Abstract
Oral squamous cell carcinoma (OSCC) is one of the leading cancers with poor disease survival rate. Herein, we explored molecular basis, in silico identification and in vitro verification of genes associated with OSCC. Five gene expression series including, GSE30784, GSE13601, GSE9844, GSE23558 and GSE37991 were screened for differentially expressed genes (DEGs). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were enriched by cluster Profiler. Further, protein–protein interaction network was analysed and hub genes were verified. A total of 6476 (up-regulated: 2848; down-regulated: 3628) DEGs were identified among OSCC patients and healthy controls. Gene Ontology analysis indicated DEGs enrichment in cellular motility, invasion and adhesion processes. KEGG analysis revealed enrichment of PI3K-Akt signalling, focal adhesion and regulation of actin cytoskeleton pathways. Subsequently, nine DEGs including APP, EHMT1, ACACB, PCNA, PLAU, FST, HMGA2, LAMC2 and SPP1 were correlated with TCGA expression data along with significant association towards patient’s survival, recognized as hub genes. This dysregulated mRNA signature of genes was validated in two OSCC cell lines with an anti-cancer agent, fisetin. Fisetin inhibited the expression of APP, EHMT1, PCNA, PLAU, FST, HMGA2, LAMC2, SPP1 and upregulated the expression of ACACB gene which were associated with growth inhibition of both the OSCC cell lines. The regulatory effect of fisetin supported crucial role of nine hub genes identified in OSCC. This study signified that hub genes and pathways might influence the aggressiveness of OSCC. Thus, the proposed hub genes could be potential diagnostic biomarker and drug targets for OSCC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02737-4.
Keywords: Gene expression profiling, Oral squamous cell carcinoma, Biomarkers, Hub genes, Survival analysis, Cytoscape
Introduction
Oral squamous cell carcinoma (OSCC), one of the most perverse type of head and neck cancer, accounts for about 354,864 new cases and 177,384 deaths worldwide every year. It is also a very frequent cancer in Southern Asia and Pacific Islands, and is the primary reason of cancer-related deaths in India and Sri Lanka amongst men (Bray et al. 2018). Customary behaviours like smoking, alcohol consumption, tobacco and betel nut chewing along with human papillomavirus infections are the key etiological factors implicated in the rise of OSCC (Nagao and Warnakulasuriya 2020). Despite the significant advancement of current therapeutics strategies, the overall disease survival rate still remains less than 65% (Siegel et al. 2019). Unfortunately, the management of patients with OSCC has not changed dramatically either, mostly due to lack of awareness for prevention and early diagnostic failures. Furthermore, the relapsing and metastatic nature of the disease accounts for poor prognosis and remains major hurdle for disease improvement. Therefore, it is extremely crucial to identify the promising early diagnostic and prognostic biomarkers that may assist to elucidate the underlying molecular mechanism of OSCC and simultaneously improve the clinical therapeutics.
In this context, microarray and high-throughput sequencing platforms emerged as promising tools to measure the differential expression level of genes between disease and healthy samples. Expression profiles from these platforms have been submitted to several database repositories, Gene Expression Omnibus (GEO) for example, is widely used database for screening cancer biomarkers and drug target discovery (Petryszak et al. 2014). Several, microarray-based gene expression profiles have been successfully performed to explore the differential expressed genes (DEGs) connected to OSCC. However, clinical translation of these findings to predict reliable biomarkers is perplexed due to variation among studies such as sample size, sample processing, tumor stage, ethnicity and types of platform and the analytical algorithms. Ding et al. (2018) studied two microarray datasets to identify pathogenic genes associated with OSCC. Zhong et al. (2019) explored several expression profiles and identified five extracellular matrix-associated genes as therapeutic target in Oral and tongue squamous cell carcinoma. Wang et al. (2020b) reported six genes with prognostic significance in OSCC. Though, such trend of transcriptome profile re-analysis provides new insight into disease pathogenesis but exhibits variations in terms of identified DEG’s and ethnicity as result of sample heterogeneity (Yuan et al. 2020). Thus, independent analysis of selected gene expression profiles with subsequent integration of DEGs across study type (microarray and RNAseq) may help in identifying the universal markers concordant across multiple OSCC studies with increased confidence and clinical applicability. In present study, 5 gene expression profiles of OSCC containing 291 tumor and 128 normal samples were screened for DEGs analysis. Further, functional annotations were enriched via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway databases. Subsequently, resulting DEGs were integrated with protein–protein interaction (PPI) network and clustering module analysis. Potential DEGs were then correlated with TCGA level 3 RNA seq data to identify high confidence hub genes primarily driving OSCC pathogenesis. The impact of dysregulated transcriptome signature of hub genes could be verified among pre- and post-drug treatment, to confirm the consequences of dysregulation. The adverse effects associated with current treatment regimens for OSCC, necessitate the use of an alternate approach such as natural agents for chemoprevention with better safety, efficacy, ease of availability, affordability, potential to overcome resistance to other traditional therapies and anticancer drugs (Choudhari et al. 2020). Fisetin, a bioactive small molecule, shown anti-cancer effects in various epithelial cancers (Sabarwal et al. 2017; Liang et al. 2020), therefore, was used to evaluate the expression pattern of altered vital genes in human OSCC cells. Overall, the differential expression signature of identified hub genes may provide a novel insight on early diagnosis and prognosis of OSCC by serving as promising biomarkers/druggable target.
Materials and methods
Gene expression profile datasets
Five human OSCC gene expression profiles, GSE30784, GSE13601, GSE9844, GSE23558 and GSE37991 were accessed from NCBI-Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) (Barrett et al. 2013). All five datasets were selected on basis of following criteria: Search term “OSCC”, Organism “Homo sapiens”, Experiment type “Expression profiles by array”, No. of samples “ > 20”, Samples “human oral tissue samples and healthy control samples”. These datasets were produced through diverse microarray platforms (Agilent, Affymetrix and Illumina) based on RNA samples extracted from oral cancer and normal tissues. The expression profile GSE23558, with 27 tumour (T) and five normal (N) oral tissue samples, is deposited by Advanced Centre for Treatment Research and Education in Cancer, India (Ambatipudi et al. 2012). GSE30784 (T = 167, N = 45), GSE13601 (T = 31, N = 26) and GSE9844 (T = 26, N = 12) are submitted by research groups from USA (Chen et al. 2008; Ye et al. 2008; Estilo et al. 2009), while GSE37991 (T = 40, N = 40) is contributed from Taiwan (Lee et al. 2015).
Screening and functional enrichment analysis of DEGs
The five transcriptome profiles were normalized by robust multi-array average method and probe level data were annotated to gene symbol. For a particular gene with several corresponding probes, the mean expression value of all probes was considered as the final expression value. Subsequently, differential gene expression analysis between tumour and normal groups for each transcriptome profile was accomplished through GEO2R tool, which is developed based on limma Bioconductor package. Genes with Benjamini and Hochberg false discovery rate (FDR) ≤ 0.05 and |log fold change|≥ 1 were considered as differentially expressed (Sun et al. 2017). Differentially expressed genes (DEGs) in OSCC were graphically represented using R v3.4.3. The biological processes, molecular functions, cellular components and metabolic pathways cognated with these DEGs were enriched with cluster Profiler (Aggarwal et al. 2017).
PPI network construction and high confidence node gene prediction
Genes differentially expressed in five transcriptome profiles were used for protein–protein interaction (PPI) data mining from STRING database. PPI probabilities (confidence score) among genes were computationally predicted by STRING based on gene neighbourhood, gene fusion, co-expression, co-occurrence, programmed text mining, annotations from curated databases and experimental evidences. A threshold confidence score ≥ 0.4 was set as cut off to retrieve interacting partners in the String App plugin of Cytoscape v3.6.1 (Cline et al. 2007). The reference organism in the String plugin was set to Homo sapiens. Network topology indices of the constructed interactome were computed using CytoHubba (Lin et al. 2008). The tool provides 12 parameters including degree, edge percolated component, maximum neighborhood component, density of maximum neighborhood component, maximal clique centrality, clustering coefficient, bottleneck, eccentricity, closeness, radiality, betweenness, and stress. These parameters are being used for identification of hub genes from protein–protein interaction networks. The degree distribution of all nodes in the network may help to explain number of first-degree interacting partner a gene has in the PPI network. The centrality indices are computed based on shortest path and are lying on the communication paths such as to control the information flow in the network. Degree, centrality indices and MCC alone or together are mostly used for hub gene prediction. The top 50 genes ranked via each of the 12 built-in centrality indices were listed out and genes featured in at least six centrality indices were considered as high confidence nodes in the PPI network (Yang et al. 2018b).
Identification of functional modules in OSCC
Modules in the PPI network were detected using MCODE (Bader and Hogue 2003). Gene ontology and pathway enrichment analysis of modules were carried out to gain insights on core biological processes and pathways involved in OSCC. These enrichments were achieved using STRING functional enrichments at a threshold FDR ≤ 0.05.
Verification of hub gene expression and survival analysis
Concordances of expression signature and prognostic significance of common genes from all five studies, high confidence nodes from PPI network were evaluated by mapping to TCGA level 3 RNA-seq data through UALCAN (http://ualcan.path.uab.edu) (Chandrashekar et al. 2017). Genes showing positive correlation in both expression as well as survival data were validated as hub genes for OSCC. The UALCAN tool is also used to monitor the association of hub gene expression with other clinical features (tumor stages and ethnicity).
Hub genes—disease association analysis
The hub genes observed to play potential role in the survival of OSCC patients, were annotated for their association with gene ontology terms, pathways and diseases. ClueGo, a Cytoscape plug-in with kappa statistics was used for deciphering dynamic functional enrichment networks of hub genes (Bindea et al. 2009). DisGeNET, prioritizes genotype–phenotype connections centred on curated genetic variations associated to human diseases from GWAS, animal studies and scientific literature, was used for gene–disease associations (Piñero et al. 2020).
Cell lines and materials
Human oral squamous cell carcinoma UM-SCC-22B and Cal33 cells were used in the study. Dulbecco’s Modified Eagle Medium (DMEM, Gibco, ThermoFisher Scientific, India), supplemented with penicillin and streptomycin, and fetal bovine serum (FBS, Gibco, ThermoFisher Scientific, India) to a final concentration of 10% were used for culturing of cells. Fisetin was purchased from Sigma-Aldrich (Sigma-Aldrich, India). Stock solutions of fisetin were prepared in dimethyl sulfoxide (DMSO) and stored at − 20 °C.
MTT assay
The assay depends on the reduction of MTT [3-(4,5- dimethylthiazolyl)-2,5-diphenyl-tetrazolium bromide] into blue formazan product via mitochondrial dehydrogenase enzymes, and hence reflects the viable cell numbers or cytotoxicity (Sabarwal et al. 2017). Cells were seeded at a density of 10,000 cells/well in a 96-well plate and then treated with DMSO (vehicle control) or fisetin (5–100 µM) for 48 h. At the end of the treatment time, the media was aspirated and 100 µl of freshly prepared MTT solution (5 mg/ml) was added into each well followed by 4 h incubation at 37 °C. The media was carefully removed after incubation period, and the formazan crystals were solubilized in 100 µl DMSO/well. Absorbance was measured at 570 nm in Thermo Scientific Varioskan Flash Spectral Scanning Multimode Reader.
Reverse transcriptase-PCR analysis of hub genes
Cells were cultured to 70–80% confluency and were treated with fisetin (25 and 50 µM) in 10% serum supplemented DMEM for 24 h. Total RNA was subjected to cDNA synthesis. Standard PCR reactions were performed using gene-specific primers (Supplementary Material 1). PCR products were visualized by agarose gel electrophoresis and imaged under low-intensity UV in Gel Doc system (Applied Biosystems). The relative expression of PCR result was normalized against GAPDH and analysed using ImageJ software (NIH, Bethesda, MD).
Results
Screening of DEGs in OSCC
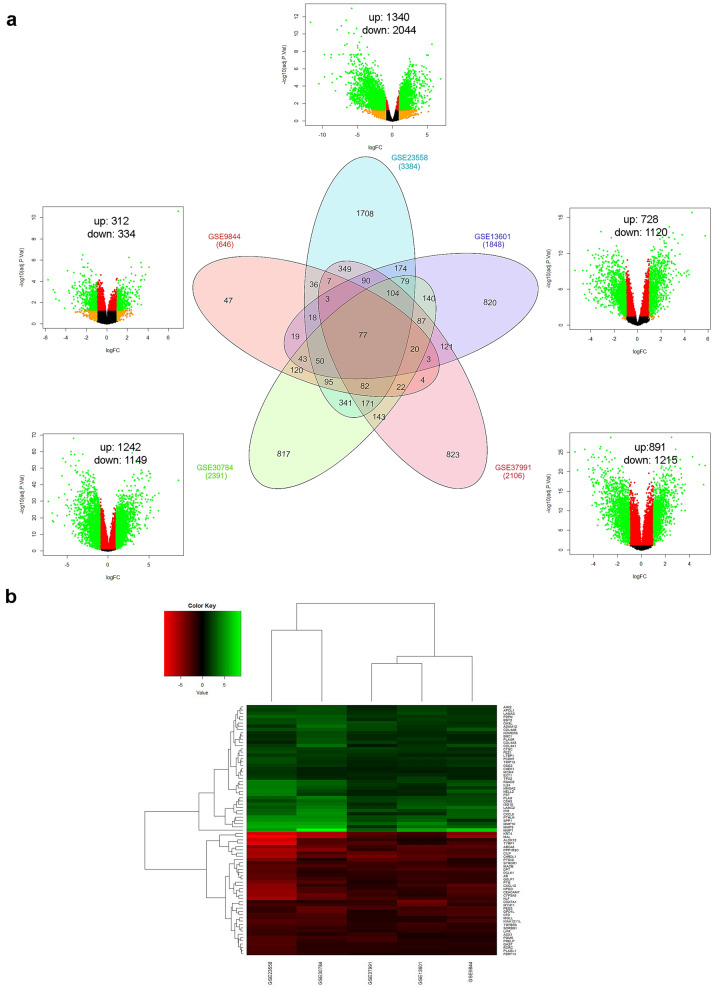
Five OSCC gene expression profiles were screened for implementing integrative analysis to recognize differentially expressed genes (DEGs) in disease condition (Fig. 1). The primary screening of expression profiles represented 291 tumor and 128 normal samples. Discrete gene expression profile based analysis via GEO2R and subsequent merging by taking union of five series exhibited 6613 dysregulated genes. Among these, 137 genes were observed to be potentially misidentified (i.e. could not be categorized into up-regulated or down-regulated groups) among GEO series and were excluded from further analysis. The final list of DEGs constituted 6476 DEGs, of which 2848 genes were up-regulated and 3628 genes were down-regulated (Fig. 2a). The top 10 up-regulated DEGs were MMP1, MMP3, CA9, MMP10, INHBA, SPP1, MMP13, PTHLH, CXCL10 and CXCL11. Conversely, the top 10 down-regulated DEGs revealed KRT76, TMPRSS11B, LOR, FDCSP, MAL, TGM3, ODAM, CRNN, FAM3B and KRT4. Integration of expression profiles also revealed that a subgroup of 77 genes was differentially expressed across all five datasets, of these; 39 were up-regulated while 38 were down-regulated (Fig. 2b). These common DEGs also perceived six genes viz. MMP1, MMP3, MMP10, SPP1, KRT4 and MAL from the list of top 20 dysregulated genes.
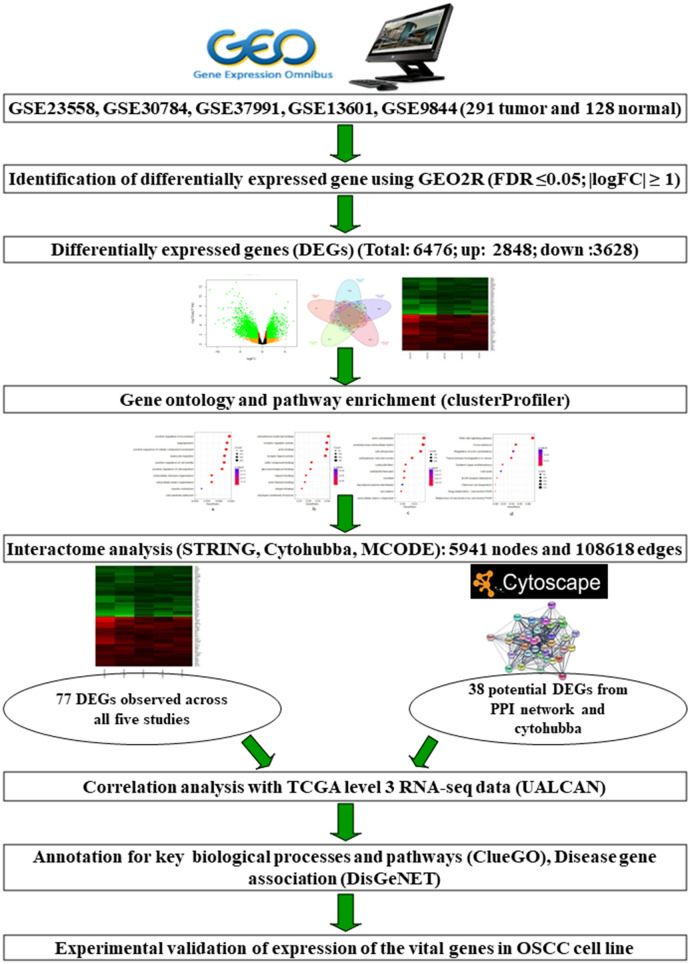
Fig. 1.
A schematic representation of systematic workflow employed in this study
Fig. 2.
Screening and identification of DEGs a Venn diagram and volcano plot exhibiting DEGs in five GEO series, x-axis in volcano plot represents log fold change while y-axis represents negative log of adj P value. Each dot symbolizes single gene, genes in green colour showed significant (|log FC|≥ 1 and adjusted P ≤ 0.05) while the genes illustrated in orange, red and black colour did not achieve significance. b Heat map of 77 common genes across all five studies (including 39 up-regulated and 38 down-regulated genes)
Functional enrichment analysis of DEGs
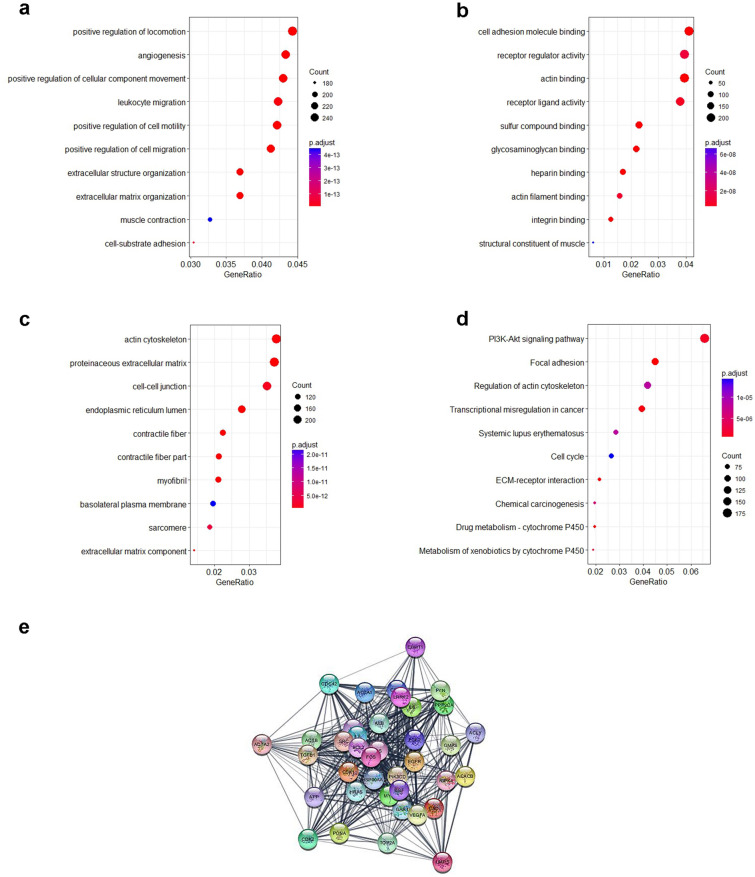
The entire sets of 6476 DEGs were enriched for their involvement in biological processes, molecular functions, cellular components and metabolic pathways. Mapping of up-regulated and down-regulated genes with various GO categories resulted in the enrichment of 1481 biological processes (BP), 154 molecular functions (MF) and 154 cellular components (CC). The observed ontology categories promote favourable ambience for developing malignant tumour in oral tissue. For instance, positive regulation of locomotion, cellular component movement, cell motility, cell migration, angiogenesis, leukocyte migration, muscle contraction, cell-substrate adhesion, extracellular matrix and structure organization are among the top enriched biological processes (Fig. 3a). Top MF included cell adhesion molecule binding, receptor regulator activity, actin binding, receptor ligand activity, sulfur compound binding, glycosaminoglycan binding, heparin binding, actin filament binding, integrin binding and structural constituent of muscle (Fig. 3b). Similarly, the top CCs involved actin cytoskeleton, proteinaceous extracellular matrix, cell–cell junction, endoplasmic reticulum lumen, contractile fibre part, myofibril, basolateral plasma membrane, sarcomere and extracellular matrix component (Fig. 3c).
Fig. 3.
Gene ontology and pathway enrichment analysis of DEGs a biological process; b molecular function; c cellular component; d metabolic pathways enriched in OSCC and e protein–protein interaction network of 38 potential hub genes
Further, complete set of DEGs were mapped to the KEGG (Kyoto Encyclopedia of Genes and Genomes) database terms. The statistical cut-off criterion FDR ≤ 0.05 acknowledged 89 over-represented pathways. Significantly high percentage of DEGs were found to be involved in PI3K-Akt signalling pathway, focal adhesion, regulation of actin cytoskeleton, transcriptional misregulation in cancer, systemic lupus erythematosus, cell cycle, ECM-receptor interaction, chemical carcinogenesis, drug metabolism—cytochrome P450 and metabolism of xenobiotics by cytochrome P450 (Fig. 3d). It signifies that the observed dysregulation in 6476 genes could potentially induce malignancy in oral tissue. Moreover, enrichment of 77 common DEGs were also detected to be over represented in pathways allied to cancer initiation and progression.
Identification of high confidence nodes from the PPI network
As the top upregulated and down-regulated genes could play important role in disease pathogenesis, but quantum of expression in disease condition is not sufficient to establish them as vital gene. Therefore, the 6476 DEGs were mapped with PPI connectivity data for construction of comprehensive network comprising of 5941 nodes and 108,618 edges. The PPI data of DEGs were reflexion of their co-occurrence, co-expression, fusion, neighbourhood and/or functional interactions. It signifies DEGs with higher centrality indices in the PPI network were likely to have significant impact on function of its interacting partners as well as their corresponding biological processes and pathways. Degree, betweeness and closeness have been the prevalent centrality indices of choice while gauging significant genes from the network. However, alongside these three parameters, other nine centrality indices being calculated by cytohubba are also shown to be imperative in prediction of high confidence nodes in the PPI network. In view of this, we have computed 12 centrality indices for the PPI network. The concatenated list of fifty top ranked genes computed through each centrality indices was screened. It was observed that 38 genes featured in the list were predicted as potential DEGs by at least 6 indices. Therefore, these 38 genes were selected as high confidence nodes in the PPI network of 5941 OSCC dysregulated transcripts (Fig. 3e).
The entire PPI network was also analysed to detect major functional modules enriched in OSCC. Top three modules were selected for subsequent analysis. Module 1 comprised of 241 genes with NBPWR1 as seed gene. Module 2 represented a network of 72 genes with AGTR1 as seed while that of module 3 was described with 183 genes with PAICS as seed (Suppl. Material 2). Cell cycle, gap junction, and platelet activation genes were enriched in all top three modules.
Verification of hub gene expression and survival analysis
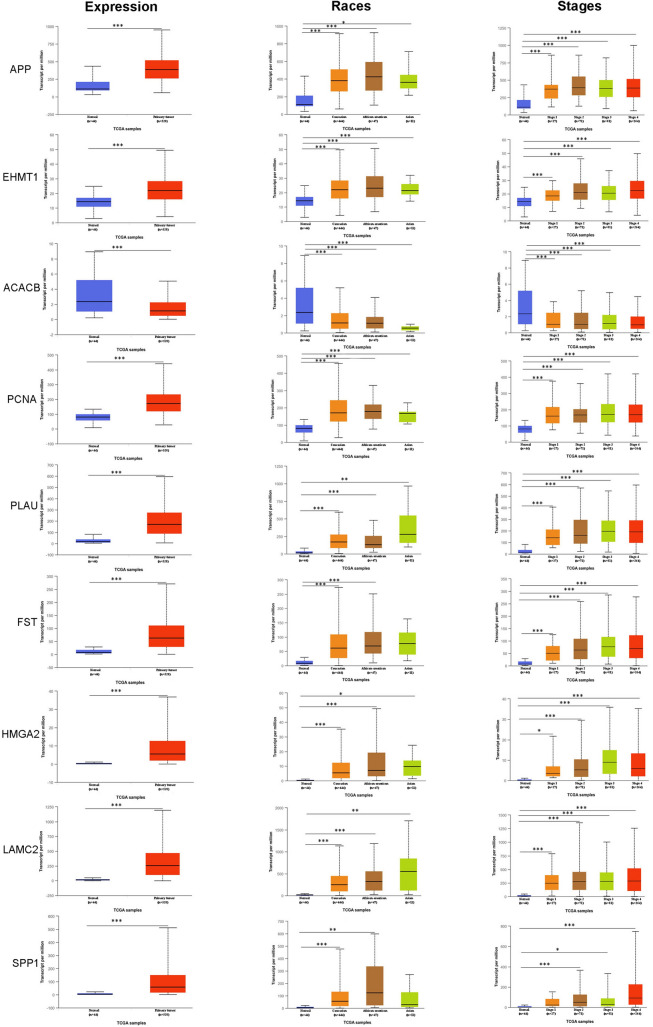
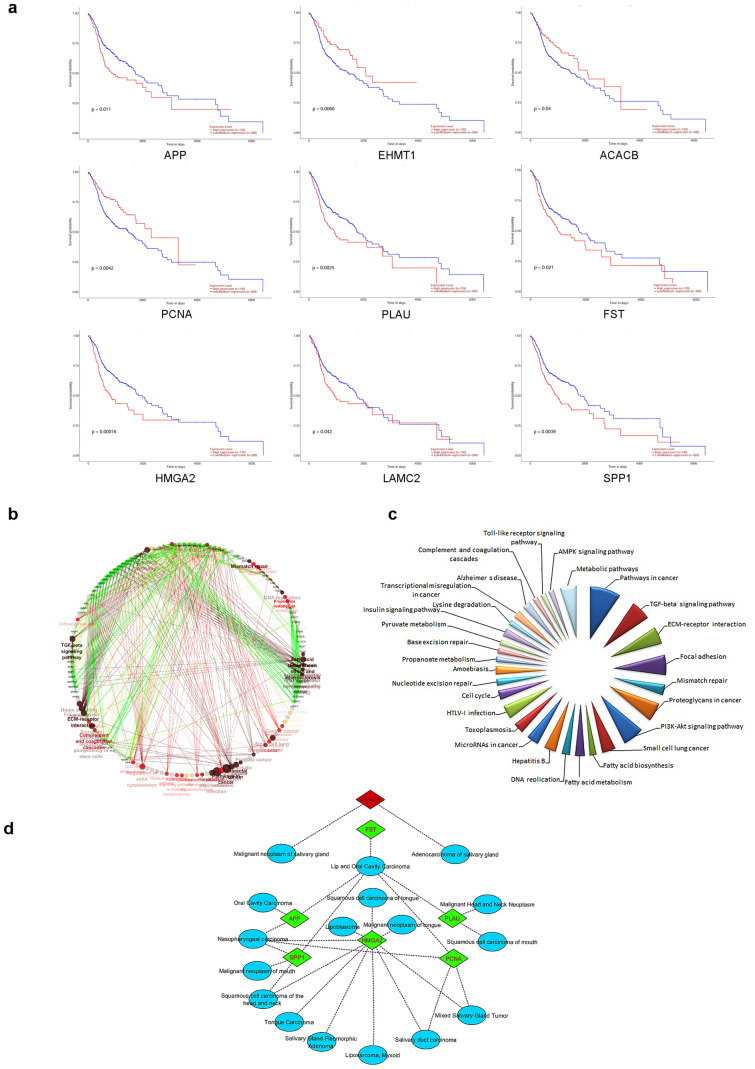
Expression signature as well as prognostic significance of 115 DEGs (77 genes differentially expressed among all five studies and 38 high confidence nodes detected from PPI network) was verified with TCGA level 3 RNA-seq data. Genes correlating with both TCGA expression and patients’ survival data were considered as hub genes and substantiated for other clinical features. Expressions of 65 genes (~ 56%) were in line with our observations made through five selected OSCC microarray gene expression profiles. It underscores, the 115 DEGs are most likely to play major role in OSCC. Subsequent, TCGA data analysis revealed that nine genes, namely, APP, EHMT1, ACACB, PCNA, PLAU, FST, HMGA2, LAMC2, and SPP1 correlated favourably with TCGA expression data (Fig. 4) and also significantly associated with worse overall survival for OSCC patients (Fig. 5a); hence, considered as hub genes. From the identified hub genes, PLAU, FST, HMGA2, LAMC2, SPP1 were emanated as differentially expressed in all five gene expression profiles. These five genes would be useful for common biomarker/target discovery. Further analysis of other clinical features showed that the differential expression signature of nine hub genes was positively connected with all tumor stages; however, no significant difference was observed between individual stages, indicating hub genes dysregulation is an early incident in OSCC development. Similarly, analysis of identified hub genes among different races/ethnicity confirmed that dysregulated signature of all nine hub genes could be used as universal biomarkers among all races/ethnic groups (Fig. 4). All these results together, validate the role of hub genes in OSCC pathogenesis.
Fig. 4.
Boxplot representing relative expression of nine hub genes in OSCC patients and healthy tissue samples, stratified based on races and tumor stages. *P < 0.05; **P < 0.01; ***P < 0.001
Fig. 5.
a Survival analysis of nine hub genes showing positive correlation with TCGA RNA-seq data. The red lines in survival plot indicate patients with high gene expression while blue lines indicate patients with a low/medium gene expression; b biological processes network of nine hub genes and their interacting partners; c metabolic pathways enriched for extended interactome by ClueGO and d annotation of nine hub genes for disease gene association network by DisGeNET
Disease association and functional analysis of hub genes
The nine hub genes linked with worst survival were subjected to functional enrichment and disease gene association analysis. ClueGo enrichment analysis revealed that hub genes significantly associated with several functions and pathways vital for cancer initiation and progression (Fig. 5b, c). Subsequently, seven genes viz. HMGA2, APP, PLAU, PCNA, SPP1, FST and ACACB were reported to be associated with OSCC along with other subtypes of HNSCC (Fig. 5d); however, their functional relevance in other subtypes need further exploration.
Fisetin inhibits cell viability of OSCC Cal33 and UM-SCC-22B cells
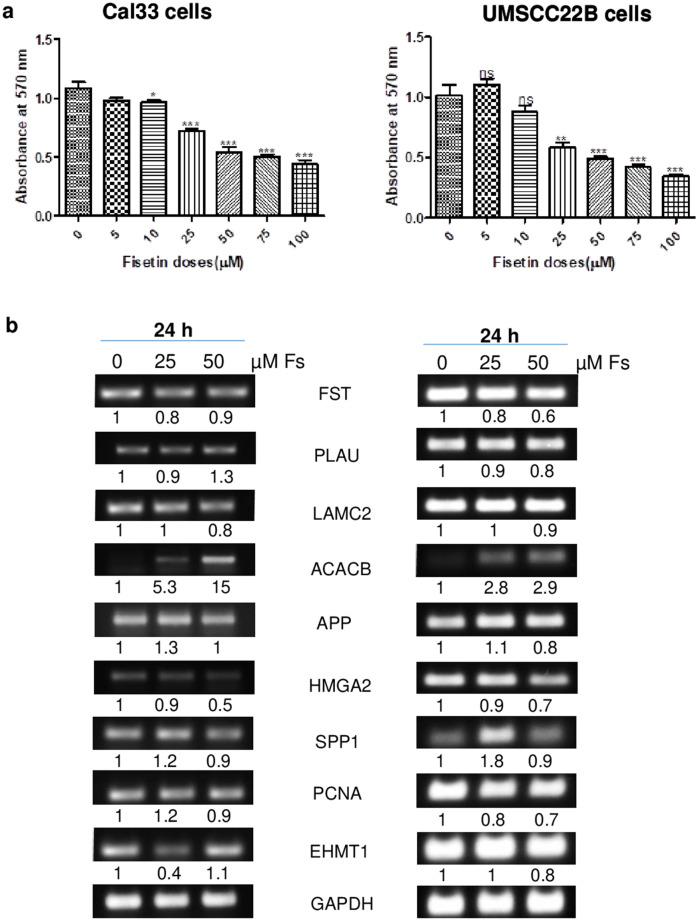
The anticancer effect of fisetin was assessed on human oral squamous cell carcinoma (OSCC) Cal33 and UM-SCC-22B cells by MTT assay (Fig. 6a). Fisetin (5-100 µM) treatment for 48 h decreased the cell viability by 10–60% (P ≤ 0.05) in Cal33 cells. In UM-SCC-22B cells, fisetin (10–100 µM) treatment resulted in 13–66% (P ≤ 0.05) decrease in cell viability. Overall, fisetin exhibited a dose-dependent decrease in cell viability of OSCC cells, suggesting for its anticancer potential against human oral cancer cells.
Fig. 6.
a Effect of fisetin on cell viability in OSCC Cal33 and UM-SCC-22B cells. Cells were seeded and treated with DMSO (control) or fisetin (5–100 µM) in DMSO. After treatment hours, cells were processed for MTT assay and absorbance was taken at 570 nm. b The anti-cancer effect of fisetin on expression analysis of hub genes in OSCC Cal33 and UM-SCC-22B cells by reverse transcriptase-PCR. *P < 0.05; **P < 0.01; ***P < 0.001. The P value was determined by comparing each treatment with control group
Fisetin modulates the expression of hub genes identified in silico
To confirm the influence of dysregulated signature of hub genes in disease condition, the mRNA expression levels were checked by reverse transcriptase-PCR after fisetin (25 and 50 µM) treatment of human oral squamous cell carcinoma UM-SCC-22B and Cal33 cells. It was observed that fisetin treatment resulted in decreased expression of FST, PLAU, LAMC2, APP, HMGA2, SPP1, PCNA and EHMT1 genes while the expression of ACACB gene was increased in both OSCC cell lines. The effect was more prominent at 50 µM concentration of fisetin (Fig. 6b). The regulating effect of fisetin on identified genes further strengthen our in silico observations by confirming their role as potential biomarkers or likely targets for OSCC. Moreover, it also draws attention towards the need for exploring the pivotal anti-cancer role of fisetin in OSCC.
Discussion
Though significant progress has been made in current treatment methods, OSCC remains one of the most prevalent cancers worldwide with increasing number of deaths. The primary causes ascribed to OSCC growth are lack of early diagnostic means, high metastatic tendency and drug resistance (Choi et al. 2019). Therefore, exploration of reliable early diagnostic and prognostic biomarkers of OSCC is urgently needed. In recent decades, with the increasing knowledge of bioinformatics tools, databases pertaining to microarray and high throughput sequencing data have emerged as a comprehensive resources to study association of altered gene expression in the disease (Altman 2016). In this regards, several microarray gene expression profiles have been explored in OSCC; however, the knowledge of precise molecular mechanisms underlying the progression is still not understood completely. In the current study, five gene expression profiles were selected for DEGs analysis among oral cavity cancer and normal control samples by a series of systematic bioinformatics approaches. A total of 6476 (2848 up-regulated and 3628 down-regulated) DEGs were identified in OSCC. Fold change analysis of DEGs unveiled MMP1 as highly up-regulated gene and KRT76 as most down-regulated gene. This immense change in the expression pattern of these two genes is suggestive of their concrete contribution in tumor driving events such as key role in initial cleavage of the extracellular matrix (Stott-Miller et al. 2011) and potential association with hyperproliferation (Ambatipudi et al. 2013), respectively. A subgroup of 77 DEGs identified in all five studies from different regions is likely to hold significance in genesis of OSCC and might be prioritized for common biomarker detection among different races.
GO enrichment analysis specified that the DEGs aids in the discrepancy of cellular motility (Stuelten et al. 2018), invasion and adhesion (Lyons and Jones 2007) which are critical parameters involved in the metastatic progression of oral cancer cells and the major cause of patient’s death. Similarly, pathway analysis confirmed frequent dysregulation in PI3K-Akt signalling, focal adhesion and regulation of actin cytoskeleton pathways. PI3K-Akt signalling pathway is considered as a master regulator of normal physiological processes such as cell proliferation, survival, metabolism and apoptosis. However, hyper-activation of this pathway has been significantly correlated with growth, survival, metastatic potential and therapeutic resistance across various human cancers (Yang et al. 2019; Hou et al. 2020). Dysregulation of focal adhesion perceived among DEGs is indicative of cellular invasion and metastasis of head and neck squamous cell carcinomas via regulation of matrix metalloproteinase expression (Chiu et al. 2016). Accumulating evidences suggest that altered activity of actin cytoskeletal proteins also stimulate tumorigenic development. For example, RHOAY42C gain of function mutation promotes focal adhesion kinase activation which further mediate PI3K/AKT-β-catenin and YAP activity to drive invasion and metastasis in diffuse gastric cancer (Zhang et al. 2020). Thus, both GO and pathway enrichments suggest involvement of the DEGs in migratory and invasive events of cancer development. Subsequently, integration of gene expression pattern with PPI network and cytohubba revealed 38 potential DEGs. Furthermore, top three significant clustering modules were enriched in cell cycle, gap junction and platelet activation pathways which also constitute vital components in OSCC. A list of 115 potential DEGs was then correlated for expression and prognostic value with transcriptome data from TCGA, revealed nine genes (APP, EHMT1, ACACB, PCNA, PLAU, FST, HMGA2, LAMC2, and SPP1) positively correlated with differential expression in OSCC patients and significantly associated with worse overall survival. Therefore, these nine DEGs were regarded as high confidence hub genes.
APP, a cell membrane protein, was identified to regulate MAPK pathway for increasing growth, migration and invasion in breast cancer cells (Wu et al. 2020a) and chemotherapeutic drugs resistance, cell survival in hepatocellular carcinoma (Wu et al. 2020b). EHMT1 was found to be over expressed in gastric cancer (Yang et al. 2018a) and acute lymphoblastic leukemia (Silva-Carvalho et al. 2020). On the other side, down-regulation of ACACB, a fatty acid synthesis gene with malonyl-CoA production facilitates direct inhibition of carnitine-palmitoyl-CoA transferase I and hence fatty acid oxidation, is consistent with its association to one of the hallmarks of cancer i.e.; reprogramming of energy metabolism (Camarda et al. 2016; Klintman et al. 2016). Zhou et al. (2018) reported significant correlation between high PCNA expression and poor colorectal cancer prognosis. PLAU is a key serine protease, well reported for involvement in fibrin dissolution, extracellular matrix degradation, EMT, invasion, metastasis, angiogenesis and immune escape of tumor cells (He et al. 2012; Zhao et al. 2020). PLAU involvement with transcriptional misregulation in cancer pathway is suggestive of its dominating role in cancer progression. Elevated serum FST level was associated with histological types, tumor progression and disease recurrence in lung cancer (Zhang et al. 2018). Another study by Janik et al., also supported that high serum FST levels influences angiogenesis and disease outcome in thymic epithelial tumors (Janik et al. 2019). HMGA2 is a chromatin-associated protein, usually expressed during embryonic stem cells development and remains absent in normal somatic cells. Interestingly, HMGA2 dysregulation stimulate its re-expression in several tumors and was found to be related with cell survival; cell cycle regulation, EMT process, drug resistance and decreased patient survival (Xu et al. 2018, p. 2; Naghizadeh et al. 2019). In present study, differential expression of LAMC2 and SPP1 genes was found to be involved in both PI3K-Akt signalling and focal adhesion pathways. The laminin component LAMC2, was associated with various tumor growth and development processes. Wang et al. (2020a, p. 2) reported that overexpression of LAMC2 modulates microenvironment acidification to initiate EMT in pancreatic cancer cells through Akt/NHE1 signaling. Similarly, SPP1 silencing inhibited ovarian cancer progression by impeding ITGβ1/FAK/AKT signalling pathway, confirming its role in carcinogenesis (Zeng et al. 2018). Subsequent, tumor stage analysis showed hub genes dysregulation is an early event in OSCC progression. Since, ethnic differences can also contribute for tumor heterogeneity, to remove such disparities hub genes were verified for patient’s race/ethinicity analysis which disclosed dysregulated signature of identified hub genes across all three races (Caucasian, African-amercian and Asian). It confirmed significance of nine hub genes as universal biomarker for OSCC. Also emphasizes that nine genes could serve as both early diagnostic and universal prognostic biomarker for OSCC. To further validate the impact in disease state, hub genes were targeted with anti-cancer agent. Herein, we used fisetin as an anti-cancer agent for abrogating the burden of OSCC by addressing hub genes. Fisetin is a dietary flavonoid mostly found in fruits and vegetables and has been shown to target multiple cancerous traits such as cell survival, apoptotic evasion, neo-angiogenesis, invasion and metastasis with no or minimal effects over normal cells (Park et al. 2019; Li et al. 2020). We observed anti-cancer effect of fisetin in OSCC cells by cell viability assay. Fisetin dose-dependently regulate the expression of most of all nine hub genes in OSCC cell lines in favour of its anticancer efficacy; however, FST, HMGA2, PCNA and ACACB genes altered mostly in response to fisetin treatment and the effect was more noticeable at higher doses of fisetin. Thus, reverse transcriptase-PCR results verified our bioinformatics analysis by confirming the prospective application of nine gene signature in OSCC diagnosis and target discovery. Nevertheless, our study is based on public dataset and validation in OSCC cell line, the results required further validation in large scale patient cohorts prior to its translational use at the point of care. Overall, we believe that altered expression and pathways represented by these nine genes would be of significant interest in pathogenesis and metastasis of OSCC.
In conclusion, our bioinformatics approach identified hub genes and key pathways that would serve as intriguing candidates for early detection and treatment for OSCC.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to CIF, JNU and ICMR-AIIMS Computational Genomics Centre, New Delhi for helping in present work.
Author contributions
Execution of research method, bioinformatics analysis, result interpretation, experimental validation, and original draft preparation were performed by Monika Yadav. Dr. Pradhan has done bioinformatics analysis, result interpretation, and visualization. Conceptualization, supervision, reviewing, and final editing of manuscript were accomplished by Dr. Singh.
Funding
The work is supported by DPRP, Department of Science and Technology (DST), India, and DST-PURSE, UGC-RN, UPE-2 from Jawaharlal Nehru University, New Delhi. M.Y. is supported by a fellowship from DST-INSPIRE (IF160971), New Delhi, India.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not include any human or animal samples.
References
- Aggarwal S, Nayek A, Pradhan D, 2017. dbGAPs: a comprehensive database of genes and genetic markers associated with psoriasis and its subtypes. Genomics. [DOI] [PubMed]
- Altman R. Current progress in bioinformatics 2016. Brief Bioinform. 2016;17:1–1. doi: 10.1093/bib/bbv105. [DOI] [PubMed] [Google Scholar]
- Ambatipudi S, Gerstung M, Pandey M, et al. Genome-wide expression and copy number analysis identifies driver genes in gingivobuccal cancers. Genes Chromosom Cancer. 2012;51:161–173. doi: 10.1002/gcc.20940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambatipudi S, Bhosale PG, Heath E, et al. Downregulation of keratin 76 expression during oral carcinogenesis of human, hamster and mouse. PLoS ONE. 2013;8:e70688. doi: 10.1371/journal.pone.0070688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Camarda R, Zhou Z, Kohnz RA, et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Méndez E, Houck J, et al. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer EpidemiolBiomarkPrev. 2008;17:2152–2162. doi: 10.1158/1055-9965.EPI-07-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y-W, Liou L-Y, Chen P-T, et al. Tyrosine 397 phosphorylation is critical for FAK-promoted Rac1 activation and invasive properties in oral squamous cell carcinoma cells. Lab Invest. 2016;96:296–306. doi: 10.1038/labinvest.2015.151. [DOI] [PubMed] [Google Scholar]
- Choi HS, Kim Y-K, Yun P-Y. Upregulation of MDR- and EMT-related molecules in cisplatin-resistant human oral squamous cell carcinoma cell lines. Int J MolSci. 2019;20:3034. doi: 10.3390/ijms20123034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhari AS, Mandave PC, Deshpande M, et al. Phytochemicals in cancer treatment: from preclinical studies to clinical practice. Front Pharmacol. 2020 doi: 10.3389/fphar.2019.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Liu P, Zhang S, et al. Screening pathogenic genes in oral squamous cell carcinoma based on the mRNA expression microarray data. Int J Mol Med. 2018;41:3597–3603. doi: 10.3892/ijmm.2018.3514. [DOI] [PubMed] [Google Scholar]
- Estilo CL, O-charoenrat P, Talbot S, et al. Oral tongue cancer gene expression profiling: identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Chen H, Probst-Kepper M, et al. PLAU inferred from a correlation network is critical for suppressor function of regulatory T cells. MolSystBiol. 2012;8:624. doi: 10.1038/msb.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Li H, Huo W. THBS4 silencing regulates the cancer stem cell-like properties in prostate cancer via blocking the PI3K/Akt pathway. Prostate. 2020;80:753–763. doi: 10.1002/pros.23989. [DOI] [PubMed] [Google Scholar]
- Janik S, Bekos C, Hacker P, et al. Follistatin impacts tumor angiogenesis and outcome in thymic epithelial tumors. Sci Rep. 2019;9:17359. doi: 10.1038/s41598-019-53671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klintman M, Buus R, Cheang MCU, et al. Changes in expression of genes representing key biologic processes after neoadjuvant chemotherapy in breast cancer, and prognostic implications in residual disease. Clin Cancer Res. 2016;22:2405–2416. doi: 10.1158/1078-0432.CCR-15-1488. [DOI] [PubMed] [Google Scholar]
- Lee C-H, Chang JS-M, Syu S-H, et al. IL-1β promotes malignant transformation and tumor aggressiveness in oral cancer. J Cell Physiol. 2015;230:875–884. doi: 10.1002/jcp.24816. [DOI] [PubMed] [Google Scholar]
- Li Y, Jia S, Dai W. Fisetin modulates human oral squamous cell carcinoma proliferation by blocking pak4 signaling pathways. Drug Des DevelTher. 2020;14:773–782. doi: 10.2147/DDDT.S229270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Kong D, Zhang Y, et al. Fisetin inhibits cell proliferation and induces apoptosis via JAK/STAT3 signaling pathways in human thyroid TPC 1 cancer cells. BiotechnolBioproc E. 2020;25:197–205. doi: 10.1007/s12257-019-0326-9. [DOI] [Google Scholar]
- Lin C-Y, Chin C-H, Wu H-H, et al. Hubba: hub objects analyzer—a framework of interactome hubs identification for network biology. Nucleic Acids Res. 2008;36:W438–W443. doi: 10.1093/nar/gkn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons AJ, Jones J. Cell adhesion molecules, the extracellular matrix and oral squamous carcinoma. Int J Oral MaxillofacSurg. 2007;36:671–679. doi: 10.1016/j.ijom.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Nagao T, Warnakulasuriya S. Screening for oral cancer: future prospects, research and policy development for Asia. Oral Oncol. 2020;105:104632. doi: 10.1016/j.oraloncology.2020.104632. [DOI] [PubMed] [Google Scholar]
- Naghizadeh S, Mansoori B, Mohammadi A, et al. Effects of HMGA2 gene downregulation by siRNA on lung carcinoma cell migration in A549 cell lines. J Cell Biochem. 2019;120:5024–5032. doi: 10.1002/jcb.27778. [DOI] [PubMed] [Google Scholar]
- Park B-S, Choi N-E, Lee JH, et al. Crosstalk between fisetin-induced apoptosis and autophagy in human oral squamous cell carcinoma. J Cancer. 2019;10:138–146. doi: 10.7150/jca.28500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Burdett T, Fiorelli B, et al. Expression Atlas update—a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–D932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarwal A, Agarwal R, Singh RP. Fisetin inhibits cellular proliferation and induces mitochondria-dependent apoptosis in human gastric cancer cells. MolCarcinog. 2017;56:499–514. doi: 10.1002/mc.22512. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- Silva-Carvalho AÉ, Alencar APD, Resende MR, et al. Epigenetic priming by EHMT1/EHMT2 in acute lymphoblastic leukemia induces TP53 and TP73 overexpression and promotes cell death. Toxicol In Vitro. 2020;69:104992. doi: 10.1016/j.tiv.2020.104992. [DOI] [PubMed] [Google Scholar]
- Stott-Miller M, Houck JR, Lohavanichbutr P, et al. Tumor and salivary matrix metalloproteinase levels are strong diagnostic markers of oral squamous cell carcinoma. Cancer EpidemiolBiomarkPrev. 2011;20:2628–2636. doi: 10.1158/1055-9965.EPI-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuelten CH, Parent CA, Montell DJ. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat Rev Cancer. 2018;18:296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Yuan Q, Wu D, et al. Identification of core genes and outcome in gastric cancer using bioinformatics analysis. Oncotarget. 2017;8:70271–70280. doi: 10.18632/oncotarget.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cai J, Du S, et al. LAMC2 modulates the acidity of microenvironments to promote invasion and migration of pancreatic cancer cells via regulating AKT-dependent NHE1 activity. Exp Cell Res. 2020;391:111984. doi: 10.1016/j.yexcr.2020.111984. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang Y, Kong F, et al. Identification of a six-gene prognostic signature for oral squamous cell carcinoma. J Cell Physiol. 2020;235:3056–3068. doi: 10.1002/jcp.29210. [DOI] [PubMed] [Google Scholar]
- Wu X, Chen S, Lu C. Amyloid precursor protein promotes the migration and invasion of breast cancer cells by regulating the MAPK signaling pathway. Int J Mol Med. 2020;45:162–174. doi: 10.3892/ijmm.2019.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chen Y, Kong W, Zhao Z. Amyloid precursor protein regulates 5-fluorouracil resistance in human hepatocellular carcinoma cells by inhibiting the mitochondrial apoptotic pathway. J Zhejiang UnivSci B. 2020;21:234–245. doi: 10.1631/jzus.B1900413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wang Y, Deng H, et al. HMGA2 enhances 5-fluorouracil chemoresistance in colorectal cancer via the Dvl2/Wnt pathway. Oncotarget. 2018;9:9963–9974. doi: 10.18632/oncotarget.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shen J, Yan D, et al. Euchromatic histone lysine methyltransferase 1 regulates cancer development in human gastric cancer by regulating E-cadherin. OncolLett. 2018;15:9480–9486. doi: 10.3892/ol.2018.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhong Z, Ding Y, et al. Bioinformatic identification of key genes and pathways that may be involved in the pathogenesis of HBV-associated acute liver failure. Genes Dis. 2018;5:349–357. doi: 10.1016/j.gendis.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X, et al. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019;18:26–26. doi: 10.1186/s12943-019-0954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Yu T, Temam S, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Kensler KH, Hu Z, et al. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet. 2020;16:e1008641. doi: 10.1371/journal.pgen.1008641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng B, Zhou M, Wu H, Xiong Z. SPP1 promotes ovarian cancer progression via Integrin β1/FAK/AKT signaling pathway. Onco Targets Ther. 2018;11:1333–1343. doi: 10.2147/OTT.S154215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Ruan Y, Xiao J, et al. Association of serum follistatin levels with histological types and progression of tumor in human lung cancer. Cancer Cell Int. 2018;18:162. doi: 10.1186/s12935-018-0664-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schaefer A, Wang Y, et al. Gain-of-function RHOA mutations promote focal adhesion kinase activation and dependency in diffuse gastric cancer. CancerDiscov. 2020;10:288–305. doi: 10.1158/2159-8290.CD-19-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Liu Z, Ren Z, et al. Triptolide inhibits pancreatic cancer cell proliferation and migration via down-regulating PLAU based on network pharmacology of Tripterygiumwilfordii Hook F. Eur J Pharmacol. 2020;880:173225. doi: 10.1016/j.ejphar.2020.173225. [DOI] [PubMed] [Google Scholar]
- Zhong P, Liu L, Shen A, et al. Five extracellular matrix-associated genes upregulated in oral tongue squamous cell carcinoma: an integrated bioinformatics analysis. OncolLett. 2019;18:5959–5967. doi: 10.3892/ol.2019.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Huang T, Xiong Y, et al. The prognostic value of proliferating cell nuclear antigen expression in colorectal cancer. Medicine (Baltimore) 2018 doi: 10.1097/MD.0000000000013752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.