Abstract
Biogenic silver nanoparticles (bio-AgNPs) are increasingly recognized as an antibiofilm and antivirulence strategy against P. aeruginosa, a bacterium that causes chronic infections in immunocompromised and cystic fibrosis patients. This study aimed to investigate the effects of subinhibitory concentrations of bio-AgNPs on motility and biofilm formation in P. aeruginosa. Bio-AgNPs were synthesized via reduction of ionic silver catalyzed by cell-free culture filtrate from Fusarium oxysporum. A total of 17 P. aeruginosa isolates and strains were evaluated for swarming, swimming, and twitching motility in the presence and absence (control) of bio-AgNPs, including 10 clinical isolates from patients with and without cystic fibrosis, 5 environmental isolates obtained from the public water supply system, and 2 reference strains (PAO1 and PA14). Isolates were identified by biochemical and molecular methods. Minimum inhibitory concentrations (MICs) were determined by the broth microdilution method. Swarming, swimming, and twitching motility assays were performed in Petri dishes. Biofilm formation capacity was assessed quantitatively by the crystal violet method. MIC values ranged from 15.62 to 62.50 µM. The results showed that subinhibitory concentrations of bio-AgNPs (½ MIC, 7.81–31.25 µM) significantly increased (p < 0.05) swarming, swimming, and twitching motility in 40.0, 40.0, and 46.7% of isolates, respectively. Subinhibitory bio-AgNP treatment enhanced (p < 0.05) biofilm formation capacity in PA14 and a cystic fibrosis isolate (P11). It is concluded that subinhibitory concentrations of bio-AgNPs increased biofilm formation and swarming, swimming, and twitching motility in PA14 and some P. aeruginosa isolates. These virulence factors are directly involved with quorum-sensing systems. Further research should investigate the effects of AgNPs on P. aeruginosa quorum sensing to help elucidate their mechanism of action at subinhibitory concentrations.
Keywords: antibiofilm, antivirulence, virulence factor, metal nanoparticle, quorum sensing
Introduction
Silver nanoparticles (AgNPs) are one of the most widely studied nanomaterials (Guo et al., 2019; Kawish et al., 2020; Saeed et al., 2020) because of their chemical stability, malleability, flexibility, high electrical and thermal conductivity, catalytic activity, relatively low cost, and, mainly, potent antimicrobial action (Durán et al., 2019). These unique properties have led AgNPs to find application in the medical industry (wound dressing, surgical instrument, and bone prosthesis production), textile industry (garment production), food industry (food packaging, countertop, and cutting board materials), and wastewater treatment (McNamara and Tofail, 2017; Gadkari et al., 2018).
AgNPs were shown to have antimicrobial properties against several pathogenic microorganisms (Scandorieiro et al., 2016; Figueiredo et al., 2019), including Pseudomonas aeruginosa (Liao et al., 2019; D’Lima et al., 2020). This bacterium can be found in various environments, such as hospitals, natural resources, and water supply systems (Dobias and Bernier-Latmani, 2013; Anversa et al., 2019; Esmaeili et al., 2019). P. aeruginosa may express different virulence factors and is closely related to healthcare-related infections and diseases in immunocompromised, cystic fibrosis, and severely burned patients. Because of these factors and its multi-antimicrobial resistance, P. aeruginosa constitutes a public health problem (Matar, 2018; Ahmed et al., 2019).
Chronic P. aeruginosa infection is established mainly through biofilm formation. Motility plays an essential role in the initial adhesion of bacterial cells to biotic and abiotic surfaces (Khan et al., 2020). Biofilm formation contributes to the survival of P. aeruginosa under adverse environmental conditions, provides protection against the host’s immune system, and confers antimicrobial resistance. This specific bacterial response is triggered by proximity to surfaces and involves complex chemical signaling networks, including quorum sensing (QS) (Khan et al., 2019; Olivares et al., 2020).
Biofilm development can be divided into four steps: (1) reversible adhesion of bacterial cells to a surface; (2) sessile growth and increased production, release, and detection of autoinducer molecules by QS systems; (3) biofilm maturation with the formation of mushroom-like structures enveloped by exopolysaccharides; and (4) disruption through the release of cells (Saxena et al., 2019). These important steps in P. aeruginosa biofilm formation depend on appendages such as flagella and type IV pili, both of which are necessary for bacterial motility (Otton et al., 2017).
P. aeruginosa is capable of three types of movement: swarming, swimming, and twitching. Swarming is performed on semi-solid surfaces (0.5 to 0.7% agar) via multicellular motion dependent on rotating helical flagella and typically involves type IV pili and a class of surfactants known as rhamnolipids (Köhler et al., 2000; Kearns, 2010). Swimming is another type of motility whereby bacteria swim on a low-viscous (0.3% agar) liquid surface using rotating flagella (Kearns, 2010; Sun et al., 2018). Twitching motility is mediated by type IV pili, occurring on solid surfaces or media with moderate viscosity (1% agar) by extension and retraction of pili (Kearns, 2010; Otton et al., 2017).
Due to the increase in multidrug resistance to antimicrobials, several studies have been carried out searching for new alternatives to interfere in the regulation of bacterial virulence and control the spread of multidrug-resistant bacteria (Shah et al., 2019; Ahmed et al., 2020; Piewngam et al., 2020). A major concern is that in these studies, subinhibitory concentrations of the compounds are used and we do not know for sure the bacterial response to these nanometals. The present study aimed to assess the effect of subinhibitory concentrations of biogenic AgNPs (bio-AgNPs) on the motility and biofilm formation capacity of P. aeruginosa.
Materials and Methods
Bacterial Isolates and P. aeruginosa Identification
This study was conducted at the Laboratory of Food Bacteriology and Microbiology of Adolfo Lutz Institute, Presidente Prudente, São Paulo, Brazil. A total of 15 isolates of P. aeruginosa from different origins were analyzed. Environmental isolates (E, P1–P5, n = 5) were obtained from the public water supply system of São Paulo State, Brazil. Clinical isolates from patients without cystic fibrosis (non-FC, P6–P10, n = 5) were obtained from the Presidente Prudente Regional Hospital, São Paulo State, Brazil. Cystic fibrosis isolates (FC, P11–P15, n = 5) were obtained from the University of São Paulo Clinics Hospital, Brazil. Two P. aeruginosa reference strains were used, PAO1 and UCBPP-PA14 (hereafter referred to as PA14). Information on bacterial isolates is summarized in Table 1 .
Table 1.
Pseudomonas aeruginosa strains and bacterial isolates (Environmental, non-CF, and CF isolates), Minimum inhibitory concentration, and 1/2 MIC of bio-AgNP used in this study.
| Reference strains or isolates | Type | Origin | bio-AgNP (µM) | |
|---|---|---|---|---|
| MIC | 1/2 MIC | |||
| PAO1 | C | Wound patient | 62.50 | 31.25 |
| PA14 | C | Burn patient | 62.50 | 31.25 |
| P1 | E | Water from the public supply system | 15.62 | 7.81 |
| P2 | E | Water from the public supply system | 31.25 | 15.62 |
| P3 | E | Water from the public supply system | 15.62 | 7.81 |
| P4 | E | Water from the public supply system | 31.25 | 15.62 |
| P5 | E | Water from the public supply system | 31.25 | 15.62 |
| P6 | non-CF | Tracheal secretion | 31.25 | 15.62 |
| P7 | non-CF | Pleural fluid | 31.25 | 15.62 |
| P8 | non-CF | Knee wound | 31.25 | 15.62 |
| P9 | non-CF | Tracheal secretion | 31.25 | 15.62 |
| P10 | non-CF | Sacral area | 31.25 | 15.62 |
| P11 | CF | Sputum and oropharyngeal | 15.62 | 7.81 |
| P12 | CF | Sputum and oropharyngeal | 15.62 | 7.81 |
| P13 | CF | Sputum and oropharyngeal | 15.62 | 7.81 |
| P14 | CF | Sputum and oropharyngeal | 15.62 | 7.81 |
| P15 | CF | Sputum and oropharyngeal | 15.62 | 7.81 |
C, clinical isolates.
E, environmental isolates.
Non-CF, clinical isolates obtained from non-cystic fibrosis patients.
CF, clinical isolates obtained from cystic fibrosis patients.
MIC, Minimum inhibitory concentration.
Isolates were seeded on Cetrimide agar (Acumedia, USA) and M-PA-C Agar (BBL, USA) and incubated at 37 and 42°C, respectively. Identification was achieved by analysis of colony morphology and biochemical characteristics (catalase, oxidase, and glucose fermentation tests). Stock cultures were maintained at −80°C in Luria–Bertani (LB) broth (Neogen, USA) containing 20% (v/v) glycerol (Synth, Brazil).
Molecular Confirmation of P. aeruginosa Isolates
All isolates biochemically identified as P. aeruginosa were confirmed by polymerase chain reaction (PCR) using a set of primers (forward, 5′-AATTCGGCAAATTTGCTGCG-3′; reverse, 5′-GGAGCTGTCGTACTCGAAGT-3′) specific to the oprL gene (209 bp) (Wong et al., 2014).
Bacterial DNA was extracted by the thermal lysis method. PCR was performed in a final volume of 20 µL containing 6.4 µL of ultrapure water, 0.8 µL of each primer (10 µM), 10 µL of GoTaq® Colorless Master Mix (Promega), and 2 µL of DNA. Amplification consisted of initial denaturation at 95°C for 10 min, followed by 30 cycles of 95°C for 1 min, 52°C for 30 s, and 72°C for 1 min, and final extension at 72°C for 5 min on a Veriti thermal cycler (Applied Biosystems, IL, USA). The amplified products were subjected to electrophoresis in agarose gel (1.5%) stained with 0.1 µL/mL SYBR Safe (Invitrogen, USA) for 40 min at 100 V. Gels were visualized under ultraviolet light (296 nm) using a transilluminator (Syngene®, DigiGenius, MD, USA), and images were captured and processed using EOS Utility® software (Canon, USA).
Preparation of Bio-AgNPs
Bio-AgNPs were synthesized according to a method previously described by Durán et al. (2005; 2006) (Patent, 2006, PI 0605681-4A2; http://www.inpi.gov.br). Bio-AgNPs were prepared by reduction of silver nitrate catalyzed by a cell-free enzyme preparation from Fusarium oxysporum (strain 551). The fungal inoculum was obtained from the culture collection of the Laboratory of Molecular Genetics, Luiz de Queiroz College of Agriculture (ESALQ), University of São Paulo, Piracicaba, São Paulo State, Brazil. First, F. oxysporum was grown for 7 days at 28°C on medium containing 0.5% (w/v) yeast extract (Neogen, USA), 2% (w/v) malt extract (Neogen, USA), 2% (w/v) agar (Acumedia, USA), and distilled water. Then, the fungal biomass (0.1 g/mL) was mixed with sterile distilled water and incubated at 28°C for 72 h under constant stirring (150 rpm). After filtration, the cell-free filtrate was mixed with 0.01 M silver nitrate (AgNO3, Nuclear, Brazil) and incubated for 15 days at 28°C in the dark. Bio-AgNPs were washed with distilled water, centrifuged at 27 000 × g and 4°C for 30 min, and incubated in an ultrasonic bath for 30 min. Washing steps were repeated three times. Quantification of Ag was performed on an EDX-7000 energy dispersive X-ray fluorescence spectrometer (Shimadzu, Japan). Particle diameter and zeta potential were determined by photon correlation spectroscopy using a Zetasizer Nano ZS (Malvern Panalytical, United Kingdom). Transmission electron microscopy was performed at the State University of Maringá, Paraná, Brazil.
Minimum Inhibitory Concentration (MIC) Determination
The MIC of bio-AgNPs for P. aeruginosa isolates was determined by the broth microdilution method in 96-well plates, as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2015). Bio-AgNPs were diluted in Mueller–Hinton broth (Difco, USA) to concentrations of 1000 to 7.81 µM. After incubation at 37°C for 24 h, MIC values were defined as the lowest concentration of bio-AgNPs capable of preventing visible microbial growth. Positive (P. aeruginosa incubated in the absence of bio-AgNPs) and negative (bio-AgNPs and broth only) controls were used.
Virulence Factor Analysis
Swarming Motility
P. aeruginosa isolates were grown in LB broth (Neogen, USA) for 24 h at 30°C. Then, swarming agar plates containing 1% glucose (Synth, Brazil), 0.5% peptone (Acumedia, USA), 0.2% yeast extract (Bacto, Difco, USA), and 0.5% agar (Acumedia, USA) were equilibrated to room temperature and inoculated at the center with 10 µL of a P. aeruginosa suspension containing 108 colony-forming units (CFU)/mL in the presence and absence (control) of subinhibitory concentrations (½ MIC) of bio-AgNPs. Plates were incubated without inversion for 24 h at 30°C (Norizan et al., 2013).
Swimming Motility
P. aeruginosa isolates were seeded on LB agar (Neogen, USA) and incubated at 37°C for 24 h. Then, one colony of each isolate was inoculated, in the presence and absence (control) of ½ MIC bio-AgNPs, on the surface of swimming agar plates containing 1.0% tryptone (Acumedia, USA), 0.5% sodium chloride (Casa Americana, Brazil), and 0.3% agar (Acumedia, USA), previously equilibrated to room temperature. Plates were incubated without inversion for 24 h at 30°C (Inoue et al., 2008).
Twitching Motility
P. aeruginosa isolates were seeded on LB agar (Neogen, USA) and incubated at 37°C for 24 h. Then, one colony of each isolate was inoculated, in the presence and absence (control) of ½ MIC bio-AgNPs, to the bottom of twitching agar plates containing 1.0% tryptone (Acumedia, USA), 0.5% yeast extract (Bacto, Difco, USA), 1.0% sodium chloride (Synth, Brazil), and 1.0% agar (Acumedia, USA). Plates were inverted and incubated at 37°C for 24 h. Subsequently, the agar was carefully removed, and the motility zone was measured to the nearest millimeter after staining with 2% crystal violet (Laborclin, Brazil) for 2 h (Otton et al., 2017). As a negative control, each isolate was inoculated in tryptone soy agar (Difco, USA) under the same conditions.
Biofilm Formation
Biofilm formation was analyzed on 96-well polystyrene microtiter plates by the modified crystal violet method described by Ramos-Vivas et al. (2019). P. aeruginosa isolates were grown on LB agar (Neogen, USA) at 37°C for 24 h. Then, 180 µL of LB broth (Neogen, USA) and 20 µL of P. aeruginosa suspension (initial concentration of 1.5 × 106 CFU/mL) were added to each well and incubated at 37°C for 24 h in the presence and absence (control) of ½ MIC bio-AgNPs. After incubation, the supernatants were discarded, and the wells were washed three times with phosphate-buffered saline (pH 7.2), fixed with 250 µL of methanol (Merck, Germany) for 10 min, and stained with a 1.0% crystal violet aqueous solution (Merck, Germany) for 15 min. Subsequently, the crystal violet was discarded and the wells washed three times with purified water. Adhered cells were resuspended in 250 µL of 33% (v/v) glacial acetic acid (Merck, Germany). Absorbance was read spectrophotometrically (Multiskan™ FC, Thermo Fisher Scientific, USA) at 620 nm.
Statistical Analysis
All experiments were conducted in triplicate on three separate occasions. Statistical analyses were performed using RStudio software version 1.2.5001. Graphical representations were constructed using GraphPad Prism version 8.4.2 (GraphPad Software Inc., USA).
Results
P. aeruginosa Isolates
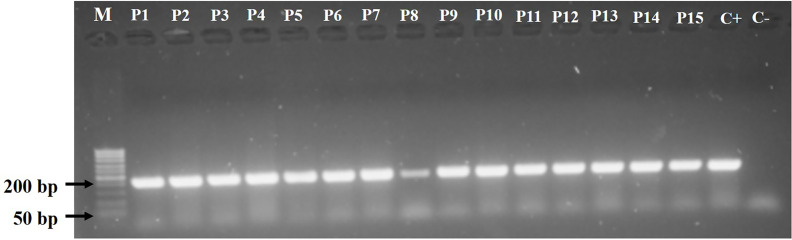
P. aeruginosa isolates were identified as oxidase-positive, catalase-positive, non-fermenter Gram-negative rods capable of growth on Cetrimide and M-PA-C agar. Isolates were confirmed to be P. aeruginosa by oprL gene detection ( Figure 1 ).
Figure 1.
Agarose gel electrophoresis (1.0%) of Pseudomonas aeruginosa isolates showing amplified PCR products for the oprL gene (209 bp). Lane 1 (M), molecular weight marker; P1–P5, environmental isolates; P6–P10, clinical isolates obtained from non-cystic fibrosis patients; P11–P15, clinical isolates obtained from cystic fibrosis patients.
Characterization of Bio-AgNPs
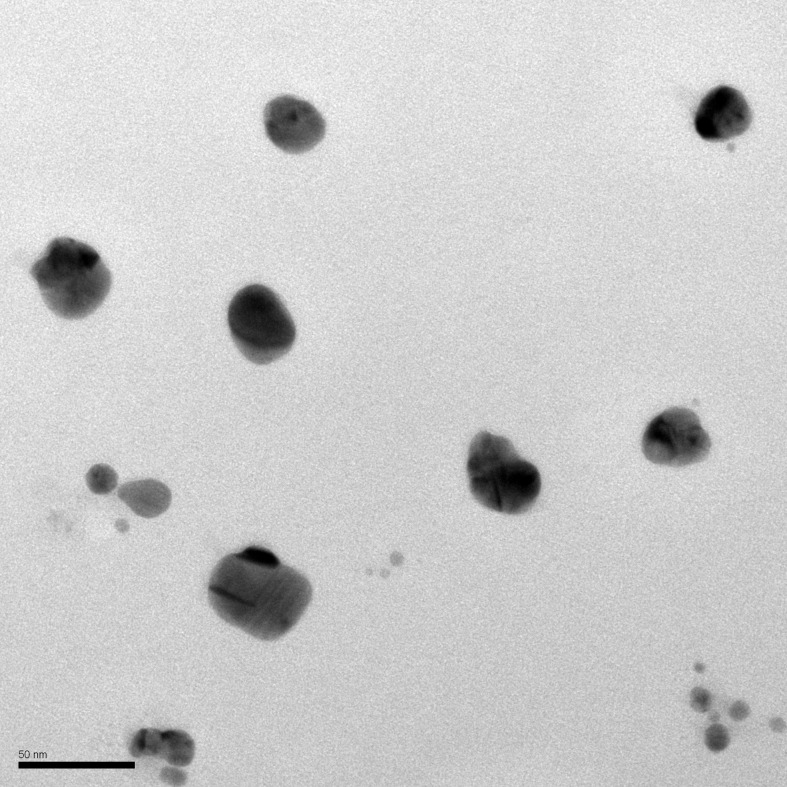
The X-ray fluorescence spectra of bio-AgNPs showed a plasmonic absorption band in the visible region (near 440 nm). Nanoparticles had a spherical shape and mean size of 73.04 nm, as demonstrated by transmission electron microscopy ( Figure 2 ). The surface charge (zeta potential) of bio-AgNPs was −18.77 mV.
Figure 2.
Characterization by transmission electron microscopy (TEM) of bio-AgNP synthesized by Fusarium oxysporum (300,000×).
MIC of Bio-AgNPs
The MIC of bio-AgNPs for P. aeruginosa PAO1 and PA14 was 62.5 µM. MICs for environmental and clinical isolates are described in Table 1 . Bio-AgNPs were used at ½ MIC (31.25, 15.62, or 7.81 µM) to test their action against P. aeruginosa motility and biofilm formation.
Effect of Bio-AgNPs on P. aeruginosa Motility
We tested the sub-MIC effects of bio-AgNPs on the three types of P. aeruginosa motility (swarming, swimming, and twitching), measured in diameter (mm). According to Table 2 , bio-AgNPs exerted distinct effects on the motility of reference strains PAO1 and PA14. In PAO1, bio-AgNPs significantly reduced swarming (p = 0.092) and swimming (p < 0.001). In PA14, however, nanoparticles enhanced swarming (p = 0.002) and twitching (p = 0.008) motility.
Table 2.
Swarming, swimming, and twitching motility in reference strains (PAO1 and PA14), environmental isolates, and clinical isolates of Pseudomonas aeruginosa in the presence and absence of biogenic silver nanoparticles (bio-AgNPs) at ½ MIC.
| Reference strainsor isolates | Type | Swarming (mm) | Swimming (mm) | Twitching (mm) | |||
|---|---|---|---|---|---|---|---|
| Control | bio-AgNP | Control | bio-AgNP | Control | bio-AgNP | ||
| PAO1 | C | 58.9 ± 10.4 | 45.7 ± 19.1** | 41.0 ± 2.5 | 29.3 ± 3.6*** | 11.6 ± 0.5 | 11.0 ± 0.8 |
| PA14 | C | 13.9 ± 4.8 | 45.0 ± 20.8** | 34.7 ± 4.0 | 36.1 ± 3.6 | 13.7 ± 1.7 | 19.1 ± 1.1** |
| P1 | E | 38.8 ± 16.1 | 47.3 ± 22.2 | 43.9 ± 2.5 | 46.3 ± 5.2 | 4.2 ± 2.1 | 4.4 ± 1.9 |
| P2 | E | 19.0 ± 8.2 | 21.4 ± 5.4 | 38.7 ± 6.0 | 48.8 ± 10.6** | 5.6 ± 1.0 | 5.2 ± 1.3 |
| P3 | E | 10.1 ± 0.6 | 15.6 ± 4.8** | 41.2 ± 3.4 | 46.3 ± 10.6 | 5.0 ± 1.6 | 5.2 ± 1.3 |
| P4 | E | 17.0 ± 5.2 | 25.6 ± 14.9 | 34.1 ± 7.3 | 34.8 ± 9.8 | 5.0 ± 1.1 | 6.3 ± 1.1** |
| P5 | E | 39.1 ± 20.4 | 44.2 ± 25.0 | 41.1 ± 3.6 | 49.7 ± 13.9 | 4.9 ± 0.9 | 5.3 ± 1.0** |
| P6 | non-CF | 10.9 ± 0.8 | 32.1 ± 29.2** | 41.6 ± 7.6 | 43.3 ± 11.1 | 9.1 ± 1.9 | 12.1 ± 5.6* |
| P7 | non-CF | 8.7 ± 0.7 | 13.2 ± 1.2** | 49.6 ± 5.0 | 46.2 ± 8.9 | 3.5 ± 0.9 | 5.0 ± 1.3* |
| P8 | non-CF | 13.3 ± 2.4 | 17.8 ± 4.2** | 33.8 ± 3.6 | 44.3 ± 4.0*** | 5.1 ± 0.3 | 6.3 ± 3.3* |
| P9 | non-CF | 14.0 ± 1.5 | 28.6 ± 8.2** | 41.2 ± 3.2 | 53.4 ± 10.6* | 7.1 ± 3.3 | 8.1 ± 4.9 |
| P10 | non-CF | 16.4 ± 6.4 | 47.8 ± 8.7*** | 25.2 ± 7.3 | 36.2 ± 6.0** | 2.8 ± 1.3 | 3.9 ± 0.9 |
| P11 | CF | 16.4 ± 6.9 | 20.6 ± 6.1 | 42.2 ± 2.5 | 48.6 ± 9.2* | 7.1 ± 0.8 | 6.2 ± 1.7 |
| P12 | CF | 32.7 ± 21.6 | 25.8 ± 13.2 | 42.6 ± 5.4 | 48.9 ± 8.1* | 5.6 ± 1.1 | 5.3 ± 1.6 |
| P13 | CF | 68.3 ± 15.5 | 73.0 ± 13.3 | 38.4 ± 5.1 | 43.0 ± 10.7 | 4.0 ± 0.6 | 3.6 ± 1.9 |
| P14 | CF | 53.6 ± 30.0 | 52.9 ± 30.5 | 40.3 ± 8.7 | 39.3 ± 15.4 | 4.1 ± 0.7 | 4.8 ± 0.6** |
| P15 | CF | 43.9 ± 30.6 | 41.2 ± 27.8 | 45.8 ± 6.5 | 46.9 ± 9.1 | 4.1 ± 0.9 | 4.4 ± 1.2** |
The results were obtained by the mean ± standard deviation of three independent experiments.
*P < 0.05, ** < 0.01, and *** < 0.001, comparing the control bacteria (without treatment) with the treated (bio-AgNP).
Bio-AgNP, silver nanoparticles.
C, clinical isolates.
E, environmental isolates.
Non-CF, clinical isolates obtained from non-cystic fibrosis patients.
CF, clinical isolates obtained from cystic fibrosis patients.
Most P. aeruginosa isolates showed an increase in swarming motility after bio-AgNP treatment, except for P12, P14, and P15 (all obtained from CF). The swarming motility diameters of six isolates (40.0%) were significantly higher (p < 0.050) in the presence of bio-AgNPs ( Table 2 ). A similar result was observed for swimming motility ( Table 2 ). This parameter increased in all isolates with bio-AgNP treatment, except P7 and P14 (non-FC and FC, respectively). Swimming motility diameters were significantly higher (p < 0.050) in the presence of bio-AgNPs in six isolates (40.0%). Twitching motility diameters increased (p < 0.050) with bio-AgNP treatment in seven isolates (46.7%).
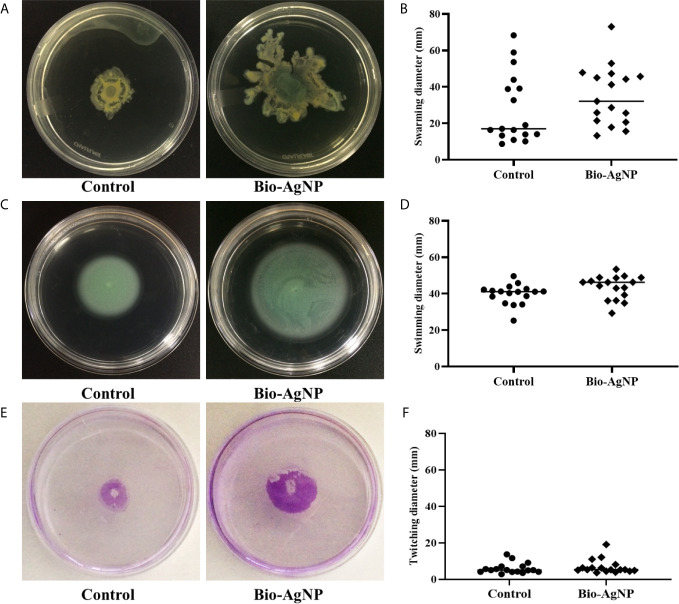
As shown in Figure 3A , the swarming motility zone of P2 in the absence of bio-AgNPs was 19.0 ± 8.2 mm, which increased to 21.4 ± 5.4 mm after bio-AgNPs were added to the culture medium. Among the tested isolates, the increase in swarming motility ranged from 6.9 to 194.5%.
Figure 3.
Pseudomonas aeruginosa motility in the absence (control) and presence of biogenic silver nanoparticles (bio-AgNPs). (A) Swarming motility of P2 isolate, (B) mean swarming motility, (C) swimming motility of P2 isolate, (D) mean swimming motility, (E) twitching motility of P6 isolate, and (F) mean twitching motility.
The swimming motility zone of P2 was 38.7 ± 6.0 mm without bio-AgNP treatment and 48.8 ± 10.6 mm with treatment ( Figure 3C ). Bio-AgNPs increased swimming motility in P. aeruginosa isolates by 2.4 to 43.7%.
P6 showed a twitching motility of 9.1 ± 1.9 mm in the absence of bio-AgNPs and 12.1 ± 5.6 mm in the presence of bio-AgNPs ( Figure 3E ). Bio-AgNP treatment increased twitching motility in P. aeruginosa isolates by 4.0 to 42.9%. The mean swarming, swimming, and twitching diameters of P. aeruginosa isolates are detailed in Figures 3B, D, F , respectively.
Effect of Bio-AgNPs on P. aeruginosa Biofilm Formation
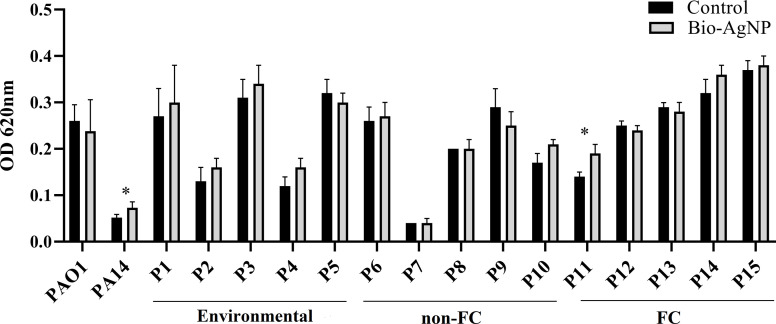
Bio-AgNP treatment promoted biofilm formation in most P. aeruginosa isolates (73.3%), except P5, P9 (non-FC), P12, P13 (FC isolates) ( Figure 4 ). Similar effects were exerted on PAO1 and PA14. However, a significant increase in biofilm formation was only observed in the PA14 reference strain (p = 0.040) and the P11 isolate (p = 0.034), obtained from a patient with cystic fibrosis ( Figure 4 ).
Figure 4.
Biofilm formation capacity of reference strains, environmental isolates, and clinical isolates of Pseudomonas aeruginosa in the absence (control) and presence of biogenic silver nanoparticles (bio-AgNPs). Results are the mean ± standard deviation of three independent experiments. *p < 0.05 compared with the control.
Discussion
The antibacterial activity of bio-AgNPs against Klebsiella pneumoniae, Staphylococcus aureus, Escherichia coli, Acinetobacter baumannii, and Salmonella Enteritidis at inhibitory concentrations was previously described by Scandorieiro et al. (2016) and Figueiredo et al. (2019). Few studies, however, have investigated the subinhibitory effects of AgNPs on clinical isolates or environmental microorganisms (Yang and Alvarez, 2015; Garuglieri et al., 2016; Grün et al., 2018).
Motility is fundamental for bacterial colonization of certain surfaces; it allows microorganisms to move toward more favorable environments and away from unfavorable conditions. Motility is also important for pathogenicity, as it is involved in host cell adhesion, colonization, biofilm formation, and survivability (Sun et al., 2018).
In the present study, the PAO1 reference strain showed a significant reduction in swarming and swimming motility with bio-AgNP treatment. In contrast, PA14 showed a significant increase in swarming and twitching motility. These reference strains differ in origin and genomic characteristics. P. aeruginosa PAO1, isolated from a wound, is a moderately virulent strain belonging to a relatively rare clonal group. PA14 is highly virulent in several infection models and is part of the most common clonal group worldwide. Among other factors, differences between strains may be due to the presence of two pathogenicity islands (PAPI-1 and PAPI-2) in PA14 but not in PAO1 (Harrison et al., 2010; Mikkelsen et al., 2011). PAPI-1 encodes several virulence-associated factors, including type IVb pili (pilS2 gene) (Bahramian et al., 2019) and PvrR, a response regulator involved in the synthesis of bacterial biofilms (Guła et al., 2019). PAPI-2 encodes ExoU cytotoxin, a potent phospholipase (Harrison et al., 2010). Researchers have shown that exclusion of one or both pathogenicity islands affects the virulence of PA14 in murine models of acute pneumonia and bacteremia (Harrison et al., 2010). According to Mikkelsen et al. (2011), even though PA14 has pathogenicity islands that are not found in PAO1, the presence or absence of specific gene clusters is not predictive of virulence. The referred study found that PA14 has an acquired mutation in the ladS gene that has a deleterious impact on biofilm formation but promotes the activity of type III secretion systems and cytotoxicity in mammalian cells. This could explain why PA14 had a lower biofilm-forming capacity than PAO1 in the present study.
Our results showed that about 40% of isolates increased swarming, swimming, and twitching activity after treatment with bio-AgNPs at sub-MIC. Villa et al. (2012) demonstrated that sublethal concentrations of oxidizing biocides increase swarming and swimming motility in the soil bacterium Azotobacter vinelandii, representing a strategy to mitigate adverse conditions. Ortega-Calvo et al. (2011) also presented evidence that AgNPs induce a repellent response in Pseudomonas putida. The mechanism that causes repellence remains unknown but it is believed to involve physiological changes caused by accumulation of AgNPs in the cell membrane and/or within cells, modifying microbial performance. The size of AgNPs also seems to influence repellence. Another study revealed that subinhibitory concentrations of AgNPs stimulated swarming and swimming in E. coli and Bacillus subtilis under aerobic and anaerobic conditions (Garuglieri et al., 2016).
Singh et al. (2019) investigated the effects of AgNPs and silver ions on P. aeruginosa PAO1. Treatment with 3.12 µg/mL AgNP increased the expression of motility-related genes, such as bswR (swarming motility regulator) and pilG (twitching motility protein), and the activation of flagellum-related genes (flgC, fliG, fliH, and fliP).
In this study, after 24 h of incubation with ½ MIC bio-AgNPs, there was an increase in biofilm production in the reference strain PA14 and isolate P11. Such an increase can be explained by one of the mechanisms of action of AgNPs in bacterial cells: formation of reactive oxygen species (ROS), which cause oxidative stress (Wang et al., 2017). The high ROS concentrations occurring under oxidative stress may lead to adverse modifications in cellular components and damage to proteins, DNA, and lipids (Su et al., 2019). Villa et al. (2012) reported that oxidative stress in A. vinelandii increased with oxidizing biocide treatment. It is probable that high oxidative stress exerted selective pressure at the initial stage of biofilm development, enhancing biofilm formation capacity.
Pseudomonas biofilms are composed of at least three types of polysaccharides: Psl, Pel, and alginate. Research has shown that pslA expression decreased in the presence of AgNPs and that expression of pslD (a gene encoding an outer membrane protein) decreased in the presence of silver ions. On the other hand, AgNPs and silver ions increased the expression of genes associated with glycosyltransferases, pslC, pslL, and pslE (Wzz/Wzc-like protein) (Singh et al., 2019).
Guo et al. (2019) observed that subinhibitory doses of AgNPs enhanced polysaccharide and protein production in P. aeruginosa CCM 3955 compared with the control, significantly altering biofilm structure. The major form of protection of bacteria against low AgNP concentrations was to increase exopolysaccharide secretion.
According to Yang and Alvarez (2015), exposure of P. aeruginosa PAO1 to sublethal AgNP concentrations improved biofilm development and increased expression of genes related to QS (lasI and lasR). P. aeruginosa has three known QS systems: las, rhl, and PQS (Lee and Zhang, 2015; Cornelis, 2020). QS systems allow communication between microorganisms through the production and diffusion of chemical or signaling molecules that control a variety of physiological functions, such as biofilm formation and motility activity (Saeki et al., 2020). Rasamiravaka et al. (2015) reported that the three QS systems of P. aeruginosa play important roles in biofilm formation and development.
Contrasting results were obtained by Hussain et al. (2019) when testing the effects of AgNPs (10–50 nm) on different microorganisms, including P. aeruginosa PAO1. At subinhibitory concentrations (1/16, 1/8, 1/4, and 1/2 MIC), there was a 42–81% reduction in swarming motility and a 22–79% reduction in biofilm formation. Another study showed that subinhibitory concentrations of gold nanoparticles (mean size of 53 nm) significantly reduced (p < 0.01) swarming, swimming, twitching, and biofilm formation (Khan et al., 2019). These differences may be related to nanoparticle size. Liao et al. (2019) showed that larger nanoparticles have lower antibacterial activity than smaller nanoparticles. The bio-AgNPs prepared in this study had a mean size of 75 nm. Other studies have shown that size may not be a determinant of virulence factor regulation. Garuglieri et al. (2016) showed that treatment with AgNPs sized 14 ± 0.3 nm increased motility and Grün et al. (2018) found that treatment with 30–70 nm AgNPs enhanced biofilm formation.
Currently, there are several commercial AgNP products widely used in the medical and food industries and water treatment plants (McNamara and Tofail, 2017; Gadkari et al., 2018; Jaiswal et al., 2019). This wide use implies that metal compounds are constantly released into the aquatic environment through sewage and wastewater disposal (Das et al., 2012; Singh et al., 2019). The release of AgNPs, even at subinhibitory concentrations, is a cause of environmental concern (Yang and Alvarez, 2015; Grün et al., 2018).
Overall, it can be inferred that the increase in phenotypic expression of biofilm and bacterial motility after exposure to bio-AgNPs may be associated with chemical signaling, as these virulence factors are regulated by the QS system. Therefore, further research is needed on the action of AgNPs in the QS system of P. aeruginosa for a better understanding of their effects at subinhibitory concentrations. Moreover, it is necessary to explore other forms of AgNP use with the aim of reducing antimicrobial resistance as well as environmental impacts.
Upon exposure to subinhibitory concentrations of bio-AgNPs, the PA14 reference strain and some P. aeruginosa isolates showed increased swarming, swimming, and twitching motility and biofilm formation. Unfortunately, some studies have also described that low doses of nanoparticles can significantly induce biofilm formation and virulence factors in vitro. This increase in bacterial exposure to nanoparticles is a cause for concern, given all the adverse effects that have not been fully clarified. Therefore, the use of subinhibitory concentrations bio-AgNPs especially in therapeutic applications must be performed with precaution.
These results are an initial contribution to broaden our understanding of the effects of subinhibitory concentrations of bio-AgNPs on bacterial behavior, an important but little-studied phenomenon. However, it is still necessary to elucidate the regulation of virulence factors in bio-AgNP-treated bacteria. Further research on the interference mechanisms of subinhibitory concentrations of bio-AgNPs in P. aeruginosa QS systems is recommended.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
EKS designed the study, interpreted the clinical data, and wrote the paper. AYY and LAA analyzed the virulence factors of Pseudomonas aeruginosa. LA performed the genomics experiments. DOG and RLBS discussed the results and implications and wrote the manuscript. RKTK and GN interpreted the clinical data. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Ahmed S. A. K. S., Rudden M., Smyth T. J., Dooley J. S. G., Marchant R., Banat I. M. (2019). Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl. Microbiol. Biotechnol. 103 (8), 3521–3535. 10.1007/s00253-019-09618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T., Pattnaik S., Khan M. B., Ampasala D. R., Busi S., Sarma V. V. (2020). Inhibition of quorum sensing-associated virulence factors and biofilm formation in Pseudomonas aeruginosa PAO1 by Mycoleptodiscus indicus PUTY1. Braz. J. Microbiol. 51 (2), 467–487. 10.1007/s42770-020-00235-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anversa L., Stancari R. C. A., Garbelotti M., Ruiz L. S., Pereira V. B. R., Nascentes G. A. N., et al. (2019). Pseudomonas aeruginosa in public water supply. Water Pract. Technol. 14 (3), 732–737. 10.2166/wpt.2019.057 [DOI] [Google Scholar]
- Bahramian A., Khoshnood S., Shariati A., Doustdar F., Chirani A. S., Heidary M. (2019). Molecular characterization of the pilS2 gene and its association with the frequency of Pseudomonas aeruginosa plasmid pKLC102 and PAPI-1 pathogenicity island. Infect. Drug Resist. 12, 221–227. 10.2147/IDR.S188527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2015). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition. CLSI document M07-A10. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cornelis P. (2020). Putting an end to the Pseudomonas aeruginosa IQS controversy. Microbiologyopen 9 (2), e962. 10.1002/mbo3.962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P., Williams C. J., Fulthorpe R. R., Hoque M. E., Metcalfe C. D., Xenopoulos M. A. (2012). Changes in Bacterial Community Structure after Exposure to Silver Nanoparticles in Natural Waters. Environ. Sci. Technol. 46 (16), 9120–9128. 10.1021/es3019918 [DOI] [PubMed] [Google Scholar]
- Dobias J., Bernier-Latmani R. (2013). Silver release from silver nanoparticles in natural waters. Environ. Sci. Technol. 47 (9), 4140–4146. 10.1021/es304023p [DOI] [PubMed] [Google Scholar]
- D’Lima L., Phadkea M., Ashokb A. D. (2020). Biogenic silver and silver oxide hybrid nanoparticles: a potential antimicrobial against multi drug-resistant Pseudomonas aeruginosa . New J. Chem. 12 (44), 4935–4941. 10.1039/c9nj04216d [DOI] [Google Scholar]
- Durán N., Marcato P. D., Alves O. L., Souza G. I. H., Esposito E. (2005). Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J. Nanobiotechnol. 3 (8), 1–7. 10.1186/1477-3155-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán N., Alves O. L., Esposito E., de Souza G. I., Marcato P. D. (2006). Processo de produção de nanopartículas de prata estabilizadas por proteínas na produção de produtos têxteis antibacterianos e o tratamento dos efluentes produzidos (Campinas: BR. Instituto Nacional da Propriedade Industrial (INPI; ). Braz. patent PI 0605681-4 A. [Google Scholar]
- Durán N., Rolim W. R., Durán M., Fávaro W. J., Seabra A. B. (2019). Nanotoxicologia de nanopartículas de prata: toxicidade em animais e humanos. Quím. Nova 42 (2), 206–213. 10.21577/0100-4042.20170318 [DOI] [Google Scholar]
- Esmaeili D., Daymad S. F., Neshani A., Rashki S., Marzhoseyni Z., Khaledi A. (2019). Alerting prevalence of MBLs producing Pseudomonas aeruginosa isolates. Gene Rep. 16:100460. 10.1016/j.genrep.2019.100460 [DOI] [Google Scholar]
- Figueiredo E. P., Ribeiro J. M., Nishio E. K., Scandorieiro S., Costa A. F., Cardozo V. F., et al. (2019). New Approach For Simvastatin As An Antibacterial: Synergistic Effect With Bio-Synthesized Silver Nanoparticles Against Multidrug-Resistant Bacteria. Int. J. Nanomed. 14, 7975–7985. 10.2147/IJN.S211756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadkari R. R., Ali S. W., Alagirusamy R., Das A. (2018). “Silver Nanoparticles in Water Purification: Opportunities and Challenges,” in Modern Age Environmental Problems and their Remediation. Eds. Oves M., Zain Khan M., M.I. Ismail I. (Cham: Springer; ). 10.1007/978-3-319-64501-8_13 [DOI] [Google Scholar]
- Garuglieri E., Cattò C., Villa F., Zanchi R., Cappitelli F. (2016). Effects of sublethal concentrations of silver nanoparticles on Escherichia coli and Bacillus subtilis under aerobic and anaerobic conditions. Biointerphases 11 (4), 04B308. 10.1116/1.4972100 [DOI] [PubMed] [Google Scholar]
- Grün A. Y., App C. B., Breidenbach A., Meier J., Metreveli G., Schaumann G. E., et al. (2018). Effects of low dose silver nanoparticle treatment on the structure and community composition of bacterial freshwater biofilms. PloS One 13 (6), p.e0199132. 10.1371/journal.pone.0199132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guła G., Dorotkiewicz-Jach A., Korzekwa K., Valvano M. A., Drulis-Kawa Z. (2019). Complex Signaling Networks Controlling Dynamic Molecular Changes in Pseudomonas aeruginosa Biofilm. Curr. Med. Chem. 26 (11), 1979–1993. 10.2174/0929867325666180912110151 [DOI] [PubMed] [Google Scholar]
- Guo J., Qin S., Wei Y., Liu S., Peng H., Li Q., et al. (2019). Silver nanoparticles exert concentration-dependent influences on biofilm development and architecture. Cell Prolif. 52 (4), p.e12616. 10.1111/cpr.12616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E. M., Carter M. E., Luck S., Ou H., He X., Deng Z., et al. (2010). Pathogenicity islands PAPI-1 and PAPI-2 contribute individually and synergistically to the virulence of Pseudomonas aeruginosa strain PA14. Infect. Imm. 78 (4), 1437–1446. 10.1128/IAI.00621-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A., Alajmi M. F., Khan M. A., Pervez S. A., Ahmed F., Amir S., et al. (2019). Biosynthesized Silver Nanoparticle (AgNP) From Pandanus odorifer Leaf Extract Exhibits Anti-metastasis and Anti-biofilm Potentials. Front. Microbiol. 10:8 (8). 10.3389/fmicb.2019.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Shingaki R., Fukui K. (2008). Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol. Lett. 281 (1), 81–86. 10.1111/j.1574-6968.2008.01089.x [DOI] [PubMed] [Google Scholar]
- Jaiswal L., Shankar S., Rhim J.-W. (2019). “Chapter 3 - Applications of nanotechnology in food microbiology,” In: Gurtler V., Ball A. S., Soni S. (eds) Methods in Microbiology. Australia: Academic Press. 10.1016/bs.mim.2019.03.002 [DOI] [Google Scholar]
- Kawish M., Ullah F., Ali H. S., Saifullah S., Ali I., ur Rehman J., et al. (2020). “Chapter 10 - Bactericidal potentials of silver nanoparticles: novel aspects against multidrug resistance bacteria,” In: Shah M. R., Imran M., Ullah S. (eds). Micro and Nano Technologies,Metal Nanoparticles for Drug Delivery and Diagnostic Applications. Pakistan: Elsevier: Elsevier. 10.1016/B978-0-12-816960-5.00010-0 [DOI] [Google Scholar]
- Kearns D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8 (9), 634–644. 10.1038/nrmicro2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F., Manivasagan P., Lee J. W., Pham D. T. N., Oh J., Kim Y. M. (2019). Fucoidan-Stabilized Gold Nanoparticle-Mediated Biofilm Inhibition, Attenuation of Virulence and Motility Properties in Pseudomonas aeruginosa PAO1. Mar. Drugs 17 (4), 208. 10.3390/md17040208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F., Pham D. T. N., Oloketuyi S. F., Kim Y. M. (2020). Regulation and controlling the motility properties of Pseudomonas aeruginosa . Appl. Microbiol. Biotechnol. 104 (1), 33–49. 10.1007/s00253-019-10201-w [DOI] [PubMed] [Google Scholar]
- Köhler T., Curty L. K., Barja F., van Delden C., Pechère J. C. (2000). Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182 (21), 5990–5996. 10.1128/jb.182.21.5990-5996.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhang L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa . Protein Cell. 6 (1), 26–41. 10.1007/s13238-014-0100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S., Zhang Y., Pan X., Zhu F., Jiang C., Liu Q., et al. (2019). Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa . Int. J. Nanomed. 14, 1469–1487. 10.2147/IJN.S191340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matar G. M. (2018). Editorial: Pseudomonas and Acinetobacter: From drug resistance to pathogenisis. Front. Cell. Infect. Microbiol. 68 (8), 1–3. 10.3389/fcimb.2018.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara K., Tofail S. A. M. (2017). Nanoparticles in biomedical applications. Adv. Phys. 2 (1), 54–88. 10.1080/23746149.2016.1254570 [DOI] [PubMed] [Google Scholar]
- Mikkelsen H., McMullan R., Filloux A. (2011). The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PloS One 6 (12), p.e29113. 10.1371/journal.pone.0029113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norizan S. N., Yin W. F., Chan K. G. (2013). Caffeine as a potential quorum sensing inhibitor. Sensors 13 (4), 5117–5129. 10.3390/s130405117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares E., Badel-Berchoux S., Provot C., Prévost G., Bernardi T., Jehl F. (2020). Clinical Impact of Antibiotics for the Treatment of Pseudomonas aeruginosa Biofilm Infections. Front. Microbiol. 9, 2894 (10):2894. 10.3389/fmicb.2019.02894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Calvo J. J., Molina R., Jimenez-Sanchez C., Dobson P. J., Thompson I. P. (2011). Bacterial tactic response to silver nanoparticles. Environ. Microbiol. Rep. 3 (5), 526–534. 10.1111/j.1758-2229.2011.00252.x [DOI] [PubMed] [Google Scholar]
- Otton L. M., Campos M. S., Meneghetti K. L., Corção G. (2017). Influence of twitching and swarming motilities on biofilm formation in Pseudomonas strains. Arch. Microbiol. 199 (5), 677–682. 10.1007/s00203-017-1344-7 [DOI] [PubMed] [Google Scholar]
- Piewngam P., Chiou J., Chatterjee P., Otto M. (2020). Alternative approaches to treat bacterial infections: targeting quorum-sensing. Expert Rev. Anti Infect. Ther. 18 (6), 499–510. 10.1080/14787210.2020.1750951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Vivas J., Chapartegui-González I., Fernández-Martínez M., González-Rico C., Fortún J., Escudero R., et al. (2019). Biofilm formation by multidrug resistant Enterobacteriaceae strains isolated from solid organ transplant recipients. Sci. Rep. 9 (1), 8928. 10.1038/s41598-019-45060-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasamiravaka T., Labtani Q., Duez P., El Jaziri M. (2015). The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. BioMed. Res. Int. 2015 (759348), 1–17. 10.1155/2015/759348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S., Iqbal A., Ashraf M. A. (2020). Bacterial-mediated synthesis of silver nanoparticles and their significant effect against pathogens. Environ. Sci. Pollut. R. 27, 37347–37356. 10.1007/s11356-020-07610-0 [DOI] [PubMed] [Google Scholar]
- Saeki E. K., Kobayashi R. K. T., Nakazato G. (2020). Quorum sensing system: Target to control the spread of bacterial infections. Microb. Pathog. 142:104068. 10.1016/j.micpath.2020.104068 [DOI] [PubMed] [Google Scholar]
- Saxena P., Joshi Y., Rawat K., Bisht R. (2019). Biofilms: Architecture, Resistance, Quorum Sensing and Control Mechanisms. Indian J. Microbiol. 59, 3–12. 10.1007/s12088-018-0757-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandorieiro S., de Camargo L. C., Lancheros C. A., Yamada-Ogatta S. F., Nakamura C. V., Oliveira A. G., et al. (2016). Synergistic and Additive Effect of Oregano Essential Oil and Biological Silver Nanoparticles against Multidrug-Resistant Bacterial Strains. Front. Microbiol. 7, 760. 10.3389/fmicb.2016.00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Gaikwad S., Nagar S., Kulshrestha S., Vaidya V., Nawani N., et al. (2019). Biofilm inhibition and anti-quorum sensing activity of phytosynthesized silver nanoparticles against the nosocomial pathogen Pseudomonas aeruginosa . Biofouling 35 (1), 34–49. 10.1080/08927014.2018.1563686 [DOI] [PubMed] [Google Scholar]
- Singh N., Paknikar K. M., Rajwade J. (2019). RNA-sequencing reveals multitude effects of silver nanoparticles on Pseudomonas aeruginosa biofilms. Environ. Sci.: Nano. 6 (6), 1812–1828. 10.1039/C8EN01286E [DOI] [Google Scholar]
- Su L., Zhang J., Gomez H., Murugan R., Hong X., Xu D., et al. (2019). Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell. Longev. 2019:5080843. 10.1155/2019/5080843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun E., Liu S., Hancock R. E. W. (2018). Surfing Motility: a Conserved yet Diverse Adaptation among Motile Bacteria. J. Bacteriol. 200 (23), p.e00394–18. 10.1128/JB.00394-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa F., Remelli W., Forlani F., Gambino M., Landini P., Cappitelli F. (2012). Effects of chronic sub-lethal oxidative stress on biofilm formation by Azotobacter vinelandii.v. Biofouling 28 (8), 823–833. 10.1080/08927014.2012.715285 [DOI] [PubMed] [Google Scholar]
- Wang L., Hu C., Shao L. (2017). The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int. J. Nanomed. 12, 1227–1249. 10.2147/IJN.S121956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y. P., Chua K. H., Thong K. L. (2014). One-step species-specific high resolution melting analysis for nosocomial bacteria detection. J. Microbiol. Methods 107, 133–137. 10.1016/j.mimet.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Yang Y., Alvarez P. J. J. (2015). Sublethal Concentrations of Silver Nanoparticles Stimulate Biofilm Development. Environ. Sci. Technol. Lett. 2 (8), 221–226. 10.1021/acs.estlett.5b00159 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.