Abstract
The identification of Tunneling Nanotubes (TNTs) and TNT‐like structures signified a critical turning point in the field of cell‐cell communication. With hypothesized roles in development and disease progression, TNTs’ ability to transport biological cargo between distant cells has elevated these structures to a unique and privileged position among other mechanisms of intercellular communication. However, the field faces numerous challenges—some of the most pressing issues being the demonstration of TNTs in vivo and understanding how they form and function. Another stumbling block is represented by the vast disparity in structures classified as TNTs. In order to address this ambiguity, we propose a clear nomenclature and provide a comprehensive overview of the existing knowledge concerning TNTs. We also discuss their structure, formation‐related pathways, biological function, as well as their proposed role in disease. Furthermore, we pinpoint gaps and dichotomies found across the field and highlight unexplored research avenues. Lastly, we review the methods employed to date and suggest the application of new technologies to better understand these elusive biological structures.
Keywords: actin protrusions, cell communication, cell signaling, tunneling nanotubes
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Development & Differentiation; Signal Transduction
Challenges from the emerging concept of cell‐to‐cell communication via tunneling nanotubes, as well as the current understanding of their formation, structure and physiological functions, are reviewed here together with nomenclature and methodological suggestions.

An overview of tunneling nanotubes
Cell‐to‐cell communication occurs through a variety of mechanisms which enable a diverse collection of biological messages to be shared between cells. These mechanisms include gap junctions (GJs), immunological and neural synapses, soluble factors, and extracellular vesicles. Stepping into the spotlight in 2004, thin, tubular membranous conduits rich in actin termed Tunneling nanotubes (TNTs) were first described and progressively gained recognition for allowing direct transport of cytoplasmic molecules, proteins, organelles, and even pathogens between distant cells. Although similar cellular structures called plasmodesmata had been described in plants (Sager & Lee, 2018), the identification of TNTs in the cells of animals was revolutionary and has incited great debate among the scientific community (Baker, 2017).
Since TNTs were first observed in cell culture (Rustom et al, 2004), the field of TNT‐mediated intercellular communication has expanded rapidly to reveal their existence in a wide variety of cell types, primarily in vitro. In two‐dimensional (2D) cell culture, TNTs are defined as: (I) thin (20–700 nm) and straight membranous protrusions hovering over the substrate and directly connecting two (or more) cells of both homotypic (same) and heterotypic (different) kind. (II) Internally, the presence of F‐actin cytoskeletal filaments enables (III) intercellular transfer of cargo. The last and most distinctive property of bona fide TNTs are (IV) open‐ended cell‐cell contact sites at each end, allowing cytoplasmic continuity (Fig 1); however, few studies have successfully demonstrated this feature. Interestingly, a subset of TNTs described as “close‐ended” protrusions facilitate electrical coupling between cells through GJs at the tip of the protrusion and recipient cell (Wang et al, 2010; Wang et al, 2012; Lock et al, 2016; Okafo et al, 2017). Notably, the functional ability to transport cargo is what makes other TNTs unique and distinct from this class of close‐ended protrusion that, given the size and selective permeability of GJs, does not allow intercellular transfer of large cargoes (e.g., mitochondria). Considering that cellular processes, including TNTs, are dynamic protrusions, it is conceivable that close‐ended TNTs eventually result in open‐ended TNTs, indicating that at some point in their lifespan, these structures will enable large intercellular cargo transport through open ends. However, whether close‐ended TNTs are different structures—and not a “time point” in their lifetime—has not yet been investigated.
Figure 1. Distinctive properties of TNTs.
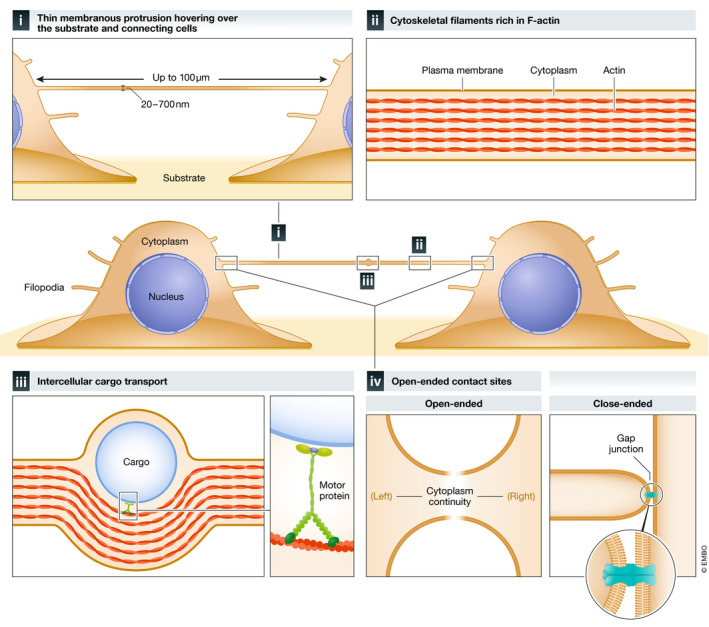
TNTs are (I) thin (20–700 nm) and long reaching (up to 100 µm in thin TNTs; < 700 nm in diameter) membranous protrusions that hover over the substrate and connect distant cells. Thick TNTs (diameter of > 700 nm) (not shown) can extend up to 250 µm in length. TNTs are (II) rich in F‐actin cytoskeletal filaments, which enable (III) internal intercellular cargo transport using motor proteins. Bona fide TNTs feature (IV) open‐ended contact sites at each end allowing for cytoplasmic continuity. A subset of TNTs contains one close‐ended contact site that facilitates electrical coupling between cells through gap junctions (GJ) at the tip of the protrusion and recipient cell.
One distinguishing characteristic of TNTs appears to be that they contain only actin. This fact was demonstrated through two key findings: (I) microtubule‐disrupting and microtubule‐stabilizing agents (nocodazole and paclitaxel, respectively) did not alter induction of TNTs (Wang et al, 2011), and (II) F‐actin depolymerizing toxins, (latrunculin, cytochalasin B, and cytochalasin D) blocked TNT formation (Bukoreshtliev et al, 2009; Schiller et al, 2013; Takahashi et al, 2013; Dupont et al, 2018). In addition to thin “canonical” TNTs fulfilling the characteristics outlined above, “thick” (> 0.7 μm in diameter), microtubule‐bearing TNTs have been found in macrophages (Onfelt et al, 2006; Souriant et al, 2019; Dupont et al, 2020), urothelial cells (Onfelt et al, 2006; Veranic et al, 2008; Resnik et al, 2018), B cells (Osteikoetxea‐Molnár et al, 2016); PC12 cells (Wang & Gerdes, 2015); astrocytes and cardiac myoblasts (Austefjord et al, 2014). These structures appear to be more resistant compared to thin TNTs, presumably from the rigidity provided by their thick architecture and microtubule cytoskeletal backbone and can reach up to 250 μm as described recently in macrophages (Dupont et al, 2020). They have been involved in the transfer of organelles (Eugenin et al, 2009; Hashimoto et al, 2016; Dupont et al, 2018) and HIV (Dupont et al, 2020); however, their specific structure and arrangement of their internal cytoskeleton, and whether they are open‐ended has not been assessed yet.
While identifying straight membranous protrusions with no substratum contact is achievable in a sparse, 2D cell culture landscape, identifying TNTs in 3D models such as organoids (Zhang et al, 2018; Pulze et al, 2020; Whitehead et al, 2020), ex vivo explants (Chinnery et al, 2008), and tissues (Lou et al, 2012), is considerably more challenging. In these and/or other ecosystems in which cells are embedded in a natural or artificial matrix, stromal elements, such as neighboring cells, can both overshadow TNTs and force them to adopt convoluted shapes compared to those seen in vitro (Korenkova et al, 2020). In these complex circumstances, TNT detection relies solely on evaluation of intercellular cargo or signal transfer, a feat of extreme technical difficulty.
In certain cases in which structures meet only some of the criterion detailed above, we suggest authors employ the term TNT‐like structure rather than TNT in order to establish a common language‐base in the field and to avoid the use of different terms for similar TNT‐like structures identified in different cell types, such as: T‐cell nanotubes in Jurkat T cells (Sowinski et al, 2008) and epithelial bridges in epithelial cells (Zani et al, 2010). In other cases, TNT synonyms, such as: bridging conduits in macrophages (Kadiu & Gendelman, 2011), nanoscale bridges in endothelial cells (Connor et al, 2015), and intercellular membranous nanotubes in urothelial cells (Lokar et al, 2012) have also been utilized. This heterogeneity in nomenclature has reportedly “created confusion in terminology” (Ariazi et al, 2017; Baker, 2017; Yamashita et al, 2018; Korenkova et al, 2020) and ultimately resulted in widespread contention over differing language ascribed to what may or may not be the same biological structure.
In spite of this dissidence, TNTs and TNT‐like structures still represent unique biological phenomena thought to play a role in a wide‐range of physiological processes and pathological conditions. These respective roles have been carefully examined in specialized reviews spanning various TNT research domains (Table 1). However, despite remarkable advances in TNT biology over the last decade, formidable challenges in this discipline remain. The absence of a TNT‐specific marker and technological limitations in the process of structural characterization have hampered our ability to (I) catalog TNTs unambiguously and determine potential existence of subclassifications, and (II) verify their existence in vivo and ascertain if they have the same functions as observed in vitro.
Table 1.
Classification of specialized reviews spanning various tunneling nanotube research domains.
| Topic | References |
|---|---|
| Molecular machineries involved TNT formation, regulation, and architecture | Abounit and Zurzolo (2012) |
| Gerdes et al (2007) | |
| Gurke et al (2008) | |
| Stability and elasticity | Drab et al (2019) |
| Morphological diversity | Austefjord et al (2014) |
| Gerdes and Carvalho (2008) | |
| Disease and therapeutic applications | Mittal et al (2019) |
| Challenges of studying them in development, health, and disease conditions | Ariazi et al (2017) |
| Jash et al (2018) | |
| Distinctions with cytonemes, and TNT‐like membranous protrusions | Korenkova et al (2020) |
| Sherer et al (2007) | |
| Yamashita et al (2018) | |
| Role in organelle transfer | Kiriyama and Nochi (2017) |
| Murray and Krasnodembskaya (2019) | |
| Vignais et al (2017) | |
| Role in neural processes and/or neurodegenerative diseases | Marzo et al (2012) |
| Victoria and Zurzolo (2017) | |
| Vargas et al (2019) | |
| Nussenzveig (2019) | |
| Role in cancer | Hekmatshoar et al (2018) |
| Matejka and Reindl (2019) | |
| Roehlecke and Schmidt (2020) | |
| Asencio‐Barría et al (2019) | |
| Pinto et al (2020) | |
| Jash et al (2018) | |
| Role in immunology | Dupont et al (2018) |
| Lin et al (2019) | |
| Onfelt et al (2005) | |
| Role in Virology | Jansens et al (2020) |
Here, we review methodologies previously and currently employed to study TNTs and suggest the future implementation of cutting‐edge technologies to enhance and further the field. We also summarize, discuss, and critically appraise existing research to outline and define the principles on which the next generation of TNT research should be based both in vitro and in vivo.
Methodology for the study of TNTs
Labeling
Given the current scarcity of a TNT‐specific biomarker, the prevailing method in TNT labeling is the staining of the entire plasma membrane with fluorescently conjugated proteins which bind glycolipids and glycoproteins, such as WGA (Abounit et al, 2015), FM1‐43 (McCoy‐Simandle et al, 2016), and MemBright (Collot et al, 2019). Alternatively, overexpressing proteins associated with the plasma membrane, such as H‐Ras, CAAX, and Lyn also serve a similar purpose (Caneparo et al, 2011; Rainy et al, 2013; Hanna et al, 2019).
Although the plasma membrane composition of TNTs remains largely unexplored (Delage & Zurzolo, 2013), previous studies identified an enrichment of specific lipids, notably cholesterol and sphingolipids, in the membrane of TNTs connecting urothelial cells (Lokar et al, 2012), and, through indirect evidence, a role of the phosphatidylinositol triphosphate (PIP3) in TNT formation in astrocytes (Wang et al, 2011) and neuronal CAD cells (Gousset et al, 2013) (see (Delage & Zurzolo, 2013) for review). In HeLa cells, PIP3 and phosphatidylinositol biphosphate (PIP2) both appear to recruit M‐Sec (Kimura et al, 2016), a protein that shares homology with Sec6, a component of the exocyst complex, that is known to be an inducer of de novo TNTs (Hase et al, 2009). Interestingly, PIP2 was localized on M‐Sec‐positive TNTs, whereas PIP3 instead accumulated in the plasma membrane ruffles at the cell periphery. Taken together, these discoveries expose a promising avenue of research which should be further explored as it may uncover important structure/function information concerning TNT membrane composition and transport role, and TNT‐specific membrane labeling hits.
Another complementary TNT‐labeling approach is the targeting of their actin backbone through either fluorescently conjugated Phalloidin staining in fixed cells (Abounit et al, 2015), expressing fluorescently conjugated actin monomers (Tardivel et al, 2016), or using different actin dyes in living cells (Lifeact fused to GFP or mCherry or the jasplakinolide‐analog dye, SiR‐actin) (Sun et al, 2019). Additionally, a recent report has shed light on a novel approach to simultaneously label actin‐, intermediate‐, and microtubule‐filaments of TNTs of urothelial cells (Resnik et al, 2019), with potential future application for co‐cytoskeletal‐distribution studies. The reproducibility of this approach in TNTs in other cell types has yet to be established, but it could be used to distinguish conduits containing only actin, microtubules, or both (Resnik et al, 2018).
Efforts made to identify TNT‐specific probes include the identification of an Aptamer, M17A2, which effectively labels TNT‐like structures in a specific breast cancer cell line (Zhang et al, 2016); however, its application and reproducibility in other cells has yet to be confirmed. In another study, laser capture microdissection and mass spectroscopy enabled Gousset and colleagues to isolate and compare membranous protrusions (including TNTs) formed by neuronal CAD cells (Gousset et al, 2019). This study revealed interesting and unexpected proteins, such as microtubule‐nucleating proteins in cells with TNTs typically deficient of microtubules. Such studies have provided our field with protein candidates to be further explored which is especially important for in vivo studies where the demand for a marker is much higher.
Detection by microscopy
Laser scanning confocal microscopy (LSCM) has revealed that the long (up to 100 μm) and thin (20–700 nm) topological nature of TNTs make them fragile and vulnerable to shearing forces (Sartori‐Rupp et al, 2019) (Box 1). The preservation of TNTs through classical fixation methods has been met with countless obstacles as TNTs’ heightened susceptibility to low‐concentrated chemical fixatives (e.g., paraformaldehyde) and long‐light exposures often results in breakage or bending.
Box 1. Detection by microscopy.
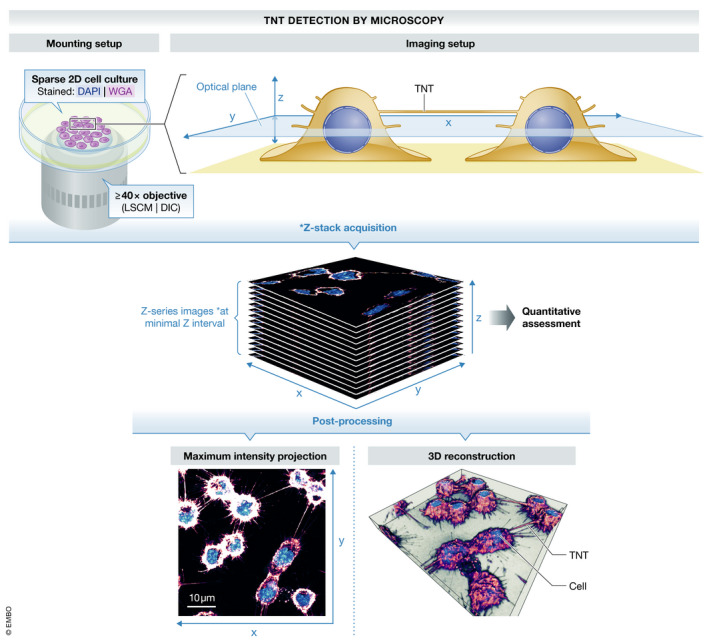
The detection of TNTs is achieved primarily through high‐magnification differential interference contrast (DIC) imaging and 3D LSCM in fixed cells (Hurtig et al, 2010), and spinning disk microscopy in living cells. However, these methods do not grant the optical resolution necessary to image these structures.
Studies using time‐gated continuous wave stimulated emission depletion (gCW‐STED) (Bénard et al, 2015) in fixed TNTs and live‐cell STED nanoscopy in living cells (Reindl et al, 2019), with a sub‐diffraction resolution of < 100 nm, are pioneering TNT microscopy methods but have yet to reach widespread implementation in the field. Structured illumination microscopy (SIM) has also enabled us and others to observe fixed TNTs in more detail (Delage et al, 2016; Halász et al, 2018; Hanna et al, 2019; Vargas et al, 2019b). Unfortunately, without a robust membrane dye or a TNT‐specific protein marker, Photo‐activated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM) do not offer any advantages.
The complexity of the matter only increases with the consideration of another outstanding obstacle—imaging these fragile structures in 3D cell cultures and tissues. One possible solution to this problem is to employ a lattice light sheet microscopy (LLSM) imaging system as done by Smith and colleagues in Ca2+‐signaling TNTs connecting human breast cancer cells (Parker et al, 2017). Even more recent developments in LLSM host a remarkable capacity to observe dynamic membranous protrusions in 4D with minimal bleaching and proved that even highly dynamic cellular protrusions, such as filopodia, can be observed at small time intervals without significant quenching (Chen et al, 2014). Implementation of tools borrowed from physics and astronomy, such as adaptive optics, may also be combined with LLSM, to allow for observation of intracellular molecules (Liu et al, 2018), an endeavor which would tremendously aid TNT research.
To circumvent the obstacle of preserving TNTs, we and others have implemented a two‐step fixation protocol leveraging extensive crosslinking via glutaraldehyde prior to paraformaldehyde fixation (Abounit et al, 2015; Carter et al, 2020). The fragility of TNTs has rendered investigations concerning membrane permeabilization experimentally cumbersome, as pores on the membrane weaken the scaffolding structure that builds TNTs. Presently, access to the lumen of TNTs is achieved through the use of milder membrane detergents (e.g., saponin) at low concentrations (Rupp et al, 2011; Costanzo et al, 2013; Zhu et al, 2018); however, the identification of inner TNT components with this method remains unsatisfactory due to its intrinsic inefficiency and damage it inflicts on the membrane. Expression of fluorescent proteins residing or getting transported within the lumen of TNTs has been used to image these structures using live microscopy, but this approach also generates complications due to the potential detrimental effects levied on the cell by protein overexpression. Moreover, this technique is indirect and cannot be used in all cell types given the existing variation in transfection susceptibility and ranging difficulties in protein expression.
To properly identify TNTs by microscopy, it is crucial to distinguish them from other membranous protrusions such as filopodia. Unlike peripheral filopodia (substrate‐adherent filopodia), TNTs hover over the substrate and even over other cells, and unlike dorsal filopodia, TNTs must connect two or more distant cells sufficiently spaced apart in order to be visualized (Fig 1) (Abounit et al, 2015). This requires cells in culture to be seeded in a medium‐confluent monolayer ecosystem warranting sufficient physical space between cells (Zhu et al, 2018). An interesting study actually found this stipulation could be overcome by cellular protease treatment (e.g., trypsin). In their study, Rustom and colleagues singularized cells by detaching them and turning them spherical, allowing for unobstructed TNT visualization (Staufer et al, 2018). However, it is currently unknown whether protease treatment of cells induces TNT formation via a stress response, for example (Zhang, 2011; Matejka & Reindl, 2019), and more intriguingly, if the observable connections are capable of intercellular cargo transport (Staufer et al, 2018). This study also highlights an intriguing conundrum in the field—the correlation between cell shape and respective capacity to form TNTs. If round cells exhibit a low membrane tension compared to a state in which cells are spreading with an increased membrane tension exerted by lamellipodial protrusions (Pontes et al, 2017), it is reasonable to think that round cells have a higher propensity to form TNTs. The more readily available membrane reservoir in round cells could also provide the extra membrane area necessary to give rise to new membranous protruding structures (Gauthier et al, 2012). However, efforts to assess the biophysical properties that mediate this relationship have not been undertaken.
Quantitative assessment
Another aspect of TNT research not sufficiently addressed, yet vital to the reproducibility of findings, is the framework employed in quantitative analysis (see Box 2). The application of machine‐learning (ML) approaches was previously proposed to identify drug targets in a single‐result algorithm application using a database of inputs obtained from not only TNTs but also GJs and hemichannels (Valdebenito et al, 2018). Using morphological and structural features that characterize TNTs, such as: height above substrate; TNT length, diameter, and straightness; actin composition; and cargo transport (Fig 1, Box 1), we propose another ML‐based approach to automate the quantification of TNTs in vitro by distinguishing them from other membranous protrusions (e.g., filopodia and lamellipodia) (Box 2). We withhold there is now sufficient ground truth data (LSCM micrographs) published in the field to tentatively train a neural network described above, but the achievement of this necessitates an agreement on a common TNT definition, data sharing, and the collaboration of numerous labs. International meetings held over recent years joining the academic leaders in the TNT field have shown TNT researchers are indeed ready to have these critical discussions and undertake an immense collaborative effort of this scale (Ariazi et al, 2017).
Box 2. Quantitative assessment.
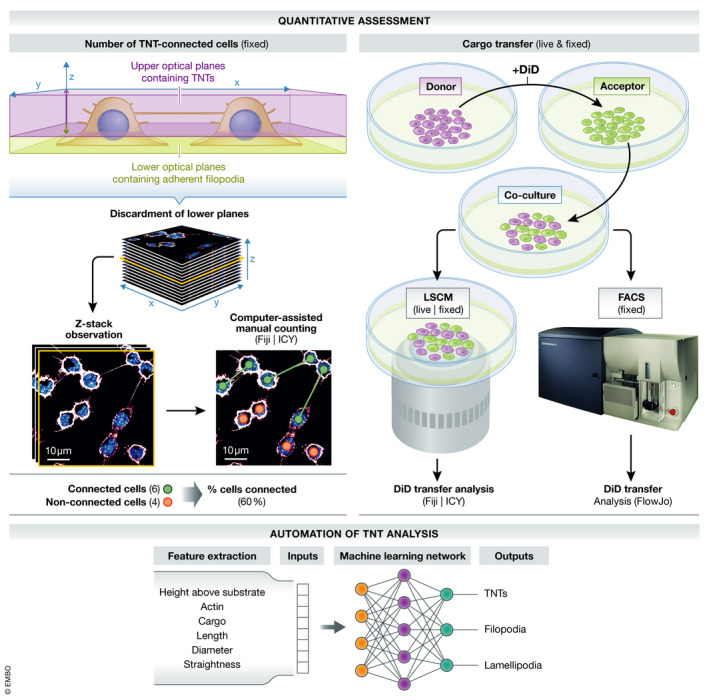
Relevant biological information from experiments is currently extracted through quantification of connected cells via TNTs using a computer‐assisted manual approach in Fiji/ImageJ or ICY (via TNT‐specific Annotator plugin) (Delage et al, 2016). However, the introduction of an effective and extensible tool to automatically and accurately count the (I) number of cells connected in a given region of interest (ROI) and (II) the number of individual tubes would improve the versatility, throughput, and reproducibility across these experiments. Recent advances in artificial intelligence (AI) and ML‐based approaches have made it possible to automatically detect and quantify dendritic spines in neurons (Smirnov et al, 2018), but no attempts have yet been made to integrate these tools in the study of TNTs.
Accurate characterization of TNTs requires the systematic accompaniment of functional assays (i.e., transport of cargo). Cargo transfer is assessed by observing organelles (e.g., lysosomes), vesicles (or endosomes) labeled with fluorescent lipophilic cationic indocarbocyanine dyes (e.g., DiI and DiD) transfer between cells in a donor and acceptor cell co‐culture system (Abounit et al, 2015; Sharma & Subramaniam, 2019). The amount of transfer is quantified by (I) live imaging microscopy or in fixed conditions by (II) LSCM or Fluorescence Activated Cell Sorting (FACS) (Abounit et al, 2015). In order to examine whether the intercellular transport of vesicles in these functional assays is TNT‐dependent and not occurring via long‐range EV‐mediated mechanisms, these experiments must be supplemented by three controls: (I) a transwell filter that reduces (or eliminates) cell‐to‐cell contact; (II) assessment of supernatant transferred from donor to acceptor cells; and (III) a mixture of non‐co‐cultured cells to obtain a background noise threshold (Gousset et al, 2009; Abounit et al, 2015; Thayanithy et al, 2017).
Quantitative analysis of TNTs could also benefit from use of controlled devices where cellular position and growth‐direction of TNTs are confined to microchannels in a micro‐fluidic chip (similarly to those routinely used for axonal growth studies (Neto et al, 2016)). This assay could in theory enhance reproducibility, throughput, and versatility of TNT studies; however, it is challenging in practice and has only been employed once thus far (Yang et al, 2016). As an alternative, we are establishing patterned surface arrays where cellular distance can be controlled in order to generate conditions which favor TNT growth in confined spaces. This method will facilitate live imaging and quantitative analysis and could be used to characterize different transport modalities inside TNTs. A collection of TNTs from cells growing on such a functionalized pattern would allow us to characterize them with greater ease as opposed to TNTs derived from sparse culture conditions. This would improve the measurement of specific cargo transfer parameters (e.g., velocity, direction, persistence), as well as the identification of the motors involved (via overexpression and knockdown studies).
Ultra‐structure of TNTs
Methods and open questions
Ultrastructural methods such as scanning and transmission electron microscopy (SEM and TEM, respectively), focused ion beam SEM (FIB‐SEM), and cryo‐EM, offer invaluable insights unobtainable through standard optical‐based imaging approaches. Thorough structural analyses of TNTs using these imaging approaches can answer central questions concerning the morphological properties which enable their function. For example, does cargo described as “being transferred” by light microscopy get shuttled within TNTs, or on the outside of the membrane? Does the cytoskeletal backbone of TNTs contain actin, microtubules, or both? How is the cytoskeleton organized to allow cargo transfer? Are TNTs open at both ends of the structure, or are they closed, filopodia‐like protrusions that mediate cargo transport via a trogocytosis‐like event, (in which an entire patch of plasma membrane gets engulfed by another cell) (Dance, 2019)? These questions would be difficult to address without the implementation of EM‐based methods (Figs 2 and 3).
Figure 2. Structural features of open‐ and close‐ended TNTs.
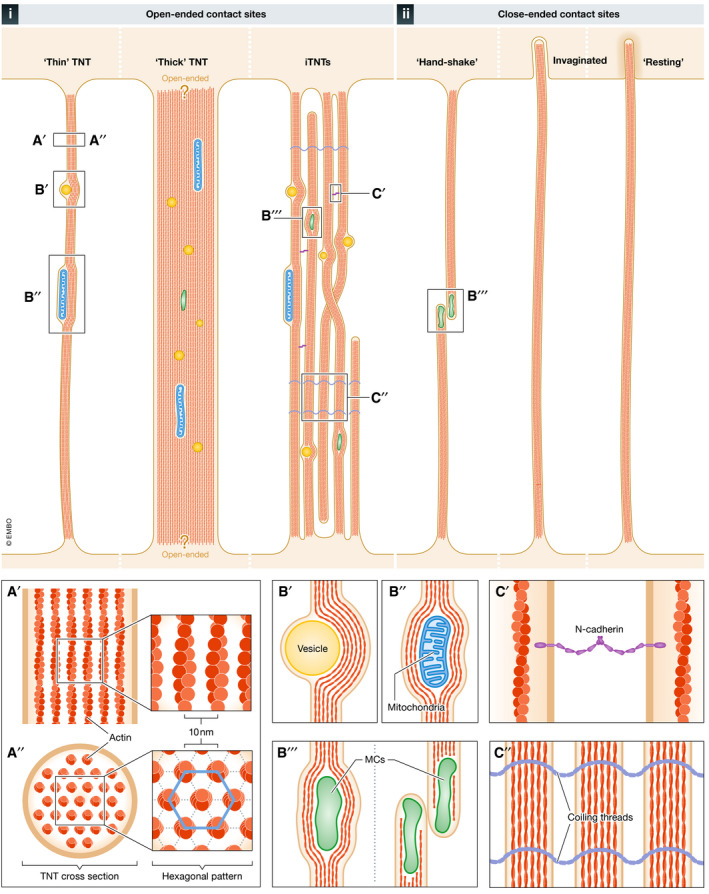
Schematic representation of the 6 main types of open‐ and close‐ended TNTs observed in vitro using electron microscopy (EM) approaches. i) Open‐ended: “Thin” TNTs, observed in photoreceptor and stem cells via SEM and in neuronal cells via cryo‐TEM/ET; “Thick” (> 700 nm) TNTs, observed in epithelial and neuronal cells with contact sites displaying questions marks to illustrate they were not demonstrated to be open; and a bundle of individual TNTs (iTNTs), a bundle of thin parallel nanotubes that get twisted together or go over and under each other, together forming one group of intertwined tubes that cannot be resolved using fluorescence microscopy; observed in neuronal and stromal cells using cryo‐TEM/ET. ii) Close‐ended (presumably facilitating cell–cell communication through gap junctions, which are not visible by EM approaches): “hand‐shake” TNTs observed in lymphocytes via ssTEM; invaginated TNTs, observed in lymphocytes via ssTEM and in neuronal cells via FIB‐SEM; and “resting” TNTs, observed in neuronal cells via FIB‐SEM. A) Top‐ (A′) and cross‐sectional (A′′) views of a TNT showing its internal actin arrangement: hexagonal actin arrays with a 10 nm distance between the center of adjacent actin filaments. B) Three types of cargoes observed within TNTs by EM: vesicles (B′), mitochondria (B′′), and membranous components (MCs) (B′′′), which could represent unidentified organelles. (C) Types of iTNT‐iTNT interlinkage processes identified by immunogold labeling and cryo‐TEM/ET: N‐Cadherin adhesion molecules between iTNTs (C′) and coiling threads (C′′), whose protein and/or lipid composition are currently known.
Figure 3. Observation of TNTs by various electron microscopy‐based approaches.
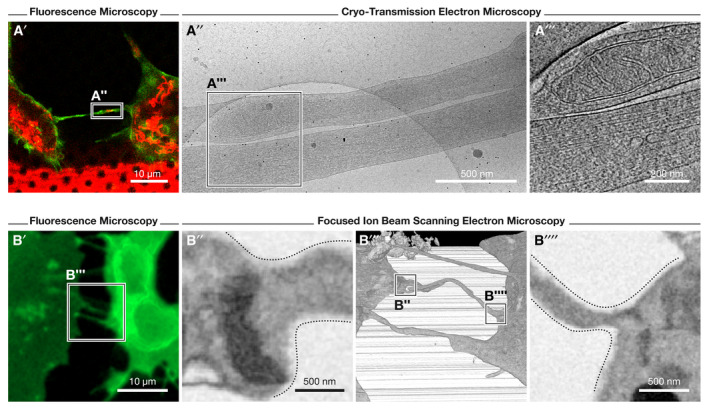
(A) Cryo‐correlative electron microscopy (cryo‐CLEM) and focused ion beam scanning EM (FIB‐SEM) approaches employed to study TNTs of SH‐SY5Y cells; images modified from (Sartori‐Rupp et al, 2019). (A′) Identification of TNT‐connected cells is first performed through fluorescence microscopy of WGA (green)‐ and mitochondria (red)‐labeled cells (seeded on EM finder grids). After vitrification of cells, the recorded position of TNTs acquired by fluorescence microscopy is imaged by cryo‐transmission EM (cryo‐TEM) and electron tomography (cryo‐ET) at medium‐ and high‐magnification (A″ and A‴, respectively). A mitochondrion can be observed within an iTNT in A‴. (B) Correlative, FIB‐SEM method employed to visualize TNTs using fluorescence microscopy (cells labeled with WGA, green) and FIB‐SEM to assess open‐ended contact sites (B″ and B′′′′, respectively) through high‐resolution 3D, volume imaging (B‴).
Morphological features and cargo
Although EM has confirmed width and length measurements of TNTs obtained by light microscopy were not far off (Table 2), EM micrographs demonstrate that the diameter of TNTs varies along their length; membrane bulges, presumably created by organelles and membranous compartments (MCs) moving through TNTs can easily expand their perimeter (Reichert et al, 2016; Scholkmann et al, 2018). Serial‐sectioning TEM (ssTEM) revealed MCs at the tips of close‐ended TNTs in Jurkat T cells (Fig 2) (Sowinski et al, 2008). Given that TNTs in these cells were shown to be close‐ended at the given time point in which they were fixed and observed, these observations suggest that MCs are unlikely to be transported intercellularly and may instead be used to deliver different molecules (including signaling molecules) at the end of the protrusion. By ssTEM, mitochondria in macrophages (Okafo et al, 2017) and ribosomes in epithelial cells (Kumar et al, 2017) were purportedly imaged within TNTs; however, due to the low magnification of the produced images, the veracity of this claim is not easily assessable.
Table 2.
Key tunneling nanotubes’ structural features obtained through electron microscopy approaches.
| Cell Model | Method | Diameter (nm) | Conformation | Open/Closed | Cytoskeleton | Cargo | References |
|---|---|---|---|---|---|---|---|
| PC12 (neuronal) | SEM, ssTEM | 50–200 | Single | Open | N/A | N/A | Rustom et al (2004) |
| Jurkat (lymphocyte) | CL‐ssTEM | 180–380 | Single | Closed | N/A | MCs | Sowinski et al (2008) |
| ARPE‐19 (photoreceptor) | SEM | 50–300 | Single | Open a | N/A | N/A | Wittig et al (2012) |
| HSPC (stem) | SEM | < 100 | Single | Open a | N/A | N/A | Reichert et al (2016) |
| KG1a (leukemic) | SEM | < 100 | Single | N/A | N/A | N/A | Reichert et al (2016) |
| A549 (epithelial) | SEM, ssTEM | 250–1,370 | Single | Open | N/A | Mitochondria & ribosomes a | Kumar et al (2017) |
| MDCK (epithelial) | SEM, ssTEM | N/A | Single | Open | Fibers a | N/A | Kumar et al (2017) |
| Macrophages (phagocyte) | SEM, ssTEM | N/A | Single | Closed | Actin a | Mitochondria a | Okafo et al (2017) |
| T24 & RT4 (bladder) | SEM | 100–200 | Single | Open | N/A | N/A | Lu et al (2017) |
| 661 W (photoreceptor) | SEM | 68–110 | Single | N/A | N/A | N/A | Scholkmann et al (2018) |
| CAD (neuronal) | SEM, cryo‐CLEM, cryo‐ET, FIB‐SEM | Individual TNT Avg. 109 | Single & bundle | Both | Actin, no microtubules | Vesicles, MCs | Sartori‐Rupp et al (2019) |
| SH‐SY5Y (neuronal) | SEM, cryo‐CLEM, cryo‐ET, FIB‐SEM | Individual TNT Avg. 120 | Single & bundle | Both | Actin, no microtubules | Vesicles, MCs, mitochondria | Sartori‐Rupp et al (2019) |
| HS‐5 (stromal) | SEM, cryo‐CLEM, cryo‐ET | ~200 | Single & bundle a | Both | Actin a | Vesicles | Kolba et al (2019) |
| STHdh (neuronal) | SEM, ssTEM | < 200 | Single | Open | N/A | N/A | Sharma and Subramaniam (2019) |
| Macrophages (phagocyte) | SEM | N/A | Single | N/A | N/A | N/A | Hanna et al (2019) |
MCs, Membranous compartments.
Not assessable or confirmed due to technique and/or insufficient image resolution employed.
Through vitrification of cells, cryo‐TEM enabled us and Piwocka and colleagues to detect vesicles inside neuronal and stromal TNTs, respectively (Kolba et al, 2019; Sartori‐Rupp et al, 2019). Furthermore, through cryo‐correlative EM (cryo‐CLEM), (identification of TNT‐connected cells using fluorescence microscopy prior to freezing and EM) and cryo‐electron tomography (cryo‐ET) (an imaging modality of TEM that produces multi‐angle, three‐dimensional volume reconstructions), we found that mitochondria also transfer within the lumen of TNTs (Fig 2) (Sartori‐Rupp et al, 2019). These tomograms demonstrated membrane bulges surrounding the cargo within, which often have a diameter equal to‐ or higher than the TNT containing them. These bulges not only contain vesicles and mitochondria, but also the actin cytoskeleton, which is likely responsible for cargo transportation (Fig 2).
Cytoskeletal architecture
Cross‐sectional analysis proved the actin in neuronal TNTs was assembled in a similar fashion as in filopodia (Aramaki et al, 2016)—hexagonally arranged, with an inter‐filament distance of 10 nm (Fig 2A). Unlike filopodia, where actin filaments can be branched, in TNTs, they generally appear to be straight (Sartori‐Rupp et al, 2019). Straight actin filaments are likely accountable for TNTs’ straight appearance. Given the distinct polarity of actin filaments, it is difficult to discern the underlying mechanisms of bidirectional cargo exchange unless actin filaments with opposite polarity coexist in the same actin bundle, or in the case of TNT‐like structures, harbor both actin and microtubules. It is therefore essential researchers increase the resolution of cryo‐TEM and ‐ET technologies (e.g., TITAN Krios cryo‐TEM) in order to correctly assess the arrangement and polarity of actin filament bundles within TNTs. The application of novel computational approaches elegantly developed by Medalia and colleagues to study actin filament polarity in cryo‐EM datasets will assist in this endeavor (Martins et al, 2021).
An intriguing discovery made by our group actually has the potential to expound upon the exact mechanisms of bidirectional TNT transport. Although historically defined as individual connections on the bases of LSCM observations, cryo‐CLEM revealed that TNTs in two neuronal cell lines, (mouse CAD and human SH‐SY5Y cells), generally appeared to be comprised of a bundle of several thin, individual TNTs (iTNTs) braided together (Figs 2 and 3) (Sartori‐Rupp et al, 2019). Contrary to a single TNT, iTNTs could in reality host opposite actin polarities allowing for a bidirectional exchange of cargoes. Interestingly, iTNTs have also been observed in stromal cells, suggesting that non‐neuronal TNTs could share this feature (Fig 2) (Kolba et al, 2019). Furthermore, even TNTs of macrophages exhibit lose‐hanging, coiled, threads that could have once formed part of a bundle which partially snapped during SEM sample preparation (Hanna et al, 2019). Similarly, the identification of multiple processes at the edge of TNTs in another report additionally supports this notion (Okafo et al, 2017). SsTEM in Jurkat T cells, however, clearly depicts TNTs as singular structures (Sowinski et al, 2008). Intriguingly, our observations in neuronal cells have also revealed the existence of single tubes with a larger diameter (average of 500 nm). The relationship between iTNT bundles and single tubes remains unclear and will require further inquiry.
Another open question that would benefit from the use of ultrastructural methods concerns the molecular motors that drive cargo transport in TNTs, which remain largely unexplored even by light microscopy approaches. Cryo‐CLEM actually enabled us to identify MyosinX (MyoX)‐positive vesicles within iTNTs (Sartori‐Rupp et al, 2019), proving correlative studies could tremendously aid in the characterization of these molecular motors.
Structural outlook
While EM has given researchers a much clearer understanding of these structures, the question of whether or not intercellular cargo transfer does indeed occur through open‐ended TNTs unfortunately cannot be addressed via cryo‐EM. Additionally, although SEM micrographs have previously suggested seamless surface transitions between TNTs and cells (Reichert et al, 2016), the images do not provide sufficient resolution to conclude whether TNTs are open‐ or close‐ended. On the contrary, qualitative ssTEM did reveal an open TNT between PC12 cells (Rustom et al, 2004), but the single example shown by the authors exhibits a short and bent shape, which differs greatly from PC12 TNTs typically observed by SEM or LSCM. In immune cells, ssTEM revealed two types of close‐ended TNTs: (I) a single protrusion invaginating into an opposing cell and (II) two filopodia‐like protrusions meeting in the middle, making a “hand‐shake” (Sowinski et al, 2008), or in a synapse‐like conformation (Okafo et al, 2017). To test whether iTNTs in neuronal cells were in fact open‐ended, we employed correlative FIB‐SEM and found most iTNTs do openly connect cells, establishing cytoplasmic continuity. In addition, closed nanotubes were either observed resting on the membrane‐ or invaginating into other cells (Sartori‐Rupp et al, 2019). Given that these observations were obtained using fixed cells, we cannot discriminate the possibility that closed structures invaginating into apposing cells represent pre‐TNT fusion events. Further studies employing high‐resolution, live imaging microscopy will be required to fully investigate this point (see Box 1).
Immunogold labeling of proteins on the membrane of TNTs could also pinpoint proteins involved in membrane fusion and interaction between close‐ended TNTs. Using this approach, we identified N‐Cadherin decorating strands between iTNTs and proposed it could play a multi‐faceted role in maintaining the stability of the overall bundle through its homophilic interactions (Fig 2) (Sartori‐Rupp et al, 2019).
The transient and fragile nature of TNTs has ultimately rendered in‐depth ultrastructural analyses technically challenging, as traditional EM involves specimen‐preparation steps which can seriously injure the morphology of TNTs (Sartori‐Rupp et al, 2019). The few structural studies available (Table 2) do detail interesting considerations but overall lack sufficient sample size to draw any quantitative conclusions. This lack of statistical power impedes our judgement of whether or not the cell‐specific differences in TNT size, cargo, cytoskeletal architecture, or overall topology illustrated above actually hold substantial biological meaning. Given most of our current TNT‐related knowledge stems from LSCM, a correlative approach, such as the one employed by Davis and colleagues for the study of Jurkat T cells (Sowinski et al, 2008) and by us in neuronal cells (Sartori‐Rupp et al, 2019), is recommended.
Mechanisms of TNT formation
Biogenesis
It is now well‐established that TNTs emerge from one of two physicomechanical origins: the first being a cell dislodgement mechanism, in which a thin actin‐bearing membranous tube forms after two adjacent cells fuse transiently and move apart; and the second being an actin‐driven protrusion mechanism, by which the tip of a filopodium‐like structure extrudes from one cell to fuse with another, onto either its soma or the tip of a filopodium emanating from it (Rustom et al, 2004; Hase et al, 2009; Wang et al, 2010).
Live imaging has revealed cell dislodgement to be the primary TNT biogenesis mechanism in a variety of cell types (Table 3). Interestingly, whole‐cell patch clamping demonstrated some dislodgment‐derived TNTs to be electrically coupled by Connexin 43 (Cx43) GJs (Wang et al, 2010). This observation is consistent with the findings of several other groups, in which immune cells known to interact by immune synapses (Finetti et al, 2017) form close‐ended TNTs (Onfelt et al, 2004; Watkins & Salter, 2005; Onfelt et al, 2006; Davis & Sowinski, 2008; Dustin et al, 2010). Whether dislodged TNTs require adhesion molecules (e.g., Cadherins) for their formation is currently unknown. The precise mechanism of actin backbone incorporation during dislodgement remains another open question. However, recent studies have demonstrated the actin‐bundling protein, Fascin, found in filopodia (Vignjevic et al, 2006) and certain neuronal dendrites (Nagel et al, 2012), is absent in the dislodged TNTs of epithelial lung cells (Dubois et al, 2018).
Table 3.
Tunneling nanotubes' preferred biogenesis mechanism.
| Cell type | Cell dislodgement | Actin‐driven | Reference |
|---|---|---|---|
| squamous cell carcinomas (SCC) | 100% | – | Sáenz‐de‐Santa‐María et al (2017) |
| neuronal crest cells (NCCs) | 100% | – | Sáenz‐de‐Santa‐María et al (2017) |
| NRK cells | 100% | – | Sáenz‐de‐Santa‐María et al (2017) |
| HEK 293 | 82% | 18% | Wang et al (2010) |
| human primary, hematopoietic progenitors | Most* | N/A | Reichert et al (2016) |
| leukemic KG1a cells | Most* | N/A | Reichert et al (2016) |
| PC12 | 7% | 93% | Bukoreshtliev et al (2009) |
In contrast to dislodgement, the actin‐driven protrusion mechanism is typical of relatively immobile cells (e.g., neurons and epithelial cells) (Bukoreshtliev et al, 2009; Gousset et al, 2013). This suggests these TNTs are formed by the action of actin protein networks which promote actin filament growth, similar to other actin‐based protrusions such as filopodia, or filopodia‐like structures (see section below). Also in this case, TNTs do not contain fascin, where evidence in neuronal CAD cells shows that fascin overexpression has an inhibitory effect on TNT formation (Gousset et al, 2013). Live imaging assays additionally showed 93% of TNTs in HeLa cells originated from filopodia‐like protrusion interplay (Bukoreshtliev et al, 2009), reinforcing this hypothesis and underlining the importance of investigating both cellular and molecular players in filopodia formation. To explore this further, here we discuss several studies focused on actin‐related proteins in the context of TNT formation.
Actin cytoskeleton and actin protein regulators
Although there exists variety in both approaches and models used to study TNT formation mechanisms, there is a consensus that these mechanisms involve proteins regulating (I) actin polymerization, (II) membrane supply and recycling, and (III) stress/inflammation and survival responses. Studies also indicate different mechanisms may operate in different cellular contexts, supporting the argument that these mechanisms are cell‐specific. Here, we will focus on proteins and signaling pathways linked to dynamic actin networks which function in the formation and regulation of other common membranous protrusions, such as filopodia (Ljubojevic et al, 2021). Molecules deployed to form TNTs via membrane recycling (e.g., different Ras‐related proteins in brain (Rabs) and M‐Sec, are described in detail elsewhere (Hase et al, 2009; Marzo et al, 2012; Dubois et al, 2018; Dupont et al, 2018; Zhu et al, 2018; Bhat et al, 2020)).
The Rho family of small GTPases is well‐characterized in the formation and rearrangement of the actin cytoskeleton in filopodia and lamellipodia (Heasman & Ridley, 2008). CDC42 was first observed at the base of TNTs connecting immune Jurkat T cells (Lachambre et al, 2014) where secramine A, a CDC42‐specific inhibitor, was shown to reduce TNT formation (Arkwright et al, 2010). In HeLa cells, inhibition of CDC42 by transfecting cells with a dominant negative construct of CDC42 also decreased TNT formation. Conversely, inhibition of Rac1 by overexpression of its dominant negative form had no significant effect (Hase et al, 2009). Treating macrophages with specific inhibitors for CDC42 and Rac1, Cox and colleagues confirmed TNTs in immune cells require both CDC42 and Rac GTPases for their formation (Hanna et al, 2017). Interestingly, our observations in neuronal CAD cells seem to indicate the opposite—that inhibition of CDC42 and Rac1 significantly increases the number of cells connected by TNTs (Delage et al, 2016). Moreover, direct inhibition of the common downstream effector of Wiskott–Aldrich syndrome protein (WASP) and WASP family verprolin‐homologous 2 (WAVE2), Arp2/3, resulted in a decrease in the formation of TNTs in macrophages (Hanna et al, 2017) but an increase in neuronal TNTs and TNT‐mediated vesicle transfer (Sartori‐Rupp et al, 2019).
Taken together, our combined results in neuronal cells disagree with those collected in macrophages (Hanna et al, 2019) and suggest a cell‐specific underlying pathway which is activated to generate TNTs. This was supported by other studies, most notably as seen for M‐Sec, which was shown to be involved in the formation of TNTs in immune (Hanna et al, 2019) but not neuronal cells (Gousset et al, 2013).
Furthermore, recent findings indicate filopodia and TNTs use distinct mechanisms of formation. More specifically, we demonstrated that the same actin regulators have opposing effects on the two structures: (I) CDC42–VASP–IRSp53 network, which induces filopodia formation serves as a negative regulator of TNTs while the (II) epidermal growth factor receptor pathway substrate 8 (Eps8), which reduces filopodia formation, induces TNT formation and increases cargo transfer (Delage et al, 2016). The discovery that these same actin regulators may have antagonistic effects relative to filopodia and TNT formation could be explained by a sort of “switch” that shifts the formation of one structure over the other (Delage et al, 2016). However, when and how this paradigm occurs is not yet known. One possible approach to investigate this further is to perform structural studies and examine the role these actin regulators have on the cytoskeletal anatomy of TNTs and filopodia of both neuronal and immune cells. This approach could also help us understand whether the structures containing both microtubules and actin—or only actin—are similar or different in their ultrastructural architecture, open‐ or close‐ended properties, and functional capabilities.
Wnt pathway
More recently, we have explored the Wingless‐related integration site (Wnt) pathway due to its function in actin cytoskeleton remodeling, filopodia formation, and neuronal growth cone guidance (Onishi et al, 2013). Our observations show the activation of the Wnt/Ca2+ pathway allows Wnt ligands to regulate the formation and stability of TNTs in neuronal cells and primary neurons (Vargas et al, 2019b). The Wnt/Ca2+ pathway achieves this through the interactional modulation of actin and the β isoform of Ca2+/calmodulin‐dependent protein kinase II (BCaMKII). Intriguingly, activation of the Wnt pathway also influences the formation of cytonemes, a specialized type of filopodia that transport signaling proteins between distant cells in development (Kornberg & Roy, 2014; Yamashita et al, 2018; Korenkova et al, 2020); however, unlike cytonemes, TNT formation is not affected by Wnt‐mediated activation of c‐Jun N‐terminal kinase (JNK) and B‐catenin (Mattes et al, 2018), suggesting TNTs and cytonemes experience differential regulation (Vargas et al, 2019b). Future studies are necessary to accurately assess whether Wnt‐induced TNTs, like cytonemes, can also transport components of the Wnt pathway (e.g., Frizzled receptor) (Stanganello & Scholpp, 2016).
MyosinX
The myosins are a superfamily of actin‐based motor proteins that “walk” on actin filaments. Of all the myosins, MyoX (an unconventional myosin motor) is the most broadly distributed within cells and is a potent filopodial inducer (Sousa & Cheney, 2005). Our group additionally showed MyoX is a critical inducer of neuronal TNTs (Gousset et al, 2013). We demonstrated that neuronal TNT formation requires the motor and tail domains of MyoX, specifically the F2 lobe of the tail protein‐ezrin‐radixin‐moesin (FERM) domain as essential. Given MyoX increases adherent‐ and dorsal filopodia formation, we postulated neuronal TNTs could arise from a specific subset of MyoX‐driven dorsal filopodia, independent of integrins, (transmembrane receptors required for the formation of adherent filopodia). Consistent with our hypothesis and subsequent data (Delage et al, 2016), MyoX indeed acts downstream of CDC42 and induces TNT formation independent of VASP, another potent inducer of dorsal filopodia formation, which conversely gets transported to the tip of filopodia by this motor (Bohil et al, 2006; Kerber & Cheney, 2011). Together, these findings demonstrate the existence of different mechanisms for dorsal filopodia formation and support a distinct role of Myo10 in TNTs.
Overall, these studies indicate that differential mechanisms operate in disparate cellular contexts, reinforcing the argument for cell specificity. Additionally, the fact filopodia and TNTs do not share the same mechanisms of formation supports the notion that they are in fact distinctive cellular structures.
Transfer function of TNTs
The principal function of TNTs being intercellular cargo transport makes them unique from other membranous protrusions, such as filopodia. The transported cargo includes various organelles, proteins, but also viruses, and bacteria. Here, we focus expressly on the role of TNTs in the transfer of mitochondria, lysosomes, amyloid aggregates, and related function in disease. For reviews that describe the involvement of TNTs in bacterial and viral transmission, please refer to Table 1.
Mitochondria
Observations derived from varying cell types have demonstrated TNTs capability for intercellular mitochondrial transfer (Vignais et al, 2017). The biological significance of this transfer is not yet fully understood. Observations in mesenchymal stem cells of TNT transfer of healthy mitochondria to cells harboring DNA defects strongly suggest an underlying protective function of mitochondrial transfer (Lin et al, 2019). In addition, mitochondria transfer through TNTs has frequently been observed in the context of different cancers (Lou et al, 2012; Antanavičiūtė et al, 2014; Thayanithy et al, 2014; Desir et al, 2018), where they are proposed to sustain tumor progression and invasion (Pinto et al, 2020).
Mitochondria are cells‘ chief energy producing organelles and thus mitochondrial dysfunction can lead to an accumulation of Reactive Oxygen Species (ROS), highly charged particles long known to pose a threat to cellular integrity and viability (Dixon & Stockwell, 2014). Intriguingly, ROS have been proven to be an effective inducer of TNTs via the major form of endogenous ROS, hydrogen peroxide (H2O2) (Liang, 2018). In rat hippocampal astrocytes, neurons, and HEK293 cells, H2O2 significantly increases the number of (homotypic and heterotypic) cells connected by TNT‐like structures compared to untreated cells (Wang et al, 2011). Notably, upon co‐culturing non‐stressed and stressed cells, the authors observed that H2O2‐treated cells were the initiators of TNT formation. Subsequent studies from our group showed that H2O2‐induced stress also increases TNT formation in neuronal cells by 70% (Gousset et al, 2013), indicating a ubiquitous effect on TNTs across distinct cell types.
Combined, these and other observations led Rustom to propose a mechanistic model of ROS‐dependent TNT formation (Rustom, 2016). In his review, Rustom describes TNT formation directly linked to the increase of ROS level in stressed cells. The underlying reason why this occurs is not yet known; however, according to the proposed model, TNTs are formed by unstressed cells in order to establish an open communication with stressed cells as a “call for help”. This event is followed by (I) the TNT‐mediated transport of mitochondria to rescue the apoptotic cell or (II) removal of cells with excessive levels of ROS. This model posits TNTs as a communication tool in cellular organization and survival during stress (Rustom, 2016).
The exact mechanism underlying ROS‐mediated TNT induction is not well understood; however, a study on TNTs in astrocytes indicated H2O2 triggers phosphorylation of the p38 mitogen‐activated protein kinase (MAPK) (Zhu et al, 2005), a kinase responsive to stress stimuli which has previously been shown to induce actin cytoskeletal remodeling in fibroblasts (Hoffman et al, 2017). Other reviews have further explicated the strict relationship between physiological levels of ROS and actin‐related proteins (Wilson & González‐Billault, 2015) and regulated actin flow at the leading edge of cells (Taulet et al, 2012). These observations support the established notion that H2O2 affects actin cytoskeletal organization and induces the formation of actin‐rich protrusions (Huot et al, 1998). Future studies will be required to pinpoint the effect redox imbalance can inflict on actin‐associated proteins and, specifically, interrogate any correlation between ROS synthesis and actin filament oxidation. As previously reported by Wilson and González‐Billault, studying this spatiotemporal relationship is a challenging quest given that both elements are short‐lived and dynamic proteins (Wilson & González‐Billault, 2015); however, novel genetic (Mishin et al, 2015) and bioluminescence‐based tools (Ishimoto & Mori, 2019) might be able to help us in these endeavors.
Another unexplored area of TNT‐mediated mitochondrial transport is the molecular motors responsible for shuttling them. Agrawal and colleagues found that the mitochondrial Rho‐GTPase, Miro1, an adaptor protein that attaches mitochondria to KIF5 motor proteins, allows for mitochondrial transport through TNTs containing microtubules (Ahmad et al, 2014). Our most recent correlative structural analysis identified mitochondria traveling through actin, not microtubules, suggesting that actin‐driven motor proteins must also be involved (Sartori‐Rupp et al, 2019). Furthermore, we found mitochondria travel through TNTs at an average velocity of 50 nm per second; however, their speed is not often uniform and instead appears to accelerate in straight TNT segments (Wang & Gerdes, 2015; Sartori‐Rupp et al, 2019).
Lysosomes
Lysosomes are membrane‐bound organelles whose foremost function is to digest macromolecules and break down cellular waste. Lysosomes were first observed in TNTs connecting macrophage cells, moving at a speed of 1 μm/s (Onfelt et al, 2006). Since then, the study of TNT‐mediated intercellular transport of lysosomes has revealed they can facilitate the transport of viruses (Eugenin et al, 2009) and proteins linked to various neurodegenerative diseases (NDs) (Victoria & Zurzolo, 2017), which will be detailed below. Interestingly, TNTs also exhibit a protective role in allowing (I) hematopoietic stem cell (HSC)‐derived macrophages to supply cystinosin‐deficient cells with functional lysosomes and (II) pulling cystinosin‐bearing lysosomes away from diseased cells (Naphade et al, 2015; Goodman et al, 2019). Additional studies have linked the direct transfer of lysosomes to lysosomal gradient preservation and endothelial cell viability (Yasuda et al, 2011). Finally, in tumor cells derived from squamous cell carcinomas, Chiara and colleagues observed that focal adhesion kinase (FAK)‐deficient cells transfer lysosomes and autophagosomes to FAC‐proficient cells via TNTs, supporting the relevance of a mechanism necessary to adapt to stress evoked by impaired FAK signaling (Sáenz‐de‐Santa‐María et al, 2017).
Amyloid proteins
Major NDs, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and Huntington’s disease (HD) all share the defining feature of proteinaceous aggregates which accumulate in the central nervous system (CNS) and cause neuronal cell death (Jucker & Walker, 2018). A growing body of literature supports the hypothesis linking the progression of these diseases to the spreading of these assemblies, which have the unique prion‐specific ability to imprint their misfolding on soluble cellular protein (Luk et al, 2009; Goedert, 2015; Hasegawa et al, 2017; Jansen et al, 2017). Recent data from our laboratory and others have revealed TNT‐mediated spread of amyloid protein assemblies associated with various NDs. This has led to our proposal that TNTs represent a major route for the intercellular amyloid spreading and are thus involved in the progression of these diseases (Victoria & Zurzolo, 2017; Vargas et al, 2019a).
In a seminal discovery, our group demonstrated that the infectious form of prion protein (PrPSc) is able to spread to other cells by hijacking TNTs (Gousset et al, 2009). We have also shown prions spread through TNTs within organelles of the endocytic pathway, (most likely recycling endosomes) (Zhu et al, 2015). TNT‐mediated transport of prions occurs between neuronal cells as well as between dendritic cells and neurons, and primary neurons and astrocytes, together accounting for the progression of the prion infection from peripheral tissue to the brain, and within the brain (Gousset et al, 2009). Amyloid proteins’ prion‐like behavior of inducing misfolding of normal proteins led our group and others to investigate the role of TNTs in the spreading of amyloid proteins involved in more common diseases, namely α‐synuclein with PD, huntingtin with HD, and Tau/amyloid beta (Aβ) with AD.
Alpha‐synuclein
One of our studies previously documented that fibrillar α‐syn could also induce cells to form TNTs and “hijack” them to efficiently spread intercellularly using lysosomal vesicles as shuttles (Abounit et al, 2016a). Follow‐up studies from our and other groups have since shown that TNT‐mediated aggregated‐α‐syn transfer occurs in human and mouse astrocytes (Loria et al, 2017; Rostami et al, 2017), SH‐SY5Y neuronal and SW‐13 epithelial cells (Dieriks et al, 2017; Dilsizoglu Senol et al, 2019), and primary brain pericytes (Dieriks et al, 2017). We have also recently observed α‐syn‐carrying TNT‐like structures in primary mouse cortical neurons (Vargas et al, 2019b) and human neuronal precursor cells (hNPCs) derived from healthy subjects and patients carrying the early‐onset PD‐associated A53T mutation in the α‐syn gene (Grudina et al, 2019).
Although our experiments in hNPCs revealing the post‐internalization preferential localization of α‐syn fibrils in lysosomes (Grudina et al, 2019), confirmed our initial demonstration in murine neuronal cells by showing that lysosomes transport α‐syn aggregates through TNTs, the mechanism and pathogenetic implication of this transfer remains to be explained (Victoria & Zurzolo, 2017). The mechanism used by α‐syn aggregates to recruit and induce the aggregation of soluble α‐syn proteins in the cytosol of acceptor cells following TNT‐mediated transfer within lysosomes also represents an exciting research avenue. Exploring this event and identifying the compartments α‐syn gets directed to when they reach “acceptor” cells hold tremendous scientific and therapeutic value (Abounit et al, 2016a).
Dismantling TNTs to diminish or even halt the spreading of intercellular α‐syn also represents a promising therapeutic avenue (Victoria & Zurzolo, 2017). Although latrunculin B was previously shown to abrogate TNT formation and intercellular α‐syn transport (Rostami et al, 2017), actin depolymerizing toxins risk collapsing the entire actin scaffold of cells. In one of our recent studies, we showed that nM concentrations of the actin‐targeting cyanobacterial macrolide, tolytoxin, significantly decreased the number of TNT‐mediated transfers of α‐syn fibrils without affecting the global actin architecture of cells (Dilsizoglu Senol et al, 2019).
Identifying inhibitory molecular targets for TNT‐related α‐syn transfer, while also avoiding effects on the actin cytoskeleton, will require further research. For example, studying the kinetics of TNT‐mediated transport of α‐syn through live imaging microscopy and particle tracking (Sartori‐Rupp et al, 2019) could provide useful insights into the speed and dynamicity of α‐syn spreading, including the distribution of potential molecular motors. Our study on the Wnt pathway, which demonstrates that α‐syn transfer through TNTs can be modulated by the Wnt/Ca2+ pathway, also represents an important future area of study and is worth characterizing (Vargas et al, 2019b). A more in‐depth look at the topological landscape governing the transfer α‐syn using CLEM approaches could also teach us about the interaction between α‐syn, lysosomes, actin, and the TNTs containing them.
Huntingtin
In the case of HTT, several mechanisms have been proposed, including extracellular vesicles (Tang, 2018) and transneuronal (synaptic) spreading (Pecho‐Vrieseling et al, 2014). Our early work has also shown the intercellular propagation of mHTT through TNTs (Costanzo et al, 2013). Primary cerebellar granule neurons could also transport mHTT in a contact‐dependent manner but we did not identify any TNT‐like structures due to the hectic dendritic landscape of primary neuron cell culture. It is likely that the co‐labeling approach recently employed by us to identify TNT‐like structures in primary cortical neurons could shed light on this intriguing possibility (Vargas et al, 2019b).
The precise molecular mechanism governing TNT‐mediated intercellular transport of mHTT is currently unknown, and little attention has been paid to it since our initial observations. In an attempt to learn more about the molecular pathway of mHTT spreading in a brain region seriously affected by HD—the striatum (Kassubek et al, 2004)—Sharma and Subramaniam focused their studies on the striatum‐enriched GTPase protein, Rhes. Using mouse normal striatal neuronal cells, the authors found Rhes induced the biogenesis of TNT‐like structures and enhanced intercellular transfer of mHTT between donor and acceptor cell populations (Sharma & Subramaniam, 2019). Consistent with studies in PC12 (Bukoreshtliev et al, 2009) and MSCs (Zhang et al, 2018), TNT‐like structures were abrogated with the F‐actin depolymerizing toxin, cytochalasin D; however, much remains unclear about the precise molecular mechanism that underlies the TNT‐mediated transport of mHTT compared to α‐syn. Future studies are needed to explore the depth and specificity involved in mHTT spreading by TNTs and how this mechanism competes with others previously described (Tang, 2018). Interestingly, Rhes also mediates Tau pathology in AD mouse models, making studies with Tau mutants an interesting research avenue.
Amyloid beta and Tau
The work by Zhang and colleagues, which previously reported TNT‐mediated Aβ transport in rat astrocytes and neurons, supports this notion (Wang et al, 2011); however, no further evidence describing the underlying mechanism or pathways involved has surfaced since then. Meanwhile, we and others have shown that extracellular monomeric and fibrillar Tau species induce the formation of TNTs, which facilitate their transfer between neuronal cells (Tardivel et al, 2016; Abounit et al, 2016b). We have also recently shown that endogenously formed Tau aggregates are found in TNTs (Chastagner et al, 2020). Together, these data indicate that proteins involved in Tauopathies and AD could hijack TNTs and use them as a means of spread, as previously described for Prion, α‐syn, and mHTT (Victoria & Zurzolo, 2017).
Molecule and Signal transfer
Major histocompatibility complexes
Studies focused on immune cells have also demonstrated that TNTs can transport major histocompatibility complexes (MHC), molecules whose main role is to present antigens at the cell surface of certain immune cells to be recognized by T cells. Transfer or association of MHC‐I class proteins with TNTs has been observed in B cells (Onfelt et al, 2004); HeLa (Schiller et al, 2013); and thymic epithelial and dendritic cells (Watkins & Salter, 2005; Millet et al, 2008). In NK cells, MHC class I chain‐related protein A was observed to transfer to the tip of a close‐ended TNT (or immune synapse) where it is postulated to interact‐ and activate the target cell (Chauveau et al, 2010). Even though it is currently unclear whether MHC molecules retain their function when transferred, or how they behave during their transfer, these studies propose an intriguing role for TNTs, including the initiation and implication of signals that can trigger an immune response. For further reading of TNT‐mediated transfer of MHC complexes, we would like to point our readership in the direction of a specialized review by Bonaccorsi and colleagues (Campana et al, 2015).
Calcium transfer
Shortly after the discovery of TNTs, numerous studies demonstrated that Calcium (Ca2+), too, could be transported via TNTs. First evidenced by observations in dendritic cells and THP‐1 monocytes, and contrary to classical cellular electrical conductivity, intercellular Ca2+ flux propagation between cells via TNTs was not driven by an action potential (Watkins & Salter, 2005). Through GJ‐specific inhibitors, the authors also showed that TNTs could transport Ca2+ independent of Gjs; however, the underlying mechanism of TNT‐mediated Ca2+ transfer was only reassessed years later, when TNTs of SH‐SY5Y and HEK293 cells were shown to involve IP3 receptors for the active propagation of Ca2+ (Smith et al, 2011). Since then, Ca2+ flux transfer through TNTs has been observed in several other cell types, including Raw cells (Hase et al, 2009), immature neurons (electrically coupled to distant astrocytes) (Wang et al, 2012), and ARPE‐19 cells (Wittig et al, 2012). Some, but not all contained Gjs, indicating that—unless TNTs can transition between an open‐ended to a GJ‐mediated, close‐ended state—Ca2+ can be transported between cells using two distinct types of TNTs. Whether and how TNTs can exist and transport Ca2+ signals between functional neurons connected by synapses has not yet been explored and future investigations will have to address this possibility.
Presence of TNT‐like structures in vivo
The data outlined above demonstrate that TNTs play a role in the intercellular communication between various cells in culture (including primary cells). This has allowed researchers to speculate on their physiopathological role; however, definitive evidence stating that TNTs exist within more complex biological contexts is currently missing.
Development
Various studies have established that during gastrulation of marine animals such as Sea Urchins, actin‐containing filopodial protrusions extend from ectodermal cells, and between primary and secondary mesenchyme cells (Miller et al, 1995). Although sightings of thin filopodia were previously made in Sea Urchins and were hypothesized to play a role in locomotion (Gustafson & Wolpert, 1963), McClay and colleagues suggest that the developmental timing, location, and rich cytoskeletal apparatus of filopodia owe their regulation to signaling organelles enabling communication of migratory cues (McClay, 1999).
During zebrafish gastrulation, long actin‐rich Intercellular Bridges (IBs) are generated from dividing epiblast cells (Caneparo et al, 2011) which do not separate by abscission as daughter cells normally do during cytokinesis. The observation of fluorescent membrane‐linked Dendra2 moving along IBs at a rate only possible through an active process led the authors to hypothesize that IBs could function as TNTs for intercellular signaling during embryonic patterning and development. However, recently published studies described similar protrusions as “retraction fiber projections” that host Migrasomes (Jiang et al, 2019), vesicular structures at the tips of projections that release contents into the environment through an EV‐like mechanism called “migracytosis” (Ma et al, 2015; Huang et al, 2019; Tavano & Heisenberg, 2019). Although IBs and migrasome‐carrying fibers resemble each other morphologically, it is currently unclear whether migrasomes are derived from IBs, or whether these two structures coexist in the complex zebrafish gastrular micro‐environment as two separate processes.
The formation of stable IBs through incomplete cytokinesis was also shown in various germline and somatic tissues (Haglund et al, 2011). For example, actin‐rich germline bridges called Ring Canals (RCs) are rims that connect germ cells in cysts of Xenopus laevis (Kloc et al, 2004), Drosophila (Haglund et al, 2010), mice (Pepling & Spradling, 1998), and several other animal models (Haglund et al, 2011). The function of these connections remains unknown; however, numerous reports have proposed they promote the exchange of cytoplasm and organelles, in a similar fashion to TNTs, with the important exception that TNTs do not form from cell division (Caneparo et al, 2011). Whether any of the structures previously identified as ring canals or IBs (such as those identified by Caneparo et al, in zebrafish) are instead TNTs forming between two different cell populations in vivo, which enable intercellular communication, synchronization of cell division, differentiation, or migration is currently unknown (Korenkova et al, 2020).
During development of the imaginal disk of Drosophila, specialized filopodia called Cytonemes were identified and are sometimes classified as the first demonstration of TNT‐like structures in vivo (Ramírez‐Weber & Kornberg, 1999). However, distinct from TNTs, cytonemes are close‐ended, specialized filopodia (Kornberg & Roy, 2014) that play an important role in the signaling of morphogens, (e.g., Bone morphogenetic protein, BMP; Sonic hedgehog, Shh; Wingless‐related integration site, Wnt, and Fibroblast growth factor, Fgf), through ligands and receptors that are currently under investigation (Kornberg, 2017; Zhang & Scholpp, 2019). Interestingly, recent evidence from our group indicates Wnt signaling is involved in the regulation of TNT formation of primary neurons (Vargas et al, 2019b). However, Wnt‐dependent TNT formation is mediated by activation of the downstream calcium‐calmodulin signaling pathway, while the formation of cytonemes appears to be mediated by the downstream activation of beta catenin (Mattes et al, 2018). Thus, while activation of the Wnt receptor is involved in both cytoneme and TNT formation, their downstream pathways differ, suggesting cytonemes and TNTs are in fact different structures that might have disparate developmental functions.
Prospective role of TNTs and TNT‐like structures in brain development
Over the past decade, our studies in neuronal cells revealed that TNT formation and stabilization requires a variety of specific proteins and ligands. Interestingly, many of these molecules are enriched during pre‐ and postnatal brain development. MyoX, for example, is regulated during early postnatal development of the CNS in mice (Sousa et al, 2006) and is highly expressed in a brain region of active cell proliferation and migration: the external‐ and internal granule cell layers of the cerebellum (Raines et al, 2012).
During neocortical brain development, MyoX is also known to form a complex with surface N‐Cadherin adhesion molecules in order to allow neuronal radial migration (Lai et al, 2015). Interestingly, we previously showed that MyoX can give rise to neuronal TNTs independent of its binding to N‐Cadherin (Gousset et al, 2013) and hypothesized that N‐Cadherin played a role in the formation and stability of TNTs through its’ homophilic interactions with N‐Cadherin molecules sitting on adjacent TNTs, “iTNTs” (Sartori‐Rupp et al, 2019). Whether N‐Cadherin, whose primary functions were classically thought to govern essential cell‐to‐cell interactions during neuronal morphogenesis and correct synaptic pairing by bringing apposed cell membranes into contact (Takeichi, 2007), also serves as an essential player for TNT formation during cortical neuron migration and polarity establishment will require further investigation.
Our most recent discovery showing that TNT‐like structures can form between primary neurons at an early stage of development, prior to the formation of synapses, supports the notion undifferentiated neurons could utilize TNTs as a mode of intercellular communication where synapses have not been established (Vargas et al, 2019b). Whether this is the case in the brain requires further inquiry. Interestingly, there are numerous instances when pre‐synaptic TNT‐mediated communication could be employed during brain development. For example, granule cells in the cerebellum are neuronal progenitors with spherical somas and a few protruding neurites that undertake two distinctive migratory events prior to synapse formation (Komuro & Kumada, 2005). The underlying pathways that govern these events have been attributed to Bergmann glia, but continue to be investigated (Galas et al, 2017). The prospect of TNTs/TNT‐like structures playing a role in neuronal migratory processes via the propagation of depolarization signals, to synchronize actin remodeling was previously proposed but never demonstrated (Gerdes et al, 2013).
Wnt ligands, (Wnt5a/7a), which induce neuronal TNTs in culture (Vargas et al, 2019b), participate in the maturation and function of pre‐synaptic terminals and dendritic maintenance (Oliva et al, 2013), and, in the case of Wnt7a, get secreted by cerebellar granule cells to regulate synaptic differentiation (Hall et al, 2000). Formation of dendritic spines is also regulated by Eps8 (Menna et al, 2013; Stamatakou et al, 2013), another neuronal TNT inducer (see above) (Delage et al, 2016). In the brain, Eps8 is localized post‐synaptically in the dendritic articulations of cerebellar granule neurons (Offenhäuser et al, 2006) and in axons of cultured hippocampal neurons, where it regulates the formation of filopodia (Menna et al, 2009).
Taken together, these studies provide evidence that TNT/TNT‐like structures involve proteins with an elevated activity in developing brain areas, rich in neuronal precursors that are non‐polarized and/or are in the process of migrating. Although these proteins were classically thought to give rise to and maintain neuronal processes (e.g., dendritic spines and axonal growth cones), our in vitro data proposes a novel molecular function in which these molecules also regulate neuronal TNTs, which are presumably engaged during neurodevelopmental processes (e.g., neuronal precursor migration). The discovery of TNT‐like structures during migratory and signaling events during early developmental processes of non‐mammalian, vertebrate organisms supports this notion (Miller et al, 1995; Caneparo et al, 2011), but our hypothesis on neurodevelopmental TNTs has never been tested.
Owing to the difficulties of identifying TNTs in vivo—given lack of a TNT‐specific marker and limitations in the resolution of light microscopy—our goal moving forward should be to focus on the implementation of long‐range, structural mapping technologies (e.g., FIB‐SEM) to detect TNTs in the developing brain without the need for a TNT‐specific marker. The technical and conceptual strategies optimized by these investigations will serve as a stepping stone that will eventually enable us to study the presence‐ and physiological function of neuronal TNTs in developed mouse models commonly used to study neurodegenerative disease conditions, where we hypothesize to find an exacerbation of TNTs (Victoria & Zurzolo, 2017).
Others
In other studies, MHC class II + cells in the mouse cornea were observed to connect via TNT‐like structures called membrane nanotubes (MNTs), < 800 nm in thickness and > 300 μm in length (Chinnery et al, 2008; Seyed‐Razavi et al, 2013). The authors hypothesize isolated (dendritic) cells may use TNTs to communicate and form an immunological syncytium; however, this hypothesis has yet to be explored.
Intervascular bridges (IVBs) are actin‐bearing fibrous strands joining neighboring capillaries in the retina of humans and mice (Mendes‐Jorge et al, 2012). The expression of common pericyte markers (NG2 and PDGFR‐b) led the authors to hypothesize IVBs are of pericyte nature. Interestingly, these bridges express Cx43, suggesting these connections allow intercellular communication. However, the precise signals transported between cells via these bridges has not yet been explored. In another more recent study, Di Polo and colleagues identified TNT‐like processes (termed interpericyte TNTs or IP‐TNTs) that connect pericytes on retinal capillaries and form a functional syncytium (Alarcon‐Martinez et al, 2020). The authors show that IP‐TNTs contain mitochondria and actin, and propagate intercellular calcium waves; however, their close‐ended tip characterized by the presence of GJs may limit the intercellular transport of larger cargoes (Zurzolo, 2020).
Various malignant tumors, including those of lung‐, ovarian‐, and laryngeal cancer, mesothelioma, and neuroblastoma, were shown to form TNT‐like connections (Antanavičiūtė et al, 2014, 2015; Ady et al, 2016; Desir et al, 2016). Mesothelioma cells forming TNTs of > 70 μm in length contained microvesicles labeled with MitoTracker (Lou et al, 2012). Human glioblastoma tumors implanted into a mouse brain and labeled with an astrocytoma cell GFP reporter form long (> 500 μm) and thick (> 1 μm) connections called Tumor Microtubes (TMs) between glioblastoma cells (Osswald et al, 2015). However, it is not clear whether TMs allow cargo exchange between cells similarly to TNTs (Pinto et al, 2020, 2021).
Finally, a report by Cherqui and colleagues identified TNT‐like extensions connecting HSC‐derived macrophages with kidney proximal tubular cells, crossing the dense basement membranes of the kidney to deliver cystinosin‐containing vesicles into diseased tubular cells (Naphade et al, 2015; Cherqui & Courtoy, 2017).
Collectively, these findings show that TNT‐like structures exist in vivo under certain physiological and/or pathological conditions. Future studies will need to demonstrate whether these different structures feature the TNT criteria described above (Fig 1).
Discussion and future perspectives
The growing number of TNT and TNT‐like structure discoveries in vitro and in vivo usher in an exciting time for the field of TNT‐mediated cell‐to‐cell communication research. We envision a new era in TNT biology in which the successful deployment of sophisticated labeling, imaging, and analytical technologies for the comprehensive characterization of TNTs will serve as an important benchmark for testing our in vitro findings in vivo, and most importantly, as a conduit between the bench and the promising therapeutic value that TNTs possess.
One of the primary objectives moving forward should be further refinement of the up‐ and downstream pathways regulating the formation of TNTs, (i.e., proteins modulating actin polymerization, membrane supply and recycling, and stress response). The outcome of these studies not only has the potential to yield a TNT‐specific marker, which the field still severely lacks, but also heralds a promising therapeutic candidate that could target TNTs during disease. A rigorous characterization of TNTs’ molecular architecture will undoubtedly benefit from 3D, correlative structural investigations peering into the cytoskeletal and membranous anatomy of TNTs at the nanoscale resolution.
Thorough molecular and structural studies will also shed light on the diversity found among TNTs (e.g., open‐ versus close‐ended) and address existing dichotomies between species / cellular models. Generating a comprehensive catalog of distinct types of TNTs will enable us to label and classify them accordingly. For the time being, we encourage authors to consider employing the terms “TNT” and “TNT‐like” in order to foster nomenclature consistency and eschew the creation of analogous‐ or cell‐specific terms, which have previously led to discrepancy and confusion (Ariazi et al, 2017; Yamashita et al, 2018). Other essential aspects which remain to be uncovered regarding the operation of these complex biological machines include the kinetics and underlying biophysical mechanisms that lead to TNT fusion during formation (Abounit & Zurzolo, 2012; Dupont et al, 2018; Drab et al, 2019).
The opportunities facing us are tremendous, but to thrive, the field will have to ascertain the presence of TNTs in vivo using animal models, verify the functional features they exhibit in culture translate to complex biological organisms, and determine their functional role during an organism’s proper or improper development. Visually accessible models like the developing zebrafish have proven to be effective systems to interrogate the presence of TNT‐like structures in vivo, during embryogenesis. However, further studies will be required to test whether these structures also exist in a mammalian model and in the development of specific biological systems such as the CNS, which is the focus of our group.
We currently face a number of limitations, but the future prospects of TNT‐mediated cell‐to‐cell communication during development and in disease are nonetheless very exciting.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank Harshavardhan Khare for his helpful insights on our machine‐learning strategy. We are grateful to Rio McLellan, Harshavardhan Khare, Stephen O’Shea, Olga Korenkova, and Anna Pepe for helpful comments on the manuscript. TNT research in the Zurzolo laboraotory is supported by Neurotunn (ANR‐16‐CE160019), Cancéropôle Ile‐de‐France (2018‐1‐PL‐03‐IP‐1), INCEPTION (ANR‐16‐CONV‐0005) and Institut de Convergence Q‐Life awards (ANR‐17‐CONV‐0005) grants to CZ. D.C.C. is supported by INCEPTION (ANR‐16‐CONV‐0005) and an Institut de Convergence Q‐Life award (ANR‐17‐CONV‐0005).
The EMBO Journal (2021) 40: e105789.
References
- Abounit S, Bousset L, Loria F, Zhu S, de Chaumont F, Pieri L, Olivo‐Marin J‐C, Melki R, Zurzolo C (2016a) Tunneling nanotubes spread fibrillar α‐synuclein by intercellular trafficking of lysosomes. EMBO J 35: 2120–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abounit S, Delage E, Zurzolo C (2015) Identification and characterization of tunneling nanotubes for intercellular trafficking. Curr Protoc Cell Biol 67: 12.10.1‐21 [DOI] [PubMed] [Google Scholar]
- Abounit S, Wu JW, Duff K, Victoria GS, Zurzolo C (2016b) Tunneling nanotubes: a possible highway in the spreading of tau and other prion‐like proteins in neurodegenerative diseases. Prion 10: 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abounit S, Zurzolo C (2012) Wiring through tunneling nanotubes–from electrical signals to organelle transfer. J Cell Sci 125: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Ady J, Thayanithy V, Mojica K, Wong P, Carson J, Rao P, Fong Y, Lou E (2016) Tunneling nanotubes: an alternate route for propagation of the bystander effect following oncolytic viral infection. Mol Ther Oncolytics 3: 16029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad T, Mukherjee S, Pattnaik B, Kumar M, Singh S, Kumar M, Rehman R, Tiwari BK, Jha KA, Barhanpurkar AP et al (2014) Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J 33: 994–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon‐Martinez L, Villafranca‐Baughman D, Quintero H, Kacerovsky JB, Dotigny F, Murai KK, Prat A, Drapeau P, Di Polo A (2020) Interpericyte tunnelling nanotubes regulate neurovascular coupling. Nature 585: 91–95 [DOI] [PubMed] [Google Scholar]
- Antanavičiūtė I, Ereminienė E, Vysockas V, Račkauskas M, Skipskis V, Rysevaitė K, Treinys R, Benetis R, Jurevičius J, Skeberdis VA (2015) Exogenous connexin43‐expressing autologous skeletal myoblasts ameliorate mechanical function and electrical activity of the rabbit heart after experimental infarction. Int J Exp Pathol 96: 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antanavičiūtė I, Rysevaitė K, Liutkevičius V, Marandykina A, Rimkutė L, Sveikatienė R, Uloza V, Skeberdis VA (2014) Long‐distance communication between laryngeal carcinoma cells. PLoS One 9: e99196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramaki S, Mayanagi K, Jin M, Aoyama K, Yasunaga T (2016) Filopodia formation by crosslinking of F‐actin with fascin in two different binding manners. Cytoskelet Hoboken NJ 73: 365–374 [DOI] [PubMed] [Google Scholar]
- Ariazi J, Benowitz A, De Biasi V, Den Boer ML, Cherqui S, Cui H, Douillet N, Eugenin EA, Favre D, Goodman S et al (2017) Tunneling nanotubes and gap junctions‐their role in long‐range intercellular communication during development, health, and disease conditions. Front Mol Neurosci 10: 333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkwright PD, Luchetti F, Tour J, Roberts C, Ayub R, Morales AP, Rodríguez JJ, Gilmore A, Canonico B, Papa S et al (2010) Fas stimulation of T lymphocytes promotes rapid intercellular exchange of death signals via membrane nanotubes. Cell Res 20: 72–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asencio‐Barría C, Defamie N, Sáez JC, Mesnil M, Godoy AS (2019) Direct intercellular communications and cancer: a snapshot of the biological roles of connexins in prostate cancer. Cancers 11: 1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austefjord MW, Gerdes H‐H, Wang X (2014) Tunneling nanotubes: diversity in morphology and structure. Commun Integr Biol 7: e27934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M (2017) How the Internet of cells has biologists buzzing. Nature 549: 322–324 [DOI] [PubMed] [Google Scholar]
- Bénard M, Schapman D, Lebon A, Monterroso B, Bellenger M, Le Foll F, Pasquier J, Vaudry H, Vaudry D, Galas L (2015) Structural and functional analysis of tunneling nanotubes (TnTs) using gCW STED and gconfocal approaches. Biol Cell 107: 419–425 [DOI] [PubMed] [Google Scholar]
- Bhat S, Ljubojevic N, Zhu S, Fukuda M, Echard A, Zurzolo C (2020) Rab35 and its effectors promote formation of tunneling nanotubes in neuronal cells. Sci Rep 10: 16803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE (2006) Myosin‐X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA 103: 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukoreshtliev NV, Wang X, Hodneland E, Gurke S, Barroso JFV, Gerdes H‐H (2009) Selective block of tunneling nanotube (TNT) formation inhibits intercellular organelle transfer between PC12 cells. FEBS Lett 583: 1481–1488 [DOI] [PubMed] [Google Scholar]
- Campana S, De Pasquale C, Carrega P, Ferlazzo G, Bonaccorsi I (2015) Cross‐dressing: an alternative mechanism for antigen presentation. Immunol Lett 168: 349–354 [DOI] [PubMed] [Google Scholar]
- Caneparo L, Pantazis P, Dempsey W, Fraser SE (2011) Intercellular bridges in vertebrate gastrulation. PLoS One 6: e20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KP, Segall JE, Cox D (2020) Microscopic methods for analysis of macrophage‐induced tunneling nanotubes. Methods Mol Biol Clifton NJ 2108: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastagner P, Loria F, Vargas JY, Tois J, Diamond IM, Okafo G, Brou C, Zurzolo C(2020) Fate and propagation of endogenously formed Tau aggregates in neuronal cells. EMBO Mol Med 12: e12025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauveau A, Aucher A, Eissmann P, Vivier E, Davis DM (2010) Membrane nanotubes facilitate long‐distance interactions between natural killer cells and target cells. Proc Natl Acad Sci USA 107: 5545–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B‐C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, Liu Z et al (2014) Lattice light‐sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346: 1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherqui S, Courtoy PJ (2017) The renal Fanconi syndrome in cystinosis: pathogenic insights and therapeutic perspectives. Nat Rev Nephrol 13: 115–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnery HR, Pearlman E, McMenamin PG (2008) Cutting edge: membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J Immunol 180: 5779–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collot M, Ashokkumar P, Anton H, Boutant E, Faklaris O, Galli T, Mély Y, Danglot L, Klymchenko AS (2019) MemBright: a family of fluorescent membrane probes for advanced cellular imaging and neuroscience. Cell Chem Biol 26: 600–614.e7 [DOI] [PubMed] [Google Scholar]
- Connor Y, Tekleab S, Nandakumar S, Walls C, Tekleab Y, Husain A, Gadish O, Sabbisetti V, Kaushik S, Sehrawat S et al (2015) Physical nanoscale conduit‐mediated communication between tumour cells and the endothelium modulates endothelial phenotype. Nat Commun 6: 8671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Abounit S, Marzo L, Danckaert A, Chamoun Z, Roux P, Zurzolo C (2013) Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci 126: 3678–3685 [DOI] [PubMed] [Google Scholar]
- Dance A (2019) Core Concept: Cells nibble one another via the under‐appreciated process of trogocytosis. Proc Natl Acad Sci USA 116: 17608–17610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DM, Sowinski S (2008) Membrane nanotubes: dynamic long‐distance connections between animal cells. Nat Rev Mol Cell Biol 9: 431–436 [DOI] [PubMed] [Google Scholar]
- Delage E, Cervantes DC, Pénard E, Schmitt C, Syan S, Disanza A, Scita G, Zurzolo C (2016) Differential identity of Filopodia and Tunneling Nanotubes revealed by the opposite functions of actin regulatory complexes. Sci Rep 6: 39632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delage E, Zurzolo C (2013) Exploring the role of lipids in intercellular conduits: breakthroughs in the pipeline. Front Plant Sci 4: 504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desir S, Dickson EL, Vogel RI, Thayanithy V, Wong P, Teoh D, Geller MA, Steer CJ, Subramanian S, Lou E (2016) Tunneling nanotube formation is stimulated by hypoxia in ovarian cancer cells. Oncotarget 7: 43150–43161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desir S, O’Hare P, Vogel RI, Sperduto W, Sarkari A, Dickson EL, Wong P, Nelson AC, Fong Y, Steer CJ et al (2018) Chemotherapy‐induced tunneling nanotubes mediate intercellular drug efflux in pancreatic cancer. Sci Rep 8: 9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieriks BV, Park TI‐H, Fourie C, Faull RLM, Dragunow M, Curtis MA (2017) α‐synuclein transfer through tunneling nanotubes occurs in SH‐SY5Y cells and primary brain pericytes from Parkinson’s disease patients. Sci Rep 7: 42984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilsizoglu Senol A, Pepe A, Grudina C, Sassoon N, Reiko U, Bousset L, Melki R, Piel J, Gugger M, Zurzolo C (2019) Effect of tolytoxin on tunneling nanotube formation and function. Sci Rep 9: 5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Stockwell BR (2014) The role of iron and reactive oxygen species in cell death. Nat Chem Biol 10: 9–17 [DOI] [PubMed] [Google Scholar]
- Drab M, Stopar D, Kralj‐Iglič V, Iglič A (2019) Inception mechanisms of tunneling nanotubes. Cells 8: 626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F, Jean‐Jacques B, Roberge H, Bénard M, Galas L, Schapman D, Elie N, Goux D, Keller M, Maille E et al (2018) A role for RASSF1A in tunneling nanotube formation between cells through GEFH1/Rab11 pathway control. Cell Commun Signal CCS 16: 66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont M, Souriant S, Lugo‐Villarino G, Maridonneau‐Parini I, Vérollet C (2018) Tunneling nanotubes: intimate communication between myeloid cells. Front Immunol 9: 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont M, Souriant S, Balboa L, Vu Manh T‐P, Pingris K, Rousset S, Cougoule C, Rombouts Y, Poincloux R, Ben Neji M et al (2020) Tuberculosis‐associated IFN‐I induces Siglec‐1 on tunneling nanotubes and favors HIV‐1 spread in macrophages. Elife 9: e52535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin ML, Chakraborty AK, Shaw AS (2010) Understanding the structure and function of the immunological synapse. Cold Spring Harb Perspect Biol 2: a002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugenin EA, Gaskill PJ, Berman JW (2009) Tunneling nanotubes (TNT) are induced by HIV‐infection of macrophages: a potential mechanism for intercellular HIV trafficking. Cell Immunol 254: 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finetti F, Cassioli C, Baldari CT (2017) Transcellular communication at the immunological synapse: a vesicular traffic‐mediated mutual exchange. F1000Res 6: 1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas L, Bénard M, Lebon A, Komuro Y, Schapman D, Vaudry H, Vaudry D, Komuro H (2017) Postnatal migration of cerebellar interneurons. Brain Sci 7: 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier NC, Masters TA, Sheetz MP (2012) Mechanical feedback between membrane tension and dynamics. Trends Cell Biol 22: 527–535 [DOI] [PubMed] [Google Scholar]
- Gerdes HH, Bukoreshtliev NV, Barroso JFV (2007) Tunneling nanotubes: a new route for the exchange of components between animal cells. FEBS Lett 581: 2194–2201 [DOI] [PubMed] [Google Scholar]
- Gerdes HH, Carvalho RN (2008) Intercellular transfer mediated by tunneling nanotubes. Curr Opin Cell Biol 20: 470–475 [DOI] [PubMed] [Google Scholar]
- Gerdes H‐H, Rustom A, Wang X (2013) Tunneling nanotubes, an emerging intercellular communication route in development. Mech Dev 130: 381–387 [DOI] [PubMed] [Google Scholar]
- Goedert M (2015) Neurodegeneration. Alzheimer’s and Parkinson’s diseases: the prion concept in relation to assembled Aβ, tau, and α‐synuclein. Science 349: 1255555 [DOI] [PubMed] [Google Scholar]
- Goodman S, Naphade S, Khan M, Sharma J, Cherqui S (2019) Macrophage polarization impacts tunneling nanotube formation and intercellular organelle trafficking. Sci Rep 9: 14529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousset K, Gordon A, Kumar Kannan S, Tovar J (2019) A novel microproteomic approach using laser capture microdissection to study cellular protrusions. Int J Mol Sci 20: 1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gousset K, Marzo L, Commere P‐H, Zurzolo C (2013) Myo10 is a key regulator of TNT formation in neuronal cells. J Cell Sci 126: 4424–4435 [DOI] [PubMed] [Google Scholar]
- Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de Chaumont F, Martino A, Enninga J et al (2009) Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol 11: 328–336 [DOI] [PubMed] [Google Scholar]
- Grudina C, Kouroupi G, Nonaka T, Hasegawa M, Matsas R, Zurzolo C (2019) Human NPCs can degrade α‐syn fibrils and transfer them preferentially in a cell contact‐dependent manner possibly through TNT‐like structures. Neurobiol Dis 132: 104609 [DOI] [PubMed] [Google Scholar]
- Gurke S, Barroso JFV, Gerdes HH (2008) The art of cellular communication: tunneling nanotubes bridge the divide. Histochem Cell Biol 129: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson T, Wolpert L (1963) The cellular basis of morphogenesis and sea urchin development. Int Rev Cytol 15: 139–214 [DOI] [PubMed] [Google Scholar]
- Haglund K, Nezis IP, Lemus D, Grabbe C, Wesche J, Liestøl K, Dikic I, Palmer R, Stenmark H (2010) Cindr interacts with anillin to control cytokinesis in Drosophila melanogaster . Curr Biol 20: 944–950 [DOI] [PubMed] [Google Scholar]
- Haglund K, Nezis IP, Stenmark H (2011) Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun Integr Biol 4: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halász H, Ghadaksaz AR, Madarász T, Huber K, Harami G, Tóth EA, Osteikoetxea‐Molnár A, Kovács M, Balogi Z, Nyitrai M et al (2018) Live cell superresolution‐structured illumination microscopy imaging analysis of the intercellular transport of microvesicles and costimulatory proteins via nanotubes between immune cells. Methods Appl Fluoresc 6: 45005 [DOI] [PubMed] [Google Scholar]
- Hall AC, Lucas FR, Salinas PC (2000) Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT‐7a signaling. Cell 100: 525–535 [DOI] [PubMed] [Google Scholar]
- Hanna SJ, McCoy‐Simandle K, Miskolci V, Guo P, Cammer M, Hodgson L, Cox D (2017) The role of Rho‐GTPases and actin polymerization during macrophage tunneling nanotube biogenesis. Sci Rep 7: 8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna SJ, McCoy‐Simandle K, Leung E, Genna A, Condeelis J, Cox D (2019) Tunneling nanotubes, a novel mode of tumor cell–macrophage communication in tumor cell invasion. J Cell Sci 132: jcs223321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K, Kimura S, Takatsu H, Ohmae M, Kawano S, Kitamura H, Ito M, Watarai H, Hazelett CC, Yeaman C et al (2009) M‐Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat Cell Biol 11: 1427–1432 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Nonaka T, Masuda‐Suzukake M (2017) Prion‐like mechanisms and potential therapeutic targets in neurodegenerative disorders. Pharmacol Ther 172: 22–33 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Bhuyan F, Hiyoshi M, Noyori O, Nasser H, Miyazaki M, Saito T, Kondoh Y, Osada H, Kimura S et al (2016) Potential role of the formation of tunneling nanotubes in HIV‐1 spread in macrophages. J Immunol 196: 1832–1841 [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ (2008) Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol 9: 690–701 [DOI] [PubMed] [Google Scholar]
- Hekmatshoar Y, Nakhle J, Galloni M, Vignais M‐L (2018) The role of metabolism and tunneling nanotube‐mediated intercellular mitochondria exchange in cancer drug resistance. Biochem J 475: 2305–2328 [DOI] [PubMed] [Google Scholar]
- Hoffman L, Jensen CC, Yoshigi M, Beckerle M (2017) Mechanical signals activate p38 MAPK pathway‐dependent reinforcement of actin via mechanosensitive HspB1. Mol Biol Cell 28: 2661–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zucker B, Zhang S, Elias S, Zhu Y, Chen H, Ding T, Li Y, Sun Y, Lou J et al (2019) Migrasome formation is mediated by assembly of micron‐scale tetraspanin macrodomains. Nat Cell Biol 21: 991–1002 [DOI] [PubMed] [Google Scholar]
- Huot J, Houle F, Rousseau S, Deschesnes RG, Shah GM, Landry J (1998) SAPK2/p38‐dependent F‐actin reorganization regulates early membrane blebbing during stress‐induced apoptosis. J Cell Biol 143: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtig J, Chiu DT, Onfelt B (2010) Intercellular nanotubes: insights from imaging studies and beyond. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2: 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto T, Mori H (2019) A new bioluminescence‐based tool for modulating target proteins in live cells. Sci Rep 9: 18239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AHP, Batenburg KL, Pecho‐Vrieseling E, Reits EA (2017) Visualization of prion‐like transfer in Huntington’s disease models. Biochim Biophys Acta Mol Basis Dis 1863: 793–800 [DOI] [PubMed] [Google Scholar]
- Jansens RJJ, Tishchenko A, Favoreel HW (2020) Bridging the gap: virus long‐distance spread via tunneling nanotubes. J Virol 94: e02120‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jash E, Prasad P, Kumar N, Sharma T, Goldman A, Sehrawat S (2018) Perspective on nanochannels as cellular mediators in different disease conditions. Cell Commun Signal 16: 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Jiang Z, Lu D, Wang X, Liang H, Zhang J, Meng Y, Li Y, Wu D, Huang Y et al (2019) Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat Cell Biol 21: 966–977 [DOI] [PubMed] [Google Scholar]
- Jucker M, Walker LC (2018) Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat Neurosci 21: 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiu I, Gendelman HE (2011) Macrophage bridging conduit trafficking of HIV‐1 through the endoplasmic reticulum and Golgi network. J Proteome Res 10: 3225–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassubek J, Juengling FD, Kioschies T, Henkel K, Karitzky J, Kramer B, Ecker D, Andrich J, Saft C, Kraus P et al (2004) Topography of cerebral atrophy in early Huntington’s disease: a voxel based morphometric MRI study. J Neurol Neurosurg Psychiatry 75: 213–220 [PMC free article] [PubMed] [Google Scholar]
- Kerber ML, Cheney RE (2011) Myosin‐X: a MyTH‐FERM myosin at the tips of filopodia. J Cell Sci 124: 3733–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Yamashita M, Yamakami‐Kimura M, Sato Y, Yamagata A, Kobashigawa Y, Inagaki F, Amada T, Hase K, Iwanaga T et al (2016) Distinct roles for the N‐ and C‐terminal regions of M‐Sec in plasma membrane deformation during tunneling nanotube formation. Sci Rep 6: 33548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriyama Y, Nochi H (2017) Intra‐ and intercellular quality control mechanisms of mitochondria. Cells 7: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Dougherty MT, Brey EM, Etkin LD (2004) Formation, architecture and polarity of female germline cyst in Xenopus. Dev Biol 266: 43–61 [DOI] [PubMed] [Google Scholar]
- Kolba MD, Dudka W, Zaręba‐Kozioł M, Kominek A, Ronchi P, Turos L, Chroscicki P, Wlodarczyk J, Schwab Y, Klejman A et al (2019) Tunneling nanotube‐mediated intercellular vesicle and protein transfer in the stroma‐provided imatinib resistance in chronic myeloid leukemia cells. Cell Death Dis 10: 817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro H, Kumada T (2005) Ca2+ transients control CNS neuronal migration. Cell Calcium 37: 387–393 [DOI] [PubMed] [Google Scholar]
- Korenkova O, Pepe A, Zurzolo C (2020) Fine intercellular connections in development: TNTs, cytonemes, or intercellular bridges? Cell Stress 4: 30–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg TB (2017) Distributing signaling proteins in space and time: the province of cytonemes. Curr Opin Genet Dev 45: 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg TB, Roy S (2014) Cytonemes as specialized signaling filopodia. Dev Camb Engl 141: 729–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kim JH, Ranjan P, Metcalfe MG, Cao W, Mishina M, Gangappa S, Guo Z, Boyden ES, Zaki S et al (2017) Influenza virus exploits tunneling nanotubes for cell‐to‐cell spread. Sci Rep 7: 40360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachambre S, Chopard C, Beaumelle B (2014) Preliminary characterisation of nanotubes connecting T‐cells and their use by HIV‐1. Biol Cell 106: 394–404 [DOI] [PubMed] [Google Scholar]
- Lai M, Guo Y, Ma J, Yu H, Zhao D, Fan W, Ju X, Sheikh MA, Malik YS, Xiong W et al (2015) Myosin X regulates neuronal radial migration through interacting with N‐cadherin. Front Cell Neurosci 9: 326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D (2018) A salutary role of reactive oxygen species in intercellular tunnel‐mediated communication. Front Cell Dev Biol 6: 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T‐K, Chen S‐D, Chuang Y‐C, Lan M‐Y, Chuang J‐H, Wang P‐W, Hsu T‐Y, Wang F‐S, Tsai M‐H, Huang S‐T et al (2019) Mitochondrial transfer of wharton’s jelly mesenchymal stem cells eliminates mutation burden and rescues mitochondrial bioenergetics in rotenone‐stressed MELAS fibroblasts. Oxid Med Cell Longev 2019: 9537504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T‐L, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J et al (2018) Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360: eaaq1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubojevic N, Henderson JM, Zurzolo C (2021) The ways of actin: why tunneling nanotubes are unique cell protrusions. Trends Cell Biol 31: 130–142 [DOI] [PubMed] [Google Scholar]
- Lock JT, Parker I, Smith IF (2016) Communication of Ca2+ signals via tunneling membrane nanotubes is mediated by transmission of inositol trisphosphate through gap junctions. Cell Calcium 60: 266–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokar M, Kabaso D, Resnik N, Sepčić K, Kralj‐Iglič V, Veranič P, Zorec R, Iglič A (2012) The role of cholesterol‐sphingomyelin membrane nanodomains in the stability of intercellular membrane nanotubes. Int J Nanomedicine 7: 1891–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria F, Vargas JY, Bousset L, Syan S, Salles A, Melki R, Zurzolo C (2017) α‐Synuclein transfer between neurons and astrocytes indicates that astrocytes play a role in degradation rather than in spreading. Acta Neuropathol 134: 789–808 [DOI] [PubMed] [Google Scholar]
- Lou E, Fujisawa S, Morozov A, Barlas A, Romin Y, Dogan Y, Gholami S, Moreira AL, Manova‐Todorova K, Moore MAS (2012) Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One 7: e33093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zheng X, Li F, Yu Y, Chen Z, Liu Z, Wang Z, Xu H, Yang W (2017) Tunneling nanotubes promote intercellular mitochondria transfer followed by increased invasiveness in bladder cancer cells. Oncotarget 8: 15539–15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk KC, Song C, O’Brien P, Stieber A, Branch JR, Brunden KR, Trojanowski JQ, Lee VM‐Y (2009) Exogenous alpha‐synuclein fibrils seed the formation of Lewy body‐like intracellular inclusions in cultured cells. Proc Natl Acad Sci USA 106: 20051–20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y, Chen L, Yan X, Du Y, Yu L (2015) Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res 25: 24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins B, Sorrentino S, Chung W‐L, Tatli M, Medalia O, Eibauer M (2021) Unveiling the polarity of actin filaments by cryo‐electron tomography. Structure 10.1016/j.str.2020.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzo L, Gousset K, Zurzolo C (2012) Multifaceted roles of tunneling nanotubes in intercellular communication. Front Physiol 3: 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matejka N, Reindl J (2019) Perspectives of cellular communication through tunneling nanotubes in cancer cells and the connection to radiation effects. Radiat Oncol Lond Engl 14: 218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes B, Dang Y, Greicius G, Kaufmann LT, Prunsche B, Rosenbauer J, Stegmaier J, Mikut R, Özbek S, Nienhaus GU et al (2018) Wnt/PCP controls spreading of Wnt/β‐catenin signals by cytonemes in vertebrates. Elife 7: e36953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClay DR (1999) The role of thin filopodia in motility and morphogenesis. Exp Cell Res 253: 296–301 [DOI] [PubMed] [Google Scholar]
- McCoy‐Simandle K, Hanna SJ, Cox D (2016) Exosomes and nanotubes: control of immune cell communication. Int J Biochem Cell Biol 71: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes‐Jorge L, Llombart C, Ramos D, López‐Luppo M, Valença A, Nacher V, Navarro M, Carretero A, Méndez‐Ferrer S, Rodriguez‐Baeza A et al (2012) Intercapillary bridging cells: immunocytochemical characteristics of cells that connect blood vessels in the retina. Exp Eye Res 98: 79–87 [DOI] [PubMed] [Google Scholar]
- Menna E, Disanza A, Cagnoli C, Schenk U, Gelsomino G, Frittoli E, Hertzog M, Offenhauser N, Sawallisch C, Kreienkamp H‐J et al (2009) Eps8 regulates axonal filopodia in hippocampal neurons in response to brain‐derived neurotrophic factor (BDNF). PLoS Biol 7: e1000138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna E, Zambetti S, Morini R, Donzelli A, Disanza A, Calvigioni D, Braida D, Nicolini C, Orlando M, Fossati G et al (2013) Eps8 controls dendritic spine density and synaptic plasticity through its actin‐capping activity. EMBO J 32: 1730–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Fraser SE, McClay D (1995) Dynamics of thin filopodia during sea urchin gastrulation. Development 121: 2501–2511 [DOI] [PubMed] [Google Scholar]
- Millet V, Naquet P, Guinamard RR (2008) Intercellular MHC transfer between thymic epithelial and dendritic cells. Eur J Immunol 38: 1257–1263 [DOI] [PubMed] [Google Scholar]
- Mishin AS, Belousov VV, Solntsev KM, Lukyanov KA (2015) Novel uses of fluorescent proteins. Curr Opin Chem Biol 27: 1–9 [DOI] [PubMed] [Google Scholar]
- Mittal R, Karhu E, Wang J‐S, Delgado S, Zukerman R, Mittal J, Jhaveri VM (2019) Cell communication by tunneling nanotubes: implications in disease and therapeutic applications. J Cell Physiol 234: 1130–1146 [DOI] [PubMed] [Google Scholar]
- Murray LMA, Krasnodembskaya AD (2019) Concise review: intercellular communication via organelle transfer in the biology and therapeutic applications of stem cells. Stem Cells 37: 14–25 [DOI] [PubMed] [Google Scholar]
- Nagel J, Delandre C, Zhang Y, Förstner F, Moore AW, Tavosanis G (2012) Fascin controls neuronal class‐specific dendrite arbor morphology. Development 139: 2999–3009 [DOI] [PubMed] [Google Scholar]
- Naphade S, Sharma J, Gaide Chevronnay HP, Shook MA, Yeagy BA, Rocca CJ, Ur SN, Lau AJ, Courtoy PJ, Cherqui S (2015) Brief reports: lysosomal cross‐correction by hematopoietic stem cell‐derived macrophages via tunneling nanotubes. Stem Cells 33: 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto E, Leitão L, Sousa DM, Alves CJ, Alencastre IS, Aguiar P, Lamghari M (2016) Compartmentalized microfluidic platforms: the unrivaled breakthrough of in vitro tools for neurobiological research. J Neurosci Off J Soc Neurosci 36: 11573–11584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzveig HM (2019) Are cell membrane nanotubes the ancestors of the nervous system? Eur Biophys J 48: 593–598 [DOI] [PubMed] [Google Scholar]
- Offenhäuser N, Castelletti D, Mapelli L, Soppo BE, Regondi MC, Rossi P, D’Angelo E, Frassoni C, Amadeo A, Tocchetti A et al (2006) Increased ethanol resistance and consumption in Eps8 knockout mice correlates with altered actin dynamics. Cell 127: 213–226 [DOI] [PubMed] [Google Scholar]
- Okafo G, Prevedel L, Eugenin E (2017) Tunneling nanotubes (TNT) mediate long‐range gap junctional communication: Implications for HIV cell to cell spread. Sci Rep 7: 16660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva CA, Vargas JY, Inestrosa NC (2013) Wnts in adult brain: from synaptic plasticity to cognitive deficiencies. Front Cell Neurosci 7: 224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Önfelt B, Nedvetzki S, Yanagi K, Davis DM (2004) Cutting edge: membrane nanotubes connect immune cells. J Immunol 173: 1511–1513 [DOI] [PubMed] [Google Scholar]
- Önfelt B, Purbhoo MA, Nedvetzki S, Sowinski S, Davis DM (2005) Long‐distance calls between cells connected by tunneling nanotubules. Sci STKE 2005: pe55 [DOI] [PubMed] [Google Scholar]
- Önfelt B, Nedvetzki S, Benninger RKP, Purbhoo MA, Sowinski S, Hume AN, Seabra MC, Neil MAA, French PMW, Davis DM (2006) Structurally distinct membrane nanotubes between human macrophages support long‐distance vesicular traffic or surfing of bacteria. J Immunol 177: 8476–8483 [DOI] [PubMed] [Google Scholar]
- Onishi K, Shafer B, Lo C, Tissir F, Goffinet AM, Zou Y (2013) Antagonistic functions of Dishevelleds regulate Frizzled3 endocytosis via filopodia tips in Wnt‐mediated growth cone guidance. J Neurosci Off J Soc Neurosci 33: 19071–19085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M et al (2015) Brain tumour cells interconnect to a functional and resistant network. Nature 528: 93–98 [DOI] [PubMed] [Google Scholar]
- Osteikoetxea‐Molnár A, Szabó‐Meleg E, Tóth EA, Oszvald Á, Izsépi E, Kremlitzka M, Biri B, Nyitray L, Bozó T, Németh P et al (2016) The growth determinants and transport properties of tunneling nanotube networks between B lymphocytes. Cell Mol Life Sci 73: 4531–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker I, Evans KT, Ellefsen K, Lawson DA, Smith IF (2017) Lattice light sheet imaging of membrane nanotubes between human breast cancer cells in culture and in brain metastases. Sci Rep 7: 11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecho‐Vrieseling E, Rieker C, Fuchs S, Bleckmann D, Esposito MS, Botta P, Goldstein C, Bernhard M, Galimberti I, Müller M et al (2014) Transneuronal propagation of mutant huntingtin contributes to non‐cell autonomous pathology in neurons. Nat Neurosci 17: 1064–1072 [DOI] [PubMed] [Google Scholar]
- Pepling ME, Spradling AC (1998) Female mouse germ cells form synchronously dividing cysts. Development 125: 3323–3328 [DOI] [PubMed] [Google Scholar]
- Pinto G, Brou C, Zurzolo C (2020) Tunneling nanotubes: the fuel of tumor progression? Trends Cancer 6: 874–888 [DOI] [PubMed] [Google Scholar]
- Pinto G, Saenz‐de‐Santa‐Maria I, Chastagner P, Perthame E, Delmas C, Toulas C, Moyal‐Jonathan‐Cohen E, Brou C, Zurzolo C (2021) Patient‐derived glioblastoma stem cells transfer mitochondria through tunneling nanotubes in tumor organoids. Biochem J 478: 21–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes B, Monzo P, Gole L, Le Roux A‐L, Kosmalska AJ, Tam ZY, Luo W, Kan S, Viasnoff V, Roca‐Cusachs P et al (2017) Membrane tension controls adhesion positioning at the leading edge of cells. J Cell Biol 216: 2959–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulze L, Congiu T, Brevini TAL, Grimaldi A, Tettamanti G, D’Antona P, Baranzini N, Acquati F, Ferraro F, de Eguileor M (2020) MCF7 Spheroid development: new insight about spatio/temporal arrangements of TNTs, amyloid fibrils, cell connections, and cellular bridges. Int J Mol Sci 21: 5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines AN, Nagdas S, Kerber ML, Cheney RE (2012) Headless Myo10 is a negative regulator of full‐length Myo10 and inhibits axon outgrowth in cortical neurons. J Biol Chem 287: 24873–24883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainy N, Chetrit D, Rouger V, Vernitsky H, Rechavi O, Marguet D, Goldstein I, Ehrlich M, Kloog Y (2013) H‐Ras transfers from B to T cells via tunneling nanotubes. Cell Death Dis 4: e726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez‐Weber FA, Kornberg TB (1999) Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell 97: 599–607 [DOI] [PubMed] [Google Scholar]
- Reichert D, Scheinpflug J, Karbanová J, Freund D, Bornhäuser M, Corbeil D (2016) Tunneling nanotubes mediate the transfer of stem cell marker CD133 between hematopoietic progenitor cells. Exp Hematol 44: 1092–1112.e2 [DOI] [PubMed] [Google Scholar]
- Reindl J, Shevtsov M, Dollinger G, Stangl S, Multhoff G (2019) Membrane Hsp70‐supported cell‐to‐cell connections via tunneling nanotubes revealed by live‐cell STED nanoscopy. Cell Stress Chaperones 24: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik N, Erman A, Veranič P, Kreft ME (2019) Triple labelling of actin filaments, intermediate filaments and microtubules for broad application in cell biology: uncovering the cytoskeletal composition in tunneling nanotubes. Histochem Cell Biol 152: 311–317 [DOI] [PubMed] [Google Scholar]
- Resnik N, Prezelj T, De Luca GMR, Manders E, Polishchuk R, Veranič P, Kreft ME (2018) Helical organization of microtubules occurs in a minority of tunneling membrane nanotubes in normal and cancer urothelial cells. Sci Rep 8: 17133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehlecke C, Schmidt MHH (2020) Tunneling nanotubes and tumor microtubes in cancer. Cancers 12: 857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostami J, Holmqvist S, Lindström V, Sigvardson J, Westermark GT, Ingelsson M, Bergström J, Roybon L, Erlandsson A (2017) Human astrocytes transfer aggregated alpha‐synuclein via tunneling nanotubes. J Neurosci Off J Soc Neurosci 37: 11835–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp I, Sologub L, Williamson KC, Scheuermayer M, Reininger L, Doerig C, Eksi S, Kombila DU, Frank M, Pradel G (2011) Malaria parasites form filamentous cell‐to‐cell connections during reproduction in the mosquito midgut. Cell Res 21: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A (2016) The missing link: does tunnelling nanotube‐based supercellularity provide a new understanding of chronic and lifestyle diseases? Open Biol 6: 160057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustom A, Saffrich R, Markovic I, Walther P, Gerdes H‐H (2004) Nanotubular highways for intercellular organelle transport. Science 303: 1007–1010 [DOI] [PubMed] [Google Scholar]
- Sáenz‐de‐Santa‐María I, Bernardo‐Castiñeira C, Secades P, Bernaldo‐de‐Quirós S, Rodrigo JP, Astudillo A, Chiara M‐D (2017) Clinically relevant HIF‐1α‐dependent metabolic reprogramming in oropharyngeal squamous cell carcinomas includes coordinated activation of CAIX and the miR‐210/ISCU signaling axis, but not MCT1 and MCT4 upregulation. Oncotarget 8: 13730–13746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager RE, Lee J‐Y (2018) Plasmodesmata at a glance. J Cell Sci 131: jcs209346 [DOI] [PubMed] [Google Scholar]
- Sartori‐Rupp A, Cordero Cervantes D, Pepe A, Gousset K, Delage E, Corroyer‐Dulmont S, Schmitt C, Krijnse‐Locker J, Zurzolo C (2019) Correlative cryo‐electron microscopy reveals the structure of TNTs in neuronal cells. Nat Commun 10: 342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller C, Diakopoulos KN, Rohwedder I, Kremmer E, von Toerne C, Ueffing M, Weidle UH, Ohno H, Weiss EH (2013) LST1 promotes the assembly of a molecular machinery responsible for tunneling nanotube formation. J Cell Sci 126: 767–777 [DOI] [PubMed] [Google Scholar]
- Scholkmann F, Seidel D, Funk RHW, Roehlecke C (2018) Ultrastructural features (bulges) of membrane nanotubes between cone‐like photoreceptor cells: an investigation employing scanning electron microscopy. Matters 10.19185/matters.201802000011 [DOI] [Google Scholar]
- Seyed‐Razavi Y, Hickey MJ, Kuffová L, McMenamin PG, Chinnery HR (2013) Membrane nanotubes in myeloid cells in the adult mouse cornea represent a novel mode of immune cell interaction. Immunol Cell Biol 91: 89–95 [DOI] [PubMed] [Google Scholar]
- Sharma M, Subramaniam S (2019) Rhes travels from cell to cell and transports Huntington disease protein via TNT‐like protrusion. J Cell Biol 218: 1972–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Lehmann MJ, Jimenez‐Soto LF, Horensavitz C, Pypaert M, Mothes W (2007) Retroviruses can establish filopodial bridges for efficient cell‐to‐cell transmission. Nat Cell Biol 9: 310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov MS, Garrett TR, Yasuda R (2018) An open‐source tool for analysis and automatic identification of dendritic spines using machine learning. PLoS One 13: e0199589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IF, Shuai J, Parker I (2011) Active generation and propagation of Ca2+ signals within tunneling membrane nanotubes. Biophys J 100: L37–L39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souriant S, Balboa L, Dupont M, Pingris K, Kviatcovsky D, Cougoule C, Lastrucci C, Bah A, Gasser R, Poincloux R et al (2019) Tuberculosis exacerbates HIV‐1 infection through IL‐10/STAT3‐dependent tunneling nanotube formation in macrophages. Cell Rep 26: 3586–3599.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AD, Berg JS, Robertson BW, Meeker RB, Cheney RE (2006) Myo10 in brain: developmental regulation, identification of a headless isoform and dynamics in neurons. J Cell Sci 119: 184–194 [DOI] [PubMed] [Google Scholar]
- Sousa AD, Cheney RE (2005) Myosin‐X: a molecular motor at the cell’s fingertips. Trends Cell Biol 15: 533–539 [DOI] [PubMed] [Google Scholar]
- Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Köhler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C et al (2008) Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV‐1 transmission. Nat Cell Biol 10: 211–219 [DOI] [PubMed] [Google Scholar]
- Stamatakou E, Marzo A, Gibb A, Salinas PC (2013) Activity‐dependent spine morphogenesis: a role for the actin‐capping protein Eps8. J Neurosci Off J Soc Neurosci 33: 2661–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanganello E, Scholpp S (2016) Role of cytonemes in Wnt transport. J Cell Sci 129: 665–672 [DOI] [PubMed] [Google Scholar]
- Staufer O, Hernandez BJE, Rustom A (2018) Protease‐resistant cell meshworks: an indication of membrane nanotube‐based syncytia formation. Exp Cell Res 372: 85–91 [DOI] [PubMed] [Google Scholar]
- Sun YY, Bradley JM, Keller KE (2019) Phenotypic and functional alterations in tunneling nanotubes formed by glaucomatous trabecular meshwork cells. Invest Ophthalmol Vis Sci 60: 4583–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Kukita A, Li Y, Zhang J, Nomiyama H, Yamaza T, Ayukawa Y, Koyano K, Kukita T (2013) Tunneling nanotube formation is essential for the regulation of osteoclastogenesis. J Cell Biochem 114: 1238–1247 [DOI] [PubMed] [Google Scholar]
- Takeichi M (2007) The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci 8: 11–20 [DOI] [PubMed] [Google Scholar]
- Tang BL (2018) Unconventional secretion and intercellular transfer of mutant huntingtin. Cells 7: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardivel M, Bégard S, Bousset L, Dujardin S, Coens A, Melki R, Buée L, Colin M (2016) Tunneling nanotube (TNT)‐mediated neuron‐to neuron transfer of pathological Tau protein assemblies. Acta Neuropathol Commun 4: 117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulet N, Delorme‐Walker VD, DerMardirossian C (2012) Reactive oxygen species regulate protrusion efficiency by controlling actin dynamics. PLoS One 7: e41342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavano S, Heisenberg C‐P (2019) Migrasomes take center stage. Nat Cell Biol 21: 918–920 [DOI] [PubMed] [Google Scholar]
- Thayanithy V, Dickson EL, Steer C, Subramanian S, Lou E (2014) Tumor‐stromal cross talk: direct cell‐to‐cell transfer of oncogenic microRNAs via tunneling nanotubes. Transl Res J Lab Clin Med 164: 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayanithy V, O’Hare P, Wong P, Zhao X, Steer CJ, Subramanian S, Lou E (2017) A transwell assay that excludes exosomes for assessment of tunneling nanotube‐mediated intercellular communication. Cell Commun Signal CCS 15: 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdebenito S, Lou E, Baldoni J, Okafo G, Eugenin E (2018) The novel roles of connexin channels and tunneling nanotubes in cancer pathogenesis. Int J Mol Sci 19: 1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JY, Grudina C, Zurzolo C (2019a) The prion‐like spreading of α‐synuclein: From in vitro to in vivo models of Parkinson’s disease. Ageing Res Rev 50: 89–101 [DOI] [PubMed] [Google Scholar]
- Vargas JY, Loria F, Wu Y‐J, Córdova G, Nonaka T, Bellow S, Syan S, Hasegawa M, van Woerden GM, Trollet C et al (2019b) The Wnt/Ca2+ pathway is involved in interneuronal communication mediated by tunneling nanotubes. EMBO J 38: e101230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veranic P, Lokar M, Schütz GJ, Weghuber J, Wieser S, Hägerstrand H, Kralj‐Iglic V, Iglic A (2008) Different types of cell‐to‐cell connections mediated by nanotubular structures. Biophys J 95: 4416–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria GS, Zurzolo C (2017) The spread of prion‐like proteins by lysosomes and tunneling nanotubes: Implications for neurodegenerative diseases. J Cell Biol 216: 2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M‐L, Caicedo A, Brondello J‐M, Jorgensen C (2017) Cell connections by tunneling nanotubes: effects of mitochondrial trafficking on target cell metabolism, homeostasis, and response to therapy. Stem Cells Int 2017: 6917941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG (2006) Role of fascin in filopodial protrusion. J Cell Biol 174: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bukoreshtliev NV, Gerdes H‐H (2012) Developing neurons form transient nanotubes facilitating electrical coupling and calcium signaling with distant astrocytes. PLoS One 7: e47429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Gerdes H‐H (2015) Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ 22: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Veruki ML, Bukoreshtliev NV, Hartveit E, Gerdes H‐H (2010) Animal cells connected by nanotubes can be electrically coupled through interposed gap‐junction channels. Proc Natl Acad Sci USA 107: 17194–17199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cui J, Sun X, Zhang Y (2011) Tunneling‐nanotube development in astrocytes depends on p53 activation. Cell Death Differ 18: 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SC, Salter RD (2005) Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity 23: 309–318 [DOI] [PubMed] [Google Scholar]
- Whitehead J, Zhang J, Harvestine JN, Kothambawala A, Liu G, Leach JK (2020) Tunneling nanotubes mediate the expression of senescence markers in mesenchymal stem/stromal cell spheroids: TNTs mediate senescence in MSC spheroids. Stem Cells 38: 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, González‐Billault C (2015) Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: implications for neuronal development and trafficking. Front Cell Neurosci 9: 381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig D, Wang X, Walter C, Gerdes H‐H, Funk RHW, Roehlecke C (2012) Multi‐level communication of human retinal pigment epithelial cells via tunneling nanotubes. PLoS One 7: e33195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Inaba M, Buszczak M (2018) Specialized intercellular communications via cytonemes and nanotubes. Annu Rev Cell Dev Biol 34: 59–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Borg TK, Ma Z, Xu M, Wetzel G, Saraf LV, Markwald R, Runyan RB, Gao BZ (2016) Biochip‐based study of unidirectional mitochondrial transfer from stem cells to myocytes via tunneling nanotubes. Biofabrication 8: 15012 [DOI] [PubMed] [Google Scholar]
- Yasuda K, Khandare A, Burianovskyy L, Maruyama S, Zhang F, Nasjletti A, Goligorsky MS (2011) Tunneling nanotubes mediate rescue of prematurely senescent endothelial cells by endothelial progenitors: exchange of lysosomal pool. Aging 3: 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani BG, Indolfi L, Edelman ER (2010) Tubular bridges for bronchial epithelial cell migration and communication. PLoS One 5: e8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Scholpp S (2019) Cytonemes in development. Curr Opin Genet Dev 57: 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Whitehead J, Liu Y, Yang Q, Leach JK, Liu G‐Y (2018) Direct observation of tunneling nanotubes within human mesenchymal stem cell spheroids. J Phys Chem B 122: 9920–9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bing T, Shen L, Song R, Wang L, Liu X, Liu M, Li J, Tan W, Shangguan D (2016) Intercellular connections related to cell‐cell crosstalk specifically recognized by an aptamer. Angew Chem Int Ed Engl 55: 3914–3918 [DOI] [PubMed] [Google Scholar]
- Zhang Y (2011) Tunneling‐nanotube: A new way of cell‐cell communication. Commun Integr Biol 4: 324–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Tan KS, Zhang X, Sun AY, Sun GY, Lee JC‐M (2005) Hydrogen peroxide alters membrane and cytoskeleton properties and increases intercellular connections in astrocytes. J Cell Sci 118: 3695–3703 [DOI] [PubMed] [Google Scholar]
- Zhu S, Bhat S, Syan S, Kuchitsu Y, Fukuda M, Zurzolo C (2018) Rab11a–Rab8a cascade regulates the formation of tunneling nanotubes through vesicle recycling. J Cell Sci 131: jcs215889 [DOI] [PubMed] [Google Scholar]
- Zhu S, Victoria GS, Marzo L, Ghosh R, Zurzolo C (2015) Prion aggregates transfer through tunneling nanotubes in endocytic vesicles. Prion 9: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurzolo C (2020) Evidence that tunnelling nanotube‐like structures connect cells in mice. Nature 585: 32–33 [DOI] [PubMed] [Google Scholar]


