Abstract
Smad2 and Smad3 (Smad2/3) are structurally similar proteins that primarily mediate the transforming growth factor-β (TGF-β) signaling responsible for driving cell proliferation, differentiation, and migration. The dynamics of the Smad2/3 phosphorylation provide the key mechanism for regulating the TGF-β signaling pathway, but the details surrounding this phosphorylation remain unclear. Here, using in vitro kinase assay coupled with mass spectrometry, we identified for the first time that nemo-like kinase (NLK) regulates TGF-β signaling via modulation of Smad2/3 phosphorylation in the linker region. TGF-β-mediated transcriptional and cellular responses are suppressed by NLK overexpression, whereas NLK depletion exerts opposite effects. Specifically, we discovered that NLK associates with Smad3 and phosphorylates the designated serine residues located in the linker region of Smad2 and Smad3, which inhibits phosphorylation at the C terminus, thereby decreasing the duration of TGF-β signaling. Overall, this work demonstrates that phosphorylation on the linker region of Smad2/3 by NLK counteracts the canonical phosphorylation in response to TGF-β signals, thus providing new insight into the mechanisms governing TGF-β signaling transduction.
Keywords: NLK, Smad2, Smad3, TGF-β signaling, linker phosphorylation
Abbreviation: ATF-3, activating transcription factor 3; BMP, bone morphogenic protein; CDK8/9, cyclin-dependent kinases 8 and 9; CTGF, connective tissue growth factor; ECM, extracellular matrix; FN, fibronectin; GSK3, glycogen synthase kinase 3; NLK, nemo-like kinase; p15, cyclin-dependent kinase inhibitor 2B; p21, cyclin-dependent kinase inhibitor 1A; PAI-1, plasminogen activator inhibitor type 1; qRT-PCR, quantitative real-time PCR; R-Smads, receptor-regulated Smad proteins; Smad, small mother against decapentaplegic; TGF-β, transforming growth factor-β; TMEPAI, transmembrane prostate androgen-induced RNA
Transforming growth factor-β (TGF-β) represents a large family of secreted polypeptide cytokines that play pivotal roles in regulating a broad spectrum of cellular processes from cell proliferation, differentiation, apoptosis, migration, to extracellular matrix (ECM) remodeling (1, 2). Aberrant TGF-β signal transductions often associate with various severe diseases, ranging from skeletal diseases, fibrosis, to cancer (3).
There are two receptors of TGF-β, TβRI and TβRII, forming a heteromeric complex responsible for transduction of TGF-β signal via the canonical path. The TGF-β ligand binds to TβRII, a constitutively active serine/threonine kinase, which further recruits and phosphorylates TβRI (4). As a result, TβRI is activated, then phosphorylates downstream transcription factors, primarily receptor-regulated Smad proteins (R-Smads), Smad2 and Smad3 at the C-terminal SXS motif. This phosphorylation event is required for the formation of ternary complex of R-Smads and Smad4 (5, 6, 7, 8) that is translocated into the nucleus and regulates the transcription of target genes in cooperation with nuclear cofactors (5, 6, 7, 8).
Smad2 and Smad3 are relatively conserved in their structures, both consist of N-terminal MH1 domain, C-terminal MH2 domain, and a linker region connecting MH1 and MH2 (9). Following the contextual cues, the linker region undergoes phosphorylation by different kinases, including MAPKs, Erk, Jnk and p38, PI3K, CDKs, ROCK, Araf, and GSK3 (10, 11, 12, 13). Proline-directed kinases prefer to phosphorylate four SP/TP sites in the linker regions, i.e., T220, S245, S250, and S255 in Smad2; T179, S204, S208, and S214 in Smad3 (14). Phosphorylation events in response to the TGF-β antagonists generally lead to the inhibition of the activation of Smad3, a critical counterbalance mechanism in the TGF-β signaling (15, 16). In contrast, agonist-induced Smad3 linker phosphorylation by cyclin-dependent kinases 8 and 9 (CDK8/9) not only enhances Smad-dependent transcription activation but also primes the linker region for the subsequent phosphorylation by glycogen synthase kinase 3 (GSK3) (17, 18, 19), which creates docking sites for the ubiquitin protein ligase NEDD4L and leads to proteasome-mediated degradation of the activated Smad3 as an mechanism of signal transduction termination (20). In addition, phosphorylation that occurred on the linker region can be reversed through dephosphorylation (21).
Homologous to Nemo in Drosophila, Nemo-like kinase (NLK) represents an evolutionarily conserved family of serine/threonine kinases (22, 23). NLK is involved in a wide variety of developmental events (24, 25, 26, 27) and also linked to several types of human cancers (28, 29, 30, 31). NLK regulates a broad spectrum of signaling pathways in mammals, including the Wnt/β-catenin, activin, YAP, mTORC1, NF-κB, NGF, and Notch signaling pathways (32, 33, 34, 35, 36). In Drosophila, Nemo antagonizes bone morphogenic protein (BMP) signaling by phosphorylating the BMP effector MAD to promote MAD export from the nucleus (37). However, the question remains as to whether NLK plays any roles in TGF-β signal transduction.
Here we demonstrate that NLK suppresses Smad2/3 mediated TGF-β signaling. NLK interacts with Smad2/3 and phosphorylates Smad2 and Smad3 in the linker region. The phosphorylation of Smad3 by NLK leads to the protein degradation of Smad3, thereby proposing NLK as a negative regulator of TGF-β signaling.
Results
NLK inhibits TGF-β-induced cell migration and transcriptional responses
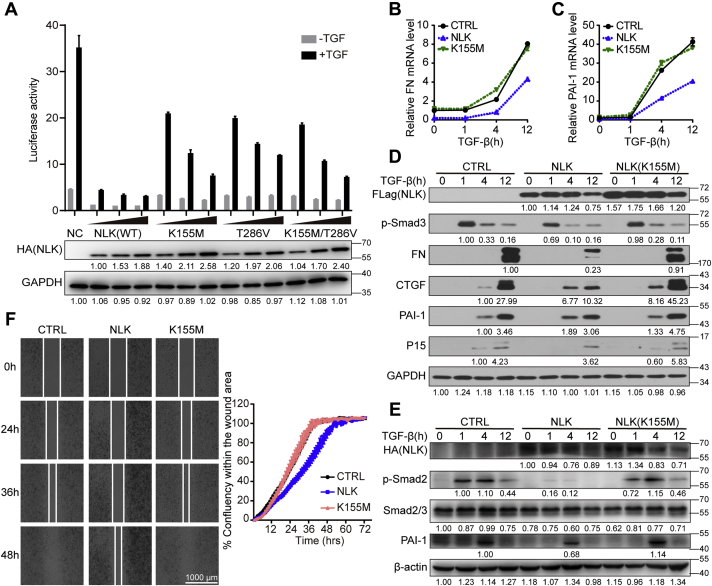
We first evaluated NLK’s functions in TGF-β signaling by measuring its impact on the Smad-driven transcription. As shown in Figure 1A, the transcription of the (SBE)4 reporter gene was robustly inhibited with increased levels of NLK in a dose-dependent manner. In clear contrast, this transcriptional inhibition by NLK was significantly alleviated when K155M, T286V, or K155M/T286V was introduced to diminish the NLK kinase activity. These data over all suggest that NLK kinase activity is essential for its inhibition of the Smad-dependent gene transcription in response to TGF-β. Furthermore, we validated this notion in HaCaT cells stably expressing exogenous NLK. The transcription of a series of established TGF-β target genes including (38, 39, 40) fibronectin (FN), plasminogen activator inhibitor type 1 (PAI-1), connective tissue growth factor (CTGF), cyclin-dependent kinase inhibitor 2B (P15), Smad7, and transmembrane prostate androgen-induced RNA (TMEPAI) was measured in the context of TGF-β signaling. We observed similar inhibition of the target gene transcription by NLK, which was compromised with NLK K155M. Interestingly, this inhibitory effect coincides with the decrease in the phosphorylation of Smad3 at C terminus that primarily mediates the transcriptional activation by TGF-β (Fig. 1, B–D and Fig. S1, A–D). We then tested HeLa cells to clarify whether this NLK’s inhibition of TGF-β signaling is cell-type-dependent. There was no significant discrepancy in the observation. The elevated level of NLK inhibits the transcription of PAI-1 induced by TGF-β, accompanied by the reduction in both the total level of Smad2/3 and the phosphorylation of the Smad2 C terminus (Fig. 1E). Furthermore, we investigated further downstream cellular activities upregulated by the TGF-β signaling. As demonstrated in the wound-healing assay, both HeLa cells (Fig. 1F) and HaCaT cells (Fig. S1E) exhibited a lower motility as the result of impaired TGF-β signaling when the wild-type NLK was overexpressed, whereas no significant difference with the kinase-inactive NLK.
Figure 1.
NLK inhibits TGF-β-induced cell migration and transcriptional responses.A, NLK affects the SBE4-luc response. The SBE4-luc reporter was cotransfected with different doses (25 ng, 50 ng, and 100 ng) of NLK (WT) or its inactive kinase forms NLK(K155M), NLK(T286V), and NLK(K155M/T286V) in HEK293 cells treated with or without 100 pM TGF-β for 24 h. The data were measured in triplicate. Values and bars represent the means and the standard deviation. The protein levels of NLK were shown below, and the relative band intensity was shown under each panel (GAPDH: relative values of band intensity; HA: relative values of the band intensity ratio of target protein to GAPDH). B and C, quantitative real-time PCR (qRT-PCR) analysis of mRNA. NLK (stably expressing HA-NLK) cells, K155M (stably expressing HA-NLK(K155M)) cells, and parental HaCaT cells were stimulated with TGF-β at the indicated times before total RNA was extracted. The data were measured in triplicate. Values and bars represent the means and the standard deviation. D, TGF-β-induced expression of PAI-1, CTGF, FN, and P15 was inhibited by NLK. HaCaT cells stably expressing FLAG-NLK or FLAG-NLK K155M were stimulated with 100 pM TGF-β for the indicated times. The relative band intensity was shown under each panel (GAPDH: relative values of band intensity; others: relative values of the band intensity ratio of target protein to GAPDH). E, TGF-β-induced expression of C-tail phosphorylated Smad2, Smad2/3, and PAI-1 was inhibited by NLK. HeLa cells stably expressing HA-NLK or HA-NLK K155M were stimulated with 100 pM TGF-β for the indicated times. The relative band intensity was shown under each panel (β-actin: relative values of band intensity; others: relative values of the band intensity ratio of target protein to β-actin). F, HeLa cells prepared as in D were tested in a wound-healing assay. Wounds were performed by seeding cells into the Culture-Insert 2 Well. Cells were treated with 100 pM TGF-β and allowed to migrate for 72 h. The data were measured in triplicate. Values and bars represent the means and the standard deviation.
Depletion of NLK enhances TGF-β-induced transcriptional responses and compromises cell growth
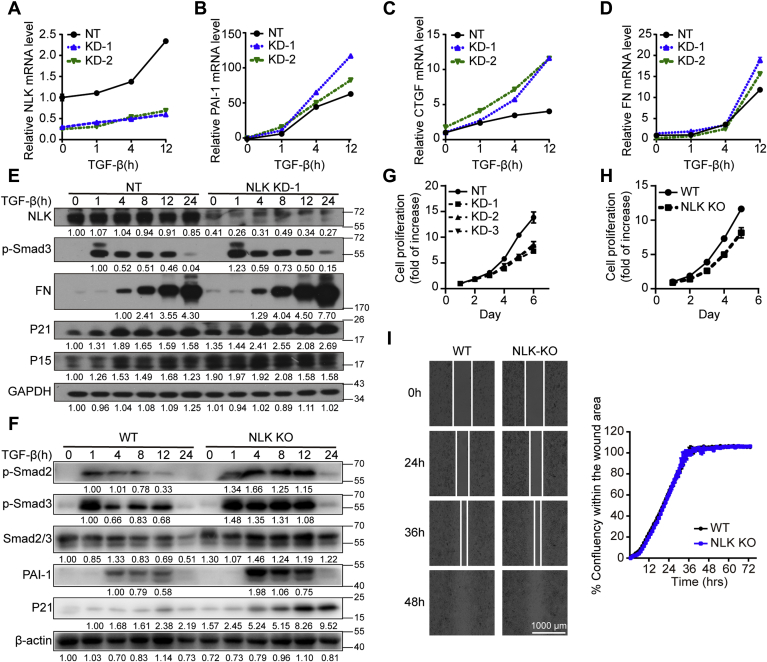
Next, we examined the function of NLK in regulating TGF-β signaling in a more physiological setting. The NLK siRNAs specifically targeting designated KD-1 and KD-2, as well as nontargeting control NT, were introduced into HaCaT cells respectively, and the efficiency of the targets was shown in Figure 2A, which also showed that TGF-beta could facilitate the mRNA expression of NLK with treatment time prolonged. A cohort of Smad3 target genes represented by PAI-1, CTGF, FN, P15, Smad7, and TMEPAI demonstrated significant reduction in their expressions (Fig. 2, B–D and Fig. S2, A–C). This impact was more evident at protein level with examination of P15 and P21 in addition to FN in HaCaT cells stably expressing NLK shRNA (NLK KD-1) (Fig. 2E). Similar results were also recorded in HeLa cells with the depletion of NLK in the genome (NLK KO), where the expression of PAI-1 and P21 was reduced together with the phosphorylation of Smad2/3 at the C terminus as well as the total levels of Smad2/3 (Fig. 2F). More importantly, the impact of the NLK depletion on the expressions of target genes was well reflected in changes in the cell proliferation with significant lower cell growth observed upon both HaCaT NLK KD cells (Fig. 2G) and HeLa NLK KO cells (Fig. 2H). However, NLK depletion did not show significant effect on cell migration (Fig. 2I and Fig. S2D). Since HaCaT and HeLa cells could migrate relatively rapidly, we hypothesized that the cell-free gap might be too narrow to exhibit the enhanced migration ability of NLK depleted cells. Collectively, these data suggest that NLK functions as a negative modulator of TGF-β signaling.
Figure 2.
Depletion of NLK enhanced TGF-β-induced transcriptional responses and compromises cell growth.A–D, qRT-PCR analysis of mRNA. HaCaT cells were transfected with NLK siRNA (KD-1 and KD-2) and nontargeting siRNA (NT), then stimulated with TGF-β at the indicated times before total RNA was extracted. The data were measured in triplicate. Values and bars represent the means and standard deviation values. E, western blot analysis of P21, P15, FN, and p-Smad3. Whole-cell lysates from HaCaT cells stably expressing NLK shRNA (NLK KD-1) and nontargeting siRNA (NT) were immunoblotted with antibodies as shown on the left. The relative band intensity was shown under each panel (GAPDH: relative values of band intensity; others: relative values of the band intensity ratio of target protein to GAPDH). F, western blot analysis of C-tail-phosphorylated Smad2/3, total Smad2/3, PAI-1, and P21 in the HeLa cells. Whole-cell lysates from the NLK-KO and WT cells were immunoblotted with antibodies as shown on the right. The relative band intensity was shown under each panel (β-actin: relative values of band intensity; others: relative values of the band intensity ratio of target protein to β-actin). G, proliferation of the HaCaT cells stably expressing NLK shRNA (KD-1; KD-2; KD-3) or nontargeting shRNA (NT) was examined by using the CCK-8 method. The data were measured in triplicate. Values and bars denote the means and standard deviation values. H, proliferation of NLK-knockout HeLa (NLK KO) cells or wild-type HeLa (WT) cells was examined by using the CCK-8 method. The data were measured in triplicate. Values and bars denote the means and standard deviation values. I, NLK-knockout HeLa (NLK KO) cells or wild-type HeLa (WT) cells were tested in a wound-healing assay. Wounds were performed by seeding cells into the Culture-Insert 2 Well. Cells were treated with 100 pM TGF-β and allowed to migrate for 72 h. The data were measured in triplicate. Values and bars represent the means and the standard deviation.
NLK interacts with Smad2/3
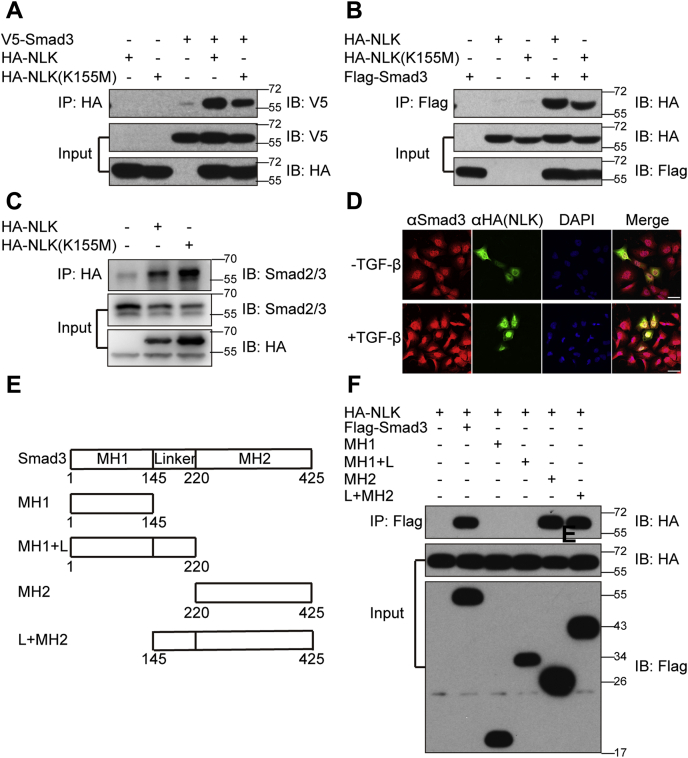
Nemo antagonizes BMP signaling by binding to and phosphorylating the BMP effector Mad (37). We tested whether NLK follows the same mechanistic trend as Nemo. Smad3 under epitopic expression was co-immunoprecipitated with NLK (Fig. 3, A and B), which is further supported by the fact that NLK was copurified by IP with endogenous Smad2/3 (Fig. 3C) confirming the physical association between NLK and Smad2/3 as its putative substrates. The kinase activity is not required for such interaction as the catalytically inactive mutant NLK K155M retained the ability to interact with Smad3. Moreover, we stained the U2OS cells for NLK and Smad3 (Fig. 3D). In the absence of TGF-β, Smad3 participated in both the nucleus and the cytoplasm, while NLK was colonized with Smad3 in the nucleus to certain extent. However, the TGF-β stimulation led to a nuclear accumulation of Smad3 that coincides with more overlapping between Smad3 and NLK, suggesting a role of NLK in regulating Smad2/3-mediated TGF-β signaling. We also looked into the structure of Smad3 responsible for interacting with NLK by introducing several truncated Smad3 proteins into HEK293T cells respectively (Fig. 3E). As shown in Figure 3F, the deletion of MH2 domain disrupted the association of NLK with Smad3, indicating that MH2 domain mediates the interaction between NLK and Smad3.
Figure 3.
NLK interacted with Smad3.A–C, co-immunoprecipitation of NLK with Smad2/3. HEK293T cells were transfected with expression plasmids as indicated. The immunoprecipitates were immunoblotted with the corresponding epitope antibodies. D, subcellular colocalization of NLK and Smad3 in the U2OS cells. U2OS cells were transfected with HA-tagged NLK for 24 h, treated with TGF-β for 1 h, and fixed. The localization of endogenous Smad3 was examined by indirect immunostaining with anti-Smad3 antibody (red). Cells treated with HA-NLK were observed through immunostaining with anti-HA antibody (green). E, schematic representation of Smad3 and its deletion mutants used in F. F, map of the region in Smad3 that interacts with NLK. Wild-type Smad3 and its deletion mutants were coexpressed with NLK in HEK293T cells. Immunoprecipitation and immunoblotting were performed as described in A–C.
NLK phosphorylates Smad2 and Smad3
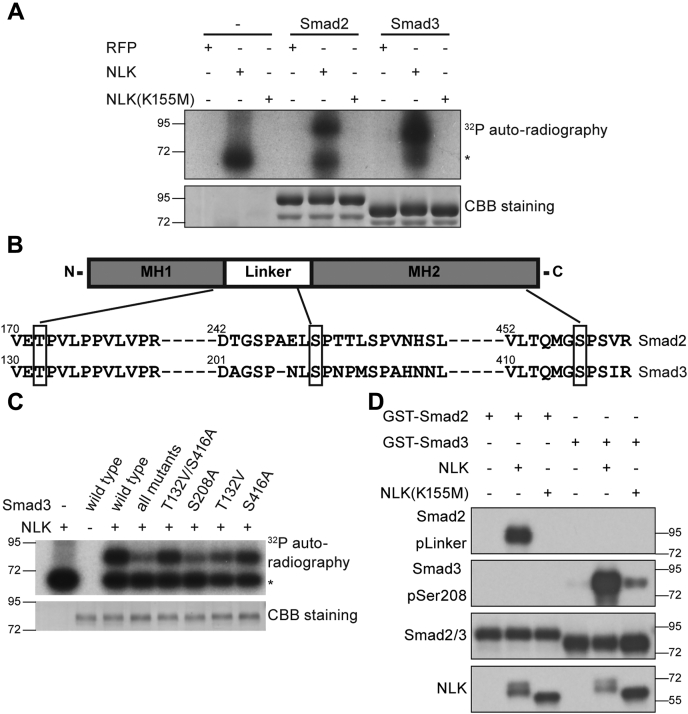
We went on to test whether Smad2/3 are indeed the substrates of NLK. GST-Smad2 and GST-Smad3 were expressed and purified from bacteria (Fig. S3). Both of them were phosphorylated by NLK, with no reaction detected with the catalytically inactive mutant NLK K155M (Fig. 4A). The potential phosphorylation sites, as shown in Figure 4B, were suggested by the mass spectrometry analysis (Tables S1-S4) and then confirmed by substituting the S/T (serine/threonine) with A/V (alanine/valine) (Fig. S4). We found sharply decreased phosphorylation with S208A and T132V/S208A/S416A substitutions but not with S416A nor with T132V/S416A (Fig. 4C), suggesting that Ser208 was the major phosphorylation site by NLK. Considering the conservation in the linker region between Smad2 and Smad3, we suspected that NLK can phosphorylate both structures in the same manner. The antibodies specifically recognizing pSer208 (phosphor-Smad3-S208) or phosphorylation on the linker region of Smad2 were raised and used in the vitro kinase assay as shown in Figure 4D. NLK phosphorylated Smad3 at Ser208 as indicated by the pSer208 signal while Smad2 was phosphorylated in the linker region by NLK. Taken together, the results confirm that NLK phosphorylates Smad3 at Ser208 and Smad2 at the analogous region.
Figure 4.
NLK phosphorylated Smad3 at Ser208 and Smad2 at the analogous site in vitro.A, NLK phosphorylated Smad2 and Smad3 in vitro. Flag-NLK, the catalytically inactive mutant Flag-NLK (K155M), or Flag-RFP immunoprecipitated was from HEK23T cells, and bacterially purified GST-Smad2 or GST-Smad3 was subjected to in vitro kinase reactions in the presence of [γ-32P] ATP. Samples were resolved by SDS-PAGE. Phosphorylated proteins were visualized by autoradiography. The asterisk denotes autophosphorylated NLK. B, schematic diagram of Smad2 and Smad3 phosphorylation sites as identified by mass spectrometry. GST-Smad2 and GST-Smad3 proteins phosphorylated by NLK were gathered form gel described in A and used in mass spectrometry analysis. C, the substitution of Ser208 with alanine in Smad3 abolished the phosphorylation of Smad3 by NLK. Bacterially purified GST-Smad3 and its point mutants were incubated with or without NLK in the presence of [γ-32P] ATP. Phosphorylated proteins were visualized by autoradiography. The asterisk denotes significant NLK autophosphorylation. CBB staining showed equalized Smad3 proteins in different reaction systems. D, an in vitro kinase assay was performed as described in A, and phosphorylated Smad2 and Smad3 were detected by the indicated phospho-specific antibodies. Equal substrate loading was confirmed by blotting with an anti-Smad2/3 antibody.
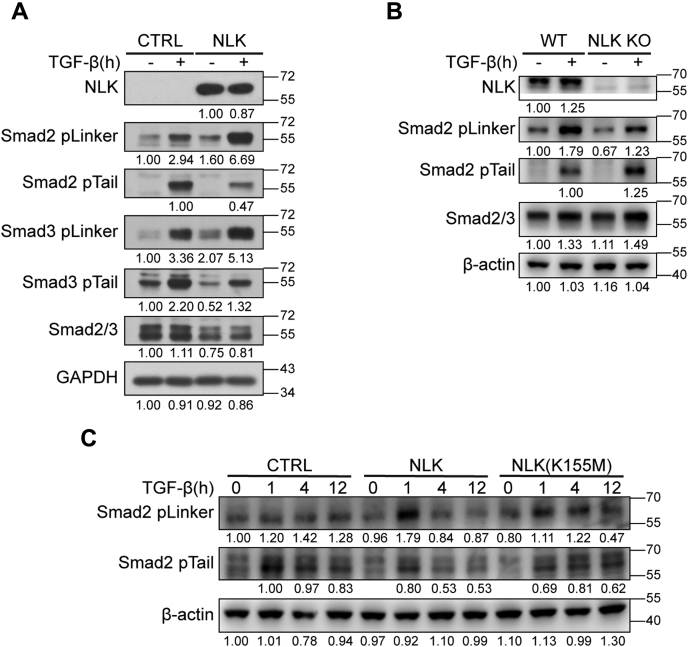
We then set to reveal the physiological relevance of Smad2/3 phosphorylation by NLK. As shown in Figure 5A, the phosphorylation of Smad2/3 at the C terminus was significantly compromised upon overexpression of NLK in the HaCaT cells while the phosphorylation on the linker regions, indicated by pSer208 in Smad3 and pSer250 in Smad2, was enhanced. Conversely, depleting NLK in HeLa cells inhibited pSer250 phosphorylation induced by NLK, while the phosphorylation of Smad2 C terminus was increased along with the level of Smad2 (Fig. 5B). Furthermore, the loss of pSer250 in Smad2 in the NLK-KO HeLa was restored by the overexpression of NLK wild type but not by the inactive form, indicating that pSer250 in Smad2 is phosphorylated by NLK under physiological conditions (Fig. 5C).
Figure 5.
NLK is required for the Smad2/3 linker phosphorylation in cells.A, HaCaT stable cells as described in Figure 1D were stimulated with TGF-β for 1 h. Whole-cell lysates were prepared for western blot analysis with the indicated antibodies. The relative band intensity was shown under each panel (GAPDH: relative values of band intensity; others: relative values of the band intensity ratio of target protein to GAPDH). B, wild-type (WT) and NLK knockout (NLK KO) HeLa cells were stimulated with TGF-β for 1 h. Whole-cell lysates were prepared for western blot analysis with the indicated antibodies. The relative band intensity was shown under each panel (β-actin: relative values of band intensity; others: relative values of the band intensity ratio of target protein to β-actin). C, NLK knockout (NLK-KO) HeLa cells were infected with lentivirus expressing NLK or the NLK mutant (K155M), then stimulated with 100 pM TGF-β for the indicated times. Whole-cell lysates were prepared for western blot analysis with the indicated antibodies. The relative band intensity was shown under each panel (β-actin: relative values of band intensity; others: relative values of the band intensity ratio of target protein to β-actin).
The linker region phosphorylation destabilizes Smad2/3 and inhibits TGF-β signaling
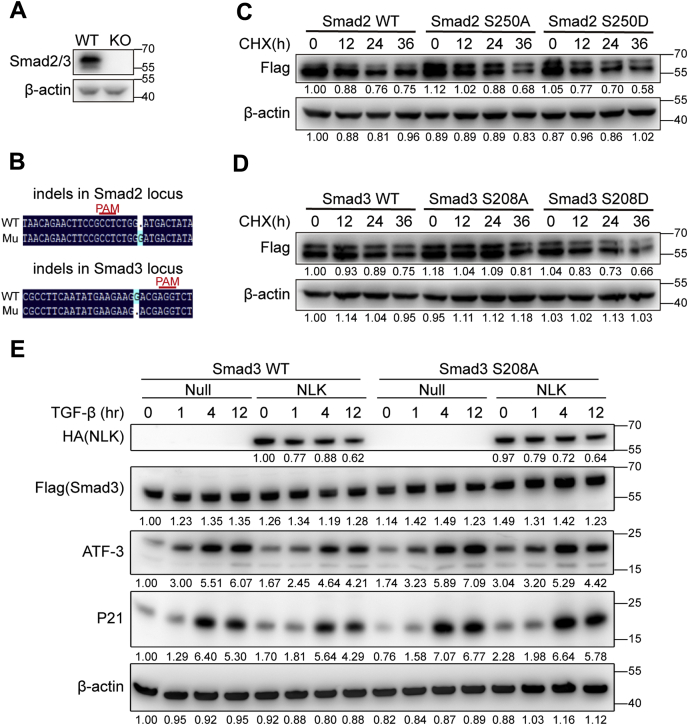
The degradation of phosphorylated Smad2/3 upon TGF-β stimulation is critical for the termination of the signaling and maintaining the dynamic kinetics of the signaling transduction. We therefore intended to check whether the linker region phosphorylation is involved in this process. Either the wild-type or the phosphorylation diminishing (S/A) or phosphorylation mimicking (S/D) Smad2/3 proteins was introduced to the HeLa cells in which endogenous Smad2 and Smad3 were depleted (Fig. 6, A and B). This setup allowed us to specifically measure the impact of the linker phosphorylation by NLK on the stability of Smad2/3. The fastest turnover of protein occurred with Smad2 S250D, followed by the wild-type protein, and the slowest with Smad2 S250A, suggesting that pSer250 by NLK decreases the stability of Smad2 (Fig. 6C). The same principle appeared also apply to Smad3 (Fig. 6D). In sum, the linker region phosphorylation by NLK prompts Smad2/3 for degradation. To further investigate the role of linker phosphorylation by NLK in TGF-β signaling, we overexpressed NLK along with either wild-type Smad3 or S/A mutant in Smad2/3-deficient HeLa cells described in Figure 6A. As shown in Figure 6E, when the linker region phosphorylation was compromised by S208 A mutation introduced to Smad3, the inhibition of TGF-β signal by NLK was impaired, leading to a higher level of transcriptional activation by TGF-β compared with the wild-type Smad3 as control. Overall, this work proposes the linker region phosphorylation of Smad2/3 by NLK as part of the crucial regulatory mechanisms of TGF-β signaling.
Figure 6.
The linker region phosphorylation destabilizes Smad2/3 and inhibits TGF-β signaling.A, validation of Smad2/3 KO in HeLa cells by western blot. B, validation of Smad2/3 KO in HeLa cells by Sanger sequencing. C and D, Smad2/3-KO HeLa cells were infected with lentivirus expressing wild-type or mutant Smad2/3, then treated with 100 μg/ml CHX for the indicated times. Whole-cell lysates were prepared for western blot analysis with the indicated antibodies. The relative band intensity was shown under each panel (β-actin: relative values of band intensity; Flag: relative values of the band intensity ratio of target protein to β-actin). E, NLK inhibits TGF-β signaling via the phosphorylation of the linker region. Smad2/3-deficient HeLa cells were infected with lentivirus expressing NLK and either Smad3(WT) or Smad3(S208A). HeLa cells were stimulated with 100 pM TGF-β for the indicated times. Subsequently, the total cell lysates were prepared for western blotting. The relative band intensity was shown under each panel (β-actin: relative values of band intensity; others: relative values of the band intensity ratio of target protein to β-actin).
Discussion
TGF-β/Smad signaling is one of the prominent elements in the cellular signal transduction network. The sensitivity and balance of this pathway are critical for lives in different forms, which is ensured by a plethora of regulatory proteins (8). Various diseases arise from the imbalance in TGF-β signaling (5). Here, we identified NLK as a novel regulator of TGF-β signaling through phosphorylating the activated Smad2/3 in the linker region and thereby attenuating the levels of activated Smad2/3.
As an atypical MAPK, NLK has been proposed as a multifaceted cellular signaling regulator (28, 30). NEMO, the homolog of NLK in Drosophila, antagonizes BMP signaling (37). To our knowledge, our work provides the first evidence for the regulatory role of NLK in TGF-β signaling. Interestingly, NLK interacts with Smad2/3 and phosphorylates in the linker region, which has been proposed as hub for integrating various inputs of regulation (14). TGF-β/BMP trigger the linker phosphorylation that often leads to the rapid turnover of activated Smads, thus serving as a negative regulatory mechanism for balancing the signal activation. Such phosphorylation is normally mediated by the transcriptional cyclin-dependent kinases CDK8/9 (17, 20) and GSK3 (18). Our work extends this kinase spectrum by revealing the role of NLK in the phosphorylation of Smad2/3 in the linker region. However, the question remains as to how CDK 8/9 and NLK cooperate in keeping TGF-β signaling on check. NLK might in collaboration with CDK8/9 phosphorylate Ser208 in Smad3. Alternatively, CDK8/9 mainly participates in the phosphorylation of the T179 [PY] motif, to create a docking site for the Pin1 cofactor, which is necessary for the peak activation of Smad3-dependent transcription (41), whereas pSer208 by NLK might serve as a priming site for subsequent Ser204 phosphorylation by GSK3 (18, 19). The timely manner featuring the latter possibility fits better our observation of the sequential events in TGF-β signaling, i.e., the Smad2/3 activation prior to the degradation of the activated proteins. Another important notion is that Pin1 prefers the pThr179 [PY] motif binding in the absence of the pS204-pS208 motif, leading to the peak activation of Smad3-dependent transcription, whereas pS204-pS208 motif shifts this interaction toward the ubiquitin ligase Nedd4L for the subsequent protein degradation (41). There, pSer208 phosphorylation signals the activated Smad3 for destruction, which is proceeded by the phosphorylation of the Thr179 [PY] motif during peak of the activation. Future work would bring more insights into the interplay between NLK and other kinases in regulating TGF-β signaling as well as the physiological and even possibly clinical relevance of these mechanisms.
Experimental procedures
Plasmids, antibodies, and other reagents
The coding regions of human NLK, Smad2, or Smad3 from a HEK293 cDNA library were amplified by RT-PCR with the corresponding primers (for NLK, the forward primer was 5’-CGCGGATCCACCATGGCGGCTTACAATGGCGGTACATCTGC-3’, and the reverse primer was 5’-TCCGCTCGAGTCACTCCCACACCAGAGGAGATG-3’; for Smad2, the forward primer was 5’-CCGGAATTCATGTCGTCCATCTTGCCATTCAC-3’, and the reverse primer was 5’-TCCGCTCGAGTTATGACATGCTTGAGCAACGCAC-3’; for Smad3, the forward primer was 5’-CGCGGATCCATGTCGTCCATCCTGCCTTTC-3’, and the reverse primer was 5’-TCCGCTCGAGTCAAGACACACTGGAACAGCGGA-3’). The PCR products with an N-terminal Flag tag, N-terminal HA tag, or N-terminal V5 tag were cloned into a modified pcDNA6/myc-HisB vector between the BamHI and XhoI sites for NLK and Smad3. Smad3 deletion mutants were generated by PCR using pcDNA6-Flag-Smad3 as the template and were subcloned into a modified pcDNA6/myc-HisB vector between the BamHI and XhoI sites with an N-terminal Flag tag. The primers for the deletion mutants of Smad3 were as follows: for the MH1 deletion mutant, 5’-CGCGGATCCATGTCGTCCATCCTGCCTTTC-3’ and 5’-TCCGCTCGAGTCACTCTGTGTGGCGTGGCACCAACAC-3’ were used; for the MH2 deletion mutant, 5’-CGCGGATCCGACCTGCAGCCAGTTACCTAC-3’ and 5’-TCCGCTCGAGTCAAGACACACTGGAACAGCGGA-3’ were used; for the MH1+linker deletion mutant, 5’-CGCGGATCCATGTCGTCCATCCTGCCTTTC-3’ and 5’-TCCGCTCGAGTCACAAGTTATTATGTGCTGGGGACATC-3’ were used; and for the linker+MH2 deletion mutant, 5’-CGCGGATCCATCCCGGCCGAGTTCCCCCCAC-3’ and 5’-TCCGCTCGAGTCAAGACACACTGGAACAGCGGA-3’ were used. Full-length GST-Smad2 and GST-Smad3 were constructed by inserting the Smad2 or Smad3 coding sequence into a pGEX-6P-1 vector (Amersham Biosciences) between the EcoRI and XhoI sites or the BamHI and XhoI sites, respectively. Point mutations were generated in all the constructs with a QuickChange site-directed mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. The primers for the NLK and Smad3 point mutations were as follows: for NLK (K155M), 5’-GTAGCGCTCATGAAGATGCCC-3’ and 5’-GGGCATCTTCATGAGCGCTAC-3’; for NLK (T286V), 5’-CGTCATATGGTTCAGGAAGTT-3’ and 5’-AACTTCCTGAACCATATGACG-3’; for Smad3 (T132V), 5’-GAGAGTAGAGGTACCAGTTCTACCTC-3’ and 5’-GAGGTAGAACTGGTACCTCTACTCTC-3’; for Smad3 (S208A), 5’-TCTCCAAACCTAGCCCCGAATCCGATG-3’ and 5’-CATCGGATTCGGGGCTAGGTTTGGAGA-3’; and for Smad3 (S416A), 5’-CAGATGGGCGCCCCAAGCATC-3’ and 5’-GATGCTTGGGGCGCCCATCTG-3’. To generate a retrovirus, the NLK coding sequence was subcloned into a modified pMSCVpuro vector with an N-terminal Flag-Strep or HA tag. To generate lentiviruses, the Smad2, Smad3, or NLK coding sequence was subcloned into a pCDH vector with an N-terminal Flag-Strep or HA tag. A lentiviral-based vector, pLentiLox 3.7, was used to express both NLK small-hairpin (shRNA) and control shRNA. In brief, chemically synthesized oligonucleotides were annealed and inserted into the pLentiLox 3.7 vector between the HpaI and XhoI sites. All the constructs were confirmed by sequencing. The shRNA targeting sequences were as follows: NLK-KD1, 5’-GCAATTCTGGAGGTTAATCATGCTTGTAC-3’; NLK-KD2, 5’-CTACTAGGACGAAGAATATTGTTTC-3’; NLK-KD3, 5’-GGAAGATAATGTACTACTGAAGATG-3’; nontargeting control, 5’-TTCTCCGAACGTGTCACGT-3’. The siRNAs used for NLK were the following: NT, 5’-TTCTCCGAACGTGTCACGT-3’; NLK KD-1, 5’-GATGCAGAGTGACCTACAT-3’; NLK KD-2, 5’-GAAGGCGCTAAGGCACATATA-3’. The sgRNAs of Smad2 and Smad3 were inserted into the lentiCRISPR v2 vector. The retrovirus and lentivirus were prepared as previously described (39).
Antibodies against the Smad2 phospho-linkers (phospho-Smad2 (Ser245/250/255)), Smad2 phospho-tails (phospho-Smad2 (Ser465/467)), Smad3 phospho-tail (phospho-Smad3 (Ser423/425)), and Smad2/3 were purchased from Cell Signaling Technology. The other antibodies were directed against: NLK (Bethyl); Flag (clone M2), HA, V5 and Flag M2-agarose (Sigma-Aldrich); Smad3 phospho-Ser208 (Abgent); Smad3 phospho-Thr179 (Bioworld); PAI-1 and P21 (BD Pharmagin); FN, CTGF and P15 (Santa Cruz Biotechnology); and GAPDH (Zhongshanjinqiao). HRP-labeled secondary antibodies were obtained from Zhongshanjinqiao.
Cell culture, transfection, and stimulation
HaCaT keratinocytes, U2OS osteosarcoma cells, HeLa cells, HEK293T cells, and HEK293 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin-streptomycin (Life Technology) and maintained at 37 °C with 5% CO2. The U2OS cells, HEK293T cells, and HEK293 cells were transfected with Neofect transfection reagent (Neo Biotech). For RNAi in HaCaT cells, siRNA oligonucleotides (at a final concentration of 20 nM) from GenePharma were transfected into cells with Lipofectamine RNAi MAX. The cells were serum-starved for 16 h prior to being treated with TGF-β1 (100 pM, R&D Systems).
Cell proliferation assay
Cell proliferation was measured with a CCK-8 cell counting kit (Dojindo Molecular Technologies) according to the manufacturer’s instructions. In brief, HaCaT cells or HeLa cells were seeded in 96-well plates. Following an overnight of attachment, the cells were exposed to TGF-β1 (100 pM) for the indicated times. Cell number was determined by measuring the absorbance of each well at 450 nm, and cell proliferation was quantified by the fold increase [(cell numbers at defined time point/original cell numbers) × 100%]. The data are represented as the mean ± SD with at least three wells for each group. Each experiment was carried out in triplicate.
Cell migration/wound-healing assay
HaCaT cells and HeLa cells were seeded into Culture-Insert 2 Well (Ibidi). After cell attachment, Culture-Insert was removed to create wounds. Cells were treated with 100 pM TGF-β and allowed to migrate in the chamber of Lionheart FX Automated Microscope (BioTek). Photos were captured once an hour automatically.
Dual-luciferase reporter assay
The SBE4-luc reporter and TK were transfected into HEK293 cells along with different doses (25 ng, 50 ng, and 100 ng) of NLK (WT) or its inactive kinase forms NLK(K155M), NLK(T286V), and NLK(K155M/T286V). Twenty-four hours after transfection, cells were treated with or without 100 pM TGF-β for 24 h. Cell lysate was prepared and luciferase activity was measured in Centro XS3 LB 960 (Berthold Technologies) with a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
In vitro kinase assay
Flag-NLK and NLK (K155M) were expressed in HEK293T cells and immunoprecipitated with anti-FLAG M2-agarose. Immunoprecipitates were washed five times with NETN buffer containing a high concentration of salt (0.5 M NaCl). The proteins were eluted by using Flag peptides (Sigma-Aldrich). Recombinant GST-tagged Smad2, GST-tagged Smad3, and GST-tagged point-mutated Smad3 were produced in Escherichia coli BL21 (DE3) cells and purified with glutathione sepharose 4B (GE Healthcare) followed by gel filtration chromatography. The purified GST-tagged Smad proteins were incubated with aliquots of immunoprecipitated NLK in 20 μl of kinase buffer containing 5 μCi [γ-32P] ATP at 30 °C for 30 min. The samples were resolved by SDS-PAGE, and the phosphorylated proteins were detected by autoradiography. The potential phosphorylation sites at Smad2 and Smad3 were identified by mass spectrometry.
Mass spectrometry
The purified GST-tagged Smad2 and Smad3 proteins were gathered, incubated with aliquots of immunoprecipitated NLK, and separated by SDS-PAGE as described above, respectively. The gel bands of interest were excised from the gel, reduced with 5 mM of DTT, and alkylated with 11 mM iodoacetamide, which was followed by in-gel digestion with sequencing grade modified trypsin (Promega) at 37 °C overnight and chymotrypsin (Promega) at 25 °C overnight. The peptides were extracted twice with 0.1% trifluoroacetic acid in 50% acetonitrile aqueous solution for 30 min and then dried in a speedvac. Peptides were redissolved in 20 μl 0.1% trifluoroacetic acid, and 6 μl of extracted peptides was analyzed by Thermo Scientific Q Exactive mass spectrometer. For LC-MS/MS analysis, the peptides were separated by a 65 min gradient elution at a flow rate 0.30 μl/min with a Thermo-Dionex Ultimate 3000 HPLC system, which was directly interfaced with a Thermo Scientific Q Exactive mass spectrometer. The analytical column was a home-made fused silica capillary column (75 μm ID, 150 mm length; Upchurch) packed with C-18 resin (300 Å, 5 μm, Varian). Mobile phase consisted of 0.1% formic acid, and mobile phase B consisted of 80% acetonitrile and 0.1% formic acid. The Q Exactive mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.0.2.1462 software, and there was a single full-scan mass spectrum in the orbitrap (350–1800 m/z, 70,000 resolution) followed by ten data-dependent MS/MS scans at 29% normalized collision energy (HCD).
The MS/MS spectra from each LC-MS/MS run were searched against the Smad2.fasta or Smad3.fasta from UniProt using an in-house Proteome Discoverer (Version PD1.4, Thermo-Fisher Scientific). The search criteria were as follows: no enzyme was chosen; two missed cleavages were allowed; carbamidomethylation (C) was set as the fixed modification; the oxidation (M) and phosphorylation were set as the variable modification; precursor ion mass tolerances were set at 20 ppm for all MS acquired in an orbitrap mass analyzer; and the fragment ion mass tolerance was set at 0.02 Da for all MS2 spectra acquired. Proteome Discoverer node ptmRS was used for analysis and mapping of peptide/protein phosphorylation sites. ptmRS Best Site Probabilities display the most likely positions of the modifications and their site score for each PSM. For each modification site, this value is an estimate of the probability (0–100%) that the site is truly modified. Any ptmRS site probabilities above 75% indicate that a site is truly modified.
Generation of CRISPR Smad2/3 knockout cells
The lentiCRISPR v2 plasmids (Addgene Plasmid #52961) containing Smad2 or Smad3 single-guide gRNAs (sgRNA sequence for Smad2: GAGTGAGTATAGTCATCCAG; sgRNA sequence for Smad3: CTTCAATATGAAGAAGGACG) were cotransfected into HeLa cells using Lipofectamine 2000 (Thermo). Single-cell clones were selected by flow cytometry. Knockout status was validated by western blot and Sanger sequencing.
Other assays
Total RNA was isolated using TRizol reagent (Ambion), and 1 μg of total RNA was used to prepare cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo) according to the manufacturer's instructions. qRT-PCR was carried out in triplicate for each target gene using LightCycler 480 SYBR Green I Master Mix (Roche) on a LightCycler 480 Real-Time system (Roche). The following primers were used.
NLK_forward: 5’-TCTTCCTGTACTCTATACCCTGTC-3’
NLK_reverse: 5’- GATATCGTAGTCGCCCTTCATC-3’
FN_forward: 5’-ACTGTACATGCTTCGGTCAG-3’
FN_reverse: 5’-AGTCTCTGAATCCTGGCATTG-3’
CTGF_forward: 5’-CTGCAGGCTAGAGAAGCAGAG-3’
CTGF_reverse: 5’- GATGCACTTTTTGCCCTTCT-3'
PAI-1_forward: 5’-ATTCAAGCAGCTATGGGATTCAA-3’
PAI-1_reverse: 5’-CTGGACGAAGATCGCGTCTG-3’
P15_forward: 5’-GGAATGCGCGAGGAGAACAA-3’
P15_reverse:5’-CATCATCATGACCTGGATCGC-3’
Smad7_forward: 5’-TTCCTCCGCTGAAACAGGG-3’
Smad7_reverse: 5’-CCTCCCAGTATGCCACCAC-3’
TMEPAI_forward: 5’-TGTCAGGCAACGGAATCCC-3’
TMEPAI_reverse: 5’-CAGGTACGGATAGGTGGGC-3’
GAPDH_forward: 5’-AATCCCATCACCATCTTCCA-3’
GAPDH_reverse: 5’-TGGACTCCACGACGTACTCA-3’
Immunoprecipitation, immunoblotting, and immunofluorescence were carried out as previously described (39).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (42) partner repository with the data set identifier PXD023957. And all the other data are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by grants from the National Key R&D Program of China (2018YFC1003500), CAMS Innovation Fund for Medical Sciences (2017-I2M-3-009, 2016-I2M-1-001, 2017-I2M-4-002), the National Natural Science Foundation of China (81672472, 81672461, 81972311, 31970794, 32000586), the State Key Laboratory Special fund from the Ministry of Science (2060204), the State Key Project on Infection Diseases of China (2017ZX10201021-007-003), the Nonprofit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT310026), and Sanming Project of Medicine in Shenzhen (SZSM202011010).
Author contributions
W. S., H. Z., and J. L. conceived the study and designed the research. J. L., Y. Z., D. W., K. L., R. H., and Y. L. performed the experiments. X. C., H. W., D. H., W. W., S. M., L. W., and M. H. analyzed the data. J. L., Y. Z., and N. Z. wrote the paper.
Edited by Alex Toker
Footnotes
Present address for Ning Zhang: Wellcome Centre for Anti-Infectives Research (WCAIR), University of Dundee, Dundee, UK.
Present address for Xiaowen Cheng: Department of Clinical Laboratory, The First Affiliated Hospital, Anhui Medical University, Hefei 230022, China.
Contributor Information
Hong Zhao, Email: zhaohong@cicams.ac.cn.
Wei Song, Email: songwei@ibms.pumc.edu.cn.
Supporting information
References
- 1.Moses H.L., Roberts A.B., Derynck R. The discovery and early days of TGF-beta: A historical perspective. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a021865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu Y., Feng X.H. TGF-beta signaling in cell fate control and cancer. Curr. Opin. Cell Biol. 2019;61:56–63. doi: 10.1016/j.ceb.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Morikawa M., Derynck R., Miyazono K. TGF-beta and the TGF-beta family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derynck R., Budi E.H. Specificity, versatility, and control of TGF-beta family signaling. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massague J. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 7.Feng X.H., Derynck R. Specificity and versatility in TGF-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 8.Massague J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 9.Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Xiong C., Jia S., Zhang Y., Chen Y.G., Wang Q., Meng A. Araf kinase antagonizes Nodal-Smad2 activity in mesendoderm development by directly phosphorylating the Smad2 linker region. Nat. Commun. 2013;4:1728. doi: 10.1038/ncomms2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon J.H., Sudo K., Kuroda M., Kato M., Lee I.K., Han J.S., Nakae S., Imamura T., Kim J., Ju J.H., Kim D.K., Matsuzaki K., Weinstein M., Matsumoto I., Sumida T. Phosphorylation status determines the opposing functions of Smad2/Smad3 as STAT3 cofactors in TH17 differentiation. Nat. Commun. 2015;6:7600. doi: 10.1038/ncomms8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J.S., Ramasamy T.S., Murphy N., Holt M.K., Czapiewski R., Wei S.K., Cui W. PI3K/mTORC2 regulates TGF-beta/activin signalling by modulating Smad2/3 activity via linker phosphorylation. Nat. Commun. 2015;6:7212. doi: 10.1038/ncomms8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamato D., Do B.H., Osman N., Ross B.P., Mohamed R., Xu S., Little P.J. Smad linker region phosphorylation is a signalling pathway in its own right and not only a modulator of canonical TGF-beta signalling. Cell. Mol. Life Sci. 2020;77:243–251. doi: 10.1007/s00018-019-03266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamato D., Burch M.L., Piva T.J., Rezaei H.B., Rostam M.A., Xu S., Zheng W., Little P.J., Osman N. Transforming growth factor-beta signalling: Role and consequences of Smad linker region phosphorylation. Cell. Signal. 2013;25:2017–2024. doi: 10.1016/j.cellsig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura I., Denissova N.G., Wang G., He D., Long J., Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura I., Wang G., He D., Liu F. Identification and characterization of ERK MAP kinase phosphorylation sites in Smad3. Biochemistry. 2005;44:12546–12553. doi: 10.1021/bi050560g. [DOI] [PubMed] [Google Scholar]
- 17.Alarcon C., Zaromytidou A.I., Xi Q., Gao S., Yu J., Fujisawa S., Barlas A., Miller A.N., Manova-Todorova K., Macias M.J., Sapkota G., Pan D., Massague J. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millet C., Yamashita M., Heller M., Yu L.R., Veenstra T.D., Zhang Y.E. A negative feedback control of transforming growth factor-beta signaling by glycogen synthase kinase 3-mediated Smad3 linker phosphorylation at Ser-204. J. Biol. Chem. 2009;284:19808–19816. doi: 10.1074/jbc.M109.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G., Matsuura I., He D., Liu F. Transforming growth factor-{beta}-inducible phosphorylation of Smad3. J. Biol. Chem. 2009;284:9663–9673. doi: 10.1074/jbc.M809281200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao S., Alarcon C., Sapkota G., Rahman S., Chen P.Y., Goerner N., Macias M.J., Erdjument-Bromage H., Tempst P., Massague J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol. Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapkota G., Knockaert M., Alarcon C., Montalvo E., Brivanlou A.H., Massague J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J. Biol. Chem. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- 22.Choi K.W., Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- 23.Brott B.K., Pinsky B.A., Erikson R.L. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc. Natl. Acad. Sci. U. S. A. 1998;95:963–968. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorpe C.J., Moon R.T. Nemo-like kinase is an essential co-activator of Wnt signaling during early zebrafish development. Development. 2004;131:2899–2909. doi: 10.1242/dev.01171. [DOI] [PubMed] [Google Scholar]
- 25.Zeng Y.A., Verheyen E.M. Nemo is an inducible antagonist of wingless signaling during Drosophila wing development. Development. 2004;131:2911–2920. doi: 10.1242/dev.01177. [DOI] [PubMed] [Google Scholar]
- 26.Ota R., Kotani T., Yamashita M. Possible involvement of Nemo-like kinase 1 in Xenopus oocyte maturation as a kinase responsible for Pumilio1, Pumilio2, and CPEB phosphorylation. Biochemistry. 2011;50:5648–5659. doi: 10.1021/bi2002696. [DOI] [PubMed] [Google Scholar]
- 27.Ota S., Ishitani S., Shimizu N., Matsumoto K., Itoh M., Ishitani T. NLK positively regulates Wnt/beta-catenin signalling by phosphorylating LEF1 in neural progenitor cells. EMBO J. 2012;31:1904–1915. doi: 10.1038/emboj.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishitani T., Ishitani S. Nemo-like kinase, a multifaceted cell signaling regulator. Cell. Signal. 2013;25:190–197. doi: 10.1016/j.cellsig.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 29.Han Y., Kuang Y., Xue X., Guo X., Li P., Wang X., Guo X., Yuan B., Zhi Q., Zhao H. NLK, a novel target of miR-199a-3p, functions as a tumor suppressor in colorectal cancer. Biomed. Pharmacother. 2014;68:497–505. doi: 10.1016/j.biopha.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Huang Y., Yang Y., He Y., Li J. The emerging role of Nemo-like kinase (NLK) in the regulation of cancers. Tumour Biol. 2015;36:9147–9152. doi: 10.1007/s13277-015-4159-7. [DOI] [PubMed] [Google Scholar]
- 31.Li S.Z., Shu Q.P., Song Y., Zhang H.H., Liu Y., Jin B.X., Liuyu T.Z., Li C., Huang X.C., Du R.L., Song W., Zhong B., Zhang X.D. Phosphorylation of MAVS/VISA by Nemo-like kinase (NLK) for degradation regulates the antiviral innate immune response. Nat. Commun. 2019;10:3233. doi: 10.1038/s41467-019-11258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishitani T., Hirao T., Suzuki M., Isoda M., Ishitani S., Harigaya K., Kitagawa M., Matsumoto K., Itoh M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat. Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- 33.Li S.Z., Zhang H.H., Liang J.B., Song Y., Jin B.X., Xing N.N., Fan G.C., Du R.L., Zhang X.D. Nemo-like kinase (NLK) negatively regulates NF-kappa B activity through disrupting the interaction of TAK1 with IKKbeta. Biochim. Biophys. Acta. 2014;1843:1365–1372. doi: 10.1016/j.bbamcr.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan H.X., Wang Z., Yu F.X., Li F., Russell R.C., Jewell J.L., Guan K.L. NLK phosphorylates Raptor to mediate stress-induced mTORC1 inhibition. Genes Dev. 2015;29:2362–2376. doi: 10.1101/gad.265116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masoumi K.C., Daams R., Sime W., Siino V., Ke H., Levander F., Massoumi R. NLK-mediated phosphorylation of HDAC1 negatively regulates Wnt signaling. Mol. Biol. Cell. 2017;28:346–355. doi: 10.1091/mbc.E16-07-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moon S., Kim W., Kim S., Kim Y., Song Y., Bilousov O., Kim J., Lee T., Cha B., Kim M., Kim H., Katanaev V.L., Jho E.H. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 2017;18:61–71. doi: 10.15252/embr.201642683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Y.A., Rahnama M., Wang S., Sosu-Sedzorme W., Verheyen E.M. Drosophila Nemo antagonizes BMP signaling by phosphorylation of mad and inhibition of its nuclear accumulation. Development. 2007;134:2061–2071. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- 38.Nakao A., Afrakhte M., Moren A., Nakayama T., Christian J.L., Heuchel R., Itoh S., Kawabata M., Heldin N.E., Heldin C.H., ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 39.Lin X., Duan X., Liang Y.Y., Su Y., Wrighton K.H., Long J., Hu M., Davis C.M., Wang J., Brunicardi F.C., Shi Y., Chen Y.G., Meng A., Feng X.H. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125:915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe Y., Itoh S., Goto T., Ohnishi E., Inamitsu M., Itoh F., Satoh K., Wiercinska E., Yang W., Shi L., Tanaka A., Nakano N., Mommaas A.M., Shibuya H., Ten Dijke P. TMEPAI, a transmembrane TGF-beta-inducible protein, sequesters Smad proteins from active participation in TGF-beta signaling. Mol. Cell. 2010;37:123–134. doi: 10.1016/j.molcel.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Aragon E., Goerner N., Zaromytidou A.I., Xi Q., Escobedo A., Massague J., Macias M.J. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (42) partner repository with the data set identifier PXD023957. And all the other data are contained within the article.