Abstract
Background
tinnitus is a heterogeneous condition associated with audiological and/or mental disorders. Chronic, severe tinnitus is reported in 1% of the population and it shows a relevant heritability, according to twins, adoptees and familial aggregation studies. The genetic contribution to severe tinnitus is unknown since large genomic studies include individuals with self-reported tinnitus and large heterogeneity in the phenotype. The aim of this study was to identify genes for severe tinnitus in patients with extreme phenotype.
Methods
for this extreme phenotype study, we used three different cohorts with European ancestry (Spanish with Meniere disease (MD), Swedes tinnitus and European generalized epilepsy). In addition, four independent control datasets were also used for comparisons. Whole-exome sequencing was performed for the MD and epilepsy cohorts and whole-genome sequencing was carried out in Swedes with tinnitus.
Findings
we found an enrichment of rare missense variants in 24 synaptic genes in a Spanish cohort, the most significant being PRUNE2, AKAP9, SORBS1, ITGAX, ANK2, KIF20B and TSC2 (p < 2E−04), when they were compared with reference datasets. This burden was replicated for ANK2 gene in a Swedish cohort with 97 tinnitus individuals, and in a subset of 34 Swedish patients with severe tinnitus for ANK2, AKAP9 and TSC2 genes (p < 2E−02). However, these associations were not significant in a third cohort of 701 generalized epilepsy individuals without tinnitus. Gene ontology (GO) and gene-set enrichment analyses revealed several pathways and biological processes involved in severe tinnitus, including membrane trafficking and cytoskeletal protein binding in neurons.
Interpretation
a burden of rare variants in ANK2, AKAP9 and TSC2 is associated with severe tinnitus. ANK2, encodes a cytoskeleton scaffolding protein that coordinates the assembly of several proteins, drives axonal branching and influences connectivity in neurons.
Keywords: Tinnitus, Extreme phenotype, Axon initial segment, Exome sequencing
Research in context.
Evidence before this study
Tinnitus is the perception of noise or ‘ringing in the ear’ in the absence of an external acoustic stimulation affecting more than 15% of population. Severe tinnitus disturbing quality of life is experienced by 1% of the population and it has a significant heritability according to twins, adoptees and familial aggregation studies [47], [48]. A systematic review of extreme phenotype strategies to search for rare variants in genetic studies of complex disorders has found evidence to support a high effectiveness to reveal rare pathogenic variants and target novel candidate genes; particularly in neurological disorders such as epilepsy or Parkinson disease. By selecting individuals with tinnitus extreme phenotype, we should expect a burden of rare variation in certain genes associated with severe tinnitus.
Added value of this study
We have found a significant enrichment of missense rare variants in synaptic genes including ANK2, TSC2 and AKAP9 in patients with tinnitus extreme phenotype in Meniere disease patients. We also replicated these findings in an independent cohort of tinnitus patients from Sweden. Gene ontology (GO) and gene-set enrichment analyses revealed several pathways and biological processes involved in severe tinnitus, the top GO terms being membrane trafficking and cytoskeletal protein binding in neurons.
Implications of all the available evidence
This is the first study reporting the association of rare variation in ANK2, TSC2 and, AKAP9 genes with severe tinnitus and supports the involvement of membrane trafficking and cytoskeletal protein binding in the pathophysiology of severe tinnitus.
Alt-text: Unlabelled box
1. Introduction
Tinnitus is the perception of noise in the absence of an external acoustic stimulation. The symptom is reported by more than 15% of the world population; however, tinnitus is considered a disorder when it is associated with emotional distress, cognitive dysfunction, and/or autonomic arousal, leading to behavioural changes and functional disability [1,2]. The main risk factor of tinnitus is hearing loss, but it is often associated with other conditions including hyperacusis, anxiety, depression, hypertension, insomnia or migraine [3]. Meniere disease (MD) is a rare inner ear disorder with a significant genetic contribution [4], characterized by episodes of vertigo, tinnitus and sensorineural hearing loss [5]. Although vertigo attacks are considered as the main symptom in the first years of the disease, persistent tinnitus is described as the most troublesome symptom by many MD patients [6,7].
Evidence for a genetic contribution to severe tinnitus is unknown since large genomic studies include individuals with self-reported tinnitus and large heterogeneity in the phenotype [8,9].The extreme phenotype (EP) strategy has been used in exome sequencing studies to investigate the genetic contribution of rare variants in rare and complex disorders [10,11]. Individuals with EP are characterized by extreme clinically relevant attributes, toxic effects, or extreme responses to a treatment. EP covers both extreme ends of a phenotype distribution in quantitative traits and a burden of rare variation is expected in certain genes in individuals with a severe tinnitus [12].
The aim of this study was to identify rare variants in synaptic genes by exome sequencing in patients with severe tinnitus. For this, we performed a gene burden analysis (GBA) in Spanish patients with MD and tinnitus EP. Candidate genes ANK2, TSC2 and AKAP9 found in the MD-EP cohort were replicated in a Swedish tinnitus cohort, but not in a third generalised genetic epilepsy cohort, overall identifying the first putative genes involved in severe tinnitus.
2. Materials
2.1. Subjects and definition of phenotype
Individuals were recruited through the Meniere disease Consortium (MeDiC), and the diagnosis of patients was performed according to the diagnostic criteria for MD stated by the Barany society [13]. The Spanish version of the Tinnitus Handicap Inventory (THI) questionnaire [14] was used to assess the tinnitus severity and the functional impact of tinnitus on daily life [15]. A total of 59 Spanish patients with chronic and persistent tinnitus were selected among 1890 individuals from the MeDiC cohort, according to percentile 90 in the THI score (extreme cases). Diagnosis and psychoacoustic characterization of chronic tinnitus in patients with MD was performed as previously reported [16].Tinnitus EP was defined in MD patients (MD-EP) with an early onset and severe persistent tinnitus according to Tinnitus Handicap Inventory (THI) score [16]. Thirty individuals with THI score ≥76 were classified as extreme phenotype (EP), 29 individuals with THI ≥ 56 and <76 were defined as almost extreme phenotype (AEP). An in-house group of patients with MD without persistent tinnitus (N =32) were used as internal controls for this study. The clinical information of patients with MD and tinnitus phenotypes is detailed in (Supplementary Tables 1 and 2). A second independent tinnitus cohort of 97 individuals from Sweden was selected as a replication cohort: the Tinnitus Swedish Tinnitus Outreach Project (STOP) [17] which originates from the LifeGene study [18]. A subgroup of 34 individuals with severe tinnitus was also selected according to the THI ≥ 56 (Supplementary Table 3). We also retrieved rare variants summary statistics data from a third cohort of patients with epilepsy, the CoGIE cohort, that consisted of 701 individuals (152 Generalised genetic epilepsy cases and 549 controls), previously reported [19]. The CoGIE cohort was select as an external control to confirm that the genes associations reported in tinnitus were not observed in a non-related neurological disorder. All cases and controls were of European ancestry.
3. Procedures
Whole exome sequencing (WES) was performed on MD-EP, MD-AEP cases and in-house MD controls. DNA was extracted from blood or saliva samples using quality controls as previously described [20]. Exon capture was done with the SureSelectXT Human All Exon V6 (Mb) kit (Agilent), and the sequencing was done using HiSeq 4000 platform (Illumina) or NovaSeq 6000 platform (llumina). Paired-end reads were generated per sample to provide an on-target coverage of 100X minimum, with a total coverage of 10GB/sample in HiSeq4000 and 18GB/sample in Novaseq 6000. Read size was 100bp on HiSeq 4000 sequenced samples and 150 bp on Novase6000 sequenced samples.
Raw reads were stored as FASTQ files for each individual. GATK best practices pipelines were utilized to generate Binary Alignment Map (BAM) and Variant Calling Format (VCF) files from raw unmapped reads [21]. Human reference genome GRCH37/hg19 was used to align the reads with the help of Burrows-Wheeler Aligner (BWA-MEM) algorithm. To filter out low quality single nucleotide variants (SNVs) the recommended hard filter was applied as "quality by depth (QD), < 2.0 || fisher strand (FS) > 60.0 || root mean square quality mapping (MQ) < 40.0 || MQRankSum < -12.5 || ReadPosRankSum < -6.0". QD is used to normalize the quality of variants to evade the inflation in the existence of deep coverage. FS determines the probability which is based on Phred-scale of the site in case there is strand biasness. This score describes if the alternate alleles are more or less on the reverse or forward strand as compared to reference allele. MQ parameter explains the mapping quality of a site.
The called variants were further filtered out by an in-house MD control dataset composed of 32 individuals to exclude variants associated with MD. The final list of remaining variants was functionally annotated using KGGSeq suite [22] v1.0 and ANNOVAR tool [23] 2019Nov04.
To search for target genes involved in tinnitus, we have used the list of genes included in SynaptomeDB (N =1886). Genes encoding synaptic components included scaffold proteins, membrane transporters, cytoskeletal/adhesion proteins, neurotransmitters and its receptors (hereafter referred as synaptic genes, SG) [24]. Additionally, hearing loss genes (N =152) from Deafness Variation Database (DVD) v.8.1(http://deafnessvariationdatabase.org) were also analyzed to separate the potential effect of rare variation in hearing loss genes on tinnitus [25].
In order to search for variants associated with tinnitus, two types of variant analysis were performed: single variant analysis (SVA) and gene burden analysis (GBA) for MD-EP and MD-AEP (Supplementary Fig. 1). A flowchart for variant analysis according to the type of variant, location in coding or non-coding regions and effect on the protein is described(Supplementary Fig. 2).We have used three independent datasets as reference population: Non-Finish European (NFE) population dataset from gnomAD.v2, NFE from gnomAD.v3 [26] and a Spanish dataset from Collaborative Spanish Variant Server (CSVS) [27].We also called small insertions and deletions (indels) from MD-EP and MD-AEP patients and filtered out by in-house controls and the filtering criteria was applied according to GATK best-practice guidelines. All variants were assessed and evaluated according to the guidelines provided by American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) [28]. The final filtered list of candidate variants was checked through IGVv.2.8.9, further validated by Sanger sequencing and represented using Illustrator for biological sequences (IBS) [29].
Gene ontology (GO) analyses and gene enrichment analyses were performed using GSEA and MsigDB (https://www.gsea-msigdb.org/gsea/index.jsp) as previously described [30]. Two gene lists generated according to the GBA for rare SNVs and indels including 24 and 31 genes were used to retrieve signalling pathways and biological processes. For Gene expression analysis, In-situ hybridization (ISH) data in the mouse brain were obtained from the Allen Brain Atlas data set (http://www.brain-map.org), methods for data collection have been described previously31(Supplementary Note 1).
3.1. Statistical analysis
NFE population datasets from gnomAD.v2 (Exomes = 56,885; Genomes = 7718), and gnomAD.v3 (Genomes = 32,399), a Spanish population dataset from CSVS (Exomes = 1942) and a Swedish population dataset from SweGen (Genomes = 1000) were used as control groups [32] to compare the minor allele frequency (MAF) and to calculate the odds ratio (OR) for Spanish MD-EP and Swedish tinnitus cohorts. For SVA the OR with 95% CI was calculated for each variant using the three control datasets and p-values were corrected by the total number of variants being compared. For GBA, total alternate alleles per gene using 2 × 2 contingency matrixes were calculated for EP, AEP and control datasets. For each gene, the OR was calculated with 95% CI and two-tailed p-value was corrected for multiple testing by the total number of genes being compared following Bonferroni-correction. A corrected p-value <0.05 was considered significant. For each gene, the Etiological Fraction (EF) was also calculated as previously described [33].
3.2. Role of the funding source
The funders of this study had no role in study design, patient recruitment, data analysis, its interpretation or writing the manuscript. The authors had full access to data used in this study with the responsibility to submit it for publication.
3.3. Ethical approval
This study has been approved by the Andalucian Ethical Review Board and written consent was obtained from all subjects to conduct genetic studies (Protocol number 722046).
4. Results
4.1. Synaptic genes in Spanish patients with tinnitus extreme phenotype
First, we performed a SVA in patients with tinnitus EP in MD. The total number of obtained variants with MAF <0.05 were 2287 for MD-EP and 1610 for MD-AEP, respectively. Two missense variants were found significantly associated in patients with MD-EP after p-correction. The first was a heterozygous variant and it was found in 3 unrelated individuals located at exon 21 in DAAM1 gene (chr14:59826182A>C; p.Asn875His; rs61740455) with MAFcsvs= 0.002 and CADD = 17.85). The associated second variant was located at exon 32 in MYH10 gene (chr17:8397065C>A; p.Ala1399Ser; rs149021341; MAFcsvs = 0.001, CADD = 22), and it was found in 2 individuals and one of the carriers was homozygous. Next, we carried out a GBA in the Spanish MD cohort with EP and AEP. For this, we selected variants with MAF<0.1 to analyze the combined effect of different common and rare variants in the same gene. The retained variants in patients were 4625 for MD-EP and 3592 for MD-AEP, respectively after filtering by MD in-house controls to rule out rare variants associated with hearing or vestibular phenotypes. The GBA of missense variants showed 24 significant genes in tinnitus MD-EP including PRUNE2, AKAP9, SORBS1, ITGAX, ANK2, KIF20B, LRPPRS, SYNPO, TSC2 (Table 1), and 18 genes for MD-AEP (Supplementary Table 4). Interestingly, none of these genes showed an enrichment of synonymous or 5`UTR variants in MD-EP (Supplementary Tables 5 and 6); additionally, the genes from synonymous analysis for AEP are detailed (Supplementary Table 7) The most significant finding for EP was an enrichment of missense variants in the ANK2 gene, against NFE population from gnomAD.v2 [OR =18.30(6.78–49.40), EF = 0.95, corrected-p =1.80E–05], gnomAD.v3 [OR =19.95(7.36–54.08), EF = 0.95, corrected-p = 7.55E–06] and Spanish population from CSVS [OR = 21.93(7.02–68.48), EF= 0.95, corrected-p = 2.02E-04]. In ANK2, four different missense rare variants were found in 3 different sporadic cases; three of the variants were novel and they have not been reported in gnomAD or CSVS databases. The variant 4:114294537G>A; exon 45 was found only in one case and two of the novel variants 4:114277102T>G; exon 38 and 4:114294509G>C; exon 45 were carried by the same patient. The third novel variant 4:114262911A>G was located at exon 33 (Supplementary Table 8 and Supplementary Fig. 3).
Table 1.
List of synaptic genes showing enrichment of missense variants in Spanish patients with Meniere disease and tinnitus extreme phenotype (MD-EP). Three reference datasets (Non-Finnish European from gnomAD.v2 or gnomAD.v3, Spanish from CSVS) were used to compare allelic frequencies for each gene in the MD cohort. Listed genes were significant when they were compared against CSVS reference dataset.
| Gene | #variants | [gnomAD.v2] OR(CI) | EF | Corrected p | [gnomAD.v3] OR(CI) | EF | Corrected p | [CSVS] OR(CI) | EF | Corrected p |
|---|---|---|---|---|---|---|---|---|---|---|
| PRUNE2 | 9 | 6.02(3.60–10.07) | 0.83 | 1.44E-08 | 5.89(3.52-9.85) | 0.83 | 2.75E-08 | 5.40(3.17-9.20) | 0.81 | 1.08E-06 |
| AKAP9 | 6 | 12.32(5.48–27.68) | 0.92 | 2.2E-06 | 13.89(6.17-31.27) | 0.93 | 4.04E-07 | 6.68(2.86-15.57) | 0.85 | 2.07E-02 |
| SORBS1 | 6 | 10.93(4.87–24.55) | 0.91 | 1.31E-05 | 11.52(5.12-25.93) | 0.91 | 6.57E-06 | 7.73(3.30-18.12) | 0.87 | 4.86E-03 |
| ITGAX | 5 | 73.02(29.68–179.66) | 0.99 | <1.00E-15 | 61.68(24.88-152.94) | 0.98 | <1.00E-15 | 14.29(5.40-37.86) | 0.93 | 1.63E-04 |
| ANK2 | 4 | 18.30(6.78–49.40) | 0.95 | 1.80E-05 | 19.95(7.36-54.08) | 0.95 | 7.55E-06 | 21.93(7.02-68.48) | 0.95 | 2.02E-04 |
| KIF20B | 4 | 7.76(3.45–17.49) | 0.87 | 1.42E-03 | 8.43(3.74-19.01) | 0.88 | 5.27E-04 | 16.57(6.71-40.92) | 0.94 | 2.15E-06 |
| TSC2 | 4 | 63.73(23.35–173.96) | 0.98 | 8.38E-13 | 53.56(19.47-147.30) | 0.98 | 2.35E-11 | 21.93(7.02-68.48) | 0.95 | 2.02E-04 |
| SPHK2 | 4 | 5.47(2.25–13.28) | 0.82 | NS | 5.51(2.27-13.39) | 0.82 | NS | 8.45(3.30-21.64) | 0.88 | 1.61E-02 |
| SYNPO | 4 | 74.87(27.35-204.94) | 0.99 | <1.00E-15 | 78.43(28.21-218.03) | 0.99 | <1.00E-15 | 32.90(9.84-110.01) | 0.97 | 2.66E-05 |
| LRPPRC | 4 | 49.75(18.29–135.32) | 0.98 | 3.73E-11 | 73.20(26.39-203.03) | 0.99 | 4.19E-13 | 21.93(7.02-68.48) | 0.95 | 2.02E-04 |
| XYLT1 | 4 | 2.00(0.74–5.38) | 0.50 | NS | 2.09(0.78-5.61) | 0.52 | NS | 10.95(3.77-31.82) | 0.91 | 2.04E-02 |
| ALCAM | 3 | 8.22(2.62–25.81) | 0.88 | NS | 9.48(3.01-29.81) | 0.89 | NS | 24.67(6.49-93.76) | 0.96 | 4.77E-03 |
| CDH13 | 3 | 12.15(4.50–32.85) | 0.92 | 1.60E-03 | 13.09(4.83-35.46) | 0.92 | 8.05E-04 | 33.08(9.87-110.87) | 0.97 | 2.69E-05 |
| DOCK7 | 3 | 52.57(16.54–167.10) | 0.98 | 3.53E-08 | 70.09(21.61-227.27) | 0.99 | 2.71E-09 | 24.67(6.49-93.76) | 0.96 | 4.77E-03 |
| BIN1 | 3 | 56.70(17.82–180.42) | 0.98 | 1.53E-08 | 73.20(22.54-237.74) | 0.99 | 1.72E-09 | 49.36(10.97-222.15) | 0.98 | 7.11E-04 |
| FLII | 3 | 26.77(8.48–84.44) | 0.96 | 3.87E-05 | 33.95(10.66-108.12) | 0.97 | 4.64E-06 | 24.67(6.49-93.76) | 0.96 | 4.77E-03 |
| HSPA4L | 3 | 17.41(5.53–54.77) | 0.94 | 1.95E-03 | 16.71(5.29-52.75) | 0.94 | 2.98E-03 | 16.44(4.60-58.77) | 0.94 | 3.11E-02 |
| IQSEC1 | 3 | 32.85(10.39–103.82) | 0.97 | 5.12E-06 | 30.49(9.59-96.94) | 0.97 | 1.32E-05 | 24.67(6.49-93.76) | 0.96 | 4.77E-03 |
| IQSEC3 | 3 | 4.16(1.33–13.04) | 0.76 | NS | 4.47(1.43-14.03) | 0.78 | NS | 16.44(4.60-58.77) | 0.94 | 3.11E-02 |
| LLGL1 | 3 | 27.29(10.06–74.03) | 0.96 | 1.57E-07 | 25.08(9.21-68.31) | 0.96 | 5.53E-07 | 13.92(4.69-41.32) | 0.93 | 4.01E-03 |
| MADD | 3 | 128.54(39.58-417.46) | 0.99 | 1.26E-12 | 73.20(22.54-237.74) | 0.99 | 1.72E-09 | 49.36(10.97-222.15) | 0.98 | 7.11E-04 |
| MBP | 3 | 170.13(51.78–559.02) | 0.99 | <1.00E-15 | 82.35(25.24-268.68) | 0.99 | 4.99E-10 | 16.44(4.60-58.77) | 0.94 | 3.11E-02 |
| MPRIP | 3 | 82.62(25.77–264.88) | 0.99 | 2.10E-10 | 78.43(24.09-255.39) | 0.99 | 8.32E-10 | 49.36(10.97-222.15) | 0.98 | 7.11E-04 |
| NRCAM | 3 | 69.68(21.82-222.56) | 0.99 | 1.49E-09 | 50.67(15.78-162.74) | 0.98 | 8.07E-08 | 49.36(10.97-222.15) | 0.98 | 7.11E-04 |
| TRAP1 | 3 | 13.72(4.37–43.13) | 0.93 | 1.39E-02 | 11.27(3.58-35.47) | 0.91 | NS | 24.67(6.49-93.76) | 0.96 | 4.77E-03 |
| VCAN | 3 | 90.37(28.13–290.35) | 0.99 | 7.41E-11 | 76.61(23.55-249.22) | 0.99 | 1.07E-09 | 24.67(6.49-93.76) | 0.96 | 4.77E-03 |
| MYO18A | 3 | 204.11(72.08–577.95) | 0.99 | <1.00E-15 | 169.90(58.69-491.88) | 0.99 | <1.00E-15 | 33.08(9.87-110.87) | 0.97 | 2.69E-05 |
| MYO5A | 3 | 13.72(4.37–43.13) | 0.93 | 1.39E-02 | 16.97(5.37-53.37) | 0.94 | 2.62E-03 | 16.44(4.60-58.77) | 0.94 | 3.11E-02 |
| PPP1R9A | 2 | 171.57(51.97–566.41) | 0.99 | <1.00E-15 | 127.78(38.15-427.99) | 0.99 | 7.12E-12 | 24.87(6.52–94.93) | 0.96 | 4.84E-03 |
| CCDC22 | 2 | 7.38(2.72–20.04) | 0.86 | NS | 8.37(3.08–22.75) | 0.88 | NS | 14.06(4.71–41.98) | 0.93 | 4.08E-03 |
| EPX | 2 | 8.17(3.01–22.19) | 0.88 | NS | 8.69(3.20–23.64) | 0.88 | 4.28E-02 | 11.61(3.95–34.11) | 0.91 | 1.54E-02 |
OR (CI)= odds ratio (95% confidence interval), EF=etiological fraction, p-corrected values obtained after Bonferroni correction, genes are ordered by number of variants, NS=Nonsignificant p-corrected values
In the next step, we selected missense variants with CADD≥20 from SG in MD-EP (561 SNV) and MD-AEP (560 SNV) for the GBA. CADD score describes the deleteriousness of SNVs and can be used to prioritize the disease causal variants explaining the underlying genetic architecture and effect size. We obtained 7 genes with significant burden of rare pathogenic variants for MD-EP (Supplementary Table 9), and 9 significant genes for MD-AEP (Supplementary Table 10), when they were compared with reference datasets.
Finally, we performed a SVA and GBA of indels in SG from Spanish patients with MD-EP and MD-AEP. Indels were further filtered out by in-house controls. A total of 1565 indels (MAF<0.05) for SVA, and 2370 indels (MAF<0.1) for the GBA were retrieved for the MD-EP, and 1404 indels for SVA (MAF<0.05) and 1693 (MAF<0.05) for GBA in the MD-AEP groups, respectively. We found an enrichment of indels in 31 genes in the MD-EP (Supplementary Table 11), including TSC2, AKAP9 and several other genes and 48 genes in the MD-AEP (Supplementary Table 12), when data were compared with European reference datasets (gnomAD.v2 and gnomAD.v3). Unfortunately, we cannot compare the allelic frequencies in MD-EP or MD-AEP for the indels using the Spanish reference data (CSVS), since the number of indels reported in CSVS dataset is low and it will overestimate the burden. We also compared rare, Loss-of-Function (LoF) variants including nonsense, splice-site and frame shift small insertions and deletions in the SG set for MD-EP and MD-AEP. We found 61 LoF variants in the MD-EP and 25 LoF variants in the MD-AEP. However, the number of nonsense or novel splice-site variants found was small, and no significant burden of LoF variants was found in MD-EP or MD-AEP.
4.2. Replication in Swedish patients with severe tinnitus
To replicate the findings in a Swedish cohort with severe tinnitus, we selected all the significant genes from MD-EP analysis. We used three different population datasets as reference controls (gnomAD.v2, gnomAD.v3 and SweGen). The observed MAF for each gene was calculated and compared with controls, whilst p-values were corrected by the total number of variants per gene. Six genes showed an enrichment of missense variants. Subsequently, we selected a subset of 34 patients with severe tinnitus (THI score ≥56) and found a burden of missense variants in ANK2, AKAP9 and TSC2 genes (Table 2). Missense variants identified in the GBA for ANK2 gene are detailed in (Supplementary Table 8); most of these variants are clustered around exons 38 to 45 across the gene sequence (Supplementary Fig. 2).Supplementary Tables 13 and 14, list missense variants found in the GBA for AKAP9 and TSC2 genes in Spanish and Swedish patients with tinnitus. Rare variants found in ANK2 and TSC2 genes were also validated by Sanger sequencing (Supplementary Figs. 4 and 5).
Table 2.
Swedish Tinnitus replication cohort, Synaptic genes showing an enrichment of missense rare variants in Swedish patients with chronic tinnitus without diagnosis of MD.
| Gene | #Variants | [gnomAD.v2] OR(CI) | EF | Corrected p | [gnomAD.v3] OR(CI) | EF | Corrected p | [SweGen] OR(CI) | EF | Corrected p |
|---|---|---|---|---|---|---|---|---|---|---|
| Non selected Tinnitus (N=97) | ||||||||||
| ANK2 | 8 | 3.20(1.92-5.33) | 0.69 | 6.25E-05 | 3.28(1.97-5.47) | 0.70 | 4.08E-05 | 2.83(1.59-5.02) | 0.65 | 3.02E-03 |
| MYO18A | 5 | 5.99(2.84-12.64) | 0.83 | 1.34E-05 | 5.94(2.81-12.57) | 0.83 | 1.61E-05 | 6.60(2.55-17.07) | 0.85 | 4.93E-04 |
| MADD | 4 | 4.99(2.23-11.17) | 0.80 | 3.78E-04 | 4.78(2.13-10.73) | 0.79 | 6.04E-04 | 3.89(1.52-9.97) | 0.74 | 1.87E-02 |
| KIF20B | 4 | 4.99(2.06-12.06) | 0.80 | 1.45E-03 | 4.89(2.02-11.87) | 0.80 | 1.77E-03 | 3.70(1.33-10.30) | 0.73 | 4.90E-2 |
| MPRIP | 3 | 35.36(11-113.70) | 0.97 | 6.53E-09 | 28.77(8.82-93.82) | 0.97 | 7.60E-08 | 15.54(2.59-93.19) | 0.94 | 8.05E-03 |
| MBP | 2 | 12.95(3.18-52.76) | 0.92 | 7.04E-04 | 35.34(8.20-152.23) | 0.97 | 3.43E-06 | 10.36(1.45-73.74) | 0.90 | 3.91E-02 |
| NRCAM | 2 | 47.15(11.13-199.77) | 0.98 | 3.37E-07 | 111.91(22.52-556.20) | 0.99 | 1.62E-08 | 20.72(1.87-229.04) | 0.95 | 2.68E-02 |
| Severe tinnitus (N=34) | ||||||||||
| AKAP9 | 3 | 4.93(2.03–12) | 0.80 | 1.29E-03 | 5.80(2.38–14.12) | 0.83 | 3.20E-04 | 3.32(1.31–8.47) | 0.70 | 3.50E-2 |
| TSC2 | 2 | 13.92(4.41–43.93) | 0.93 | 1.4E-05 | 10.97(3.47–34.66) | 0.91 | 9.02E-05 | 12.87(3.29–50.31) | 0.92 | 4.81E-04 |
| ANK2 | 2 | 11.51(3.65–36.28) | 0.91 | 6.06E-05 | 13.20(4.17–41.78) | 0.92 | 2.26E–05 | 4.73(1.38–16.17) | 0.79 | 2.60E-2 |
OR (CI)= odds ratio (95% confidence interval), EF=etiological fraction, p-corrected values obtained after Bonferroni correction, genes are ordered by number of variants
In addition, we used an independent cohort of generalized genetic epilepsy to determine if the association of ANK2, TSC2 and AKAP9 genes with severe tinnitus was a non-specific finding, since some neurological disorders such as epilepsy could also share some common genetic background with tinnitus. For this, we performed a GBA using the same SG list in this epilepsy cohort, but none of the genes showed a significant enrichment of missense variants strongly suggesting the genes captured here are tinnitus-specific.
Lastly, we performed GBA of indels in the Swedish cohort using synaptic genes with MAF<0.1. We found 2 genes in the tinnitus cohort (N = 97), and 6 genes in subgroup with severe tinnitus (N = 34), showing a significant burden of indels (Supplementary Table 15).
To investigate the association of rare missense variants in hearing loss genes with MD-EP, we performed a GBA of missense variants using hearing loss genes in patients with MD. We obtained 305 variants from EP and 313 from AEP with MAF<0.1, respectively. The 6 genes included USH1G, ILDR1, OTOA, PCDH15, CACNA1D and NARS2 were found significant in MD-EP (Supplementary Table 16), and 4 genes showed significant enrichment in MD-AEP (Supplementary Table 17). To replicate the burden of rare variants found in hearing loss genes in MD-EP patients, we selected a subset of 62 patients with self-reported hearing problems from the Swedish cohort. Then, we performed a GBA in the 6 significant hearing loss genes of MD-EP, however, none of these genes showed an enrichment of missense variants in this cohort.
4.3. Gene ontology and gene-set enrichment analysis in patients with tinnitus
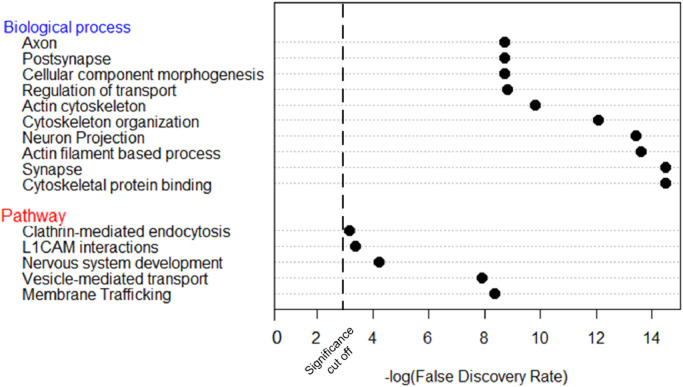
We selected 55 significant genes from MD-EP cohort to perform GO and gene-set enrichment analysis including 24 genes with enrichment of missense variants and 31 genes with enrichment of indels analysis. The most significant pathway and GO biological processes involved were the membrane trafficking and cytoskeletal protein binding (Fig. 1, Supplementary Table 18).
Fig 1.
Gene ontology (GO) and gene-set enrichment analysis This analyses were performed using 55 genes obtained in the gene burden analysis of MD-EP and GSEA tool was used to obtained molecular pathways and biological processes.
4.4. ANK2 and TSC2 gene expression profile in the mouse brain
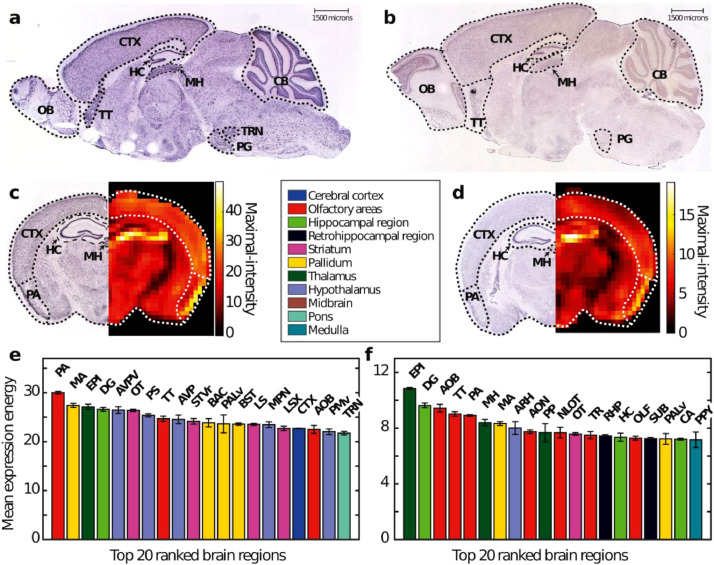
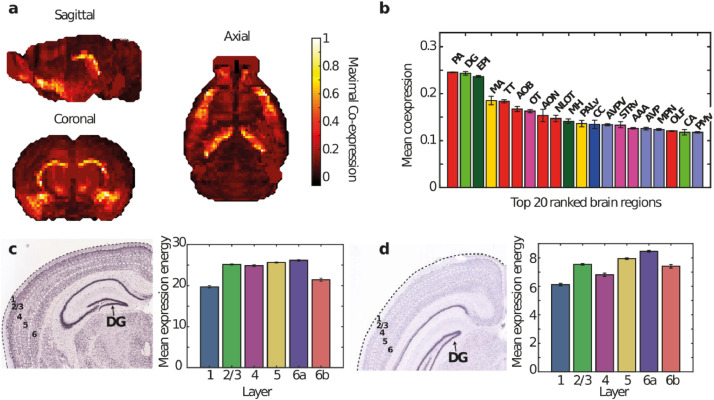
In-situ hybridization (ISH) data in the mouse (Ank2: n = 2, Tsc2: n = 2, one coronally and one sagittally sections each) obtained from the Allen Mouse Brain Atlas [31] demonstrated strong Ank2 and Tsc2 expression in a number of brain regions (Figs. 2 and 3). Visual inspection revealed strong expression of both genes in the cortex, hippocampus (pyramidal layer of CA1, CA2 and CA3 and the granule cell layer of the dentate gyrus), olfactory bulb (the granule and mitral layers), hypothalamus and cerebellum. In addition, to subregions of other brain regions, notably: Tenia tecta, the epithalamus (especially the medial habenula), piriform area (layer 2) and the magnocellular nucleus (Fig. 3). There was a marked similarity in the brain wide expression of Ank2 and Tsc2, this could potentially suggest a common mechanism or brain regions of interest. To confirm this co-expression, 4,104 genes in the mouse brain were compared (n = 4104). These data were used to build a probability distribution for deriving a given amount of coexpression, based on this coexpression of Ank2 and Tsc2 was found to be highly significant (coexpression = 0.9031, p = 0.0091), Fig. 3, (Supplementary Note 2). Somewhat comparable human data (though in a much lower quantity, i.e. Ank2: n = 3, Tsc2 = 2 brains) were also found via the BioGPS gene portal system (http://biogps.org). These data demonstrated strong expression of both Ank2 and TSC2 in cortex (occipital and parietal lobes, and prefrontal cortex), hypothalamus, cerebellum (peduncles) and the amygdala. Interestingly, the profiles of normalized brain expression of Ank2 and Tsc2 were also significantly correlated suggesting similar expression in the human brain also (Pearson's, r = 0.507, p = 0.0031). Single cell RNA-seq data from the Allen Cell Types Database also revealed similarities between human and mouse expression [34]. For both humans and mice Ank2 and Tsc2 expression was significantly differentially distributed in cortical neurons (KS test, humans: Ank2, p =1.1 × 10−6, Tsc2, p = 1.4 × 10−12, mouse: Ank2, p = 1.8 × 10−20, TSC2, p = 1.2 × 10−5), where stronger expression was commonly observed in pyramidal neurons when compared to inhibitory interneurons (Supplementary Fig. 6).
Fig 2.
Brain wide expression profiles of ANK2 and TSC2 in the mouse brain taken from in-situ hybridization data from the Allen Brain Atlas data set (http://www.brain-map.org). Sagittal sections of expression in the adult mouse (P56) brain for both (a) ANK2 and (b) TSC2. Strong expression for both genes is found in a number of brain regions, including: CTX = Cortex, HC = Hippocampus, CB = Cerebellum, MH = Medial habenula, TT = Taenia tecta. Coronal sections (c) and (d), left panels) were taken from Allen Brain Atlas in a pre-rendered to fit an annotated format (c) and (d), right panels) allowing easy identification expression in different brain regions. This was used to identify brain regions demonstrating strongest expression (e) and (f), see text for details). PA = piriform area, MA = magnocellular nucleus, EPI = epithalamus, DG = dentate gyrus, AVPV = anteroventral periventricular nucleus, OT = olfactory tubercle, PS = parastrial nucleus, TT = tenia tecta, AVP = anteroventral preoptic nucleus, STVr = Striatum, ventral region, BAC = bed nucleus of the anterior commissure, PALv = pallidum, ventral region, BST = bed nuclei of the stria terminalis, LS = lateral septal nucleus, MPN = medial preoptic nucleus, LSX = lateral septal complex, CTX = cortex, AOB = accessory olfactory bulb, PMv = ventral premammillary nucleus, TRN = tegmental reticular nucleus, PP = peripeduncular nucleus, NLOT = nucleus of the lateral olfactory tract, ARH = arcuate hypothalamic nucleus, PP = peripenducular nucleus, TR = postpiriform transition area, RHP = retrohippocampal region, PPY = Parapyramidal nucleus.
Fig 3.
Significant brain wide co-expression of ANK2 and TSC2 was found in the mouse brain (see text). 3) Mean across-section (sagittal, coronal and axial planes) co-expression of ANK2 and TSC2. Colorbar indicates strong coexpression in yellow. (b) Mean coexpression in 209 brain regions was calculated and ranked revealing the top 20 brain regions where ANK2 and TSC2 were co-expressed. Coronal sections revealing layer specific Ank2 expression in the cortex (c), left panel), mean expression for each cortical layer (across the whole brain) was calculated (c), right panel) revealing strongest expression in layers 2/3, 5 and 6a. Coronal sections revealing layer specific TSC2 expression in the cortex (d), left panel), mean expression for each cortical layer (across the whole brain) was calculated (d), right panel) revealing strongest expression in layers 2/3, 5 and 6a.
5. Discussion
The present study reports for the first time a burden of rare missense and structural variants in several SG in patients with severe tinnitus. These genes are involved in cytoskeleton organization and cytoskeleton protein binding in neurons suggesting novel mechanisms involved in tinnitus severity. In particular, a burden of missense rare and novel variants in ANK2, AKAP9 and TSC2 genes in Spanish MD patients with severe tinnitus (MD-EP), which was replicated in a Swedish cohort of individuals with severe tinnitus. Using a large generalized genetic epilepsy cohort, we could confirm the specificity of these new genes to tinnitus.
The synapse between sensory inner hair cells, primary auditory neurons and these neurons itself are potential candidates for tinnitus, but its perception and long term maintenance involves complex networks in the central nervous system, both in auditory and in non-auditory structures [35]. GO analyses suggest that membrane trafficking and cytoskeletal protein binding in neurons are involved at the molecular level. Future studies in a larger cohort of tinnitus patients will confirm these predictions.
Tinnitus is associated with hearing loss in 90% of cases, according to standard pure tone audiograms. The most accepted causative model of tinnitus is based on the reduction in the auditory input associated with hearing loss, which leads to increased gain in the auditory pathway; that is, an amplification of spontaneous activity in the auditory neurons will lead to the perception of tinnitus [36]. This change in the intrinsic neuronal excitability after sensory deprivation occurs at the axon initial segments (AISs), the site of initiation of the action potential, which increase in length, and expression of voltage-dependent Na+ channels and Ankyrin-G. A membrane scaffolding protein encoded by the ANK2 gene in the AISs.
The ANK2 (ENSG00000145362) gene, located at chromosome 4q25–q26, encodes Ankyrin-2, a large structural protein that carries death and ankyrin repeat containing domains. The Ankyrin gene has 46 exons in total and the exon 37/38 is brain specific [37,38]. It belongs to the ankyrin family that links the integral proteins to the fundamental spectrin-actin cytoskeleton and plays an important role in different activities including micrometer scale organization of plasma membranes in a broad spectrum of physiological context. ANK2 encodes two different polypeptide including Ankyrin-2 (expressed in different tissues) and giant Ankyrin-2, a neuro-specific isoform variant expressed broadly in the central nervous system, with 2133 residues encoded by exon 37 between death and spectrin-binding domains [38]. Giant Ankyrin-2 is a key protein to keep connectivity and neural activity in the central nervous system. It contributes to the development, maintenance and the refinement of neural circuits in different brain areas. The neural signals that arise at AISs site regulate the neural activity. However, the lack of auditory input can cause an increase in the length of AISs ultimately exciting the auditory neurons in avian brainstem [39]. In addition, this is accompanied with an increase in whole-cell Na+ current, membrane excitability and spontaneous firing. After auditory deprivation, the preservation of auditory function indicates that the change may have occurred at synaptic functionality level rather than at the structural level. However, the homeostatic changes occurring at AISs might play an important role to maintain the integrity of the remaining neurons in auditory circuits [39], something that may also occur in severe tinnitus.
Rare variations in ankB isoform may produce an increase of axonal branching [38]. In humans, rare variants in ANK2 gene have previously been reported in individuals with autism spectrum disorder [38] and long QT syndrome [40].
Epidemiological and genetic studies consistently support that severe tinnitus has a genetic contribution[47]1[48] and common and rare variants with epistatic effects shape the phenotype [3,41]. A recent GWAS using a broad definition for tinnitus found a small number of loci and common variants with small effect sizes [42].
Tinnitus as a neurological disorder may not only result from sensory deprivation as it probably occurs in high-frequency hearing loss or MD, or after synaptic reorganization that lead to changes on the neuronal excitability at different brain areas, but also from enhanced connectivity with non-auditory brain regions as it is often observed in tinnitus patients or individuals with severe tinnitus [43].
ANK2 is expressed in a number of distinct auditory and non-auditory brain regions within the mouse brain. We investigated ANK2 and TSC2 co-expression profile by selecting RNAseq data, confirming that both genes have a significant co-expression in the mouse. Human data (on a much grosser) scale appears to confirm this with significant correlation of these genes across the brain regions for which data was available. In addition, both genes demonstrated significantly greater expression in excitatory neurons than inhibitory neurons, potentially suggesting their importance in the function of this class of neurons.
By using https://shield.hms.harvard.edu/index.html, we have confirmed the expression of ANK2, TSC2 and AKAP9 in spiral ganglion neurons. However, it is difficult to assess quantitatively about expression throughout the auditory system as the Allen Brain ISH data are not annotated to include subregions, and not all auditory sub-nuclei are categorized. However, visual inspection shows noticeably stronger expression of ANK2 in the dorsal Inferior Colliculus. Expression of TSC2 does not stand out relative to other non-auditory nuclei.
Interestingly, we have found a burden of rare variation in AKAP9, another gene previously associated with long-QT syndrome [44]. AKAP9 encodes A-kinase anchor protein 9, a member of the A-kinase anchor protein family, whose known function is binding to the protein kinase A (PKA) regulatory subunit with the objective of enclose it to different parts of the cell where phosphorylation is needed [45].
Our study also reveals a significant enrichment of rare variants in TSC2 gene in patients with MD-EP and severe tinnitus. Tuberous Sclerosis Complex 2 (also known as TSC2 or Tuberin) is a known tumor suppressor protein part of the tuberous sclerosis complex (TSC) along with TSC1. This complex is involved in the negative regulation of mTORC1 activity. Loss of tuberin function causes constitutive activation of the mTORC1 signaling pathway leading to tuberous sclerosis tumors [46]. The regulation of the Mtorc1 pathway via the TSC complex has been found to be a key part in some age-associated diseases, including age-related hearing loss. Finally, we found highly significant co-expression of ANK2 and TSC2 across the mouse brain, potentially suggesting they are expressed in similar neuronal subtypes. Mouse brain co-expression of ANK2 and TSC2 was particularly strong in limbic brain regions (i.e. the hypothalamus, epithalamus, striatum, pallidum and hippocampus), that form a complex circuit distributed across the brain. In addition, strong expression was found across cortex particularly in cortical layers generally associated with cortical projections (i.e. layers 2/3, 5 and 6).
6. Limitations
Our study has several limitations. The EP strategy is not a representative of complete phenotypic variance observed in MD. Secondly, most of our MD patients were females and future genetic studies should consider gender differences in tinnitus. The third limitation of the study is that other neural pathways not related to synapses may be implicated in tinnitus. Our EP approach will be extended to investigate these pathways in future studies. Finally, indel analysis is highly dependent on the callers and annotation tools and most of the indels in our dataset were not found in either the gnomAD or the CSVS databases.
Our study reveals a burden of rare variants in SG, including ANK2, AKAP9 and TSC2 in patients with severe tinnitus and predicts the involvement of ANK2 in the cytoskeleton re-organization in the axonal initial segment. We thus propose that axonal branching is a likely mechanism to enhance connectivity in auditory and non-auditory brain regions ultimately leading to severe tinnitus.
Acknowledgments
Acknowledgments
This project is a part of European School of Interdisciplinary Tinnitus (ESIT) research and Sana Amanat is a PhD student in Biomedicine Program at the University of Granada. We would like to thank all patients for their participation in this study. We thank Pablo Roman-Naranjo and Marisa Flook for their assistance with DNA extraction; Alba Escalera-Balsera for her help during sample preparation for Sanger sequencing; Niklas Edvall and Natalia Trpchevska for assistance with the STOP cohort.
Data sharing
Clinical and genomic data from each individual are protected according to EU regulation on data protection. Aggregated, anonymized genomic datasets will be shared upon request according to consortium agreement regulations defined by H2020 MSCA-ITN-2016–722046, the H2020-SC1-2019-848261, and the GNP-182 GENDER-Net Co-Plus Fund.
Funding sources
This study has been funded by H2020 MSCA-ITN-2016–722046, the H2020-SC1-2019-848261, and the GNP-182 GENDER-Net Co-Plus Fund (JALE and CRC). The project leading to these results has received funding from “la Caixa” Foundation (ID 100010434), under agreement LCF/PR/DE18/52010002 (JALE). This project is a part of European School of Interdisciplinary Tinnitus (ESIT) research and Sana Amanat is a PhD student in Biomedicine Program at the University of Granada. CRC received additional funding from Svenska Läkaresällskapet (SLS-779681), Hörselforskningsfonden (503). The data handling for STOP and SweGen cohorts were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX partially funded by the Swedish Research Council through Grant agreement No. 2018-05973. The time provided by JS was funded by the University of Nottingham, Nottingham Research Fellowship. PM was supported by the BMBF Treat-ION Grant (01GM1907) and the DFG Research Unit FOR2715 (FNR INTER/DFG/17/11583046).
Declaration of Competing Interest
C.R.C. is supported by the UK National Institute for Health Research (NIHR) Biomedical Research Centre but the views expressed herein are his own and do not represent those of NIHR nor the UK Department of Health and Social Care. The other authors declare no competing interest.
Author contributions
JALE conceived the study design and developed the scientific arguments. SA and AGM performed the bioinformatics analyses for MD cohort, AGM performed bioinformatics analyses on Swedish tinnitus cohort. SA performed statistical analysis for MD and Swedish tinnitus cohorts. JS contributed to the analysis of mouse gene expression datasets. JALE, PPC, JMES, IA, ASV, ABC recruited extreme phenotype patients from the MD cohort. BC and CRC collected data for STOP cohort. PM performed bioinformatic analyses in the epilepsy cohort. JALE, SA, AGM draft the manuscript and all authors have read and approved the final draft.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103309.
Appendix. Supplementary materials
References
- 1.Vanneste S., Joos K., De Ridder D. Prefrontal cortex based sex differences in tinnitus perception: same tinnitus intensity, same tinnitus distress, different mood. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilles A., Camp G., Van de Heyning P., Fransen E. A pilot genome-wide association study identifies potential metabolic pathways involved in tinnitus. Front Neurosci. 2017;11:1–10. doi: 10.3389/fnins.2017.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Escamez J.A., Amanat S. Heritability and genetics contribution to tinnitus. Otolaryngol Clin. 2020;53:501–513. doi: 10.1016/j.otc.2020.03.003. NA. [DOI] [PubMed] [Google Scholar]
- 4.Gallego-Martinez A., Requena T., Roman-Naranjo P., Lopez-Escamez J.A. Excess of rare missence variants in hearing loss genes in sporadic Meniere disease. Front Genet. 2019;10 doi: 10.3389/fgene.2019.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Carpena P., Lopez-escamez J.A. Current understanding and clinical management of Meniere ’s disease. Semin Neurol. 2020;40(1):138–150. doi: 10.1055/s-0039-3402065. [DOI] [PubMed] [Google Scholar]
- 6.Ueberfuhr M.A., Wiegrebe L., Krause E., Gürkov R., Drexl M. Tinnitus in normal-hearing participants after exposure to intense low-frequency sound and in Ménière's disease patients. Front Neurol. 2017;7:1–11. doi: 10.3389/fneur.2016.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herraiz C., Tapia M.C., Plaza G. Tinnitus and Ménière's disease: characteristics and prognosis in a tinnitus clinic sample. Eur Arch Oto Rhino Laryngol. 2006;263:504–509. doi: 10.1007/s00405-006-0019-9. [DOI] [PubMed] [Google Scholar]
- 8.Vona B., Nanda I., Shehata-Dieler W., Haaf T. Genetics of tinnitus: still in its infancy. Front. Neurosci. 2017 doi: 10.3389/fnins.2017.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amanat S., Gallego-Martinez A., Lopez-Escamez J.A. Genetic inheritance and its contribution to tinnitus. Curr Top Behav Neurosci. 2020 doi: 10.1007/7854_2020_155. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Gracia J.L., Ruiz-Ilundain M.G., Garcia-Ribas I., Carrasco E.M. The role of extreme phenotype selection studies in the identification of clinically relevant genotypes in cancer research. Cancer. 2002;95:1605–1610. doi: 10.1002/cncr.10877. [DOI] [PubMed] [Google Scholar]
- 11.Peloso G.M., Rader D.J., Gabriel S., Kathiresan S., Daly M.J., Neale B.M. Phenotypic extremes in rare variant study designs. Eur J Hum Genet. 2016;24:924–930. doi: 10.1038/ejhg.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amanat S., Requena T., Lopez-Escamez J.A. A systematic review of extreme phenotype strategies to search for rare variants in genetic studies of complex disorders. Genes. 2020;11:987. doi: 10.3390/genes11090987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Escamez J.A., Carey J., Chung W.H., Goebel J.A., Magnusson M., Mandalà M. Diagnostic criteria for Menière's disease. J Vestib Res Equilib Orientat. 2015;25:1–7. doi: 10.3233/VES-150549. [DOI] [PubMed] [Google Scholar]
- 14.Herráiz C., Plaza G., Tapia M.C. Evaluación de la incapacidad en pacientes con acúfenos. Acta Otorrinolaringol Esp. 2001;52:534–538. doi: 10.1016/s0001-6519(01)78247-7. [DOI] [PubMed] [Google Scholar]
- 15.Newman C.W., Sandridge S.A., Jacobson G.P. Psychometric adequacy of the Tinnitus Handicap Inventory (THI) for evaluating treatment outcome. J Am Acad Audiol. 1998;9:153–160. [PubMed] [Google Scholar]
- 16.Perez-Carpena P., Martinez-Martinez M., Martínez Carranza R.A., Batuecas-Caletrio A., Lopez-Escamez J.A. A tinnitus symphony in 100 patients with Meniere's disease. Clin Otolaryngol. 2019;44:1176–1180. doi: 10.1111/coa.13438. [DOI] [PubMed] [Google Scholar]
- 17.Cederroth C.R., Lugo A., Edvall N.K., Lazar A., Bulla J., Uhlen I. Association between hyperacusis and tinnitus. J Clin Med. 2020 Jul;289(8):2412. doi: 10.3390/jcm90824122020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almqvist C., Paul H.A., Leif W.F., Erik G. LifeGene — a large prospective population-based study of global relevance. Eur J Epidemiol. 2011 Jan;26(1):67–77. doi: 10.1007/s10654-010-9521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.May P., Girard S., Harrer M., Bobbili D.R., Schubert J., Wolking S. Rare coding variants in genes encoding GABA a receptors in genetic generalized epilepsies: an exome-based case-control study. Lancet Neurol. 2018;17:699–708. doi: 10.1016/S1474-4422(18)30215-1. [DOI] [PubMed] [Google Scholar]
- 20.Szczepek A.J., Frejo L., Vona B., Trpchevska N., Cederroth C.R., Caria H. Recommendations on collecting and storing samples for genetic studies in hearing and tinnitus research. Ear Hear. Mar/Apr. 2019;40(2):219–226. doi: 10.1097/AUD.00000000000006142019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Depristo M.A., Banks E., Poplin R., Garimella K. V., Maguire J.R., Hartl C. Technical reports a framework for variation discovery and genotyping using next-generation DNA sequencing data. 2011; 43. doi:10.1038/ng.806. [DOI] [PMC free article] [PubMed]
- 22.Li M.X., Gui H.S., Kwan J.S.H., Bao S.Y., Sham P.C. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Li M., Hakonarson H. ANNOVAR : functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010 Sep;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirooznia M., Wang T., Avramopoulos D., Valle D., Thomas G., HuganirSynaptome R.L.D.B. An ontology-based knowledgebase for synaptic genes. Bioinformatics. 2012;28(6):897–899. doi: 10.1093/bioinformatics/bts040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azaiez H., Booth K.T., Ephraim S.S., Crone B., Black-ziegelbein E.A., Marini R.J. Genomic landscape and mutational signatures of deafness-associated genes. Am J Hum Genet. 2018;103:484–497. doi: 10.1016/j.ajhg.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peña-Chilet M., Roldán G., Perez-Florido J., Ortuño F.M., Carmona R., Aquino V. CSVS, a crowdsourcing database of the Spanish population genetic variability. Nucleic Acids Res. 2021;49(D1):D1130–D1137. doi: 10.1093/nar/gkaa794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards S., Aziz N., Bale S., Bick D., Das S. ACMG Standards and guidelines standards and guidelines for the interpretation of sequence variants : a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. 2015: 1-20. [DOI] [PMC free article] [PubMed]
- 29.Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z. an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015 Oct;1531(20):3359–3361. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007 Jan;445(7124):168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 32.Ameur A., Dahlberg J., Olason P. SweGen: a whole-genome data resource of genetic variability in a cross-section of the Swedish population. Eur J Hum Genet. 2017;25:1253–1260. doi: 10.1038/ejhg.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh R., Mazzarotto F., Whiffin N. Quantitative approaches to variant classification increase the yield and precision of genetic testing in Mendelian diseases: the case of hypertrophic cardiomyopathy. Genome Med. 2019;11:5. doi: 10.1186/s13073-019-0616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staining H. Allen cell types databases. Allen Inst. Brain Sci. 2015;88:1–9. [Google Scholar]
- 35.Guitton M.J. Tinnitus : pathology of synaptic plasticity at the cellular and system levels. Front System Neurosci. 2012;6:1–7. doi: 10.3389/fnsys.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaette R, Kempter R. Development of tinnitus-related neuronal hyperactivity through homeostatic plasticity after hearing loss: a computational model. Eur J Neurosci. 2006 Jun;23(11):3124-38. doi: 10.1111/j.1460-9568.2006.04774.x. PMID: 16820003. [DOI] [PubMed]
- 37.Mohler PJ, Gramolini AO, Bennett V. The ankyrin-B C-terminal domain determines activity of ankyrin-B/G chimeras in rescue of abnormal inositol 1,4,5-trisphosphate and ryanodine receptor distribution in ankyrin-B (-/-) neonatal cardiomyocytes. J Biol Chem. 2002 Mar;22277(12):10599–10607. doi: 10.1074/jbc.M110958200. [DOI] [PubMed] [Google Scholar]
- 38.Yang R, Walder-Christensen KK, Kim N, Wu D, Lorenzo DN, Badea A. ANK2 autism mutation targeting giant ankyrin-B promotes axon branching and ectopic connectivity. Proc Natl Acad Sci U S A. 2019 Jul;116(30):15262–15271. doi: 10.1073/pnas.1904348116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuba H., Oichi Y., Ohmori H. Presynaptic activity regulates Na+ channel distribution at the axon initial segment. Nature. 2010;465:1075–1078. doi: 10.1038/nature09087. [DOI] [PubMed] [Google Scholar]
- 40.Swayne LA, Murphy NP, Asuri S, Chen L, Xu X, McIntosh S. Novel Variant in the ANK2 Membrane-Binding Domain Is Associated With Ankyrin-B Syndrome and Structural Heart Disease in a First Nations Population With a High Rate of Long QT Syndrome. Circ Cardiovasc Genet. 2017 Jan;10(1) doi: 10.1161/CIRCGENETICS.116.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cederroth C.R., Pirouzifard M., Trpchevska N., Idrizbegovic E., Canlon B., Sundquist J. Association of genetic vs environmental factors in Swedish adoptees with clinically significant tinnitus. JAMA Otolaryngol Head Neck Surg. 2019;145:222–229. doi: 10.1001/jamaoto.2018.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clifford RE, Maihofer AX, Stein MB, Ryan AF, Nievergelt CM. Novel Risk Loci in Tinnitus and Causal Inference With Neuropsychiatric Disorders Among Adults of European Ancestry. JAMA Otolaryngol Head Neck Surg. 2020 Sep;24146(11):1015–1025. doi: 10.1001/jamaoto.2020.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlee W, Weisz N, Bertrand O, Hartmann T, Elbert T. Using auditory steady state responses to outline the functional connectivity in the tinnitus brain. PLoS One. 2008;3(11) doi: 10.1371/journal.pone.0003720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hedley P.L., Jørgensen P., Schlamowitz S., Wangari R., Moolman-Smook J., Brink P.A. The genetic basis of long QT and short QT syndromes: a mutation update. Hum Mutat. 2009;30:1486–1511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- 45.Chen L., Kass R.S. A-kinase anchoring protein 9 and IKs channel regulation. J Cardiovasc Pharmacol. 2011;58:459–461. doi: 10.1097/FJC.0b013e318232c80c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozcan U., Ozcan L., Yilmaz E., Düvel K., Sahin M., Manning B.D. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell. 2008;29:541–551. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trpchevska N, Bulla J, Prada Hellberg M. Sex-Dependent Aggregation of Tinnitus in Swedish Families. J Clin Med. 2020;9(12):3812. doi: 10.3390/jcm9123812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maas IL, Brüggemann P, Requena T. Genetic susceptibility to bilateral tinnitus in a Swedish twin cohort. Genet Med. 2017;19(9):100–101. doi: 10.1038/gim.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.