Abstract
Clinical targeted sequencing allows for the selection of patients expected to have a better treatment response, and reveals mechanisms of resistance to molecular targeted therapies based on actionable gene mutations. We underwent comprehensive genomic testing with either our original in-house CLHURC system or with OncoPrime. Samples from 24 patients with estrogen receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer underwent targeted sequencing between 2016 and 2018. Germline and somatic gene alterations and patients’ prognosis were retrospectively analyzed according to the response to endocrine therapy. All of the patients had one or more germline and/or somatic gene alterations. Four patients with primary or secondary endocrine-resistant breast cancer harbored germline pathogenic variants of BRCA1, BRCA2, or PTEN. Among somatic gene alterations, TP53, PIK3CA, AKT1, ESR1, and MYC were the most frequently mutated genes. TP53 gene mutation was more frequently observed in patients with primary endocrine resistance compared to those with secondary endocrine resistance or endocrine-responsive breast cancer. Recurrent breast cancer patients carrying TP53-mutant tumors had significantly worse overall survival compared to those with TP53-wild type tumors. Our 160-gene cancer panel will be useful to identify clinically actionable gene alterations in breast cancer in clinical practice.
Subject terms: Breast cancer, Cancer genomics
Introduction
Although recent advances in therapeutic strategies have improved survival in breast cancer, metastatic breast cancer is an incurable disease. Genomic alterations in tumors are acquired during the development from early to late stages1–3. While there are numerous reports of genomic features in early and metastatic breast cancers4–7, they have not conclusively demonstrated the clinical usefulness of genomic sequencing for treatment decisions. Clinical targeted sequencing facilitated by next-generation sequencing (NGS), identifies genetically-based drivers and pathways to help develop and apply valid molecular-targeted therapies. Furthermore, it might reveal mechanisms of resistance to molecular-targeted therapies8.
Breast cancer is the most common malignancy in women. Almost 80% of breast cancers are classified into the estrogen receptor (ER)-positive subtype9. Endocrine therapy for ER-positive breast cancer was one of the earliest targeted treatments. The clinical benefit rate for highly endocrine-responsive disease can be up to 70–80%10. Furthermore, endocrine therapy causes no severe side effects and maintains or improves quality of life. However, the resistance to endocrine therapy is an unavoidable problem especially in metastatic breast cancer. We previously reported that endocrine responsiveness after recurrence was a key issue for improved post-relapse survival in ER-positive recurrent breast cancer11. Recently, inhibitors of molecules within well-defined signaling pathways, including mammalian target of rapamycin (mTOR), cyclin-dependent kinase 4/6 (CDK4/6), phosphatidylinositol 3 kinase (PI3K), and akt murine thymoma viral oncogene (AKT), have been combined with endocrine agents to overcome resistance to endocrine therapy12,13. Understanding predictive factors for the response to endocrine therapy will facilitate the selection of patients to be treated with specific targeted therapies in ER-positive breast cancer. The 4th European School of Oncology (ESO)-European Society for Medical Oncology (ESMO) International Consensus Guidelines for Advanced Breast Cancer (ABC 4) defines endocrine resistance as primary and secondary endocrine resistances14. Since the molecular characteristics of tumors differ according to endocrine responsiveness15, the investigation of actionable gene alterations based on response to endocrine therapy will help to select appropriate treatments in ER-positive metastatic breast cancer.
In April 2016, an original clinical sequencing system named ‘CLHURC’ for patients with cancer was started at our hospital16–20. Targeted sequencing of 160 cancer-related genes using genomic DNA from both tumor tissue and blood was performed under the CLHURC framework. We previously reported the clinical utility of ‘CLHURC’ genomic testing for patients with meningioma16, thyroid carcinoma17, sinonasal papilloma18, pancreatic cancer19, and various other cancers such as colorectal, breast, stomach, and lung cancers20. The ‘CLHURC’ system has been proven to be safe and secure, and could be feasible and promising for application in clinical practice. Here, we report our experience of clinical targeted sequencing in ER-positive, human epidermal growth factor receptor 2 (HER2)-negative advanced breast cancer. We sought to verify the clinical utility of our 160-gene cancer panel for identifying clinically actionable gene alterations in patients with endocrine-responsive and endocrine-resistant breast cancer.
Results
Characteristics of patients
Characteristics of patients at the time of clinical sequencing are summarized in Table 1, and detailed information including disease-free and overall survivals according to endocrine responsiveness is shown in Table 2. Of the 24 patients with ER-positive, HER2-negative metastatic breast cancer, ten (42%) patients had a family history of breast and/or ovarian cancers. Eighteen (75%) patients had recurrent breast cancer and four (17%) patients presented with Stage IV disease. Tumor samples were obtained from the primary tumors of 13 (54%) patients and from the metastatic tumors of 11 (46%) patients. At the time of clinical sequencing, 15 (63%) patients were undergoing endocrine therapy; six (25%) as first-line, four (17%) as second-line, and five (21%) as third-line or later. Endocrine responsiveness in metastatic breast cancer was classified as primary endocrine resistance in seven (29%) patients, secondary endocrine resistance in eight (33%) patients, and endocrine-responsive breast cancer in nine (33%) patients.
Table 1.
Characteristics of patients at the time of clinical sequencing.
| Characteristics | Patients (n = 24) |
|---|---|
| Age, median (range), years | 65.5 (33–73) |
| Sex | |
| Female | 24 (100%) |
| Male | 0 |
| ECOG performance status | |
| 0 | 13 (54%) |
| 1 | 7 (29%) |
| 2 | 2 (8%) |
| 3 | 2 (8%) |
| Family history | |
| Breast cancer and/or ovarian cancer | 10 (42%) |
| Other cancer | 8 (33%) |
| No family history of cancer | 6 (25%) |
| Smoking history | |
| Yes | 8 (33%) |
| No | 16 (67%) |
| Initial stage of diagnosis | |
| I | 1 (4%) |
| II | 11 (46%) |
| III | 6 (25%) |
| IV | 4 (17%) |
| Unknown | 2 (8%) |
| Histopathology | |
| Invasive ductal carcinoma | 23 (96%) |
| Invasive lobular carcinoma | 1 (4%) |
| Site of the sample used for clinical sequencing | |
| Primary tumor | 13 (54%) |
| Metastatic tumor | 11 (46%) |
| Systemic therapy for metastatic breast cancer | |
| First-line endocrine therapy | 6 (25%) |
| Second-line treatment | |
| Endocrine therapy | 4 (17%) |
| Chemotherapy | 2 (8%) |
| Third-line or later | |
| Endocrine therapy | 5 (21%) |
| Chemotherapy | 7 (29%) |
| Endocrine responsiveness in metastatic breast cancer | |
| Primary endocrine resistance | 7 (29%) |
| Secondary endocrine resistance | 8 (33%) |
| Endocrine-responsive breast cancer | 9 (38%) |
| Platform used for clinical sequencing | |
| CLHURC (in-house) | 22 (92%) |
| OncoPrime | 2 (8%) |
Table 2.
Characteristics, treatment, and survivals in 24 patients according to endocrine responsiveness.
| No | Agea | Family history | Initial Stage | ER | Adjuvant endocrine therapy | DFS | First site of metastatic disease | Ageb | Site of sample | Platform | OS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary endocrine resistance | |||||||||||
| 1 | 42 | Other | II | 100 | None | 1 | Lymph node, bone | 44 | Primary | CLHURC | 33 |
| 2 | 33 | Breast/ovary | III | 5 | None | 2 | Lymph node, lung, liver, spleen | 33 | Primary | OncoPrime | 12 |
| 3 | 67 | Other | II | 80 | AI | 3 | Chest wall | 70 | Primary | CLHURC | 42 |
| 4 | 41 | Breast/ovary | II | 90 | TAM, OFS | 16 | Lung | 44 | Primary | CLHURC | 57 |
| 5 | 48 | Other | − | − | TAM | 7 | Lymph node | 67 | Metastasis (ovary) | CLHURC | > 269 |
| 6 | 42 | Breast/ovary | II | 95 | TAM | 11 | Bone | 45 | Metastasis (chest wall) | OncoPrime | 46 |
| 7 | 61 | None | II | 100 | AI | 23 | Liver, bone | 66 | Metastasis (liver) | CLHURC | 71 |
| Secondary endocrine resistance | |||||||||||
| 8 | 61 | Other | II | > 65 | AI | 36 | − | 65 | Primary | CLHURC | − |
| 9 | 58 | Breast/ovary | III | 100 | TAM | 54 | Chest wall | 72 | Primary | CLHURC | 187 |
| 10 | 63 | Breast/ovary | II | − | TAM → AI | 60 | Chest wall | 68 | Primary | CLHURC | 78 |
| 11 | 32 | Breast/ovary | IV | > 65 | N/A | N/A | Bone | 33 | Primary | CLHURC | 20 |
| 12 | 66 | None | IV | 100 | N/A | N/A | Lymph node, pleura | 70 | Primary | CLHURC | 71 |
| 13 | 43 | Breast/ovary | III | 100 | TAM | 56 | Lymph node | 50 | Metastasis (lymph node) | CLHURC | 107 |
| 14 | 51 | Other | II | − | TAM → AI | 58 | Lymph node | 64 | Metastasis (lung) | CLHURC | > 188 |
| 15 | 28 | Breast/ovary | II | > 65 | TAM | 65 | Lymph node, bone | 38 | Metastasis (liver) | CLHURC | 114 |
| Endocrine-responsive breast cancer | |||||||||||
| 16 | 61 | Breast/ovary | I | > 65 | TAM | 97 | Lymph node, bone | 70 | Primary | CLHURC | > 159 |
| 17 | 46 | Other | III | 60 | AI | 128 | − | 57 | Primary | CLHURC | − |
| 18 | 39 | None | IV | > 65 | N/A | N/A | Bone | 46 | Primary | CLHURC | 126 |
| 19 | 72 | Other | IV | 90 | N/A | N/A | Lymph node, bone, pleura | 72 | Primary (ILC) | CLHURC | > 49 |
| 20 | 57 | None | III | 100 | AI → TAM | 74 | − | 66 | Metastasis (bone) | CLHURC | − |
| 21 | 58 | None | III | − | AI | 77 | − | 68 | Metastasis (liver) | CLHURC | − |
| 22 | 39 | Breast/ovary | − | − | TAM | 124 | Lung | 59 | Metastasis (lung) | CLHURC | > 204 |
| 23 | 59 | Other | II | 100 | TAM → AI | 148 | Brain | 73 | Metastasis (brain) | CLHURC | > 205 |
| 24 | 39 | None | II | 95 | TAM | 247 | Lymph node | 66 | Metastasis (lymph node) | CLHURC | 361 |
ER estrogen receptor (% of positive cells), DFS disease-free survival (months), OS overall survival (months), AI aromatase inhibitor, TAM tamoxifen, OFS ovarian function suppression, ILC invasive lobular carcinoma, N/A not applicable, − unknown.
aAge at the time of initial diagnosis (years).
bAge at the time of clinical sequencing (years).
Germline and somatic gene alterations
Detailed information regarding germline and somatic gene alterations according to endocrine responsiveness is shown in Table 3. All of the patients had one or more germline and/or somatic gene alterations. Germline pathogenic variants were identified in four (17%) patients: BRCA1 in one patient (No. 13), BRCA2 in two patients (No. 4 and No. 15), and PTEN in one patient (No. 11). All four patients harboring germline pathogenic variants had family history of breast and/or ovarian cancers. Somatic gene alterations were not detected in one patient (No. 11) who instead has a germline PTEN pathogenic variant. Germline pathogenic variants were not detected in patients with endocrine-responsive breast cancer.
Table 3.
Germline and somatic gene alterations in 24 patients according to endocrine responsiveness.
| No | Age | Germline pathogenic variant | Somatic gene alteration | ||
|---|---|---|---|---|---|
| Actionable gene mutation | Copy-number variant | ||||
| Gain | Loss | ||||
| Primary endocrine resistance | |||||
| 1 | 44 | TP53 T253Nfs*11, PIK3CA V105_R108del | |||
| 2 | 33 | TP53 Y220C, BRCA1 L63* | |||
| 3 | 70 | TP53 A161D, TERT V299M | ESR1, MYC, CDK4, ECT2L, TNFAIP3 | ||
| 4 | 44 | BRCA2 T630Nfs*6 | DICER1 A518V | ||
| 5 | 67 | PIK3CA H1047L, TSC1 A84T | FAM46C, NRAS | ||
| 6 | 45 | TP53 E11Q, STK11 T189l, CEBPA S190P | |||
| 7 | 66 | TP53 E339*, PIK3CA H1047R, ESR1 D538G, MAP2K4 R75Tfs*7 | |||
| Secondary endocrine resistance | |||||
| 8 | 65 | AKT1 E17K, ESR1 D538G | |||
| 9 | 72 | TP53 Q192*, PIK3CA C420R, ESR1 G415_C417del, BRAF R389C, ATM S1863F | MYC, GNAS | ||
| 10 | 68 | CRLF2, SMO | |||
| 11 | 33 | PTEN R130* | |||
| 12 | 70 | RB1 R621S | |||
| 13 | 50 | BRCA1 Q1447Rfs*22 | AKT1 E17K, CREBBP Q1259*, NF1 W561* | CDKN2A | |
| 14 | 64 | ESR1 G442R | |||
| 15 | 38 | BRCA2 D252Vfs*24 | ERBB2 G727A | MYC, TERT | BRCA2, ARID1A, RB1 |
| Endocrine-responsive breast cancer | |||||
| 16 | 70 | PIK3CA H1047R, CSF1R A245T, ARID1A Q1334del, FANCA P1411L, SF3B1 K700E | |||
| 17 | 57 | PIK3R1 R534Q, AKT1 E17K, PTEN L325F | |||
| 18 | 46 | PIK3CA E453K, KIT V540L | |||
| 19 | 72 | PIK3CA H1047R | |||
| 20 | 66 | AKT1 E17K, ROS1 D2213E, FLT3 S454L, DICER1 S678F, MAP3K1 N1305D, MAP3K1 N749Ifs*12 | |||
| 21 | 68 | ESR1 E380Q, HIST1H3B D82Y | PRKAR1A, BRIP1, CD79B | ||
| 22 | 59 | AKT1 E17K | |||
| 23 | 73 | TP53 E11Q, ASXL1 E1033V | HIST1H3B, PRKAR1A, DAXX | ||
| 24 | 66 | AKT1 E17K, DDR2 T283I | KLF6 | ||
Age age at the time of clinical sequencing (years).
Somatic gene alterations were identified in 23 (96%) of tumors (Table 3): TP53 (7 tumors, 29%), PIK3CA (7 tumors, 29%), AKT1 (6 tumors, 25%), and ESR1 (6 tumors, 25%) were the most frequently mutated genes. The median numbers of germline and/or somatic gene alterations were three (range 2–7) in primary endocrine resistance, two (range 1–7) in secondary endocrine resistance, and three (range 1–6) in endocrine-responsive breast cancer. RB1 gene alterations were detected in two tumors with secondary endocrine resistance (RB1 R621S in the primary tumor of No. 12 and RB1 loss in the metastatic tumor of No. 15). A patient (No. 2) whose primary tumor carried a TP53 missense mutation (TP53 Y220C) and a BRCA1 nonsense mutation (BRCA1 L63*) did not harbor germline BRCA1 pathogenic variants.
Among somatic gene alterations, missense mutation was the most frequently detected mutation type. Mutation types found within the TP53 gene were missense in four tumors (Y220C in No. 2, A161D in No. 3, and E11Q in No. 6 and No. 23), nonsense in two tumors (E339* in No. 7, and Q192* in No. 9), and frameshift in one tumor (T253Nfs*11 in No. 1). Mutation types found within the PIK3CA gene were missense in six tumors (H1047L in No. 5, H1047R in No. 7, 16, and 19, C420R in No. 9, E453K in No. 18) and in-frame deletion in one tumor (V105_R108del in No. 1). All six tumors with AKT1 gene alterations had the E17K missense mutation. Mutation types found within the ESR1 gene were missense in four tumors (D538G in No. 7 and No. 8, G442R in No. 14, and E380Q in No. 21), in-frame deletion in one tumor (G415_C417del in No. 9), and amplification in one tumor (No. 3).
Copy number variations such as copy number gain and copy number loss were identified as somatic mutations in nine tumors. Copy number gain was observed in seven (29%) tumors. All MYC gene alterations in three tumors corresponded to copy number gain (No. 3, 9, and 15). Copy number loss was identified in three (13%) tumors (No. 10, 13, and 15). A patient (No. 10) whose tumor had copy number loss of the CRLE2 and SMO genes did not carry other somatic gene mutations nor germline pathogenic variants.
TP53 gene mutation either in a primary or metastatic site was more frequently observed in patients with primary endocrine resistance compared to those with secondary endocrine resistance or endocrine-responsive breast cancer (P = 0.014, Table 4). In contrast, the frequencies of alterations in the PIK3CA, AKT1, ESR1, and MYC genes were not correlated with a particular endocrine-responsive status (Table 4). In addition, the frequencies of alterations in the TP53, PIK3CA, AKT1, ESR1, and MYC genes were similar in samples taken from primary or metastatic sites (Table 5).
Table 4.
Number of patients harboring somatic gene alterations according to endocrine responsiveness.
| Primary resistance (n = 7) | Secondary resistance (n = 8) | Responsive (n = 9) | P value | |
|---|---|---|---|---|
| TP53 | 5 (71%) | 1 (13%) | 1 (11%) | 0.014* |
| PIK3CA | 3 (43%) | 1 (13%) | 3 (33%) | 0.41 |
| AKT1 | 0 | 2 (25%) | 4 (44%) | 0.13 |
| ESR1 | 2 (29%) | 3 (38%) | 1 (11%) | 0.44 |
| MYC | 1 (14%) | 2 (25%) | 0 | 0.29 |
*P < 0.05 is considered significant.
Table 5.
Number of patients harboring somatic gene alterations according to the site of the sample used for clinical sequencing.
| Primary tumor (n = 13) | Metastatic tumor (n = 11) | P value | |
|---|---|---|---|
| TP53 | 4 (31%) | 3 (27%) | 0.85 |
| PIK3CA | 5 (38%) | 2 (18%) | 0.28 |
| AKT1 | 2 (15%) | 4 (36%) | 0.24 |
| ESR1 | 3 (23%) | 3 (27%) | 0.81 |
| MYC | 2 (15%) | 1 (9%) | 0.64 |
Overall survival according to genetic alterations
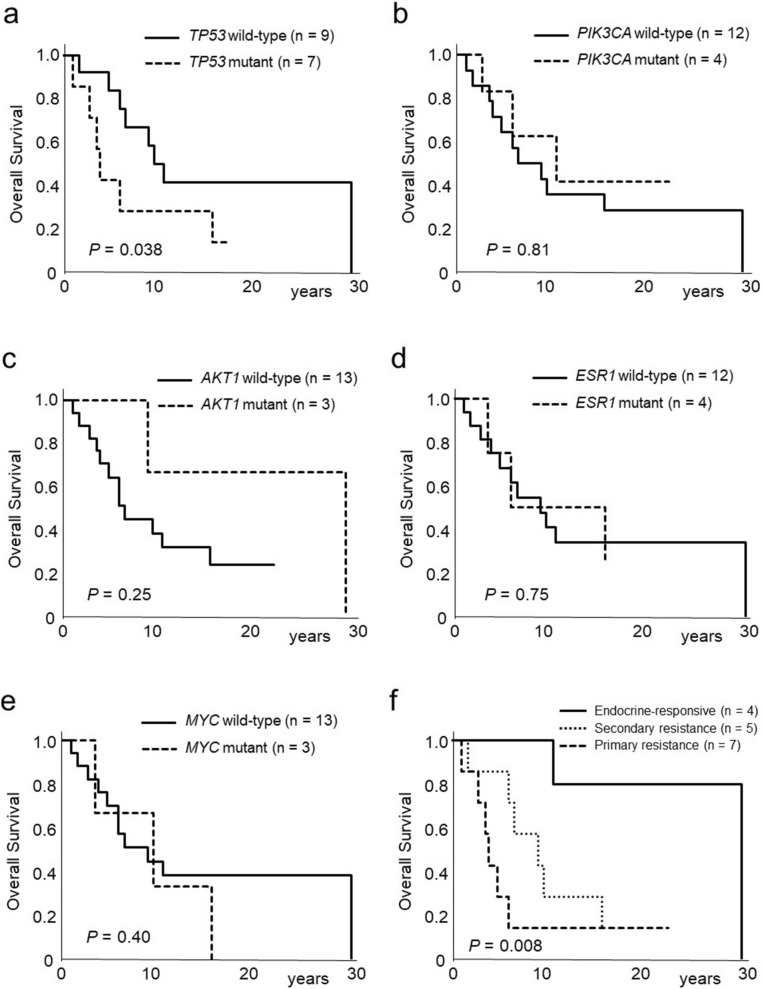
We then determined whether particular sets of genetic alterations correlated with overall survival (time from the diagnosis of primary breast cancer to death) in recurrent breast cancer patients. Kaplan − Meier analysis showed that recurrent breast cancer patients with TP53-mutant tumors had significantly worse overall survival compared to those with TP53-wild type tumors (P = 0.038, Fig. 1a). The presence of alterations in the PIK3CA, AKT1, ESR1, and MYC genes was not correlated with overall survival (Fig. 1b–e). Recurrent breast cancer patients with endocrine-responsive disease had significantly longer overall survival compared to those with primary or secondary endocrine-resistant breast cancers (P = 0.008, Fig. 1f). Univariate analysis using clinical factors (age, family history, smoking history, initial stage of diagnosis, and endocrine responsiveness) and gene mutation status (TP53, PIK3CA, AKT1, ESR1, and MYC) demonstrated that endocrine responsiveness was the only significant factor affecting overall survival in recurrent breast cancer patients (P = 0.0059).
Figure 1.
Kaplan–Meier curves of the effect of TP53 (a), PIK3CA (b), AKT1 (c), ESR1 (d), and MYC (e) gene alterations for overall survival in recurrent breast cancer patients. (f) Kaplan–Meier curves of the effect of endocrine responsiveness for overall survival in recurrent breast cancer patients.
Discussion
Since 2019 in Japan, NGS-based panel testing has been covered by insurance for metastatic cancer patients who have not had (or do not respond to) standard treatments. In the present study, we describe our experience of clinical targeted sequencing in ER-positive metastatic breast cancer. The results demonstrated that all of the 24 patients had one or more germline and/or somatic gene alterations. Four (17%) patients with primary or secondary endocrine-resistant breast cancer harbored germline pathogenic variants of BRCA1, BRCA2, or PTEN. TP53 gene mutations either in the primary or metastatic sites were associated with primary endocrine resistance and poor overall survival.
Large-scale genomic sequencing studies have revealed that ER-positive breast cancer is heterogeneous regarding gene expression, mutation, and copy number variations4,6. Somatic gene mutations of PIK3CA, GATA3, and MAP3K1 in luminal A subtype, and those of TP53, MDM2, PIK3CA, PTEN, and cyclin D1 in luminal B subtype were identified in early breast cancer4. On the other hand, gene alterations of TP53, ESR1, GATA3, KMT2C, NCOR1, AKT1, NF1, RIC8A, and RB1 were more frequently observed in metastatic breast cancer compared with those in early breast cancer in hormone receptor-positive, HER2-negative disease7. This latter report also showed that TP53, RB1, and NF1 gene alterations correlated with poor prognosis in hormone receptor-positive, HER2-negative metastatic disease. Furthermore, a study on endocrine-resistant advanced breast cancers revealed that mutations in the mitogen-activated protein kinase (MAPK) pathway, transcriptional regulators of ER (MYC, CTCF, FOXA1, and TBX3), and ESR1 mutations were present in post-endocrine therapy tumors21. Identification of the mechanisms of resistance to endocrine therapy in each patient will help inform the use of specific molecular-targeted therapies in order to optimize the therapeutic regimen.
The TP53 gene is the most frequently mutated gene in all cancers, and is also the most frequent mutation found in breast cancer4. A TP53 gene mutation was present in 12% of luminal A and 32% of luminal B subtypes, although the frequency of TP53 gene alterations in luminal subtypes is lower than that in basal-like (84%) or HER2-positive (75%) subtypes4. Approximately 90% of TP53 gene alterations encode missense mutant proteins that span ~ 190 different codons located in the DNA-binding domain22. Our recent analysis of TP53 gene alterations in the primary tumors of 56 ER-positive breast cancers showed that all of the 22 mutations existed at different sites23. Ellis and colleagues examined somatic gene alterations in pretreatment tumors from patients who were treated with neoadjuvant aromatase inhibitor therapy24. They reported that mutations in the TP53 signaling pathway including TP53, ATR, APAF1, or THBS1 were present in 38% of the aromatase inhibitor-resistant tumors (11 of 29 tumors). We previously reported that p53 protein accumulation in the nucleus, a surrogate marker for TP53 missense mutation, was a prognostic marker for disease-free survival in patients treated with aromatase inhibitors as adjuvant endocrine therapy25. Patients treated with adjuvant endocrine therapy who had TP53 wild-type tumors also showed longer 5-year overall survival26. Furthermore, TP53 mutations in the primary or metastatic tumors were correlated with poor prognosis in ER-positive metastatic breast cancer27,28. Similarly, we showed in the present study that TP53 mutations either in the primary or metastatic sites affected primary endocrine resistance and poor outcome in ER-positive metastatic breast cancer.
The activation of PI3K/AKT/mTOR signaling pathway is involved in endocrine resistance in ER-positive breast cancer12,13. Mutations of the PIK3CA gene are frequently observed in luminal A (49%) and luminal B (32%) subtypes4,29. Approximately 95% of mutations exist in the helical domain (exon 9, commonly E542 and E545) and the kinase domain (exon 20, commonly H1047)30.We and others previously reported that PIK3CA mutations were a positive prognostic marker in ER-positive, HER2-negative early breast cancer31–33. In contrast, activation of the PI3K pathway is suggested to cause endocrine resistance in ER-positive advanced breast cancer, and therefore inhibitors for PI3K, AKT, and mTOR have potential therapeutic utility in patients with this tumor subtype12,13. Furthermore, Mosele and colleagues recently reported that patients with PIK3CA-mutated metastatic tumors presented with a poor prognosis and resistance to chemotherapy in hormone receptor-positive, HER2-negative metastatic breast cancer34. However, the present study indicated that PIK3CA mutations were present in seven tumors (29%) either in the primary or metastatic sites, and that PIK3CA mutations were not associated with endocrine responsiveness or overall survival.
On the other hand, somatic RB1 gene alterations were identified in two tumors in the present study. Loss of RB protein is one of the key mechanisms of CDK4/6 inhibitor resistance35. Because a CDK4/6 inhibitor combined with endocrine therapy is recommended as first- or second-line treatments in ER-positive, HER2-negative metastatic breast cancer36, information regarding the presence of RB1 gene alterations was essential for selecting an appropriate treatment strategy.
A survey of real-world outcomes of studies that employed targeted sequencing of small panels (48 to 50 genes) in metastatic breast cancer patients revealed that few patients were admissible to clinical trials with molecular-targeted agents37. In contrast, clinical targeted sequencing also led to germline secondary findings. The rate of actionable secondary findings was shown to be from 2 to 5%37–41. In the present study, four (17%) out of 24 patients harbored germline pathogenic variants including BRCA1, BRCA2, or PTEN. All four patients with germline pathogenic variants had family members with breast and/or ovarian cancers, whereas germline pathogenic variants were not detected in patients with endocrine-responsive disease. Patients in whom standard treatments are not effective, or whose family members had breast cancer, are likely to undergo targeted sequencing in clinical practice. Furthermore, one patient in the present study with no germline BRCA1 pathogenic variants had a primary tumor with a somatic BRCA1 gene mutation. De novo somatic BRCA1/2 mutations in breast cancer are considered to be rare: the prevalence of somatic BRCA1 gene mutations in primary tumors is only 1.55%, and that of somatic BRCA2 gene mutations is 1.68%42. Although the clinical success of PARP inhibitors for women with germline BRCA1/2 pathogenic variants is clear, the utility of using PARP inhibition to treat breast cancers with somatic BRCA1/2 mutations is not known.
The present study has several limitations. First, the size of our cohort was too small for a valid statistical analysis and to provide reliable real-world evidence. Furthermore, the site of samples used for clinical sequencing was either the primary or metastatic tumor, because it is often difficult to obtain samples from metastatic sites in clinical practice. However, we do show details of the characteristics, treatments and gene alterations of each patient (Tables 2 and 3). Because 24 samples were all the cases that we experienced during the study period, it is difficult to set an independent dataset to replicate the original findings. It is necessary to construct a prediction model for endocrine therapy based on the results of gene mutations43. Using the deep learning method will help to predict endocrine resistance and the efficacy of targeted therapies for precision medicine in ER-positive breast cancer44.
Second, our clinical targeted sequencing with 160 cancer-related genes is not breast cancer-specific. Thus, some genes essential for breast cancer progression, especially germline variants, might not be covered in this system. Third, the estimation of alterations of each gene has been improving over time. Consequently, we reanalyzed the sequencing results with reference to the newest versions (the Ensembl Variant Effect Predictor (version 95) for somatic variants and ClinVar for germline variants).
In this study, we report our clinical experience of targeted sequencing in ER-positive, HER2-negative metastatic breast cancer. Germline and somatic gene alterations as well as patients’ prognosis were retrospectively analyzed, and we determined whether mutation patterns were correlated with a particular response to endocrine therapy. Our study demonstrated that 17% of patients with primary or secondary endocrine-resistant breast cancer harbored germline pathogenic variants of BRCA1, BRCA2, or PTEN. TP53 gene mutations were affected by primary endocrine resistance and poor overall survival. Our 160-gene cancer panel will be useful to identify clinically actionable gene alterations in patients with endocrine-responsive and endocrine-resistant breast cancer in clinical practice. Accumulation of real-world data on clinical targeted sequencing from individual patients will help to select appropriate therapies in endocrine-resistant breast cancer.
Methods
Patients and clinical targeted sequencing
Between April 2016 and March 2018, 24 patients with ER-positive, HER2-negative metastatic breast cancer underwent comprehensive genomic testing with either our original in-house CLHURC system16–20 or with OncoPrime (EA Genomics, Morrisville, NC, USA). In the CLHURC system, genomic DNA was extracted both from tumor tissue and peripheral blood mononuclear cells obtained from cancer patients, who provided consent to undergo comprehensive genomic testing. After checking the quality of the DNA based on the DNA integrity number (DIN) score calculated using the Agilent 2000 TapeStation (Agilent Technologies, Waldbronn, Germany), we performed a targeted amplicon exome sequencing for 160 cancer-related genes using the Illumina MiSeq sequencing platform (Illumina, San Diego, CA, USA). The list of 160 cancer-related genes included in the CLHURC comprehensive cancer panel was shown in the previous study19,20. The minimum amount of DNA was 50 ng and the minimum quality of DNA had a DIN score over 3.1. This study was approved by the Ethics Committee of the Hokkaido University Graduate School of Medicine (No. 18–049), and conformed to the guidelines of the 1996 Declaration of Helsinki. Informed consent was obtained from each patient.
Bioinformatics analysis
Data from the MiSeq runs were processed initially using the MiSeq platform-specific software MiSeq Reporter (Illumina) to generate sequence reads as a FASTQ file. The generated FASTQ files were then processed with Trim-galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) for trimming adapter sequences and filtering and remove poor signal-profile reads. Burrows-Wheeler Aligner (BWA)45 was used to align the paired-end reads to the hg19 human reference genome to generate BAM format files. BAM files were coordinately sorted and indexed with SAMtools (v1.6)46. Indels in sequence alignment files were left-aligned, and local realignment around Indels was done with the Genome Analysis ToolKit (GATK 3.4–46) RealignerTargetCreator and IndelRealigner47. Base quality score recalibration was performed with GATK tools. For checking sequence run quality, Illumina InterOp library (https://illumina.github.io/interop/index.html) was used. FASTQ files were quality controlled using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (v0.11.6), and BAM files were evaluated those qualities with GATK DepthOfCoverage and Qualimap 2 (v2.2.2)48. VarScan 249 was used to identify single nucleotide variants and small insertions/deletions. If the sample had the normal control data, tumor-normal pair analysis was conducted for calling variants. Copy number variants were identified using CNVkit50 with matched normal. If without normal, Panel of Normal dataset was used as the reference. For calling somatic variant, possible germline single nucleotide polymorphisms (SNPs) were excluded using Single Nucleotide Polymorphism Database (dbSNP) (https://www.ncbi.nlm.nih.gov/snp/), the gnomAD database (https://gnomad.broadinstitute.org/) and the ToMMo 4.7KJPN database (https://jmorp.megabank.tohoku.ac.jp/ijgvd/). Variant filtering was conducted using bcftools51 with alternate allele count, variant allele frequency and read depth and normal pool dataset created before this study, followed by manual review with Integrative Genomics Viewer (Version2.3.57)52 and BAF-LogR profiles. We have used the Ensembl Variant Effect Predictor (version 95)53 for genomic and functional annotation of the variants.
Germline variants were classified according to the American College of Medical Genetics and Genomics (ACMG) guidelines38, and retained as pathogenic with conflicting interpretation in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/)54. The results of germline secondary findings were disclosed to patients only who agreed to disclose germline information.
Endocrine responsiveness in metastatic breast cancer
We defined endocrine responsiveness according to ABC 414 as follows: Primary endocrine resistance was defined as relapse while on the first 2 years of adjuvant endocrine therapy, or progressive disease (PD) within first 6 months of first-line endocrine therapy for advanced breast cancer, while on endocrine therapy. Secondary endocrine resistance was defined as relapse while on adjuvant endocrine therapy but after the first 2 years, or relapse within 12 months of completing adjuvant endocrine therapy, or PD within 24 months of first-line endocrine therapy for advanced breast cancer. Endocrine-responsive breast cancer was defined as relapse after 12 months of completing adjuvant endocrine therapy, or PD ≥ 24 months of first-line endocrine therapy for advanced breast cancer.
Statistical analysis
The significance of differences among three groups was evaluated using the chi-squared test. P values < 0.05 were considered significant. Estimation of overall survival (time from the diagnosis of primary breast cancer to death) was performed using the Kaplan–Meier method, and differences between survival curves were assessed using the log-rank test. The Cox proportional hazards model was used for prognostic analysis. Statistical analysis was performed using Excel software (Microsoft corp., Albuquerque, MX, USA).
Acknowledgements
This work was partially supported by a grant from Japan Society for the Promotion of Science KAKENHI Grant Number 17K10531 from the Ministry of Education, Science, Sports and Culture, Japan.
Author contributions
Study concept and design (K.H., H.Y.), acquisition and interpretation of data (K.H., T.A., H.H., T.T., T.O., J.K., Y.O., I.Y., I.K., H.N., H.Y.), analysis of data (K.H., T.A., H.Y.), drafting of the manuscript (K.H., H.Y.). All authors read and approved the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yates LR, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32:169–184. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng CKY, et al. Genetic heterogeneity in therapy-naive synchronous primary breast cancers and their metastases. Clin. Cancer Res. 2017;23:4402–4415. doi: 10.1158/1078-0432.CCR-16-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nik-Zainal S, Morganella S. Mutational signatures in breast cancer: The problem at the DNA level. Clin. Cancer Res. 2017;23:2617–2629. doi: 10.1158/1078-0432.CCR-16-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertucci F, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569:560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 8.Tsuchida J, et al. Clinical target sequencing for precision medicine of breast cancer. Int. J. Clin. Oncol. 2019;24:131–140. doi: 10.1007/s10147-018-1373-5. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita H, et al. Estrogen receptor-positive breast cancer in Japanese women: trends in incidence, characteristics, and prognosis. Ann. Oncol. 2011;22:1318–1325. doi: 10.1093/annonc/mdq596. [DOI] [PubMed] [Google Scholar]
- 10.Iwase H. Treatment strategy for metastatic breast cancer with estrogen receptor-positive tumor. Int. J. Clin. Oncol. 2015;20:249–252. doi: 10.1007/s10147-015-0795-6. [DOI] [PubMed] [Google Scholar]
- 11.Ogiya A, et al. Post-relapse survival in patients with the early and late distant recurrence in estrogen receptor-positive HER2-negative breast cancer. Breast Cancer. 2017;24:473–482. doi: 10.1007/s12282-016-0730-3. [DOI] [PubMed] [Google Scholar]
- 12.Araki K, Miyoshi Y. Mechanism of resistance to endocrine therapy in breast cancer: The important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer. 2018;25:392–401. doi: 10.1007/s12282-017-0812-x. [DOI] [PubMed] [Google Scholar]
- 13.Brufsky AM, Dickler MN. Estrogen receptor-positive breast cancer: Exploiting signaling pathways implicated in endocrine resistance. Oncologist. 2018;23:528–539. doi: 10.1634/theoncologist.2017-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardoso F, et al. 4th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 4)dagger. Ann. Oncol. 2018;29:1634–1657. doi: 10.1093/annonc/mdy192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tryfonidis K, Zardavas D, Katzenellenbogen BS, Piccart M. Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treat. Rev. 2016;50:68–81. doi: 10.1016/j.ctrv.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Yuzawa S, et al. Clinical impact of targeted amplicon sequencing for meningioma as a practical clinical-sequencing system. Mod. Pathol. 2016;29:708–716. doi: 10.1038/modpathol.2016.81. [DOI] [PubMed] [Google Scholar]
- 17.Bandoh N, et al. Targeted next-generation sequencing of cancer-related genes in thyroid carcinoma: A single institution's experience. Oncol. Lett. 2018;16:7278–7286. doi: 10.3892/ol.2018.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasukawa S, et al. Genetic mutation analysis of the malignant transformation of sinonasal inverted papilloma by targeted amplicon sequencing. Int. J. Clin. Oncol. 2018;23:835–843. doi: 10.1007/s10147-018-1296-1. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi H, et al. Genomic testing for pancreatic cancer in clinical practice as real-world evidence. Pancreatology. 2018;18:647–654. doi: 10.1016/j.pan.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi H, et al. Clinical impact of a cancer genomic profiling test using an in-house comprehensive targeted sequencing system. Cancer Sci. 2020;111:3926–3937. doi: 10.1111/cas.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Razavi P, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34:427–438. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018;25:154–160. doi: 10.1038/cdd.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita H, et al. HER2 gene amplification in ER-positive HER2 immunohistochemistry 0 or 1+ breast cancer with early recurrence. Anticancer Res. 2020;40:645–652. doi: 10.21873/anticanres.13994. [DOI] [PubMed] [Google Scholar]
- 24.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M, et al. p53 accumulation is a strong predictor of recurrence in estrogen receptor-positive breast cancer patients treated with aromatase inhibitors. Cancer Sci. 2014;105:81–88. doi: 10.1111/cas.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ungerleider NA, et al. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 2018;20:115. doi: 10.1186/s13058-018-1044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meric-Bernstam F, et al. Survival outcomes by TP53 mutation status in metastatic breast cancer. JCO Precis. Oncol. 2018 doi: 10.1200/PO.17.002452018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim JY, et al. Clinical implications of genomic profiles in metastatic breast cancer with a focus on TP53 and PIK3CA, the most frequently mutated genes. Oncotarget. 2017;8:27997–28007. doi: 10.18632/oncotarget.15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes SA, et al. COSMIC: Mining complete cancer genomes in the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishida N, et al. PIK3CA mutation, reduced AKT serine 473 phosphorylation, and increased ERalpha serine 167 phosphorylation are positive prognostic indicators in postmenopausal estrogen receptor-positive early breast cancer. Oncotarget. 2018;9:17711–17724. doi: 10.18632/oncotarget.24845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loi S, et al. PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor-positive breast cancer. Proc. Natl. Acad. Sci. U S A. 2010;107:10208–10213. doi: 10.1073/pnas.0907011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabine VS, et al. Mutational analysis of PI3K/AKT signaling pathway in tamoxifen exemestane adjuvant multinational pathology study. J. Clin. Oncol. 2014;32:2951–2958. doi: 10.1200/JCO.2013.53.8272. [DOI] [PubMed] [Google Scholar]
- 34.Mosele F, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 2020;31:377–386. doi: 10.1016/j.annonc.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Portman N, et al. Overcoming CDK4/6 inhibitor resistance in ER-positive breast cancer. Endocr. Relat. Cancer. 2019;26:R15–R30. doi: 10.1530/ERC-18-0317. [DOI] [PubMed] [Google Scholar]
- 36.Cardoso F, et al. 5th ESO-ESMO international consensus guidelines for advanced breast cancer (ABC 5) Ann. Oncol. 2020;31:1623–1649. doi: 10.1016/j.annonc.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pezo RC, et al. Impact of multi-gene mutational profiling on clinical trial outcomes in metastatic breast cancer. Breast Cancer Res. Treat. 2018;168:159–168. doi: 10.1007/s10549-017-4580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalia, S. S. et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet. Med.19, 249–255. 10.1038/gim.2016.190 (2017). [DOI] [PubMed]
- 39.Dorschner MO, et al. Actionable, pathogenic incidental findings in 1000 participants' exomes. Am. J. Hum. Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amendola LM, et al. Actionable exomic incidental findings in 6503 participants: Challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dewey FE, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:1549. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 42.Wang L. An exploration of mutation status of cancer genes in breast cancers. Next Gen. Seq. Appl. 2014;1:1000103. [Google Scholar]
- 43.Yu H, et al. LEPR hypomethylation is significantly associated with gastric cancer in males. Exp Mol. Pathol. 2020;116:104493. doi: 10.1016/j.yexmp.2020.104493. [DOI] [PubMed] [Google Scholar]
- 44.Liu M, et al. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer's disease. Neuroimage. 2020;208:116459. doi: 10.1016/j.neuroimage.2019.116459. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okonechnikov K, Conesa A, Garcia-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao R, Yu T, Qing Y, Wang J, Shen Y. Evaluation of copy number variant detection from panel-based next-generation sequencing data. Mol. Genet. Genomic Med. 2019;7:e00513. doi: 10.1002/mgg3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landrum MJ, et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]