Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) is most aggressive among all gastrointestinal tumors. The complex intra-tumor heterogeneity and special tumor microenvironment in PDAC bring great challenges for developing effective treatment strategies. We aimed to delineate dynamic changes of tumor microenvironment components during PDAC malignant progression utilizing single-cell RNA sequencing.
Methods
A total of 11 samples (4 PDAC I, 4 PDAC II, 3 PDAC III) were used to construct expression matrix. After identifying distinct cell clusters, subcluster analysis for each cluster was performed. New cancer associated fibroblasts (CAFs) subset was validated by weighted gene co-expression network analysis, RNA in situ hybridization and immunofluorescence.
Findings
We found that ductal cells were not dominant component while tumor infiltrating immune cells and pancreatic stellate cells gradually accumulated during tumor development. We defined several new Treg and exhausted T cell signature genes, including DUSP4, FANK1 and LAIR2. The analysis of TCGA datasets showed that patients with high expression of DUSP4 had significantly worse prognosis. In addition, we identified a new CAFs subset (complement-secreting CAFs, csCAFs), which specifically expresses complement system components, and constructed csCAFs-related module by weighted gene co-expression network analysis. The csCAFs were located in the tissue stroma adjacent to malignant ductal cells only in early PDAC.
Interpretation
We systematically explored PDAC heterogeneity and identified csCAFs as a new CAFs subset special to PDAC, which may be valuable for understanding the crosstalk inside tumor.
Funding
This study was supported by The Natural Science Foundation of China (NO.81572339, 81672353, 81871954) and the Youth Clinical Research Project of Peking University First Hospital (2018CR28).
Keywords: Pancreatic cancer, scRNA-seq, Tumor microenvironment, Cancer associated fibroblasts, Intra-tumor heterogeneity
Research in context.
Evidence before this study
A growing body of studies revealed the extensive intra-tumor heterogeneity in breast cancer, liver cancer, lung cancer and pancreatic cancer using bulk-seq. However, little is known about dynamic changes of tumor microenvironment during PDAC malignant progression using single cell sequencing (scRNA-seq). CAFs play a pivotal role in facilitating tumor survival and migration, while many therapeutic strategies to deplete fibroblasts failed. It is imperative to further understand the role of CAFs.
Added value of this study
We systematically explored the dynamic changes of tumor microenvironment from early PDAC to late PDAC using scRNA-seq analysis, and identified a new cancer-associated fibroblasts (CAFs) subpopulation, named as csCAFs. Weighted gene co-expression network (WGCNA) was applied to verify their identification and biological function. More importantly, we also confirmed the existence of the new CAF subset and analyzed their location in human pancreatic cancer tissue.
Implications of all the available evidence
Together, our study may provide a valuable clue to understand the CAF heterogeneity and help develop new anti-CAF drugs for improving the prognosis of PDAC patients.
Alt-text: Unlabelled box
Introduction
Pancreatic ductal adenocarcinoma (PDAC) has the highest mortality among all gastrointestinal tumors. Based on annual cancer statistical reports, there has been only a slight improvement in the prognosis of PDAC with a 5-year survival rate of 8-9% recently [1]. This plight is mainly attributed to the lack of reliable markers for early diagnosis, low surgical resection rate and chemoradiotherapy resistance of PDAC, and only 15–20% patients with pancreatic cancer are eligible for radical excision, which is the only potential curative therapy [2,3]. The majority of PDAC patients have major vascular invasion and distant metastasis at the time of diagnosis. Even after curative resection, most of patients still suffer from local recurrence or systematic metastasis within a short time, with a 5-year survival rate of only 20-30% [4]. Despite recent paradigm shifting from the traditional "surgery first" approach to the modern "multi-disciplinary" that improved the short-term prognosis of PDAC patients, the multi-disciplinary approach did not contribute enough for their long-term survival [5]. Therefore, it is imperative to further explore pathological features of PDAC to develop effective treatment strategies.
Recently, single cell RNA-sequencing (scRNA-seq) made it increasingly possible to unveil the complex heterogeneity in PDAC microenvironment with unprecedented resolution [6], [7], [8], [9]. The scRNA-seq can be used to determine new cell types and their marker genes based on the availability of genome-wide expression data, indicate the developmental trajectories of specific cells by pseudotime analysis (Monocle2) [10], and identify gene-gene co-regulated network [11], [12], [13]. For example, scRNA-seq revealed plenty of distinct immune cell subsets with different state in breast tumor microenvironment, which provided new insight that different immune cell subsets play a different role in promoting and opposing tumor progression [14]. In fact, PDAC is characterized by complex immune microenvironment comprised of multiple types of inflammatory cells, such as macrophage, T cell and B cell. Many studies have demonstrated that the regulatory T cells (Tregs), regulatory B cells (Bregs) and tumor associated macrophage (TAMs) in PDAC play a vital role in tumorigenesis and tumor progression, which are potential therapeutic targets [15], [16], [17], [18], [19]. Moreover, there are dense stroma consisted of proliferating cancer associated fibroblasts (CAFs) and abundant extracellular matrixes in PDAC. This natural barrier impedes drugs delivery and compresses the space of blood vessels, creating a pro-tumor milieu [20], [21], [22]. However, the depletion of αSMA (+) myofibroblasts could reduce desmoplasia but enhance hypoxia, epithelial-to-mesenchymal transition (EMT), and shorten animal survival [23]. Some clinical trials and in vivo studies demonstrated that the inhibition of Hh pathway could not improve the prognosis of PDAC but even accelerate disease progression [24], [25], [26]. Therefore, it is necessary to further understand the heterogeneity and biological function of CAFs.
In this study we applied single cell RNA-sequencing approach to systematically explore dynamic changes of tumor microenvironment during PDAC malignant progression. A total of 11 samples (4 PDAC I, 4 PDAC II, 3 PDAC III referring to patients with stage I, II and III PDAC respectively) were used to construct expression matrix for subsequent analysis. We found that the proportion of ductal cell, immune cell, CAF and their gene expression profile had significant changes from early PDAC to late PDAC. Interestingly, we identified few Tregs, TAMs/M2 with unique signature genes and compared biological process and molecular function in PDAC with different clinical stages by GO and KEGG analysis. Next we identified three distinct subtypes of CAFs in PDAC – cCAFs (classical CAFs), csCAFs (complement-secreting CAFs) and PSCs (pancreatic stellate cells), and we performed weighted gene co-expression network analysis (WGCNA) to confirm the identification of complement-secreting CAFs (csCAFs) in PDAC microenvironment. The csCAF related brown module genes were significantly enriched for the components of complement system. Finally, we analyzed the location of csCAFs in human PDAC tissue by immunofluorescence (IF) and RNA in situ hybridization (RNA ISH).
Methods
Human PDAC Samples for scRNA Sequencing, RNA ISH, IF and RT-qPCR
The raw expression matrix for single-cell transcriptome analysis was from GSA (Genome Sequence Archive) database (GSA: https://bigd.big.ac.cn/gsa), under accession number CRA001160 [27]. According to the article, all patient samples were obtained from Department of General Surgery of Peking Union Medical College Hospital (PUMCH). In addition, the clinical characteristics of PDAC patients profiled by scRNA-seq were listed (Supplementary Table S1).
A total of 21 postoperative PDAC specimens for RNA ISH and IF (3 PDACI, 3 PDACII, 3 PDACIII) and RT-qPCR (4 PDACI, 4 PDACII, 4 PDACIII) were retrieved from the department of general surgery of Peking University First Hospital in china. The clinical characteristics of 9 patients for RNA ISH and IF are shown in Table S1.
This study was approved by Ethics Committee of Peking University First Hospital (Approval No. 2019-147) and conducted in accordance with ethical guidelines (Declaration of Helsinki). Written informed consent was obtained from all participants.
Single cell RNA-seq data Quality Control and Analysis
Gene-cell matrixes were filtered to remove cells (<200 transcripts/cell, >10% mitochondria genes) and genes (<10 cells/gene). Then the matrix was imported into the R package Seurat (v 3.1.2) for subsequent analysis [28]. The gene expression levels were normalized so that the number of unique molecular identifiers in each cell (UMI/cell) is equal to the median UMI and then natural-log transformed. Total 2000 highly variable genes were generated for performing PCA reduction dimension. Significant principle components were determined using Jackstraw. Finally, single cell clustering was visualized by t-SNE (t-Distributed Stochastic Neighbour Embedding) utilizing previous computed principle components 1 to 10.
Gene Ontology and KEGG analysis and PPI network
The package (clusterProfiler) was applied to analyze and visualize functional profiles (GO and KEGG) of gene clusters [29]. We selected 0.05 as the cutoff of p value and q value. The protein-protein interaction network construction in ductal cell subcluster analysis were conducted through STRING online tool (STRING: https://string-db.org). All functions were run using default parameters.
TCGA and HPA database Analysis
The differential analysis, survival analysis and correlation analysis were performed with GEPIA (Gene Express Profiling Interactive Analysis) online tool, using a standard professing pipeline [30]. These data were from the prevalent TCGA and GTEx PDAC samples. In addition, we also detected the expression level of COL12A1, SDC1 and CTHR1 in normal pancreas tissue and pancreatic cancer tissue by IHC from HPA database (The Human Protein Atlas; HPA: https://www.proteinatlas.org) [31].
Pseudotime Analysis
R package (Monocle 2) was applied to conduct cellular trajectory analysis with the assumption that one-dimensional ‘time’ can describe the high-dimensional expression values, so called pseudotime analysis of single cells. The clusters identified as T cell and fibroblast cell were loaded into R environment. And we presented cell trajectory and position with tree structure in two-dimension space after log normalization and DDDTree reduction dimension. Then we set the pattern of each cell in the plot according to specific markers expression level.
Weighted Gene Co-expression Network Analysis
We randomly picked out 30–50 cells from cCAFs, csCAFs and PSCs respectively to construct gene expression matrix for WGCNA analysis. A signed network was constructed using any genes that was expressed at UMI value of 1 or higher at least 5 samples. After constructing the adjacency matrix and selecting appropriate soft-power (β = 3), we calculated the topological overlaps matrix (TOM). Genes with high similar co-expression relationship were grouped together using average linkage hierarchical clustering upon TOM. Through Dynamic Tree Cut algorithm, we completed the construction of gene network and identification of modules. Next we proved brown module was significantly associated with csCAFs subpopulation by scatter plot of Gene Significance (GS) vs Module Membership (MM) and eigengene adjacency heatmap. Finally, the hub genes representing csCAFs in brown module were exported to perform GO and KEGG analysis and visualization by Cytoscape software (v 3.6.0).
Real-time PCR Assay
Total RNA was extracted from human PDAC tissue and adjacent normal tissue by TRIzol reagent (Invitrogen, USA). First-strand cDNAs were synthesized from the 2 ug total RNA with ReverTra Ace® qPCR RT kit (TOYOBO, Japan) according to the manufacturer's instructions. And the quantitative real-time polymerase chain reaction (RT-qPCR) was conducted using SYBR Green Realtime PCR Master Mix (TOYOBO, Japan). Each sample was tested in triplicate wells in triplicate experiments. The relative expression level of C3 and C7 were normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and calculated by the 2- ΔΔCt method. The following primers were used: C3 (Forward primer: CGGATCTTCACCGTCAACCA, Reverse primer: GATGCCTTCCGGGTTCTCAA), C7 (Forward primer: CCTCTTTGCTGGGCAGATCA, Reverse primer: AAGCCGGGCACATAAATGGA), GAPDH (Forward primer: GTATTGGGCGCCTGGTCACC, Reverse primer: CGCTCCTGGAAGATGGTGATGG).
RNA in Situ Hybridization and Immunofluorescence Assay
RNA ISH was performed on freshly prepared 4% paraformaldehyde (DEPC water) fixed paraffin-embedded tissue sections. Sections were treated by protase K (20 ug/ml) for 8 min at 37°C, then washed three times in PBS. Each section was added hybridization buffer and incubated at 37°C for 1 h, then sections were incubated with probe against C3 (TTACCAGGGTCCTCCCAGCGGTTCTTATCT) and COL1A1 (CGCAGGTGATTGGTGGGATGTCTTCGTCT) overnight. After washing, sections were stained with DAPI (Servicebio, China) for 8 min in the dark and mounted.
IF was also performed on freshly prepared 4% paraformaldehyde fixed paraffin-embedded tissue sections. Deparaffinization and antigen retrieval were conducted according to the IF protocol. The 3% H2O2 was used to block endogenous peroxidase at room temperature for 25 min, then sections were blocked with 3% BSA at temperature for 30 min. After removing blocking solution, sections were incubated with primary antibodies for C3 (Abcam, 1:100 dilution) and COL1A1 (Servicebio, 1:200 dilution) overnight at 4°C, and washed with PBS, then stained with TSA-FITC (Servicebio, China) and TSA-CY3 (Servicebio, China) secondary antibodies for 1 h at room temperature. DAPI was used as counterstain.
RNA ISH and IF imaging of fixed tissue were conducted with Nikon Eclipse CI and Nikon DS-U3. All sections were scanned and then analyzed by CaseViewer 2.0 software. A total of 5 pictures for each section were obtained randomly to calculate the relative number of csCAFs in PDAC tissue with different stages.
Statistical Analysis
All data were presented as mean values and standard deviation (SD). Comparison of three group for PDAC with different stages was performed by one-way ANOVA in RNA ISH, IF and Real-time qPCR assay. Spearman's rank correlation was used to analyze the relationship between IL2RA and Treg/Exhausted T cells signature genes. The Kaplan-Meier method and the corresponding log-rank test were performed to identify the prognostic value of marker genes. All statistical analyses were conducted using SPSS version 22.0 software (SPSS, Chicago, IL, USA). Statistical significance was defined as *p < 0.05, **p < 0.01, ***p < 0.001.
Role of Funders
The funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Single-cell analysis uncovers cellular component and evolution during PDAC progression
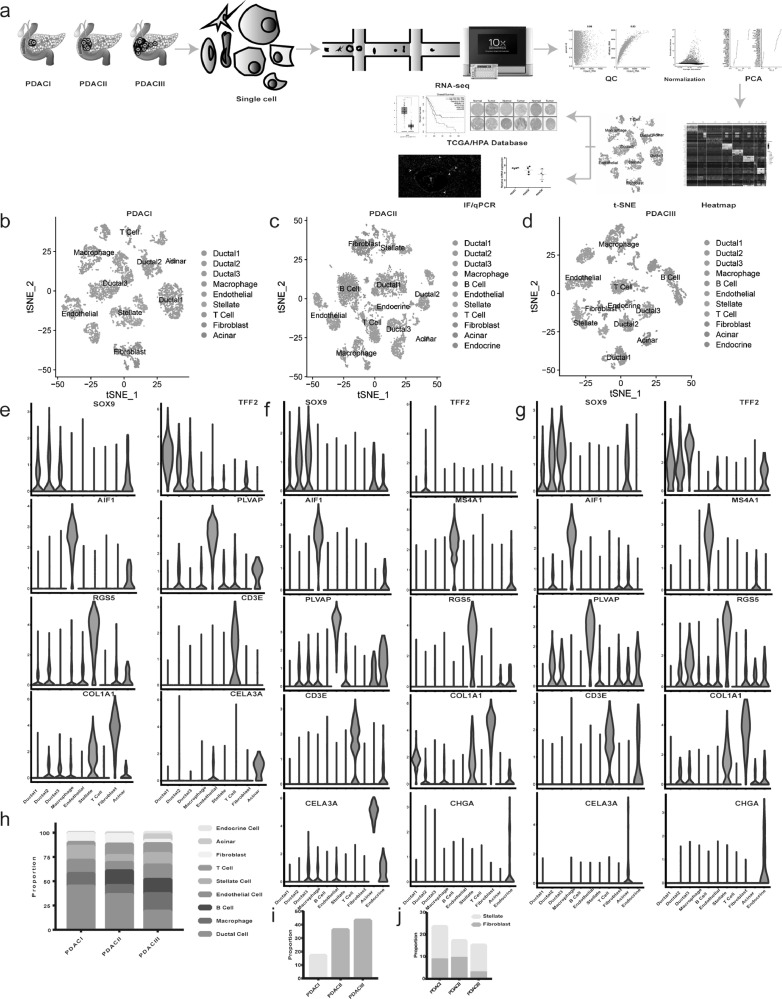
To comprehensively analyze dynamic changes in tumor microenvironment during PDAC malignant progression, 18555 cells from PDAC with different clinical stages were sequenced by single-cell RNA-seq approach (Fig. 1A and Supplementary Table S1 and 2). We performed gene filtering, normalization, principle component analysis, and finally identified 9 distinct clusters, including ductal cell, macrophage, B Cell, endothelial cell, stellate cell, T cell, fibroblast, acinar and endocrine cell (Fig. 1 B-D and Supplementary Fig.S1A-D). Signature genes for each cluster were in accordance with well-known cell markers recorded in the literature (Fig. 1 E-G and Supplementary Fig.S1E and Table S3).
Fig. 1.
scRNA-seq delineats the dynamic changes of tumor microenvironment components during PDAC progression. (a) Workflow described by flow diagram. PDAC samples with different clinical stages were dissociated into single cells, all cells were subjective to capture, library preparation, RNA-seq using 10 × Genomics platform, then the QC, normalization, PCA and subsequent bioinformatics analysis were conducted. (b-d) The t-distributed stochastic neighbor embedding (t-SNE) plot showing clustering information in PDACI, II, III respectively. (e-g) Violin plots demonstrating the identity of each cluster through analyzing the expression of well-known cell type specific markers. (h) The proportion of cells changing from PDAC I, II to PDAC III. (i) The proportion of immune cells (T cell, B cell and macrophage) gradually increased from PDAC I, II to PDAC III. (j) The bar chart showing the proportion of pancreatic stellate cells (PSCs) in all cancer associated fibroblasts (CAFs) in PDAC I, II, III separately.
Then we counted each cluster in three groups, which represented the samples with different clinical stages, and found that the majority of cells in PDAC I were ductal cells (44.88%), and identified only a small number of immune cells (17.06%). Interestingly, the number of ductal cells gradually decreased with tumor stage progression (44.88%-36.04%–18.95%) (Fig. 1 H and Supplementary Table S4), while the proportion of immune cells, including T cell, B cell and macrophage, increased gradually (17.06%-36.15%–43.14%) (Fig. 1 I). It suggested that ductal cell would not be the major component in tumor microenvironment when the tumor developed toward higher malignancy, immune cells migrated and accumulated in tumor, reflecting human immune system's response for tumor lesion. In addition, the amount of cancer-associated fibroblasts (CAFs) consisted of fibroblasts and stellate cells decreased gradually (23.49%–17.09%–15.12%), but the stellate cells evolved into predominant population in PDAC III, which could account for abundant extracellular matrix in advanced PDAC (Fig. 1 J).
Ductal cell heterogeneity during PDAC progression
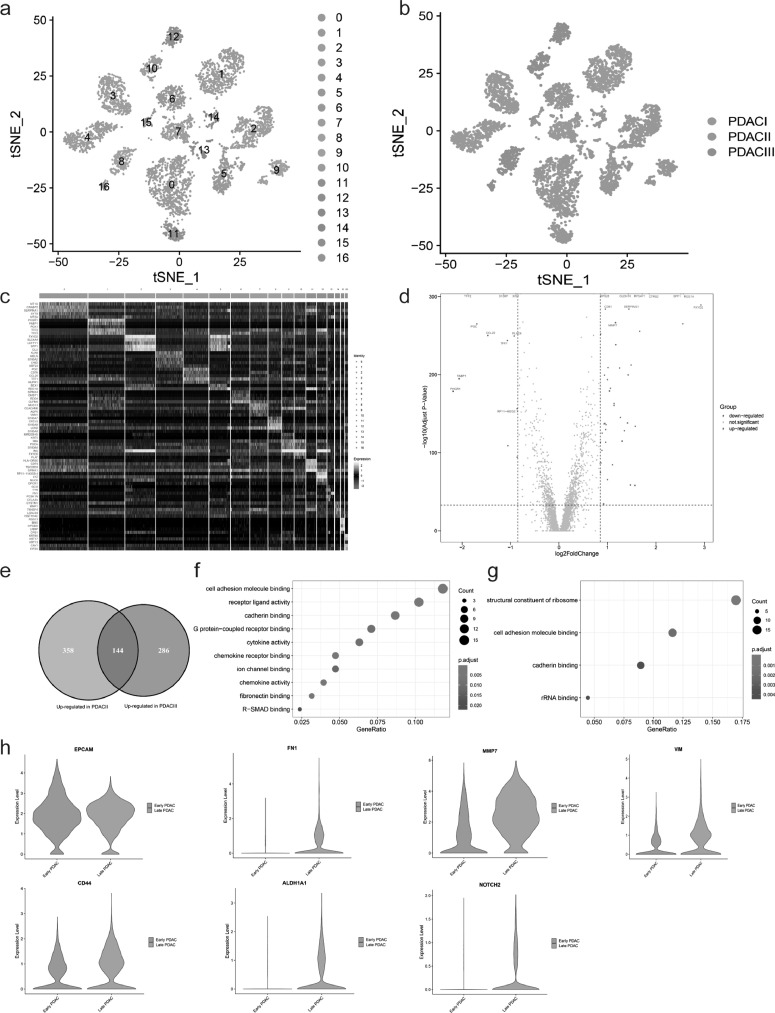
To reveal changes of ductal cells during PDAC malignant progression, we conducted the subcluster analysis for ductal cells. A total of 5800 ductal cells were analyzed, which were divided into different clusters with unique gene signatures upon t-SNE analysis (Fig. 2 A and C). Furthermore, t-SNE plot indicated that ductal cells from different tumor stage formed the markedly different clusters (Fig. 2B), suggesting gradual changes in gene expression pattern of ductal cells during PDAC malignant progression. Common ductal cell markers such as SOX9 and epithelial cell marker EPCAM were expressed in all ductal cell subpopulations, confirming ductal cell identity (Supplementary Fig.S2A).
Fig. 2.
Late PDAC is characterized by Epithelial-Mesenchymal Transition (EMT) and Cancer Stem cell (CSC) properties. (a-b) The t-SNE plots showing ductal cell subpopulations in subcluster analysis, according to clustering distribution in the a, sample source in the b. (c) Heatmap presenting the marker genes expression levels among ductal cell subpopulation. (d) Volcano plot showing the differential expression genes (DEGs) of ductal cells between PDAC I and II, red and blue dots represented the genes upregulated and downregulated respectively (PDAC II vs PDAC I). The names of top 10 up/downregulated genes were marked. (e) Venn diagram indicating the intersection between upregulated genes (PDAC II vs I) and those (PDAC III vs I). (f-g) Gene ontology analysis showing molecular function terms for genes upregulated in PDAC II and III in the f, in PDAC I in the g. (h) Violin plot demonstrating late PDAC had significantly higher expression levels of mesenchymal and cancer stem cell markers, and EPCAM (ductal cell marker) confirming their ductal cell identity.
We then compared ductal cells from PDAC II with those from PDAC I, and detected 502 upregulated and 259 downregulated genes (Fig. 2 D), and 144/502 of the upregulated genes were also significantly upregulated in ductal cells from PDAC III (Fig. 2 E and Supplementary Fig.S2B). GO analysis indicated that the upregulated genes in late PDAC (PDAC II and III) were significantly enriched for several cancer-related terms, such as cell adhesion, cytokine activity, chemokine receptor binding, chemokine activity and R-SMAD binding (Fig. 2 F and Supplementary Fig.S2C and E), suggesting increased ability of tumor cells to respond to cytokine and chemokine with tumor progression. In contrast, the upregulated genes in early PDAC (PDAC I) were mainly related to normal biological function, including structural constituent of ribosome, cell adhesion molecule binding, cadherin binding and rRNA binding (Fig. 2G and Supplementary Fig.S2D and F).
In addition, we performed gene expression analysis of epithelial markers (EPCAM), mesenchymal markers (FN1, MMP7 and VIM) and cancer stem cell (CSC) related markers (CD44, ALDH1A1 and NOTCH2). The results showed that ductal cells in early PDAC mainly exhibited epithelial expression profile, in contrast with ductal cells in late PDAC which were enriched for mesenchymal markers and had higher expression levels of CSC related genes (Fig. 2 H). These data supported the notion that epithelial-mesenchymal transition and cancer stem cell properties gradually accumulated with the progression of tumor, promoting tumor invasion and metastasis.
New subgroup of Cancer-Associated Fibroblasts was detected
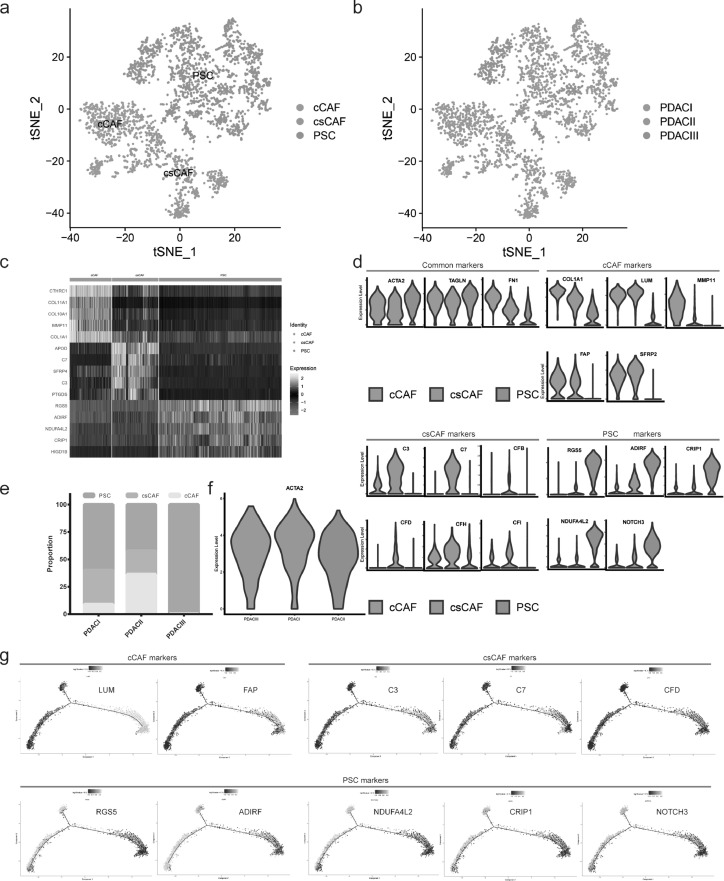
In order to further corroborate the finding that there exist three main subpopulations of CAF – myCAFs (myofibroblastic CAFs), iCAFs (inflammatory CAFs) and apCAFs (antigen-presenting CAFs) in PDAC microenvironment [9,32]. A total of 2958 mesenchymal cells from PDAC with different clinical stages were analyzed, which formed three distinct subclusters with unique gene signatures upon t-SNE analysis (Fig. 3 A-C and Supplementary Fig.S3A and Table S5). Common markers, such as ACTA2, TAGLN and FN1, were expressed in all subpopulations, confirming PDAC mesenchymal cell identity (Fig. 3D).
Fig. 3.
New cancer-associated fibroblast subset: csCAFs (complement-secreting CAFs) were detected. (a-b) The t-SNE plot showing CAF subpopulations in subcluster analysis, according to clustering distribution in the a, sample source in the b. (c) Heatmap presenting the marker genes expression levels among CAF subpopulations. (d) Violin plot demonstrating the identity of each CAF subcluster, pan-CAF common markers (top left), cCAFs markers (top right), csCAFs markers (bottom left), PSCs markers (bottom right). (e) The proportion of CAF subpopulations changing during PDAC progression. (f) Violin plot showing the ACTA2 (αSMA) expression levels of PSC in PDAC I, II, III respectively. (g) Pseudotime analysis exploring the development trajectories of CAFs, each dot corresponds to a single cell, the CAFs subtype shown according to specific markers expression level.
In addition, three subclusters were identified according to specific gene expression profile: cCAFs (classical CAFs) expressed high levels of COL1A1, LUM (extracellular matrix related components), MMP11 (matrix remodeling molecular), FAP (stromal fibroblast activeted protein) and SFRP2 (modulators of Wnt signaling), which are involved in extracellular matrix deposition; csCAFs (complement-secreting CAFs) showed highly activated transcription of complement system, including C3, C7, CFB, CFD, CFH, CFI, which may regulate immune and inflammation response within the tumor; PSCs (pancreatic stellate cells) were distinct from both cCAF and csCAF, and expressed marker genes of stellate cells, such as RGS5, ADIRF, CRIP1, NDUFA4L2, NOTCH3 and PDGFA (Fig. 3D). Then we analyzed the proportion of these subpopulations in PDAC with different clinical stages. The microenvironment in PDAC III contained only one kind of mesenchymal cell (PSCs) but not cCAFs and csCAFs. Furthermore, PSCs were predominant in PDAC I/II/III and expressed high levels of αSMA (encoded by ACTA2) (Fig. 3E-F), consistent with previous conclusion that PDAC is characterized by a dense stroma consisted of proliferating myfibroblasts (pancreatic stellate cell) and extracellular matrix components [4].
We applied the Monocle 2 algorithm to explore the developmental trajectories of three subpopulations based on transcriptional similarities [10]. The results indicated that tree structure began with cCAFs with signature genes of LUM and FAP and csCAFs with signature genes of C3, C7, CFD, and ended with PSCs with signature genes of RGS5, ADIRF, NDUFA4L2, CRIP1 and NOTCH3, which demonstrated the possibility of evolution from cCAFs and csCAFs towards PSCs (Fig. 3G). Simultaneously, we also found the development trend of cells from early PDAC to late PDAC, which was in accordance with tumor progression direction (Supplementary Fig.S3B).
To characterize csCAFs which expressed high levels of complement, we compared csCAFs and cCAFs, and detected 333 upregulated and 273 downregulated genes (Supplementary Fig.S3C). KEGG analysis indicated that the upregulated genes were significantly enriched for key signaling pathways such as MAPK, Rap1, Foxo and p53 signaling pathway (Supplementary Fig.S3D). Moreover, we analyzed a small subcluster of PSCs which only represented PDAC III (cluster 4) and found that the marker genes were enriched in biological process related to IFN gamma mediated response and presentation of peptide antigen (Supplementary Fig.S3E and Table S6). Furthermore, we utilized TCGA and HPA (The Human Protein Atlas) database to examine clinical significance of cCAFs marker genes, and found that the markers including COL12A1, SDC1 and CTHRC1 had higher expression level in PDAC compared with normal tissue and indicated worse prognosis (Supplementary Fig.S3F-G).
Landscape of tumor infiltrating immune cells in PDAC
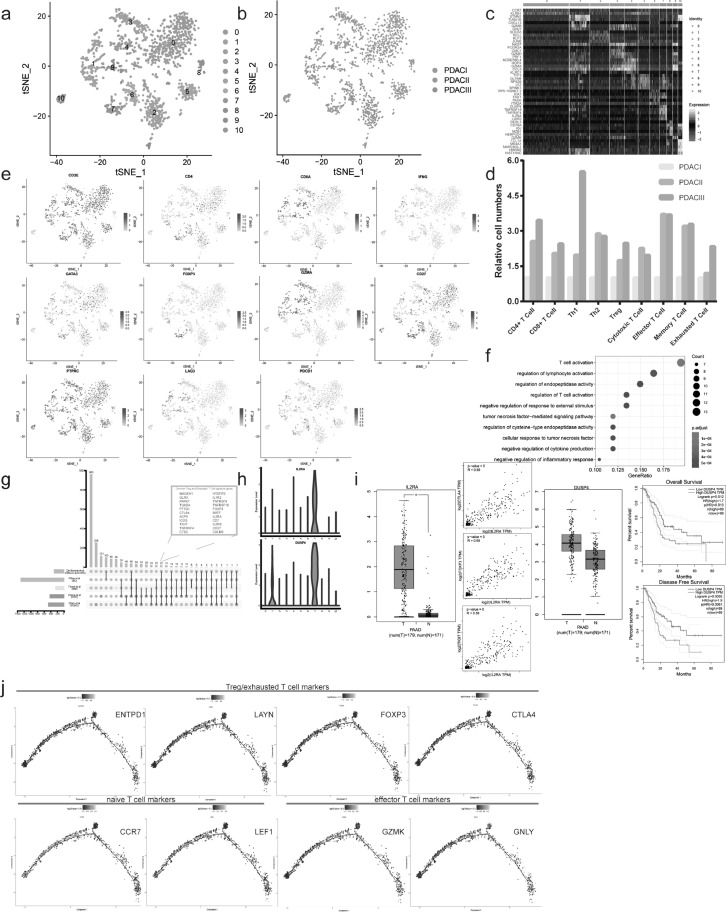
Tumor infiltrating immune cells, including Tregs, exhausted T cells, tumor associated macrophages (TAMs) and Bregs, play significant role in tumorigenesis and metastasis [33], [34], [35]. And cancer immunotherapies aiming at tumor microenvironment, such as anti-CTLA4 and anti-PD1 therapies obviously improved patients’ prognosis in some cancers. However, their therapeutic responses varied according to cancer types and patients [36]. So it's imperative to screen out candidate markers to predict the response to treatments and develop new immunotherapy strategies based on further understanding to tumor microenvironment. Therefore, we analyzed total 1521 T cells in PDAC, which formed 11 distinct subclusters with unique signature genes upon unsupervised clustering (Fig. 4A-C and Supplementary Table S7). Based on specific cell markers, the relative proportion of subsets, including CD4+T cell, CD8+T cell, Th1/2, Treg, cytotoxic T cell, effector T cell, memory T cell and exhausted T cell, were investigated. PDAC II/III presented more cytotoxic T cells, effector T cells and memory T cells than PDAC I. Simultaneously, the results showed more Tregs and exhausted T cells accumulated in the advanced PDAC as well (Fig. 4D-E). In fact, anti-tumor T cell responses arose in PDAC but may be disabled during tumor progression by the negative regulation from accumulated Tregs and exhausted T cells.
Fig. 4.
The evolution of T cell subpopulations during PDAC malignant development. (a-b) The t-SNE plots showing T cell subpopulations in subcluster analysis, according to clustering distribution in the a, sample source in the b. (c) Single-cell profiling heatmap comparing the marker genes expression level among T cell subcluster, each column and row represented a single cell and gene respectively. (d) T cell subsets proportion in PDAC with different clinical stages. (e) The t-SNE plots showing the expression level of specific T cell subsets marker gene. (f) GO analysis showing biological process terms for marker genes in Tregs (cluster7). (g) Upsetplot for Treg and exhausted T cell signature genes from different studies, the box in the upper right enumerated 22 common signature gene among all studies. (h) DUSP4 and IL2RA having specific expression pattern in Treg/exhausted T cell (clsuter7) based on violin plot. (i) TCGA database indicating the value of IL2RA and DUSP4 as diagnosis and prognosis markers. Scatter plot showing IL2RA has significant correlation with canonical Treg/exhausted T cell markers (CTLA4, FOXP3, TIGIT). (j) Pseudotime analysis exploring the development trajectories of T cell (naïve T cell – effector T cell – Treg/exhausted T cell).
In addition, we compared the cluster 2 (representing PDAC III) and cluster 0 (representing PDAC II), and found 116 upregulated genes and 114 downregulated genes. The genes with higher expression levels in PDAC II than PDAC III were mainly enriched for T cell activation, regulation of lymphocyte activation, lymphocyte differentiation and MHC class II receptor activity based on GO analysis (Supplementary Fig.S4A-C).
Interestingly, the cluster 7 had enriched expression of known regulatory and exhausted T cell markers, FOXP3, TNFRSF18, CTLA4, TIGIT, LAYN, and was therefore identified as Treg/exhausted T cell. This subpopulation was only a small part of total T cells, and total 481 genes were specifically expressed in this subcluster, and significantly enriched in some biological processes, including regulation of T cell activation, negative regulation of cytokine production, negative regulation of inflammatory response (Fig. 4F and Supplementary Table S7). There was an obvious overlap between signature genes for cluster 7 and Treg and exhausted T cell markers reported in previous studies on no-small cell lung cancer, colorectal cancer, breast cancer, melanoma and hepatocellular carcinoma [33,[37], [38], [39]]. Among the 33 common markers from previous studies, 22 genes were identified in our data (Fig. 4G).
Next we focused on two important signature genes for the diagnosis and prognosis of PDAC. IL2RA (encoding interleukin 2 receptor alpha chain), IL2RB together with IL2RG constitute the high-affinity IL2 receptor, which is involved in the regulation of immune response by controlling regulatory T cells (Tregs) [40]. The expression level of IL2RA in pancreatic cancer samples was significantly higher than that of normal tissues, and we found a positive correlation between IL2RA and classical Treg markers – CTLA4, FOXP3, TIGIT (R: 0.66, 0.68, 0.58, respectively) using TCGA database (Fig. 4H-I). Another marker, DUSP4 (a member of the dual specificity protein phosphatase subfamily), exhibited highly specific expression pattern in cluster 7 (Treg/exhausted T cell), but was not found in previous reported common markers (Fig. 4H). Based on differential expression and survival analysis by TCGA database, the patients with high expression level of DUSP4 had significantly worse prognosis than those with low expression level of DUSP4 (OS: HR = 1.7, p = 0.013; DFS: HR = 1.9, p =0.0061) (Fig. 4I). In addition, correlation analysis indicated that T cells with high expression of regulatory/exhausted T cell markers, including CTLA4, DUSP4, FOXP3, MAGEH1, THADA, TIGIT, TNFRSF4/9/18, had the lower expression level of Interferon Gamma (IFNG) based on single cell sequencing data (Supplementary Fig.S4D), suggesting that Treg/exhausted T cell may suppress activated lymphocytes by downregulating IFNG expression.
To better understand the functional states and relationship of T cells, we utilized Monocle 2 algorithm to figure out their developmental trajectories. The pseudotime analysis showed that tree structure began with naïve T cells with the signature genes of CCR7, LEF1, followed by effector T cells with the signature genes of GZMK, GNLY and Treg/exhausted T cells with the signature genes of ENTPD1, LAYN, FOXP3 and CTLA4 (Fig. 4J). The regulatory and exhausted T cells mainly were enriched at late T cell development, demonstrating T cell state transition from activation to suppression and exhaustion, consistent with previous study [39].
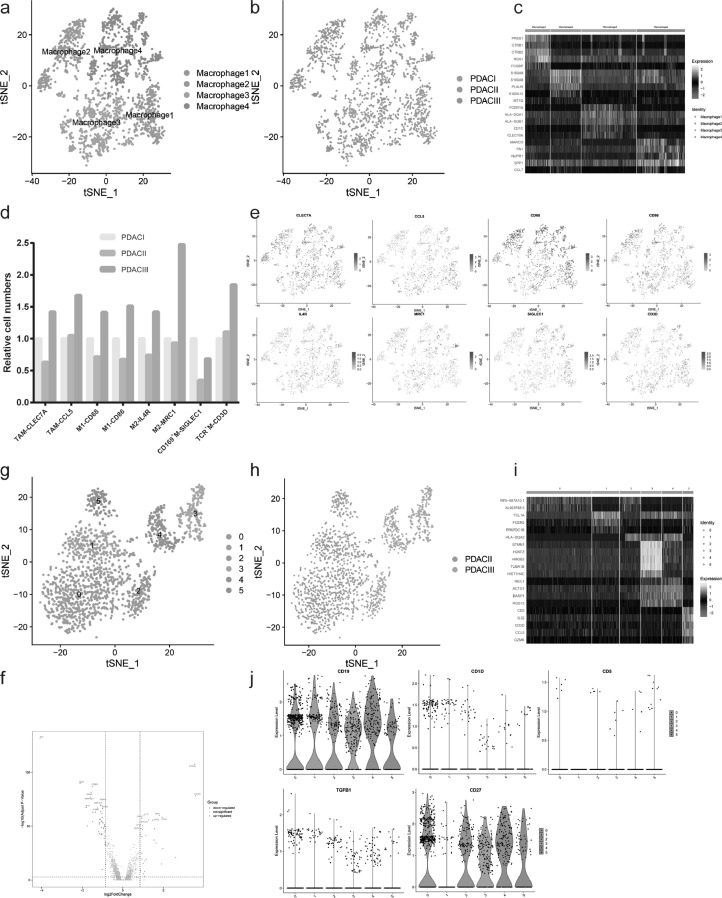
Another important subgroup of tumor infiltrating immune cells is macrophage, the heterogeneity of which remains unclear in PDAC. Next we characterized macrophage heterogeneity during PDAC progression using unsupervised clustering, and revealed 4 distinguished macrophage subpopulations (Fig. 5 A-C and Supplementary Table S8). According to the cell markers of macrophage subsets, we counted the number of these subsets in PDAC with different clinical stages, including tumor associated macrophage (TAM), M1/2, CD169+ macrophage and TCR+ macrophage. The results showed that late PDAC (PDAC III) had more M1, M2 and TAM than early PDAC (PDAC I and II) (Fig. 5 D-E). In accordance with T cell subset analysis, the anti-tumor components (M1) increased, accompanied by the infiltration and accumulation of pro-tumor components (M2 and TAM), which created immune-suppressive environment. In addition, we compared macrophage 2 (representing PDAC III) and macrophage 1 (representing PDAC I and II) and found 296 upregulated genes and 461 downregulated genes (Fig. 5F). The upregulated genes in PDAC III were chiefly enriched for neutrophil degranulation, neutrophil mediated immunity, G protein-coupled receptor binding, cytokine and chemokine activity (Supplementary Fig.S4E-F).
Fig. 5.
The evolution of macrophage and B cell subpopulations during PDAC malignant development. (a-b) The t-SNE plots showing macrophage subpopulations in subcluster analysis according to clustering distribution in the a, sample source in the b. (c) Heatmap of macrophage subclusters and marker genes. (d) Macrophage subsets proportion in PDAC with different clinical stages. (e) The t-SNE plots showing the expression level of specific macrophage subsets marker genes. (f) The DEGs between PDAC III and PDAC I in macrophage subcluster analysis shown by volcano plot, red and blue dots represented the genes upregulated and downregulated respectively (PDAC III vs PDAC I and II). (g-h) The t-SNE plots presenting B cell subpopulations in B cell subcluster analysis based on clustering distribution in the g, sample source in the h. (i) Heatmap of B cell subclusters and related marker genes. (j) Violin plot showing the expression level of common B cell marker (CD19), Breg markers (CD1D, CD5, TGFB1), plasmocyte/memory B cell marker (CD27) among B cell subclusters.
Then we analyzed B cell infiltration in PDAC. A total of 1657 B cells were analyzed upon unsupervised clustering, which formed 6 distinct subclusters with different signature genes (Fig. 5G-I and Supplementary Table S8). We found no infiltration of B cells in PDAC I, while B cells in PDAC III showed radical difference with those in PDAC II based on transcriptional analysis. Thus we speculated that B cell gradually penetrated into tumor microenvironment during PDAC malignant development and evolved into different subpopulations with unique gene expression profiles and pro-tumor or anti-tumor function. The common marker CD19 was expressed in all B cell subclusters, confirming B cell identity (Fig. 5J). We found no regulatory B cells (Bregs) in tumor microenvironment according to the expression of Breg markers, such as CD1D, CD5 and TGFB1. The main forms of B cells were plasmocyte and memory B cells, expressing high level of CD27. Simultaneously, there was a subcluster (cluster 1) in PDAC II, identified as naïve B cell with lack of CD27 expression (Fig. 5J). Moreover, we found that the upregulated genes in PDAC III were mainly enriched for neutrophil mediated immunity, neutrophil degranulation and activation by GO analysis, and B cells in PDAC III expressed high level of costimulatory molecules, such as CD40 and CD86, which may promote T cell activation by interacting with CD40L and CD28 on T cell membrane (Supplementary Fig.S4G-H).
Gene-network module of csCAF are identified by WGCNA
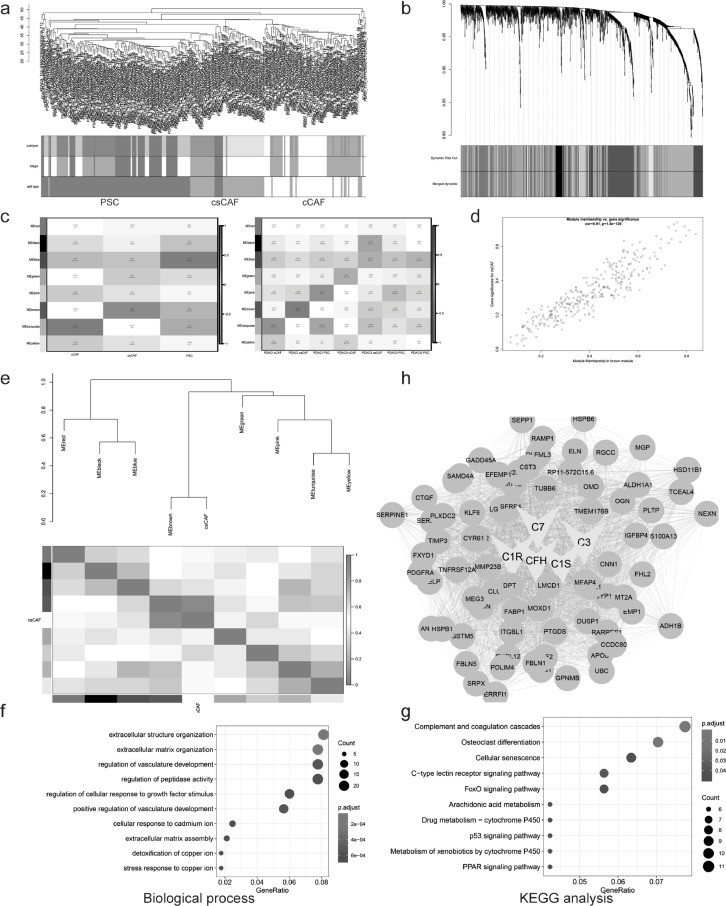
Weighted gene co-expression network analysis (WGCNA) is a system biology method for describing the correlation patterns among genes in RNA sequencing [41,42]. Here we took advantage of this powerful tool to figure out the interesting modules, consisting of a group of highly correlated genes, which represented the biological functions of subcluster. We randomly picked out 30–50 cells from cCAFs, csCAFs and PSCs respectively to construct gene expression matrix for WGCNA analysis, and their group information were listed as well (Supplementary Table S9). Then we performed cluster analysis for samples and found that the cells belonging to the same cell type were assigned to one group (Fig. 6A), which was consistent with the previous unsupervised clustering results in Fig. 3A. After choosing the soft-thresholding power, the algorithm for gene network construction and identification of modules was conducted, it showed that 8 modules were identified and there existed significant correlation between modules – MEturquoise, MEbrown, MEblue, and subpopulations – cCAFs, csCAFs, PSCs respectively (Fig. 6B-C). To better prove the correlation between Gene Significance (GS) for csCAFs and Module Membership (MM) in the brown module, a scatter plot of GS vs MM and eigengene adjacency heatmap were conducted. The results indicated that brown module could represent subpopulation csCAFs well (Fig. 6D-E).
Fig. 6.
Gene-network module of csCAF were identified by WGCNA. (a) Clustering dendrogram of samples based on Euclidean distance. (b) Clustering dendrogram of genes, with dissimilarity based on topological overlap using Dynamic Tree Cut algorithm, together with assigned merged module colors and the original module colors. (c) The heatmap of module-trait associations. Each row corresponds to a module eigengene, column to CAF subsets, based on cell type in the left panel, subtype in the right panel. In addition, each box contained the corresponding correlation and p-value. (d) A scatter plot of Gene Significance (GS) for csCAF vs Module Membership (MM) in the brown module. There is a highly significant correlation between csCAFs and brown module. (e) Visualization of eigengene network representing the relationships among modules and csCAFs. The upper panel showed a hierarchical clustering dendrogram of the eigengenes, the lower panel showed the eigengene adjacency. (f-g) Gene ontology and KEGG analysis showing biological process (f) and pathway (g) terms for genes in brown module (csCAFs hub genes). (h) Visualization of network connections among the most connected genes in the brown module representing csCAFs using Cytoscape software.
Next, to facilitate a biological interpretation, we would like to know whether the genes in brown module were significantly enriched in key gene ontologies. Thus we exported a list of gene in modules to conduct gene ontology and functional enrichment analysis (Supplementary Table S10). The genes in brown module were chiefly enriched for extracellular structure organization, regulation of vasculature development and complement and coagulation cascades (Fig. 6F-G). In addition, the brown module contained many genes that were the components of complement system, confirming the identity of csCAFs again. Finally, we exported the top 80 hub genes in brown module for the visualization of network connections among these genes by Cytoscape software [43] (Fig. 6H and Supplementary Table S10).
csCAF are detectable in early PDAC
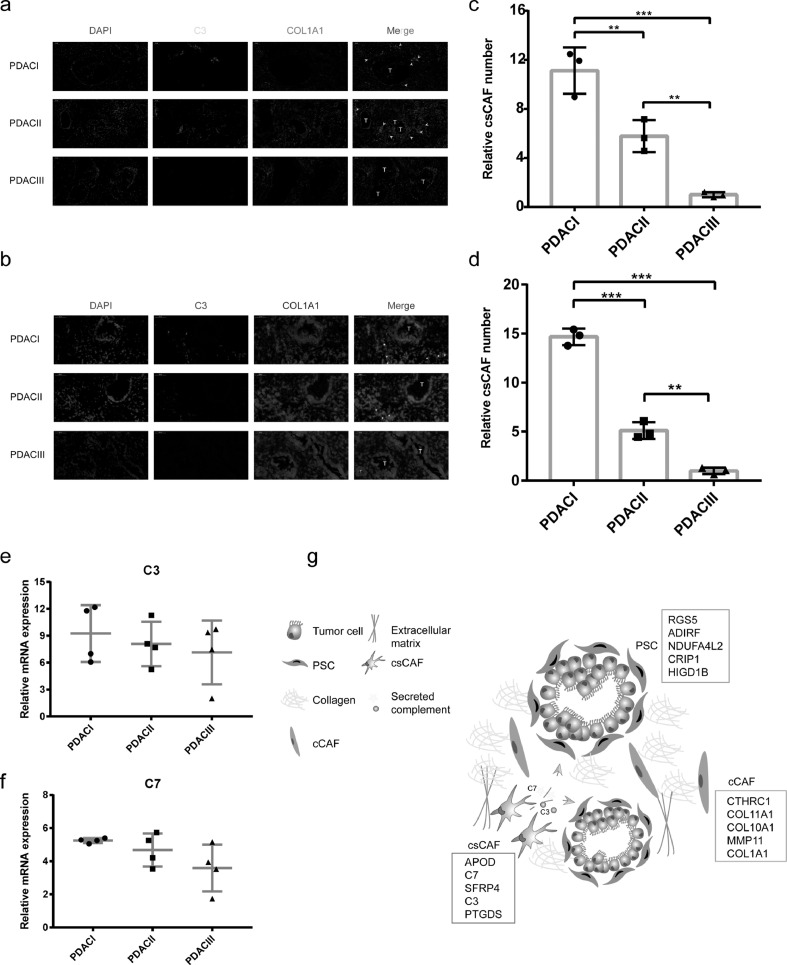
Following the identification of csCAFs by scRNA-seq and the detection of csCAF-related module by WGCNA, we decided to demonstrate the existence of these cells and figure out their location in human PDAC. We utilized the fibroblast marker COL1A1 and complement molecule C3 to stain human PDAC sections with different clinical stages by RNA ISH (RNA in situ hybridization) and IF (immunofluorescence). We further identified the presence of csCAFs and these cells were located in the tissue stroma next to malignant ductal cells only in PDACI and II (Fig. 7A-B). Then we counted the relative number of csCAFs in PDAC with different stages, and found that the PDACI had significantly more csCAF than PDACII and III (Fig. 7C-D). This suggested that the csCAFs may play a tumor-suppressive role in PDAC microenvironment and gradually decreased during the progression of tumor. In addition, we detected the relative mRNA expression levels of C3 and C7 in human PDAC tissue by RT-qPCR, and found a declining trend of C3 and C7 expression levels with the increase of tumor stages (Fig. 7E-F). Collectively, these results identified a novel CAF subpopulation – csCAFs, which is helpful for better understanding the heterogeneity of CAFs in PDAC (Fig. 7G). However, the potential value of these cells still remain to be explored.
Fig. 7.
csCAF were detected in human PDAC tissue by RNA ISH and IF. (a) The representative immunofluorescence (IF) image for co-staining of COL1A1 (red) and C3 (green) in human PDAC sections with different stages (3 PDAC I, 3 PDAC II, 3 PDAC III), the nuclear were stained with DAPI (blue), T, tumor glands, arrows were pointing to examples of csCAFs. (b) The representative RNA in situ hybridization (RNA ISH) image for co-staining of COL1A1 (green) and C3 (red) in human PDAC sections. (c-d) Bar plot showing the relative number of csCAFs in PDAC with different stages in IF in the c and in RNA ISH in the d. (e-f) The relative expression level of C3 (e) and C7 (f) in PDAC tissue with differnet stages by RT-qPCR. (g) Illustrated summary of CAF subpopulations and their location in PDAC. The marker genes of each CAF subpopulation were listed in corresponding boxes, csCAFs played a vital role in tumor microenvironment by secreting complement (C3 and C7).
Discussion
A growing body of researches have demonstrated the obvious intra-tumor heterogeneity in PDAC that brings great challenges for developing effective treatment strategies. Thus we should explore more precise targeted drugs aiming at the specific subpopulations of T cell, B cell, macrophage and CAF rather than the whole group. In this study, we systematically analyzed dynamic changes of PDAC microenvironment during tumor malignant development by single-cell sequencing. Notably, the ductal cells were not the dominant component, and tumor infiltrating immune cells, pancreatic stellate cells, and dense extracellular matrixes gradually accumulate from early PDAC to late PDAC. Based on epithelial, mesenchymal and cancer stem cell markers analysis, we found that EMT and CSC properties were key feature of advanced PDAC. Consistent with previous studies [44,45], anti-tumor immune response arose but were disabled by negative regulation from Tregs, exhausted T cells, TAMs with the development of tumor.
Previous studies identified many regulatory and exhausted T cell markers by single cell sequencing or bulk sequencing [33,[37], [38], [39]]. Compared with these data, our study found 22 common markers and several new markers, such as DUSP4, FANK1, LAIR2. Through analyzing TCGA datasets, we also found that the patients with high expression level of DUSP4 had significantly worse prognosis. Transcriptome profiling in Dusp4-deficient mice showed that DUSP4 could enhance the expression of a segment of canonical Treg signature genes [46]. In addition, a large number of studies revealed that DUSP4 could impact patients’ prognosis and chemoresistance in many types of cancers [47], [48], [49], [50], [51]. All these results suggested DUSP4 as a Treg signature gene is eligible for prognosis marker and potential therapy target.
A recent study performing cross-species single cell analysis comprehensively delineated the CAFs heterogeneity in PDAC and defined three CAF subsets – myofibroblastic CAFs (myCAFs), inflammatory CAFs (iCAFs) and antigen-presenting CAFs (apCAFs) [9]. In our study, we identified a new CAF subpopulation, complement-secreting CAFs (csCAF), but didn't find iCAFs and apCAFs. To some degree, there has similarity between iCAFs reported previously and csCAFs, because the iCAFs also had enriched expression of complement system components – C3, CFD. The divergence between two studies is probably due to tumor sample heterogeneity, different CAF enrichment method and analytical perspective. In addition, we detected that the microenvironment in PDAC III only contained one kind of CAFs (PSCs), lacking of other CAFs subpopulation. Pseudotime analysis for CAFs showed the possibility of mutual transformation between different CAF subpopulations. These findings demonstrate CAF has dynamic plasticity during PDAC progression, which is in accordance with precious research [52]. It is a noteworthy strategy to make use of CAF plasticity for the conversion of pro-tumor CAFs into anti-tumor ones.
The intricate crosstalk between CAFs and other cells in microenvironment has been explored in the past several decades. Some studies supported that CAFs could favor tumor growth, survival, migration, angiogenesis and metastasis by secreting various signaling molecules and vesicles [53], [54], [55], [56], [57]. However, other studies supported the anti-tumor function of CAFs [58], [59], [60]. In fact, there are distinct CAF subpopulations with different secretory phenotypes and functions. We utilized single cell sequencing data and WGCNA to identify a new CAF (csCAF). These CAFs specifically expressed complement components – C3, C7, C1R/S, CFD, CFH, CFI, compared with other CAFs. We also found the csCAFs subpopulation were detectable in human PDAC tissue using RNA ISH and IF. Moreover, a growing body of studies also identified the CAF subpopulation enriched for complement system components in different organs [61], [62], [63]. Complement system is not only the important component of innate immunity, but also plays the pivotal role in modulating cell-cell interaction inside tumor [64,65]. A series of experiments showed that the activation of complement system could lead to tumor-promoting inflammation [66,67]. Complement protein C3, C7, CFH could promote tumor proliferation, invasion, metastasis and drug resistance by regulating complement-complement receptors mediated biological processes [68], [69], [70], [71]. However, in this study, we did not further explore the potential function of csCAFs for PDAC by isolating these cells. We believe that it is considerably valuable to study the crosstalk between csCAFs and cancer cells, macrophages, T cells, endothelial cells and other CAFs. Understanding CAFs heterogeneity, we can develop new effective strategies and make it more accurate in the process of anti-cancer associated fibroblast treatment. In addition, there are other limitations in this study, such as the relative low coverage of 3’end sequencing and limited sample size. In the section of ductal cell subcluster analysis, we could not distinguish malignant ductal cells from normal ductal cells, likely resulting in gene expression bias.
In conclusion, our findings may provide a new insight for delineating the dynamic changes of tumor microenvironment components during PDAC malignant progression. The further studies about a new CAF subset (csCAF) are worthwhile for understanding PDAC microenvironment and improving patients’ prognosis.
Funding sources
This study was supported by The Natural Science Foundation of China (NO.81572339, 81672353, 81871954) and the Youth Clinical Research Project of Peking University First Hospital (2018CR28).
Contributors
Conceptualization, KC, XDT; Resources and Formal Analysis, KC; Validation, KC, QW, MZL, HHG, WKL, FW and XDT; Investigation and Visualization, KC, QW; Methodology, KC, HHG and MZL; Writing – original draft, KC; Project administration, YMY; Writing – review & editing XDT; Supervision, XDT and YMY. All authors read and approved the final version of the manuscript.
Data Sharing Statement
Correspondence and requests for materials should be addressed to KC, XDT and YMY.
Declaration of Competing Interest
We declare there are no any competing financial interests in relation to this work.
Acknowledgements
We thank Zebin Mao for suggestions and discussion, Lin Wo for technical support in single cell sequencing analysis, Yongsu Ma, Xudong Zhao, Shouge Zang for collecting pancreatic specimens, Baofa Sun and Dali Han for original scRNA-seq data. We thank the members in the Central of Medical and Health Analysis in Peking University for their assistance with fluorescence images. We thank all individuals who provided written consent.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103315.
Contributor Information
Xiaodong Tian, Email: tianxiaodong@pkufh.com.
Yinmo Yang, Email: yangyinmosci@163.com.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Ryan DP, Hong TS, adenocarcinoma Bardeesy N.Pancreatic. New Engl J Med. 2014;371(22):2140–2141. doi: 10.1056/NEJMc1412266. [DOI] [PubMed] [Google Scholar]
- 3.Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378(9791):607–620. doi: 10.1016/S0140-6736(10)62307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrido-Laguna I, Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat Rev Clin Oncol. 2015;12(6):319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y. Current status and future prospect of surgical treatment for pancreatic cancer. Hepatobiliary Surg Nutr. 2020;9(1):89–91. doi: 10.21037/hbsn.2019.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosein AN, Huang H, Wang Z, Parmar K, Du W, Huang J. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight. 2019;5 doi: 10.1172/jci.insight.129212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard V, Semaan A, Huang J, San Lucas FA, Mulu FC, Stephens BM. Single-cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin Cancer Res. 2019;25(7):2194–2205. doi: 10.1158/1078-0432.CCR-18-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29(9):725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9(8):1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rostom R, Svensson V, Teichmann SA, Kar G. Computational approaches for interpreting scRNA-seq data. FEBS Lett. 2017;591(15):2213–2225. doi: 10.1002/1873-3468.12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541(7637):331–338. doi: 10.1038/nature21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015;25(10):1499–1507. doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174(5):1293–1308. doi: 10.1016/j.cell.2018.05.060. e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Lazarus J, Steele NG, Yan W, Lee HJ, Nwosu ZC. Regulatory T-cell depletion alters the tumor microenvironment and accelerates pancreatic carcinogenesis. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-19-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keenan BP, Saenger Y, Kafrouni MI, Leubner A, Lauer P, Maitra A. A Listeria vaccine and depletion of T-regulatory cells activate immunity against early stage pancreatic intraepithelial neoplasms and prolong survival of mice. Gastroenterology. 2014;146(7):1784–1794. doi: 10.1053/j.gastro.2014.02.055. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das S, Bar-Sagi D. BTK signaling drives CD1d(hi)CD5(+) regulatory B-cell differentiation to promote pancreatic carcinogenesis. Oncogene. 2019;38(17):3316–3324. doi: 10.1038/s41388-018-0668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74(18):5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Huang L, Ding G, Huang H, Cao G, Sun X. Interferon gamma inhibits CXCL8-CXCR2 axis mediated tumor-associated macrophages tumor trafficking and enhances anti-PD1 efficacy in pancreatic cancer. J Immunother Cancer. 2020;8(1) doi: 10.1136/jitc-2019-000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helms E, Onate MK, Sherman MH. Fibroblast Heterogeneity in the Pancreatic Tumor Microenvironment. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-19-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dauer P, Zhao X, Gupta VK, Sharma N, Kesh K, Gnamlin P. Inactivation of cancer-associated-fibroblasts disrupts oncogenic signaling in pancreatic cancer cells and promotes its regression. Cancer Res. 2018;78(5):1321–1333. doi: 10.1158/0008-5472.CAN-17-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol. 2017;10(1):76. doi: 10.1186/s13045-017-0448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR. Depletion of Carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2015;28(6):831–833. doi: 10.1016/j.ccell.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111(30):E3091–E3100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim EJ, Sahai V, Abel EV, Griffith KA, Greenson JK, Takebe N. Pilot clinical trial of hedgehog pathway inhibitor GDC-0449 (vismodegib) in combination with gemcitabine in patients with metastatic pancreatic adenocarcinoma. Clin Cancer Res. 2014;20(23):5937–5945. doi: 10.1158/1078-0432.CCR-14-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Jesus-Acosta A, Sugar EA, O'Dwyer PJ, Ramanathan RK, Von Hoff DD, Rasheed Z. Phase 2 study of vismodegib, a hedgehog inhibitor, combined with gemcitabine and nab-paclitaxel in patients with untreated metastatic pancreatic adenocarcinoma. Br J Cancer. 2020;122(4):498–505. doi: 10.1038/s41416-019-0683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Song F, Zhu J, Zhang S, Yang Y, Chen T. GSA: Genome Sequence Archive<sup/>. Genomics Proteomics Bioinformatics. 2017;15(1):14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 32.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Simone M, Arrigoni A, Rossetti G, Gruarin P, Ranzani V, Politano C. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity. 2016;45(5):1135–1147. doi: 10.1016/j.immuni.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H. Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets. Cancer Cell. 2019;35(4):588–602. doi: 10.1016/j.ccell.2019.02.009. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balkwill F, Montfort A, Capasso M. B regulatory cells in cancer. Trends Immunol. 2013;34(4):169–173. doi: 10.1016/j.it.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EV. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity. 2016;45(5):1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X. Landscape of infiltrating t cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356. doi: 10.1016/j.cell.2017.05.035. e16. [DOI] [PubMed] [Google Scholar]
- 40.Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, Cooper JD. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009;41(9):1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 42.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munn DH, Bronte V. Immune suppressive mechanisms in the tumor microenvironment. Curr Opin Immunol. 2016;39:1–6. doi: 10.1016/j.coi.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arina A, Corrales L, Bronte V. Enhancing T cell therapy by overcoming the immunosuppressive tumor microenvironment. Semin Immunol. 2016;28(1):54–63. doi: 10.1016/j.smim.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Yan D, Farache J, Mingueneau M, Mathis D, Benoist C. Imbalanced signal transduction in regulatory T cells expressing the transcription factor FoxP3. Proc Natl Acad Sci U S A. 2015;112(48):14942–14947. doi: 10.1073/pnas.1520393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du F, Yu L, Wu Y, Wang S, Yao J, Zheng X. miR-137 alleviates doxorubicin resistance in breast cancer through inhibition of epithelial-mesenchymal transition by targeting DUSP4. Cell Death Dis. 2019;10(12):922. doi: 10.1038/s41419-019-2164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen X, Tan W, Li W, Li W, Zhu S, Zhong J. miR-1226-3p promotes sorafenib sensitivity of hepatocellular carcinoma via downregulation of DUSP4 expression. J Cancer. 2019;10(12):2745–2753. doi: 10.7150/jca.31804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M, Zhang J, Berger AH, Diolombi MS, Ng C, Fung J. Compound haploinsufficiency of Dok2 and Dusp4 promotes lung tumorigenesis. J Clin Invest. 2019;129(1):215–222. doi: 10.1172/JCI99699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X, Gao F, Wang J, Tao L, Ye J, Ding L. MiR-122-5p inhibits cell migration and invasion in gastric cancer by down-regulating DUSP4. Cancer Biol Ther. 2018;19(5):427–435. doi: 10.1080/15384047.2018.1423925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hijiya N, Tsukamoto Y, Nakada C, Tung Nguyen L, Kai T, Matsuura K. Genomic loss of DUSP4 contributes to the progression of intraepithelial neoplasm of pancreas to invasive carcinoma. Cancer Res. 2016;76(9):2612–2625. doi: 10.1158/0008-5472.CAN-15-1846. [DOI] [PubMed] [Google Scholar]
- 52.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y. IL1-Induced JAK/STAT signaling is antagonized by TGFbeta to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9(2):282–301. doi: 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albrengues J, Meneguzzi G, Gaggioli C. [Carcinoma-associated fibroblasts in cancer: the great escape] Med Sci (Paris) 2014;30(4):391–397. doi: 10.1051/medsci/20143004012. [DOI] [PubMed] [Google Scholar]
- 54.Mezawa Y, Orimo A. The roles of tumor- and metastasis-promoting carcinoma-associated fibroblasts in human carcinomas. Cell Tissue Res. 2016;365(3):675–689. doi: 10.1007/s00441-016-2471-1. [DOI] [PubMed] [Google Scholar]
- 55.Yamamura Y, Asai N, Enomoto A, Kato T, Mii S, Kondo Y. Akt-Girdin signaling in cancer-associated fibroblasts contributes to tumor progression. Cancer Res. 2015;75(5):813–823. doi: 10.1158/0008-5472.CAN-14-1317. [DOI] [PubMed] [Google Scholar]
- 56.Fang Y, Zhou W, Rong Y, Kuang T, Xu X, Wu W. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp Cell Res. 2019;383(1) doi: 10.1016/j.yexcr.2019.111543. [DOI] [PubMed] [Google Scholar]
- 57.Eiro N, Gonzalez L, Martinez-Ordonez A, Fernandez-Garcia B, Gonzalez LO, Cid S. Cancer-associated fibroblasts affect breast cancer cell gene expression, invasion and angiogenesis. Cell Oncol (Dordr) 2018;41(4):369–378. doi: 10.1007/s13402-018-0371-y. [DOI] [PubMed] [Google Scholar]
- 58.Yeh CR, Slavin S, Da J, Hsu I, Luo J, Xiao GQ. Estrogen receptor alpha in cancer associated fibroblasts suppresses prostate cancer invasion via reducing CCL5, IL6 and macrophage infiltration in the tumor microenvironment. Mol Cancer. 2016;15:7. doi: 10.1186/s12943-015-0488-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue T, Adachi K, Kawana K, Taguchi A, Nagamatsu T, Fujimoto A. Cancer-associated fibroblast suppresses killing activity of natural killer cells through downregulation of poliovirus receptor (PVR/CD155), a ligand of activating NK receptor. Int J Oncol. 2016;49(4):1297–1304. doi: 10.3892/ijo.2016.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trimboli AJ, Cantemir-Stone CZ, Li F, Wallace JA, Merchant A, Creasap N. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461(7267):1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M, Yang H, Wan L, Wang Z, Wang H, Ge C. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol. 2020;73(5):1118–1130. doi: 10.1016/j.jhep.2020.05.039. [DOI] [PubMed] [Google Scholar]
- 62.Sebastian A, Hum NR, Martin KA, Gilmore SF, Peran I, Byers SW. Single-cell transcriptomic analysis of tumor-derived fibroblasts and normal tissue-resident fibroblasts reveals fibroblast heterogeneity in Breast Cancer. Cancers. 2020;12(5) doi: 10.3390/cancers12051307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krenkel O, Hundertmark J, Ritz TP, Weiskirchen R, Tacke F. Single Cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells. 2019;8(5) doi: 10.3390/cells8050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afshar-Kharghan V. The role of the complement system in cancer. J Clin Invest. 2017;127(3):780–789. doi: 10.1172/JCI90962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mamidi S, Hone S, Kirschfink M. The complement system in cancer: ambivalence between tumour destruction and promotion. Immunobiology. 2017;222(1):45–54. doi: 10.1016/j.imbio.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 66.Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 2015;160(4):700–714. doi: 10.1016/j.cell.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 67.Bulla R, Tripodo C, Rami D, Ling GS, Agostinis C, Guarnotta C. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun. 2016;7:10346. doi: 10.1038/ncomms10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kwak JW, Laskowski J, Li HY, McSharry MV, Sippel TR, Bullock BL. Complement Activation via a C3a Receptor Pathway Alters CD4(+) T Lymphocytes and mediates lung cancer progression. Cancer Res. 2018;78(1):143–156. doi: 10.1158/0008-5472.CAN-17-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Sun SN, Liu Q, Yu YY, Guo J, Wang K. Autocrine complement inhibits IL10-Dependent T-cell-mediated antitumor immunity to promote tumor progression. Cancer Discov. 2016;6(9):1022–1035. doi: 10.1158/2159-8290.CD-15-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zha H, Wang X, Zhu Y, Chen D, Han X, Yang F. Intracellular activation of complement C3 Leads to PD-L1 antibody treatment resistance by modulating tumor-associated macrophages. Cancer Immunol Res. 2019;7(2):193–207. doi: 10.1158/2326-6066.CIR-18-0272. [DOI] [PubMed] [Google Scholar]
- 71.Seol HS, Lee SE, Song JS, Rhee JK, Singh SR, Chang S. Complement proteins C7 and CFH control the stemness of liver cancer cells via LSF-1. Cancer Lett. 2016;372(1):24–35. doi: 10.1016/j.canlet.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.