK cells relate to the dissemination of SARS-CoV-2 to other organs, and to their trafficking into lungs, which accounts for the inflammatory condition, ARDS, and disease progression. Some opposite reports indicated that the frequency of CD16+ CD56+ NK cells had no significant discrepancy between mild and severe COVID-19 cases [16,91]. Since various results indicate that the number of NK cell is heterogeneous in COVID-19 patients as well as in different stages of the disease, further studies are required to determine the exact role of these cells in immunogenicity and immunopathogenesis of SARS-CoV-2 infection.
3.2. Neutrophil responses
Neutrophils, as other innate immune cells, play a critical role against infections like SARS-CoV-2. Since neutrophils are gradually upregulated in the peripheral blood circulation of COVID-19 patients during the disease progression and severity, they can be considered as a disease severity predictor [92]. In a study on 44 mild- and 17 severe or critical COVID-19 cases, a significant increase in the absolute number of blood neutrophils and neutrophil-to-lymphocyte ratio (NLR) was detected in severe patients that those in mild stages [84]. It has also been found that the elevated level of NLR is concerned with poor prognosis and severe condition of patients, which could be considered as a precise predictor factor of disease severity and prognosis [16,84,93].
Of note, neutrophils producing Neutrophil extracellular traps (NETs), which are known as extracellular webs of DNA/histones, also intensify the inflammatory responses in severe cases of COVID-19. NETs have been demonstrated to play a role in cytokine release syndrome, coagulopathy, excessive thrombosis, cystic fibrosis, respiratory failure, ARDS, and multi-organ injury. Myeloperoxidase (MPO)-DNA and NETosis with cell-free DNA were frequently found in severe SARS-CoV-2 illness [94,95]. Moreover, Liu et al. [16] and Li et al. [96] found that the severe or critical COVID-19 patients had a considerable higher count of neutrophils and NLR, increased pro-inflammatory cytokines, and lymphopenia compared to mild cases or healthy subjects. Similarly, another study reported the upregulated levels of activated neutrophils in ARDS and ventilated-COVID-19 patients when compared with unventilated cases and healthy subjects. Of interest, overexpression of chemoattractants for neutrophil recruitment in the course of COVID-19 suggests the importance of granulocytes in disease pathogenesis and severity [29]. It is worth noting that greater levels of CXCL-2 and CXCL-8 lead to neutrophil attraction into infection sites, which subsequently exacerbate the pulmonary inflammation [97]. Some other related studies also reported the increased levels of total neutrophils or NLR in cases with severe illness compared to non-severe patients and controls. In line, an augmented number of neutrophils were also reported in severe or lethal SARS-CoV or MERS-CoV patients [18,19].
3.3. Monocyte responses
Dysregulation of monocyte and macrophage populations have also been reported in the course of the disease. Due to the ACE2 expression on monocytes and macrophages, they are infected by SARS-CoV-2 leading to the activation of proinflammatory genes; whereas, ACE2 expression is downregulated in peripheral blood monocytes [98,99]. In COVID-19 infected patients, activated monocytes in peripheral blood are responsible for the disease severity and poor prognosis, characterized by the expression of CD11b, CD14, CD16, CD68, CD80, CD163, and CD206 markers, as well as producing the IL-6, IL-10, and TNF-α cytokines [98]. Intriguingly, the considerable association was discovered between these peripheral activated monocytes and hemophagocytic lymphohistiocytosis syndrome in COVID-19 patients, which may elicit the cytokine storm and disease progression [100].
According to a published report, scRNA-seq analysis in PBMC samples provided evidence of the monocyte alterations and revealed an elevated ratio of classical CD14+ monocytes in the early recovery stage of 2019-nCoV patients compared to healthy controls. However, it was normal in late recovery stage patients. Additionally, a higher level of CD14++ IL-1β + and IFN-activated monocytes was discovered in early and late recovery patients compared to control group [56]. In this context, Zhou et al. [99] demonstrated the significant greater counts of IL-6-producing CD14+ CD16+ inflammatory monocytes in ICU-admitted COVID-19 patients with severe condition. Some other studies illustrated that CD14+ monocyte expressing the HLA class II genes was considerably downregulated in severe COVID-19 patients [67,84,85,101]. Additionally, Zhang et al. revealed that the frequency of classical monocytes (CD14++ CD16-) was decreased in COVID-19 patients compared to healthy individuals; whereas, intermediate (CD14++ CD16+) and non-classical (CD14+ CD16++) monocytes were found in elevated levels. Their findings also showed a significantly higher level of activated IL-6 and TNF-α secreting monocytes in severe COVID-19 patients, which increased the ICU admission risk. Contrarily, better prognosis and recovery were observed in patients with a normal proportion of monocytes [98].
The role of peripheral blood monocytes in inflammation and progression of SARS-CoV-2 disease have also been evaluated. In this regard, Guo et al. [102] reported the noticeable enriched presence of a unique subpopulation of monocytes expressing PHLDA2, ETS2, and NFIL3 inflammatory genes in severe COVID-19 cases compared to the recovery stage cases and healthy controls, using the scRNA-seq data. Moreover, it has been found that monocytes are involved in inflammatory storm and disease progression. On the other hand, monocytes could interact with CD4+ T cell and plasma cells, which strongly induce the inflammation during the severe stage of the disease. Similarly, increased frequency of inflammatory monocytes producing IL-6 and GM-CSF cytokines were detected in COVID-19 patients with the severe condition [58]. By contrast, Wilk et al. [84] showed a remarkable decreased levels of CD16+ monocytes, as well as a strong shift in cell phenotype into CD14+ monocyte, in severe cases, mostly in ARDS patients. However, no significant higher expression levels of proinflammatory cytokines by peripheral blood monocytes were found in severe COVID-19 cases. To accurately determine the monocyte alterations in the course of SARS-CoV-2 infection, further information is required.
3.4. Macrophage responses
Macrophages are important innate immune cells producing the inflammatory cytokines such as IL-1, IL-6, and TNF-α in COVID-19, which result in cytokine storm and lung injury in severe illness. Macrophages show variations during the various stages of the COVID-19. In addition to monocytes expressing ACE2, CD68+ and CD169+ macrophages also express ACE2, which have been found in the spleen and lymph nodes of 2019-nCoV cases. These macrophages and other ACE2 positive myeloid cells can be targeted by SARS-CoV-2 [64].
Thus far, contradictory results of macrophage alteration have been reported in COVID-19 cases. According to Autopsy reports of COVID-19 cases, the accumulation of inflammatory macrophages was evidenced in lung tissue [103]. As an example, Zhou et al. indicated the non-significant enhance of macrophages (M0, M1, and M2) in the lungs of 2019-nCoV patients compared to healthy subjects [104]. Contrarily, Liao et al. reported that COVID-19 patients in severe stages with ARDS had enriched levels of inflammatory monocyte-derived FCN1+ macrophage in the bronchoalveolar lavage fluid than those in mild stages. Additionally, they found that in severe cases, inflammatory monocyte-derived macrophages and SPP1+ macrophages were greater than mild ones. In return, alveolar macrophages were predominantly greater in mild cases and controls [45]. Based on the evaluation of mononuclear cells in BAFL and peripheral blood of SARS-CoV-2 cases, increased production of IFN-induced protein 10 (IP-10) and Monocyte Chemoattractant Protein-1 (MCP-1) revealed the tendency of macrophages to traffic toward the sites of infection in lungs [97]. In the same subject, it was found that increased levels of inflammatory macrophages and their hyperactivity resulted in hyper-inflammation in severe stage, which led to impaired tissue repair and fibrosis in the lungs [105]. Based on the results, mononuclear phagocyte (MNP) in the severe stages of COVID-19 was specified by a notable increase in inflammatory monocyte-derived macrophages as well as the lower population of tissue-resident alveolar macrophages. Elevated levels of neutrophil and the monocyte-macrophages influx have been also documented in the severe or lethal patients of SARS-CoV and MERS-CoV [18,19].
In recent studies, it was shown that macrophages play a substantial role in eliciting the cytokine storm and inflammation in severe and poor prognosis COVID-19 cases by producing the inflammatory factors like CXCL10 [106,107]. Besides, abundant proinflammatory monocyte-derived macrophages were found in BAFL of severe COVID-19 patients compared to mild cases and healthy subjects, which can be replaced damaged infected alveolar macrophages in the severe stage. These increased amounts of macrophages lead to cytokine storm and ARDS that are known as hallmarks of severe cases [87].
3.5. Dendritic cell responses
Dendritic cells (DCs) are important antigen-presenting cells essential for activating the innate and adaptive immune responses. There is little information about DC cell variations in SARS-CoV-2 infection, and no definitive findings have been reported. In this context, Wilk et al. demonstrated the reduced proportion of both conventional DCs and plasmacytoid DCs in COVID-19 cases when compared with healthy subjects [84]. In another study, in comparison to healthy controls, resting DCs were found to decrease in COVID-19 patients versus the increased number of activated DCs in the lungs [104]. Similarly, Liao et al. investigated the alterations of DC cells in BALFs of diverse severity of COVID-19 patients by single-cell RNA sequencing. As a result, severe or critical COVID-19 had decreased proportions of myeloid dendritic cells (mDCs) and pDCs compared to moderate cases [45]. In another investigation conducted by Wang et al. [49], the counts and activation status of DC were assessed in mild, severe, and extremely severe COVID-19 illness. Their findings indicated the lower activation of DCs in extremely severe patients and non-significant difference in all of the severities.
3.6. Immune therapies in severe COVID-19
Understanding the immunological changes that occur in severe COVID-19 is critical for designing better immunotherapies for these patients [108]. Since accumulating evidence suggests that severe COVID-19 forms a hyperinflammation syndrome, immunomodulation may be potentially useful as a complementary therapy to supportive care measures. The reduction of hyper-inflammatory reaction may improve the outcome in patients with severe COVID-19 infection in order to prevent mortality. This is of particular interest, especially when keeping in mind that no specific antiviral treatment has been developed for SARS-CoV-2. There are different treatment modalities being tried or under trial against COVID-19 [109]. Currently available cytokine blockers were developed when COVID-19 was considered a predominantly immunoinflammatory disease [110]. In China, an anti-human IL-6 receptor monoclonal antibody is approved for severe cases of COVID-19 affected by pulmonary complications [111]. Anakinra, an anti-IL-1, is another modality used against severe forms of COVID-19 patients [112]. Inhibition of inflammatory responses via blockade of IL-1 may play a role in the treatment of severe or refractory COVID-19 patients [113]. In the same line, interference with other cytokines such as IL-2, and TNF may be other therapeutic options for the management of COVID-19 [114]. Blockade of IL-2/IL-2R pathway has been shown to participate in regulation of T cell-mediated immunity in vivo in critical patients with COVID-19 pneumonia. Using checkpoint inhibitors is another therapeutic strategy in patients with the novel infectious disease COVID-19. Immune checkpoint inhibitors, that control the balance of stimulatory and inhibitory signals toward a state of immune activation, have shown clinical activity in the treatment of a variety of cancers [115]. Recent publications propose that immune checkpoint inhibitors can be safely employed in cancer patients with a COVID-19 infection. Accordingly, Luo et al. demonstrated that PD-1 inhibitor use is not associated with an increased risk of severity and mortality from SARS-CoV-2 infection in lung cancer patients [116]. In addition, passive immunization with hyperimmune intravenous immunoglobulin (HIVIG) has been tried for the treatment of COVID-19 disease [117]. IVIG that may contain neutralizing antibodies is also known to interrupt the storm of inflammatory cytokines [118]. Moreover, immunosuppressive drugs including corticosteroids exert inhibitory effects on cytokine production and on a broad range of innate and adaptive immune responses [119,120]. However, unlike other respiratory viral infections, the use of systemic corticosteroids in the setting of COVID-19 is still controversial [121]. Taken together, clinical experience suggests that immunosuppressors play a major role in maintaining remission and as earlier the immunosuppressive therapy is started, better is the improvement in severe COVID-19 patients.
4. Conclusion
Through the available data, we tried to elaborate further on how alterations in different leukocyte populations reflect the immune system dynamic responses during COVID-19 infection. These dynamic properties should be taken into consideration, since it can affect the data interpretation. When considering therapeutic aspects, it is crucial to differentiate between COVID-19 patients in the early course of illness who respond better to antiviral pharmacotherapy and those in advanced stages who are likely to benefit from anti-inflammatory therapy [122]. Mounting an early and effective induction of the adaptive immune response to the virus may retard or help prevent progression to the critical stage of the illness. This review highlights the immune correlates of severe clinical disease and protection against the disease caused by SARS-CoV-2 in patients. A disease-associated decline in the numbers of T-cells, both of the CD4+ and the CD8+ subsets, increased neutrophil counts, neutrophil-to-lymphocyte ratio and neutrophil-to-CD8+ T cell ratio have been proposed to be the immune risk indicators associated with an increased risk of adverse clinical events. In support of Siddiqu and Mehra [105] who proposed a clinical staging system by developing a three-stage classification model that recognizes early infection (stage I), pulmonary involvement without and with hypoxia (stage II), and systemic hyperinflammation (stage III), findings from this review support for an abnormal development of immune response at systemic level and also provides diagnostic clues for the three grades of increasing severity of COVID-19 disease which correspond with the underlying immune pathology. A relatively low frequency of CD4+ and CD8+ T cell subsets at hospital admission can identify patients at risk for severe COVID-19 disease. Moreover, aberrant CD8+ T cell responses have shown to be linked to a high risk of death in COVID-19 pneumonia cases. Taken together, immunological parameters for disease severity include elevated level of neutrophils, increased inflammatory T cell subsets (Th17 and Th1 cells) and their related cytokines, decreased number of peripheral NK cells, and reduced total number of CD4+ and CD8+ T cells. The data of Table 1 summarize the studies now available presenting information about the comparative analysis of immune response to SARS-CoV-2 virus in mild and severe stages. Prospective randomized trials and longer observation periods, however, are needed to confirm these pilot results.
Table 1.
Alterations of innate and adoptive immune cells in mild to severe stages of COVID-19 infection.
| Study | No. of patients | Disease stage | Overall changes of innate immune cells in COVID-19 patients | Overall changes of adoptive immune cells in COVID-19 patients | Comparison of immune cell alteration in severe vs. mild stages | Ref |
|---|---|---|---|---|---|---|
| Huang et al. | 41 | ICU: 13 Non-ICU: 28 |
– | ↓ Lymphocyte (26/41) | ↑ Neutrophil ↑ Lymphocytes | [11] |
| Qin et al. | 452 | Severe: 286 Non-severe: 166 |
↓ NK cell | ↑ B cells ↑ T helper cells ↑ T suppressor cells ↑ Treg cells | ↑ Neutrophils ↑ NLR ↓ Lymphocytes; Monocytes, Eosinophils, NK cells; Basophils; Th cell; Treg cells | [123] |
| Chen et al. | 21 | Mild: 10 Severe: 11 |
↓ NK cell (8/14) | ↑ B cells (7/14) ↓ Lymphocyte (9/21) ↓ Total T cells (13/14) ↓ T CD4+ cells (12/14) ↓ T CD8+ cells (14/14) | ↑ Neutrophil ↓ Lymphocyte ↓ B cells ↓ T CD4+ cells ↓ T CD8+ cells | [77] |
| Liu et al. | 40 | Mild: 27 Severe: 13 |
– | ↓ Lymphocyte | ↑ Neutrophils ↓ Lymphocyte ↓ CD3+ T cells ↓ CD8+ T cells | [16] |
| Diao et al. | 262 | Mild: 151 Severe: 40 Critical: 13, 8 Perished: 8 Control: 40 |
– | ↑PD1+ CD4+ T cells; PD1+ CD8+ T cells ↓Total T cells (166/222); CD4+ T cells (166/222); CD8+ T cells (156/222) | ↑ PD1+ CD4+ T cells ↑ PD1+ CD8+ T cells ↓ Total T cells ↓ CD4+ T cells ↓ CD8+ T cells | [124] |
| Liu et al. | 80 | Severe: 69 Non-severe: 11 |
– | ↓ Lymphocyte (60/80) | ↑ Neutrophils ↑ NK cells ↓ Lymphocytes | [125] |
| Chen et al. | 29 | Mild: 15 Severe: 9 Critical: 5 |
– | ↓ Lymphocyte (20/29) | No difference | [126] |
| Wang et al. | 69 | sPO2 ≥ 90%: 55 sPO2 < 90%: 14 |
↑ Neutrophil (41/67) ↑ Basophil (49/69) |
↓ lymphocyte (29/69) | ↑ Neutrophil ↓ Lymphocytes |
[127] |
| Wang et al. | 138 | ICU P: 36 Non-ICU: 102 |
– | ↓ lymphocyte | ↑ Neutrophil ↓ Lymphocytes |
[27] |
| Ouyang et al. | 11 | Mild: 5 Severe: 6 |
↑ Neutrophil | ↓ Lymphocyte | ↑ Neutrophil | [128] |
| Li et al. | 548 | Severe: 269 Non-severe: 279 |
↑ Neutrophil (118/542) | ↓ Lymphocytes (118/542) | ↑ Neutrophil ↓ Lymphocytes |
[129] |
| Wan et al. | 123 | Mild: 102 Severe: 21 |
↓ NK cell (45/123) | ↓ CD4+ T cells (74/123) ↓ CD8+ T cells (42/123) ↓ B cells (32/123) | ↓ CD4+ T cells ↓ CD8+ T cells |
[130] |
| Shi et al. | 56 | Mild:31 Severe: 25 |
↑ Neutrophils ↑ NLR ↓ NK cell |
↑ Treg cells ↓ CD4+ T cells ↓ CD8+ T cells ↓ B cells |
↑ Neutrophil ↓ Lymphocytes |
[82] |
| Zheng et al. | 16 | Mild: 10 Severe: 6 |
– | ↓ T cells | ↑ TIGIT+CD8+ T cells ↓ Granulocytes; CD4+ T cells |
[42] |
| Yang et al. | 53 | Mild: 19 Severe: 34 |
– | ↓ CD4+ T cells ↓ CD8+ T cells |
↑ Neutrophil ↓ CD4+ T cells ↓ CD8+ T cells |
[106] |
| Gong et al. | 100 | Mild: 34 Severe: 34 Critical: 32 |
↓ Eosinophils | – | ↑ Neutrophils ↓ Eosinophils ↓ Lymphocytes |
[133] |
| Chen et al. | 48 | Mild: 21 Severe: 10 Critical: 17 |
– | ↓ Lymphocytes | ↑ Neutrophils ↓ Lymphocytes |
[134] |
| Qi et al. | 267 | Severe: 50 Non-severe: 217 |
– | ↓ Lymphocytes (231/267) ↓ CD3+ T cells (51/96) ↓ CD4+ T cells (74/96) |
↑ Neutrophils ↓ CD3+ T cells ↓ CD4+ T cells |
[135] |
| Li et al. | 110 | Mild: 57 Severe: 39 Critical: 14 |
– | ↓ CD4+ T cells ↓ CD8+ T cells |
↓ CD4+ T cells ↓ CD8+ T cells |
[136] |
| Xu et al. | 155 | Mild: 125 Severe: 30 |
– | ↓ CD4+ T cells ↓ CD8+ T cells |
↓ CD4+ T cells ↓ CD8+ T cells |
[137] |
| Zhou et al. | 43 | ICU: 12 Non-ICU: 21 Control: 10 |
↑CD14 + CD16+ monocytes; GM-CSF+ CD14+ monocytes |
↑ Tim-PD-1-CD4+ T cells ↑ Tim-PD-1- CD8+ T cells ↑ GM-CSF-CD4+ T cells ↑ IL6-CD4+ T cells ↓ CD4 T+ cells ↓ CD8+ T cells |
↑ Tim-PD-1-CD4+ T cells ↑ IFNγ-GM-CSF-CD4+ T cells ↑ GM-CSF-CD4+ T cells ↑ IL-6-CD4+ T cells ↑ CD14-CD16+ Monocytes ↑ GM-CSF-CD14 + Monocytes ↑ IL-6 + CD14+ Monocytes ↓ CD8+ T cells |
[138] |
Abbreviations: COVID-19, coronavirus disease 2019; N, number of cases; ICU, intensive care unit; NK, natural killer; Th, T helper cell; Treg, regulatory T cells; SpO2, blood oxygen saturation; NLR, neutrophil-to-lymphocyte ratio; PD-1, programmed cell death 1; Tim, T cell Ig and mucin domain; TIGIT, T cell immunoglobulin and ITIM domain; GM-CSF; Granulocyte-macrophage colony-stimulating factor; IFN-γ, Interferon gamma; IL, Interleukin.
Author contributions
Mahnaz Ghaebi: Conceptualization, Writing - original draft preparation Safa Tahmasebi: Writing - original draft preparation Maryam Jozghorbani: Visualization Alireza Sadeghi: Review & editing Abdolreza Esmaeilzadeh: Conceptualization, Review & editing, Supervision.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Research data not shared.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgements
The authors would like to dedicate this study to healthcare workers, which are struggling with the novel coronavirus. The authors received no specific funding for this work.
References
- 3.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses: Springer. 2015:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30(7):1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gattinoni L., Chiumello D., Caironi P., Busana M., Romitti F., Brazzi L., et al. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Springer. 2020;46:1099–1102. doi: 10.1007/s00134-020-06033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Control CfD . 2020. Prevention. Novel Coronavirus, Wuhan, China. Prevention. [Google Scholar]
- 8.Adhikari S.P., Meng S., Wu Y.-J., Mao Y.-P., Ye R.-X., Wang Q.-Z., et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9(1):1–12. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [Internet]; 2020 [cited 2020 Mar 21]. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghaebi M., Osali A., Valizadeh H., Roshangar L., Ahmadi M. Vaccine development and therapeutic design for 2019-nCoV/SARS-CoV-2: challenges and chances. J. Cell. Physiol. 2020;235(12):9098–9109. doi: 10.1002/jcp.29771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marofi F., Vahedi G., Biglari A., Esmaeilzadeh A., Athari S.S. Mesenchymal stromal/stem cells: a new era in the cell-based targeted gene therapy of cancer. Front. Immunol. 2017;8:1770. doi: 10.3389/fimmu.2017.01770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahmasebi S., Khosh E., Esmaeilzadeh A. The outlook for diagnostic purposes of the 2019-novel coronavirus disease. J. Cell. Physiol. 2020;235(12):9211–9229. doi: 10.1002/jcp.29804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esmaeilzadeh A., Elahi R. Immunobiology and immunotherapy of COVID-19: a clinically updated overview. J. Cell. Physiol. 2021;236(4):2519–2543. doi: 10.1002/jcp.30076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F, Zhao S, Yu B. A new coronavirus associated with human respiratory disease in China [published online ahead of print February 3, 2020]. Nature.10.
- 18.Dandekar A.A., Perlman S. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C., Chen X., Cai Y., Zhou X., Xu S., Huang H., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu L., Chen S., Fu Y., Gao Z., Long H., H-w Ren, et al. Risk factors associated with clinical outcomes in 323 COVID-19 patients in Wuhan, China. Medrxiv. 2020;71(16):2089–2098. doi: 10.1093/cid/ciaa539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K.D., Zhao J., Auh S., Yang X., Du P., Tang H., et al. Adaptive immune cells temper initial innate responses. Nat. Med. 2007;13(10):1248–1252. doi: 10.1038/nm1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esmaeilzadeh A., Tahmasebi S., Athari S.S. Chimeric antigen receptor-T cell therapy: applications and challenges in treatment of allergy and asthma. Biomed. Pharmacother. 2020;123 doi: 10.1016/j.biopha.2019.109685. [DOI] [PubMed] [Google Scholar]
- 26.Elahi R., Khosh E., Tahmasebi S., Esmaeilzadeh A. Immune cell hacking: challenges and clinical approaches to create smarter generations of chimeric antigen receptor T cells. Front. Immunol. 2018;9:1717. doi: 10.3389/fimmu.2018.01717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang D., Lin M., Wei L., Xie L., Zhu G., CSD Cruz, et al. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. Jama. 2020;323(11):1092–1093. doi: 10.1001/jama.2020.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., et al. Neutrophil-to-lymphocyte ratio predicts severe illness patients with 2019 novel coronavirus in the early stage. MedRxiv. 2020;18(206) doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peteranderl C., Herold S. The impact of the interferon/TNF-related apoptosis-inducing ligand signaling axis on disease progression in respiratory viral infection and beyond. Front. Immunol. 2017;8:313. doi: 10.3389/fimmu.2017.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamming I., Timens W., Bulthuis M., Lely A., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet. 2020;395(10235):1517–1520. doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Xu W., Hu G., Xia S., Sun Z., Liu Z., et al. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of immune response in patients with COVID-19 in Wuhan. Clin. l. Infect. Dis. Chin. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohlmeier J.E., Cookenham T., Roberts A.D., Miller S.C., Woodland D.L. Type I interferons regulate cytolytic activity of memory CD8+ T cells in the lung airways during respiratory virus challenge. Immunity. 2010;33(1):96–105. doi: 10.1016/j.immuni.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J., Yamane H., Paul W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 2009;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kheirandish H. Corona virus and salt intake. J. Med. Chem. Sci. 2021;4(1):1–7. [Google Scholar]
- 40.Ghaebi M., Abdolmohammadi-Vahid S., Ahmadi M., Eghbal-Fard S., Dolati S., Nouri M., et al. T cell subsets in peripheral blood of women with recurrent implantation failure. J. Reprod. Immunol. 2019;131:21–29. doi: 10.1016/j.jri.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 41.Ganji A., Farahani I., Khansarinejad B., Ghazavi A., Mosayebi G. Increased expression of CD8 marker on T-cells in COVID-19 patients. Blood Cell Mol. Dis. 2020;83 doi: 10.1016/j.bcmd.2020.102437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du R.-H., Liang L.-R., Yang C.-Q., Wang W., Cao T.-Z., Li M., et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur. Respir. J. 2020;55(5) doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020 In preparation. [Google Scholar]
- 46.Liu Z., Long W., Tu M., Chen S., Huang Y., Wang S., et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Inf. Secur. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J., Qi T., Liu L., Ling Y., Qian Z., Li T., et al. Clinical progression of patients with COVID-19 in Shanghai, China. J. Inf. Secur. 2020;80(5):e1–e6. doi: 10.1016/j.jinf.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F., Hou H., Luo Y., Tang G., Wu S., Huang M., et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5(10) doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salomé B., Magen A. Nature Publishing Group; 2020. Dysregulation of Lung Myeloid Cells in COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong C., Lam C., Wu A., Ip W., Lee N., Chan I., et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin H.-S., Kim Y., Kim G., Lee J.Y., Jeong I., Joh J.-S., et al. Immune responses to Middle East respiratory syndrome coronavirus during the acute and convalescent phases of human infection. Clin. Infect. Dis. 2019;68(6):984–992. doi: 10.1093/cid/ciy595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44(6):1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53(3):368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moghadam S., Erfanmanesh M., Esmaeilzadeh A. Interleukin 35 and hepatocyte growth factor; as a novel combined immune gene therapy for multiple sclerosis disease. Med. Hypotheses. 2017;109:102–105. doi: 10.1016/j.mehy.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020;7(6):998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian S., Hu W., Niu L., Liu H., Xu H., Xiao S.-Y. Pulmonary pathology of early phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 2020;15(5):700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neidleman J., Luo X., Frouard J., Xie G., Gurjot G., Stein E.S., et al. SARS-CoV-2-specific T cells exhibit unique features characterized by robust helper function, lack of terminal differentiation, and high proliferative potential. bioRxiv. 2020;1(6) doi: 10.1016/j.xcrm.2020.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M., Endeman H., et al. Phenotype of SARS-CoV-2-specific T-cells in COVID-19 patients with acute respiratory distress syndrome. medRxiv. 2020 doi: 10.1101/2020.04.11.20062349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmadi M., Yousefi M., Abbaspour-Aghdam S., Dolati S., Aghebati-Maleki L., Eghbal-Fard S., et al. Disturbed Th17/Treg balance, cytokines, and miRNAs in peripheral blood of patients with Behcet’s disease. J. Cell. Physiol. 2019;234(4):3985–3994. doi: 10.1002/jcp.27207. [DOI] [PubMed] [Google Scholar]
- 64.Lee Y.K., Landuyt A.E., Lobionda S., Sittipo P., Zhao Q., Maynard C.L. TCR-independent functions of Th17 cells mediated by the synergistic actions of cytokines of the IL-12 and IL-1 families. PLoS One. 2017;12(10) doi: 10.1371/journal.pone.0186351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoeve M.A., Savage N.D., de Boer T., Langenberg D.M., de Waal Malefyt R., Ottenhoff T.H., et al. Divergent effects of IL-12 and IL-23 on the production of IL-17 by human T cells. Eur. J. Immunol. 2006;36(3):661–670. doi: 10.1002/eji.200535239. [DOI] [PubMed] [Google Scholar]
- 66.Zheng Y., Huang Z., Ying G., Zhang X., Ye W., Hu Z., et al. Comparative study of the lymphocyte change between COVID-19 and non-COVID-19 pneumonia cases suggesting uncontrolled inflammation might not be the main reason of tissue injury. medRxiv. 2020 ID: ppmedrxiv-20024885. [Google Scholar]
- 67.Liu Y., Du X., Chen J., Jin Y., Peng L., Wang H.H., et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Inf. Secur. 2020;81(1):e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional http://science.sciencemag.org/ Downloaded from exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol.10. [DOI] [PMC free article] [PubMed]
- 69.Liu Y., Zhang C., Huang F., Yang Y., Wang F., Yuan J., et al. Elevated plasma level of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl. Sci. Rev. 2020;7(6):1006–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu Y, Zhang C, Huang F, Yang Y, Wang F, Yuan J, et al. Novel Coronavirus (2019-nCoV) Infections Trigger an Exaggerated Cytokine Response Aggravating Lung Injury. 2020f. 2020.
- 71.Faure E., Poissy J., Goffard A., Fournier C., Kipnis E., Titecat M., et al. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One. 2014;9(2):e88716. doi: 10.1371/journal.pone.0088716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., et al. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4(3) doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ait Addi R., Benksim A., Cherkaoui M. Pregnancy and COVID-19: what we need to know. Electron. J. Gen. Med. 2020;17(6) (2020) [Google Scholar]
- 74.Roncati L., Nasillo V., Lusenti B., Riva G. Signals of Th2 immune response from COVID-19 patients requiring intensive care. Ann. Hematol. 2020;1 doi: 10.1007/s00277-020-04066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei X.-S., Wang X., Niu Y.-R., Ye L.-L., Peng W.-B., Wang Z.-H., et al. 2020. Clinical Characteristics of SARS-CoV-2 Infected Pneumonia With Diarrhea. Available at SSRN 3546120. [Google Scholar]
- 76.Spellberg B., Edwards J.E., Jr. Type 1/type 2 immunity in infectious diseases. Clin. Infect. Dis. 2001;32(1):76–102. doi: 10.1086/317537. [DOI] [PubMed] [Google Scholar]
- 77.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunologic features in severe and moderate forms of coronavirus disease. J. Clin. Invest. 2019:137244. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., et al. Single-cell transcriptomic analysis of SARS-CoV-2 reactive CD4+ T cells. bioRxiv. 2020 doi: 10.2139/ssrn.3641939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schorer M., Lambert K., Rakebrandt N., Rost F., Kao K.-C., Yermanos A., et al. Rapid expansion of Treg cells protects from collateral colitis following a viral trigger. Nat. Commun. 2020;11(1):1–13. doi: 10.1038/s41467-020-15309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duerr G., Heine A., Hamiko M., Zimmer S., Luetkens J., Nattermann J., et al. Parameters predicting COVID-19-induced myocardial injury and mortality. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang W., He J. Lie puyi, et al. The definition and risks of cytokine release syndrome-like in 11 COVID-19-infected pneumonia critically ill patients: disease characteristics and retrospective analysis. Intensive Care Cri. Care Med. 2020 doi: 10.1101/2020.02.26.20026989. [DOI] [Google Scholar]
- 82.Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., et al. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160(3):261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., JL McKechnie, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020:1–7. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.National Research Project for SARS BG The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am. J. Clin. Pathol. 2004;121(4):507–511. doi: 10.1309/WPK7Y2XKNF4CBF3R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020:1–3. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 88.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marquardt N., Kekäläinen E., Chen P., Kvedaraite E., Wilson J.N., Ivarsson M.A., et al. Human lung natural killer cells are predominantly comprised of highly differentiated hypofunctional CD69− CD56dim cells. J. Allergy Clin. Immunol. 2017;139(4):1321–1330. doi: 10.1016/j.jaci.2016.07.043. (e4) [DOI] [PubMed] [Google Scholar]
- 90.Sun D., Li H., Lu X.-X., Xiao H., Ren J., Zhang F.-R., et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center’s observational study. World J. Pediatr. 2020:1–9. doi: 10.1007/s12519-020-00354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng Y., Huang Z., Yin G., Zhang X., Ye W., Hu Z., et al. 2020. Study of the Lymphocyte Change Between COVID-19 and Non-COVID-19 Pneumonia Cases Suggesting Other Factors Besides Uncontrolled Inflammation Contributed to Multi-Organ Injury. [Google Scholar]
- 92.Chen J., Fan H., Zhang L., Huang B., Zhu M., Zhou Y., et al. Retrospective analysis of clinical features in 101 death cases with COVID-19. medRxiv. 2020 [Google Scholar]
- 93.Lagunas-Rangel F.A. Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. J. Med. Virol. 2020;92(10):1733–1734. doi: 10.1002/jmv.25819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Narasaraju T., Yang E., Samy R.P., Ng H.H., Poh W.P., Liew A.-A., et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am. J. Pathol. 2011;179(1):199–210. doi: 10.1016/j.ajpath.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., et al. Neutrophil extracellular traps (NETs) as markers of disease severity in COVID-19. medRxiv. 2020;51:446–453. [Google Scholar]
- 96.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang D., Guo R., Lei L., Liu H., Wang Y., Wang Y., et al. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv. 2020;109(1):13–22. doi: 10.1002/JLB.4HI0720-470R. [DOI] [Google Scholar]
- 99.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., et al. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+ CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. BioRxiv. 2020 doi: 10.1101/2020.02.12.945576. [DOI] [Google Scholar]
- 100.Mehta P., DF McAuley, Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (London, England) 2020;395(10229):1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ong E.Z., YFZ Chan, Leong W.Y., NMY Lee, Kalimuddin S., SMH Mohideen, et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020;27(6):879–882. doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guo C., Li B., Ma H., Wang X., Cai P., Yu Q., et al. Tocilizumab treatment in severe COVID-19 patients attenuates the inflammatory storm incited by monocyte centric immune interactions revealed by single-cell analysis. BioRxiv. 2020;11(1):3924. doi: 10.1101/2020.04.08.029769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yao X., Li T., He Z., Ping Y., Liu H., Yu S., et al. A pathological report of three COVID-19 cases by minimally invasive autopsies. Zhonghua bing li xue za zhi= Chinese journal of pathology. 2020;49 doi: 10.3760/cma.j.cn112151-20200312-00193. [DOI] [PubMed] [Google Scholar]
- 104.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–890. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Siddiqi H.K. Mehra MR. COVID-19 illness in native and immunosuppressed states: a clinical–therapeutic staging proposal. J. Heart Lung Transplant. 2020;39(5):405. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang Y., Shen C., Li J., Yuan J., Yang M., Wang F., et al. Exuberant elevation of IP-10. MCP-3 and IL-1ra during SARS-CoV-2 infection is associated with disease severity and fatal outcome. medRxiv. 2020;(2002):2020. [Google Scholar]
- 107.Vaninov N. Nature Publishing Group; 2020. In the Eye of the COVID-19 Cytokine Storm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marofi F., Azizi R., Motavalli R., Vahedi G., Nasimi M., Yousefi M., et al. COVID-19: our current knowledge of epidemiology, pathology, therapeutic approaches, and diagnostic methods. Anti Cancer Agents Med. Chem. 2021 doi: 10.2174/1871520621666210201101245. [DOI] [PubMed] [Google Scholar]
- 109.Reche A., Kolse R., Gupta S., Ingle A., Chhabra K.G., Nimbulkar G. Therapeutic options for COVID–19: pandemic–a review. Int. J. Res. Pharm. Sci. 2020;11(Special Issue 1) [Google Scholar]
- 110.Abdin S.M., Elgendy S.M., Alyammahi S.K., Alhamad D.W., Omar H.A. Tackling the cytokine storm in COVID-19, challenges, and hopes. Life Sci. 2020;257 doi: 10.1016/j.lfs.2020.118054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu X. 2020. A Multicenter, Randomized Controlled Trial for the Efficacy and Safety of Tocilizumab in the Treatment of New Coronavirus Pneumonia (COVID-19). Registration Number on ChiCTR: ChiCTR2000029765 of February; p. 13. [Google Scholar]
- 112.Huet T., Beaussier H., Voisin O., Jouveshomme S., Dauriat G., Lazareth I., et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2(7):e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dimopoulos G., de Mast Q., Markou N., Theodorakopoulou M., Komnos A., Mouktaroudi M., et al. Favorable anakinra responses in severe Covid-19 patients with secondary hemophagocytic lymphohistiocytosis. Cell Host Microbe. 2020;28(1):117–123. doi: 10.1016/j.chom.2020.05.007. (e1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shi H., Wang W., Yin J., Ouyang Y., Pang L., Feng Y., et al. The inhibition of IL-2/IL-2R gives rise to CD8+ T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020;11(6):1–8. doi: 10.1038/s41419-020-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Webster R.M. The immune checkpoint inhibitors: where are we now? Nat. Rev. Drug Discov. 2014;13(12):883. doi: 10.1038/nrd4476. [DOI] [PubMed] [Google Scholar]
- 116.Luo J., Rizvi H., Egger J.V., Preeshagul I.R., Wolchok J.D., Hellmann M.D. Impact of PD-1 blockade on severity of COVID-19 in patients with lung cancers. Cancer Discov. 2020;10(8):1121–1128. doi: 10.1158/2159-8290.CD-20-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu J., Chen Y., Li R., Dong X., Li Y., Xu Q., et al. 2020. Intravenous Immunoglobulin Treatment for Patients With Severe COVID-19: A Retrospective Multi-Center Study. [Google Scholar]
- 118.Nimmerjahn F., Ravetch J.V. Anti-inflammatory actions of intravenous immunoglobulin. Annu. Rev. Immunol. 2008;26:513–533. doi: 10.1146/annurev.immunol.26.021607.090232. [DOI] [PubMed] [Google Scholar]
- 119.Heming N., Sivanandamoorthy S., Meng P., Bounab R., Annane D. Immune effects of corticosteroids in sepsis. Front. Immunol. 2018;9:1736. doi: 10.3389/fimmu.2018.01736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stern A., Skalsky K., Avni T., Carrara E., Leibovici L., Paul M. Corticosteroids for pneumonia. Cochrane Database Syst. Rev. 2017;(12) doi: 10.1002/14651858.CD007720.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng C., Wang J., Guo H., Lu Z., Ma Y., Zhu Y., et al. Risk-adapted treatment strategy for COVID-19 patients. Int. J. Infect. Dis. 2020;94:74–77. doi: 10.1016/j.ijid.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Benegiamo M., Sammarro A., Rella E., D’Addona A., Garcia-Godoy F. Using antiseptic mouthrinses to reduce Sars-Cov2 oral viral load. Electron. J. Gen. Med. 2021;18(1):em273. 2021. [Google Scholar]
- 123.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of immune response in patients with COVID-19 in Wuhan. China Clin. Infect. Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. covidwho-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang J.C., Zhang X., Wu G., Yi J. 2019. The Potential Role of IL-6 in Monitoring Coronavirus Disease. [Google Scholar]
- 126.Chen L., Liu H., Liu W., Liu J., Liu K., Shang J., et al. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua jie he he hu xi za zhi= Zhonghua jiehe he huxi zazhi= Chinese journal of tuberculosis and respiratory diseases. 2020;43 doi: 10.3760/cma.j.issn.1001-0939.2020.0005. [DOI] [PubMed] [Google Scholar]
- 127.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020;71(15):769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ouyang Y., Yin J., Wang W., Shi H., Shi Y., Xu B., et al. Down-regulated gene expression spectrum and immune responses changed during the disease progression in COVID-19 patients. Clin. Infect. Dis. 2020;71(16):2052–2060. doi: 10.1093/cid/ciaa462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li X., Xu S., Yu M., Wang K., Tao Y., Zhou Y., et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020;146(1):110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wan S., Yi Q., Fan S., Lv J., Zhang X., Guo L., et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019-nCoV pneumonia (NCP) medRxiv. 2020 [Google Scholar]
- 133.Gong J., Dong H., Xia S.Q., Huang Y.Z., Wang D., Zhao Y., et al. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. MedRxiv. 2020 doi: 10.1186/s12879-020-05681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis. 2020;71(8):1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qi D., Yan X., Tang X., Peng J., Yu Q., Feng L., et al. 2020. Epidemiological and Clinical Features of 2019-nCoV Acute Respiratory Disease Cases in Chongqing Municipality, China: A Retrospective, Descriptive, Multiple-Center Study. [Google Scholar]
- 136.Guohua L., Ling L., Min H., Haibiao L., Peifeng K., Zishao Z. Value of various inflammatory markers combined with lymphocyte subsets on clinical diagnosis of different clinical types of COVID-19. J. Chong. Med. Univ. 2020:10. [Google Scholar]
- 137.Xu J., Han M., Zhao F., Zhang T., Ma L. Clinical manifestations and sero-immunological characteristics of 155 patients with COVID-19. Chin. J. Nosocomiol. 2020;30(15):961–965. [Google Scholar]
- 138.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., Sun R., Tian Z., Xu X., Wei H. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+ CD16+ monocytes in severe pulmonary syndrome patients of a new coronavirus. bioRxiv. 2020 doi: 10.1101/2020.02.12.945576. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data not shared.