Abstract
Fungus-growing termites are engaged in a tripartite mutualism with intestinal microbes and a monocultivar (Termitomyces sp.) in the fungus garden. The termites are often plagued by entomopathogen (Metarhizium anisopliae) and fungus garden is always threatened by competitors (Xylaria spp.). Here, we aim to understand the defensive role of intestinal microbes, the actinomycetes which were isolated from the gut of Macrotermes barneyi. We obtained 44 antifungal isolates, which showed moderate to strong inhibition to Xylaria sp. HPLC analysis indicated that different types of polyenes (tetraene, pentene, and heptaene) existed in the metabolites of 10 strong antifungal Streptomyces strains. Two pentene macrolides (pentamycin and 1′14-dihydroxyisochainin) were firstly purified from Streptomyces strain HF10, both exhibiting higher activity against Xylaria sp. and M. anisopliae than cultivar Termitomyces. Subsequently, tetraene and heptaene related gene disruption assay showed that the mutant strains lost the ability to produce corresponding polyenes, and they also had significantly decreased activities against Xylaria sp. and M. anisopliae compared to that of wild type strains. These results indicate that polyene-producing Streptomyces from the guts of M. barneyi have strong inhibition to competitor fungus and polyenes contribute to inhibitory effects on Xylaria sp.
Keywords: fungus-growing termite, Macrotermes barneyi, Streptomyces, polyene, Xylaria
Introduction
Many fungus-growing insects (ants, termites, and southern pine beetles) actively cultivate one or two symbiotic fungi (Basidiomycota or Ascomycota) in an obligatory association (Chapela et al., 1994; Aanen et al., 2002; Hofstetter et al., 2006). The fungi serve as food for the insects, and the insects provide nutrients for fungus growth. However, the fungi cultivated by fungus-growing ants (Attini), fungus-growing termites (Macrotermitinae), and fungus-growing beetles (Dendroctonus frontalis) are plagued by specialized pathogens Escovopsis spp., Xylaria spp., and Ophiostoma minus, respectively (Currie et al., 1999a; Hofstetter et al., 2006; Ju and Hsieh, 2007). In addition, entomopathogenic fungi, such as Beauveria bassiana and Metarhizium anisopliae, are abundant in soil, and can be infectious to fungus-growing insects (Strasser et al., 2000; Mburu et al., 2009).
The multilateral symbioses systems of fungus-growing insects are compared to human agricultural systems. The symbiotic interactions in fungus-growing ant and beetle systems have been documented (Ramadhar et al., 2014). Microbial symbionts of fungus-growing ants and southern pine beetles, mainly Pseudonocardia and Streptomyces (Currie et al., 1999b; Cafaro et al., 2011), protect their fungal cultivars from competitor fungi via production of antifungal compounds, including dentigerumycin, gerumycins, antimycins, 9-methoxyrebeccamycin, cyphomycin, candicidin, mycangimycin, and macrolactam frontalamides (Scott et al., 2008; Haeder et al., 2009; Oh et al., 2009a, b; Blodgett et al., 2010; Schoenian et al., 2011; Sit et al., 2015; van Arnam et al., 2015; Chevrette et al., 2019b). Leaf-cutting ant-associated Pseudonocardia and Amycolatopsis isolates with antifungal activities protect hosts from entomopathogenic fungi (Sen et al., 2009; Mattoso et al., 2012).
Fungus-growing termites (Termitidae: Macrotermitinae), a group of higher termites, are abundant in tropical and subtropical regions of Asia and Africa (Liu et al., 2013). They have a significant effect on plant decomposition and element cycling (Li et al., 2017, 2021). The bacteria in termite mound soils could improve the fertility of the soil and suppress soil borne plant pathogens through the production of antibiotics and nutrient fixation, thus they might help reduce the farm use of chemical fertilizers and pesticides (Enagbonma and Babalola, 2019). Actinobacteria from nests of termite species, including Macrotermes natalensis, Microtermes sp., and Odontotermes sp., show higher antifungal activity against cultivar Termitomyces than Xylaria (Visser et al., 2012), while other Actinobacteria strains, or chemicals such as Actinomycin D and macrotermycins isolated from termite-associated actinomycetes exhibit selective antifungal activity against competitors (Pseudoxylaria or Xylaria) over Termitomyces (Beemelmanns et al., 2017; Yin et al., 2019). The role of actinobacteria in fungus-growing termites remains to be further explored.
Macrotermes barneyi, is a fungus-growing termite in the subfamily of Macrotermitinae, and widely distributed in southern China (Wu et al., 2012). M. barneyi lives in obligate symbiosis with a specialized fungal cultivar Termitomyces sp. (Basidiomycotina), which is the only visible fungus in active nests. Xylaria sp. thrives in abandoned termite nests and is the competitor of Termitomyces sp. Here, we assess the antifungal activity of actinomycetes through strain isolation, paired bioassay, HPLC analysis, compound identification, gene disruption and bioactivity assay. The results indicated that M. barneyi associated antifungal Streptomyces produced different types of polyenes, which contributes to the inhibitory activity against the antagonistic fungi.
Materials and Methods
Sample Collection and Tested Fungi
The workers and soldiers of M. barneyi were collected in July 2017 and June 2018 from termite nests in Hunan (E 112° 96′, N 26° 58′) and Guangdong (E 113° 60′, N 24° 82′), China. The termites were transferred into sterilized tubes and stored on dry ice. Gut dissection was performed within 48 h. The insect-pathogenic fungi M. anisopliae ACCC 30103 and B. bassiana ACCC 30730 were purchased from the Agricultural Culture Collection of China, Beijing, China. Termitomyces sp. and Xylaria sp. were isolated from the fungus gardens of M. barneyi. Strain identity was verified by PCR amplification and sequencing of the ITS gene using the primers ITS1 and ITS4 (Heine et al., 2018).
Gut Dissection and DNA Extraction
Termite workers were surface sterilized by successive soaking in sterile water and phosphate buffered saline, and then rinsed with 70% ethanol and sterile water (Ramadhar et al., 2014). 150 workers and 50 soldiers were dissected. The workers guts were aseptically removed with forceps and divided into foregut, midgut, and hindgut, which were immersed in PBS buffer (NaCl 8 g/L, KCl 0.2 g/L, Na2HPO4 1.42 g/L, KH2PO4 0.27 g/L, pH 7.4) (Schmitt-Wagner et al., 2003). The samples were homogenized and transferred into sterile tubes to be used for actinomycetes isolation.
Strain Isolation and Phylogenetic Analysis
Actinomycetes were isolated by the serial dilution method on Gause’s No.1, M2 (Mincer et al., 2002), M4 (Mincer et al., 2002), PY-CMC (Min et al., 1994), HVA (Subramani and Aalbersberg, 2013), and Chitin media (Benndorf et al., 2018). Fifty workers and soldiers were dissected to obtain gut samples. After shaking at 150 rpm for 30 min, gut suspensions of 10–1 to 10–3-fold dilutions were plated onto six isolation media with or without inhibitors (50 μg/mL potassium dichromate, 50 μg/mL cycloheximide, and 20 μg/mL nalidixic acid) (Malviya et al., 2014). The plates were incubated at 30°C for 7–15 days. Colonies with distinct morphological characteristics were transferred and purified on yeast extract-malt extract (ISP2) agar plates (Shirling and Gottlieb, 1966). Isolates were kept in 20% glycerol at −80°C for long-term preservation.
For genomic DNA extraction, actinomycetes were grown in nutrient-rich liquid Tryptic Soy Broth (TSB) medium at 30°C for 4 days. Cells were then harvested, and the genomic DNA was extracted using a bacterial DNA Extraction Kit (Omega, BioTek, United States). The 16S rRNA gene was amplified by PCR using general primers 27F and 1492R (Liu et al., 2013). Amplification reactions were standardized in a total volume of 50 μl containing 2× EasyTaq master mix (Takara, Dalian, China), 100 ng genomic DNA of isolated actinomycetes, 10 μM of each primer and sterile water. Cycle parameters for PCR were 10 min at 95°C (initial denaturation); 35 cycles at 95°C for 30 s, 58°C for 30 s and 72°C for 2 min; 7 min at 72°C (final elongation). The amplified 16S rRNA fragments were cloned into the pMD19-T vector. After sequencing, these complete 16S rRNA gene sequences were compared with available sequences in the GenBank database using the BLAST program in NCBI. The phylogenetic tree was constructed via the neighbor-joining tree algorithm using MEGA version 7.0 (Kumar et al., 2016). The confidence values of nodes in the trees were evaluated by 1000 bootstrap replicates (Felsenstein, 1985).
Actinobacteria–Fungi Paired Challenge Assays
Actinobacteria-fungi paired bioassays were performed according to a previously reported method (Benndorf et al., 2018). Strains were grown at 30°C for 3 days in ISP2 or TSB. Aliquots of 20 μl liquid culture were used to inoculate on the centers of Potato Dextrose Agar (PDA) plates. Then, plates were inoculated at the edges with two agar pieces covered with fungal mycelium (Xylaria sp.). All assays were performed in triplicate. Plates were incubated for 7–10 days at 30°C and checked daily until a clear and stable zone of inhibition (ZOI) appeared (normally after 7 days). The ZOI values were given by measuring the distance of two inoculated Xylaria. The final ZOI is the average value of three replicates. The categories of strong inhibition (ZOI > 2 cm), moderate inhibition (ZOI 0.5–2 cm), and little or no inhibition (ZOI < 0.5 cm) were defined (Scott et al., 2008).
Antifungal Assay of Crude Extracts
Actinomycetes isolates with strong antifungal activity against Xylaria sp. in Actinobacteria-fungi paired bioassays were cultivated in 40 ml TSB, MS (2% D mannitol, 2% soybean meal, pH 7.2) and ISP2 liquid media for 4 days at 30°C with shaking at 150 rpm. The liquid broth of 21 actinomycetes isolates was centrifuged at 12000 × g for 10 min. Culture supernatants were dried in vacuum rotary evaporator at 38°C and dissolved in 2 ml methanol, which was used as crude extracts. The assay was conducted using the agar diffusion method (Haeder et al., 2009). First, sterilized stainless-steel Oxford cups (10 × 6 × 8 mm) were placed on PDA plates. Next, PDA agar medium with fungal spores and mycelium was introduced into the upper plates. After solidification, the Oxford cups were removed, the crude extracts (100 μl) were added into each well. The diameters of the inhibition zones around the wells were observed after incubation for 1 days at 30°C. The active crude extracts were subjected to HPLC analysis.
HPLC Analysis of the Active Crude Extracts
The active crude extracts were applied to HPLC (DIONEX Ultimate 3000 instrument) analysis. HPLC was operated at a flow rate of 1 mL/min with a C18 column (YMC-Pack Pro, 250 × 4.6 mm, 5 μm). Water-0.1% formic acid (solvent A) and acetonitrile-0.1% formic acid (solvent B) were used as the mobile phases. The column was eluted for a conventional separation by using elution gradient of 20% B, increased to 35% B in 3 min, 45% B in 10 min, 90% B in 20 min, and 100% B in 22 min (held for 3 min). Afterward, the elution gradient was reduced to 20% B in 27 min and sustained for 3 min.
Genome Sequencing and Analysis
The Streptomyces strains were cultivated in 20 ml TSB liquid medium for 2 days at 30°C with shaking at 150 rpm. The culture broth (20 ml) was centrifuged at 12000 × g for 10 min, culture supernatant was removed and cell pellets were harvested. Genomic DNA was isolated from cell pellets that were physically ground in liquid N2 and then extracted using a bacterial DNA Extraction Kit (Omega, BioTek, United States). Whole-genome sequencing was performed using PacBio SMRT sequencing technology by the Novogene sequencing company. Biosynthetic gene clusters (BGCs) of secondary metabolites were predicted by antiSMASH 4.0 (Blin et al., 2017). The annotation of the polyene BGCs was performed using Blastp (non-redundant proteins).
Isolation and Identification of the Active Compounds From Streptomyces sp. HF10
To isolate the antifungal compounds from Streptomyces sp. HF10, we set out a large-scale culture (20 L ISP7 broth) (Shirling and Gottlieb, 1966) for 4 days at 30°C with shaking at 160 rpm. The culture was centrifuged to obtain the supernatant followed by absorption using macroporous resin D101 overnight. The resin was loaded on a column and washed with water and then eluted with 50% methanol and 100% methanol, respectively. The antifungal activity of each fraction was tested. The active fractions were dried by evaporation to obtain the crude extract.
The crude extract was loaded on a middle-pressure liquid chromatography column (MPLC; 80 g RP-18 silica gel; 20, 40, 60, and 100% acetonitrile containing 0.1% formic acid, 200 ml for each gradient) to yield Fr.1-4. Fr 2 was subjected to Sephadex LH-20 to yield Fr.2a and 2b. Fr.2a and Fr.2b were subjected to MPLC (25%, 30% acetonitrile containing 0.1% formic acid) to yield Fr.2a1 and Fr.2b2. Fr.2a1 and Fr.2b2 were purified by semi-preparative reverse-phase HPLC (DIONEX Ultimate 3000 instrument, YMC-Pack Pro C18, 10 × 250 mm, 5 μm, flow rate 4 mL/min, UV detection at 350 nm) to yield 1 (10 mg, 38% acetonitrile containing 0.1% formic acid, tR = 9.5 min) and 2 (9 mg, 29% acetonitrile containing 0.1% formic acid, tR = 8.5 min), respectively. Subsequently, compounds 1 and 2 dissolved in methanol were subjected to high resolution mass spectrometry (HRMS) for determination of molecular mass. Compounds 1 and 2 were dissolved in DMSO-d to measure 1H, 13C-NMR and two-dimensional NMR spectra on a 600 MHz spectrometer.
In vitro Antifungal Assay and Determination of Minimal Inhibitory Concentrations
Antifungal activity of the purified compounds was monitored by the paper disc diffusion method (Um et al., 2013). An agar piece covered with fungal mycelium (Xylaria sp. or Termitomyces sp.) was inoculated at the center of the PDA plates. Plates were incubated for 7 days at 30°C until a clear colony was apparent. Sterile discs were placed around the colony covered with different amounts (2.5, 5, and 10 μg) of purified pentamycin or 1′14-dihydroxyisochainin. DMSO was used as a negative control. All of the tested discs were observed on a daily basis for 3 days. A liquid antifungal assay was performed to measure minimal inhibitory concentrations (MICs) of compounds. To prepare mycelia or spore suspensions, Xylaria sp., Termitomyces sp. and two entomopathogens were cultivated on PDA plates 5–7 days, then mycelia or spores were collected and homogenized in potato dextrose broth medium to maintain the absorbance of OD600 approximately 0.5. The suspension was transferred into the wells of 96-well microplate followed by adding 2-fold serial dilution of purified compounds or amphotericin with a final concentration of 128, 64, 32, 16, 8, 4, 2, and 1 μg/mL. Subsequently, in order to determine the exact difference in antifungal activity of purified compounds against tested fungi, different concentrations of pentamycin (30, 25, and 20 μg/mL), 1′14-dihydroxyisochainin (150 145, and 140 μg/mL) or amphotericin (60, 55, 50, 45, and 40 μg/mL) were applied against Termitomyces sp. The above suspension without adding compound used as negative control was simultaneously cultured. The plates were incubated at 30°C and monitored for inhibition. The MIC value was calculated as the lowest concentration showing complete inhibition of the tested strain. All of the assays were performed in triplicate.
Construction of Natamycin Biosynthetic Gene Disruption Mutant of Streptomyces sp. GS7
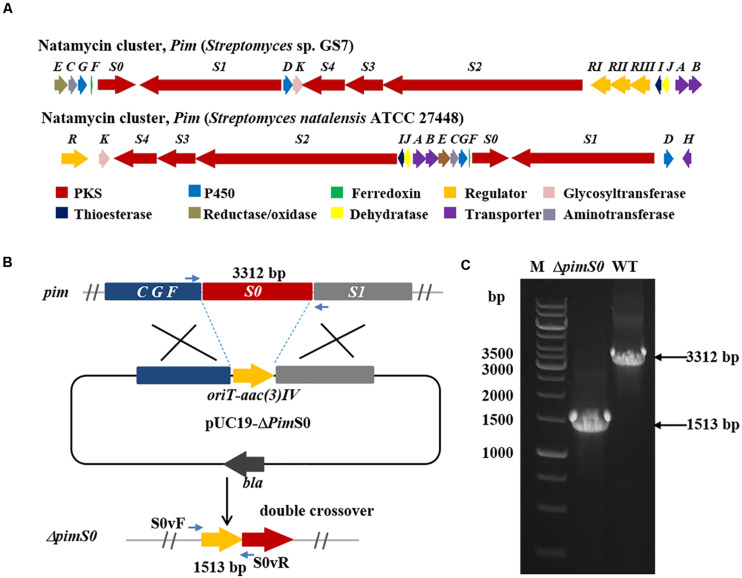
To inactivate the BGC of natamycin (pim) in Streptomyces sp. GS7, the polyketide synthase gene (pimS0) was replaced with a gene disruption cassette by homologous recombination. The gene disruption cassette containing a selectable apramycin resistance gene aac(3)IV and an origin of transfer gene oriT were jointly amplified by PCR from plasmid PIJ773 (Gust et al., 2003) with the primers (aac(3)IV-P4F/aac(3)IV-P4R). Two homologous arms flanking pimS0 were amplified from the genomic DNA of Streptomyces sp. GS7 with the primers (dP4LF/dP4LR, dP4RF/dP4RR). The above three fragments were assembled into pUC19 by Gibson assembly (Gibson et al., 2009), to yield pUC19-ΔpimS0. The constructed plasmid pUC19-ΔpimS0 was transformed into Streptomyces sp. GS7 by conjugation and the exconjugants were selected on MS agar medium with apramycin (Sangon Biotech, Shanghai, China). After three rounds of non-selective growth, the desired double cross-over mutants were confirmed by PCR. Amplification reactions were performed in a final volume of 25 μl containing 2× Rapid Taq master mix (Vazyme Biotech Co., Nanjing, China), 50 ng genomic DNA of mutants or wide-type strains, 10 μM of each primer (S0vF/S0vR), 5 μl PCR enhancer (Vazyme Biotech Co., Nanjing, China) and sterile water. PCR amplification program involved an initial DNA denaturation at 95°C for 10 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s and elongation at 72°C for 3 min 30s, which followed by a final extension at 72°C for 7 min. All primers were listed in Supplementary Table 7.
Construction of the Candicidin Biosynthetic Gene Disruption Mutant of Streptomyces sp. GF20
To inactivate the BGC of candicidin (fsc) in Streptomyces sp. GF20, the polyketide synthase gene (fscA) was replaced with a gene disruption cassette by homologous recombination. The gene disruption cassette oriT-aac(3)IV was amplified by PCR from plasmid pSET152 (Bierman et al., 1992) with the primers (aac(3)IV-XbaI-F/aac(3)IV-XbaI-F) and digested with XbaI. Two homologous arms flanking fscA were amplified from the genomic DNA of Streptomyces sp. GF20 with the primers (dP7-PstI-LF/dP7-XbaI-LR, dP7-XbaI-RF/dP7-HindIII-RR). Homologous arms were digested with PstI/XbaI or HindIII/XbaI, respectively. The three fragments were ligated into pSPRam, which is pOJ260 (Bierman et al., 1992) derivative containing the reporter melanin gene mel (Wang et al., 2018) and resistance marker aadA (spectinomycin-resistance), to yield pSPRam-ΔfscA. The constructed plasmid pSPRam-ΔfscA was transformed into Streptomyces sp. GF20 by conjugation and exconjugants were selected on MS agar medium with apramycin. Then the cells were cultured on MS medium plates containing 5 mg/L copper sulfate and 100 mg/L tyrosine, white colonies as double cross-over mutants were picked (Wang et al., 2018). The double cross-over mutants were confirmed by PCR. The reaction systems were same as described early. Program parameters for PCR were 10 min at 98°C (initial denaturation); 35 cycles at 95°C for 30 s, 58°C for 30 s and 72°C for 6 min; 7 min at 72°C (final elongation). All used primers were listed in Supplementary Table 7.
Results
Isolation and Phylogenetic Analysis of Actinomycetes Strains
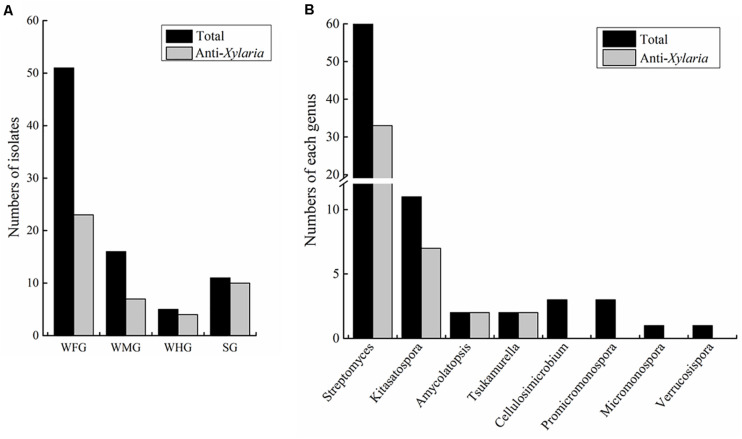
Actinobacteria strains were isolated from the foregut, midgut and hindgut of workers as well as the gut of soldiers. According to distinct morphological features of actinomycetes and 16S rRNA gene sequence analysis, in total, we obtained 83 strains of actinomycetes belonging to eight genera from two termite samples collected from Hunan and Guangdong (Figure 1 and Supplementary Table 1). Among these, 72 strains were isolated from workers, and 11 from soldiers. Overall, 60 Streptomyces strains were isolated from the foregut, midgut and hindgut of workers and the gut of soldiers. 11 Kitasatospora strains were isolated from the foregut of workers and the gut of soldiers (Figure 1). Defined by a threshold of <98.65% sequence similarity, seven putative new Actinobacteria species (Streptomyces, Kitasatospora, and Amycolatopsis) were obtained from termite workers (Supplementary Table 2). Among them, six strains (HF5, HF17, GF5, GF6, GF15, and GF18) were isolated from the foregut and one strain GM8 from the midgut of M. barneyi. Phylogenetic analysis of all of the isolates based on 16S rRNA sequences revealed that the isolates formed two clades within the Actinobacteria phylum (Figure 2A), Streptomyces and Kitasatospora isolates clustering into one clade and the remaining 12 isolates clustering into another clade.
FIGURE 1.
Statistics of Actinobacteria isolated from intestines of Macrotermes barneyi. (A) Total numbers of Actinobacteria isolates and those with anti-Xylaria activity from the worker foregut (WFG), worker midgut (WMG), worker hindgut (WHG), and the soldier gut (SG), respectively. (B) Numbers of Actinobacteria isolates and those with anti-Xylaria activity in each genus. The anti-Xylaria activity was examined by the paired challenge assay.
FIGURE 2.
Phylogenetic and anti-Xylaria activity analyses of the isolated actinomycetes. (A) Phylogenetic tree of all the isolated strains. The tree was constructed with Mega 7.0 by the Neighbor-Joining method. The branch values indicate bootstrap support (>30 are given) of 1000 replicates. The ability to inhibit the growth of Xylaria sp. is indicated with black squares (strong inhibition, ZOI > 2 cm), gray squares (moderate inhibition, ZOI = 0.5–2 cm), and open squares (little or no inhibition, ZOI < 0.5 cm) (Scott et al., 2008). HF: Hunan foregut, HM: Hunan midgut, HH: Hunan hindgut, GF: Guangdong foregut, GM: Guangdong midgut, GH: Guangdong hindgut, GS: Guangdong soldier. (B) Examination of the anti-Xylaria activity of the isolated actinomycetes by paired challenge assay. Inhibition of representative Streptomyces against Xylaria sp. is shown. (i–v) Streptomyces sp. strains against Xylaria sp. (i–iii) HF10, GS7, and GF20, strong inhibition; (iv) GM6, moderate inhibition; (v) GM1, no inhibition; (vi) control, Xylaria sp.
The Isolated Actinomycetes Exhibited Antifungal Activity Against Xylaria sp.
To explore the antifungal activity of the isolated strains, all the Actinobacteria isolates were challenged against fungal garden competitor Xylaria sp. Forty-four actinomycetes isolates, which belongs to four genera (Streptomyces, Kitasatospora, Amycolatopsis, and Tsukamurella), exhibited moderate to strong antifungal activity against Xylaria sp. (Figure 2A). Among these, 23, 7, and 4 strains were isolated from foregut, midgut, and hindgut of workers, respectively, and 10 strains were isolated from the gut of soldiers. Twenty-one isolates with strong antifungal activity belong to two genera (Streptomyces and Kitasatospora) and cluster into one clade (Figure 2A). It was noted that Streptomyces strains GF2 and GM11 have the same closest type strain (Streptomyces drozdowiczii NRRL B-24297), HF10 and GM9 have the same type strain (Streptomyces misionensis JCM 4497), GF26 and HM2 have the same type strain (Streptomyces sampsonii ATCC 25495) in the blast search in NCBI (Supplementary Table 1), GF2, HF10, and GF26 presented antifungal activity while GM11, GM9, and HM2 had no antifungal activity (Figure 2A), which was supported by the study that taxonomic and metabolic incongruence exists in Streptomyces (Chevrette et al., 2019a).
Paired challenge assays of five representative strains were shown in Figure 2B. As shown in this figure, three Streptomyces isolates (HF10, GS7, and GF20) exhibited strong inhibition against Xylaria sp. (Figure 2Bi–iii). Streptomyces sp. GM6 exhibited moderate inhibition against Xylaria sp. and Streptomyces sp. GM1 displayed no inhibition against Xylaria sp. (Figure 2Biv,v).
Polyene Compounds Were Detected in the Metabolites of Majority of the Streptomyces Strains
To identify putative compounds responsible for the antifungal activity, 21 actinomycetes strains with strong antifungal activity against Xylaria sp. were selected and cultivated in TSB, MS, and ISP2 liquid media, respectively. Agar diffusion assay showed that the fermentation broth of 15 strains had antifungal activity against M. anisopliae. Here M. anisopliae was used instead of Xylaria sp. owing to the instability of crude extracts. The metabolites of these strains were analyzed by HPLC and the profiles of 10 out of the 15 Streptomyces strains, GS7, GS11, GM12, HF10, GF21, GF25, HM3, GF20, GF26, and GH2, showed peaks with typical UV/vis spectra of four type of polyenes (Table 1 and Supplementary Figures 1–10). Among these strains, GS7, GS11, and GM12 showed tetraene peaks (292, 305, and 320 nm) (Supplementary Figures 1–3; Mendes et al., 2001); HF10, GF21, and GF25 showed pentene peaks (325, 340, and 358 nm) (Supplementary Figures 4–6; Xiong et al., 2012); HM-3 showed linear heptaene peaks (354, 272, and 393 nm) (Supplementary Figure 7; Oh et al., 2009b); GF20, GF26, and GH2 showed heptaene peaks (360, 381, and 406 nm) (Supplementary Figures 8–10; Haeder et al., 2009). While no obvious polyene peaks were observed in the metabolite profiles of the remaining five strains (Kitasatospora sp. HF13, HF15, and GS2, Streptomyces sp. HH1 and GS5).
TABLE 1.
Predicted polyenes from antifungal crude extracts of isolated actinomycetes.
| Strains | Isolated resource | Antifungal activitya | Absorption peaks (nm) | Polyene type | References |
| Streptomyces GS7 | Solider gut | +++ | 293,306,320 | Cyclic tetraene | Mendes et al., 2001 |
| Streptomyces GS11 | Solider gut | +++ | 293,306,320 | Cyclic tetraene | Mendes et al., 2001 |
| Streptomyces GM12 | Worker midgut | +++ | 294,306,321 | Cyclic tetraene | Mendes et al., 2001 |
| Streptomyces HF10 | Worker foregut | +++ | 324,340,358 | Cyclic pentene | Xiong et al., 2012 |
| Streptomyces GF21 | Worker foregut | ++ | 325,340,358 | Cyclic pentene | Xiong et al., 2012 |
| Streptomyces GF25 | Worker foregut | ++ | 325,340,358 | Cyclic pentene | Xiong et al., 2012 |
| Streptomyces HM3 | Worker midgut | +++ | 353,372,393 | Linear heptaene | Oh et al., 2009b |
| Streptomyces GF20 | Worker foregut | +++ | 360,381,400 | Cyclic heptaene | Haeder et al., 2009 |
| Streptomyces GF26 | Worker foregut | ++ | 360,382,401 | Cyclic heptaene | Haeder et al., 2009 |
| Streptomyces GH2 | Worker hindgut | + | 360,382,401 | Cyclic heptaene | Haeder et al., 2009 |
| Kitasatospora HF13 | Worker foregut | +++ | – | – | – |
| Kitasatospora HF15 | Worker foregut | +++ | – | – | – |
| Streptomyces HH1 | Worker hindgut | ++ | – | – | – |
| Kitasatospora GS2 | Solider gut | ++ | – | – | – |
| Streptomyces GS5 | Solider gut | + | – | – | – |
aThe antifungal activity against Metarhizium anisopliae. IZD: Inhibition zone diameter excluding width of Oxford cups. +++: IZD ≥ 1.5 cm. ++: IZD ≥ 1 cm. +: IZD ≥ 0.5 cm.
Prediction of BGC and Purification of Antifungal Polyene Compounds From Streptomyces sp. HF10 and Bioactivity Assays of Purified Compounds
To identify polyene BGCs, whole genome sequencing of strain HF10 was performed. Comparative analysis revealed that the genetic organization of the target gene cluster in strain HF10 was almost identical to the reported pentamycin gene cluster in Streptomyces sp. S816 (Figure 3A and Supplementary Table 3; Zhou et al., 2019).
FIGURE 3.
Streptomyces sp. strain HF10 produces antibiotics pentamycin and 1′14-dihydroxyisochainin. (A) The genetic organization of the pentamycin BGC of strain HF10 compared with the reported one in Streptomyces sp. 816. The BGC of pentamycin in strain HF10 was identified by whole genome sequencing and bioinformatic analysis. (B) HPLC profiles of metabolites from strain HF10 (left) and UV/vis spectra of compounds 1 and 2 (right). (C) The chemical structures of pentamycin (1) and 1′14-dihydroxyisochainin. (2). (D) Antifungal activity of pentamycin and 1′14-dihydroxyisochainin by disc diffusion assay. Pentamycin and 1′14-dihydroxyisochainin were purified from strain HF10 as described in section “Materials and Methods.” Pentamycin (middle) and 1′14-dihydroxyisochainin (right) against Xylaria sp. Pentamycin (middle) and 1′14-dihydroxyisochainin (right) against Termitomyces sp. Left, schematic map showing sample arrangement in experiments. A gradient amount (2.5, 5.0, and 10 μg) of pentamycin or 1′14-dihydroxyisochainin was used. An agar piece covered with fungal mycelium (Xylaria sp. or Termitomyces sp.) was inoculated at the center of the PDA plate for 7 days at 30°C. Sterile discs containing the compounds dissolved in DMSO were placed around the colony. The plates were cultured at 30°C for 3 days and observed on daily basis.
To identify the antifungal compounds produced by strain HF10 which had the strongest activity against Xylaria sp. as observed in the paired bioassay (Figure 2B), we performed a scale-up fermentation of strain HF10 and obtained 10 mg of pure compound 1 and 9 mg of pure compound 2 after a series of column chromatography purifications (Figure 3B). The relative molecular masses of compounds 1 and 2 were detected at m/z 670.3936 (1) and m/z 642.3630 (2) by HRMS, respectively (Supplementary Figures 11, 12). The 1H- and 13C-NMR data of 1 and 2 were summarized in Supplementary Table 4 (Supplementary Figures 13–20; Li et al., 1989). Based on these results, the chemical structures of 1 and 2 were determined to be pentamycin and 1′14-dihydroxyisochainin, respectively (Figure 3C).
In paper disc diffusion assays, pentamycin (1) exhibited stronger activity against Xylaria sp. (Figure 3D middle) than Termitomyces sp. (Figure 3D middle). With the tested concentrations, 1′14-dihydroxyisochainin (2) also selectively suppressed Xylaria sp. (Figure 3D right) rather than Termitomyces sp. (Figure 3D right). In the MIC determination assays, the pentamycin (1) exhibited the lowest MIC (4 μg/mL) against Xylaria sp., while the MIC of amphotericin B against Xylaria sp. was 16 μg/mL (Table 2). The pentamycin consistently inhibited the growth of Xylaria sp., M. anisopliae, and B. bassiana more strongly than that of Termitomyces sp. While, 1′14-dihydroxyisochainin selectively suppressed the growth of pathogens but not Termitomyces sp. Thus, the antifungal activities of the two polyene compounds against Xylaria sp. and entomopathogens were stronger than that of the cultivar.
TABLE 2.
Minimal inhibitory concentrations (MIC, μg/mL) of pentamycin and 1′14-dihydroxyisochainin.
| Strains | MIC (μg/mL) |
||
| Pentamycin | 1′14-dihydroxyisochainin | Amphotericin B | |
| M. anisopliae ACCC 30103 | 8 | 64 | 16 |
| B. bassiana ACCC 30730 | 8 | 64 | 32 |
| Xylaria sp. | 4 | 32 | 16 |
| Termitomyces sp. | 20 | >150 | 60 |
Amphotericin B was used as a positive control.
Mutants Completely Abolished the Polyene Production and the Activities of the Mutants Against Antagonistic Fungi Were Greatly Weakened
Two Streptomyces strains, GS7 and GF20 (Figure 2B), producing tetraene and heptaene compounds (Supplementary Figures 1, 8) revealed by HPLC analysis, were subjected to genome sequencing. The sequence alignment showed that the putative tetraene cluster in GS7 was homologous to the natamycin synthesis cluster of Streptomyces natalensis ATCC 27448 and showed 77% sequence identity (Figure 4A; Mendes et al., 2001). The putative heptaene cluster in GF20 was homologous to the candicidin synthesis cluster of Streptomyces sp. FR-008 and showed 100% sequence identity (Figure 5A; Chen et al., 2003). Potential ORFs responsible for natamycin and candicidin biosynthesis were shown in Supplementary Tables 5,6.
FIGURE 4.
Disruption of a key natamycin biosynthetic gene pimS0 in Streptomyces sp. GS7. (A) Comparison of the genetic organization of natamycin BGCs in Streptomyces sp. GS7 and S. natalensis ATCC 27448. (B) Schematic representation for disruption of pimS0. (C) Verification of the pimS0-disrupted mutant by PCR. M, DNA marker. The fragments were amplified using primers of S0vF/S0vR.
FIGURE 5.
Disruption of candicidin biosynthetic gene fscA in Streptomyces sp. GF20. (A) Comparison of the genetic organization of candicidin BGCs in Streptomyces sp. GF20 and Streptomyces sp. FR-008. (B) Schematic representation for disruption of fscA in Streptomyces sp. GF20. (C) Verification of the fscA-disrupted mutant by PCR. M, DNA size marker. The fragments were amplified using primers of AvF/AvR.
The 3.2 kb fragment of pimS0 and 5.7 kb fragment of fscA, corresponding to the polyketide synthase gene of polyenes BGCs pim (natamycins) and fsc (candicidins) in strains GS7 and GF20, respectively, have been replaced with the gene disruption cassette (Figures 4B, 5B). Two mutants GS7ΔpimS0 and GF20ΔfscA generated by double cross-over were obtained and verified by PCR (Figures 4C, 5C) with primers S0vF/S0vR and AvF/AvR, respectively (Supplementary Table 7). HPLC analysis showed that two mutant strains were completely unable to produce compounds 3, 4 (Figure 6A), and 5 (Figure 7A). Streptomyces-fungi paired bioassays showed that both mutants (GS7ΔpimS0 and GF20ΔfscA) had significantly less inhibitory effects on the growth of Xylaria sp. and M. anisopliae than the corresponding wild type strains (Figures 6B,C, 7B,C). In addition, both wild type and mutant strains, exhibited stronger inhibition to Xylaria sp. than M. anisopliae. The results suggested that tetraene and heptaene, produced by Streptomyces sp. GS7 and GF20, respectively, are major antifungal compounds against the pathogens (Xylaria sp. and M. anisopliae), and compared to M. anisopliae, Streptomyces tends to inhibit Xylaria sp. more strongly than M. anisopliae.
FIGURE 6.
The deletion mutant of natamycins in Streptomyces sp. GS7 had weaker antifungal activities against Xylaria sp. and Metarhizium anisopliae. (A) HPLC profiles of the metabolites of Streptomyces sp. GS7 wild type and the GS7ΔpimS0 mutant. The triangle symbol (Δ) denotes a homolog of natamycin. (B,C) Paired challenge assays of GS7 and the deletion mutant (GS7ΔpimS0) against Xylaria sp. (Xa, B) and M. anisopliae (Ma, C). (D,E) HRMS spectra of compounds 3 and 4 from GS7. (F,G) The predicted chemical structures of natamycin/pimaricin (3) and de-epoxynatamycin/de-epoxypimaricin (4).
FIGURE 7.
The deletion mutant of candicidins in Streptomyces sp. GF20 had weaker antifungal activities against Xylaria sp. and M. anisopliae. (A) HPLC profiles of the metabolites of wide type Streptomyces sp. GF20 and mutant GF20 ΔfscA. The asterisk symbols (*) denote homologs of candicidin. (B,C), paired challenge assays of GF20 and the deletion mutant (GF20 ΔfscA) against Xylaria sp. (Xa, B) and M. anisopliae (Ma, C). (D) HRMS spectrum of compound 5 from GF20. (E) The predicted structure of candicidin D (5).
The relative molecular masses of compounds 3, 4, and 5 were m/z 665.3123, 649.1131, and 1108.5778, respectively (Figures 6D,E, 7D). UV/Vis spectra of compound 3 and 4 were identical to pimaricin and de-epoxypimaricin from S. natalensis ATCC 27448, and compound 5 was identical to candicidin D from Streptomyces sp. FR-008 (Mendes et al., 2001; Chen et al., 2003). The identity of BGC, UV/Vis spectrum and relative molecular mass indicated that compounds 3, 4, and 5 are probably natamycin/pimaricin, de-epoxynatamycin/de-epoxypimaricin, and candicidin D, respectively (Figures 6F,G, 7E).
Discussion
The success of termite fungiculture depends on multiple factors, such as the control of pathogens within termite fungus farms, termite antimicrobial peptides and gut bacteria with antimicrobial properties (Um et al., 2013; Poulsen, 2015; Bodawatta et al., 2019). The results in this study revealed that fungus-growing termite M. barneyi-associated actinomycetes produced different type of polyenes, which greatly inhibited competitor fungus Xylaria sp.
Actinobacteria Isolation From the Digestive Tract of M. barneyi
The actinomycetes were isolated from M. barneyi workers and soldiers. Owing to relative smaller size of soldier guts than that of workers, also a fewer numbers of soldier collected than workers, only worker guts were separated into the different gut sections (foregut, midgut, and hindgut) (Figure 1A). By a culture-based approach, we obtained 83 Actinobacteria strains from the guts of M. barneyi, which belongs to 8 genera but mainly Streptomyces and Kitasatospora (Figure 1B). Previously, actinomycetes of 4 genera including Streptomyces, Cellulosimicrobium, Promicromonospora, and Micromonospora have been isolated from workers intestines of M. natalensis and Odontotermes formosanus (Benndorf et al., 2018), and genus Kitasatospora strains have been isolated from the cuticle of fungus-growing termite (Visser et al., 2012). To our knowledge, this is the first time to obtain actinomycetes isolates of three genera Amycolatopsis, Tsukamurella, and Verrucosispora from fungus-growing termites.
Based on 16S rRNA sequence analysis, seven strains were predicted to be potential novel actinomycetes (Supplementary Table 1). Recently, a number of new actinomycetes have been isolated and identified from the worker gut of M. natalensis (Benndorf et al., 2020a, b; Schwitalla et al., 2020). The intestinal microflora of fungus-growing termite represents a promising resource of novel actinomycetes.
It was noted there were relatively higher numbers of strains obtained from the foregut of worker than other gut sections. The possible explanation is: the variety and abundance of hindgut microflora are highest among three gut sections (Chew et al., 2018), and actinomycetes are at very low level in termite guts (Otani et al., 2014), thus on the plates spreading with hindgut sample, a large number of bacteria overgrew and slow-growing actinomycetes were difficult to grow. Therefore, there was few actinomycetes obtained from the hindgut. Pre-treatments of hindgut samples by drying heating (Bredholt et al., 2008) or adding inhibitor such as chloramine-T (Hayakawa et al., 1997) may facilitate the selective isolation of actinomycetes from the hindgut. Although majority of actinomycetes were isolated from the foregut, considering the foregut is the first gut section of ingested food, we could not preclude these actinomyces originate from the nest environment, where many actinobacteria strains exist (Visser et al., 2012).
Antifungal Actinobacteria Strains From the Digestive Tract of M. barneyi Worker and Soldier
Plate paired assay revealed that 53% of the actinomycetes, mainly Streptomyces and Kitasatospora genus possessed antifungal activity against antagonistic fungus Xylaria. Majority of these antifungal actinomycetes were isolated from worker intestines, which could be explained by the fact that the workers have a greater chance of being exposed to pathogens than that of soldiers, since workers are responsible for transporting and digesting external plant material (Li et al., 2017). Additionally, the defensive microbes (most likely Actinobacteria and Bacillus) in the gut is helpful to selectively inhibit the potential antagonists when the substrate first passes through the worker gut and avoid the entry of pathogens into fungal comb (Um et al., 2013; Poulsen, 2015).
Soldiers mainly play a defensive role in the colony by using their large and strong mandibles as well as by chemical substances secreted from a frontal gland on the head (He et al., 2018). In the present study, we also obtained Streptomyces with antifungal activity against Xylaria sp. from soldier guts. Previous study of gut bacterial metagenomic analysis by Poulsen revealed that the soldiers have nearly similar or a bit higher relative abundance of Streptomyces than workers in M. natalensis (Poulsen et al., 2014). To our knowledge, this is the first time to obtain the Streptomyces isolates from the soldier, considering the relatively high percentage of antifungal strains, which suggest the potential role of soldier-associated Streptomyces in fungus comb against antagonistic fungus Xylaria.
Polyenes-Producing Streptomyces Contribution to Inhibition of Xylaria
A key strategy of insects coping with environmental threats is the use of molecular defenses from symbiotic microbes (van Arnam et al., 2018), especially from insect-associated Streptomyces and Pseudonocardia (Scott et al., 2008; Haeder et al., 2009; Oh et al., 2009a, b; Blodgett et al., 2010; Schoenian et al., 2011; Sit et al., 2015; van Arnam et al., 2015; Chevrette et al., 2019b). To identify putative compounds responsible for the antifungal activity, the metabolites of antifungal strains were analyzed by HPLC, and the results revealed that the ten antifungal Streptomyces strains produce four type of polyenes with different number of conjugated double bonds. Several polyene compounds, including candicidin (Haeder et al., 2009; Barke et al., 2010), nystatin P1 (Barke et al., 2010), selvamicin (van Arnam et al., 2016), filipins (Gao et al., 2014), and mycangimycin (Oh et al., 2009b) have been reported from the symbiont actinomycetes of fungus-farming ants and southern pine beetles. Bacillus sp. from M. natalensis produced a polyene polyketide, bacillaene, which selectively inhibits antagonistic fungus of Termitomyces (Um et al., 2013), However, as far as we know, polyene compounds have not previously been isolated from fungus-growing termites-associated actinomycetes (Bi et al., 2011; Carr et al., 2012; Zhang et al., 2013, 2020; Kim et al., 2014; Benndorf et al., 2018; Klassen et al., 2019; Guo et al., 2020). Three strains with strong antifungal activities and potential polyene products corresponding to tetraene, pentene and heptaene, respectively, were subjected to bulk culture, probably due to chemical instability of polyenes (Worthen et al., 2001), finally only two pentene compounds (pentamycin and 1′14-dihydroxyisochainin) were purified from Streptomyces sp. HF10 (Figure 3C). Pentamycin (also called fungichromin) and 1′14-dihydroxyisochainin belong to polyene macrolides, containing the antifungal antibiotics amphotericin B (Sun et al., 2015) and nystatin (Fjaervik and Zotchev, 2005). Previously, fungichromin have been isolated from endophytic actinomycetes (Human et al., 2016) and lower termite-associated actinomycetes (Mevers et al., 2017). The compound 1′14-dihydroxyisochainin is an analog of chainin, which was first isolated from a soil actinomycete with antifungal activity against phytopathogens (Thirumalachar, 1955; Gopalkrishnan et al., 1968).
Although two purified pentene compounds are not novel products, they represent the first report on polyene compound from fungus-growing termite-associated Streptomyces. Bioactivity assays showed that the competitor fungus Xylaria sp. was the most susceptible to inhibition of two pentene compounds, compared with entomopathogen (B. bassiana and M. anisopliae) and fungal cultivar Termitomyces sp., which was similar to the studies in leaf-cutting ants and the southern pine beetle (Haeder et al., 2009; Oh et al., 2009b). Candicidin identified from ant-associated Streptomyces is highly active against pathogenic fungus Escovopsis sp. (Haeder et al., 2009). Our results suggest the potential role of the pentamycin in protecting the fungus comb of M. barneyi against competitor fungus Xylaria sp.
Since we failed to obtain tetraene and heptaene compounds from cultures of Streptomyces sp. GS7 and GF20, disruptive mutants of tetraene and heptaene BGCs were constructed (Figures 4, 5). HPLC analysis revealed that the corresponding polyene peaks disappeared and paired challenge assays showed the mutants had obviously weaker activities against antagonistic fungus Xylaria than the wild type strains, suggesting that tetraene and heptaene compounds produced by GS7 and GF20 contribute to the inhibition against the antagonistic fungi. Furthermore, both the wild type and mutant strains exhibited stronger inhibitory effects on antagonistic fungus (Xylaria sp.) than entomopathogen (M. anisopliae) (Figures 6B,C, 7B,C), suggesting tetraene and heptaene produced by Streptomyces firstly selectively inhibited the fungus comb antagonistic fungus Xylaria. Interestingly, the recent study by Bodawatta et al. (2019) showed that M. natalensis foraging workers significantly avoided the mycopathogen-exposed substrates, and did not show any preference between entomopathogen-exposed and control substrate. Overall, the present study by pentene compound purification, tetraene and heptaene BGCs gene disruption and bioactivity assays suggest that polyenes produced by M. barneyi-associated Streptomyces greatly contribute to inhibition of antagonistic fungi.
It was noted that mutants retained slight activity against Xylaria sp. after disruption of tetraene and heptaene BGCs (Figures 6B, 7B). Thus, except for polyenes, some other active compounds also inhibit the antagonistic fungi. In leaf-cutting ants, different antifungal secondary metabolites exhibited strong synergistic effects against pathogenic fungi (Schoenian et al., 2011). We inferred that in termite guts multiple compounds including polyenes and non-polyenes complement and reinforce the activities against pathogens.
The complex web of interactions involving insects (Poulsen et al., 2014), their fungal crops (Wang et al., 2015; Otani et al., 2019), specialized pathogens (Guo et al., 2016), symbiotic fungus (Xu et al., 2020), and symbiotic bacteria has become both a model system for chemical ecology and a source of naturally occurring small molecules. We are still a long way from identifying additional antifungal compounds in this system and understand thoroughly the chemical basis of symbiotic or antagonistic associations among termites, fungal cultivar, cultivar competitors, entomopathogenic fungi and antibiotic-producing actinomycetes.
Conclusion
In conclusion, through actinomycetes isolation, bioactivity assays, active product purification, and BGCs gene disruption analysis, we show that Streptomyces isolated from the gut of fungus-growing termite M. barneyi are capable of producing a variety of polyenes, which significantly inhibit antagonistic fungus Xylaria over entomopathogenic fungi and fungal cultivar Termitomyces. The results indicate that potential role of different type of polyenes produced by Streptomyces in protection of fungus comb against the antagonistic fungus.
Data Availability Statement
The complete 16S rRNA genes of 83 isolated actinomycetes in this study were deposited in GenBank (MN826234-MN826316). The GenBank accession numbers for the genomes of the three strains are CP047144-CP047145 (Streptomyces sp. HF10), CP047146 (Streptomyces sp. GS7), and CP047147 (Streptomyces sp. GF20).
Author Contributions
JL, MS, YJ, and JW performed the experiments. JN, YL, and JL designed the experiments. JL and JN analyzed the data and wrote the manuscript. YL, YS, and QH helped to revision of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Haoxin Wang from Shandong University for suggestions and supports regarding the study.
Footnotes
Funding. This study was supported by grants from National Key R&D Program of China (No. 2020YFA0906800), the National Natural Science Foundation of China (31970119), and the Young Scholars Program of Shandong University (2016WLJH31).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.649962/full#supplementary-material
References
- Aanen D. K., Eggleton P., Rouland-Lefevre C., Guldberg-Froslev T., Rosendahl S., Boomsma J. J. (2002). The evolution of fungus-growing termites and their mutualistic fungal symbionts. Proc. Natl. Acad. Sci. U.S.A. 99 14887–14892. 10.1073/pnas.222313099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barke J., Seipke R. F., Gruschow S., Heavens D., Drou N., Bibb M. J., et al. (2010). A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 8:109. 10.1186/1741-7007-8-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemelmanns C., Ramadhar T. R., Kim K. H., Klassen J. L., Cao S. G., Wyche T. P., et al. (2017). Macrotermycins A-D, glycosylated macrolactams from a termite-associated Amycolatopsis sp M39. Org. Lett. 19 1000–1003. 10.1021/acs.orglett.6b03831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benndorf R., Guo H. J., Sommerwerk E., Weigel C., Garcia-Altares M., Martin K., et al. (2018). Natural products from Actinobacteria associated with fungus-growing termites. Antibiotics 7:83. 10.3390/antibiotics7030083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benndorf R., Martin K., Kufner M., de Beer Z. W., Vollmers J., Kaster A. K., et al. (2020a). Actinomadura rubteroloni sp. nov. and Actinomadura macrotermitis sp. nov., isolated from the gut of the fungus growing-termite Macrotermes natalensis. Int. J. Syst. Evol. Microbiol. 70 5255–5262. 10.1099/ijsem.0.004403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benndorf R., Schwitalla J. W., Martin K., de Beer Z. W., Vollmers J., Kaster A. K., et al. (2020b). Nocardia macrotermitis sp. nov. and Nocardia aurantia sp. nov., isolated from the gut of the fungus-growing termite Macrotermes natalensis. Int. J. Syst. Evol. Microbiol. 70 5226–5234. 10.1099/ijsem.0.004398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi S. F., Li F., Song Y. C., Tan R. X., Ge H. M. (2011). New acrylamide and oxazolidin derivatives from a termite-associated Streptomyces sp. Nat. Prod. Commun. 6 353–355. [PubMed] [Google Scholar]
- Bierman M., Logan R., Obrien K., Seno E. T., Rao R. N., Schoner B. E. (1992). Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia Coli to Streptomyces spp. Gene 116 43–49. 10.1016/0378-1119(92)90627-2 [DOI] [PubMed] [Google Scholar]
- Blin K., Wolf T., Chevrette M. G., Lu X. W., Schwalen C. J., Kautsar S. A., et al. (2017). antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45 W36–W41. 10.1093/nar/gkx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blodgett J. A. V., Oh D. C., Cao S. G., Currie C. R., Kolter R., Clardy J. (2010). Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. U.S.A. 107 11692–11697. 10.1073/pnas.1001513107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodawatta K. H., Poulsen M., Bos N. (2019). Foraging Macrotermes natalensis fungus-growing termites avoid a mycopathogen but not an entomopathogen. Insects 10:185. 10.3390/insects10070185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredholt H., Fjaervik E., Johnsen G., Zotchev S. B. (2008). Actinomycetes from sediments in the Trondheim fjord, Norway: diversity and biological activity. Mar. Drugs 6 12–24. 10.3390/Md6010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro M. J., Poulsen M., Little A. E. F., Price S. L., Gerardo N. M., Wong B., et al. (2011). Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc. R. Soc. B Biol. Sci. 278 1814–1822. 10.1098/rspb.2010.2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr G., Poulsen M., Klassen J. L., Hou Y. P., Wyche T. P., Bugni T. S., et al. (2012). Microtermolides A and B from termite-associated Streptomyces sp. and structural revision of vinylamycin. Org. Lett. 14 2822–2825. 10.1021/ol301043p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapela I. H., Rehner S. A., Schultz T. R., Mueller U. G. (1994). Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science 266 1691–1694. 10.1126/science.266.5191.1691 [DOI] [PubMed] [Google Scholar]
- Chen S., Huang X., Zhou X., Bai L., He J., Jeong K. J., et al. (2003). Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem. Biol. 10 1065–1076. 10.1016/j.chembiol.2003.10.007 [DOI] [PubMed] [Google Scholar]
- Chevrette M. G., Carlos-Shanley C., Louie K. B., Bowen B. P., Northen T. R., Currie C. R. (2019a). Taxonomic and metabolic incongruence in the ancient genus Streptomyces. Front. Microbiol. 10:2170. 10.3389/Fmicb.2019.02170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrette M. G., Carlson C. M., Ortega H. E., Thomas C., Ananiev G. E., Barns K. J., et al. (2019b). The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 10:516. 10.1038/s41467-019-08438-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew Y. M., Lye S., Salleh M. M., Yahya A. (2018). 16S rRNA metagenomic analysis of the symbiotic community structures of bacteria in foregut, midgut, and hindgut of the wood-feeding termite Bulbitermes sp. Symbiosis 76 187–197. 10.1007/s13199-018-0544-5 [DOI] [Google Scholar]
- Currie C. R., Mueller U. G., Malloch D. (1999a). The agricultural pathology of ant fungus gardens. Proc. Natl. Acad. Sci. U.S.A. 96 7998–8002. 10.1073/pnas.96.14.7998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C. R., Scott J. A., Summerbell R. C., Malloch D. (1999b). Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398 701–704. 10.1038/19519 [DOI] [Google Scholar]
- Enagbonma B., Babalola O. O. (2019). Potentials of termite mound soil bacteria in ecosystem engineering for sustainable agriculture. Ann. Microbiol. 69 211–219. 10.1007/s13213-019-1439-2 [DOI] [Google Scholar]
- Felsenstein J. (1985). Confidence-limits on phylogenies - an approach using the bootstrap. Evolution 39 783–791. 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Fjaervik E., Zotchev S. B. (2005). Biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei. Appl. Microbiol. Biotechnol. 67 436–443. 10.1007/s00253-004-1802-4 [DOI] [PubMed] [Google Scholar]
- Gao H., Gruschow S., Barke J., Seipke R. F., Hill L. M., Orivel J., et al. (2014). Filipins: the first antifungal “weed killers” identified from bacteria isolated from the trap-ant. RSC Adv. 4 57267–57270. 10.1039/c4ra09875g [DOI] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., Smith H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6 343–345. 10.1038/Nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gopalkrishnan K. S., Narasimhachari N., Joshi V. B., Thirumalachar M. J. (1968). Chainin: a new methylpentaene antibiotic from a species of Chainia. Nature 218 597–598. 10.1038/218597a0 [DOI] [PubMed] [Google Scholar]
- Guo H., Schwitalla J. W., Benndorf R., Baunach M., Steinbeck C., Gorls H., et al. (2020). Gene cluster activation in a bacterial symbiont leads to halogenated angucyclic maduralactomycins and spirocyclic actinospirols. Org. Lett. 22 2634–2638. 10.1021/acs.orglett.0c00601 [DOI] [PubMed] [Google Scholar]
- Guo H. J., Kreuzenbeck N. B., Otani S., Garcia-Altares M., Dahse H. M., Weigel C., et al. (2016). Pseudoxylallemycins A-F, cyclic tetrapeptides with rare allenyl modifications isolated from Pseudoxylaria sp. X802: a competitor of fungus-growing termite cultivars. Org. Lett. 18 3338–3341. 10.1021/acs.orglett.6b01437 [DOI] [PubMed] [Google Scholar]
- Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100 1541–1546. 10.1073/pnas.0337542100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeder S., Wirth R., Herz H., Spiteller D. (2009). Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. U.S.A. 106 4742–4746. 10.1073/pnas.0812082106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa M., Iino H., Takeuchi S., Yamazaki T. (1997). Application of a method incorporating treatment with chloramine-T for the selective isolation of Streptosporangiaceae from soil. J. Ferment. Bioeng. 84 599–602. 10.1016/S0922-338x(97)81919-8 [DOI] [Google Scholar]
- He S., Johnston P. R., Kuropka B., Lokatis S., Weise C., Plarre R., et al. (2018). Termite soldiers contribute to social immunity by synthesizing potent oral secretions. Insect Mol. Biol. 27 564–576. 10.1111/imb.12499 [DOI] [PubMed] [Google Scholar]
- Heine D., Holmes N. A., Worsley S. F., Santos A. C. A., Innocent T. M., Scherlach K., et al. (2018). Chemical warfare between leafcutter ant symbionts and a co-evolved pathogen. Nat. Commun. 9:2208. 10.1038/S41467-018-04520-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter R. W., Cronin J. T., Klepzig K. D., Moser J. C., Ayres M. P. (2006). Antagonisms, mutualisms and commensalisms affect outbreak dynamics of the southern pine beetle. Oecologia 147 679–691. 10.1007/s00442-005-0312-0 [DOI] [PubMed] [Google Scholar]
- Human Z. R., Moon K., Bae M., de Beer Z. W., Cha S., Wingfield M. J., et al. (2016). Antifungal Streptomyces spp. associated with the infructescences of Protea spp. in South Africa. Front. Microbiol. 7:1657. 10.3389/Fmicb.2016.01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Y. M., Hsieh H. M. (2007). Xylaria species associated with nests of Odontotermes formosanus in Taiwan. Mycologia 99 936–957. 10.3852/mycologia.99.6.936 [DOI] [PubMed] [Google Scholar]
- Kim K. H., Ramadhar T. R., Beemelmanns C., Cao S., Poulsen M., Currie C. R., et al. (2014). Natalamycin A, an ansamycin from a termite-associated Streptomyces sp. Chem. Sci. 5 4333–4338. 10.1039/C4SC01136H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen J. L., Lee S. R., Poulsen M., Beemelmanns C., Kim K. H. (2019). Efomycins K and L from a termite-associated Streptomyces sp. M56 and their putative biosynthetic origin. Front. Microbiol. 10:1739. 10.3389/fmicb.2019.01739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yelle D. J., Li C., Yang M., Ke J., Zhang R., et al. (2017). Lignocellulose pretreatment in a fungus-cultivating termite. Proc. Natl. Acad. Sci. U.S.A. 114 4709–4714. 10.1073/pnas.1618360114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Young S. E., Poulsen M., Currie C. R. (2021). Symbiont-mediated digestion of plant biomass in fungus-farming insects. Annu. Rev. Entomol. 66 297–316. 10.1146/annurev-ento-040920-061140 [DOI] [PubMed] [Google Scholar]
- Li Z., Rawlings B. J., Harrison P. H., Vederas J. C. (1989). Production of new polyene antibiotics by Streptomyces cellulosae after addition of ethyl (Z)-16-phenylhexadec-9-enoate. J. Antibiot. 42 577–584. 10.7164/antibiotics.42.577 [DOI] [PubMed] [Google Scholar]
- Liu N., Zhang L., Zhou H. K., Zhang M. L., Yan X., Wang Q., et al. (2013). Metagenomic insights into metabolic capacities of the gut microbiota in a fungus-cultivating termite (Odontotermes yunnanensis). PLoS One 8:e69184. 10.1371/journal.pone.0069184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malviya N., Yandigeri M. S., Yadav A. K., Solanki M. K., Arora D. K. (2014). Isolation and characterization of novel alkali-halophilic actinomycetes from the Chilika brackish water lake, India. Ann. Microbiol. 64 1829–1838. 10.1007/s13213-014-0831-1 [DOI] [Google Scholar]
- Mattoso T. C., Moreira D. D. O., Samuels R. I. (2012). Symbiotic bacteria on the cuticle of the leaf-cutting ant Acromyrmex subterraneus subterraneus protect workers from attack by entomopathogenic fungi. Biol. Lett. 8 461–464. 10.1098/rsbl.2011.0963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mburu D. M., Ochola L., Maniania N. K., Njagi P. G. N., Gitonga L. M., Ndung’u M. W., et al. (2009). Relationship between virulence and repellency of entomopathogenic isolates of Metarhizium anisopliae and Beauveria bassiana to the termite Macrotermes michaelseni. J. Insect Physiol. 55 774–780. 10.1016/j.jinsphys.2009.04.015 [DOI] [PubMed] [Google Scholar]
- Mendes M. V., Recio E., Fouces R., Luiten R., Martin J. F., Aparicio J. F. (2001). Engineered biosynthesis of novel polyenes: a pimaricin derivative produced by targeted gene disruption in Streptomyces natalensis. Chem. Biol. 8 635–644. 10.1016/s1074-5521(01)00033-3 [DOI] [PubMed] [Google Scholar]
- Mevers E., Chouvenc T., Su N. Y., Clardy J. (2017). Chemical interaction among termite-associated microbes. J. Chem. Ecol. 43 1078–1085. 10.1007/s10886-017-0900-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H. K., Choi Y. J., Ha J. K., Cho K. K., Lee S. S. (1994). Isolation and identification of anaerobic rumen bacterium, Actinomyces sp. 40 and enzymatic properties of β-1,4-endoglucanase. Asian Austral. J. Anim. 7 373–382. 10.5713/ajas.1994.373 [DOI] [Google Scholar]
- Mincer T. J., Jensen P. R., Kauffman C. A., Fenical W. (2002). Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl. Environ. Microb. 68 5005–5011. 10.1128/Aem.68.10.5005-5011.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. C., Poulsen M., Currie C. R., Clardy J. (2009a). Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol. 5 391–393. 10.1038/nchembio.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. C., Scott J. J., Currie C. R., Clardy J. (2009b). Mycangimycin, a polyene peroxide from a mutualist Streptomyces sp. Org Lett. 11 633–636. 10.1021/ol802709x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani S., Challinor V. L., Kreuzenbeck N. B., Kildgaard S., Christensen S. K., Larsen L. L. M., et al. (2019). Disease-free monoculture farming by fungus-growing termites. Sci. Rep. 9:8819. 10.1038/S41598-019-45364-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani S., Mikaelyan A., Nobre T., Hansen L. H., Kone N. A., Sorensen S. J., et al. (2014). Identifying the core microbial community in the gut of fungus-growing termites. Mol. Ecol. 23 4631–4644. 10.1111/mec.12874 [DOI] [PubMed] [Google Scholar]
- Poulsen M. (2015). Towards an integrated understanding of the consequences of fungus domestication on the fungus-growing termite gut microbiota. Environ. Microbiol. 17 2562–2572. 10.1111/1462-2920.12765 [DOI] [PubMed] [Google Scholar]
- Poulsen M., Hu H. F., Li C., Chen Z. S., Xu L. H., Otani S., et al. (2014). Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc. Natl. Acad. Sci. U.S.A. 111 14500–14505. 10.1073/pnas.1319718111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadhar T. R., Beemelmanns C., Currie C. R., Clardy J. (2014). Bacterial symbionts in agricultural systems provide a strategic source for antibiotic discovery. J. Antibiot. 67 53–58. 10.1038/ja.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt-Wagner D., Friedrich M. W., Wagner B., Brune A. (2003). Axial dynamics, stability, and interspecies similarity of bacterial community structure in the highly compartmentalized gut of soil-feeding termites (Cubitermes spp.). Appl. Environ. Microb. 69 6018–6024. 10.1128/Aem.69.10.6018-6024.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenian I., Spiteller M., Ghaste M., Wirth R., Herz H., Spiteller D. (2011). Chemical basis of the synergism and antagonism in microbial communities in the nests of leaf-cutting ants. Proc. Natl. Acad. Sci. U.S.A. 108 1955–1960. 10.1073/pnas.1008441108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla J. W., Benndorf R., Martin K., Vollmers J., Kaster A. K., de Beer Z. W., et al. (2020). Streptomyces smaragdinus sp. nov., isolated from the gut of the fungus growing-termite Macrotermes natalensis. Int. J. Syst. Evol. Microbiol. 70 5806–5811. 10.1099/ijsem.0.004478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. J., Oh D. C., Yuceer M. C., Klepzig K. D., Clardy J., Currie C. R. (2008). Bacterial protection of beetle-fungus mutualism. Science 322:63. 10.1126/science.1160423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R., Ishak H. D., Estrada D., Dowd S. E., Hong E., Mueller U. G. (2009). Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc. Natl. Acad. Sci. U.S.A. 106 17805–17810. 10.1073/pnas.0904827106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirling E. B., Gottlieb D. (1966). Methods for characterizing Streptomyces species. Int. J. Syst. Bacteriol. 16 313–340. 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- Sit C. S., Ruzzini A. C., Van Arnam E. B., Ramadhar T. R., Currie C. R., Clardy J. (2015). Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc. Natl. Acad. Sci. U.S.A. 112 13150–13154. 10.1073/pnas.1515348112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser H., Vey A., Butt T. M. (2000). Are there any risks in using entomopathogenic fungi for pest control, with particular reference to the bioactive metabolites of Metarhizium, Tolypocladium and Beauveria species? Biocontrol Sci. Technol. 10 717–735. 10.1080/09583150020011690 [DOI] [Google Scholar]
- Subramani R., Aalbersberg W. (2013). Culturable rare actinomycetes: diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 97 9291–9321. 10.1007/s00253-013-5229-7 [DOI] [PubMed] [Google Scholar]
- Sun J., Liu X. H., Jiang G. S., Qi Q. G. (2015). Inhibition of nucleic acid biosynthesis makes little difference to formation of amphotericin B-tolerant persisters in Candida albicans biofilm. Antimicrob. Agents Chemother. 59 1632–1638. 10.1128/Aac.03765-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalachar M. J. (1955). Chainia, a new genus of the Actinomycetales. Nature 176 934–935. 10.1038/176934b0 [DOI] [PubMed] [Google Scholar]
- Um S., Fraimout A., Sapountzis P., Oh D. C., Poulsen M. (2013). The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp that inhibit potentially antagonistic fungi. Sci. Rep. 3:3250. 10.1038/Srep03250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arnam E. B., Currie C. R., Clardy J. (2018). Defense contracts: molecular protection in insect-microbe symbioses. Chem. Soc. Rev. 47 1638–1651. 10.1039/c7cs00340d [DOI] [PubMed] [Google Scholar]
- van Arnam E. B., Ruzzini A. C., Sit C. S., Currie C. R., Clardy J. (2015). A rebeccamycin analog provides plasmid-encoded niche defense. J. Am. Chem. Soc. 137 14272–14274. 10.1021/jacs.5b09794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arnam E. B., Ruzzini A. C., Sit C. S., Horn H., Pinto-Tomas A. A., Currie C. R., et al. (2016). Selvamicin, an atypical antifungal polyene from two alternative genomic contexts. Proc. Natl. Acad. Sci. U.S.A. 113 12940–12945. 10.1073/pnas.1613285113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser A. A., Nobre T., Currie C. R., Aanen D. K., Poulsen M. (2012). Exploring the potential for Actinobacteria as defensive symbionts in fungus-growing termites. Microb. Ecol. 63 975–985. 10.1007/s00248-011-9987-4 [DOI] [PubMed] [Google Scholar]
- Wang J., Li W., Wang H., Lu C. (2018). Pentaketide ansamycin microansamycins A-I from Micromonospora sp. reveal diverse post-PKS modifications. Org. Lett. 20 1058–1061. 10.1021/acs.orglett.7b04018 [DOI] [PubMed] [Google Scholar]
- Wang L., Feng Y., Tian J., Xiang M., Sun J., Ding J., et al. (2015). Farming of a defensive fungal mutualist by an attelabid weevil. ISME J. 9 1793–1801. 10.1038/ismej.2014.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthen D. R., Jay M., Bummer P. M. (2001). Methods for the recovery and purification of polyene antifungals. Drug Dev. Ind. Pharm. 27 277–286. 10.1081/Ddc-100103727 [DOI] [PubMed] [Google Scholar]
- Wu Y., Chi S., Yun C., Shen Y., Tokuda G., Ni J. (2012). Molecular cloning and characterization of an endogenous digestive beta-glucosidase from the midgut of the fungus-growing termite Macrotermes barneyi. Insect Mol. Biol. 21 604–614. 10.1111/j.1365-2583.2012.01164.x [DOI] [PubMed] [Google Scholar]
- Xiong Z. Q., Zhang Z. P., Li J. H., Wei S. J., Tu G. Q. (2012). Characterization of Streptomyces padanus JAU4234, a producer of actinomycin X-2, fungichromin, and a new polyene macrolide antibiotic. Appl. Environ. Microb. 78 589–592. 10.1128/Aem.06561-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Shao M., Yin C., Mao Z., Shi J., Yu X., et al. (2020). Diversity, bacterial symbionts, and antimicrobial potential of termite-associated fungi. Front. Microbiol. 11:300. 10.3389/fmicb.2020.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Jin L., Li S., Xu X., Zhang Y. (2019). Diversity and antagonistic potential of actinobacteria from the fungus-growing termite Odontotermes formosanus. 3 Biotech 9:45. 10.1007/s13205-019-1573-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Song T., Wu J., Zhang S. X., Yin C. P., Huang F., et al. (2020). Antibacterial and cytotoxic metabolites of termite-associated Streptomyces sp. BYF63. J. Antibiot. 73 766–771. 10.1038/s41429-020-0334-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y. L., Li S., Jiang D. H., Kong L. C., Zhang P. H., Xu J. D. (2013). Antifungal activities of metabolites produced by a termite-associated Streptomyces canus BYB02. J. Agric. Food Chem. 61 1521–1524. 10.1021/jf305210u [DOI] [PubMed] [Google Scholar]
- Zhou S., Song L., Masschelein J., Sumang F. A. M., Papa I. A., Zulaybar T. O., et al. (2019). Pentamycin biosynthesis in Philippine Streptomyces sp. S816: cytochrome P450-catalyzed installation of the C-14 hydroxyl group. ACS Chem. Biol. 14 1305–1309. 10.1021/acschembio.9b00270 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete 16S rRNA genes of 83 isolated actinomycetes in this study were deposited in GenBank (MN826234-MN826316). The GenBank accession numbers for the genomes of the three strains are CP047144-CP047145 (Streptomyces sp. HF10), CP047146 (Streptomyces sp. GS7), and CP047147 (Streptomyces sp. GF20).