Abstract
Objective
To investigate the expression and clinical value of the E-selectin gene (SELE) in colorectal cancer (CRC).
Methods
Using gene expression profiles and clinicopathological data for patients with CRC from The Cancer Genome Atlas, and tumor and adjacent normal tissues from 31 patients with CRC from Xianyang Central Hospital, we studied the correlation between SELE gene expression and clinical parameters using Kaplan–Meier and Cox proportional hazards regression analyses.
Results
Higher expression of SELE was significantly associated with a poorer prognosis and shorter survival in patients with CRC. The median expression level of SELE was significantly higher in CRC tissues compared with healthy adjacent tissue. Cox regression analysis showed that the prognosis of CRC was significantly correlated with the expression of SELE. Immunohistochemical analysis also showed that positive expression of E-selectin increased significantly in line with increasing TNM stage.
Conclusion: This study confirmed that SELE gene expression is an independent prognostic factor in patients with CRC.
Keywords: Colorectal cancer, SELE, The Cancer Genome Atlas, transcript profiling, clinical prognosis, survival
Introduction
Colorectal cancer (CRC) is a major threat to human health, with about two million new cases of CRC worldwide in 2018.1 CRC is the third most common malignant tumor globally, accounting for 690,000 deaths, and the fourth most common malignant tumor in China. The incidence of CRC is increasing rapidly in the eastern developed region of China, accounting for 18.6% of the total number of cases worldwide. The 5-year survival rate of CRC in China is also significantly lower than that in developed countries in Europe and North America, mainly due to the difficulty in early diagnosis and the presence of distant metastases. Early diagnosis, prevention, and treatment of metastases are therefore key to improving the prognosis and survival rate of patients with CRC. Gene testing aids targeted therapy, and the prompt selection of targeted drugs with specific efficacy represents an economic saving for the patient, as well as avoiding delays with disease treatment.
The prognostic values of KRAS and BRAF gene2 mutations have remained controversial. In addition, the SELE gene encodes E-selectin, which is found in cytokine-stimulated endothelial cells and is thought to be responsible for the accumulation of leukocytes at sites of inflammation by mediating the adhesion of cells to the vascular wall. Its structure includes lectin- and epidermal growth factor-like domains followed by short consensus repeat domains containing six conserved cysteine residues. E-selectin is part of the selectin family of cell adhesion molecules, which are glycoprotein sugar chains or sheath sugars found on the surface of white blood cells and tumor cells. Selectin ligands include saliva-acidized Lewis oligosaccharides (sLeX and sLeA) and other molecules with similar structures,3 and the interaction between selectin and these ligands and the subsequent adhesion response have been shown to play an essential role in tumor invasion and metastasis.4 Numerous studies5–7have shown that sLeX and sLeA antigens are expressed on the surface of tumor cells, and can combine with selectins on the surface of vascular endothelial cells to mediate the adhesion of tumor cells to endothelial cells. Stronger expression is therefore associated with more intense adhesion and a greater ability to form metastatic foci in vitro.8,9 Studies4,10have shown that selectin and ligand-mediated adhesion play an essential role in tumor cell metastasis. An in-depth study of selectin expression will therefore aid the early diagnosis of CRC, evaluation of its prognosis, and the search for new therapeutic targets. The current study analyzed the prognostic role of SELE in CRC based on the analysis of data from The Cancer Genome Atlas (TCGA) database.
Materials and Methods
Transcript profiling of SELE gene
We analyzed data for patients with colon cancer (TCGA-COAD) and rectal cancer (TCGA-READ) from the TCGA database11 (https://portal.gdc.cancer.gov/). We successively selected patients with adenomas and adenocarcinomas, with quantitative transcript data suitable for assessing gene expression. Finally, we chose Fragments per kilobase of exon model per million mapped fragments (FPKM) as the workflow type to plot single gene expression. After downloading and collating the data, we used the R package “limma” to extract the data and obtain the mean value for any repeated genes. The “Clinical” option was selected to extract clinical data.
Expression of SELE gene using correlation function by Gene Expression Profiling Interactive Analysis (GEPIA)
GEPIA12 contains genotype–tissue expression and TCGA data (http: //gepia.cancer-pku.cn/). We evaluated the expression of SELE and other characteristic genes in CRC (TP53, ERCC1, KRAS, NRAS, BRAF, SMAD4, APC, SOX9, MRC1, MSH2, NDRG4, BMP3, VIM, KDR, EGFR, TFPI2) using the correlation function of GEPIA.
Immunohistochemical validation
Study subjects
We collected information on patients with colon cancer treated at Xianyang Central Hospital in Shaanxi between January 2019 and June 2019. Patients with postoperative pathological information including tumor size, differentiation degree, TNM stage, and immunohistochemical detection of E-selectin were followed-up from postoperative discharge for 4 to 6 months, until January 2020. The pathology was reviewed independently by three pathologists without access to the clinical data. The study was approved by Xianyang Central Hospital ethics committee (Approval number 20190037), and all patients signed informed consent.
Immunohistochemistry
Ethanol, phosphate-buffered saline (PBS), 3% H2O2, 10% normal goat serum, 0.01 M citrate buffer, xylene, and antibodies were provided by Maxim Biotechnology Co. Ltd. (Fuzhou, China). DAB chromogenic solution (Catalog number AR1022) was purchased from Boster Biological Technology Co. Ltd. (Wuhan, China). Rabbit anti-E-selectin antibody (1:500; Catalog number-1273R) was purchased from Beijing Biosynthesis Biotechnology Co. Ltd. (Beijing, China). Horseradish peroxidase-conjugated Affinipure goat anti-rabbit IgG (1:2000; Catalog number sa00001-2) was purchased from Proteintech (Wuhan, China).
Paraffin-embedded tissue slices were dewaxed and subjected to antigen repair using citrate buffer solution in a pressure cooker with air-injection for 2 minutes, and cooled to room temperature. Endogenous peroxidase activity was blocked by incubation at room temperature with 3% H2O2 for 10 minutes, followed by the addition of 5% bovine serum albumin (BSA) to the slices for 30 minutes at room temperature to block non-specific antigen reactions. The primary antibody was then added to the working solution at a dilution of 1:500, 1% BSA, incubated overnight at 4°C, and then washed three times with PBS for 3 minutes each. Each section was then incubated with two drops of horseradish peroxidase-conjugated Affinipure goat anti-rabbit IgG (dilution ratio 1:2000, 1% BSA dilution) for 30 minutes at room temperature, rinsed three times in PBS for 3 minutes each, and DAB color was developed, followed by hematoxylin staining and covering with a neutral resin seal. The slides were observed and photographed under a light microscope. E-selectin-positive signals were indicated by brownish yellow color. Less than 10% of positive cells was considered as a negative result, and ≥10% was considered positive.
Statistical analysis
Data were analyzed using SPSS Statistics for Windows, Version 22.0 (IBM Corp., Armonk, NY, USA) and R (version 3.6) software (www.R-project.org), and graphs were produced using GraphPad Prism (version 5.0) (GraphPad Prism Inc., La Jolla, CA, USA). We carried out single-gene data analysis using R (www.R-project.org) and the Bioconductor package13 (www.bioconductor.org). We analyzed SELE expression using t-tests and Kruskal–Wallis tests. Clinicopathological correlation analysis was conducted using logistic regression analysis, and survival was evaluated by Kaplan–Meier analysis and log-rank test. Multivariate analysis was carried out using a Cox proportional risk regression model, and Pearson’s correlation analysis was used to assess correlations between genes and related factors. A P-value <0.05 was considered significant.
Results
SELE gene expression in CRC and adjacent healthy tissues
Data for 31 patients with colon cancer (18 men, 13 women; median age 72 years, range 46–88 years), including four cases of metastasis, treated at Xianyang Central Hospital between January 2019 and June 2019 were collected, according to a random number table. Seventy-two samples including carcinoma and healthy adjacent tissue were available. We compared SELE expression levels between CRC and healthy tissues (Figure 1a). Compared with healthy adjacent tissues, the median level of SELE gene expression was significantly higher in the tumor samples (P< 0.001). Matched pair analysis between CRC and healthy tissues (Figure 1b) in the same patient showed that SELE was significantly overexpressed in CRC samples (P < 0.001).
Figure 1.
Differential expression of E-selectin gene (SELE) in relation to survival and clinical stage in patients with colorectal cancer (CRC). (a) Scatter plot of SELE gene expression in CRC and healthy adjacent tissues. (b) SELE expression in paired CRC and healthy adjacent tissues in the same patient. (c) Survival analysis in relation to SELE gene expression based on The Cancer Genome Atlas data. (d) Differential expression of SELE gene in patients with different stages of CRC (Kruskal–Wallis test).
Kaplan–Meier survival analysis
We initially identified 475 patients with colon or rectal adenoma/adenocarcinoma from TCGA database (https://portal.gdc.cancer.gov). After excluding patients with missing data, we obtained 332 samples with enough data to conduct survival analysis (average follow-up 2.8 years, longest 12 years). After analyzing and collating the data, we obtained survival times and survival status and evaluated the correlation between OS and SELE gene expression (Figure 1c). Survival gradually declined over time. We also divided cases into low and high SELE expression according to the median value (0.51), then assessed survival according to high or low SELE expression. The 5-year OS rate was significantly higher in the low-expression group (0.704, 95% confidence interval [CI] 0.577–0.860) compared with the high-expression group (0.526, 95% CI 0.385–0.718) (P=0.017). Patient survival time could thus be predicted according to the expression level of the SELE gene, with patients with low expression levels more likely to survive for >5 years, and patients with high levels more likely to survive for <5 years.
Clinical correlation analysis
We investigated SELE gene expression in relation to clinical stage using Kruskal–Wallis and logistic regression analyses (Figure 1d). There was no significant correlation between CRC stage and SELE gene expression.
SELE gene expression had stronger prognosis prediction ability than TNM stage or other clinical features in CRC
We analyzed SELE gene, age, sex, clinical stage, primary lesion, increased involvement of adjacent tissues (T), distant metastasis (M), and regional lymph node involvement (N) as independent factors related to clinical survival using the survival package in single-factor Cox regression analysis (Table 1). Univariate analysis identified SELE gene expression, age, clinical stage, primary tumor, increased involvement of adjacent tissues (T), distant metastasis (M), and regional lymph node involvement (N) as independent prognostic factors (all P < 0.05). However, sex had no significant effect on the prognosis of CRC.
Table 1.
Univariate and multivariate Cox analyses of clinical parameters in relation to overall survival.
| Variable |
Univariate Cox analysis |
Multivariate Cox analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | P value | HR | 95%CI | P value | |||
| Age | 1.029 | 1.005 | 1.055 | 0.017 | 1.040 | 1.015 | 1.066 | 0.001 |
| Sex | 1.126 | 0.668 | 1.899 | 0.655 | 0.995 | 0.582 | 1.699 | 0.984 |
| Stage | 2.502 | 1.850 | 3.382 | <0.001 | 1.614 | 0.672 | 3.875 | 0.283 |
| T | 2.926 | 1.742 | 4.917 | <0.001 | 1.453 | 0.769 | 2.748 | 0.249 |
| M | 5.226 | 3.065 | 8.911 | <0.001 | 1.699 | 0.517 | 5.587 | 0.382 |
| N | 2.175 | 1.608 | 2.941 | <0.001 | 1.353 | 0.800 | 2.286 | 0.258 |
| SELE | 1.287 | 1.062 | 1.559 | 0.009 | 1.312 | 1.077 | 1.597 | 0.0067 |
HR, hazard ratio; CI, confidence interval.
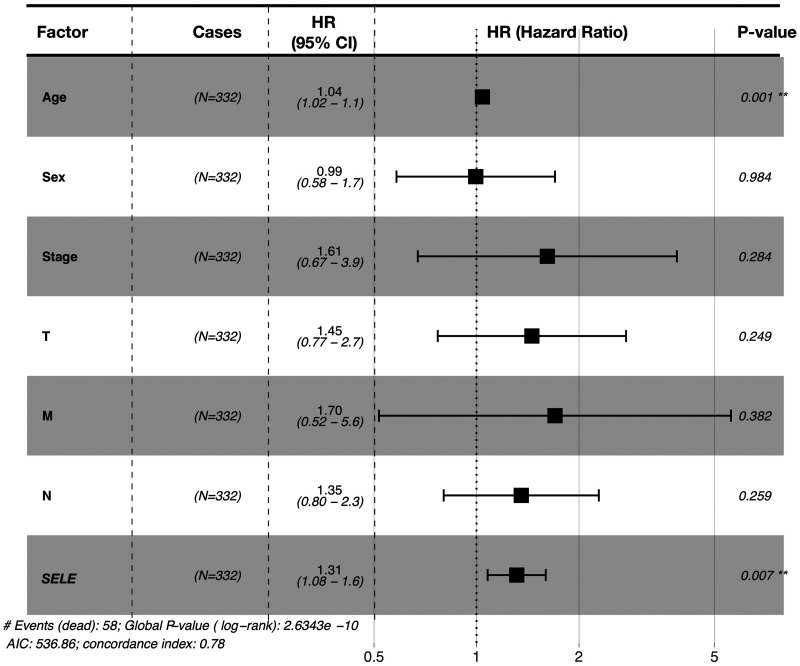
We then carried out multivariate Cox regression analysis using the R package “survminer” (Figure 2). This indicated that SELE gene expression (P = 0.0067) and age (P = 0.0014), but not clinical stage or TNM stage, were significantly and independently correlated with a prognosis of CRC.
Figure 2.
Multivariate analysis of clinicopathological factors affecting overall survival in patients with colorectal cancer (**P < 0.05).
AIC, Akaike information criterion.
Association between SELE and key genes in CRC
We also analyzed the correlations between expression levels of SELE and mismatch repair genes using the correlation analysis function in the GEPIA database. We analyzed the correlations between expression levels of SELE and MLH1, MSH2, MSH6, and PMS2 mismatch repair genes separately (Figure 3). SELE was significantly correlated with MSH2 expression (P<0.05). The American Cancer Society recommends targeted therapy and chemotherapy as the two main drug therapies for CRC. Based on the NCCN guidelines of the United States,14 experts in the field strongly recommended that all patients diagnosed with stage IV metastasis should undergo RAS (KRAS, NRAS) and BRAF gene-status detection in cancer tissues. We therefore analyzed the expression levels of SELE, RAS (KRAS, NRAS), and BRAF genes,15 and expression of the vascular endothelial growth factor receptor gene (KDR), ERCC1, and UGT1A1*28/*6 gene polymorphism.16 SELE was significantly correlated with expression of NRAS BRAF, and KDR17 (P<0.05). We also analyzed the correlations between SELE expression and expression of a series of CRC-related genes including SMAD4, APC, TP53, NDRG4, MRC1, BMP3, VIM, TFPI2, and SOX9. SMAD4, APC, TP53, NDRG4, MRC1, VIM, TFPI2, and SOX9 were significantly correlated with SELE gene expression (all P<0.05).
Figure 3.
Correlations between SELE and key genes in colorectal cancer. SELE gene expression was significantly correlated with expression of MSH2, NRAS, BRAF, KDR, SMAD4, APC, TP53, NDRG4, MRC1, VIM, TFPI2, and SOX9 genes (P < 0.05).
TPM, transcripts per million kilobases.
Immunohistochemical validation
Positive E-selectin staining indicated by brown-yellow granules was located in the cytoplasm and cell membrane in both cancer cells and vascular endothelial cells, but not in the negative control group (0.1 mol/L PBS) (Figure 4). The incidence of positive E-selectin expression was significantly higher in cancer cells compared with healthy intestinal mucosal glandular epithelial cells (P<0.05, rank sum test). The incidence of positive E-selectin expression, calculated according to the product of the number of positive cells and staining intensity, changed significantly with increasing TNM stage (P<0.05). We also analyzed the correlations between E-selectin and clinical features and prognosis, which confirmed the result of TCGA analysis. SELE gene expression levels were higher in CRC compared with healthy adjacent tissues, and increased with increasing stage, with a higher SELE expression level indicating a worse prognosis.
Figure 4.
Positive E-selectin signals were detected in the cytoplasm and/or cell membrane, presenting as brownish-yellow granules, in cancer cells and vascular endothelial cells. The incidence of E-selectin positivity was significantly higher in cancer cells compared with healthy intestinal mucosal glandular epithelial cells (P < 0.05). E-selectin expression increased significantly with increasing TNM stage (P < 0.05).
Discussion
We analyzed transcript profiling and related clinical data for patients with CRC in TCGA database, to investigate the correlation between SELE gene expression and prognosis in CRC. We showed that SELE gene expression levels were significantly higher in CRC tissues compared with adjacent healthy tissues. We also analyzed survival, clinicopathological, and SELE gene expression data in relation to CRC using TCGA database. Univariate Cox regression analysis suggested that SELE gene expression levels differed significantly between colon cancer tissues in relation to tumor size, invasion depth, and distant metastasis, with higher expression associated with increased tumor differentiation degree, lymph node metastasis, and clinical T, N, and M stages. These results suggest that high expression of the SELE gene may promote malignant biological behavior in colon cancer.
Various factors affect the prognosis of CRC. The current study showed that the poorer prognosis in patients with high expression of SELE was associated with shorter survival, determined by multiple regression and Kaplan–Meier analyses. The value of SELE gene expression was independent of T, N, and M stages and other clinical data, indicating that is was an essential prognostic factor in CRC. The application of anti-tumor drugs significantly down-regulated SELE expression levels in different colon cancer cell lines.18 Overall, these results suggest that SELE may act as an oncogene.
Myeloid cells include macrophages, neutrophils, acidic granulocytes, mast cells, and dendritic cells. Myeloid cells exist in peripheral circulating blood and are recruited into tumor tissue to promote angiogenesis in the tumor microenvironment.19 Granulocytes, myeloid cells, tumor cells, and vascular endothelial cell membranes all secrete E-selectin. In addition, considering the crucial role of tumor angiogenesis in tumor development, cancer cells have been shown to secrete inflammatory cytokines such as interleukin-1β and tumor necrosis factor-α, to induce SELE at sites of distant metastasis, and inhibit the anti-tumor adaptive immune response.20 The current results showed that increased expression of SELE led to a poorer clinical outcome and shorter survival.
This study provided conclusive evidence for increased expression of SELE as a clinical adverse prognostic factor in CRC. Survival analysis and univariate and multivariate Cox regression analyses showed consistent results, implicating SELE as an oncogene for CRC, as supported by other related studies.10 Numerous studies21–23 confirmed that SELE was strongly linked to the generation and invasion of CRC in small samples. Other researchers24 detected preoperative serum E-selection and CA19-9 in 152 patients with CRC and 28 healthy volunteers, and showed that patients with higher serum expression levels of E-selection and CA19-9 had a significantly lower 5-year overall survival rate. Researchers have also used E-selectin as a marker of hematogenous metastasis and prognosis in CRC. SELE expression was analyzed in 202 patients with different clinical stages of colon cancer with lymphatic vessel invasion and intestinal perforation treated with oxaliplatin-assisted chemotherapy, using sequence analysis.25 Positive results occurred in patients with phase II and III colon cancer, suggesting that clinicians could use SELErs3917412 as a predictor for intestinal cancer. Although some studies found that the average level of selectin expression was significantly higher in CRC patients than in healthy subjects, E-selectin expression levels decreased in a stepwise manner in patients with liver metastasis, patients with lymph node metastasis, and patients without metastasis.10 The current study analyzed transcriptome profiling and clinically relevant data for CRC from TCGA database, which provided more reliable results compared with the above small-sample study.
The possible signaling pathways underlying these mechanisms are not clear. Some studies suggested that the interaction between HT29 colon cancer cells expressing DR3 and SELE induced and activated the phosphoinositide 3-kinase (PI3K)/Akt pathway,26 which is the mechanism of activation of colon cancer cells. DR3 stimulation increased the viability of colon cancer cells by activating the PI3K/nuclear factor-kappa B pathway. SELE gene expression was closely related to tumor-related circulating endothelial cells in 55 healthy donors and 81 metastatic tumors27 (including colon and rectal cancers). Our results also indicated that SELE might be a biomarker of poor prognosis and survival in patients with CRC. The mechanism may involve Toll-like receptor,28 JAK-STAT,29 and transforming growth factor-β pathway pathways,30 pathways in cancer,31 and Nod-like receptor signaling28 and other signaling pathways related to CRC.32 In addition, through the GEPIA database, our results confirmed that SELE expression was significantly correlated with the expression of other CRC-related genes, further supporting the scientific value of SELE gene expression in CRC.
Although our study showed encouraging results, there were some limitations. The study was mainly based on bioinformatics methods and immunohistochemical validation in a small cohort. More external verification is therefore required before clinical application. In addition, further studies are needed to clarify the detailed mechanism of SELE in CRC.
In summary, our study confirmed the importance of SELE and SELE-related genes in the occurrence and development of CRC, thus laying the foundation for further research into the mechanism of SELE in CRC.
Footnotes
Declaration of conflicting interest: The authors declare no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD: Na Li https://orcid.org/0000-0001-6821-2518
References
- 1.Ferlay J, Colombet M, Soerjomataram I, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2019; 144: 1941–1953. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 3.Koi M, Okita Y, Carethers JM. Fusobacterium nucleatum infection in colorectal cancer: linking inflammation, DNA mismatch repair and genetic and epigenetic alterations. J Anus Rectum Colon 2018; 2: 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paschos KA Canovas D, andBird NC.. The engagement of selectins and their ligands in colorectal cancer liver metastases. J Cell Mol Med et al. ; 14: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paavonen T, Renkonen R. Selective expression of sialyl-Lewis x and Lewis a epitopes, putative ligands for L-selectin, on peripheral lymph-node high endothelial venules. Am J Pathol et al. ; 141: 1259–1264. [PMC free article] [PubMed] [Google Scholar]
- 6.Borentain P, Carmona S, Mathieu S, et al. Inhibition of E-selectin expression on the surface of endothelial cells inhibits hepatocellular carcinoma growth by preventing tumor angiogenesis. Cancer Chemother Pharmacol et al. ; 77: 847–856. [DOI] [PubMed] [Google Scholar]
- 7.Starzonek S, Maar H, Labitzky V, et al. Systematic analysis of the human tumor cell binding to human vs. murine E-and P-selectin under static vs. dynamic conditions. Glycobiology et al. ; 30: 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nosho K, Sukawa Y, Adachi Y, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol 2016; 22: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korphaisarn K, Pongpaibul A, Limwongse C, et al . Deficient DNA mismatch repair is associated with favorable prognosis in Thai patients with sporadic colorectal cancer. World J Gastroenterol 2015; 21: 926–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korniluk A, Kamińska J, Kiszło P, et al. Lectin adhesion proteins (P-, L-and E-selectins) as biomarkers in colorectal cancer. Biomarkers et al. ; 22: 629–634. [DOI] [PubMed] [Google Scholar]
- 11.Reilly NM, Novara L, Di Nicolantonio F, et al . Exploiting DNA repair defects in colorectal cancer. Mol Oncol 2019; 13: 681–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45: W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sepulveda JL. Using R and Bioconductor in clinical genomics and transcriptomics. J Mol Diagn 2020; 22: 3–20. [DOI] [PubMed] [Google Scholar]
- 14.Provenzale D, Gupta S, Ahnen DJ, et al. NCCN Guidelines Insights: Colorectal Cancer Screening, Version 1.2018. J Natl Compr Canc Netw 2018; 16: 939–949. [DOI] [PubMed] [Google Scholar]
- 15.Cremolini C, Rossini D, Dell'Aquila E, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol 2019; 5: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C, Tang X, Qu Y, et al. UGT1A1 gene polymorphism is associated with toxicity and clinical efficacy of irinotecan-based chemotherapy in patients with advanced colorectal cancer. Cancer Chemother Pharmacol 2016; 78: 119–130. [DOI] [PubMed] [Google Scholar]
- 17.Price TJ, Tang M, Gibbs P, et al. Targeted therapy for metastatic colorectal cancer. Expert Rev Anticancer Ther 2018; 18: 991–1006. [DOI] [PubMed] [Google Scholar]
- 18.Lee CY, Hsieh SL, Hsieh S, et al . Inhibition of human colorectal cancer metastasis by notoginsenoside R1, an important compound from Panax notoginseng. Oncol Rep 2017; 37: 399–407. [DOI] [PubMed] [Google Scholar]
- 19.Murdoch C, Muthana M, Coffelt SB, et al. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer 2008; 8: 618–631. [DOI] [PubMed] [Google Scholar]
- 20.Pang X, Wang Y, Liu M. M1-macrophage polarization is upregulated in deep vein thrombosis and contributes to the upregulation of adhesion molecules. Hum Immunol 2019; 80: 883–889. [DOI] [PubMed] [Google Scholar]
- 21.Turpin A, Labreuche J, Fléjou JF, et al. Prognostic factors in patients with stage II colon cancer: Role of E-selectin gene polymorphisms. Dig Liver Dis et al. ; 51: 1198–1201. [DOI] [PubMed] [Google Scholar]
- 22.Deschepper FM, Zoppi R, Pirro M, et al. L1CAM as an E-selectin ligand in colon cancer. Int J Mol Sci et al. ; 21: 8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoppi R. Validation of novel biomarkers for colorectal cancer detection and production of novel antibodies against E-selectin ligands et al. http://hdl.handle.net/10362/100622.
- 24.Sato H, Usuda N, Kuroda M, et al. Significance of serum concentrations of E-selectin and CA19-9 in the prognosis of colorectal cancer. Jpn J Clin Oncol 2010; 40: 1073–1080. [DOI] [PubMed] [Google Scholar]
- 25.Custodio A, Moreno-Rubio J, Aparicio J, et al. Pharmacogenetic predictors of outcome in patients with stage II and III colon cancer treated with oxaliplatin and fluoropyrimidine-based adjuvant chemotherapy. Mol Cancer Ther 2014; 13: 2226–2237. [DOI] [PubMed] [Google Scholar]
- 26.Porquet N, Poirier A, Houle F, et al. Survival advantages conferred to colon cancer cells by E-selectin-induced activation of the PI3K-NFκB survival axis downstream of Death receptor-3. BMC Cancer 2011; 11: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smirnov DA, Foulk BW, Doyle GV, et al. Global gene expression profiling of circulating endothelial cells in patients with metastatic carcinomas. Cancer Res 2006; 66: 2918–2922. [DOI] [PubMed] [Google Scholar]
- 28.Wan M, Liu JR, Wu D, et al. E-selectin expression induced by Porphyromonas gingivalis in human endothelial cells via nucleotide-binding oligomerization domain-like receptors and Toll-like receptors. Mol Oral Microbiol 2015; 30: 399–410. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang X, Song Q, et al. Characterization of the chromatin accessibility in an Alzheimer's disease (AD) mouse model. Alzheimers Res Ther 2020; 12: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrascal MA, Silva M, Ramalho JS, et al. Inhibition of fucosylation in human invasive ductal carcinoma reduces E-selectin ligand expression, cell proliferation, and ERK1/2 and p38 MAPK activation. Mol Oncol 2018; 12: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi K, Li N, Yang M, et al. Identification of key genes and pathways in female lung cancer patients who never smoked by a bioinformatics analysis. J Cancer 2019; 10: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Wang YB. Systematic large-scale meta-analysis identifies miRNA-429/200a/b and miRNA-141/200c clusters as biomarkers for necrotizing enterocolitis in newborn. Biosci Rep 2019; 39: BSR20191503. [DOI] [PMC free article] [PubMed] [Google Scholar]