Abstract
Background
Sphingolipid metabolism is among the top dysregulated pathways in non-small cell lung carcinomas (NSCLC). However, the molecular control of sphingolipid metabolic reprogramming in cancer progression remains unclear.
Methods
We first determined the correlation between sphingolipid metabolic gene expression and patient prognosis. We then carried out sphingolipidomics analysis of health individual and NSCLC patient sera as well as B3GNT5 and GAL3ST1 genetically perturbed NSCLC cell lines. We used these cell lines to perform tumorigenesis study to determine the cellular role of B3GNT5 and GAL3ST1 in cancer growth and progression.
Findings
The expression of B3GNT5 and GAL3ST1 among sphingolipid metabolic enzymes is most significantly associated with patient prognosis, whilst sphingolipidomics analysis of healthy individual and NSCLC patient sera identifies their metabolites, lacto/neolacto-series glycosphingolipid and sulfatide species, as potential biomarkers that were more effective than current clinical biomarkers for staging patients. Further network analysis of the sphingolipidomes reveals a circular network of coregulated sphingolipids, indicating that the lacto/neolacto-series glycosphingolipid/sulfatide balance functions as a checkpoint to determine sphingolipid metabolic reprograming during patient progression. Sphingolipidomics analysis of B3GNT5/GAL3ST1 genetically perturbed NSCLC cell lines confirms their key regulatory role in sphingolipid metabolism, while B3GNT5 and GAL3ST1 expression has an opposite role on tumorigenesis.
Interpretation
Our results provide new insights whereby B3GNT5 and GAL3ST1 differentially regulate sphingolipid metabolism in lung cancer growth and progression.
Funding
This work was supported by the Natural Science Foundation of China (81872142, 81920108028); Guangzhou Science and Technology Program (201904020008); Guangdong Science and Technology Department (2020A0505100029, 2019A1515011802, 2020A1515011280, 2020B1212060018, 2020B1212030004); China Postdoctoral Science Foundation (2019M650226, 2019M650227).
Keywords: B3GNT5: GAL3ST1, Lung cancer, Sphingolipidomics, Tumor growth
Research in Context.
Evidence before this study
Sphingolipid metabolism reprogramming is among the top dysregulated pathways in NSCLC patients, while the molecular control of sphingolipid metabolic reprogramming in cancer progression remains unknown. Furthermore, it remains unclear whether the alteration in NSCLC patients’ serum sphingolipidomes correlates with patient progression.
Added value of this study
Here, we first show that the expression of B3GNT5 and GAL3ST1 among sphingolipid metabolic enzymes is most significantly correlated with patient prognosis, while their metabolites, lacto/neolacto-series glycosphingolipid and sulfatide, function as markers for stratifying NSCLC patients. The balance of lacto/neolacto-series glycosphingolipid/sulfatide level works as a checkpoint to determine sphingolipid metabolic reprograming in cancer progression. At the cellular level, we show that expression of B3GNT5 or GAL3ST1 regulates sphingolipid metabolism and tumorigenesis.
Implications of all the available evidence
All the available evidence show that the change in tumor sphingolipid metabolic enzyme expression directly orchestrates the serum levels of their metabolites in NSCLC patients, indicating that our sphingolipid profiling approach could be a valuable diagnostic tool.
Alt-text: Unlabelled box
Introduction
Lung cancer is the leading cause of cancer-related death in the world and has a poor prognosis because most of the patients are not diagnosed until at advanced or metastatic stages [1]. In particular, non-small cell lung carcinoma (NSCLC), which accounts for 85–90% of newly diagnosed lung cancers, is often diagnosed at the metastatic stage, with median survival of just 1 year [2,3]. Due to lack of an effective method and biomarker for early diagnosis and cancer staging, together with poor understanding of its progression mechanism, its 5-year survival rate remains <15% across all stages of NSCLC [4]. It is therefore a clinically unmet need to identify effective biomarkers and therapeutic targets for NSCLC patients.
Genome-wide studies have ranked sphingolipid metabolism as one of the top dysregulated pathways in pathophysiological states of the lung cancer patients, suggesting that harnessing and restoring sphingolipid homeostasis in the patients could be an effective therapeutic approach to tackle the disease [5]. Sphingolipids are a class of lipids containing a backbone of sphingoid bases, which can be either phosphorylated, acylated, glycosylated or sulfated by specific metabolic enzymes, in order to produce different subclasses and species of the sphingolipids [6]. They are known to collaboratively regulate cell membrane biology, while some of them can function as signal molecules to regulate cell survival, proliferation and migration [7]. For instance, ceramide (Cer), the key subclass of sphingolipid metabolism, is shown to regulate cell apoptosis, while ceramide-1-phosphate (C1P), sphingosine-1-phosphate (S1P), glycosphingolipid (GSL) (including lacto/neolacto-series glycosphingolipid) and sulphated galactoceramide (also known as sulfatide), have been implicated in cell proliferation and survival [6,8,9]. Although the term “sphingolipid rheostat” was proposed to describe the ability of ceramide and S1P to differentially regulate cell growth and survival [10], administration of C2 and C6 ceramides showed limited benefits in a phase II cutaneous breast cancer study [11]. Alternatively, small inhibitors were developed to target S1P metabolic enzyme, sphingosine kinase type 1 (SPHK1) in order to reduce S1P production, but they have shown mixed results in preclinical animal models [12]. Collectively, these findings prompt us to re-examine sphingolipid metabolic pathway in order to identify the key metabolic enzymes that are responsible for sphingolipid metabolic reprogramming in cancer growth and progression.
In this study, we sought to explore the molecular control of sphingolipid metabolic reprogramming in cancer growth and progression. We first provided clinical evidence that NSCLC patients with high beta-1,3-N-acetylglucosaminyltransferase 5 (B3GNT5) or low galactose-3-O-sulfotransferase 1 (GAL3ST1) expression correlated strongly with poor prognosis, while sphingolipidomics analysis of healthy individual and NSCLC patient sera showed that their sphingolipid metabolites were more specific and effective than currently available biomarkers in staging NSCLC patients. Network analysis revealed the circular organization of a sphingolipid coregulatory network, indicating that the level of lacto/neolacto-series glycosphingolipids and sulfatide species dictates sphingolipid metabolic reprogramming in cancer progression. Finally, combining sphingolipidomics with genetic perturbation of B3GNT5/GAL3ST1 in NSCLC cell lines confirmed their regulatory roles in sphingolipid metabolism, while B3GNT5 and GAL3ST1 expression differentially regulated cancer cell migration, invasion, tumor sphere formation and in vivo tumor growth.
Collectively, our data uncover the lacto/neolacto-series glycosphingolipid/sulfatide balance as a checkpoint to determine sphingolipid metabolic reprogramming in cancer growth and progression, and that is controlled by B3GNT5/GAL3ST1 expression.
Methods
Human clinical specimens and ethnics
Serum samples were collected from healthy individuals and treatment naive NSCLC patients (age-matched and body-mass index–matched) with informed consent, and the research program and all the related procedures were approved by the Ethics Committee of Sun Yat-sen Memorial Hospital (Guangzhou, China) and stored according to standard operating procedures for Good Clinical Laboratory Practice standards (Ref no.: 2020-108). The 8th Edition Lung Cancer Stage Classification system was used to classify the NSCLC patients with adenocarcinoma in our cohort. A total of 100 healthy individuals and 100 NSCLC patients (subset of stage Ⅰ patients (n = 63), subset of stage Ⅱ/Ⅲ patients (n = 37)) samples were analyzed as a training dataset in this study. A total of 100 healthy individuals and 100 NSCLC patients (subset of stage Ⅰ patients (n = 70), subset of stage Ⅱ/Ⅲ patients (n = 30)) samples were analyzed as a validation dataset in this study. The clinicopathological features of NSCLC patients and healthy individuals were shown in Supplementary Table 1. Power calculation was performed for the training cohort using the assumptions given by Gpower software (https://stats.idre.ucla.edu/other/gpower/) [13].: α of 0.05; power (1-β) of 0.80; and effect size of 0.3. Test family, t test, statistical test, Correlation:Point biserial model. The calculated total sample size was 82.
Clinical study using online databases from GEO, TCGA and Hou lung datasets
To identify all relevant datasets, we searched GEO for NSCLC expression profiling studies. Studies were included in the systematic review if (i) they were gene profiling studies in patients with NSCLC; (ii) they used NSCLC tissue and normal lung tissue for comparison; (iii) they used the same platform; (iv) they contained more than three samples meeting the quality control standard in experimental and controlled group. As a result, 4 panels of NSCLC gene expression datasets were included: GSE19188, GSE27262, GSE18842 and GSE30219 [14], [15], [16], [17]. Effectivity measures considered included: objective response, 1- and 2-year survival. Maentel-Haenszel combined odds ratio (OR) was estimated in the metanalysis. The statistical analysis of effectiveness across categories was performed using a one-way analysis of variance (ANOVA). Differences were statistically considered for p values = 0.01. All results obtained were weighted according to number of patients [18]. For the Hou lung dataset, it was obtained by using assession code GSE19804 [19]. The Cancer Genome Atlas (TCGA) employs high-throughput techniques to assist the understanding of the genetic basis of diseases. These databases mainly provide 3 types of transcriptional expression data, including Counts, the Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) and the upper quartile FPKM(FPKMUQ). Among these three types of data, Count is the raw data without any pre-processing operation. FPKM and FPKM-UQ data are obtained by using different normalization methods. We used the Count data for differential gene calculation. In order to eliminate the influence of gene length, we mainly used FPKM data rather than raw Counts for NSCLC subtype classification. We excluded samples without sufficient expression data in the study [20].
RNA isolation and RT-PCR condition
RNA was isolated using RNAasy Kit (Cat# AM9738, Invitrogen-Thermo Fisher Scientific, USA) and was reverse transcribed using 5×All-In-One MasterMix (with AccuRT Genomic DNA Removal kit) (Cat# G492, Applied Biological Materials, Canada) following the procedure our lab published before [21]. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using LightCycler 480 (Roche, Switzerland) in technical triplicates analyzed on LightCycler 480 Software Version 1.5.1 from Roche. Results were normalized to the gene β-actin. To verify the gene expression of online database, we tested 10 couples of normal and cancer tissues from non-small cell lung carcinoma patients. ASAH1 (N-acylsphingosine amidohydrolase 1); ACER3 (alkaline ceramidase 3); B3GNT5 (UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5); B4GALT6 (beta-1,4-galactosyltransferase 6); CERK (ceramide kinase); CERS1 (ceramide synthase 1); CERS2 (ceramide synthase 2); CERS3 (ceramide synthase 3); CERS4 (ceramide synthase 4); CERS5 (ceramide synthase 5); CERS6 (ceramide synthase 6); CERT1 (Ceramide transporter 1); GAL3ST1 (galactose-3-O-sulfotransferase 1); GAL3ST2 (galactose-3-O-sulfotransferase 2); GAL3ST4 (galactose-3-O-sulfotransferase 4); GALC (galactosylceramidase); GBA (glucosylceramidase beta); GBA2 (glucosylceramidase beta 2); SGPL1 (sphingosine-1-phosphate lyase 1); SGPP1 (sphingosine-1-phosphate phosphatase 1); SGPP2 (sphingosine-1-phosphate phosphatase 2); SMPD1 (sphingomyelin phosphodiesterase 1); SMPD2 (sphingomyelin phosphodiesterase 2); SMPD3 (sphingomyelin phosphodiesterase 3); SMPD4 (sphingomyelin phosphodiesterase 4); SPHK1 (sphingosine kinase 1); SPHK2 (sphingosine kinase 2); SPTLC1 (serine palmitoyltransferase long chain base subunit 1); SPTLC2 (serine palmitoyltransferase long chain base subunit 2); UGCG (UDP-glucose ceramide glucosyltransferase); UGT8 (UDP glycosyltransferase 8) primers were purchased from Gene WIZ (Suzhou, China). mRNA expression was measured by RT-PCR. The primer sequences were given below:
The sequences of RT-PCR primers.
| Gene | Direction | Primer set |
|---|---|---|
| ASAH1 | F | AACTCGATGCTAAGCAGGGTA |
| R | GCGATCATCAAGGAAGAAGGG | |
| ACER3 | F | GTGCCTTTAACCCCAGAGTCT |
| R | GTGCAGTCAGCTCGTCCAA | |
| B3GNT5 | F | TTCAAGACTTTTGGATTGGTCGT |
| R | CGGCTGTGTAGTCAGGGTAAG | |
| B4GALT6 | F | TCATTCCTTTCCGTAATCGCC |
| R | GAAAAGCATCGCACGGTTAAAA | |
| CERK | F | TATCAACCCGTTTGGAGGAAAAG |
| R | ATGGAGGCTAAGGTGAACAGT | |
| CERS1 | F | TGTGGGCATCCTTGTGCTC |
| R | GAGGCGGAACCAGAACCAG | |
| CERS2 | F | GGTAGAGCGTTGGTTCCGTC |
| R | GGCAATGAAGGCAATCAGGTAA | |
| CERS3 | F | AACATTCCACAAGGCAACCAT |
| R | GACTCCTAAACCATCTTTCCACC | |
| CERS4 | F | CTGTCCAGTTTCAACGAGTGG |
| R | TCCCGGTCTTCTAGCTCTGTC | |
| CERS5 | F | TCGCCATCGGAGGAATCAG |
| R | CCAAGGTGACGACCAGAGAAA | |
| CERS6 | F | GGACCACAAATTGCTCCGC |
| R | GGCTTCTCCTGATTGCGTCT | |
| CERT1 | F | AAAGGCCACAGTTTACGTGAG |
| R | CTTCTGTAGCGTGTCAACTTGT | |
| GAL3ST1 | F | CAAGACCCGGATCGCTACTAC |
| R | TGTCATAGCCCAGGTCGAAGA | |
| GAL3ST2 | F | GGCGGCTTGCAGAGATACTT |
| R | GCGGTAGAGGATGTTGAGCAC | |
| GAL3ST4 | F | CTACGCTCGCAACTTACTATGG |
| R | ACAGTGGAAACAGGATGGATGA | |
| GALC | F | TATTTCCGAGGATACGAGTGGT |
| R | CCAGTCGAAACCTTTTCCCAG | |
| GBA | F | GCTGTTGAGTGGATACCCCTT |
| R | GGCGGACATTGTGGTGAGT | |
| GBA2 | F | CAAGCTAACAACGTCTCCCTAAG |
| R | GATCATGTCGATGAAAGGTGTCT | |
| SGPL1 | F | CCTAGCACAGACCTTCTGATGT |
| R | ACTCCATGCAATTAGCTGCCA | |
| SGPP1 | F | CCATTTGTGGACCTGATTGACA |
| R | ACTTCCTAGTATCTCGGCTGTG | |
| SGPP2 | F | TCACCGCACTCCTCATCGT |
| R | CCGGGTTGGGCTGTAGTAATC | |
| SMPD1 | F | CCAGGTTACATCGCATAGTGC |
| R | TGATGGCGGTGAATAGACCTTT | |
| SMPD2 | F | GCTCCATCTAAGTGGCATGGT |
| R | TCTTCTGGGTGCATGTTGAGG | |
| SMPD3 | F | GCTGCCCTTTGCGTTTCTC |
| R | TCCAGCCGTGAATAGATGTAGG | |
| SMPD4 | F | GACAGTCCTCTGTACCACAACA |
| R | CGAACGGATTCAGGGCCAA | |
| SPHK1 | F | GGCTGCTGTCACCCATGAA |
| R | TCACTCTCTAGGTCCACATCAG | |
| SPHK2 | F | ATGGCATCGTCACGGTCTC |
| R | CTCCCAGTCAGGGCGATCTA | |
| SPTLC1 | F | GGTGGAGATGGTACAGGCG |
| R | TGGTTGCCACTCTTCAATCAG | |
| SPTLC2 | F | TGCTCACGTATGTGGGGTATG |
| R | GATTGGCCGATTCCAGTTGTC | |
| UGCG | F | GAATGGCCGTCTTCGGGTT |
| R | AGGTGTAATCGGGTGTAGATGAT | |
| UGT8 | F | CTGTTGGGATAGCGAAGGCTG |
| R | GTGGCCTCTCTCGTGCAAG |
Clinical study performed by using the Kaplan–Meier plotter database
The prognostic significance of the mRNA expression of topoisomerase family genes in NSCLC was evaluated using the Kaplan-Meier plotter (www.kmplot.com), an online database including gene expression data and clinical data. With the purpose to assess prognostic value of a specific gene, the patient samples were divided into two cohorts according to the median expression of the gene (high vs. low expression). We analyzed the overall survival (OS) of NSCLC patients by using a Kaplan-Meier survival plot. Briefly, the 24 genes were uploaded into the database respectively to obtain the Kaplan-Meier survival plots, in which the number-at-risk was shown below the main plot. Log-rank p-value and hazard ratio (HR) with 95% confidence intervals were calculated and displayed on the webpage. Then, we exported plot data as text and plotted the Kaplan-Meier survival curve using Graphpad Prism software. The desired Affymetrix ID of each gene in NSCLC was valid. In this study, “array quality control” was selected “exclude outlier arrays”. Affymetrix ID: ASAH1 (210979_at), ACER3 (222687_s_at), B3GNT5 (1554835_a_at), B4GALT6 (235333_at), CERK (218421_at), CERS1 (229448_at), CERS2(222212_s_at), CERS3 (1554252_a_at), CERS4 (218922_s_at), CERS5 (239491_at), CERS6 (212446_s_at), CERT1 (219625_s_at), GAL3ST1(205670_at), GAL3ST2 (1553046_s_at), GAL3ST4(219815_at), GALC, GBA, GBA2 (223921_s_at), SGPP2(226560_at), SMPD1, SMPD2 (205622_at), SMPD3 (219695_at), SMPD4 (207856_s_at), SPHK1(219257_s_at), SPHK2 (209857_s_at), SPTLC1 (202277_at), SPTLC2 (203127_s_at), and UGT8 (208358_s_at).
Chemicals
Water (LC-MS grade) (Cat# 85189, Thermo Scientific, USA), methanol (MeOH, LC-MS grade) (Cat# AA47192K2, Alfa Aesar-Fisher Chemical, USA), isopropanol (IPA, LC-MS grade) (Cat# MB1241, Fisher Chemical, USA), chloroform (CHCl3, HPLC grade) (Cat# C607SK-4, Fisher Chemical, USA), formic acid (LC-MS grade) (Cat# 28905, Thermo Fisher Scientific), acetic acid (LC-MS grade) (Cat# 984303, Thermo Fisher Scientific), ammonium acetate (purity ≥ 98%) (Cat# AM9070G, Thermo Fisher Scientific) were purchased from Thermo Fisher Scientific (USA). Potassium hydroxide (KOH, purity ≥ 85%) (Cat# 221473) were purchased from Sigma-Aldrich (Germany). HyperSep silica gel SPE column (200 mg, 3 mL) (Cat# 60108-410, Thermo Fisher Scientific) were purchased from Thermo Fisher Scientific. The LIPID SEARCH internal standard cocktail (Cer/Sph Mixture Ⅱ, 25 μM each of 9 compounds in ethanol) (Cat# LM6005, Avanti Polar Lipids, USA) was purchased from Avanti Polar Lipids. It was composed of uncommon sphingolipids which included 17-carbon chain length sphingoid base analogs C17-sphingosine [So (d17:1)], C17-sphinganine [Sa (d17:0)], C17-sphingosine-1-phosphate [S1P (d17:1)], C17-sphinganine-1-phosphate [Sa1P (d17:0)], C18-ceramide [Cer (d18:1/12:0)], C18-ceramide-1-phosphate [C1P (d18:1/12:0)], C18-sphingomyelin [SM (d18:1/12:0)], C18-glucosylceramide (d18:1/12:0), C18-lactosylceramide [LacCer (d18:1/12:0)]. Sulfatide (d18:1/24:0) (Cat# 860578, Avanti Polar Lipids), C16 Galactosyl (α) Dihydroceramide (d18:0/16:0) (Cat# 860730, Avanti Polar Lipids), C17:0 globotriaosylceramide (GB3) (Cat# 860699, Avanti Polar Lipids).
Serum sample collection and processing
The blood samples were collected and processed by strictly following standard operating procedures for serum and plasma collection procedure [22]. Briefly, the blood samples were collected from patients at a fasting state, which were added into serum separator tubes and gently inverted the tubes 8–10 times. The blood samples were then allowed to clot after leaving tube undisturbed at room temperature for 20 min. The tubes were soon after centrifuged at 3000 × g for 30 min at 4 °C. Afterwards, the liquid components (serum) were transferred into sterile Eppendorf tube and frozen at -80 ℃ for next step experiments. To avoid the hemolyzed serum samples, spectrophotometer (NanoDrop™ One, Thermo Fisher Scientific) was used to measure the oxyhemoglobin absorbance of our processed samples at k = 414 nm. For those hemolyzed serum samples that showed a peak at 414 nm, they would not be used for next step analysis.
Serum sphingolipid extraction
Total sphingolipid extract was prepared following the procedures developed in previous studies [23]. Briefly, 100 μl serum of NSCLC patient or healty individuals was accurately weighed into a glass bottle, in which 1.5 ml of chloroform (CHCl3) /MeOH (1:2, v/v) and internal standard C17 Ceramide (d18:1/17:0) (Cat# 860517, Avanti Polar Lipids), sphinganine-D7 (Cat# 860658, Avanti Polar Lipids) and sphingosine1-phosphate-D7 (Cat# 860641, Avanti Polar Lipids) was added. The mixture was vortex 15 min and incubated at 48 °C for 12 h. Then, 75 μl of KOH in MeOH (1M) was added and were incubated at 37 °C for 2 h to cleave potentially interfering glycerolipids. After neutralization by formic acid and centrifuged at 3800 rpm for 8 min, the supernatant was collected and dried by nitrogen blowing (Savant SPD1010 SpeedVac Concentrator) (Thermo Fisher Scientific, USA).
Chromatographic conditions
Chromatographic separation was performed using a Dionex Ultimate 3000 RS UHPLC system (Thermo Fisher Scientific, USA), equipped with a binary solvent delivery system and a standard auto-sampler. A Hyperil Gold C18 column (100 × 2.1 mm, 1.9 um; Thermo Fisher Scientific, USA) was used to separate sphingolipids from the patients’ serums. The mobile phase consisted of (A) MeOH (methanol)/H2O/HCOOH (formic acid) (60:40:0.2, v/v/v) and (B) MeOH/IPA (isopropyl alcohol)/HCOOH (60:40:0.2, v/v/v), both containing 10 mM NH4OAc. A linear gradient was optimized as follows (flow rate, 0.3 mL/min): 0min, 0%B; 3 min, 10% B; 5 min, 40% B; 5.3 min, 55% B; 8 min, 60% B; 8.5 min, 80% B; 10.5 min, 80% B; 16 min, 90% B; 19 min, 90% B; 22 min, 100% B. The injection volume was 1 μl, and the column temperature was maintained at 40 ℃ for each run. A typical run time was 25 min. Qualitative and quantitative analysis was performed using Q Extractive mass spectrometer (Thermo Fisher Scientific, USA). Parameters included full scan/data dependent ms2 Top 20 analysis mode, ESI mode for positive analysis. Spray voltage + 3500V / -3000V, sheath gas is 40 arb, auxiliary gas is 10 arb, ion transfer tube temperature is 320 ℃ auxiliary gas heating temperature is 350 ℃ full scan first stage full scan resolution is 70,000, mass range is 100–1500 m/z; Data Dependent Analysis, Data depends on ms2 secondary resolution of 17,500, top 10, collision energy of 10, 20, 40, mass range of 50–3000 m/z.
Lipid nomenclature
Lipid nomenclature in this study follows the classification system used in LIPID MAPS (Lipidomics Gateway), annotation of ceramide moiety of glycosphingolipid denotes hydroxyl-group number (d or t means two or three hydroxyl groups) in sphingoid backbone, carbon number of sphingoid backbone, double bond number in sphingoid backbone, carbon number of N-acyl chain, double bond number in N-acyl chain, and hydroxyl-group number in N-acyl chain (e.g., in d18:1/24:0(OH), OH means one hydroxyl group); and annotation of headgroup denotes sugar residues composition and sequence (e.g., Fuc, Hex and GalNAc mean fucose, glucose/galactose and N-acetyl-D-galactosamine, respectively; Cer means ceramide) [24].
Sphingolipidomics analysis
Serum sphingolipidomics data was applied to run our processed serum samples according to the manufacturer's instruction and software program [23]. Briefly, sphingolipids were identified using internal standard LipidSearch database (Thermo Fisher Scientific, USA) and quantified by using internal standard, based on their accurate precursor ion mass and fragment features. After performing untargeted lipidomic analysis for identifying sphingolipids, the standard curve was used to calibrate the linear range of these identified sphingolipids, and used targeted MS/MS to verify our idenitified sphingolipids (Supplementary Table 2). The results, including sphingolipid names, sample names, and concentrations, were imported into SIMCA-P+ 14.1 software (Sartorius, Sweden) for multivariate statistical analysis. Principal component analysis (PCA) was used to visualize general clustering between controls and models. Partial least-squares discriminant analysis (PLS-DA) and OPLS-DA was carried out to identify the differentially expressed sphingolipids responsible for two groups (NSCLC patients vs healthy individuals or early stage (stage Ⅰ) vs advanced stage (stage Ⅱ/Ⅲ) NSCLC patients)
Annotation enrichment analysis
Annotation enrichment analysis was performed on the differentially expressed genes (at p < 0.05 and an absolute log2 (fold-change) in expression ≥ 1.5, student's t test) among tumors and normal tissues. Annotation enrichment analysis was calculated using the web service DAVID (https://david.ncifcrf.gov/) with default settings as previously described [25].
Sphingolipidomics data normalization
The normalization of sphingolipidomics results were based on internal standard. Values were averaged over the three technical replicates, and log2 transformed against the corresponding average abundance measured in NSCLC patients or healthy individuals.
Sphingolipid coregulation network analysis
Sphingolipid coregulation circular network analysis was done as previously described [26]. Briefly, the sphingolipid coregulation study was done by using an R package in order to carry out weight correlation network analysis (WGCNA). The gradient method was used to test the independence and average connectivity of different modules with different power values (ranging from 1 to 20). An appropriate power value of 7 is determined when the degree of independence is 0.7. Once the power value was determined, module construction was continued by the WGCNA algorithm, which also identifies the common representation module. The threshold of 0.7 was tested and confirmed to be enough to leave out the single data that most contributed to any one correlation value, and not determined by any single serum sample. By using the implementation of Cytoscape together with various natural network layout algorithms, the circularity of sphingolipid coregulatory network was confirmed to be accurate in the selected network layout optimization algorithm. The network of “edge” and “node” generated from WGCNA were visualized complex networks and integrating these with any type of attribute data by Cytoscape.
Western blot analysis
Western blots were performed as previously described [27]. Briefly, cell lines were harvested and lysed with RIPA lysis buffer (Cat# 89900, Thermo Fisher Scientific, USA). Protein concentration was determined by using the Pierce BCA Protein assay kit (Cat# 23225, Thermo Fisher Scientific). 10–30 μg of protein from each sample was loaded onto 8-12% polyacrylamide gels. The protein was then transferred to a nitrocellulose membrane and incubated 1h in 5% milk Tris-buffered saline with 0.1% Tween-20 (TBS-T), followed by an overnight incubation of primary antibody diluted at 1:250 in 2.5% milk in TBS-T for 1 h at room temperature. The following primary antibodies were used: rabbit polyclonal anti-B3GNT5 (Cat# Ab238819, Abcam, USA), rabbit polyclonal anti-GAL3ST1 (Cat# Ab232758, Abcam, USA), mouse monoclonal anti-GAPDH (6C5) (Cat# sc-32233, Santa Cruz Biotechnology, USA). Chemiluminescence was detected by using the ECL detection system (Bio-Rad, USA).
Cell lines and transfection
A549 (Human lung adenocarcinoma cell) (Cat# CRL-7909, RRID:CVCL_0023), and H1299 (Human lung large cell carcinoma cell) (Cat# CRL-5803, RRID:CVCL_0060) cell lines were obtained from the American Type Culture Collection (ATCC, USA). PC9 (Human lung adenocarcinoma cell) (Cat# 90071810) cell line was obtained from Sigma-Aldrich (Germany). HCC827 (Human lung adenocarcinoma cell) (Catl# TCHu153) cell line was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell lines were cultured in DMEM (Cat# C11995500BT, Gibco-Thermo Fisher Scientific, USA) or RPMI1640 (Cat# C11875500BT, Gibco) supplemented with 10% fetal bovine serum (Cat# 11054020, Gibco) and 1% Penicillin and Streptomycin (Cat# 15140122, Gibco) at 37 °C in a 5% CO2 incubator. To generate stable B3GNT5 or GAL3ST1 expressing cells, cancer cells were transfected with either pspax2-B3GNT5 vector (Cat# LPP-CS-W0127-Lv205-01-400; Genechem, Shanghai, China), pspax2-GAL3ST1 vector (Cat# LPP-CS-A6425-Lv205-01-400, Genechem, Shanghai, China), or the empty vector alone, using Lipofectamine 3000 (Cat# L3000015, Invitrogen-Thermo Fisher Scientific, USA) according to the manufacturer's instruction, which were then subjected to puromycin(Cat# ant-pr-1, InvivoGen, USA) selection (5 μg/mL) for 2 weeks. To generate stable B3GNT5 or GAL3ST1 knock-down cell lines, cancer cells were transfected with pSuper-retro-neo-shB3GNT5 or pSuper-retro-neo-shGAL3ST1 (Genechem, Shanghai, China) or the scramble control vector, using Lipofectamine 3000 and were selected with neomycin (0.5 mg/mL) (Cat# N1142, Sigma-Aldrich, Germany) for 2 weeks. For siRNA transfection, cancer cells were transiently transfected with either 100 nM non-silencing control (NSC) or B3GNT5/GAL3ST1 targeted siRNA molecules (a mixture of 3 siRNA targeting sequences for each gene; purchased from GenePharma) using Lipofectamine RNAiMAX transfection reagent (Cat# 13778150, Invitrogen-Thermo Fisher Scientific, USA) according to the manufacturer's instruction for 48 h. The transfected cells were then used for a range of in vitro assays.
Cell proliferation, colony formation, migration, invasion assays and sphere formation assay
Cell proliferation assay was performed by using Cell Counting Kit-8 (CCK-8) (Cat# CK04, Dojindo Molecular Technologies, Japan) according to the manufacturer's instructions. Briefly, cells were seeded into 96-well plates at a density of 1 × 103 cells/well with 100 μl culture medium. After culturing for 24 h, 10 μl CCK-8 reagent was added to each well. Similarly, 100 μl complete medium containing 10 μl CCK-8 solution was added to respective wells at different time points (12, 24, 36, 48 and 60 h). The plates were incubated in dark at 37 °C for 2 h, and absorbance at 450 nm wavelength was measured. Independent experiments were repeated for five times.
For colony formation assays, cancer cells (500 cells/well) were plated in each well of a six-well cell culture plate. At day 14, the plates were fixed in 4% paraformaldehyde, stained with 1% crystal violet, and the numbers of colonies were then counted under a AID vSpot Spectrum microscope (AID, Germany).
For wound-healing assays, the wounds were made by dragging a plastic pipette tip across the cell surface after cells grew to 90% confluence in 6-well plates. The remaining cells were washed by using PBS, and then incubated at 37 °C with serum-free medium. Migrating cells at the wound front were photographed and compared at indicated time using Image J software.
For Invasion assays, the assays were performed using a chamber system with membrane pre-coated with matrixgel (8 μm pore size) (Cat# 354480, BioCoat-Corning, USA). 3 × 104 cells were placed in the upper chamber with 0.1 ml of serum-free medium, whereas the lower chamber of a 24-well plate was loaded with 0.5 ml of medium containing 10% fetal bovine serum. After 24 h incubation, the cells were fixed with 4% paraformaldehyde (Cat# G1101, Servicebio, Wuhan, China) and then counterstained with 0.1% crystal violet (Cat# G1063, Solarbio, Beijing, China), which were then counted under a microscope.
For sphere formation assays, a total of 500 single cells were plated in 6-well clear flat bottom ultra-low attachment plates (Cat# 3471, Corning, USA). The cells were grown in Dulbecco's modified Eagle medium/F12 (Cat# 12634010, Gibco, USA) supplemented with 20 ng/mL both EGF (Cat# 315-09, PeproTech, USA) and bFGF (Cat# 450-33, PeproTech, USA), and 1 x B-27 supplement (Cat# 17504044, Gibco, USA). After 5 days, the formed tumor spheres were imaged under a microscope and the sizes of tumor spheres were measured by image J.
Xenograft tumor model
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Sun Yat-sen University (Approved No. SYSU-IACUC-2020-B0362) and met the ARRIVE guildlines. Female nude mice (BALB/c nu/nu, 4 weeks old), which were purchased from the Laboratory Animal Center of Sun Yat-Sen University (Guangzhou, China), were injected with 3×106 of either stable B3GNT5 expressing A549 cells, empty vector transfected A549 cells, stable GAL3ST1 expressing PC9 cells or empty vector transfected PC9 cells. Tumor volumes were monitored by using calipers every 3-4 days. Tumor volume (V) was calculated: V = (Length x width2) x 0.52. After 31 to 49 days, animals were culled, tumors excised and fixed in formalin overnight and then paraffin-embedded for further analysis.
General statistics
All violin plots and bar charts are presented as mean ± s.e.m. Statistical significances were calculated using GraphPad Prism (GraphPad Software, USA). p-values were calculated with student's t test, unless otherwise indicated. Correlation values given were Pearson's linear correlation coefficients. The quality or performance of the predicted models were evaluated by developed in previous studies[28]. In details, we used the receiver operating characteristic (ROC) curve, which was obtained by calculating the sensitivity and specificity of the test at every possible cut-off point, and plotting sensitivity (the proportion of true positive results) against 1-specificity (the proportion of false positive results) as described previously [29]. Briefly, the area under the ROC curve (AUC) was a reflection of how good the potential biomarker was at distinguishing NSCLC patients and healthy individuals. The AUC served as a single measure, independent of prevalence, that summarized the discriminative ability of a test across the full range of cut-offs. The closer the AUC to 1, the better the overall diagnostic performance of the biomarker, and the closer it to 0.5, the poorer the test. A rough guide for classifying the accuracy of a diagnostic test is the traditional academic point system: AUC values between 0.90–1.0 is excellent, 0.80–0.90 is good, 0.70–0.80 is fair, 0.60–0.70 is poor, 0.50–0.60 is failed.
Role of the funding source
The corresponding author confirmed that the funders had no role in study design, data collection and interpretation or writing of report, while he possessed full access to all the data generated in the study and held the full responsibility for the paper submission.
Results
Altered expression pattern of sphingolipid metabolic pathway genes correlates strongly with poor prognosis in NSCLC patients
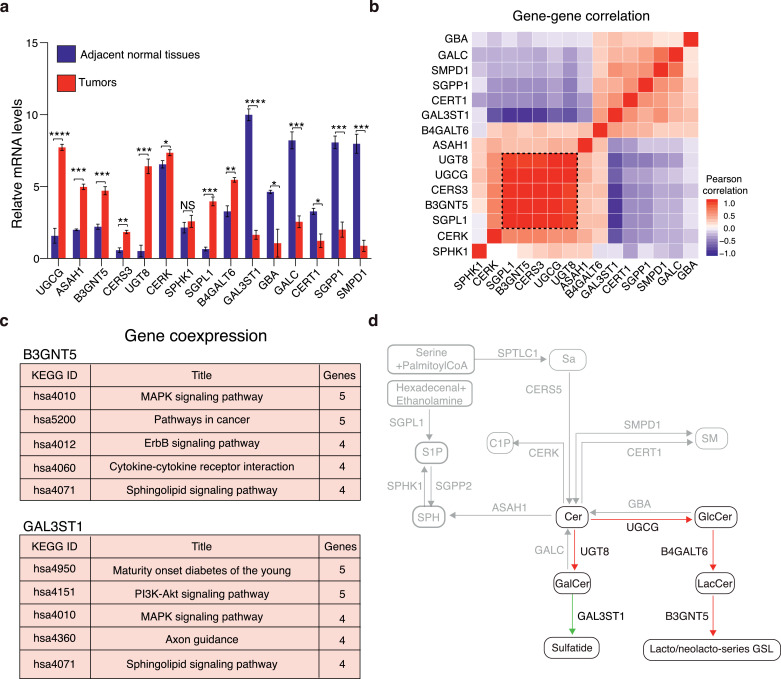
In this study, sphingolipids and shorthand are named using common, officially accepted terms and builds upon the LIPID MAPS terminology [30,31]. To determine the clinical relevance of sphingolipid metabolic gene expression in NSCLC patients, we selected 31 key metabolic genes based on the sphingolipid metabolic pathway (https://www.kegg.jp/kegg-bin/show_pathway?map00600), particularly focusing on ceramide metabolism and its downstream sphingomyelin (SM), ceramide-1-phosphate (C1P), sphingosine-1-phosphate (S1P), galactosylceramides (GalCer), sulfatide, glucosylceramide (GlcCer), lactosylceramide (LacCer) and lacto/neolacto-series glycosphingolipid (lacto/neolacto-series GSL) synthesis network, including several poorly studied genes and their related metabolites [6,32]. We analyzed mRNA expression from three different studies, including the Cancer Genome Atlas (TCGA), Gene Expression Omnibus dataset (GEO) and Hou lung dataset. Mapping our expression results onto the known sphingolipid metabolic pathway indicated that 15 of these genes were significantly dysregulated in cancer tissues compared to normal lung tissues (between p < 0.01–p < 0.0001, student's t test) (Fig. 1 a, Supplementary figure 1 a). Apart from ceramide synthase 3 (CERS3), ceramide kinase (CERK), sphingosine kinase 1 (SPHK1), N-acylsphingosine amidohydrolase 1 (ASAH1), sphingosine-1-phosphate lyase 1 (SGPL1), sphingosine-1-phosphate phosphatase 1(SGPP1) that involved in the synthesis or hydrolysis of sphingolipids (Cer, C1P, S1P, SPH etc.), most of the up-regulated sphingolipid metabolic genes in cancer tissues were related to the metabolism of glycosylated- or sulfated-sphingolipids, for instance, GlcCer by UDP-glucose ceramide glucosyltransferase (UGCG), LacCer by beta-1,4-galactosyltransferase 6 (B4GALT6), lacto/neolacto-series glycosphingolipid by beta-1,3-N-acetylglucosaminyltransferase 5 (B3GNT5), GalCer by UDP glycosyltransferase 8 (UGT8) etc. In contrast, genes involved in synthesis/degradation of either sulfatide by galactose-3-O-sulfotransferase 1 (GAL3ST1), S1P by SGPP1 or sphingomyelin (SM) by sphingomyelin phosphodiesterase 1 (SMPD1) were significantly down-regulated in cancer tissues as compared to normal lung tissues. Importantly, gene-gene expression correlation study showed that the down-regulated gene, GAL3ST1, was negatively correlated to either of the following up-regulated genes, including UGCG, B3GNT5, B4GALT6 or UGT8, whilst these up-regulated genes were positively correlated to each other (Fig. 1 b), suggesting that these correlated genes may coregulate sphingolipid metabolism in NSCLC patients.
Fig. 1.
Alteration of sphingolipid metabolic gene expression that involved in the metabolism of bioactive sphingolipids strongly correlates with poor prognosis in NSCLC patients. (a) Metabolic network shows selected sphingolipid metabolic reactions (Kyoto Encyclopedia of Genes and Genomes pathway, KEGG), together with main metabolites (rounded rectangles) and 15 selected sphingolipid metabolic genes (rectangles). Heatmap analysis of the RNA-seq datasets obtained from TCGA, GEO and Hou lung indicates a consistent alteration of 15 sphingolipid metabolic gene expression in NSCLC patient tissues as compared to the normal lung tissues, across all three databases. (GEO: n = 70 normal lung tissues, n = 1251 tumor tissues; TCGA database: n = 59 normal lung tissues, n = 535 tumor tissues; Hou lung database: n=65 normal lung tissues, n = 91 tumor tissues). Table shows the average fold change and statistical analysis of 15 sphingolipid metabolic gene expression between tumor tissues and normal lung tissues obtained from the three databases. (b) Pearson correlation matrix of individual sphingolipid metabolic genes. Rows and columns correspond to the 15 measured enzymes. Each colored square illustrates the gene-gene interaction; color code: red, positive correlation; blue, negative correlation; the color intensity represents the correlation strength. The values range between -1.0 and 1.0. A correlation of -1.0 shows a perfect negative correlation, while a correlation of 1.0 demonstrates a perfect positive correlation. A correlation of 0.0 indicates no relationship between the two genes. (c) Violin plots show the relative mRNA expression level of CERS3, ASAH1, SPHK1, CERK, UGT8, GAL3ST1, B4GALT6 or B3GNT5 in the tumors derived from early stage (stage I) and advanced stage (stage Ⅱ/Ⅲ) respectively (n=223 early stage (stage I) NSCLC patients, n = 179 advanced stage (stage Ⅱ/Ⅲ) NSCLC patients). The datasets are obtained from TCGA database. Values are shown as log2 fold-change relative to normal lung tissues. Violin plots represent means ± s.e.m. (d) Kaplan-Meier curves of overall survival in relation to the CERS3, ASAH1, SPHK1, CERK, UGT8, GAL3ST1, B4GALT6 and B3GNT5 gene expression. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. NS, no significant difference. (a, c) Student's t test. (d) Log-rank test.
To confirm the clinical significance of our finding, we investigated any correlation between the expression of these sphingolipid metabolic genes and patient prognosis. Interestingly, our data showed that the expression of lacto/neolacto-series glycosphingolipid and GalCer synthesis pathway genes, including B4GALT6, B3GNT5 and UGT8, was increased (between p < 0.05–p < 0.0001, student's t test), while the sulfatide metabolic gene GAL3ST1 was down-regulated in advanced stage patients (stage Ⅱ/Ⅲ) as compared to early stage patients (stage I) (p<0.001, student's t test) (Fig. 1 c). Nevertheless, there was no significant difference in the expression of CERS3, ASAH1, CERK and SPHK1 between early stage and advanced stage patients (NS, student's t test) (Fig. 1 c). Indeed, the role of SPHK1 and its metabolite S1P in cancer progression still remains controversial [33]. In addition, high expression of either SPHK1, UGT8, B4GALT6, B3GNT5 was correlated with poor patient survival (between p < 0.01–p < 0.0001, log-rank test), while low expression of GAL3ST1 stratified patients into those with poor survival (p < 0.0001, log-rank test) (Fig. 1 d). In contrast, expression of sphingolipid metabolic genes that mainly involved in ceramide metabolic pathway such as CERK, CERS1-6, ASAH1, SMPD1-4 or SPTLC1-2 etc, was not correlated with overall survival, implying that the glycosylated- or sulfated-sphingolipids, such as lacto/neolacto-series glycosphingolipid and sulfatide, may have more significant roles in lung cancer progression (Fig. 1 d, Supplementary figure 1 b). Together, our data revealed the alteration of metabolic gene expression that involved in lacto/neolacto-series glycosphingolipid and sulfatide synthesis associated with poor prognosis in NSCLC patients.
Tumor exhibits a distinct sphingolipid metabolic gene expression profile compared with the normal adjacent lung tissue
To confirm the result of our RNA-seq database analysis, we collected and evaluated the expression of sphingolipid metabolic genes in paired tumor tissues and adjacent normal lung tissues from NSCLC patients. The clinicopathological features of NSCLC patients and healthy individuals included in this study were summarized in supplementary Table 1. RT-PCR data confirmed that the expression pattern of sphingolipid metabolic genes in our collected patient samples was consistent with the result from our RNA-seq database analysis (Fig. 2 a, Supplementary figure 2 a). Again, the expression levels of UGCG, B4GALT6, B3GNT5 and UGT8 etc., were up-regulated (between p < 0.05–p < 0.0001, student's t test), while GAL3ST1 level was significantly decreased in cancer tissues compared with the adjacent normal lung tissues (p < 0.0001, student's t test). To further analyze any correlation between these genes, we performed a hierarchical clustering of the correlation matrix of the significantly altered genes by using Pearson correlation method. As expected, GAL3ST1 was negatively associated to either UGCG, UGT8, B3GNT5 or B4GALT6, while these up-regulated genes were positively correlated to each other. This hierarchical view on gene expression was a sub-optimal representation of sphingolipid coregulation, indicating that these genes were highly likely to coregulate sphingolipid metabolism in NSCLC patients (Fig. 2 b, Supplementary figure 2 b). By perfoming gene co-expression studies using COXPRESdb v7 co-expression database (34), our result indicated that both B3GNT5 and GAL3ST1 genes co-expressed with the genes that involved in sphingolipid signaling and MAPK signaling pathways (Fig. 2 c). Overall, our result indicated that average of lacto/neolacto-series glycosphingolipid synthesis pathway and their upstream ceramide hydrolysis pathway gene expression was up-regulated, while sulfatide synthesis pathway gene expression was down-regulated in tumor tissues as compared with the adjacent normal lung tissues (Fig. 2 d).
Fig. 2.
Tumor shows an altered pattern of sphingolipid metabolic enzyme expression as compared with the adjacent normal lung tissue. (a) RT-PCR analysis shows the relative expression of sphingolipid metabolic genes in matched NSCLC patient tumors and adjacent normal lung tissues (n = 10 NSCLC patients). (b) Heatmap represents the Pearson correlation matrix of individual sphingolipid metabolic genes. Rows and columns correspond to the sphingolipid metabolic genes. (c) Gene co-expression study, using COXPRESdb v7 databse, reveals that both B3GNT5 and GAL3ST1 genes coexpressed with the genes that involved in MAPK signaling and sphingolipid signaling pathways. (d) Schematic representation of the sphingolipid metabolic network shows up-regulated (highlight in red) and down-regulated (highlight in green) sphingolipid metabolic genes in NSCLC tumors as compared to the adjacent normal lung tissues. Bar charts represent means ± s.e.m. *p< 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. N.S., no significant difference. (a) Student's t test.
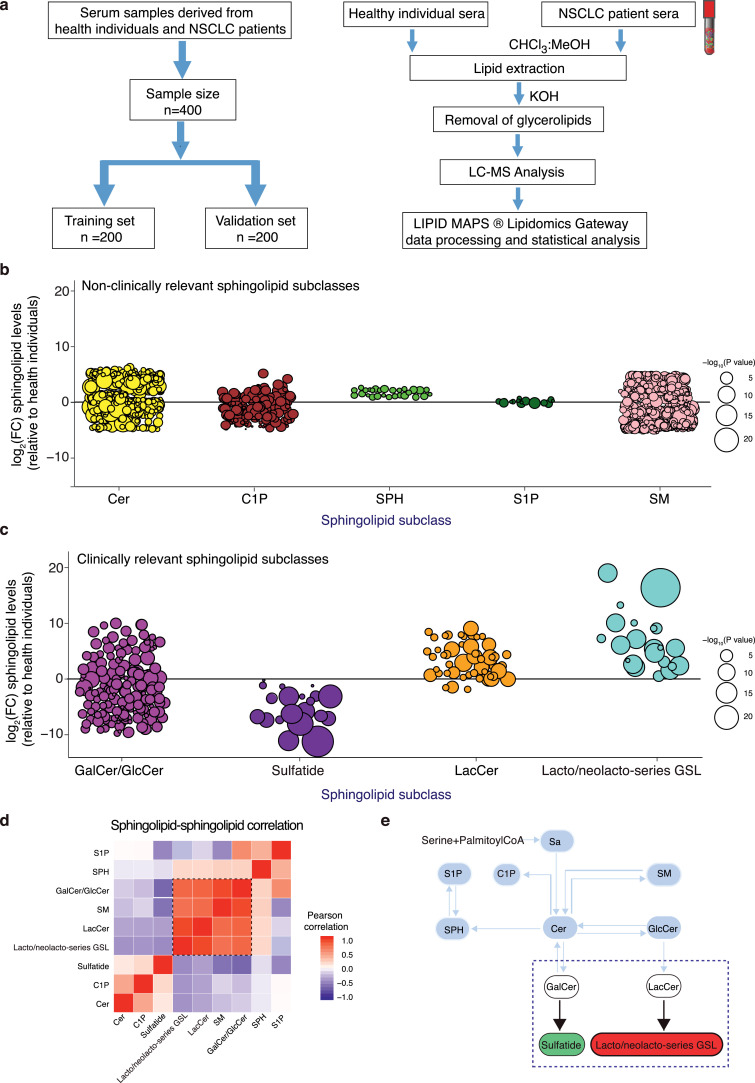
Serum sphingolipidomics analysis reveals strongly altered sphingolipid metabolic state in NSCLC patients
To study any correlation between serum sphingolipidomes and tumor sphingolipid metabolic gene expression, we performed a high resolution LC-MS based sphingolipidomics approach using the serum samples derived from NSCLC patients and healthy individuals. In our patient cohort, the serum samples from 100 healthy individuals and 100 treatment naïve NSCLC patients were used as a training dataset, while another 100 healthy individuals and 100 treatment naïve NSCLC patient samples were collected for a validation dataset (Fig. 3 a, Supplementary table 1). By performing the LC-MS based sphinglipidomics approach, we successfully detected approximately 847 unique ion masses that were previously characterized sphingolipid compositions, while majority of them belonged to 9 different sphingolipid subclasses (which were named according to the LIPID MAPS terminology), including Cer, C1P, SPH, S1P, SM, GalCer/GlcCer (abbreviation: Hex1Cer), sulfatide, LacCer (abbreviation: Hex2Cer) and lacto/neolacto series-glycosphingolipid (abbreviation: HexnHexNAcnFucn-Cer, n refers to number of sugars). In this study, the sphingolipid compositions were primarily assigned based on their high-resolution MS data. The presence of sulfate group was confirmed by the observation of diagnostic fragment ion originating from in-source neutral loss of H2SO4. The structures of the “net” sulfate part were verified by high-resolution MS/MS data. In addition, these fragments provided key information for the location of sulfate group. By using a sulfatide species, (3′-sulfo)Galβ-Cer (d18:1/16:0), as an example, based on neutral loss m/z 682.5544 [M+H-H2SO4]+ from its accurate mass 779.5234, the sphingolipid was matched with a sulfated sphingolipid candidate with the formula of C40H77O11NS and one double bond. In addition, the structures of “net” sphingolipid part were verified by our high-resolution MS/MS data. MS/MS experiments on the [M+H]+ ion at m/z 682.5544 provided characteristic fragments which revealed some aspects of the structure of glycosphingolipid. Firstly, one 162 and 180 gap in the pattern of these fragmentations of m/z 682.5544 to m/z 520.5022 and 502.4923, was observed as a result of losing one hexosyl group, indicating that this sphingolipid belonged to sulfated glycosphingolipid. Secondly, the breakage of a glycosidic bond produced neutral loss of m/z 504.5076 from 682.5544, suggested that there were glucosyl or galactosyl group present in this sphingolipid. Thirdly, a cleavage in ceramide moiety gave rise to the ion at m/z 282.2779 [M-SPH(d18:1)+H-H2O] which yielded ions at m/z 264.2684 and m/z 252.2675, reflecting a C18 sphingosine backbone. Finally, the ions at m/z 272.2576 [Amide(16:0+O)+H] were yielded from a C16 fatty acid chain. Based on fragment clues mentioned above, the structure of (3′-sulfo)Galβ-Cer (d18:1/16:0) was elucidated. For lacto/neolacto series-glycosphingolipid species, they expressed more loss of sugar fragment ions. Using Hex6HexNAc3Fuc2-Cer (d18:1/16:0) as an example, the fragment ion at m/z 1116.5676 referred to Hex6HexNAc3Fuc2-Cer (d18:1/16:0) with loss of a hexosyl group; the ion at m/z 1034.5333 reflected Hex6HexNAc3Fuc2-Cer (d18:1/16:0) with loss of a hexosyl and a pentosyl group; the ion at m/z 944.5017 referred to the loss of two galactoses and one pentosyl from Hex6HexNAc3Fuc2-Cer (d18:1/16:0), while all of these fragment ions got two hydrogen adduct.
Fig. 3.
Serum sphingolipidomics analysis indicates altered sphingolipid metabolic states in NSCLC patients as compared with healthy individuals. (a) Schematic representation of the workflow for sample allocation as well as the extraction and identification procedure of serum sphingolipids from NSCLC patients and healthy individuals. (b, c) Manhattan plots reveal the relative levels of non-clinically relevant sphingolipid subclasses, including ceramide (Cer), ceramide-1-phosphate (C1P), sphingosine (SPH), sphingosine-1-phosphate (S1P), sphingomyelin (SM) and the relative levels of clinically relevant sphingolipid subclasses, such as galactosylceramide (GalCer)/glucosylceramide (GlcCer), sulfatide, lactosylceramide (LacCer), lacto/neolacto-series glycosphingolipid (lacto/neolacto-series GSL) in NSCLC patients as compared to healthy individuals. Values are shown as log2 fold-change relative to healthy individuals. Each dot represents a sphingolipid species, color codes per sphingolipid subclass. Dot size indicates significance (-log10 value). Serum sphingolipidomics data are combined of 100 NSCLC patients and 100 healthy individuals from the training dataset and represented as mean. (d) Analysis of correlations between different sphingolipid subclasses, as function of correlation strength. Black dotted line represents strongly correlated sphingolipids. (e) Overview of different sphingolipid levels in NSCLC patients. lacto/neolacto-series glycosphingolipid and sulfatide species are indicated in a close-up of the sphingolipid metabolic network. Red color represents up-regulated sphingolipids and green for down-regulated sphingolipids. Blue dotted line represents p < 0.05. All data are obtained from the training dataset. (b, c) Student's t test.
We have already shown that the expression of metabolic enzymes that involved in the synthesis of Cer, C1P, SPH, S1P and SM, were not correlated with patient prognosis, while the expression of glycosylated- or sulfated-sphingolipid metabolic enzymes that involved in GalCer/GlcCer, sulfatide, LacCer and lacto/neolacto-series glycosphingolipid synthesis, associated strongly with patient overall survival. We therefore performed Manhattan plot analysis of the relative serum abundance of these sphingolipid subclasses and their species in two separate groups according to the clinical relevance of their metabolic enzyme expression. In comparison with the non-clinically relevant sphingolipid subclasses (Fig. 3 b), the relative serum level of many glycosylated- or sulfated-sphingolipid species, including GalCer/GlcCer, sulfatide, LacCer and lacto/neolacto-series glycosphingolipid species, was highly altered in NSCLC patients as compared with healthy individuals (Fig. 3 c and Supplementary Table 3). In particular, the serum level of many LacCer species and its downstream metabolite lacto/neolacto-series glycosphingolipid species was up-regulated, while the level of majority sulfatide species was decreased in NSCLC patients as compared with healthy individuals (each dot represents a sphingolipid species, circle size represents -log10 P-value, bigger means more significant difference) (Fig. 3 c and Supplementary Table 3). Furthermore, Pearson correlation analysis showed that the serum levels of GalCer/GlcCer, LacCer and lacto/neolacto-series glycosphingolipid were positively correlated, while the sulfatide level was negatively associated to all these positively correlated sphingolipids in NSCLC patients (Fig. 3 d). Based on our serum sphingolipidomes and gene expression profiling data, we purposed that the regulation of lacto/neolacto-series glycosphingolipid and sulfatide balance by their metabolic enzymes B3GNT5 and GAL3ST1 functioned as a checkpoint to determine sphingolipid metabolism in NSCLC patients (Fig. 3 e). Collectively, our data indicated that the alteration of sphingolipid metabolic gene expression in tumor tissues directly reflected into the serum level of their metabolites in NSCLC patients, implying that our serum sphingolipid profiling approach could be a diagnostic tool for cancer patients.
Identification of lacto/neolacto-series glycosphingolipid and sulfatide species as prominent biomarkers of NSCLC patients
To test whether the change in serum sphingolipid abundance can distinguish NSCLC patients from healthy individuals, we employed principle least squares (PLS) and orthogonal PLS (OPLS) discriminant analysis methods [35] to interpret their serum sphingolipidomes (Supplementary fig. 3 a and b), allowing us to identify which predictor variables were influencing the most the classification by calculating and examining the variable importance in projection (VIP) scores. The criterion of VIP score with greater than 1 was used as a cut-off point for our variable selection. By calculating the VIP indices of our training dataset spectra, 24 sphingolipids have been selected as potential sphingolipid biomarkers (Supplementary Table 4). Among 24 sphingolipids, majority of them were glycosylated-sphingolipid (i.e. GalCer/GlcCer, LacCer and lacto/neolacto-series glycosphingolipid) and sulfated-sphingolipid (sulfatide) species. To validate these potential biomarkers, the relative abundance of individual sphingolipids was employed as variations to predict the grouping of individuals. We assessed their classification performance (including specificity and sensitivity) using the area under the curve (AUC) of the receiver operator characteristic (ROC) curve method in order to distinguish NSCLC patients from healthy individuals. As shown in supplementary Fig. 4 a-c, when compared to group 1 (including Cer, S1P, C1P and SM) and group 2 (including GalCer/GlcCer and LacCer), group 3 (including sulfatide and lacto/neolacto-series glycosphingolipid) was changed most dramatically in NSCLC patients as compared with healthy individuals. For instance, we observed 30-fold increase in Hex7HexNAc3Fuc1-Cer (d18:1/24:0) and more than 50-fold decrease in (3′-sulfo)Galβ-Cer (d18:1/24:0) levels in NSCLC patients as compared to healthy individuals (Supplementary Fig. 4 a-c, Supplementary Table 4). Furthermore, we used a prediction model to calculate the sensitivity and specificity of serum sphingolipids to diagnose NSCLC patients. Among 9 sphingolipid subclasses, lacto/neolacto-series glycosphingolipid and sulfatide species, including Hex7HexNAc3Fuc1-Cer(d18:1/24:0), Hex7HexNAc3Fuc1- Cer (d18:1/24:1), Hex6HexNAc3Fuc2-Cer (d18:1/18:0) and Hex6HexNAc3Fuc2-Cer (d18:1/16:0), (3′-sulfo)Galβ-Cer (d18:1/24:0), (3′-sulfo)Galβ-Cer (d18:1/24:1), (3′-sulfo)Galβ-Cer (d18:0/16:0), and (3′-sulfo)Galβ-Cer (d18:1/16:1) had the highest AUC values, sensitivity and specificity in both training and validation datasets as compared with other sphingolipid subclasses, indicating that they were the most effective biomarkers of distinguishing NSCLC patients from heathy individuals (Supplementary Fig. 4, Supplementary Table 5). In addition, their chemical structures, chromatograms and MS/MS data were defined and given in Supplementary Fig. 5 a-d and Supplementary Fig. 6 a-d. To prove the accuracy of our finding, we examined the specificity of currently used clinical diagnostic tumor biomarkers in our serum samples, such as carcinoembryonic antigen (CEA), carbohydrate antigen 125 (CA125), cytokeratin 19 fragment (CYFRA21-1), and neuron-specific enolase (NSE), and previously reported sphingolipid biomarker, sphingomyelin (SM(d14:1/22:0)) species (but not the most abundant species of sphingomyelin (i.e. SM (d18.1/22:0)) correlated to patient diagnosis) [36,37] indicating that the serum level of SM (d14:1/22:0), CA125, and CEA, could also differentiate NSCLC patients from healthy individuals (Supplementary Fig. 4 d-e).
Evaluation of lacto/neolacto-series glycosphingolipid and sulfatide species as serum biomarkers of lung cancer progression
To evaluate the clinical significance of our findings, we performed a correlation study between the serum level of lacto/neolacto-series glycosphingolipid/sulfatide species and cancer progression. The normalized intensity data for selected species from lacto/neolacto-series glycosphingolipid and sulfatide subclasses were summarized as a heatmap in Fig. 4 a. To further confirm the efficacy of these biomarkers, we also performed principle component analysis (PCA) with our sphingolipidomics data, which showed a clear separation of healthy individuals, early stage (stage I) and advanced stage (stage Ⅱ/Ⅲ) patients by using these biomarkers (R2X[1]=0.531, PCA) (Fig. 4 b). Our result indicated that NSCLC patients with high serum lacto/neolacto-series glycosphingolipid level and low serum sulfatide level were associated strongly with advanced disease (stage Ⅱ/Ⅲ) in both training and validation datasets (between p < 0.05–p < 0.0001, student's t test) (Supplementary figure 7 a and b). To test whether these sphingolipid biomarkers can effectively stratify NSCLC patients, we calculated the ROC values of these sulfatide and lacto/neolacto-series glycosphingolipid species for differentiating early stage patients from advanced stage patients. Among them, Hex7HexNAc3Fuc1-Cer (d18:1/24:0) and (3′-sulfo)Galβ-Cer (d18:1/24:0) yielded the highest individual AUC (0.9775 and 0.9632 respectively), as well as the highest individual sensitivity (86% and 100% respectively) and specificity (100% and 96% respectively), indicating that they were the best sphingolipid biomarkers of NSCLC stage classification (between p<0.05 – p<0.0001, student's t test) (Fig. 4 c and d). Coincidently, recent studies suggested that the cellular level of these sphingolipid subclasses affected breast cancer progression [8,38].
Fig. 4.
Analysis of serum sphingolipid abundance reveals a circular sphingolipid coregulatory network, identifying lacto/neolacto-series glycosphingolipid and sulfatide species as biomarkers of NSCLC stage classification. (a) Heatmap shows a comparison of the serum levels of lacto/neolacto-series glycosphingolipid and sulfatide among healthy individuals, early stage (stage I) and advanced stage (stage Ⅱ/Ⅲ) NSCLC patients. Using a color scale, the serum concentration of sphingolipids was log2-transformed relative to average abundance of healthy individuals. (b) Principle Component Analysis (PCA) was performed to test whether healthy individuals, early stage (stage Ⅰ) or advanced stage (stage Ⅱ/Ⅲ) NSCLC patients could be separated into different groups according to their serum lacto/neolacto-series glycosphingolipids and Sulfatide levels as mentioned in A. Healthy individual (blue), early stage patient (red) and advanced stage patient (green) groups are shown in separated clusters on the score plot. (c, d) ROC curves represent individual lacto/neolacto-series glycosphingolipid and sulfatide species for stratifying early stage (stage Ⅰ) patients from advanced stage (stage Ⅱ/Ⅲ) patients. (e) Network visualization of the serum sphingolipid-sphingolipid correlations. The serum sphingolipid-sphingolipid correlation matrix was transformed into a network where nodes represent individual sphignolipid species and edges for correlations of 0.7 or higher. Node sizes represent significance. Color represents each sphingolipid subclass. In particular, lake blue circle represents for lacto/neolacto-series glycosphingolipid and purple circle for sulfatide. (f) Network close-up showed that lacto/neolacto-series glycosphingolipid and sulfatide species were strongly correlated and mainly distributed at the center of the sphingolipid coregulatory network. (g) Nodes of the network are color coded based on the relative fold change of sphingolipid abundance for healthy individuals, stage Ⅰ and Ⅱ/Ⅲ NSCLC patients. Red color represents an increased in relative fold change in sphingolipid abundance and green color codes for down-regulation in the sphingolipid coregulatory network. All results are obtained from the training dataset (n = 100 healthy individuals, n = 63 stage Ⅰ NSCLC patients, n = 37 stage Ⅱ/Ⅲ NSCLC patients). (a-g) *p< 0.05, **p< 0.01, ***p< 0.001, ****p< 0.0001. (c, d) Student's t test.
To prove the importance of our finding, we studied the specificity of currently used diagnostic tumor biomarkers, such as CEA, CA125, CYFRA21-1 and NSE, and previously reported sphingolipid biomarker, SM species [36,37] in our sample cohort. Strikingly, there was no significant difference in the serum levels of SM (22:0) species (including SM (d14:1/22:0), SM (d15:1/22:0), SM (d15:2/22:0) and SM (d18:0/22:0)), and also CEA and NSE between early stage and advanced stage patients, indicating these biomarkers were not specific for NSCLC stage classification in both training and validation datasets (Supplementary Fig. 7 c and d). Although the serum level of CA125 and CYFRA21-1 showed a modest difference between early stage and advanced stage NSCLC patients in the validation dataset (Supplementary Fig. 7 d), their AUC values were less than 0.7 (CA125: 0.5799; CYFRA21-1: 0.6484), and led to poor sensitivity (CA125:61.90%; CYFRA21-1:70.00%) and specificity (CA125: 45.16%; CYFRA21-1: 51.61%) of a diagnostic test (Supplementary Table 5). Overall, our result identified lacto/neolacto-series glycosphingolipid and sulfatide species as potential biomarkers that were more effective and specific than the currently used clinical markers for staging NSCLC patients.
Analysis of serum sphingolipid abundance reveals the circular organization of sphingolipid coregulatory network in NSCLC patients
Given the change in serum sphingolipid compositions in NSCLC patients as compared to healthy individuals, we next analyzed the correlation of sphingolipid abundance at the level of individual sphingolipid species to expose the larger organizational principles that reflect sphingolipid metabolism in NSCLC patients. We transformed the sphingolipid-sphingolipid correlation matrix into a network where nodes represented individual sphingolipid species, node sizes represented -log10 P-value (bigger means more significant difference), and edges represented correlations of 0.7 or higher. Remarkably, this correlation network displayed a circular network of coregulated sphingolipids (Fig. 4 e) and indicated the distribution of our 8 newly identified modified sphingolipid biomarkers, including Hex7HexNAc3Fuc1-Cer (d18:1/24:0), Hex7HexNAc3Fuc1-Cer (d18:1/24:1), Hex6HexNAc3Fuc2-Cer (d18:1/18:0), Hex6HexNAc3Fuc2-Cer (d18:1/16:0), (3′-sulfo)Galβ-Cer (d18:1/24:1), (3′-sulfo)Galβ-Cer (d18:1/24:1), (3′-sulfo)Galβ-Cer (d18:0/16:0), and (3′-sulfo)Galβ-Cer (d18:1/16:1), at the center of the network, showing that they had the strongest correlation with each other and also with different sphingolipid subclasses, such as sphingoid base, glycosphingolipid and ceramide etc (Fig. 4 f). When we transformed the color coding of each node according to the log2 relative fold change in SPL abundance, it revealed different SPL metabolic states for healthy individuals, early stage and advanced stage patients (Fig. 4 g), indicating that the change in the serum level of lacto/neolacto-series glycosphingolipid and sulfatide species directly orchestrated SPL metabolic reprogramming in cancer progression. Indeed, previous work showed that sulfatide could sensitize cancer cells to microenvironmental stress factors such as hypoxia and chemotherapy(8), implying that the serum level of sulfatide may be an indicator of patient response to therapies. Moreover, the dysregulated synthesis of lacto/neolacto-series glycosphingolipid and sulfatide plays a role in regulating cancer cell proliferation and survival [32,39,40]. Collectively, our data demonstrated, for the first time, that a circular network of coregulated sphingolipids dictated cancer progression in NSCLC patients and that was determined by the lacto/neolacto-series glycosphingolipid/sulfatide balance.
Aberrant expression of sphingolipid metabolic enzyme B3GNT5 or GAL3ST1 regulates tumor cell growth and progression
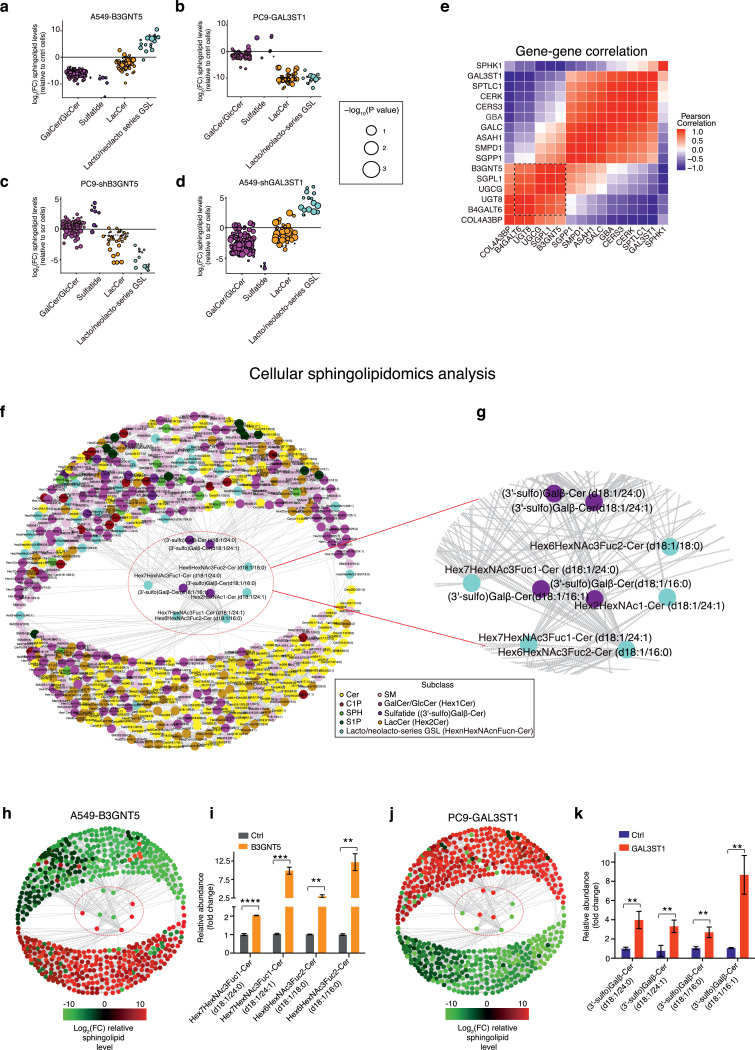
Since the cellular roles of B3GNT5 and GAL3ST1 in lung cancer cells remained unclear, we first checked the expression of B3GNT5 and GAL3ST1 in human NSCLC cell lines, indicating that A549 cells had low B3GNT5 and high GAL3ST1 expression as compared to the rest of cell lines, while H1299 and PC9 cells had an opposite expression pattern (Supplementary figure 8 a). In contrast, HCC827 cells expressed both B3GNT5 and GAL3ST1 (Supplementary figure 8 a). We therefore stably overexpressed or transiently/stably knock-down B3GNT5/GAL3ST1 in these cell lines respectively, expression of which was confirmed by western blot (Fig. 5 a, Supplementary figure 8 b). These transfected cells were then used to perform a range of in vitro experiments, including CCK8 proliferation assays, colony formation assays, scratch assays, transwell invasion assays, and tumor sphere formation assays. As expected, overexpression of B3GNT5 in A549 and HCC827 cells significantly enhanced cell proliferation, colony formation, migration, invasion, or/and tumor sphere formation as compared to empty vector transfected cells (Fig. 5 b, d, f, h and j), while knocking down B3GNT5 had the opposite effect on PC9 and H1299 cells (Supplementary Fig. 8 c, e, g, i and k). Conversely, exogenous expression of GAL3ST1 in PC9 and H1299 cells dramatically inhibited cell proliferation, colony formation, migration, invasion or/and tumor sphere formation as compared to the empty vector transfected cells (Fig. 5 c, e, g, i and k), while depleting GAL3ST1 increased the tumorigenesis in A549/HCC827 cells (Supplementary Fig. 8 d, f, h, j and l).
Fig. 5.
Aberrant expression of B3GNT5 or GAL3ST1 regulates cancer cell growth, migration, invasion and sphere formation abilities and in vivo tumor growth. (a) Western blot analysis of stable B3GNT5/GAL3ST1 expressing cells and empty vector transfected cells (ctrl) as indicated. Molecular weight markers (in kDa) are shown on the right. (b, c) CCK8 proliferation assays of stable B3GNT5/GAL3ST1 expressing cells and empty vector transfected cells. Bar charts indicate the relative proliferation of stable B3GNT5/GAL3ST1 expressing cells as compared to empty vector transfected cells. (d, e) Colony formation assays of stable B3GNT5/GAL3ST1 expressing cells and empty vector transfected cells. Representative pictures of colony formation from each group are given. (f, g) Wound healing assay of stable B3GNT5/GAL3ST1 expressing cells and empty vector transfected cells. Bar charts represent the relative wound closure of stable B3GNT5/GAL3ST1 expressing cells as compared to empty vector transfected cells. Representative images of wound healing assays from each group are given. (h, i) Transwell invasion assays. Overexpression of B3GNT5 increased the invasion ability of A549/HCC827 cells, while GALST3 overexpression inhibited PC9/H1299 cell invasion as compared to empty vector control cells. Representative images of crystal violet stained invaded cells from each group are given. (j, k) Sphere formation assays of stable B3GNT5/GAL3ST1 expressing cells and empty vector transfected cells. Bar charts indicate the number of tumor spheres per group (n = 3 experimental repeats). (l, m) Nude mice were subcutaneously injected with either stable B3GNT5 expressing cells (A549-B3GTN5), empty vector transfected A549 cells (A549-Ctrl), stable GAL3ST1 expressing PC9 cells (PC9-GAL3ST1) or empty vector transfected PC9 cells (PC9-ctrl). Tumor growth curves showed that B3GNT5 overexpression increased the tumor growth of A549 cells as compared with empty vector transfected cells, while overexpression of GAL3ST1 repressed the tumor growth of PC9 cells when compared with empty vector transfected cells (ctrl) (n = 5 mice per group). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. NS, no significant difference. (b-k) Student's t test. (l, m) Two-way ANOVA test. Scale bars in (f-k) represent 100 μm.
To confirm our observation in vivo, we subcutaneously injected stable B3GNT5/GAL3ST1 expressing cells or empty vector transfected cells into nude mice, indicating that aberrant expression of B3GNT5 in A549 cells significantly increased tumor burden as compared to empty vector transfected cells (p < 0.0001, two-way ANOVA test) (Fig. 5 l), while GAL3ST1 overexpression inhibited the tumor growth of PC9 cells as compared to empty vector control cells (p < 0.01, two-way ANOVA test) (Fig. 5 m). Overall, our results showed that expression of B3GNT5 and GAL3ST1 had an opposite effect on cancer cell migration, invasion and sphere formation as well as in vivo tumor growth.
Sphingolipidomics analysis of B3GNT5/GAL3ST1 genetically perturbed lung cancer cell lines indicate their key regulatory role in sphingolipid metabolism
To confirm our serum sphingolipidomics data, we therefore performed sphingolipidomics analysis of different NSCLC cell lines, including B3GNT5/GAL3ST1 stably expressing lung cancer cells, B3GNT5/GAL3ST1 knock-down lung cancer cells, empty vector transfected cells and scramble control cells. As expected, Manhattan plot analysis indicated that overexpression of B3GNT5 in A549 and HCC827 cells increased the relative level of lacto/neolacto-series glycosphingolipid species and down-regulated the level of sulfatide species as compared with empty vector transfected cells, while knocking down B3GNT5 in PC9 and H1299 cells had an opposite effect on their expression levels (Fig. 6 a and c, Supplementary figure 9 a and c). In contrast, GAL3ST1 overexpression up-regulated the relative level of sulfatide species and decreased the level of lacto/neolacto-series glycosphingolipid species in PC9 and H1299 cells as compared with empty vector transfected cells, while depletion of GAL3ST1 in A549 and HCC827 cells down-regulated the relative level of sulfatide species and increased the relative abundance of lacto/neolacto-series glycosphingolipid species compared with scramble control cells (Fig. 6 b and d, Supplementary figure 9 b and d). In addition, comparing the sphingolipid metabolic gene expression across all NSCLC cell lines revealed pairs of positively and negatively correlated sphingolipid metabolic enzymes, which were same as the pairs we identified in the tumor tissues derived from NSCLC patients (Fig. 6 e). Interestingly, RT-PCR analysis showed that exogenous expression of B3GNT5 reduced GAL3ST1 expression in A549 and HCC827 cells, while knock-down of B3GNT5 in H1299 cells enhanced GAL3ST1 expression (Supplementary figure 9 e-g). In addition, GAL3ST1 overexpression significantly decreased B3GNT5 expression in H1299 cells, while depletion of GAL3ST1 increased B3GNT5 expression in A549 and HCC827 cells (Supplementary figure 9 h-j), indicating that the expression of B3GNT5 and GAL3ST1 was negatively correlated in some NSCLC cell lines.
Fig. 6.
Sphingolipidomics analysis of B3GNT5/GAL3ST1 genetically perturbed lung cancer cells confirms the circular sphingolipid coregulatory network. (a-d) Manhattan plot analysis of the relative levels of clinically relevant sphingolipid subclasses, including galactosylceramide (GalCer)/glucosylceramide (GlcCer), sulfatide, lactosylceramide (LacCer), lacto/neolacto-series glycosphingolipid in B3GNT5/GAL3ST1 stably overexpressed A549/PC9 cells or B3GNT5/GAL3ST1 stably knock-down A549/PC9 cells. Data are normalized to the levels of these sphingolipid subclasses in empty vector transfected cells or scramble control transfected cells. (e) Heatmap represents the Pearson correlation matrix of individual sphingolipid metabolic genes. Rows and columns correspond to the sphingolipid metabolic genes. Black dotted boxes show clusters of strongly positively correlated sphingolipid metabolic genes. (f) Network visualization of the sphingolipid-sphingolipid correlations in cancer cells. Nodes represent each sphingolipid species. Colors represent sphingolipid subclasses. Edges are correlations of r ≥ 0.7. (g) Network close-up reveals the strongly correlated lacto/neolacto-series glycosphingolipid and sulfatide species distributed at the center of the sphingolipid coregulatory network. (h, j) Nodes of the network are color-coded based on the relative fold change of sphingolipid abundance for B3GNT5/GAL3ST1 stably expressing cells after normalized to empty vector transfected cells. (i, k) Bar charts indicate that overexpression of B3GNT5/GAL3ST1 in A549 or PC9 cells increases the relative abundance levels of lacto/neolacto-series glycosphingolipid and sulfatide species as compared to empty vector transfected cells (ctrl) (n = 3 experimental repeats). Bar charts represent means ± s.e.m. **p<0.01; ***p<0.001; ****p<0.0001. (a-d, i, k) Student's t test.
To better represent the correlation between these sphingolipids, we again transformed the sphingolipid-sphingolipid correlation matrix into a network where nodes represented individual sphingolipid species and edges represented positive correlation of 0.7 or higher. Strikingly, our network study confirmed the existence of circular sphingolipid coregulatory network in lung cancer cells (Fig. 6 f). Consistent with our finding, our newly identified sphingolipid biomarkers, including Hex7HexNAc3Fuc1-Cer (d18:1/24:0), Hex7HexNAc3Fuc1-Cer (d18:1/24:1), Hex6HexNAc3Fuc2-Cer (d18:1/18:0), Hex6HexNAc3Fuc2-Cer (d18:1/16:0)), (3′-sulfo)Galβ-Cer (d18:1/24:0), (3′-sulfo)Galβ-Cer (d18:1/24:1), (3′-sulfo)Galβ-Cer (d18:1/16:0), and (3′-sulfo)Galβ-Cer (d18:1/16:1) again distributed at the center of this network, indicating that they possessed the strongest correlation with other sphingolipid subclasses, including sphingoid base, glycosphingolipid and ceramide etc (Fig. 6 f and g). To characterize the effect of B3GNT5/GAL3ST1 expression on sphingolipid metabolism, we transformed the color coding of each node in the network according to the log2 fold change in sphingolipid abundance between B3GNT5/GAL3ST1 stably expressing cells and empty vector transfected cells. Strikingly, our result revealed an almost completely opposite pattern of increased and decreased sphingolipids between B3GNT5 and GAL3ST1 stably expressing cells (Fig. 6 h and j). In addition, overexpression of B3GNT5 or GAL3ST1 up-regulated the relative levels of its related metabolites in A549 cells or PC9 cells as compared to empty vector transfected cells (Fig. 6 i and k). Taken together, our data confirmed that the lacto/neolacto-series glycosphingolipid/sulfatide balance, which is controlled by their metabolic enzymes B3GNT5 and GAL3ST1, works as a checkpoint to determine sphingolipid metabolic reprogramming during cancer progression (Fig. 7).
Fig. 7.
Schematics diagram represents the role of “lacto/neolacto-series glycosphingolipid/sulfatide balance” in determining sphingolipid metabolism and cancer progression. Schematics representation of the correlation between the lacto/neolacto-series glycosphingolipid/sulfatide balance and cancer progression. Tai chi graphs indicate that the synthesis cycle of sulfatide (Yin) and lacto/neolacto-series GSL (Yang) determines sphingolipid metabolic states in cancer progression, while the Yin-Yang balance is controlled by B3GNT5/GAL3ST1 expression. When the synthesis of lacto/neolacto-series glycosphingolipid species increases, it favors cancer progression and vice versa.
Discussion
NSCLC has a dismal prognosis mostly due to an advanced stage at the time of diagnosis. One way to improve prognosis would be to diagnose NSCLC at an earlier stage. However, the current diagnostic tools are unable to perform accordingly and no effective biomarker (s) for NSCLC are yet available. Although sphingolipid metabolism has been shown to be the top dysregulated pathway in NSCLC patients [41], the molecular control of sphingolipid metabolism still remains poorly studied. In addition, it is unclear whether the change in serum level of sphingolipids could effectively stratify NSCLC patients at different clinical stages.
Recent studies suggest that increased expression of SPHK1, which catalyses the conversion of pro-survival metabolite S1P from ceramide, is observed in multiple cancers [33,42,43]. However, the simple concept that the balance between ceramide and S1P synthesis, often termed as the “sphingolipid rheostat”, controls cell survival is somewhat controversial due to the fact that new SPHK1 inhibitors do not affect cancer cell proliferation or apoptosis [44,45], and some studies demonstrating higher ceramide levels observed in metastatic breast cancer [46]. When we examined mRNA expression data obtained from different databases (i.e. TCGA, GEO, Hou lung) as well as our own patient cohort, we demonstrated that the alteration of metabolic gene expression pathway involved in the synthesis of glycosphingolipid, such as lacto/neolacto-series glycosphingolipid and sulfatide, but not ceramide and S1P, was most significantly associated with poor prognosis in NSCLC patients. Indeed, lacto/neolacto-series glycosphingolipid and sulfatide are the key components of cellular membranes, which are able to associate other sphingolipids and cholesterol to form lipid rafts [47], and therefore involve in many important cellular signaling pathways [9]. Although our clinical studies demonstrated the importance of lacto/neolacto-series glycosphingolipid and sulfatide species in cancer progression, knowledge about the prognostic and/or predictive role of these sphingolipids in human cancer is still very limited. By collecting a cohort of serum samples from NSCLC patients and healthy individuals, we performed serum sphingolipidomics analysis with these samples to identify the potential sphingolipid biomarkers of NSCLC patients. Interestingly, our data indicated that lacto/neolacto-series glycosphingolipid and sulfatide were the serum sphingolipid biomarkers of NSCLC patients, which were more effective than currently available tumor biomarkers in staging NSCLC patients. Overall, we provided first evidence that the alteration in sphingolipid metabolic gene expression B3GNT5 and GAL3ST1 in tumors correlated with the serum levels of their metabolites, lacto/neolacto-series glycosphingolipid and sulfatide, in NSCLC patients. Since it is not practical to obtain sufficient tumor biospies from NSCLC patient at different stages for sphingolipidomics analysis and diagnostic purpose, it therefore highlights the importance of our serum sphingolipidomics study.
Our approach also led to discovery of a circular organization of sphingolipid coregulatory network in NSCLC patients. Due to the fact that the circular sphingolipid network reflected the correlation among individual sphingolipids, sphingolipid metabolic changes, and the spatial organization of sphingolipids, it provided us a landscape of the sphingolipid metabolic reprogramming in cancer progression, indicating that the lacto/neolacto-series glycosphingolipid/sulfatide balance acted like a checkpoint to dictate sphingolipid metabolic change in cancer progression. In other words, our network analysis successfully identified B3GNT5 and GAL3ST1 as the key regulators of sphingolipid metabolism in lung cancer progression. At the cellular level, we showed that B3GNT5 and GAL3ST1 differentially regulated cancer cell proliferation, migration, invasion, sphere formation as well as in vivo tumor growth. Consistent with this finding, the intracellular level of lacto/neolacto-series glycosphingolipid and sulfatide has been recently implicated in controlling cancer cell properties [40,48]. Interestingly, sphingolipidomics analysis of B3GNT5/GAL3ST1 genetically perturbed lung cancer cell lines confirmed their regulatory role in sphingolipid metabolism, indicating that their expression differentially regulated the synthesis of lacto/neolacto-series glycosphingolipid and sulfatide in order to determine sphingolipid metabolic reprogramming in lung cancer cells. Importantly, we showed that the expression of B3GNT5 and GAL3ST1 was negatively correlated in human tumor tissues and some NSCLC cell lines. Further study revealed that B3GNT5 overexpression/depletion in A549/HCC827/H1299 cell lines caused a significant change in GAL3ST1 expression, while overexpression/knock-down of GAL3ST1 altered the expression of B3GNT5 in the cell lines. This result may explain why we observed up-regulation of lacto/neolacto-series GSL and down-regulation of sulfatide in B3GNT5 overexpressing cells/GAL3ST1 depleted cells and vice versa. Further investigation is required to understand the potential regulatory mechanism of their expression and develop specific small inhibitors/activators targeting these two important sphingolipid metabolic enzymes in the future.
Our work provides novel insight whereby the lacto/neolacto-series glycosphingolipid/sulfatide balance works as a checkpoint to determine sphingolipid metabolic reprogramming in lung cancer growth and progression, and that is controlled by B3GNT5/GAL3ST1 expression. Importantly, we identify potential serum sphingolipid biomarkers for NSCLC stage classification and new therapeutic targets for an improved lung cancer treatment.
Contributors
Q.M., X-T.H., X-B.Z. and X.K. carried out majority of the experiments and analyzed and interpreted the data, and contributed equally to the paper; Y-M.M., Y.C., L.S., X.J., X.Q., C.H. assisted with the experiments; C.L., and M.W., provided clinical samples; C.L., M.W. and P.-P.W. conceived the study. Q.M., X.K. and P.-P.W. verified the underlying data. Q.M. and P.-P.W. wrote the manuscript. All authors reviewed and approved the final manuscript.
Data sharing statement
All data are available in this manuscript and supplementary files.
Declaration of Competing Interests
The authors have no relevant conflicts to report.
Acknowledgements
This work was supported by the Natural Science Foundation of China (81872142, 81920108028); Guangzhou Science and Technology Program (201904020008); Guangdong Science and Technology Department (2020A0505100029, 2020B1212060018, 2020B1212030004); Guangdong Natural Science Foundation (Grant no. 2019A155011802, 2020A1515011280) and China Postdoctoral Science Foundation (2019M650226, 2019M650227).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103301.
Contributor Information
Chao Liu, Email: liuchao3@mail.sysu.edu.cn.
Minghui Wang, Email: wmingh@mail.sysu.edu.cn.
Ping-Pui Wong, Email: huangbp3@mail.sysu.edu.cn.
Appendix. Supplementary materials
Reference
- 1.Torre LA, Freddie B, Siegel RL, Jacques F, Joannie LT, Ahmedin J. Global cancer statistics, 2012. Ca A Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.R R, TG B, N K. Genetics and biomarkers in personalisation of lung cancer treatment. J Lancet. 2013;382(9893):720–731. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 3.Reck M, Rabe KF. Precision diagnosis and treatment for advanced non–small-cell lung cancer. N Engl J Med. 2017;377(9):849–861. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 4.Galvan-Femenia I, Guindo M, Duran X, Calabuig-Farinas S, Mercader JM, Ramirez JL. Genomic profiling in advanced stage non-small-cell lung cancer patients with platinum-based chemotherapy identifies germline variants with prognostic value in SMYD2. Cancer Treat Res Commun. 2018;15:21–31. doi: 10.1016/j.ctarc.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Petrache I, Berdyshev EV. Ceramide signaling and metabolism in pathophysiological states of the lung. Annu Rev Physiol. 2015;78(1):463. doi: 10.1146/annurev-physiol-021115-105221. [DOI] [PubMed] [Google Scholar]
- 6.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18(1):33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2017;19(3):175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suchanski J, Grzegrzolka J, Owczarek T, Pasikowski P, Piotrowska A, Kocbach B. Sulfatide decreases the resistance to stress-induced apoptosis and increases P-selectin-mediated adhesion: a two-edged sword in breast cancer progression. Breast Cancer Res. 2018;20(1):133. doi: 10.1186/s13058-018-1058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrill AH., Jr Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev. 2011;111(10):6387–6422. doi: 10.1021/cr2002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuvillier O, Pirianov G, Kleuser B, Vanek P, Coso O, Gutkind S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 1996;381(6585):800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 11.Jatoi A, Suman V, Schaefer P, Block M, Loprinzi C, Roche P. A phase II study of topical ceramides for cutaneous breast cancer. Breast Cancer Res Treat. 2003;80(1):99–104. doi: 10.1023/A:1024409123726. [DOI] [PubMed] [Google Scholar]
- 12.Shaw J, Costa-Pinheiro P, Patterson L, Drews K, Spiegel S, Kester MJAicr. Novel Sphingolipid-Based Cancer Therapeutics in the Personalized Medicine Era. 2018;140:327-66. [DOI] [PMC free article] [PubMed]
- 13.Faul F, Erdfelder E, Lang A-G, Buchner A.G* Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 14.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5(186):186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129(2):355–364. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]
- 16.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19(10):2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 17.Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC, Chou HY. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 2012;103(9):1640–1650. doi: 10.1111/j.1349-7006.2012.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferriols LF, Pitarch MJ, Magraner GJ. 2006. Pharmacoeconomic assessment of taxanes as first-line therapy for advanced or metastatic non-microcytic lung cancer. [DOI] [PubMed] [Google Scholar]
- 19.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5(4):e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.YM M, J L, C W, J X, DN Z, XJ Y. Monocytes/Macrophages promote vascular CXCR4 expression via the ERK pathway in hepatocellular carcinoma. Oncoimmunology. 2018;7(3) doi: 10.1080/2162402X.2017.1408745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Liu J, Chen J, Wang H, Yang L, Chen F. A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat Commun. 2018;9(1):1614. doi: 10.1038/s41467-018-03537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JR, Zhang H, Yau LF, Mi JN, Lee S, Lee KC. Improved sphingolipidomic approach based on ultra-high performance liquid chromatography and multiple mass spectrometries with application to cellular neurotoxicity. Anal Chem. 2014;86(12):5688–5696. doi: 10.1021/ac5009964. [DOI] [PubMed] [Google Scholar]
- 24.Mi J, Han Y, Xu Y, Kou J, Li WJ, Wang JR. Deep profiling of immunosuppressive glycosphingolipids and sphingomyelins in wild cordyceps. J Agric Food Chem. 2018;66(34):8991–8998. doi: 10.1021/acs.jafc.8b02706. [DOI] [PubMed] [Google Scholar]
- 25.Zou R, Zhang D, Lv L, Shi W, Song Z, Yi B. Bioinformatic gene analysis for potential biomarkers and therapeutic targets of atrial fibrillation-related stroke. J Transl Med. 2019;17(1):45. doi: 10.1186/s12967-019-1790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MS K, B S, LX H, CL B, A F, GI V. A conserved circular network of coregulated lipids modulates innate immune responses. Cell. 2015;162(1):170–183. doi: 10.1016/j.cell.2015.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong PP, Demircioglu F, Ghazaly E, Alrawashdeh W, Stratford MR, Scudamore CL. Dual-action combination therapy enhances angiogenesis while reducing tumor growth and spread. Cancer Cell. 2015;27(1):123–137. doi: 10.1016/j.ccell.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 28.Wang JR, Gao WN, Grimm R, Jiang S, Liang Y, Ye H. A method to identify trace sulfated IgG N-glycans as biomarkers for rheumatoid arthritis. Nat Commun. 2017;8(1):631. doi: 10.1038/s41467-017-00662-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Z Y, H C, Y Z, J A, Y L, W G. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: a pilot study. Oncol Lett. 2018;16(1):761–768. doi: 10.3892/ol.2018.8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54(6):1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K F, Y O, Y O, RH B, P Z, O T. New era of research on cancer-associated glycosphingolipids. Cancer Sci. 2019;110(5):1544–1551. doi: 10.1111/cas.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Don AS, Lim XY, Couttas TA. Re-configuration of sphingolipid metabolism by oncogenic transformation. Biomolecules. 2014;4(1):315–353. doi: 10.3390/biom4010315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obayashi T, Kagaya Y, Aoki Y, Tadaka S, Kinoshita K. COXPRESdb v7: a gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2018;47(D1):D55–D62. doi: 10.1093/nar/gky1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biancolillo A, De Luca S, Bassi S, Roudier L, Bucci R, Magri AD. Authentication of an Italian PDO hazelnut ("Nocciola Romana") by NIR spectroscopy. Environ Sci Pollut Res Int. 2018;25(29):28780–28786. doi: 10.1007/s11356-018-1755-2. [DOI] [PubMed] [Google Scholar]
- 36.Yu Z, Chen H, Ai J, Zhu Y, Li Y, Borgia JA. Global lipidomics identified plasma lipids as novel biomarkers for early detection of lung cancer. Oncotarget. 2017;8(64) doi: 10.18632/oncotarget.22391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Q, Zhang P, Wu R, Lu K, Zhou H. Identifying the best marker combination in CEA, CA125, CY211, NSE, and SCC for lung cancer screening by combining ROC curve and logistic regression analyses: is it feasible? Dis Markers. 2018;2018 doi: 10.1155/2018/2082840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang PK, Hsiao M, Hsu TL, Chang CF, Wu CY, Chen BR. Signaling pathway of globo-series glycosphingolipids and beta1,3-galactosyltransferase V (beta3GalT5) in breast cancer. Proc Natl Acad Sci USA. 2019;116(9):3518–3523. doi: 10.1073/pnas.1816946116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.S I, K A, T M, M A, SI N, structure HHJC. Glycosphingolipid GM2 Induces Invasiveness in Irradiation-tolerant. Lung Cancer Cells. 2018;43(2):177–185. doi: 10.1247/csf.18026. [DOI] [PubMed] [Google Scholar]
- 40.Dong YW, Wang R, Cai QQ, Qi B, Wu W, Zhang YH. Sulfatide epigenetically regulates miR-223 and promotes the migration of human hepatocellular carcinoma cells. J Hepatol. 2014;60(4):792–801. doi: 10.1016/j.jhep.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Alberg AJ, Armeson K, Pierce JS, Bielawski J, Bielawska A, Visvanathan K. Plasma sphingolipids and lung cancer: a population-based, nested case-control study. Cancer Epidemiol Biomarkers Prev. 2013;22(8):1374–1382. doi: 10.1158/1055-9965.EPI-12-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosa R, Marciano R, Malapelle U, Formisano L, Nappi L, D'Amato C. Sphingosine kinase 1 overexpression contributes to cetuximab resistance in human colorectal cancer models. Clin Cancer Res. 2013;19(1):138–147. doi: 10.1158/1078-0432.CCR-12-1050. [DOI] [PubMed] [Google Scholar]
- 43.Hart PC, Chiyoda T, Liu X, Weigert M, Curtis M, Chiang CY. SPHK1 is a novel target of metformin in ovarian cancer. Mol Cancer Res. 2019 doi: 10.1158/1541-7786.MCR-18-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rex K, Jeffries S, Brown ML, Carlson T, Coxon A, Fajardo F. Sphingosine kinase activity is not required for tumor cell viability. PLoS One. 2013;8(7):e68328. doi: 10.1371/journal.pone.0068328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnute ME, McReynolds MD, Kasten T, Yates M, Jerome G, Rains JW. Modulation of cellular S1P levels with a novel, potent and specific inhibitor of sphingosine kinase-1. Biochem J. 2012;444(1):79–88. doi: 10.1042/BJ20111929. [DOI] [PubMed] [Google Scholar]
- 46.Moro K, Kawaguchi T, Tsuchida J, Gabriel E, Qi Q, Yan L. Ceramide species are elevated in human breast cancer and are associated with less aggressiveness. Oncotarget. 2018;9(28):19874–19890. doi: 10.18632/oncotarget.24903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ho MY, Yu AL, Yu J. Glycosphingolipid dynamics in human embryonic stem cell and cancer: their characterization and biomedical implications. Glycoconj J. 2017;34(6):765–777. doi: 10.1007/s10719-016-9715-x. [DOI] [PubMed] [Google Scholar]
- 48.Ishihara S, Aoki K, Mizutani T, Amano M, Nishimura S-I, Haga H. Glycosphingolipid GM2 Induces Invasiveness in Irradiation-tolerant Lung Cancer Cells. Cell Struct Function. 2018;43(2):177–185. doi: 10.1247/csf.18026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.