Abstract
Background & Aims:
The mechanisms by which the I148M mutant variant of the patatin-like phospholipase domain-containing 3 (PNPLA3I148M) drives development of nonalcoholic steatohepatitis (NASH) is not known. The aim of this study was to obtain insights on mechanisms underlying PNPLA3I148M induced acceleration of NASH.
Approach & Results:
Hepatocyte-specific overexpression of empty vector (Luc), human wild-type PNPLA3 (PNPLA3WT), or PNPLA3I148M was achieved using adeno-associated virus (AAV)-8 in DIAMOND mice followed by chow diet or high fat Western diet with ad lib administration of sugar in drinking water (WDSW) for 8 weeks. Under WDSW, PNPLA3I148M overexpression accelerated steatohepatitis with increased steatosis, inflammation ballooning and fibrosis (p< 0.001 vs other groups for all). Silencing PNPLA3I148M after its initial overexpression abrogated these findings. PNPLA3I148M caused 22:6n3 docosahexanoic acid depletion and increased ceramides under WDSW in addition to increasing triglycerides and diglycerides especially enriched with unsaturated fatty acids. It also increased oxidative stress and ER-stress. Increased total ceramides was associated with STAT3 activation with downstream activation of multiple immune-inflammatory pathways at a transcriptomic level by network analyses. Silencing PNPLA3I148M reversed STAT3 activation. Conditioned media from HepG2 cells overexpressing PNPLA3I148M increased procollagen mRNA expression in LX2 cells; this was abrogated by hepatocyte STAT3 inhibition.
Conclusions:
Under WDSW, PNPLA3I148M overexpression promotes steatosis and NASH by metabolic reprogramming characterized by increased triglycerides and diglycerides, n3 PUFA depletion and increased ceramides with resultant STAT3 phosphorylation and downstream inflammatory pathway activation driving increased stellate cell fibrogenic activity.
Keywords: nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, DIAMOND model, PNPLA3, ceramide, innate immune response, STAT3, inflammation, fibrosis
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD), especially its aggressive variant nonalcoholic steatohepatitis (NASH), is a major cause of liver related mortality (1). NAFLD is a heterogeneous disorder driven by complex gene-environment interactions that produce variable clinical and histological phenotypes of the disease that progress at variable rates towards cirrhosis (2, 3). Understanding the basis for such heterogeneity is critical for development of highly effective therapeutics for affected individuals.
The patatin-like phospholipase domain containing 3 (PNPLA3) gene was the first major gene associated with NASH (4, 5). Patients with NAFLD who carry the PNPLA3 rs738409 C/G mutation encoding an isoleucine to methionine substitution at the amino acid position 148 (PNPLA3I148M) are more likely to develop steatohepatitis and fibrosis (6). PNPLA3 is a triglyceride hydrolase whose function is lost in the mutant protein (7). PNPLA3I148M is the principal genetic risk factor for disease progression and delinks disease severity from underlying insulin resistance (8, 9).
In initial studies, overexpression of the mutant human PNPLA3 in C57Bl6J mice led to increased steatosis with accumulation of PNPLA3 protein on lipid droplets but without development of steatohepatitis and fibrosis (10). This was subsequently shown to be related to proteosomal dysfunction and reduced turnover of the mutant protein which accumulated on lipid droplets (11). While this explains worsened steatosis, these data however do not provide information on the cellular or molecular basis for PNPLA3I148M-associated development of steatohepatitis and fibrosis.
In an elegant study, the human mutation (I148M) was introduced into the mouse PNPLA3 gene by homologous recombination (12). Mice were then fed a 70% sucrose diet to induce NAFLD. Subsequent knockdown of PNPLA3 with a GalNAc-conjugated anti-sense oligonucleotide decreased steatosis and inflammation. However, a limitation of this study is that a clear phenotype of NASH with fibrosis was not established by introduction of the I148M mutation. Furthermore, this study evaluated the impact of the human mutation in the mouse gene rather than the function of the human mutant gene itself. In other studies, global targeted deletion of PNPLA3 did not induce NASH in a murine model (13).
We have recently established and validated a diet-induced animal model of NAFLD (DIAMOND) using an isogenic inbred C57Bl6J/s129svlmJ mice that sequentially develop a fatty liver then steatohepatitis and progressive fibrosis on a high-fat (Western) diet with ad lib provision of sugars in drinking water (WDSW) (14, 15). The first aim of the current study was to leverage this sequential development of lesions to test the hypothesis that overexpression of PNPLA3I148M would cause development of NASH with fibrosis at a time point (8 weeks) when the model would normally only develop a fatty liver thus demonstrating disease acceleration by PNPLA3I148M. The second aim was to confirm the specificity of these findings by rescuing the phenotype by silencing PNPLA3I148M after its initial overexpression. The third aim was to take advantage of this accelerated disease phenotype to define the metabolomic, transcriptomic and molecular basis of PNPLA3I148M-related acceleration of steatohepatitis and fibrosis. Finally, to confirm the effects of hepatocyte-specific PNPLA3 expression on stellate cell activation, procollagen gene expression in stellate cells was measured after exposure to conditioned media from hepatocytes with and without overexpressed PNPLA3I148M.
MATERIALS AND METHODS
Diet-induced animal model of NAFLD
The previously reported and well-characterized DIAMOND mice were used (14–17). This mouse strain develops a fatty liver (2–8 weeks) and then steatohepatitis (by 16 weeks) followed by progressive fibrosis on a WDSW diet. This diet (42% calories from fat, 0.1% cholesterol [Harlan TD.88137]) is similar in composition to the average American adult diet (cdc.gov/nchs/fastats/diet.htm). Fructose-glucose (23.1g/L d-fructose + 18.9 g/L d-glucose) was added to drinking water and provided ad libitum. The rationale for using this model was that it sequentially develops steatosis, steatohepatitis and then fibrosis allowing us to test if more advanced lesions could be induced earlier in the course of the disease thus providing proof of concept of disease acceleration by PNPLA3I148M.
Hepatocyte-specific expression of PNPLA3
An adeno-associated virus (AAV)-8 construct with a thyroxine-binding globulin promoter was used for hepatocyte-specific overexpression of PNPLA3 (18). Three AAV constructs, AAV-Luc empty vector, AAV-PNPLA3WT and AAV-PNPLA3I148M were obtained from the Viral Vector Core at the University of Massachusetts (Supplemental Figure 1). A pilot study was initially conducted using two different doses of AAV (1×1011 GC/mouse and 3×1011 GC/mouse) administered via the retro-orbital route in order to determine the dose of AAV-PNPLA3 required for optimal expression. The siRNA of placebo lipid nanoparticles and si-hPNPLA3 were obtained from our collaborators at Merck.
Study Design
Based on this pilot study, approximately 8-week old male DIAMOND mice (age range 6 – 10 weeks old) were injected with 3×1011 GC/mouse of Luc, PNPLA3WT or PNPLA3I148M. Mice were then randomized to chow diet/normal water (CDNW) or to WDSW for 8 weeks. These studies were performed in two cohorts with eight to ten mice per group. All mice were housed in a 12 hour light–12 hour dark cycle in a 21–23 °C facility.
To test the specificity of findings attributed to PNPLA3I148M, 8-week old male mice were injected with AAV-8 carrying PNPLA3I148M and started on WDSW. After four weeks, half the mice received si-hPNPLA3I148M at 0.5 mg/kg body weight via tail vein injection while the other half received scrambled sequences, given once per week for a total of 4 weeks. Mice were then euthanized and tissues collected for analysis. All procedures were performed according to protocols approved by the Animal Care and Use Committee of Virginia Commonwealth University.
For in vitro studies, hepatocytes and stellate cells were isolated from mice liver as previously described (19) and assayed to determine the site of human PNPLA3 expression. In vitro studies were also performed using HepG2 hepatoma cells and LX2 stellate cells. PNPLA3I148M was induced in HepG2 cells by addition of high glucose to the medium containing 10% fetal bovine serum, penicillin (100IU/ml) and Streptomycin (100μg/ml). LX2 cells were exposed overnight to conditioned media from HepG2 cells with or without induction of PNPLA3I148M before harvesting for procollagen mRNA measurement. To determine STAT3 effect on induction of fibrosis in LX2 cells, HepG2 cells were pretreated with a STAT3 inhibitor (Stattic, Sigma Co.) prior to collection of conditioned media.
Laboratory and histological analysis, RNA, protein and metabolite analyses
Protein and mRNA extraction followed by RNA Seq, PCR or Western blot were performed on tissues as previously described (14) (Supplemental methods). Metabolomic analysis were also performed using the One-Way-Lipidomics (OWL) platform as described previously (20) (Supplemental materials). Similarly, histological assessment followed established methods and was performed in a masked manner by a dedicated senior hepato-pathologist (PB) using hematoxylin/eosin and Sirius Red stained sections scanned in using the Aperio 3.0 system (14).
Analytical Approach
Descriptive and across group comparisons for weight, histological scores and other parameters was performed using SPSS and plotted on Graphpad Prism software. T-test or ANOVA were used as appropriate for continuous variables and Fishers Exact test was used for comparison of proportions. A p value of 0.05 was used to establish statistical significance.
Metabolomic analyses were performed using Metabo-analyst 4.0 software using the statistical and enrichment modules as described previously by us (21, 22). Similarly, transcriptomic analyses were performed using Genespring GX 12.5 (14). Pathway enrichment was analyzed and visualized using Gene Ontology and Metacores curated pathways (23). Integrated analysis of gene expression and metabolites to evaluate pathway status was performed using the Joint pathway module of Metabo-analyst 4.0. Network analyses of metabolites was also performed using the network module of Metabo-analyst 4.0.
RESULTS
Liver-specific expression of human wildtype and mutant PNPLA3 in mouse liver using adeno-associated virus
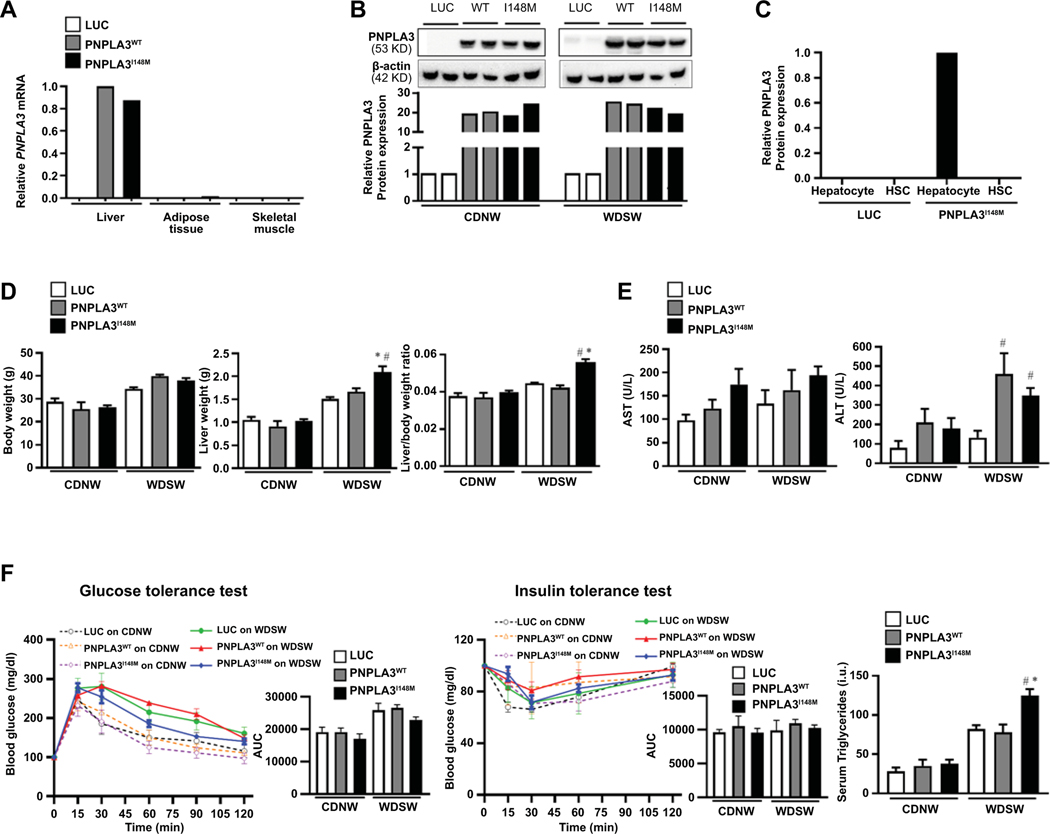
In pilot studies, DIAMOND mice were given retro-orbital injections of AAV with empty luciferase vector Luc, PNPLA3WT or PNPLA3I148M. Real time PCR using primers for human PNPLA3 confirmed liver-specific expression in mice injected with PNPLA3WT and PNPLA3I148M, with little or no expression in adipose tissue and skeletal muscle in these mice (Figure 1A). Mice injected with Luc showed no PNPLA3 mRNA expression (Figure 1A). Over-expression of human PNPLA3 protein was confirmed by Western blot in both PNPLA3WT and PNPLA3I148M mice, while no PNPLA3 protein was noted with Luc (Figure 1B). In pilot studies, two weeks following PNPLA3 administration, hepatocytes and stellate cells were isolated from the liver; PNPLA3 mRNA was overexpressed solely in hepatocytes and not in hepatic stellate cells (Figure 1C).
Figure 1: AAV-mediated PNPLA3 expression is liver-specific and its effects are dependent on a high fat high sugar diet.
(A) Liver-specific overexpression of human PNPLA3 gene in syngeneic C57Bl6J/129S1svlmJ mice (n=3 mice per group). Adeno associated virus (AAV) containing empty luciferase (luc), PNPLA3WT, or PNPLA3I148M was introduced by retro-orbital injection. Real-time PCR demonstrated high liver expression of both PNPLA3 forms with virtually no expression in other organs. (B) Mice were randomized to CDNW or WDSW ad libitum for 8 weeks after retro-orbital injection of the three AAV constructs. Western blot analysis was performed in liver lysates to determine PNPLA3 protein expression, which was normalized to AAV-luc on CDNW or WDSW. (C) Hepatocytes and stellate cells were isolated from the liver 2 weeks after injection of the AAV constructs. PNPLA3 was expressed in hepatocytes but not stellate cells confirming hepatocyte specific expression. (D) Mice were randomized to CDNW or WDSW after injection of the three AAV constructs. All mice on WDSW gained more weight than mice on WDSW. The liver weight and liver/body weight ratio was significantly increased in PNPLA3I148M mice compared to other groups on WDSW diet. (E) Plasma collected at the time of sacrifice was used to determine aminotransferase levels, with ALT reaching significance in PNPLA3WT and PNPLA3I148M mice on WDSW compared to luc. (F) There were no differences across the AAV-groups with respect to glucose homeostasis and insulin resistance measured by glucose tolerance test and insulin tolerance tests, however PNPLA3I148M mice had significant hypertriglyceridemia on WDSW compared to other groups. The mice data shown represent 8–10 mice per group unless previously indicated. # represents p<0.05 compared to luc on WDSW; * represents p<0.05 compared to PNPLA3WT on WDSW.
After establishing feasibility in these pilot studies, mice were injected with AAV-Luc, AAV-PNPLA3WT or AAV-PNPLA3I148M and randomized to ad libitum feeding on CDNW or WDSW for 8 weeks. As expected, body weight was higher in all mice groups on WDSW compared to those on CDNW (Fig. 1D; left panel). While PNPLA3WT mice on WDSW gained slightly more weight compared to Luc mice, the liver weight (Figure 1D; middle panel) and liver/body weight ratio (Figure 1D, right panel) were similar in these two groups of mice. In contrast, PNPLA3I148M on WDSW showed increased liver weight and liver/body weight ratio compared to Luc and PNPLA3WT on an identical diet. There was a trend for increased transaminases in both PNPLA3 groups versus Luc which reached significance for ALT in both PNPLA3WT and PNPLA3I148M under WDSW conditions (Figure 1E).
PNPLA3 did not alter systemic insulin resistance but led to dyslipidemia under conditions of Western diet
Mice on WDSW showed a trend towards increased blood glucose when assessed by glucose tolerance test (Figure 1F; left panel), however there were no statistically significant differences in insulin sensitivity between Luc, PNPLA3WT or PNPLA3I148M on WDSW diet as measured by insulin tolerance test (Figure 1F; middle panel). Under CDNW conditions, there was no difference in circulating triglyceride levels among mice groups (Figure 1F; right panel). In contrast, PNPLA3I148M mice showed increased circulating triglyceride levels compared to mice with Luc or PNPLA3WT on WDSW diet.
Human PNPLA3I148M expression specifically accelerated steatohepatitis and led to liver fibrosis in DIAMOND mice under high fat-sugar water diet conditions
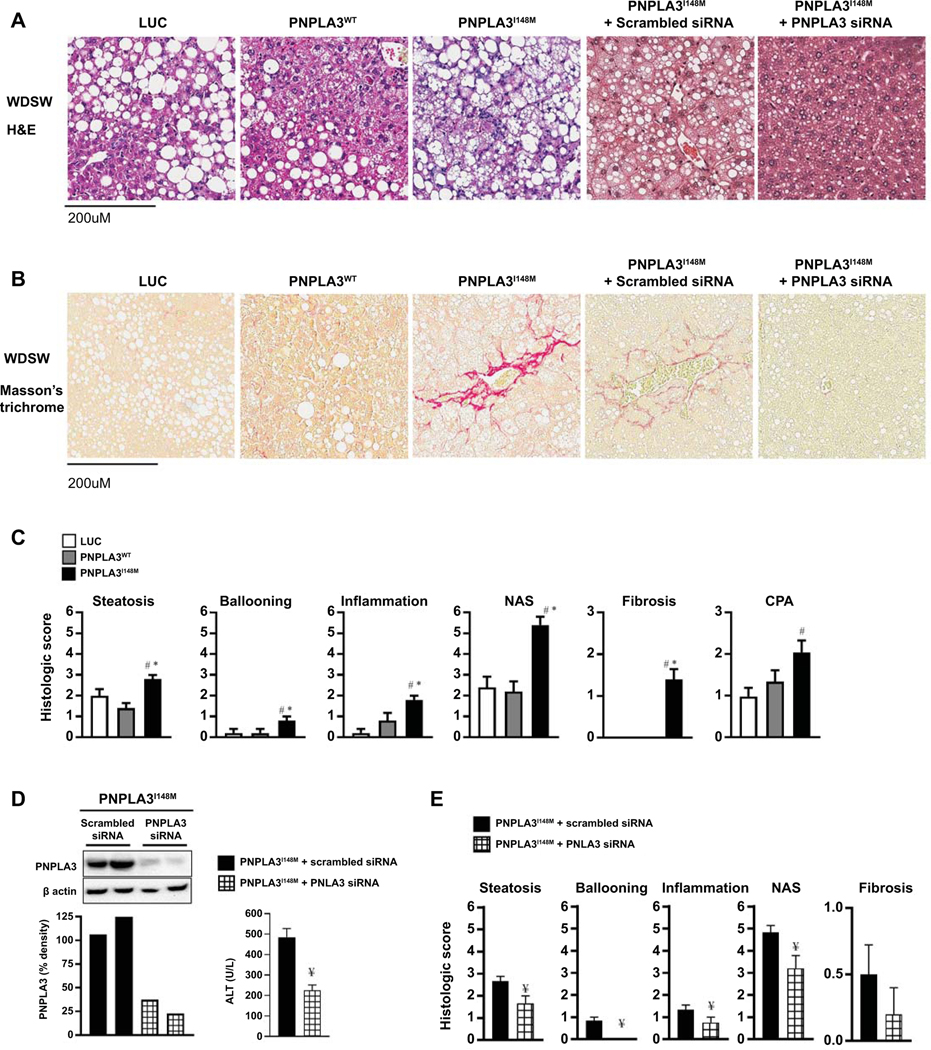
Under chow diet conditions, all three groups had essentially normal liver histology (supplemental Figure 2A and 2B). As expected, after 8 weeks of WDSW diet, Luc mice developed profound fatty liver but without any evidence of steatohepatitis (Figure 2A) or fibrosis (Figure 2B). Similarly, mice with PNPLA3WT mainly had fatty liver with some mice demonstrating mild inflammation but no fibrosis. In sharp contrast, the majority PNPLA3I148M mice developed florid steatohepatitis with fibrosis (p< 0.0001 vs other groups) (Figure 2A and B).
Figure 2: PNPLA3I148M expression aggravates steatohepatitis and fibrosis which is reversed by PNPLA3I148M silencing.
(A) Liver tissue collected at the time of sacrifice were sectioned for histological analysis; representative Hematoxylin and eosin (H&E) stained sections of liver tissue for mice on WDSW are shown. Whereas Luc and PNPLA3WT mice had a fatty liver, PNPLA3I148M mice developed steatohepatitis with fibrosis. The specificity of these findings were confirmed through siRNA-mediated silencing of PNPLA3I148M which resulted in only fatty liver despite feeding on WDSW, in contrast to PNPLA3I148M mice who received scrambled siRNA in which steatohepatitis and fibrosis were re-demonstrated. (B) Sirius Red staining showed that fibrosis developed only in PNPLA3I148M mice with or without scrambled siRNA. (C) Histological scoring of H&E sections showed that the severity of steatosis, ballooning, lobular inflammation, and NAFLD activity score were all significantly higher in PNPLA3I148M mice compared to Luc and PNPLA3WT. Similarly, scoring of Sirius Red stained sections showed that fibrosis stage and collagen proportional area (CPA) were increased only in PNPLA3I148M mice. (D) A Western blot demonstrating effective silencing of PNPLA3 I148M after its initial overexpression in liver tissue is shown. Silencing PNPLA3I148M led to significant reduction in liver inflammation as demonstrated by reduced ALT. (E) Silencing PNPLA3I148M also reduced the severity of steatosis, ballooning, lobular inflammation, NAFLD activity score and fibrosis stage. Histological sections are representative sections and the graphical data represent a total of 8–10 in each group for Luc, PNPLA3WT and PNPLA3I148M and 6 mice each for the experiment with scrambled sequence or siRNA administration. # represents p<0.05 compared to luc on WDSW; * represents p<0.05 compared to PNPLA3WT on WDSW; ¥ represents p<0.05 compared to scrambled siRNA-PNPLA3I148M
In addition to acceleration of steatohepatitis and fibrosis, PNPLA3I148M significantly worsened the severity of individual histological lesions of NASH including steatosis, hepatocellular ballooning and lobular inflammation (Figure 2C). This translated into a significantly higher NAFLD activity score (p< 0.001 vs both Luc and PNPLA3WT) (Figure 2C). Only PNPLA3I148M mice developed fibrosis with some mice developing bridging fibrosis (Figure 2C). This was reflected in a higher collagen proportionate area as well.
The specificity of the findings was tested by comparison of mice overexpressing PNPLA3I148M who received siRNA directed against placebo lipid nanoparticles or PNPLA3I148M. Silencing was confirmed by Western blot (Figure 2D; left panel), with accompanying decrease in biochemical inflammation as evidenced by decreased ALT (Figure 2D; right panel). These mice did not develop steatohepatitis or fibrosis (Figure 2A and B). In comparison to PNPLA3I148M mice who received scrambled siRNA, those with PNPLA3I148M silencing showed decreased steatosis and lobular inflammation whereas ballooning disappeared entirely and fibrosis score was decreased (Figure 2E).
Mechanisms underlying PNPLA3I148M-associated acceleration of NASH and fibrosis
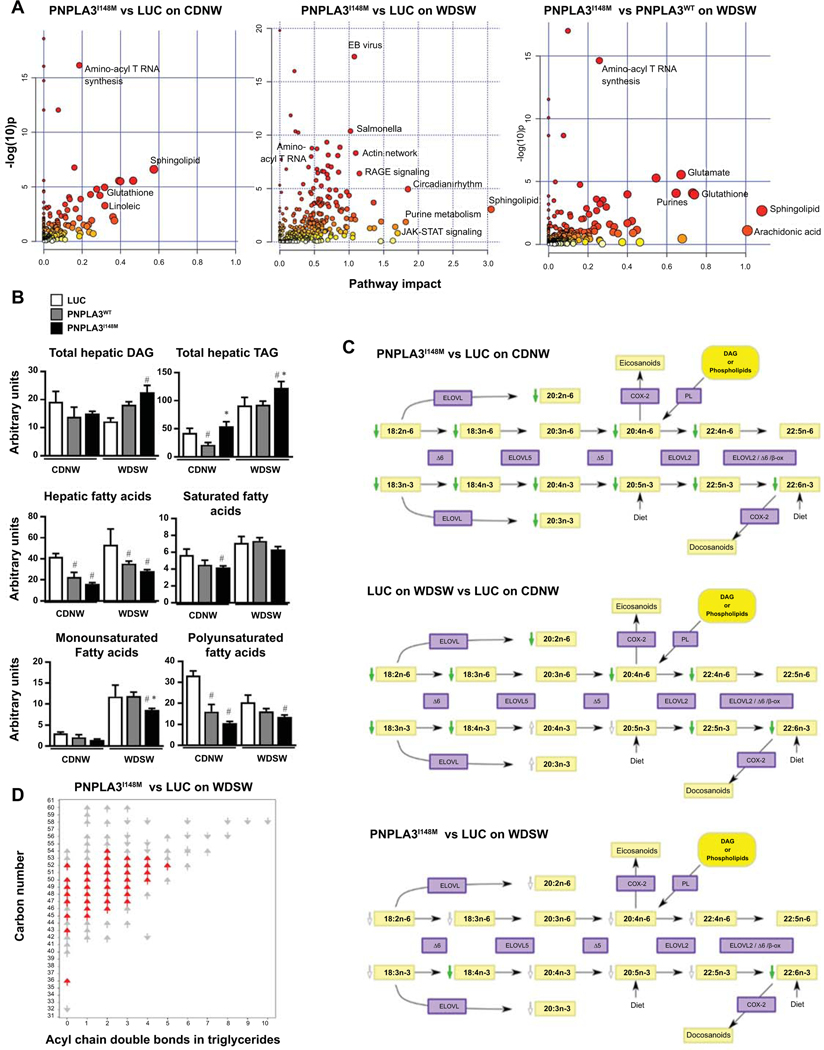
We hypothesized that, based on the known triglyceride hydrolysis inhibition and transacylase functions of PNPLA3I148M (7, 24), overexpression of the mutant gene would remodel the lipidome which in turn would alter cell physiology and gene expression resulting in greater injury and inflammation which would drive more fibrosis. An integrated pathway analysis including both differentially expressed genes and metabolites was performed to identify the metabolic reprogramming signature of PNPLA3I148M. The pathway significance (based on differential expression with weightage based on proportion of differentially expressed metabolites versus genes) versus impact (based on the number of other significantly altered pathways that connect with differentially expressed nodes) was plotted (Figure 3A; supplemental Excel Tables 1–4).
Figure 3: Metabolic reprogramming induced by overexpression of PNPLA3I148M.
(A) Integrated analysis of metabolites and gene-expression data obtained from metabolomic and RNA seq analysis of liver tissue was performed. Under chow diet, the pathways most significantly altered (largest number of significantly altered metabolites and genes) by PNPLA3I148M (vs Luc) when plotted as a function of pathway impact (number of significantly altered pathways intersecting with differentially expressed genes and metabolites in a specific pathway on network analysis) were PUFA and sphingolipid metabolism (left panel). Under WDSW diet, there was a larger set of pathways, mainly metabolic, that were both significantly altered and had a high impact score (middle panel). The specificity of these changes for PNPLA3I148M was tested by comparison to PNPLA3WT under WDSW diet (right panel). These confirmed that the main impact attributable to mutation of the PNPLA3 gene was a metabolic reprogramming with main effects on PUFA, glutathione and sphingolipid pathways. (B) Total DAG, TAG, FFA, and SFA, MUFA and PUFA are shown and demonstrate an increase in DAG and TAG, and a decrease in all fatty acid species in PNPLA3I148M mice on WDSW compared to the other groups on the same diet. (C) Significantly altered PUFA (shown by green arrows) in PNPLA3I148M vs Luc under chow diet are shown (top panel). Luc mice on WDSW diet vs CDNW demonstrated a major depletion of PUFA by a WDSW diet (middle panel). Additional changes in PUFA in PNPLA3I148M (vs Luc mice) on WDSW diet over and on top of changes induced by the diet itself were seen (bottom panel). Several PUFA were further decreased (shown in non-colored arrows) reaching significance for docosahexanoic acid (green arrow). (D) The depletion of PUFAs was accompanied by an increasing number of triglycerides double bonds. Data for 4–5 mice in each group are shown. # represents p<0.05 compared to luc on the same diet; * represents p<0.05 compared to PNPLA3WT on the same diet.
Under chow-diet conditions, the principal pathways affected by PNPLA3I148M included sphingolipids, polyunsaturated fatty acids (PUFA) and glutathione metabolism. In contrast, there was a larger impact under high fat sugar water diet with multiple additional metabolic pathways involved. Comparison of PNPLA3I148M with PNPLA3WT further confirmed that the principal effect attributable to the mutant gene was a metabolic reprogramming of the liver with dominant effects on glutathione and sphingolipids.
A: Metabolic reprogramming due to PNPLA3I148M overexpression
A1: Effects on fatty acid metabolism:
Total triglycerides (TAGs) were increased by PNPLA3I148M versus Luc and PNPLA3WT under both diets whereas DAGs were increased under WDSW diet only (Figure 3B). The hepatic free fatty acid pool was decreased by both PNPLA3 types under chow diet conditions driven mainly by a decrease in PUFAs including 18:3n3 linolenic acid, 22:6n3 docosahexanoic acid and 20:4n6 arachidonic acid (Figure 3B-C; Supplemental Figure 3A). WDSW diet itself led to significant depletion of multiple PUFAs (Figure 3C); under these conditions, there was a further decrease in specific PUFAs for both PNPLA3 groups, reaching statistical significance for 22:6n3 docosahexanoic acid for PNPLA3I148M (Figure 3C; Supplemental Figure 3A). There was a corresponding increase in unsaturation of TAGs (Figure 3D) and DAGs.
Based on integrated pathway analyses and molecular analyses, there were no major effects of PNPLA3I148M on the de novo lipogenesis pathway (Supplemental Figure 3B) although mild increase in protein levels of total SREBP, acyl CoA carboxylase, and fatty acid synthetase was seen (Supplemental Figure 3C). Of note, CGI58 which has been linked to PNPLA3 TAG hydrolytic function (25) was induced by overexpression of PNPLA3I148M but did not decrease significantly after silencing PNPLA3 suggesting that co-factors may maintain its overexpression (Supplemental Figure 3D).
A2: Effects on sphingolipid metabolism:
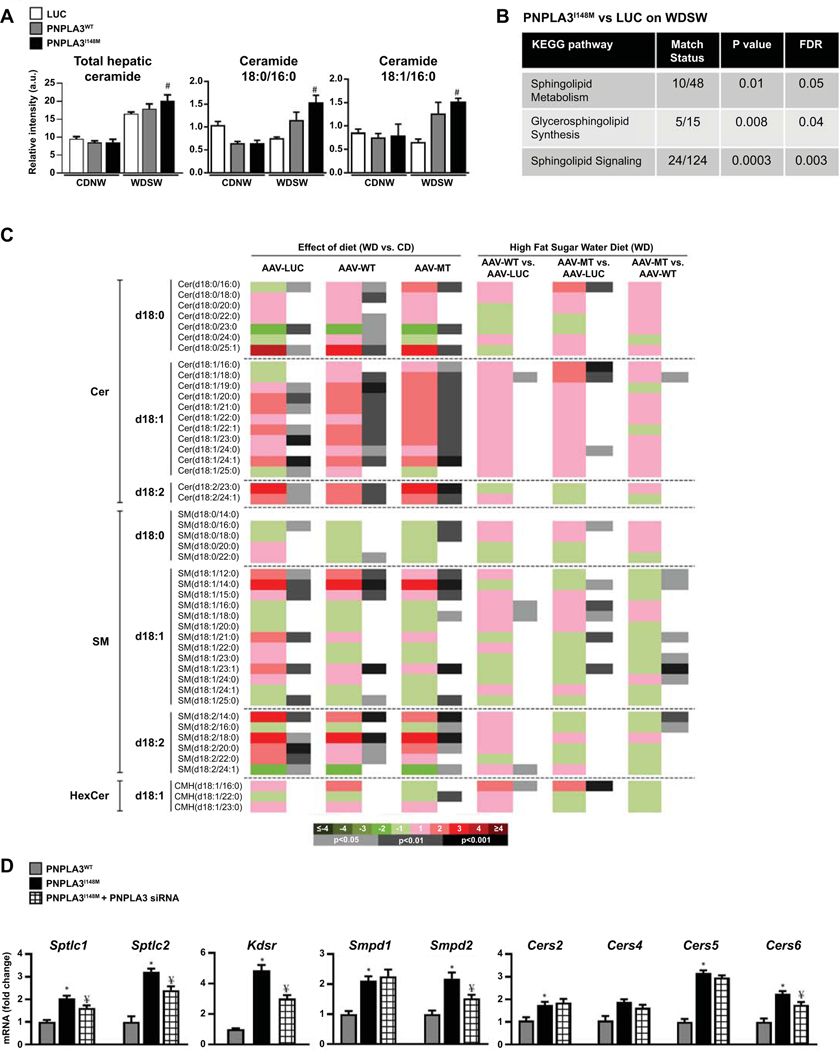
Total ceramides were not significantly different across groups under chow diet conditions but were increased significantly by WDSW in all groups (Figure 4A). This was significantly greater in PNPLA3I148M vis-à-vis luc and PNPLA3WT groups. Ceramide upregulation in PNPLA3I148M mice was accompanied by significant changes in sphingolipid signaling and metabolism pathways at a transcriptomic and metabolomic level respectively (Figure 4B-C).
Figure 4: PNPLA3I148M upregulates ceramides signaling.
(A) Total ceramides were increased in all groups on WDSW diet, with PNPLA3I148M on WDSW demonstrating significantly increased in total ceramides including unsaturated and saturated ceramides compared to other groups. (B) KEGG pathway analysis between PNPLA3I148M and luc on WDWS demonstrate upregulation of sphingolipid signaling pathways. (C) A heat-map of altered sphingolipids is shown with effect of diet (WDSW vs CDNW) in Luc, PNPLA3WTand PNPLA3I148M mice (left three columns). Pairwise comparisons of groups on WDSW are also shown (right three columns). Both diet and PNPLA3 expression remodel the sphingolipidome, with the greatest increase in ceramides seen in PNPLA3I148M mice on WDSW diet. (D) Real time PCR analysis of hepatic expression of genes involved in ceramide synthesis shows upregulation of several key genes involved in de novo synthesis, salvage pathway and sphingomyelinase in PNPLA3I148M mice compared to PNPLA3WT on WDSW. Silencing PNPLA3 I148M resulted in downregulation of these genes, indicating that the observed increments were linked to PNPLA3I148M overexpression. Data from 4–5 mice in each group are shown. # represents p<0.05 compared to luc; * represents p<0.05 compared to PNPLA3WT; ¥ represents p<0.05 compared PNPLA3I148M
PNPLA3I148M overexpression specifically increased expression of multiple genes in the ceramide synthesis, sphingomyelin and salvage pathways (Fig. 4D). These included both isoforms of serine palmitoyl CoA transferase (Sptlc) 1 and 2, the rate-limiting step in ceramide synthesis from serine and palmitic acid in the de novo synthesis pathway. The mRNA expression level of 3-ketodihydrosphingosine reductase (Kdsr), also in the de novo synthesis pathway, was also increased in PNPLA3I148M mice compared to PNPLA3WT mice on WDSW. There was also an increase in the sphingomyelinases sphingomyelin phosphodiesterase (Smpd) 1 and 2 which convert sphingomyelin to ceramide. In addition, there was an increase in the expression of ceramide synthetase (Cers) isoforms 2, 4, 5 and 6 in PNPLA3I148M mice compared to PNPLA3WT mice. Cers1 and Cers3 mRNA levels were similar among the groups on WDSW (data not shown). SiRNA-mediated silencing of PNPLA3I148M resulted in a clear reversal of Spltc 1 and 2, Kdsr, Smpd2 and Cer6 induction despite feeding on a high fat diet (Figure 4D).
B: Mutant PNPLA3 impacts key cellular processes and promotes cell death:
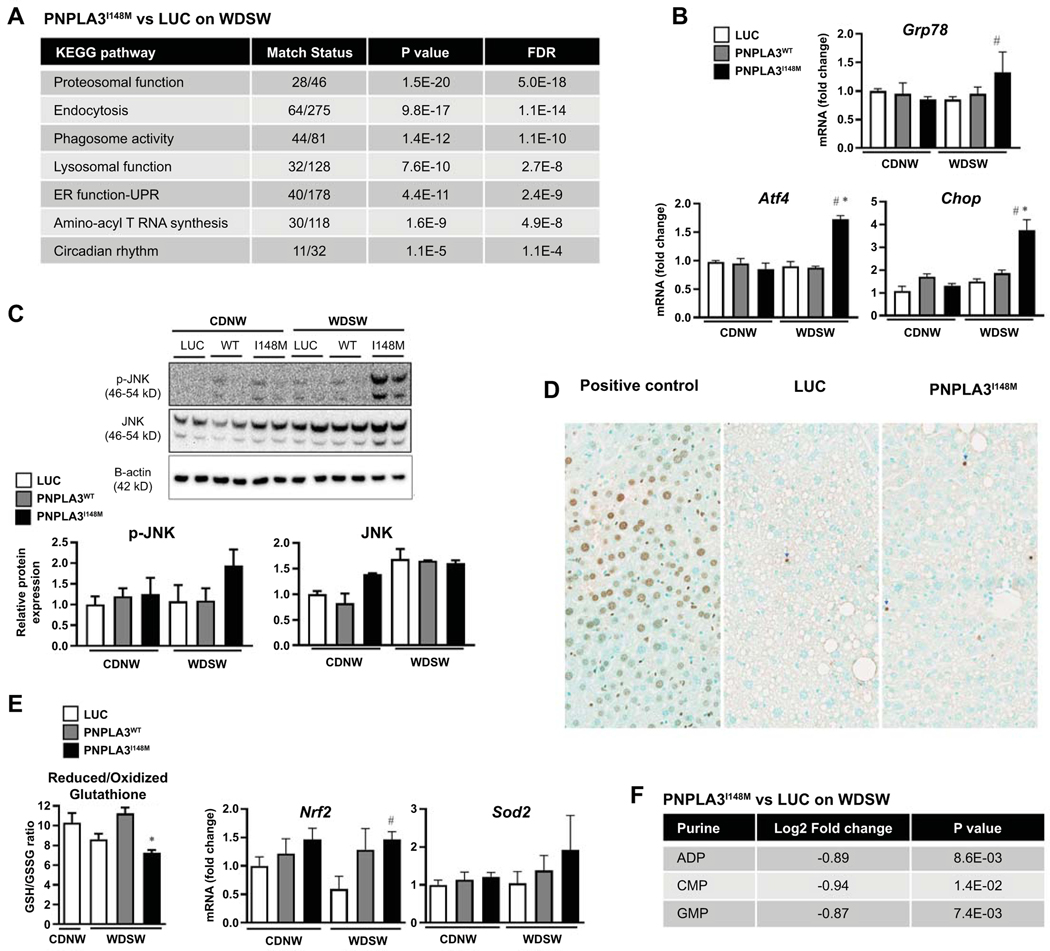
Integrated pathway analysis of PNPLA3I148M compared to AAV-luc under WDSW revealed a significant impact (False discovery rate < 0.05) on several cellular processes including proteosomal, phagosomal, and lysosomal function, amino-acyl T RNA synthesis, circadian rhythm and endoplasmic reticulum (ER) function (Figure 5A). PNPLA3I148M expression was also associated with several markers of the unfolded protein response including GRP78, ATF 4, CHOP and JNK (Figure 5B-C). This was associated with modestly increased apoptosis (Figure 5D) and activation of cell death pathways (supplemental Excel sheets).
Figure 5: PNPLA3I148M promotes ER stress, oxidative stress and cell death.
(A) KEGG pathway analysis showed significant alteration of key cellular processes involved in ER stress and cell turnover in PNPLA3I148M mice versus Luc on WDSW. (B) Real time PCR confirmed significant upregulation of hepatic genes involved in ER stress, including GRP78, ATF4, and CHOP in mice overexpressing PNPLA3I148M compared to Luc and PNPLA3WT. (C) Despite similar levels of PNPLA3 protein expression, upregulation of ER stress as demonstrated by increase in phosphorylated JNK was specific to PNPLA3I148M, indicating what this was not simply a function of excess PNPLA3 protein synthesis. (D) This was accompanied by increased cell death shown by TUNEL assay in PNPLA3I148M mice. (E) PNPLA3I148M mice on WDSW showed increase in oxidative stress as evidenced by decrease in the reduced/oxidized gluthathione ratio and increased expression of superoxide dismutase and nrf-2 genes. (F) There was also a depletion of multiple purines including ADP, CMP, and GMP in PNPLA3I148M mice compared to luc on WDSW. Data from 4–6 mice in each group are shown.
In addition, PNPLA3I148M expression resulted in increased oxidative stress as evidenced by accumulation of oxidized glutathione (Figure 5E; left panel), increased oxidized eicosanoids, superoxide dismutase and nrf2 expression and glutathione pathway activation in PNPLA3I148M versus AAV-luc on WDSW (Figure 5E and Figure 3A). Furthermore, there was a depletion of multiple purines including ADP, CMP, and GMP in PNPLA3I148M mice compared to luc on WDSW (Figure 5F).
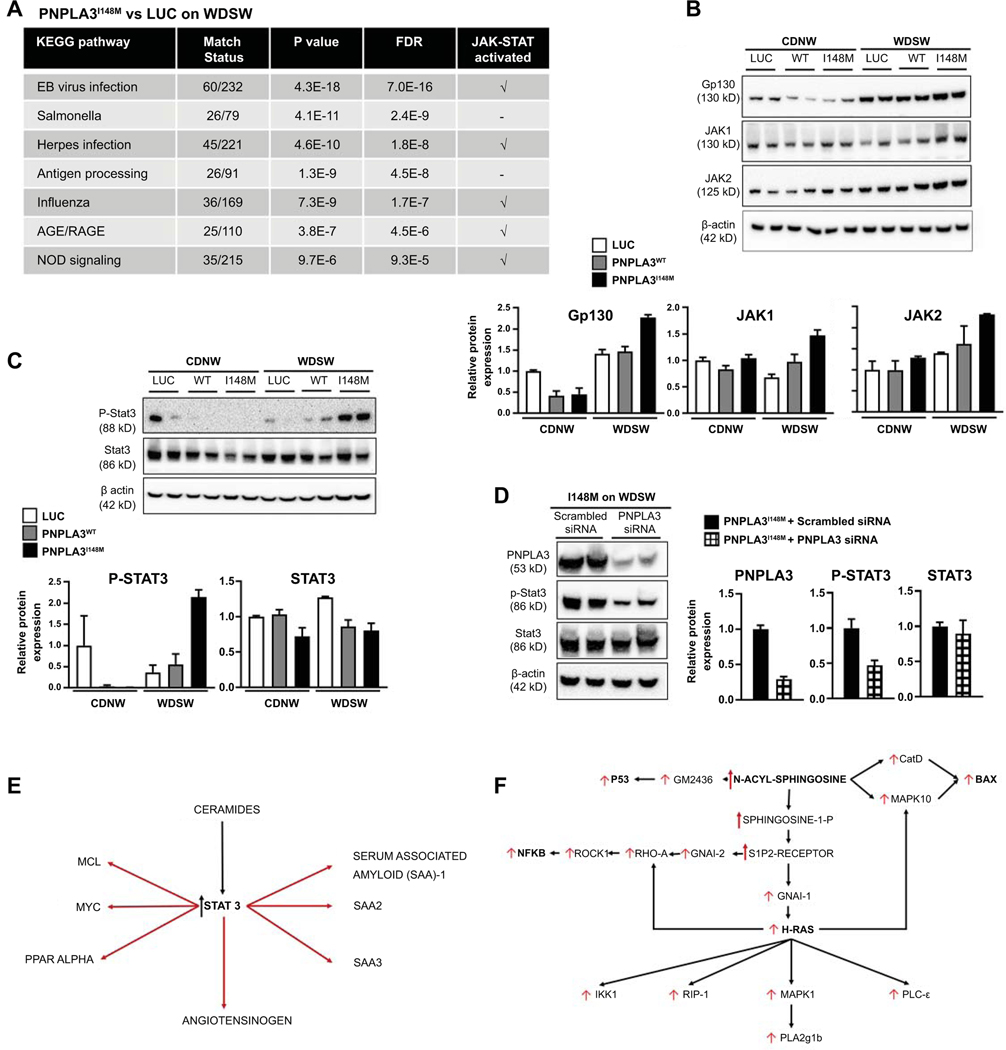
C: Sphingolipids and STAT3 activation in the inflammatory response to PNPLA3
Multiple inflammatory pathways including both the innate and adaptive immune systems were activated in the liver of mice with PNPLA3I148M versus AAV-luc on WDSW (Figure 6A). Of note, the top innate immune pathways activated all demonstrated transcriptomic activation of the JAK-STAT system. JAK and STAT3 activation were further confirmed by Western blot and its relationship to PNPLA3I148M confirmed by its deactivation upon silencing the gene (Figure 6B-D). This was accompanied by improved histological activity and fibrosis as well as reversal of steatohepatitis in mice with siRNA-mediated silencing of PNPLA3I148M (Figure 2A, B, E). Pathway analysis also revealed numerous targets of STAT3, well known to be activated by increased ceramides (26), that were also activated and linked to the transcriptomic inflammatory response to PNPLA3I148M (Figure 6E). Similarly, integrated pathway analysis revealed a strong signature of ceramide related inflammatory activation including h-ras and downstream PLA2 activation (Figure 6F). These changes were only noted under WDSW and not in chow diet conditions. Together, these provide a key role for sphingolipids in the inflammatory response to PNPLA3I148M under WDSW conditions.
Figure 6: PNPLA3I148M on high fat diet differentially upregulates innate immune system and inflammatory pathways.
(A) KEGG pathway analysis of combined metabolite-gene expression pathways showed significant upregulation of immune-inflammatory pathways, with several pathways related to the innate immune system and adaptive immune system significantly activated in PNPLA3I148M (vs Luc) on WDSW diet. Most of these pathways included JAK-STAT activation as a component. (B and C): Activation of Gp130, JAK and STAT3 signaling was confirmed by Western blot analysis which showed increased protein levels in PNPLA3I148M mice on WDSW compared to the other groups. (D) The specificity of these findings for PNPLA3I148M was further established by STAT3 inactivation following silencing of PNPLA3I148M. (E) Network analysis was performed to identify key metabolites linked to activation status of inflammatory pathways specifically activated by PNPLA3I148M overexpression under WDSW conditions (vs Luc and PNPLA3WT). A strong signal for STAT3 targets coordinately activated with STAT3 (at a transcriptomic level) and specifically linked to accelerated steatohepatitis was noted. (F) Similarly, multiple injury-inflammatory targets were activated coordinately with ceramides (specifically those immediately upstream of n-acyl sphingosine) that were linked to the accelerated histological phenotype of NASH in PNPLA3I148M. Data from 4–6 mice in each group are shown.
Unbiased transcriptomic analyses further revealed a strong signature for activation of both innate and adaptive immune-inflammatory pathways (Supplemental Figure 4A) including activation of TNF-α, IL-6, RIG-1, STAT1, serum-associated amyloid and ICAM1 (Supplemental Figure 4B and C). Based on enrichment analysis, several of the top 10 gene ontology processes and process networks were involved in immune-inflammatory response (Supplemental Figure 5A and B). Using the “analyze-networks” algorithm, networks of genes were identified that were coordinately altered by exposure to WDSW in PNPLA3I148M mice but not in Luc. A network comprised of 50 genes with 23 altered genes (p= 5.46 × 10−54) centered around complement C1q, cluster of differentiation 14 (CD-14) and STAT-1 was identified as the topmost differentially altered gene network in the PNPLA3I148M group compared to Luc on WDSW diet (Supplemental Figure 5C).
Network analysis further identified a network related to MHC class II genes as a major network to be activated by WDSW diet in PNPLA3I148M mice. These included 17 differentially altered genes (p=1.85 × 10−39) including cyclin-dependent kinase inhibitor p21, human leukocyte antigen (HLA)-DQA1, rho associated coiled-coil containing protein kinase 2 (ROCK2), paired box protein (PAX)-8, and myosin regulatory light chain (MLC)-2 (Supplemental Figure 5D).
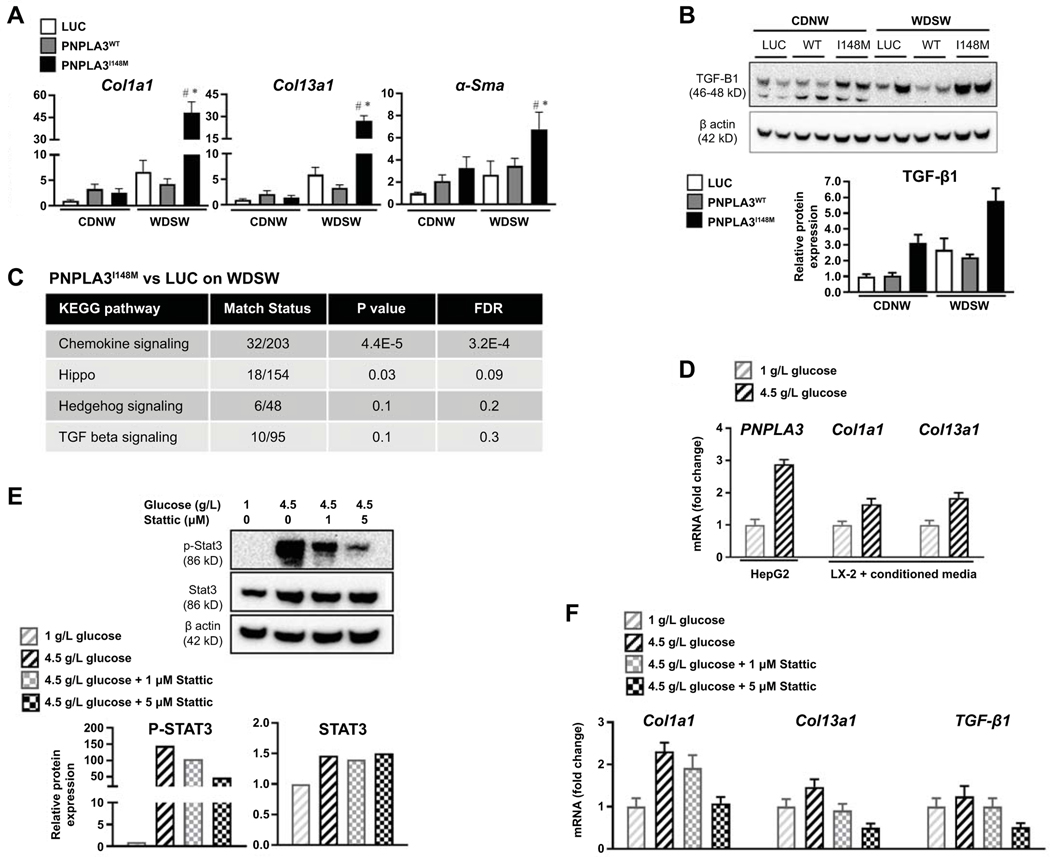
D: PNPLA3I148M related hepatocellular injury can activate stellate cells
Procollagen I, III and α-smooth muscle actin mRNA were all activated in PNPLA3I148M compared to AAV-luc and PNPLA3WT under WDSW conditions (Figure 7A). This was accompanied by increased TGF-β1 protein levels (Figure 7B) and activation of several known fibrogenic pathways (Figure 7C). To further determine cross-talk between hepatocytes overexpressing PNPLA3I148M and stellate cells, PNPLA3 gene expression was induced by glucose exposure in HepG2 cells that are known to express PNPLA3I148M (21) (Figure 7D; left panel). Conditioned media from these cells significantly increased procollagen I and III mRNA expression by LX2 cells (Figure 7D; middle and right panel). This experiment was repeated after inhibition of STAT3 in the HepG2 cells, resulting in a STAT3 inhibition dose-dependent decrease in procollagen and TGF-β1 mRNA (Figure 7E-F).
Figure 7: PNPLA3I148M upregulates fibrogenic pathways which is reversed by silencing.
(A) Realtime PCR of liver tissue showed upregulation of procollagen I, III and α-smooth muscle actin mRNA in PNPLA3I148M mice compared to luc and PNPLA3WT under WDSW conditions. (B) Western blot analysis showed upregulation of TGF-β protein levels in PNPLA3I148M mice on WDSW. (C) KEGG pathway analysis showed differential upregulation of signaling pathways related to fibrosis, with chemokine and hippo pathway signaling reaching statistical significance. (D) Cross-talk between hepatocytes overexpressing PNPLA3I148 and hepatic stellate cells was explored in vitro. HepG2 cells, known to express PNPLA3I148, were exposed to high glucose which resulted in upregulation of PNPLA3I148 (left panel). Conditioned media from HepG2 cells treated with high or low glucose resulted in dose-dependent upregulation of procollagen I and 3 mRNA by LX2 cells (middle and right panel). (E) Treatment of HepG2 cells with a selective Stat3 inhibitor, Stattic, resulted in a dose-dependent decrease in Stat3 activation despite exposure to high glucose. (F) Selective Stat3 inhibition resulted in a corresponding downregulation of procollagen 1 and 3 and TGF-β1 mRNA in HepG2 cells.
DISCUSSION
The current study provides proof of concept that development of steatohepatitis with fibrosis is accelerated and its severity worsened by overexpression of PNPLA3I148M in a murine model of NASH. This allowed us to obtain novel insights on interactions between gene expression and metabolites to dissect the roles of diet and PNPLA3 status in driving the observed acceleration of NASH with fibrosis.
Prior attempts to overexpress PNPLA3 in mice or introduce the I148M mutation in to the mouse gene have not succeeded in establishing a phenotype of steatohepatitis (10, 12). The model used in this study is distinct from prior studies by having the capacity to sequentially develop fatty liver followed by typical steatohepatitis and progressive fibrosis (14). Further, there is significant transcriptomic and cell signaling concordance with human NASH (15). These factors along with the overexpression of the entire human gene distinguish the approach taken in this study and are likely to have contributed to the development of an accelerated disease phenotype.
The principal observation in this study is that, after accounting for the effects of a WDSW and general properties of PNPLA3WT, metabolic reprogramming stood out as the hallmark feature linked to PNPLA3I148M-induced NASH acceleration (Figure 3A). This is in line with the known lipid trafficking and remodeling functions of PNPLA3 (7, 24, 27).
It has been previously reported that PNPLA3I148M increased transfer of labeled oleic acid to TAGs (24) and enhanced unsaturated fatty acid content of circulating triglycerides (28). The current study demonstrates widespread depletion of both n3 and n6 PUFAs under both chow diet and WDSW diet by both forms of PNPLA3 but with greater depletion by PNPLA3I148M under WDSW conditions which by itself also led to depletion of PUFAs (Figure 3). This was accompanied by increasing unsaturation of TAGs and DAGs indicating transfer of unsaturated fatty acids to other lipids as a core function of PNPLA3. Based on known biology (29), PUFA depletion should promote NASH and provides a potential explanation for PNPLA3 gene status as a modulator of response to n3 PUFA treatment (30).
Another novel observation is the impact of PNPLA3I148M on sphingolipid metabolism and its downstream effect on inflammation and cell death. In a prior study in humans, patients were dichotomously classified to have insulin-resistance or PNPLA3-associated NASH (31). In that study, ceramide upregulation was characteristic of insulin resistance-associated NASH but not of PNPLA3-associated NASH. This binary assessment does not permit evaluation of the role of PNPLA3I148M in the context of background obesity, the situation seen in many patients with NASH (32). In the current study, we demonstrate substantial differences in ceramides and sphingomyelins based on both diet and PNPLA3 status. Specifically, this was linked to activation of the ceramide synthesis and salvage pathways which were reversed by silencing PNPLA3I148M. These changes in ceramides were linked by pathway analysis to activation of STAT3 and several differentially activated inflammatory pathways that were directly attributable to PNPLA3I148M under WDSW conditions i.e. seen in both PNPLA3I148M vs Luc and also vs PNPLA3WT. Furthermore, STAT3 inhibition in HepG2 cells overexpressing PNPLA3 modulated stellate cell collagen gene expression establishing this as a relevant pathway connecting hepatocyte injury to increased fibrogenic drive due to PNPLA3I148M.
Yet another important observation is the broad transcriptomic foot-print indicative of activated innate and adaptive immune system, in line with recent observations in humans although altered IL32 was not noted (33). Some of the novel findings in this regard is the evidence for activation of multiple STAT3 targets and also activation of the complement system, MHC class II signaling and other elements of adaptive immune system. The reversal of STAT3 activation by silencing PNPLA3I148M even in the background of a WDSW diet further indicates the relevance of this pathway in PNPLA3I148M-NASH.
It has been reported that PNPLA3 is expressed to a greater extent in hepatic stellate cells than in hepatocytes and activation of the gene in stellate cells promotes collagen synthesis (34, 35). The current study demonstrates that, independent of stellate cell overexpression, PNPLA3I148M overexpression in hepatocytes can accelerate fibrosis in the context of NASH. This was further confirmed by the ability of conditioned media from hepatocytes overexpressing PNPLA3I148M to activate stellate cells. The two mechanisms are not mutually exclusive and it is likely that in humans with NASH, PNPLA3I148M contributes to fibrosis via both hepatocyte and stellate cell mechanisms.
As with most studies, this study too has limitations. The overexpression of PNPLA3 was not physiological. There are also differences in murine versus human lipid-metabolism especially related to lipoproteins, cholesterol and bile acids. These may explain increased circulating triglycerides seen with PNPLA3I148M in the current model whereas decreased triglycerides have been reported in humans (36). Also, the potential effects of altered murine PNPLA3 expression cannot be distinguished in these studies although hepatic PNPLA3 expression is not very high in mice. Despite these differences, the current study provides several novel and specific insights on the mechanisms underlying PNPLA3I148M-associated acceleration of NASH and fibrosis.
In summary, the current study provides proof of concept that NASH with fibrosis can be accelerated by PNPLA3I148M but only under conditions of WDSW diet. The findings establish that this gene-environmental interaction leads to metabolic reprogramming, particularly affecting PUFA and sphingolipid metabolic pathways with downstream activation of inflammatory and cell injury pathways especially via STAT3 activation. They further demonstrate the ability of hepatocyte overexpression of PNPLA3I148M to activate stellate cells. These provide insights in to the biology underlying PNPLA3I148M-mediated NASH acceleration and direction for future studies.
Supplementary Material
Acknowledgments
Financial support:
1. A grant from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA
2. RO1 DK 10596 to Dr. Arun Sanyal
3. T32 DK 07150 to Dr. Arun Sanyal
Abbreviations:
- NAFLD
Nonalcoholic fatty liver disease
- NAFL
Nonalcoholic fatty liver
- NASH
Nonalcoholic steatohepatitis
- PNPLA3
Patatin-like phospholipase domain containing 3
- DIAMOND
Diet-induced animal model of NAFLD
- AAV
Adeno-associated virus
- Luc
Luciferase
- WT
Wildtype
- CDNW
Chow diet/ normal water
- WDSW
Western diet/ sugar water
- PUFA
Polyunsaturated fatty acids
- EGTA
Ethylene glycol tetra-acetic acid
- DMEM
Dulbecco’s modified Eagle’s medium
- NAS
Nonalcoholic fatty liver disease activity score
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- DAG
Diacylglycerols
- TAG
Triacylglycerols
- Sptlc
Serine palmitoyl CoA transferase
- Kdsr
3-ketodihydrosphingosine reductase
- Smpd2
Sphingomyelin phosphodiesterase 2
- Cers
Ceramide synthetase
- IL-6
Interleukin-6
- TNF-α
Tumor necrosis factor alpha
- IRF-1
Interferon regulatory factor 1
- RIG-1
Retinoic acid-inducible gene I
- GRO-2
Growth related oncogene 2
- ICAM-1
Intercellular adhesion molecule 1
- STAT
Signature of transducer and activator of transcription
- SAA1
Serum amyloid A1
- HLA
Human leukocyte antigen
- ROCK2
Rho associated coiled-coil containing protein kinase 2
- PAX
Paired box protein
- MLC
Myosin regulatory light chain
- SREBP
Sterol regulatory element-binding transcription factor
- FAS
Fatty acid synthetase
- ACC
Acyl CoA carboxylase
- ER
Endoplasmic reticulum
- Nrf2
Nuclear factor erythroid
- Sod2
Superoxide dismutase 2
- Grp-78
Glucose related protein 78
- Atf-4
Activating transcription factor-4
- Chop
CCAAT-enhancer-binding protein homologous protein
- JNK
c-Jun N-terminal kinase
- Col-α
Collagen alpha
- α-SMA
Alpha smooth muscle actin
- TGF-β
Transforming growth factor beta
Footnotes
Disclosures:
Dr. Sanyal is President of Sanyal Bio. He has stock options in Genfit, Galmed, Exhalenz, Durect, Tiziana, Algernon and Indalo. He has served as a consultant to Intercept, Gilead, Bristol Myers Squibb, Novartis, Pfizer, Lilly, Novo Nordisk, Astra Zeneca,Medimmune, Merck, Allergan, Albireo, Boehringer Ingelhiem, Celgene, NGM, Glympse, Conatus, Genentech, Tern, Takeda, Hemoshear, Immuron, Surrozen, Poxel, Path AI, Second Genome, Zydus, Chiasma, Surrozen, Poxel, Blade, Pliant, Liposcience, Cymabay, Salix, Ferring and Teva. His institution has received grants from Intercept, Gilead, Novartis, Merck, Astra Zeneca, Malinckrodt, Pfizer, Lilly, Salix and Bristol Myers Squibb. VCU has ownership interests in Sanyal Bio. Dr. Alonso is an employee of OWL Metabolomics. All the remaining authors have no disclosures or competing interests.
REFERENCES
- 1.Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018;67:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018;24:908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleiner DE, Brunt EM, Wilson LA, Behling C, Guy C, Contos M, Cummings O, et al. Association of Histologic Disease Activity With Progression of Nonalcoholic Fatty Liver Disease. JAMA Netw Open 2019;2:e1912565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valenti L, Al-Serri A, Daly AK, Galmozzi E, Rametta R, Dongiovanni P, Nobili V, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 2010;51:1209–1217. [DOI] [PubMed] [Google Scholar]
- 6.Anstee QM, Day CP. The Genetics of Nonalcoholic Fatty Liver Disease: Spotlight on PNPLA3 and TM6SF2. Semin Liver Dis 2015;35:270–290. [DOI] [PubMed] [Google Scholar]
- 7.He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 2010;285:6706–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dongiovanni P, Donati B, Fares R, Lombardi R, Mancina RM, Romeo S, Valenti L. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol 2013;19:6969–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speliotes EK, Butler JL, Palmer CD, Voight BF, Consortium G, Consortium MI, Nash CRN, et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology 2010;52:904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smagris E, BasuRay S, Li J, Huang Y, Lai KM, Gromada J, Cohen JC, et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2015;61:108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.BasuRay S, Smagris E, Cohen JC, Hobbs HH. The PNPLA3 variant associated with fatty liver disease (I148M) accumulates on lipid droplets by evading ubiquitylation. Hepatology 2017;66:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linden D, Ahnmark A, Pingitore P, Ciociola E, Ahlstedt I, Andreasson AC, Sasidharan K, et al. Pnpla3 silencing with antisense oligonucleotides ameliorates nonalcoholic steatohepatitis and fibrosis in Pnpla3 I148M knock-in mice. Mol Metab 2019;22:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basantani MK, Sitnick MT, Cai L, Brenner DS, Gardner NP, Li JZ, Schoiswohl G, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res 2011;52:318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asgharpour A, Cazanave SC, Pacana T, Seneshaw M, Vincent R, Banini BA, Kumar DP, et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J Hepatol 2016;65:579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cazanave S, Podtelezhnikov A, Jensen K, Seneshaw M, Kumar DP, Min HK, Santhekadur PK, et al. The Transcriptomic Signature Of Disease Development And Progression Of Nonalcoholic Fatty Liver Disease. Sci Rep 2017;7:17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacana T, Cazanave S, Verdianelli A, Patel V, Min HK, Mirshahi F, Quinlivan E, et al. Dysregulated Hepatic Methionine Metabolism Drives Homocysteine Elevation in Diet-Induced Nonalcoholic Fatty Liver Disease. PLoS One 2015;10:e0136822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanyal AJ, Pacana T. A Lipidomic Readout of Disease Progression in A Diet-Induced Mouse Model of Nonalcoholic Fatty Liver Disease. Trans Am Clin Climatol Assoc 2015;126:271–288. [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Z, Yan H, Ou H. Human thyroxine binding globulin (TBG) promoter directs efficient and sustaining transgene expression in liver-specific pattern. Gene 2012;506:289–294. [DOI] [PubMed] [Google Scholar]
- 19.Cazanave SC, Wang X, Zhou H, Rahmani M, Grant S, Durrant DE, Klaassen CD, et al. Degradation of Keap1 activates BH3-only proteins Bim and PUMA during hepatocyte lipoapoptosis. Cell Death Differ 2014;21:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayo R, Crespo J, Martinez-Arranz I, Banales JM, Arias M, Minchole I, Aller de la Fuente R, et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: Results from discovery and validation cohorts. Hepatol Commun 2018;2:807–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min HK, Sookoian S, Pirola CJ, Cheng J, Mirshahi F, Sanyal AJ. Metabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in Huh-7 hepatoma cells. Am J Physiol Gastrointest Liver Physiol 2014;307:G66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr Protoc Bioinformatics 2019;68:e86. [DOI] [PubMed] [Google Scholar]
- 23.Kumar DP, Caffrey R, Marioneaux J, Santhekadur PK, Bhat M, Alonso C, Koduru SV, et al. The PPAR alpha/gamma Agonist Saroglitazar Improves Insulin Resistance and Steatohepatitis in a Diet Induced Animal Model of Nonalcoholic Fatty Liver Disease. Sci Rep 2020;10:9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruhanen H, Perttila J, Holtta-Vuori M, Zhou Y, Yki-Jarvinen H, Ikonen E, Kakela R, et al. PNPLA3 mediates hepatocyte triacylglycerol remodeling. J Lipid Res 2014;55:739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Kory N, BasuRay S, Cohen JC, Hobbs HH. PNPLA3, CGI-58, and Inhibition of Hepatic Triglyceride Hydrolysis in Mice. Hepatology 2019;69:2427–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doshi UA, Shaw J, Fox TE, Claxton DF, Loughran TP, Kester M. STAT3 mediates C6-ceramide-induced cell death in chronic lymphocytic leukemia. Signal Transduct Target Ther 2017;2:17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.BasuRay S, Wang Y, Smagris E, Cohen JC, Hobbs HH. Accumulation of PNPLA3 on lipid droplets is the basis of associated hepatic steatosis. Proc Natl Acad Sci U S A 2019;116:9521–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luukkonen PK, Nick A, Holtta-Vuori M, Thiele C, Isokuortti E, Lallukka-Bruck S, Zhou Y, et al. Human PNPLA3-I148M variant increases hepatic retention of polyunsaturated fatty acids. JCI Insight 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiktorowska-Owczarek A, Berezinska M, Nowak JZ. PUFAs: Structures, Metabolism and Functions. Adv Clin Exp Med 2015;24:931–941. [DOI] [PubMed] [Google Scholar]
- 30.Scorletti E, West AL, Bhatia L, Hoile SP, McCormick KG, Burdge GC, Lillycrop KA, et al. Treating liver fat and serum triglyceride levels in NAFLD, effects of PNPLA3 and TM6SF2 genotypes: Results from the WELCOME trial. J Hepatol 2015;63:1476–1483. [DOI] [PubMed] [Google Scholar]
- 31.Luukkonen PK, Zhou Y, Sadevirta S, Leivonen M, Arola J, Oresic M, Hyotylainen T, et al. Hepatic ceramides dissociate steatosis and insulin resistance in patients with non-alcoholic fatty liver disease. J Hepatol 2016;64:1167–1175. [DOI] [PubMed] [Google Scholar]
- 32.Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ, Nash CRN. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology 2010;52:894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baselli GA, Dongiovanni P, Rametta R, Meroni M, Pelusi S, Maggioni M, Badiali S, et al. Liver transcriptomics highlights interleukin-32 as novel NAFLD-related cytokine and candidate biomarker. Gut 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruschi FV, Claudel T, Tardelli M, Caligiuri A, Stulnig TM, Marra F, Trauner M. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepatic stellate cells. Hepatology 2017. [DOI] [PubMed] [Google Scholar]
- 35.Pirazzi C, Valenti L, Motta BM, Pingitore P, Hedfalk K, Mancina RM, Burza MA, et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet 2014;23:4077–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pirazzi C, Adiels M, Burza MA, Mancina RM, Levin M, Stahlman M, Taskinen MR, et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol 2012;57:1276–1282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.