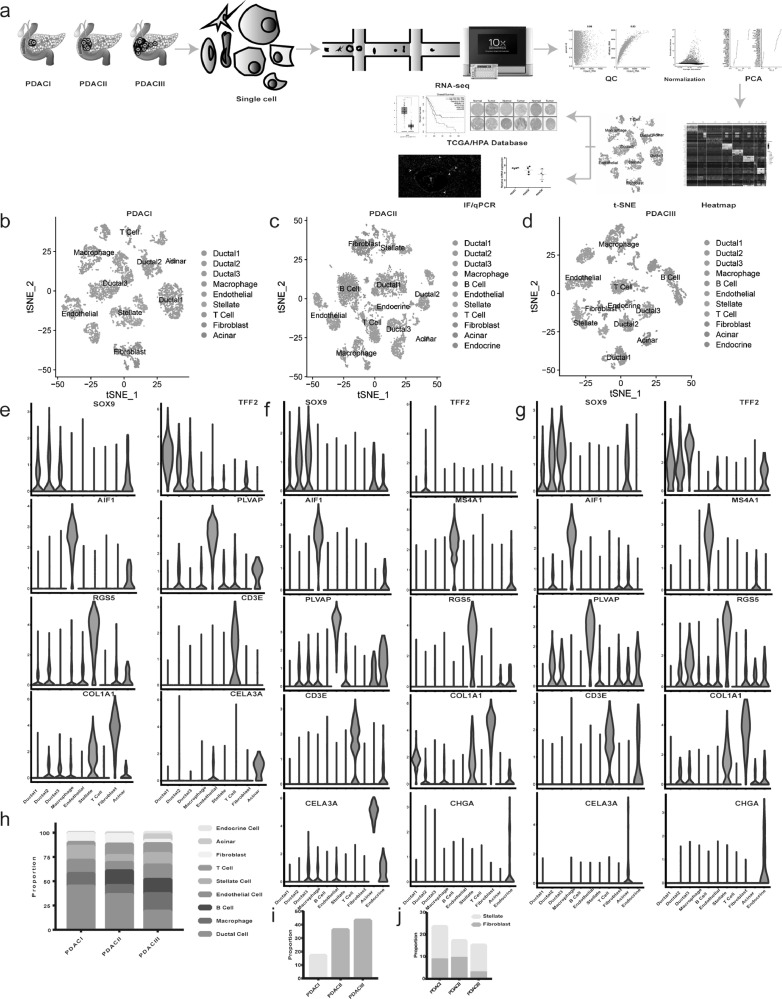
Fig. 1.
scRNA-seq delineats the dynamic changes of tumor microenvironment components during PDAC progression. (a) Workflow described by flow diagram. PDAC samples with different clinical stages were dissociated into single cells, all cells were subjective to capture, library preparation, RNA-seq using 10 × Genomics platform, then the QC, normalization, PCA and subsequent bioinformatics analysis were conducted. (b-d) The t-distributed stochastic neighbor embedding (t-SNE) plot showing clustering information in PDACI, II, III respectively. (e-g) Violin plots demonstrating the identity of each cluster through analyzing the expression of well-known cell type specific markers. (h) The proportion of cells changing from PDAC I, II to PDAC III. (i) The proportion of immune cells (T cell, B cell and macrophage) gradually increased from PDAC I, II to PDAC III. (j) The bar chart showing the proportion of pancreatic stellate cells (PSCs) in all cancer associated fibroblasts (CAFs) in PDAC I, II, III separately.