Abstract
Dynamic chemical modifications of RNA represent novel and fundamental mechanisms that regulate stemness and tissue homeostasis. Rejuvenation and wound repair of mammalian skin are sustained by epidermal progenitor cells, which are localized within the basal layer of the skin epidermis. N6‐methyladenosine (m6A) is one of the most abundant modifications found in eukaryotic mRNA and lncRNA (long noncoding RNA). In this report, we survey changes of m6A RNA methylomes upon epidermal differentiation and identify Pvt1, a lncRNA whose m6A modification is critically involved in sustaining stemness of epidermal progenitor cells. With genome‐editing and a mouse genetics approach, we show that ablation of m6A methyltransferase or Pvt1 impairs the self‐renewal and wound healing capability of skin. Mechanistically, methylation of Pvt1 transcripts enhances its interaction with MYC and stabilizes the MYC protein in epidermal progenitor cells. Our study presents a global view of epitranscriptomic dynamics that occur during epidermal differentiation and identifies the m6A modification of Pvt1 as a key signaling event involved in skin tissue homeostasis and wound repair.
Keywords: epidermal progenitor cell, lncRNA, m6A modification, Pvt1 signaling
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Development & Differentiation; RNA Biology
Mettl14‐dependent long non‐coding RNA methylation supports skin stem cell function and wound repair.
Introduction
Chemical modifications of RNA are critically involved in many biological processes. To date, over 150 different RNA modifications have been identified with a wide variety of chemical diversities (http://rna‐mdb.cas.albany.edu/RNAmods/) (Cantara et al, 2011). Besides abundant and diverse modifications on tRNA and rRNA, some of these modifications also occur on mammalian mRNA and lncRNA (long noncoding RNA). These post‐transcriptional changes are crucial to many fundamental processes including cell differentiation, tissue development, metabolism, neural functions, immune response, and viral infection. N6‐methyladenosine (m6A) is one of the most abundant modifications found in eukaryotic mRNA and lncRNA. The m6A modification is ubiquitous among all higher eukaryotes and among viruses that replicate inside host nuclei. Each mammalian mRNA contains ~3 m6A modifications within a consensus sequence of Pu[G>A]m6AC[A/C/U] (Harper et al, 1990). Antibody‐based m6A‐profiling has revealed human and mouse m6A RNA methylomes (Dominissini et al, 2012; Meyer et al, 2012), indicating that these modifications are preserved across different tissues and are subject to dynamic modulation, further suggesting the regulatory roles of m6A. The recent extensive functional characterizations of m6A in eukaryotes uncovered it as a basic mechanism to control processing, transport, translation, and stability of mRNAs and lncRNAs (Yue et al, 2015; Zhao et al, 2017; Roundtree et al, 2017a; Zhao et al, 2018). This modification is installed on mRNA by a dedicated methyltransferase complex, with Mettl3 (methyltransferase like 3) and Mettl14 (Methyltransferase like 14) forming the core catalytic complex, but also with a number of other protein cofactors (Yue et al, 2015; Wen et al, 2018; Yue et al, 2018). It can be removed by two erasers, FTO (fat mass and obesity‐associated protein/α‐ketoglutarate‐dependent dioxygenase) and ALKBH5 (AlkB homolog 5). There are a number of proteins that have been identified as reader proteins that could recognize m6A‐methylated RNA (Wang et al, 2014; Wang et al, 2015; Liu et al, 2015b; Hsu et al, 2017; Shi et al, 2017; Roundtree et al, 2017b; Huang et al, 2018), including the YTH (YT521‐B homology) domain family of proteins (Theler et al, 2014; Xu et al, 2014). Studies from the past few years suggest that m6A as an mRNA chemical mark allows cells to group hundreds to thousands of transcripts for coordinated translation and transcriptome turnover during stem cell differentiation or in response to cellular and environmental signals (Zhao et al, 2017; Roundtree et al, 2017a).
Mammalian skin protects us from many different environmental damages (Fuchs, 2008). The epidermal progenitor/stem cells that localize at the basal layer of the epidermis play a critical role in tissue homeostasis of skin. During epidermal stratification, basal progenitor cells can migrate upward from the basement membrane and differentiate to replenish the tissue with new differentiated epidermal keratinocytes (Fuchs, 2008; Lopez‐Pajares et al, 2013; Perdigoto et al, 2014). Differentiation of basal progenitor cells involves the synthesis and modification of different lipid and protein molecules of differentiated corneocytes, the controlled disruption, and degradation of subcellular organelles and the nucleus, as well as the permanent withdrawal from the cell cycle. Deregulation of skin differentiation can lead to many different skin diseases, such as inflammatory skin diseases and skin cancers.
Despite our understanding of the transcriptional changes and signal transductions involved in skin differentiation (Fuchs, 2008; Lopez‐Pajares et al, 2013; Perdigoto et al, 2014), the potential role of m6A modification in skin development and adult skin tissue homeostasis remains largely unclear. Using a mouse genetics approach, we found that conditional ablation of Mettl14 (Yue et al, 2015), the core subunit of the RNA methyltransferase in skin epidermis, can lead to diminished stemness of basal progenitor cells and aberrant epidermal differentiation in vivo. With transcriptome‐wide profiling, we further demonstrated profoundly altered RNA m6A methylomes upon epidermal differentiation. Gene Ontology (GO) term and enrichment analysis unraveled various enriched RNA clusters after skin differentiation, including both mRNAs and lncRNAs. LncRNAs represent an emerging class of noncoding RNAs that integrate a myriad of external signaling to regulate gene expression and epigenetic reprogramming of cells (Flynn & Chang, 2014). Increasing evidence have implicated lncRNAs in epidermal development and differentiation (Hombach & Kretz, 2013; Wan & Wang, 2014); however, the complexity of the signaling networks involved in these processes in skin remains poorly defined. Our data revealed that the lncRNA Pvt1 (plasmacytoma variant translocation gene 1) displayed significantly reduced m6A modification upon epidermal differentiation. In the human genome, Pvt1 is located in the well‐established cancer risk region 8q24 on chromosome 8 and is immediately adjacent to the oncogene MYC (myelocytomatosis oncogene) (Tseng et al, 2014; Colombo et al, 2015). 8q24 has frequent genetic aberrations, including translocation, amplification, and viral integration in different types of malignancies in humans. The copy number of Pvt1 was co‐amplified in more than 98% of MYC‐copy‐increase cancers, and a gain of Pvt1 expression is required for a high MYC oncogene level in 8q24‐amplifed cancer cells. It has been shown that Pvt1 can physically interact with MYC and protect it from protein degradation (Tseng et al, 2014). Adding to this, the MYC‐Pvt1 genes are syntenic not only in the human genome, but also in other mammalian species, including the mouse (chromosome 15) and rat (chromosome 7). In the skin epidermis, mounting evidence has demonstrated a central role of MYC in controlling tissue homeostasis, stemness of basal progenitor cells, and skin carcinogenesis (Arnold & Watt, 2001; Waikel et al, 2001; Frye et al, 2003; Zanet et al, 2005; Oskarsson et al, 2006; Watt et al, 2008). Interestingly, in addition to regulation of translation or RNA stability, the m6A modification can also lead to profound changes in the mRNA or lncRNA secondary structure, and thus alter their interaction with proteins, a process known as an “m6A switch” (Liu et al, 2015b; Zhou et al, 2016). For instance, the lncRNA MALAT1 (metastasis associated lung adenocarcinoma transcript 1) can undergo a conformational change to expose a U5‐tract for recognition and binding by HNRNPC (heterogeneous nuclear ribonucleoprotein C) upon m6A modification (Liu et al, 2015b; Zhou et al, 2016). Our data showed that the m6A modification on five key residues of Pvt1 is essential for its interaction with MYC and protects MYC protein stability. Overall, our results unraveled RNA methylome dynamics upon epidermal differentiation and identified an important molecular pathway whereby m6A modification of Pvt1 regulates differentiation of somatic stem cells in skin.
Results
Conditional ablation of Mettl14 leads to aberrant skin development and inhibits skin wound healing
RNA modifications can coordinate complex translation and transcriptome turnover during stem cell differentiation (Zhao et al, 2017; Roundtree et al, 2017a). Western blot analysis indicates a specific decrease in Mettl14 protein upon calcium‐induced epidermal differentiation in vitro (Appendix Fig S1A), suggesting a potentially significant role of RNA m6A modification in this process. To further investigate RNA m6A modification in skin development and tissue homeostasis, we developed a skin cKO (conditional knockout) mouse model of Mettl14. A targeting cassette with two loxP sites was engineered into the Mettl14 locus in mouse chromosome 3 by homologous recombination (Yoon et al, 2017). To conditionally delete Mettl14 in the skin epithelium, we bred Mettl14fl/fl mice with transgenic mice carrying K14‐Cre recombinase, which can efficiently excise floxed genomic DNA by embryonic day E15.5 (Vasioukhin et al, 1999; Wu et al, 2008) (Fig 1A and Appendix Fig S1B). cKO mice are born in the expected Mendelian ratio but exhibit significant perinatal lethality. The cKO animals are smaller and often have tighter, smoother, and shiny skin compared with their littermates (Fig 1A). Immunohistochemistry confirms loss of Mettl14 in skin epithelial cells in the cKO animals (Fig 1B). Mass spectrometry analysis shows a dramatic decrease of global mRNA m6A modification in cKO skin epidermis (Appendix Fig S1C).
Figure 1. Mettl14 regulates skin tissue homeostasis and wound healing.
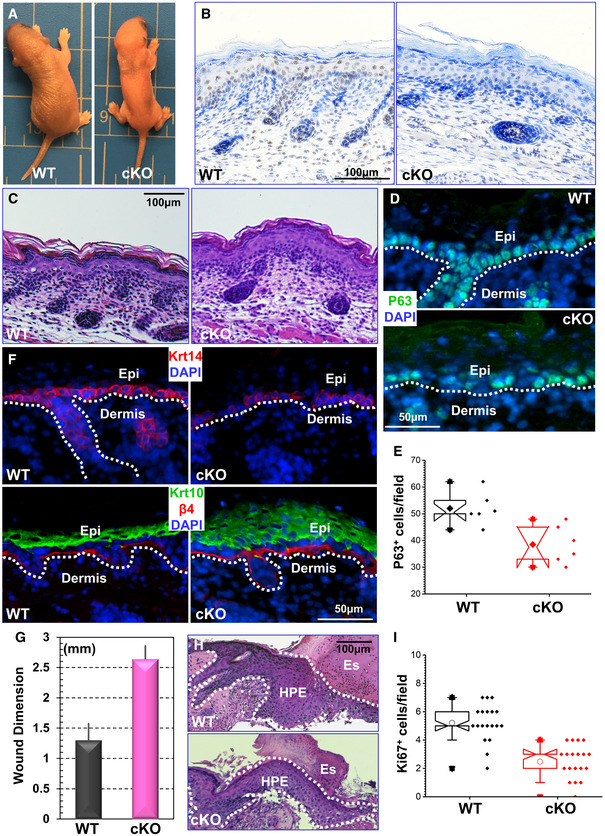
- Mettl14 cKO leads to perinatal lethality. Newborn cKO pups are smaller with tight and shiny skin.
- Immunohistochemistry confirms loss of Mettl14 expression in skin in cKO animals.
- H/E staining of newborn skin sections from WT and Mettl14 cKO mice.
- Expression of p63 in skin epidermis was examined by immunofluorescence staining in WT and cKO mice.
- Number of p63‐positive cells in WT and cKO skin was quantified and shown as box and whisker plots. The plot indicates the mean (solid diamond within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid triangles), and minimum and maximum measurements (solid squares). n = 6 (biological repeats), P < 0.05 (Student’s t‐test).
- Skin stratification in WT and cKO skin was determined by immunofluorescence staining with different antibodies as indicated. Krt14: Keratin 14; Krt10: Keratin 10; β4: β4‐integrin; DAPI for nucleus staining. The dashed line denotes the basement membrane that separates dermis and epidermis (Epi).
- Wound healing as monitored by wound size 8 days post‐injury. n = 3; P < 0.01 (Student’s t‐test). Error bar represents s.d. (standard deviation).
- Histological staining of skin sections at the wound edges. Halves of wound sections are shown. Note significant reduction of HPE (hyperproliferative epidermis) in cKO skin. Es: eschar. Dotted lines denote epidermal boundaries.
- Quantification of Ki67‐positive cells present in wound HPE. The plot indicates the mean (open circles within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid triangles), and minimum and maximum measurements (solid squares). n = 19, P < 0.01 (Student’s t‐test).
To first investigate the role of Mettl14 in skin development, we analyzed WT and cKO skin from newborn pups. Skin histology shows that the epidermis is notably thicker upon Mettl14 deletion (Fig 1C, and quantification in Appendix Fig S1D). Transcription factor p63 is a master regulator controlling epidermal morphogenesis and stemness (Botchkarev & Flores, 2014; Melino et al, 2015). Immunofluorescence staining demonstrates a significantly reduced number of p63‐positive basal cells in cKO skin (Fig 1D and quantification in Fig 1E). Along with decreased and disorientated basal progenitor cells, Mettl14 cKO skin epidermis has an expanded spinous layer as determined by staining with the early differentiation marker, Keratin 10 (Fig 1F, quantification in Appendix Fig S1E).
Wounding in skin can mobilize quiescent epidermal progenitor cells for proliferation and migration. To circumvent the issue of perinatal lethality in Mettl14 cKO animals, we bred the Metttl14l/fl mice with transgenic mice carrying the K14‐Cre‐ERT2 allele (Vasioukhin et al, 1999; Wu et al, 2008). The resultant inducible cKO animals can grow to adulthood with no apparent changes in skin epidermis or hair coat. However, when challenged to skin injury after administration of tamoxifen, Mettl14 cKO skin exhibited a significant delay in repairing full‐thickness wounds as compared to WT skin (Fig 1G, Appendix Fig S1F). Histological analysis revealed that the area of hyperproliferative epithelium that typically proliferates and migrates into the wound site was significantly diminished following injury (Fig 1H). Wound‐induced hyperproliferation of epidermal cells was also inhibited in cKO skin (Fig 1I). Together, our results strongly suggest that m6A modification of RNA mediated by Mettl14 plays a critical role in skin tissue homeostasis and wound repair in vivo.
Loss of Mettl14 impairs epidermal stemness
Although wound‐induced hyperproliferation in adult epidermis is inhibited upon ablation of Mettl14, epidermal proliferation is not significantly affected in newborn skin of the cKO mice (Appendix Fig S2A). Potential markers for long‐term epidermal progenitor cells are not clearly defined in vivo, and different models have been proposed for epidermal tissue maintenance (Lavker & Sun, 1982; Potten et al, 1982; Morris et al, 1985; Loeffler et al, 1987; Potten & Loeffler, 1987; Jones et al, 1995; Clayton et al, 2007; Mascre et al, 2012; Rompolas et al, 2016; Sada et al, 2016; Mesa et al, 2018; Dekoninck et al, 2020). However, accumulating evidence reveals the existence of distinct basal cell populations with hierarchical organization and proliferation dynamics in skin epidermis, including slow‐cycling progenitor cells and committed progenitor cells with limited proliferative potential (Lavker & Sun, 1982; Potten et al, 1982; Morris et al, 1985; Loeffler et al, 1987; Potten & Loeffler, 1987; Jones et al, 1995; Mascre et al, 2012). With classical label‐retaining analysis (Appendix Fig S2B), we found that loss of Mettl14 in skin leads to dramatic decrease in the slow cycling, label‐retaining cells in the basal layer (Fig 2A and quantification in 2B).
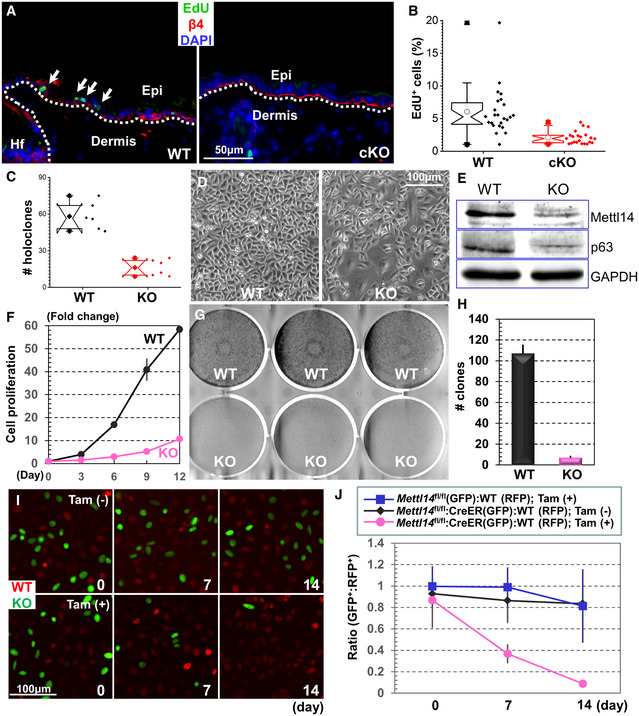
Figure 2. Loss of Mettl14 impairs epidermal stemness.
-
AEdU staining of WT or Mettl14 cKO skin after pulse‐chase labeling. Skin samples were counterstained with antibody against β4‐integrin. Note reduced EdU label‐retaining cells in cKO skin epidermis. Arrows indicate Edu‐positive cells.
-
BLabel‐retaining cells in WT or Mettl14 cKO skin were quantified and shown as box plots. The plot indicates the mean (open circle within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid triangles), and minimum and maximum measurements (solid squares). n = 6 (biological repeats), P < 0.05 (Student’s t‐test). n = 19, P < 0.01 (Student’s t‐test).
-
CNumber of holoclones derived from WT and Mettl14 cKO skin was quantified and shown as box and whisker plots. The plot indicates the mean (solid diamond within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid triangles), and minimum and maximum measurements (solid squares). n = 6 (biological repeats), P < 0.05 (Student’s t‐test). n = 24, P < 0.01 (Student’s t‐test).
-
DMorphology of primary keratinocytes isolated from WT or Mettl14 cKO skin.
-
EExpression of Mettl14 and p63 in WT and KO cells was determined by immunoblots with respective antibodies. Immunoblot for GAPDH was used as loading control.
-
FProliferation of WT and Mettl14 null cells in vitro was quantified and shown as dot plots. n = 3, P < 0.01 (Student’s t‐test) for Days 6, 9, and 12, and P < 0.05 (Student’s t‐test) for Day 3. Error bar represents s.d.
-
G, HCFE (colony formation efficiency) of WT and Mettl14 null cells was determined in vitro. Results were quantified and shown as bar graph (H). n = 19, P < 0.01 (Student’s t‐test). Error bar represents s.d.
-
IFluorescence microscopy demonstrates different survival capability of WT and Mettl14 inducible KO cells with or without Tamoxifen (TAM) treatment.
-
JRatio of WT and Mettl14‐inducible KO cells in the co‐culture model was quantified and shown as dot plots. n = 8, P < 0.01 (Student’s t‐test) for KO cells with TAM treatment compared with WT cells or KO cells without TAM stimulation at both Days 7 and 14. Error bar represents s.d.
Upon culture in vitro, primary epidermal keratinocytes will generate three different types of colonies, named holoclones, meroclones, and paraclones, with only holoclones containing undifferentiated epidermal progenitor cells (Rheinwald & Green, 1975; Green et al, 1977; Blanpain & Fuchs, 2006). Deletion of Mettl14 led to a dramatic reduction in holoclones when primary cells were cultured on 3T3 feeder layers (Fig 2C). Whereas WT holoclone cells are typically small and tightly packed with undifferentiated morphology, the KO colonies usually have cells that are large and multinucleated with senescent or differentiated morphology (Fig 2D). Immunoblots confirmed loss of Mettl14 expression in KO cells. Expression of p63 was also significantly reduced in KO cells (Fig 2E). When passaged in vitro, the KO cells proliferate slower with significantly longer doubling time (Fig 2F) and reduced EdU (5‐ethynyl‐2’‐deoxyuridine) incorporation (Appendix Fig S2C) compared with WT controls. The colony formation efficiency (CFE) was markedly decreased upon loss of Mettl14 (Fig 2G and quantification in 2H), and re‐expression of WT Mettl14 but not Mettl14 R298P mutant that is deficient in methyltransferase activity (Liu et al, 2018) can restore the CFE in vitro (Appendix Fig S2D).
To further investigate the change of epidermal stemness, we carried out a clonal competition and lineage trace analysis (Siegle et al, 2014) using our skin organoid culture and transplantation model (Yue et al, 2017; Li et al, 2019). As Mettl14‐deficient cells failed to sustain long‐term culture in vitro, we isolated primary epidermal keratinocytes from Mettl14‐inducible KO strain. Before treatment with tamoxifen, the inducible KO cells grow at a comparable rate with their WT counterparts. To monitor their epidermal regeneration capacity and trace their fate upon engraftment, we transduced the WT and inducible KO cells with lentivirus encoding either H2B‐RFP or H2B‐GFP. Fluorescently labeled cells were mixed at a 1:1 ratio to generate skin organoids. Interestingly, whereas clone size measurement with fluorescent microscopy revealed no significant changes between WT and inducible KO cells before tamoxifen treatment, induction of Cre recombination with tamoxifen led to continual loss of Mettl14 KO cells in the skin, which became barely detectible 14 days post‐tamoxifen treatment (Fig 2I and quantification in 2J).
N6‐methyladenosine modification of Pvt1 regulates stemness of epidermal progenitor cells
In order to uncover how changes in the RNA methylomes regulate self‐renewal and differentiation of epidermal progenitor cells, we performed m6A sequencing and RNA sequencing of polyadenylated RNAs isolated from undifferentiated and differentiated cells (Dataset EV1 and EV2). Our profiling identified 7,379 and 6,945 RNA species with m6A modifications in progenitor cells and differentiated cells, respectively. Among them, 260 RNAs have markedly reduced m6A modification upon differentiation (fold change > 2), whereas 1,474 RNAs have elevated levels of m6A modification. GO term enrichment analysis of RNAs with altered m6A modification revealed a variety of enriched clusters following keratinocyte differentiation (Appendix Fig S3), including annotations pertaining to RNA processing and metabolisms.
LncRNAs have emerged as an important class of regulators involved in many processes, including skin development and homeostasis (Hombach & Kretz, 2013; Wan & Wang, 2014). Our analysis revealed substantially altered m6A modifications of different lncRNAs during skin differentiation, including Gas5 (growth arrest‐specific 5), Neat1 (nuclear enriched abundant transcript 1), Malat1 (metastasis associated in lung adenocarcinoma transcript 1), Snhg18 (small nucleolar RNA host gene 18) and Pvt1 (Fig 3A). Among these, Pvt1, a potential oncogenic lncRNA associated with MYC, displayed significantly reduced m6A modification in differentiated cells. We verified this change by RT–PCR from differentiated and undifferentiated cells. Pvt1 contains five potential m6A modification sites that match the Pu[G>A]m6AC[A/C/U] consensus sequence (Harper et al, 1990). The five potential sites are distributed as two clusters (A282, A294, and A303 in cluster 1, and A446 and A452 in cluster 2) at Exon 2. To examine their potential modification, we prepared different mutants of Pvt1 and then determined their m6A methylation level by α‐m6A immunoprecipitation followed with RT–PCR. When each cluster was mutated (A–G), we saw ~50% reduction in the m6A modification (Fig 3B). When both clusters were mutated (A5G), methylation of Pvt1 was nearly abolished (Fig 3B).
Figure 3. Pvt1 regulates epidermal stemness and tissue homeostasis.
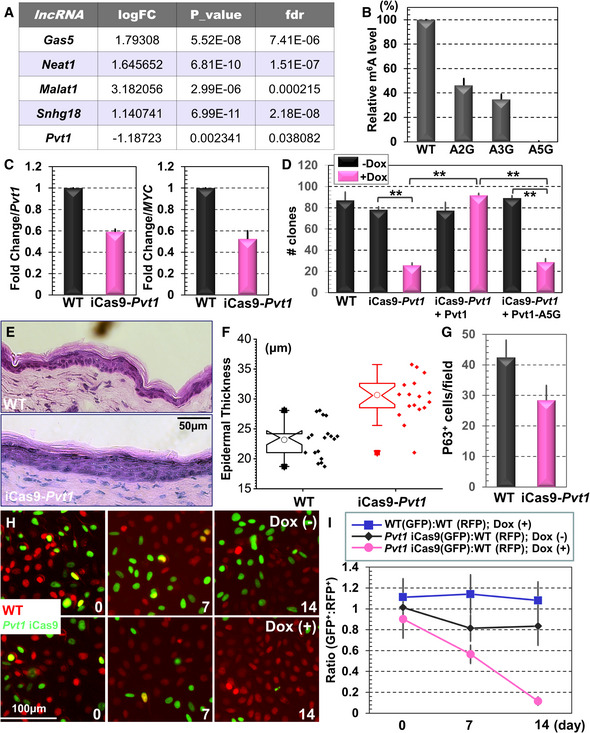
- Profiling of RNA methylome by m6A‐seq shows significant changes of lncRNA modifications upon epidermal differentiation in vitro.
- Mutations in the predicted m6A modification sites can significantly reduce Pvt1 methylation. n = 3. Error bar represents s.d.
- Deletion of endogenous Pvt1 by an inducible Cas9 (iCas9) system can significantly reduce Pvt1 RNA level (quantification from RT–PCR, left panel) and MYC protein level (quantification from immunoblot, right panel) expression. n = 3, P < 0.01 (Student’s t‐test). Error bar represents s.d.
- CFE of WT, Pvt1‐inducible KO, and Pvt1‐inducible KO cells rescued with WT or mutant Pvt1 was quantified and presented as bar graph. n = 3, **P < 0.01 (Student’s t‐test). Error bar represents s.d.
- Skin organoids derived from WT or Pvt1‐inducible KO cells were grafted to nude mice. The regenerated skin was analyzed by H/E staining.
- Epidermal thickness of WT and Pvt1 KO skin grafts was quantified and presented as box and whisker plots. The plot indicates the mean (open circle within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid triangles), and minimum and maximum measurements (solid squares). n = 6 (biological repeats), P < 0.05 (Student’s t‐test). n = 18, P < 0.01 (Student’s t‐test).
- Number of p63‐positive cells in WT and cKO skin was quantified and shown as bar graph. n = 5, P < 0.01 (Student’s t‐test). Error bar represents s.d.
- Fluorescence microscopy demonstrates different survival capability of WT and Pvt1‐inducible KO cells with or without Doxycycline (Dox) treatment.
- Ratio of WT and Pvt1‐inducible KO cells in the co‐culture model was quantified and shown as dot plots. n = 8, P < 0.01 (Student’s t‐test) for KO cells with Dox treatment compared with WT cells or KO cells without Dox stimulation at both Days 7 and 14. Error bar represents s.d.
Although MYC has been shown to play a pivotal role in skin controlling tissue homeostasis, stemness, and skin carcinogenesis, the role of Pvt1 in these processes remains unclear. Mouse Pvt1 gene locates at chromosome 15 with 9 exons. As Pvt1 is a noncoding RNA, we developed a CRISPR (clustered regularly interspaced short palindromic repeats) KO strategy that deletes the whole gene by applying two gRNAs (guide RNAs) that are complementary to the sequences at the both ends of Pvt1 gene (Appendix Fig S4A). Interestingly, the Pvt1 KO cells failed to propagate after selection, suggesting that it is an essential gene for maintaining stemness of epidermal progenitor cells in vitro. To resolve this issue, we used a doxycycline‐inducible platform to deliver the Cas9 (CRISPR‐associated protein 9) protein (iCas9) (Cao et al, 2016), together with a PiggyBac transposon vector encoding the two gRNAs required for Pvt1 deletion (Li et al, 2013). In stably transduced cells, induction with doxycycline led to a significant reduction in Pvt1 expression (Fig 3C). Consistent with previous reports, a decrease in Pvt1 RNA results in a reduction in MYC protein at a comparable level, as determined by immunoblots (Fig 3C). No significant changes in MYC mRNA level were detected in the induced KO cells (Appendix Fig S4B), suggesting that Pvt1 regulates the MYC level post‐transcriptionally.
To determine the role of Pvt1 methylation in epidermal stemness, we first examined CFE in Pvt1 inducible KO cells, and KO cells rescued with lentiviral vector expressing either WT Pvt1 or Pvt1‐mutant deficient in m6A modification (mutation of all five adenosine residues to guanidine) in a doxycycline‐inducible manner. Reduced Pvt1 expression led to a dramatic decrease in CFE, and re‐expression of WT but not mutant form of Pvt1 can restore the CFE in vitro (Fig 3D). Consistent with CRISPR‐mediated deletion of Pvt1, silencing of Pvt1 expression by siRNA also led to significant decrease in CFE (Appendix Fig S4C and D). As in silico analysis (Gruber et al, 2008) shows that A282, 294, and 446 may be involved in the secondary structure formation of Pvt1 lncRNA (Appendix Fig S4E), we prepared a Pvt1 mutant in which the three corresponding T residues were mutated to C (A5GT3C mutant) to preserve the secondary structure. When this mutant was expressed in Pvt1 KO cells, it failed to restore CFE in vitro as A5G mutant, suggesting that the defect in epidermal stemness is not due to potential secondary structural change in Pvt1 (Appendix Fig S4F).
To determine the role of Pvt1 in an in vivo setting, we took advantage of the mouse skin organoid and transplantation model that has been recently established by our group (Yue et al, 2017; Li et al, 2019). Skin organoids derived from WT and Pvt1‐inducible KO cells can both be efficiently grafted to host mice and regenerate skin. However, upon doxycycline administration, the KO skin grafts grew notably thicker with expansion of spinous cells and a decrease in p63‐positive basal cells, resembling the phenotypes of Mettl14 skin (Fig 3E–G and Appendix Fig S4G–H). We further assessed the changes in epidermal stemness by the clonal competition assay. We transduced the WT and inducible Pvt1 KO cells with lentivirus encoding either H2B‐RFP or H2B‐GFP and mixed the cells at a 1:1 ratio to generate the skin organoids. Interestingly, induction of Cas9 with doxycycline for Pvt1 deletion led to continual loss of KO cells in the skin (Fig 3H and quantification in 3I). Taken together, our results provide compelling evidence that lncRNA Pvt1 plays an important role in sustaining epidermal stemness, which requires its m6A modification.
Modification of Pvt1 regulates epidermal stemness via its interaction with MYC
MYC is critically involved in epidermal proliferation, differentiation, and carcinogenesis (Arnold & Watt, 2001; Waikel et al, 2001; Frye et al, 2003; Zanet et al, 2005; Oskarsson et al, 2006; Watt et al, 2008). Gene set enrichment analysis (GSEA) on our RNA‐seq data shows that MYC‐targeted gene sets are significantly upregulated in epidermal progenitor cells with the highest enrichment scores (Appendix Fig S5A). Hierarchical cluster analysis with another MYC‐regulated gene set also confirms that this group of genes are down‐regulated upon epidermal differentiation. Pvt1 physically associates with MYC (Tseng et al, 2014). As a critical signaling molecule, the MYC protein has a relatively short half‐life, and its degradation is mediated by phosphorylation at a key threonine residue at its N terminus. Pvt1 interaction with MYC has been shown to inhibit this phosphorylation and protect MYC from protein degradation machinery (Tseng et al, 2014; Colombo et al, 2015). We speculate that methylation of Pvt1 may act as an “m6A switch” that regulates its interaction with MYC. To examine this hypothesis, we first tested and quantified Pvt1 and MYC interaction by co‐immunoprecipitation. Treatment of Pvt1 with FTO in vitro, a demethylase specific for m6A, lead to Pvt1 demethylation (Appendix Fig S5B) and a dramatic reduction in MYC and Pvt1 interaction (Fig 4A, and Appendix Fig S5C and D). Consistent with this finding, calcium‐induced differentiation causes diminished m6A modification and MYC interaction of Pvt1. Pvt1 mutant deficient in m6A modification (A5G and A5GT3C) also binds MYC in a significantly lower level compared with WT Pvt1 (Fig 4A, and Appendix Fig S5C and D). Immunoblots show that the protein level of MYC is decreased in both differentiated cells and Mettl14 KO cells (Fig 4B). MYC protein stability is markedly reduced upon deletion of Mettl14, Pvt1, or calcium‐induced keratinocyte differentiation (Fig 4C–E and quantification in Appendix Fig S5E). Ectopic expression of MYC in Pvt1 deficient cells can significantly restore CFE in vitro (Appendix Fig 5F), suggesting that MYC is the key downstream effector of Pvt1 in controlling epidermal stemness.
Figure 4. Pvt1 methylation regulates epidermal stemness through its interaction with MYC.
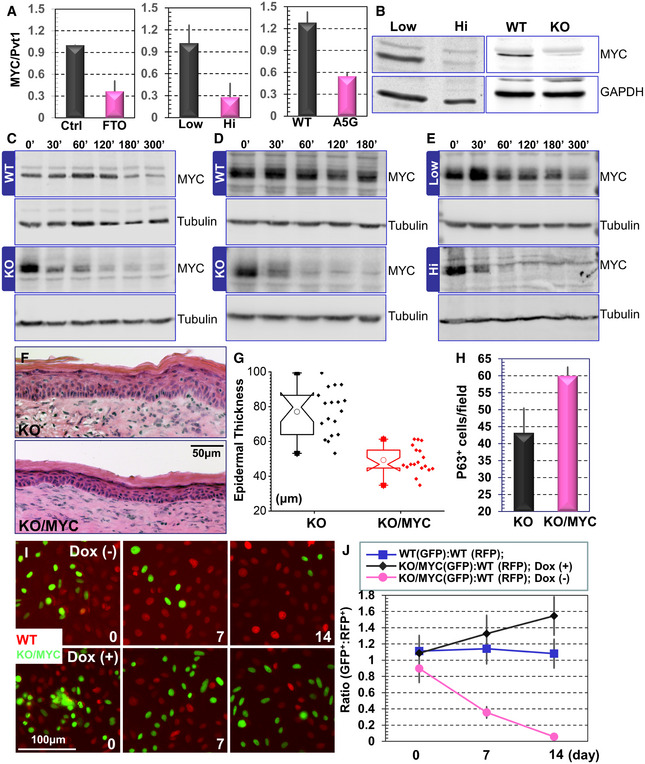
-
AInteraction between Pvt1 and MYC was determined by immunoprecipitation followed with RT–PCR. Note significant decrease in Pvt1 and MYC interaction upon FTO treatment (left panel), calcium shift‐induced differentiation (middle panel, Hi: high calcium), and Pvt1 methylation site mutations (right panel). n = 3, P < 0.01 (Student’s t‐test). Error bar represents s.d.
-
BImmunoblots show decreased MYC protein level in differentiated keratinocytes and Mettl14 KO cells.
-
C‐EUndifferentiated WT and Mettl14 KO cells (C); or undifferentiated WT and Pvt1 KO cells (D); or WT cells before and after calcium shift‐induced differentiation (E) were treated with cycloheximide. Cell lysates were collected at 0, 30, 60, 90, 120, 180, and 300 min post‐treatment and subjected to immunoblotting with different antibodies as indicated.
-
FSkin organoids derived from Mettl14 KO cells or KO cells rescued with exogenous expression of MYC were grafted to nude mice. The regenerated skin was analyzed by H/E staining.
-
GEpidermal thickness of KO and KO skin rescued with MYC expression was quantified and presented as box and whisker plots. The plot indicates the mean (solid diamond within the box), 25th percentile (bottom line of the box), median (middle line of the box), 75th percentile (top line of the box), 5th and 95th percentile (whiskers), 1st and 99th percentile (solid triangles), and minimum and maximum measurements (solid squares). n = 6 (biological repeats), P < 0.05 (Student’s t‐test). n = 18, P < 0.01 (Student’s t‐test).
-
HNumber of p63‐positive cells in KO and KO skin rescued with MYC expression was quantified and shown as bar graph. n = 5, P < 0.01 (Student’s t‐test). Error bar represents s.d.
-
IFluorescence microscopy demonstrates different survival capability of WT and Mettl14 KO cells with inducible expression of MYC.
-
JRatio of WT and Mettl14 KO cells with inducible expression of MYC in the co‐culture model was quantified and shown as dot plots. n = 8, P < 0.01 (Student’s t‐test) for KO cells without Dox treatment (no MYC expression) compared with WT cells or KO cells with Dox stimulation (exogenous MYC expression) at both Days 7 and 14. Error bar represents s.d.
To determine the relevance of the Pvt1‐MYC axis in Mettl14‐driven processes in skin, we prepared Mettl14 KO cells with an inducible expression of MYC. Treatment with doxycycline can lead to elevated MYC expression in a dose‐dependent manner (Appendix Fig S5G). To examine the significance of MYC re‐expression in vivo, we prepared skin organoids with Mettl14 KO cells or KO cells rescued with MYC expression. Upon engraftment to host animals, we found that exogenous expression of MYC in Mettl14‐deficient cells can restore normal epidermal architecture. The rescued skin grafts have increased p63‐positive basal cells, with epidermal thickness and spinous layer comparable to normal WT skin (Fig 4F–H, and Appendix Fig S5H and I). In clonal competition assay, restored MYC expression can significantly protect the KO clones in skin (Fig 4I and J). Taken together, our data suggest that Mettl14‐mediated RNA modification enhances skin epidermal stemness through Pvt1 and MYC signaling cascade.
Discussion
Defects in skin epidermal differentiation or wound repair can result in dire consequences for our survival because the skin provides an essential barrier protecting us against many different environmental damage (Blanpain & Fuchs, 2006; Fuchs, 2008). Tissue homeostasis and regeneration of our skin are driven by the epidermal basal progenitor cells and are controlled by the critical balance of cell proliferation, cell differentiation, and cell death. Stemness and differentiation of epidermal progenitor cells are controlled by a complex and choreographed signaling network; however, the complexity of the multilevel regulation of these processes remains ill defined. In this study, we uncovered a unique epitranscriptomic mechanism whereby N6‐methyladenosine modification of lncRNA Pvt1 in epidermal progenitor cells maintains stemness and regulates skin tissue homeostasis and wound repair.
Protein‐coding regions comprise only ~1–2% of our genome, and a major component of the noncoding genome are lncRNAs (Flynn & Chang, 2014; Koch, 2017). Compared with mRNAs, lncRNAs usually have a relatively lower expression level but strong tissue‐specific expression patterns. Although nearly 28,000 lncRNAs have been cataloged in human genome, most of the lncRNAs have not been functionally characterized. To date, only a few lncRNAs have been identified as playing an important role in epidermal stemness and homeostasis with cultured human primary keratinocytes or human skin tissue samples as model systems, including DANCR (differentiation antagonizing non‐protein‐coding RNA) (Kretz et al, 2012), TINCR (terminal differentiation‐induced noncoding RNA) (Kretz et al, 2013), PRANCR (progenitor renewal associated noncoding RNA) (Cai et al, 2020), SMRT‐2 (SCC mis‐regulated transcripts‐2) (Lee et al, 2018), and uc.291 (UC‐containing lncRNA) (Panatta et al, 2020). Kretz et al (2012) and Kretz et al (2013) have shown that expression of DANCR is required for maintaining epidermal stemness, whereas expression of TINCR is induced upon differentiation, and TINCR enhances epidermal differentiation via a post‐transcriptional mechanism by binding with protein STAU1 (staufen1) (Kretz et al, 2012; Kretz et al, 2013). Consistent with previous report (Kretz et al, 2012), our RNA‐seq revealed a significant decrease in ANCR transcript level upon calcium‐induced differentiation of mouse epidermal progenitor cells. However, contradictory to the role of TINCR in promoting epidermal differentiation (Kretz et al, 2013), transcription of Gm20219, the mouse homolog of TINCR, is significantly reduced in differentiated mouse keratinocytes. It is noteworthy that lncRNAs are less sequence‐conserved compared with protein‐coding RNAs, and many functional lncRNAs are human or primate specific. It is likely that Gm20219 and TINCR may function in a different manner in mouse and human epidermal cells. Neither ANCR nor TINCR shows significant changes in m6A modifications upon epidermal differentiation, suggesting their functions are regulated by different mechanisms in skin epithelial cells.
Our results revealed Pvt1, a new lncRNA, that plays a pivotal role in skin tissue homeostasis. Pvt1 associates with MYC and prevents its protein degradation (Tseng et al, 2014), and our data show that m6A methylation is an important regulator of this process. With in vitro culture and skin transplantation models, we show that RNA m6A modification mediated by Mettl3/Mettl14 complex regulates epidermal stemness by controlling Pvt1 and MYC interaction through Pvt1 methylation, uncovering a key and novel molecular mechanism underlying skin tissue homeostasis, regeneration, and wound repair.
MYC is a proto‐oncogene critically involved in many cellular processes. In the skin epidermis, deletion of endogenous MYC by the Krt5 promoter‐driven expression of Cre leads to skin hypoplasia, impaired epidermal stem cell self‐renewal, and delayed wound healing in vivo (Zanet et al, 2005), resembling the phenotypes of Mettl14 deletion and CRISPR‐mediated KO of Pvt1 in skin epidermal cells. In a separate study, when MYC is ablated by an inducible Cre‐ER system, tissue homeostasis in adult skin IFE (interfollicular epidermis) and hair follicles seems to be rather normal, but the cKO skin is more resistant to Ras‐induced carcinogenesis as a result of upregulation of p21 (Oskarsson et al, 2006). Interesting, constitutive or inducible overexpression of MYC in basal progenitor cells causes both increased keratinocyte proliferation and differentiation in vivo. Constitutive MYC overexpression driven by Krt5 promoter also leads to spontaneous skin carcinogenesis when the animals age (Watt et al, 2008). Together, it suggests that MYC plays a multifaceted role in skin tissue homeostasis and tumorigenesis, depending on the level and duration of MYC activation. MYC protein level is significantly reduced in Mettl14 null cells, and ectopic expression of MYC in Mettl14 KO cells can restore epidermal stemness, strongly suggesting that MYC is a key downstream effector of Mettl14 in regulation of skin tissue homeostasis.
Our profiling of m6A‐modified transcripts also revealed several other lncRNA targets beyond Pvt1 (Fig 3A). Among them, Gas5 has been associated with a decreased cell cycle and increased cell death and could function as a potential tumor suppressor (Goustin et al, 2019). Its upregulation after a calcium shift is consistent with suppressed cell proliferation upon epidermal differentiation. Neat1 and MALAT1 (also known as Neat2) are abundant nuclear lncRNAs that localize at paraspeckles and nuclear speckles, respectively. It has been shown that Neat1 and MALAT1 associate with many active chromatin sites with distinct gene body binding patterns (West et al, 2014). It will be interesting to pursue how m6A modifications on these lncRNAs may contribute to skin development and wound repair in the future.
Aberrant skin tissue homeostasis can contribute to the etiology of many skin diseases, including inflammatory skin disorders and skin cancers. Cutaneous SCC (squamous cell carcinoma) is one of the most common cancers in the United States (Preston & Stern, 1992; Alam & Ratner, 2001), which can be highly invasive and metastatic, leading to severe morbidity and mortality for the patients (Burton et al, 2016). Increasing evidence suggest that m6A modification of mRNA and lncRNA are involved in cancer development and progression (Yue et al, 2015; Roundtree et al, 2017a; Dai et al, 2018). Key players of the m6A pathway, including writer, eraser, and reader proteins have been shown to play diverse roles in different type of tumorigenesis. Recently, Zhou et al (2019) have shown that expression of Mettl3 is upregulated in cutaneous SCCs and that a knockdown of Mettl13 in SCC cell lines in vitro can inhibit its tumorigenicity by reducing p63 expression (Zhou et al, 2019). Understanding the molecular mechanisms underlying epidermal stemness and homeostasis provides essential basis for developing effective therapies for skin diseases, including skin SCC.
In closing, our studies provide important insights into the molecular mechanisms whereby RNA post‐transcriptional modification regulates epidermal stemness and tissue homeostasis.
Materials and Methods
Antibodies, reagents, and plasmid DNA constructions
Guinea pig α‐Krt5, rabbit α‐Krt10, and Loricrin antibodies were generous gifts from Dr. Elaine Fuchs at the Rockefeller University. Rabbit Krt 14 and Krt 10 antibodies were obtained from Covance (Princeton, NJ). Rat monoclonal β4‐integrin (CD104) was obtained from BD Pharmingen (Franklin lakes, NJ). P63 antibody was obtained from Genetex (Irvine, CA). Mettl14 antibody was obtained from Sigma (St Louis, MO). MYC, GAPDH, and α‐tubulin antibodies were obtained from ProteinTech (Rosemont, IL). Mouse monoclonal antibody against Vinculin, α‐Flag‐conjugated beads, and α‐Flag antibodies were obtained from Sigma (St. Louis, MO). Rabbit polyclonal antibodies against Ki67 were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Other chemicals or reagents were obtained from Sigma, unless otherwise indicated.
CRISPR targeting vector against Pvt1 was generated with primers: ACA CCG TGG GAC CTA TGT AGG ACC TG; AAA ACA GGT CCT ACA TAG GTC CCA CG; ACA CCG GAT GAT GCT TCT CCT GGT AG; AAA ACT ACC AGG AGA AGC ATC ATC CG. Pvt1 was cloned with primers: CCG CTC GAG CGT GAC GTC ACG GGC CAC CCG CCA GCC CCG CGC TCT ACC AGG CAG AGC GCG; CCC AAG CTT ACC GGT CTT GGT CAA ATT GTT TTA TTG AGC. Pvt1 mutants at m6A modification sites were constructed with primers: GCA GGC CGT GTC TGC TGG GGC CTC TCT GGA GGC TTC CTG AGC CCT GTG GAT TTA CAG TAA; TTA CTG TAA ATC CAC AGG GCT CAG GAA GCC TCC AGA GAG GCC CCA GCA GAC ACG GCC TGC; GAT GCC CAC TGA AAA CAA GGG CCG AAG CTA AGA GGA TTG TAT CTC TAC; GTA GAG ATA CAA TCC TCT TAG CTT CGG CCC TTG TTT TCA GTG GGC ATC. MYC was cloned from Addgene clone 74164.
RNA extraction and quantitative RT–PCR
Total RNA was isolated with TRIzol reagent (Invitrogen) using Direct‐zol kit (Zymo Research). mRNA was extracted from the total RNA using a Dynabeads® mRNA purification kit (Invitrogen). mRNA concentration was measured by ultraviolet absorbance at 260 nm.
For analysis of mRNA expression, 200–500 ng of RNA was reverse‐transcribed into cDNA using the High‐Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative real‐time PCR analysis was then performed using PerfeCTa SYBER green Supermix (Quantabio) on the StepOnePlus RT–PCR system (Thermo Fisher Scientific). GAPDH or ACTB was used as endogenous control. Each sample was run in triplicates.
Quantification of RNA m6A modification levels via mass spectrometry
Total RNA was subjected to two rounds of poly(A) selection with the Dynabeads® mRNA purification kit (Invitrogen). 25 ng of the purified mRNA was digested by nuclease P1 (1 U) in 20 μl of buffer containing 20 mM NH4OAc pH 5.5 at 42°C for 2 h, followed by the addition FastAP buffer (2.3 µl, Thermo Scientific) and alkaline phosphatase (1 U) and incubation at 37°C for 4 h. The sample was then filtered (0.22 μm pore size, 4 mm diameter, Millipore), and 5 μl of the solution was injected into the LC‐MS/MS. The nucleosides were separated by reverse‐phase ultra‐performance liquid chromatography on a C18 column (Agilent) with online mass spectrometry detection using a Sciex 6500+ triple‐quadrupole LC mass spectrometer in positive electrospray ionization mode. The nucleosides were quantified by using the nucleoside‐to‐base ion mass transitions of 282–150 (m6A) and 268–136 (A). Quantification was carried out by comparison with a standard curve obtained from pure nucleoside standards run with the same batch of samples. The ratio of m6A to A was calculated based on the calibrated concentrations.
Analysis of m6A‐sequencing
Total RNA was isolated from primary mouse keratinocyte with or without calcium shift‐induced differentiation. Polyadenylated RNA was further enriched from total RNA using a Dynabeads® mRNA purification kit (Invitrogen). RNA was fragmented using a Bioruptor ultrasonicator (Diagenode) with 30 s on/off for 30 cycles. m6A‐IP was performed using the Epimark N6‐methyladenosine enrichment kit (New England Biolabs) and library preparation was performed using the Illumina TruSeq Stranded mRNA Sample Prep Kit according to previously published protocols (Dominissini et al, 2012). Sequencing was carried out at the University of Chicago Genomics Facility on an Illumina HiSeq4000 machine in single‐read mode with 50 base pairs per read.
m6A‐seq data were analyzed according to the protocol previously described (Meng et al, 2014). Briefly, HISAT2 (version 2.1.0) (Kim et al, 2015) was used to align the sequence reads to reference genome and transcriptome (mm10). Then, the exomePeak R/Bioconductor package (version 3.7) was used to find m6A peaks and the QNB package (version 1.1.11) (Liu et al, 2017) was used to identify differentially methylated regions. Significant peaks with false discovery rate less than 0.05 were annotated to the RefSeq database (mm10). Differential gene expression was calculated using DESeq2 (version 1.18.1) using the sequencing reads from input samples (Love et al, 2014). Gene Ontology (GO) analysis on the differentially methylated genes was performed using Cytoscape app with BINGO plugin (Maere et al, 2005). Gene Set Enrichment Analysis was performed using GSEA software created by Broad Institute (Mootha et al, 2003; Subramanian et al, 2005).
Quantification of RNA m6A methylation via m6A‐immunoprecipitation
Protein G Dynabeads (Thermo Scientific) was washed and resuspended in IP buffer containing 20 mM Tris (pH 7.4), 750 mM NaCl, and 0.5% NP40. Next, m6A antibody (New England Biolabs) was added to the beads and incubated for 30 min in 4°. After incubation, the supernatant was removed and washed with IP buffer, then purified RNA was added, and incubated for 1 h in 4°. The beads were then washed with low salt buffer containing 10 mM Tris (pH 7.4), 10 mM NaCl, 0.1% NP40, followed by a second wash with high salt buffer containing 10 mM Tris (pH 7.4), 500 mM NaCl, 0.1% NP40. Finally, the RNA was eluted with RLT buffer (Qiagen) and further purified using the RNA Clean and Concentrator‐5 kit (Zymo Research). The relative level of m6A‐modified RNA was quantified via RT–PCR as described in the previous section.
Quantification of Pvt1‐myc interaction via myc‐immunoprecipitation
Cell lysate was collected from transfected 293 cells expressing MYC with HA tag. 50 µl anti‐HA agarose beads (Peirce) were added to the lysate and incubated at 4° for 1 h. After incubation, the beads were washed with RNA–protein binding buffer containing 25 mM Tris (pH 7.4) 150 mM KCl, 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, and SUPERNase (Invitrogen) 0.1 U/µl. Next, purified RNA was added to the myc‐HA beads and incubated for 2 h in 4°. Finally, the beads were washed with RNA–protein binding buffer and RNA was purified using RNA Clean and Concentrator‐5 kit (Zymo Research). The relative level of myc‐bound Pvt1 was determined by RT–PCR as described in the previous section.
FTO demethylation of RNA
Removal of m6A methylation of mRNA and lncRNA was performed as previously described (Wei et al, 2018). In brief, 200 ng of polyadenylated RNA purified from cultured cells was incubated with 2 µM purified FTO in 20 µl reaction system. The reaction buffer was composed of 50 mM HEPES buffer (pH6.5), 100 mM KCl, 2 mM MgCl2, 2 mM L‐ascorbic acid, 300 µM α‐ketoglutarate, 150 µM (NH4)2Fe(SO4)2•6H2O, and SUPERNase (Invitrogen) 0.2 U/µl. After 1‐h incubation, the reaction was quenched by added 5 mM EDTA. For control group, EDTA was added prior to incubation. The demethylated RNA was purified using RNA Clean and Concentrator‐5 kit (Zymo Research).
Skin organotypic culture and grafting
Decellularized dermis (1 cm × 1 cm) was prepared from newborn CD1 mice. 1.5 × 106 cultured keratinocytes were seeded onto the dermis in cell culture insert. After overnight attachment, the skin culture was exposed to air/liquid interphase. To transplant to nude animals (2–3 months old), two 1 cm × 1 cm wounds were introduced to the back skin. After transplantation, the wound edge was sealed with surgical glue. The grafted animals were housed separately, and the wound bandages were usually removed 1 week post‐surgery (Liu et al, 2015a; Yue et al, 2016). All the experiments had more than three biological replicates (independent skin grafts). For phenotypic analysis, at least three sections were taken from each graft for analysis and quantification by staining.
Generation of Mettl14 cKO mice
The Mettl14 conditionally targeted strain was kindly provided by Dr. Chuan He (University of Chicago). Mettl14 cKO animals were generated by breeding the strain with K14‐Cre or K14‐CreERT2. To avoid potential maternal effect, we use male mice with K14‐Cre or CreERT2 transgene for breeding. All mice used in this study were bred and maintained at the ARC (animal resource center) of the University of Chicago in accordance with institutional guidelines.
Histology and immunofluorescence
Skin or wound samples were embedded in OCT, sectioned, and fixed in 4% paraformaldehyde. Sections were subjected to hematoxylin and eosin staining or immunofluorescence staining as described (Guasch et al, 2007). Antibodies were diluted according to manufacturer’s instruction unless indicated. Measurements of stratification markers were carried out using ImageJ.
Skin wound healing
For skin wound healing assays, same sex littermates of ~12‐week‐old mice were anesthetized, and two full‐thickness excisional wounds were made on both sides of the dorsal midline (Wu et al, 2008). Mice were housed separately, and no self‐induced trauma was observed in control or cKO mice. Tissue was collected 2–6 days after wounding, and wound re‐epithelialization was evaluated by histological analyses. Hyperproliferative epidermis was identified by hematoxylin and eosin staining, and the length of HE that extended into the wounds was measured and quantified.
Cell culture
Primary mouse keratinocytes were isolated from the epidermis of newborn mice using trypsin, after prior separation of the epidermis from the dermis by an overnight dispase treatment. Keratinocytes were plated on mitomycin C–treated 3T3 fibroblast feeder cells until passage 3. Cells were cultured in E‐media supplemented with 15% serum with a final concentration of 0.05 mM Ca2+.
Protein biochemical analysis
Western blot was performed as described previously (Wu et al, 2004). Cell lysates were prepared with RIPA (radioimmunoprecipiation assay) buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 10% Glycerol, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X‐100, 1% Sodium Deoxycholate, 0.1% SDS) containing protease inhibitors and phosphatase inhibitors. Equal amounts of the cell lysates were separated using SDS‐polyacrylamide gel electrophoresis (PAGE) and electroblotted onto a NC membrane. The immunoblot was incubated with Odyssey blocking buffer (Li‐Cor) at room temperature for 1 h, followed by an overnight incubation with the primary antibody. Blots were washed three times with Tween 20/Tris‐buffered saline (TBST) and incubated with a 1:10,000 dilution of secondary antibody for 1 h at room temperature. Blots were washed three times with TBST. Visualization and quantification were carried out with the LI‐COR Odyssey scanner and software (LI‐COR Biosciences, Nebraska, USA).
For MYC protein stability test, cells were treated with 20 nM cycloheximide (CHX). Whole cell lysates were collected at 0, 30, 60, 90, 120, 180, and 300 min post‐CHX treatment and subjected to immunoblotting with MYC antibody. Band intensity is determined by densitometry.
Statistical analysis
Statistical analysis was performed using Excel, OriginPro, or SciDAVis software. Box plots were used to describe the entire population without assumptions on the statistical distribution. A Student t‐test was used to assess the statistical significance (P value) of the difference for the experiments.
Author contributions
Experiment design: XW, CH, and YM; Experiments: JL, YW, BH, YL, and JZ; Data analysis: JL, YW, BH, and XW; Manuscript writing: JL and XW. Manuscript editing: All authors.
Conflict of interest
CH is a scientific founder and a scientific advisory board member of Accent Therapeutics, Inc.
Supporting information
Review Process File
Appendix
Dataset EV1
Dataset EV2
Source Data for Appendix
Acknowledgements
We thank Dr. Elaine Fuchs at the Rockefeller University for sharing reagents. We thank Linda Degenstein from the University of Chicago transgenic core facility for her excellent technical assistance. The animal studies were carried out in the ALAAC‐accredited animal research facility at the University of Chicago. This work was supported by grants R01 AR063630, R01 OD023700, R01 DA047785, R21 AA027172 to XW, and HG008935 to CH from the National Institutes of Health. JL is supported by the University of Chicago cancer biology training grant (T32, CA 9594‐30). CH is a Howard Hughes Medical Institute Investigator.
The EMBO Journal (2021) 40: e106276.
Contributor Information
Chuan He, Email: chuanhe@uchicago.edu.
Yanlei Ma, Email: yanleima@fudan.edu.cn.
Xiaoyang Wu, Email: xiaoyangwu@uchicago.edu.
Data availability
For this study, we will make our data available to the scientific community, which will avoid unintentional duplication of research. All the research data will be shared openly and in a timely manner in accordance with the most recent NIH guidelines (http://grants.nih.gov/grants/policy/data_sharing/). The RNA sequencing results are available at GEO (accession code: GSE165912): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165912.
References
- Alam M, Ratner D (2001) Cutaneous squamous‐cell carcinoma. N Engl J Med 344: 975–983 [DOI] [PubMed] [Google Scholar]
- Arnold I, Watt FM (2001) c‐Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol 11: 558–568 [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E (2006) Epidermal stem cells of the skin. Annu Rev Cell Dev Biol 22: 339–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Flores ER (2014) p53/p63/p73 in the epidermis in health and disease. Cold Spring Harb Perspect Med 4: a015248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton KA, Ashack KA, Khachemoune A (2016) Cutaneous squamous cell carcinoma: a review of high‐risk and metastatic disease. Am J Clin Dermatol 17: 491–508 [DOI] [PubMed] [Google Scholar]
- Cai P, Otten ABC, Cheng B, Ishii MA, Zhang W, Huang B, Qu K, Sun BK (2020) A genome‐wide long noncoding RNA CRISPRi screen identifies PRANCR as a novel regulator of epidermal homeostasis. Genome Res 30: 22–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF (2011) The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 39: D195–D201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Wu L, Zhang SM, Lu M, Cheung WK, Cai W, Gale M, Xu Q, Yan Q (2016) An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res 44: e149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E, Doupe DP, Klein AM, Winton DJ, Simons BD, Jones PH (2007) A single type of progenitor cell maintains normal epidermis. Nature 446: 185–189 [DOI] [PubMed] [Google Scholar]
- Colombo T, Farina L, Macino G, Paci P (2015) PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int 2015: 304208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Wang H, Zhu L, Jin H, Wang X (2018) N6‐methyladenosine links RNA metabolism to cancer progression. Cell Death Dis 9: 124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekoninck S, Hannezo E, Sifrim A, Miroshnikova YA, Aragona M, Malfait M, Gargouri S, de Neunheuser C, Dubois C, Voet T et al (2020) Defining the design principles of skin epidermis postnatal growth. Cell 181: 604–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, Salmon‐Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob‐Hirsch J, Amariglio N, Kupiec M et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature 485: 201–206 [DOI] [PubMed] [Google Scholar]
- Flynn RA, Chang HY (2014) Long noncoding RNAs in cell‐fate programming and reprogramming. Cell Stem Cell 14: 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Gardner C, Li ER, Arnold I, Watt FM (2003) Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development 130: 2793–2808 [DOI] [PubMed] [Google Scholar]
- Fuchs E (2008) Skin stem cells: rising to the surface. J Cell Biol 180: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goustin AS, Thepsuwan P, Kosir MA, Lipovich L (2019) The Growth‐Arrest‐Specific (GAS)‐5 long non‐coding RNA: a fascinating lncRNA widely expressed in cancers. Noncoding RNA 5: 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Rheinwald JG, Sun TT (1977) Properties of an epithelial cell type in culture: the epidermal keratinocyte and its dependence on products of the fibroblast. Prog Clin Biol Res 17: 493–500 [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL (2008) The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E (2007) Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell 12: 313–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JE, Miceli SM, Roberts RJ, Manley JL (1990) Sequence specificity of the human mRNA N6‐adenosine methylase in vitro . Nucleic Acids Res 18: 5735–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombach S, Kretz M (2013) The non‐coding skin: exploring the roles of long non‐coding RNAs in epidermal homeostasis and disease. BioEssays 35: 1093–1100 [DOI] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J et al (2017) Ythdc2 is an N(6)‐methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL et al (2018) Recognition of RNA N(6)‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20: 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PH, Harper S, Watt FM (1995) Stem cell patterning and fate in human epidermis. Cell 80: 83–93 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch L (2017) Functional genomics: screening for lncRNA function. Nat Rev Genet 18: 70 [DOI] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J et al (2013) Control of somatic tissue differentiation by the long non‐coding RNA TINCR. Nature 493: 231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez‐Pajares V, Qu K, Zheng GX, Chow J, Kim GE et al (2012) Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 26: 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavker RM, Sun TT (1982) Heterogeneity in epidermal basal keratinocytes: morphological and functional correlations. Science 215: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Lee CS, Mah A, Aros CJ, Lopez‐Pajares V, Bhaduri A, Webster DE, Kretz M, Khavari PA (2018) Cancer‐associated long noncoding RNA SMRT‐2 controls epidermal differentiation. J Invest Dermatol 138: 1445–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhuang Y, Han M, Xu T, Wu X (2013) piggyBac as a high‐capacity transgenesis and gene‐therapy vector in human cells and mice. Dis Model Mech 6: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kong Q, Yue J, Gou X, Xu M, Wu X (2019) Genome‐edited skin epidermal stem cells protect mice from cocaine‐seeking behaviour and cocaine overdose. Nat Biomed Eng 3: 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yue J, Huang H, Gou X, Chen SY, Zhao Y, Wu X (2015a) Regulation of focal adhesion dynamics and cell motility by the EB2 and Hax1 protein complex. J Biol Chem 290: 30771–30782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T (2015b) N(6)‐methyladenosine‐dependent RNA structural switches regulate RNA‐protein interactions. Nature 518: 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang SW, Huang Y, Meng J (2017) QNB: differential RNA methylation analysis for count‐based small‐sample sequencing data with a quad‐negative binomial model. BMC Bioinformatics 18: 387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, Tienda SM, Chryplewicz A, Zhu AC, Yang Y et al (2018) m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol 20: 1074–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler M, Potten CS, Wichmann HE (1987) Epidermal cell proliferation. II. A comprehensive mathematical model of cell proliferation and migration in the basal layer predicts some unusual properties of epidermal stem cells. Virchows Arch B Cell Pathol Incl Mol Pathol 53: 286–300 [PubMed] [Google Scholar]
- Lopez‐Pajares V, Yan K, Zarnegar BJ, Jameson KL, Khavari PA (2013) Genetic pathways in disorders of epidermal differentiation. Trends Genet 29: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Mascre G, Dekoninck S, Drogat B, Youssef KK, Brohee S, Sotiropoulou PA, Simons BD, Blanpain C (2012) Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489: 257–262 [DOI] [PubMed] [Google Scholar]
- Melino G, Memmi EM, Pelicci PG, Bernassola F (2015) Maintaining epithelial stemness with p63. Sci Signal 8: re9 [DOI] [PubMed] [Google Scholar]
- Meng J, Lu Z, Liu H, Zhang L, Zhang S, Chen Y, Rao MK, Huang Y (2014) A protocol for RNA methylation differential analysis with MeRIP‐Seq data and exomePeak R/Bioconductor package. Methods 69: 274–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa KR, Kawaguchi K, Cockburn K, Gonzalez D, Boucher J, Xin T, Klein AM, Greco V (2018) Homeostatic epidermal stem cell self‐renewal is driven by local differentiation. Cell Stem Cell 23: 677–686.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149: 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E et al (2003) PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Fischer SM, Slaga TJ (1985) Evidence that the centrally and peripherally located cells in the murine epidermal proliferative unit are two distinct cell populations. J Invest Dermatol 84: 277–281 [DOI] [PubMed] [Google Scholar]
- Oskarsson T, Essers MA, Dubois N, Offner S, Dubey C, Roger C, Metzger D, Chambon P, Hummler E, Beard P et al (2006) Skin epidermis lacking the c‐Myc gene is resistant to Ras‐driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev 20: 2024–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panatta E, Lena AM, Mancini M, Smirnov A, Marini A, Delli Ponti R, Botta‐Orfila T, Tartaglia GG, Mauriello A, Zhang X et al (2020) Long non‐coding RNA uc.291 controls epithelial differentiation by interfering with the ACTL6A/BAF complex. EMBO Rep 21: e46734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E (2014) Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med 4: a015263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten CS, Wichmann HE, Loeffler M, Dobek K, Major D (1982) Evidence for discrete cell kinetic subpopulations in mouse epidermis based on mathematical analysis. Cell Tissue Kinet 15: 305–329 [DOI] [PubMed] [Google Scholar]
- Potten CS, Loeffler M (1987) Epidermal cell proliferation. I. Changes with time in the proportion of isolated, paired and clustered labelled cells in sheets of murine epidermis. Virchows Arch B Cell Pathol Incl Mol Pathol 53: 279–285 [PubMed] [Google Scholar]
- Preston DS, Stern RS (1992) Nonmelanoma cancers of the skin. N Engl J Med 327: 1649–1662 [DOI] [PubMed] [Google Scholar]
- Rheinwald JG, Green H (1975) Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6: 331–343 [DOI] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V (2016) Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352: 1471–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C (2017a) Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P et al (2017b) YTHDC1 mediates nuclear export of N(6)‐methyladenosine methylated mRNAs. Elife 6: e31311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, Tumbar T (2016) Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol 18: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C (2017) YTHDF3 facilitates translation and decay of N(6)‐methyladenosine‐modified RNA. Cell Res 27: 315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegle JM, Basin A, Sastre‐Perona A, Yonekubo Y, Brown J, Sennett R, Rendl M, Tsirigos A, Carucci JA, Schober M (2014) SOX2 is a cancer‐specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat Commun 5: 4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theler D, Dominguez C, Blatter M, Boudet J, Allain FH (2014) Solution structure of the YTH domain in complex with N6‐methyladenosine RNA: a reader of methylated RNA. Nucleic Acids Res 42: 13911–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC et al (2014) PVT1 dependence in cancer with MYC copy‐number increase. Nature 512: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E (1999) The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA 96: 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR (2001) Deregulated expression of c‐Myc depletes epidermal stem cells. Nat Genet 28: 165–168 [DOI] [PubMed] [Google Scholar]
- Wan DC, Wang KC (2014) Long noncoding RNA: significance and potential in skin biology. Cold Spring Harb Perspect Med 4: a015404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G et al (2014) N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature 505: 117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C (2015) N(6)‐methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Frye M, Benitah SA (2008) MYC in mammalian epidermis: how can an oncogene stimulate differentiation? Nat Rev Cancer 8: 234–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A et al (2018) Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell 71: 973–985.e975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L et al (2018) Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self‐renewal. Mol Cell 69: 1028–1038.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE (2014) The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 55: 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan JL (2004) Focal adhesion kinase regulation of N‐WASP subcellular localization and function. J Biol Chem 279: 9565–9576 [DOI] [PubMed] [Google Scholar]
- Wu X, Kodama A, Fuchs E (2008) ACF7 regulates cytoskeletal‐focal adhesion dynamics and migration and has ATPase activity. Cell 135: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10: 927–929 [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez‐Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L et al (2017) Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell 171: 877–889.e817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C (2015) RNA N6‐methyladenosine methylation in post‐transcriptional gene expression regulation. Genes Dev 29: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Zhang Y, Liang WG, Gou X, Lee P, Liu H, Lyu W, Tang WJ, Chen SY, Yang F et al (2016) In vivo epidermal migration requires focal adhesion targeting of ACF7. Nat Commun 7: 11692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Gou X, Li Y, Wicksteed B, Wu X (2017) Engineered epidermal progenitor cells can correct diet‐induced obesity and diabetes. Cell Stem Cell 21: 256–263.e254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H et al (2018) VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 4: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Pibre S, Jacquet C, Ramirez A, de Alboran IM, Gandarillas A (2005) Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J Cell Sci 118: 1693–1704 [DOI] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, He C (2017) Post‐transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Nachtergaele S, Roundtree IA, He C (2018) Our views of dynamic N(6)‐methyladenosine RNA methylation. RNA 24: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T (2016) N(6)‐methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol 428: 822–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Gao Y, Lv D, Wang C, Wang D, Li Q (2019) METTL3 mediated m(6)A modification plays an oncogenic role in cutaneous squamous cell carcinoma by regulating DeltaNp63. Biochem Biophys Res Commun 515: 310–317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Appendix
Dataset EV1
Dataset EV2
Source Data for Appendix
Data Availability Statement
For this study, we will make our data available to the scientific community, which will avoid unintentional duplication of research. All the research data will be shared openly and in a timely manner in accordance with the most recent NIH guidelines (http://grants.nih.gov/grants/policy/data_sharing/). The RNA sequencing results are available at GEO (accession code: GSE165912): https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165912.


