Abstract
Hyperuricemia (HUA), a chronic disease caused by metabolic disorders of purine, is often accompanied by other diseases such as gout, type 2 diabetes mellitus (T2DM), and hyperlipidemia. However, little is known about the relationship between HUA and these diseases on the protein level. We performed label-free liquid chromatography MS/MS spectrometry analysis of urine samples from 26 HUA patients and 25 healthy controls, attempting to establish the possible protein links between HUA and these diseases by profiling urine proteome. A total of 2119 proteins were characterized in sample proteomes. Among them, 11 were found decreased and 2 were found increased in HUA samples. Plausible pathways found by enrichment analysis of these differentially expressed proteins (DEPs) include the processes for insulin receptor recycling and lipid metabolism, suggesting potential links between HUA and T2DM and hyperlipidemia. The abundance changes of three key proteins (VATB1, CFAD, and APOC3) involved in these processes were validated by enzyme-linked immunosorbent assay (ELISA). In conclusion, our result provides proteomic evidence, for the first time, that the aberrant pathways enriched by described key DEPs are closely related to the incidence of HUA and its concomitant diseases.
Introduction
Hyperuricemia (HUA) is a chronic disease caused by metabolic disorders of purine.1 The incidence of HUA is growing rapidly worldwide, especially in developed countries and regions. Recent studies suggested that factors such as sex, age, ethnicity, specific genetic mutation, dietary habit, and environmental factors contribute to this increasing prevalence.2,3 HUA is easily ignored in the early stage due to the lack of symptoms until gouty arthritis and renal tophi occur.
HUA is known associated with higher incidences of other chronic and metabolic diseases, such as hyperlipidemia, type 2 diabetes mellitus (T2DM), hypertension, major adverse cardiovascular events, and chronic kidney disease.4−6 Until now, little is known about the relationship between HUA and these diseases. HUA has been confirmed as an independent risk factor for the occurrence and development of T2DM. Research showed that the risk of occurrence of T2DM was increased by 17% with every 60 μmol/L serum uric acid (UA) increase.7 Studies also showed that HUA contributed to the progression of diabetic kidney disease in T2DM, as well as a significant increase in the risk of diabetic vascular complications.8,9 Previous experimental studies suggested several possible mechanisms for the relationships between HUA and T2DM.10−12 Among them, the leading hypothesis was that the elevated serum UA level imbalanced proinflammatory endocrine and/or reduced endothelial nitric oxide bioavailability, which subsequently resulted in insulin resistance. Another possible mechanism for HUA/T2DM link was that intracellular urate was shown to promote hepatic gluconeogenesis by activating AMPD and inhibiting AMPK.13
Serum uric acid (UA) level was also reported to be positively associated with an elevated level of total cholesterol, triglyceride, and low-density lipoprotein cholesterol.14 Apolipoproteins were reported to play a key role in the pathophysiological process of lipid abnormalities among HUA patients.15 This process has been demonstrated to be mediated by polymorphisms in the apolipoprotein APOA1-APOC3-APOA4 gene cluster.16,17
With the development of high-throughput experimental technologies, holistic analysis approaches have been applied to the studies of HUA and gout. A number of genes associated with urate transporting and interacting proteins were identified in the genome-wide association studies.18−22 Metabolomics studies have discovered several dipeptides, essential amino acids, steroid hormones, and their metabolic pathways as important regulators of serum UA.23,24 And inflammation induced by the interleukin 6/signal transducer and activator of transcription 3 signaling pathway participated in hyperuricemia-induced kidney injury.25 Complement C3, haptoglobin, complement C4, and apolipoprotein A1 were significantly higher expressed serum proteins in the Uygur hyperuricemia group than in the normal control group by two-dimensional gel electrophoresis (2-DE) and matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry (MALDI-TOF-MS).26
Urine is one of the most valuable clinical samples for the discovery of biomarkers. Compared with blood and tissue fluid, urine protein detection has the following advantages. First, urine is a kind of sample that can be collected noninvasively and repeatedly. Second, urine contains proteins that are filtered from plasma and secreted from the urinary system and, therefore, can reflect changes in the whole body system.27,28 Thirdly, lots of studies have shown that urine is more suitable for mass spectrometry analysis than blood,29−31 as it is less affected by the high-abundance proteins such as albumin and immunoglobulins. In addition to the applications in disease diagnosis, the urine proteome might offer new insight into the pathogenesis of hyperuricemia.
Mass spectrometry (MS)-based proteomics has become a powerful tool that can routinely detect and quantify thousands of proteins, providing great opportunities in the discovery of biomarkers for a broad range of potential clinical applications. In this study, we performed label-free liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis on the urine samples from patients with HUA and healthy controls to characterize the urinary proteomic features of HUA and to establish possible protein links between HUA and its concomitant diseases.
Results
Patient Group
Twenty-six patients (20 males and 6 females) aged between 22 and 71 years (mean age, 37.9 years) with HUA were enrolled in the study as the HUA group, with 25 normal volunteers (20 males and 5 females, aged between 24 and 70 years with a mean age of 39.2 years) recruited as the control group. No statistical differences were found in the distribution of gender and age between these two groups. Individuals in both study groups have been confirmed clinically without other diseases such as hyperlipidemia, diabetes, and liver and kidney diseases. Basic information of enrolled individuals is given in Table 1. Their detailed clinical data are provided in Table S1.
Table 1. Basic Characteristics of the Individualsa.
| control (n = 25) | hyperuricemia (n = 26) | T or χ2 | p value | ||
|---|---|---|---|---|---|
| age (years) | 39.16 ± 12.43 | 37.85 ± 10.64 | –0.406 | 0.686 | |
| gender (male) | 20(80.0) | 20(76.9) | 0.071 | 0.789 | |
| serum uric acid (μmol/L) | male | 362.50 ± 51.04 | 508.85 ± 82.02 | 6.775 | 0.000 |
| female | 254.40 ± 85.76 | 483.67 ± 175.21 | 2.656 | 0.026 | |
| urea nitrogen (mmol/L) | 4.70 ± 1.15 | 4.70 ± 0.92 | 0.002 | 0.998 | |
| serum creatinine (μmol/L) | 85.84 ± 15.45 | 85.96 ± 16.64 | 0.027 | 0.979 | |
| ALT (U/L) | 24.24 ± 12.55 | 25.08 ± 12.31 | 0.240 | 0.837 | |
| AST (U/L) | 21.84 ± 5.74 | 20.58 ± 4.99 | –0.839 | 0.405 | |
| total protein (g/L) | 75.87 ± 3.27 | 77.95 ± 4.22 | 1.968 | 0.055 | |
| albumin (g/L) | 49.90 ± 2.52 | 49.45 ± 1.95 | –0.705 | 0.484 | |
| total cholesterol (mmol/L) | 4.32 ± 0.66 | 4.49 ± 0.61 | 0.992 | 0.326 | |
| triglyceride (mmol/L) | 1.08 ± 0.40 | 1.29 ± 0.45 | 1.787 | 0.08 | |
| LDL cholesterol (mmol/L) | 2.64 ± 0.60 | 2.87 ± 0.59 | 1.356 | 0.181 | |
| HDL cholesterol (mmol/L) | 1.34 ± 0.37 | 1.21 ± 0.43 | –1.147 | 0.257 | |
| fasting blood glucose (mmol/L) | 4.84 ± 0.59 | 4.95 ± 0.32 | 0.823 | 0.415 |
Values for continuous data are mean ± SD; for categorical data are count (percentage).
General Information of Proteome Results
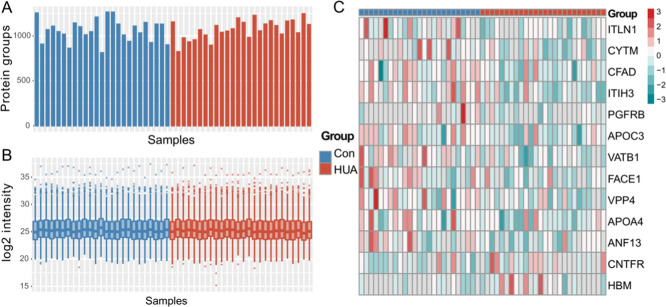
A total of 2119 proteins were identified and quantified by the label-free LC–MS/MS approach with MaxQuant software (version 1.6.2.10).32 Both peptide-spectrum match (PSM) and protein false discovery rate (FDR) were set to 0.01. Also, the minimum number of unique peptides for each protein was set to 2. As shown in Figure 1A,1B, similar numbers of proteins were identified between all samples in the two groups (Figure 1A), and protein abundance distributions among samples were nearly identical (Figure 1B). In total, 78.01% of proteins are classified as extracellular proteins (extracellular exosome, extracellular space, extracellular matrix, or extracellular region) in Gene Ontology (GO). To evaluate the comprehensiveness of our urinary proteomic data set, the 2119 proteins were compared with those identified in previous urinary proteomic studies on the concomitant diseases of HUA. Our data set includes 56.12% (463/825) of proteins with at least two unique peptides reported in a T2DM urinary proteome by Anh et al.,33 63.45% (349/550) of gene products identified in another T2DM urinary proteome by Guillén-Gómez et al.,34 and 69.00% (512/742) proteins in a urinary proteome data set from patients with hypertension.35 Besides, our proteome data set contains 78.95% (1579/2000) of the normal urinary proteome demonstrated in Leng et al.’s study,36 which employed very similar sample preparation and proteomic analysis approaches as in this study. These observations indicate good technical reproducibility and validity of our proteomic analysis. A total of 1102 proteins with valid abundance values in at least 50% of the samples in a certain group were selected for further analysis (Table S2).
Figure 1.
General information of proteome results. HUA, hyperuricemia group; Con, control group. (A) Number of proteins identified in each sample. (B) Abundance distribution. (C) Heatmap of all differentially expressed proteins (DEPs) in HUA. The color scale indicates the z-score-transformed abundance data of each DEP. Gray indicates a missing value in quantification.
Differential Urinary Proteins in Hyperuricemia
Thirteen DEPs were identified between HUA and Con groups by the t-test (p value <0.05, fold change ≥1.5, Table 2). Eleven of them were downregulated and two were upregulated in the HUA group (Figure 1C).
Table 2. Differentially Expressed Proteins in Hyperuricemia Compared to Normal Controla.
| symbol | accession | description | p value | fold change | change |
|---|---|---|---|---|---|
| ITIH3 | Q06033 | inter-α-trypsin inhibitor heavy chain H3 | 0.0015 | 0.5404 | downregulation |
| CFAD | P00746 | complement factor D | 0.0292 | 0.5322 | downregulation |
| APOC3 | P02656 | apolipoprotein C-III | 0.0140 | 0.5883 | downregulation |
| VATB1 | P15313 | V-type proton ATPase subunit B kidney isoform | 0.0052 | 0.5990 | downregulation |
| CYTM | Q15828 | cystatin-M | 0.0112 | 0.4469 | downregulation |
| FACE1 | O75844 | CAAX prenyl protease 1 homolog | 0.0351 | 0.6010 | downregulation |
| VPP4 | Q9HBG4 | V-type proton ATPase 116 kDa subunit a isoform 4 | 0.0076 | 0.6057 | downregulation |
| RNF13 | O43567 | E3 ubiquitin-protein ligase RNF13 | 0.0091 | 0.6572 | downregulation |
| APOA4 | P06727 | apolipoprotein A-IV | 0.0383 | 0.6431 | downregulation |
| ITLN1 | Q8WWA0 | intelectin-1 | 0.0422 | 0.0624 | downregulation |
| PDGFRB | P09619 | platelet-derived growth factor receptor β | 0.0462 | 0.5782 | downregulation |
| HBM | Q6B0K9 | hemoglobin subunit mu | 0.0311 | 8.5500 | upregulation |
| CNTFR | P26992 | ciliary neurotrophic factor receptor subunit α | 0.0419 | 1.7430 | upregulation |
Fold change, the fold change (HUA/control).
Gene Ontology (GO) Enrichment and Pathway Analysis
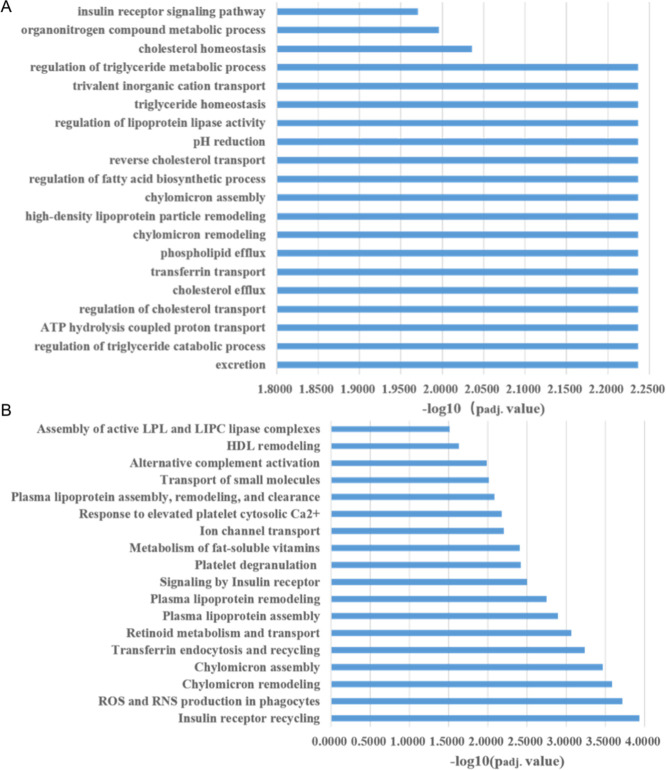
GO enrichment analysis result is outlined in Table S3. Enriched biological processes are mainly involved in lipid metabolism, adenosine 5′-triphosphate (ATP) hydrolysis coupled proton transport, and the insulin receptor signaling pathway (Figure 2A). Enriched molecular functions include cholesterol binding, enzyme inhibitor activity, proton transmembrane transporter activity, etc. Pathway analysis using Reactome (Figure 2B and Table S4) also showed enriched pathways of insulin receptor recycling as well as those related to lipid metabolism, such as plasma lipoprotein and chylomicron assembly and remodeling. Besides, pathways of platelet degranulation, metabolism of fat-soluble vitamins, and others were also enriched among the DEPs.
Figure 2.
Functional enrichment analysis of DEPs. (A) Significantly enriched biological processes in Gene Ontology. (B) Significantly enriched Reactome pathways.
Validation of DEPs
Three DEPs with interesting molecular functions that might be linked to the pathogenesis of the HUA-associated diseases were validated by enzyme-linked immunosorbent assay (ELISA). Samples used for validation include 17 HUA and 24 control samples used in the MS analysis, and 8 HUA and 7 control samples from another cohort. A one-sided t-test was performed on the logarithm-transformed protein-to-creatine ratio of the proteins between the control group and the HUA group. Downregulation of APOC3 (p = 0.0137) CFAD (p = 0.0418), and VATB1 (p = 0.0885) in the HUA group was validated at the significance level of 0.1. The results are shown in Figure 4.
Figure 4.
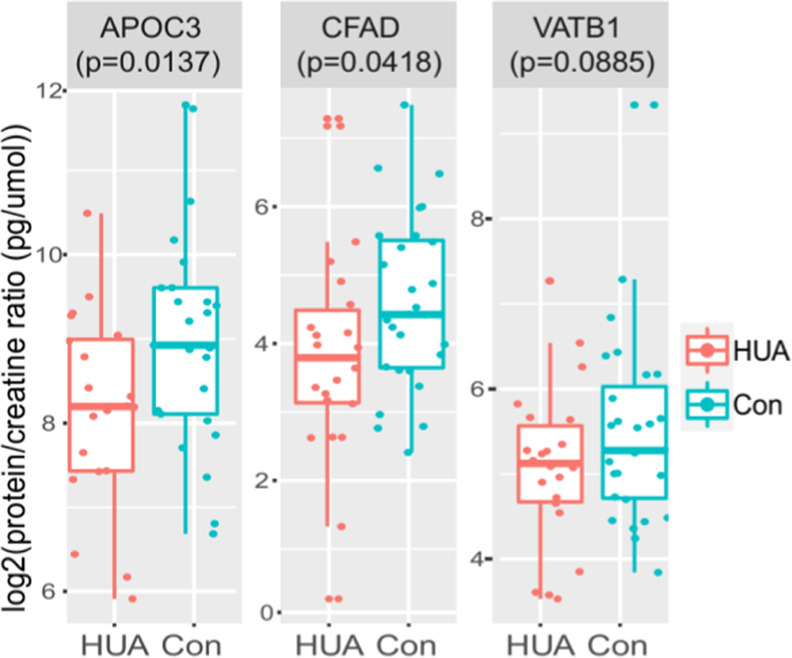
Validation of five DEPs by ELISA Three DEPs with interesting molecular functions that might be linked to the pathogenesis of the HUA-associated diseases were validated by ELISA. The one-sided t-tests were performed on the logarithm-transformed protein-to-creatine ratio of the proteins between the control and the HUA group.
Discussion
HUA is an independent risk factor for the occurrence and development of many diseases, such as gout, hyperlipidemia, T2DM, hypertension, cardiovascular disease, and chronic kidney disease. However, the specific pathogenesis is still not clear. In this study, we identified a total of 13 proteins that had significant abundance changes in the urine of HUA patients. Since a large proportion of urinary proteins come from renal filtration of plasma proteins, it can be assumed that changes of these proteins in the urine can reflect their corresponding changes in the blood. We discussed the associations between these altered proteins and reported potential molecular changes of some diseases that are closely related to HUA in the following subsections, showing that urinary proteomics may help explain their pathogenesis. Some of these urinary proteins may serve as noninvasive biomarkers for monitoring and early detection of some of these diseases in the future.
VATB1 and CFAD May Promote the Occurrence of T2DM by Affecting Insulin in HUA Patients
Previous studies have confirmed that HUA is an independent risk factor for T2DM and urate-lowering therapy can notably ameliorate insulin resistance.37,38 VATB1, together with another downregulated protein VPP4, is involved in the insulin receptor recycling pathway. The two proteins are mainly expressed in distal renal tubular epithelial cells, which is one of the main sites of insulin degradation. They are components of the proton channel of V-ATPase, which could acidify the eukaryotic endosomal lumen by pumping H+ into it from the cytoplasm.39 Physically, the acidification of the endosomal lumen triggers insulin to dissociate from its receptor and be degraded in the endosome lumen. The insulin receptor is then recycled into the plasma membrane after being dephosphorylated.39 We hypothesize that decreased VATB1 and VPP4 may lead to insufficient and untimely acidification of the endosomal lumen, disturbing the insulin receptor recycling and delaying its binding to insulin, which is a crucial feature of insulin resistance (Figure 3).
Figure 3.
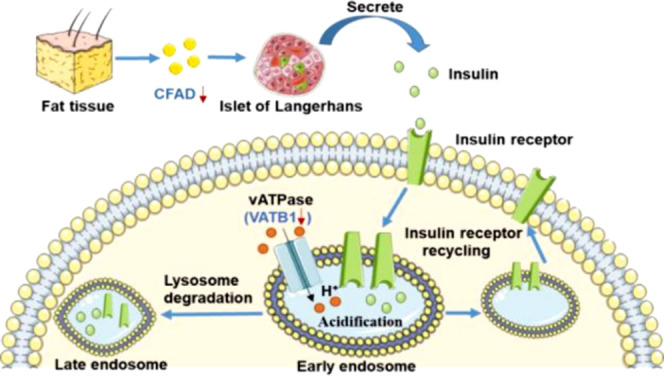
Schematic diagram of possible functional mechanisms of insulin resistance induced by VATB1 and CFAD. DEPs are indicated with blue characters and red arrows. Decreased VATB1 may lead to insufficient and untimely acidification of the endosomal lumen, which consequently disturbs the insulin receptor recycling and delays its binding to insulin, leading to insulin resistance. Downregulated CFAD may be involved in the development of T2DM by decreasing insulin secretion.
CFAD also has a close relationship with T2DM. CFAD40 is a positive regulator of the alternative complementary pathway by catalyzing the cleavage of complement factor B, a rate-limiting protease of complement activation. Historically, CFAD was known as adipsin which can regulate energy homeostasis and systemic metabolism. Research has demonstrated that T2DM patients with β cell failure are deficient in CFAD and CFAD acts, at least, in part, via C3a to potentiate insulin secretion.41 Decreased urinary CFAD in HUA patients may be involved in the development of T2DM by decreasing insulin secretion.
APOC3 May Participate in the Pathogenesis of Hypertriglyceridemia in HUA Patients
Increased serum UA is a well-known risk factor for hyperlipidemia, and the serum triglyceride level has the strongest association with the serum UA level.42 Some biological processes and pathways related to lipid metabolism were significantly enriched in patients with HUA, such as regulation of the triglyceride catabolic process (Figure 2A), plasma lipoprotein and assembly, and remodeling (Figure 2B). APOC3 is synthesized in the liver and intestine and is the most abundant C apolipoprotein in human plasma.43 It was reported that APOC3 could change triglyceride metabolism by affecting the activity of lipoprotein lipase (LPL) and clearance of triglyceride-rich lipoproteins (TRLs).44 However, it is still difficult to speculate how APOC3 participated in the development of hypertriglyceridemia in HUA patients based on the limited understanding of the molecular mechanisms of HUA. Future studies are needed to first validate their downregulation in blood and the corresponding organs and then to discover their roles in the pathogeneses of HUA-related hypertriglyceridemia.
Possible Relationships between HUA and Other DEPs
The positive correlation between serum uric acid and hemoglobin levels has long been known.45−48 HUA is also commonly occurred in patients with polycythemia. Increased red cell degradation in subjects with increased red cell mass may be the cause of this correlation.48
In vitro study in vascular smooth muscle cells showed that uric acid could induce proliferative pathways through its phosphorylation. This process is thought to play a crucial role in the development of cardiovascular diseases.49
Several proteins that are known factors of HUA-induced injuries have also been detected in our study. HUA could activate the renin–angiotensin system (RAS), which is closely related to metabolic syndromes and cardiovascular and kidney diseases.50,51 Two key factors in this system, angiotensinogen and angiotensin-converting enzyme (ACE), were quantified in our study. Our proteomic data set also includes MMP7 and fibronectin, which associate with HUA-induced renal fibrosis,50 as well as CXCL-12, an indicator of inflammation.50 However, these proteins exhibited no significant differences between HUA and control groups (Table 3). The fact that all patients were free from hypertension and any renal diseases at the time of urine collection may be the explanation of these observations.
Table 3. Known Proteins Associated with HUA-Induced Disease.
Conclusions
In conclusion, HUA is a systemic metabolic disease that may have impacts on various systems of the human body. Taking advantage of advanced proteomic techniques, the present study characterized the urinary proteomic features of HUA and explored the possible protein links between HUA and its concomitant diseases. Specifically, we identified and validated three key downregulated expressed proteins: VATB1 and CFAD, which are associated with insulin resistance and regulation of insulin secretion, and APOC3, which is related to triglyceride metabolism. We hypothesize these candidate proteins may be related to the pathogenesis of T2DM and hypertriglyceridemia associated with HUA.
These proteins may help us to monitor and detect the subsequent occurrence of the concomitant diseases in patients with HUA timely and provide evidence for intervention. Other DEPs may give us more clues for HUA-related diseases. Further work to verify the above DEPs in blood samples from HUA patients will help us to reveal the detailed relationships between HUA and its concomitant diseases.
Methods
Patients
Volunteers were divided into the control group and experiment group. In the control group, there were 25 healthy people (20 males and 5 females) aged between 24 and 70 years. The HUA group consisted of 26 patients with HUA (20 males and 6 females) aged between 22 and 71 years. Both groups did not include the cases associated with other common diseases, such as hyperlipidemia, diabetes, liver, and kidney diseases. The diagnosis criteria of HUA patients are as follows: fasting serum uric acid level ≥420 μmol/L for males and ≥360 μmol/L for females, associated with gout or not. All volunteers with informed consent documents were from the physical examination population in the physical examination center of Guangdong Provincial Hospital of Chinese Medicine. The study was approved by the Ethics Committee of Guangdong Provincial Hospital of Chinese Medicine.
Sample Collection and Storage
Approximately 10 mL of midstream urine from each volunteer was collected into one sterile urinary sediment tube according to the criteria of morning urine retention. All collected urine samples were stored at −80 °C and left standing.
Sample Preparation
One milliliter of urine sample was centrifuged at 176 000g for 70 min at 4 °C to collect the pellets. We utilized a previously described method52 with modifications to remove uromodulin (UMOD). Briefly, resuspension buffer (50 mM Tris, 250 mM sucrose, pH 8.5) and dithiotheitol (DTT) were added to the pellets and the suspension was then heated at 65 °C for 30 min. Wash buffer (10 mM tetraethylammonium (TEA), 100 mM NaCl, pH 7.4) was added to the pallets, and second ultracentrifugation was carried out at 176 000g for 40 min. Then, the supernatant was discarded. NH4HCO3 (50 mM) was added to the precipitate and heated at 95 °C for 3 min. Samples were then digested with trypsin overnight at 37 °C. Then, the supernatant was extracted by centrifugation with 100% ACN at 10× 1000g for 5 min and dried in a vacuum concentrator (Thermo Scientific); the resulting peptides were used for mass spectrometry analysis.
Liquid Chromatography–Tandem Mass Spectrometry
Peptides were loaded onto a trap column (100 μm × 2 cm, homemade; particle size, 3 μm), separated by a homemade silica microcolumn (150 μm × 12 cm, particle size, 1.9 μm) with a gradient of 5–35% mobile phase B (80% acetonitrile and 0.1% formic acid) at a flow rate of 800 nL/min for 30 min. The analysis was performed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Rockford, IL) connected to an Easy-nlc 1200 System (Thermo Fisher Scientific). Peptides were analyzed by an Orbitrap mass analyzer with a resolution of 60 000 in full scan mode and 15 000 in MS/MS mode. The MS/MS analysis was performed in a data-dependent mode. One full scan was followed by up to 15 data-dependent MS/MS scans with higher-energy collision dissociation (normalized collision energy of 27%). The dynamic exclusion time was set as 15 s.
Mass Spectrometry Data Analysis
The mass spectrometry raw files were analyzed with MaxQuant (version 1.6.2.10). A total of 51 raw files were analyzed together to exploit the match-between-run algorithm, which enables the identification of peptides that were not selected for fragmentation in one run by checking whether these peptides were sequenced in another run.53 We used the Andromeda search engine54 to search the detected features against the human Swiss-Prot reference proteome from Uniprot (downloaded on 18 March 2019, 20251 proteins). Only tryptic peptides that were at least seven amino acids in length with up to two missed cleavages were considered. N-acetylation of proteins’ N-termini and oxidation of methionine were set as variable modifications. The first search peptide tolerance was set to 4.5 ppm for the main search peptide tolerance for the Q Exactive data at the MS level and 0.05 Da at the MS/MS level. A false discovery rate (FDR) of 1% was imposed for peptide-spectrum matches (PSMs) and protein identification using a target–decoy approach. The number of minimal unique peptides was set to 2. To normalize the differences in peptide loading amounts, relative quantification was performed using the default parameters of the MaxLFQ algorithm55 with the minimum ratio count set to 2. For all other search parameters, the default settings were used.
The “proteinGroups.txt” file produced by MaxQuant was further analyzed in the R language (R version 3.5.1). Proteins from the reverse database and contaminants were removed.
Statistical Analysis
A two-sided t-test was performed on the logarithmized LFQ intensities with a p-value cutoff of 0.05 and fold change ≥1.5.
Protein Gene Ontology and Pathway Analysis
Cellular component annotation for identified proteins was performed using DAVID (https://david.ncifcrf.gov/) with the GOTERM_CC_DIRECT database. To further research the biological functions of the DEPs in hyperuricemia compared to normal control, functional annotation was performed. Gene Ontology enrichment analysis was performed using STRING (version 11.0, https://string-db.org/), an open-source software tool, for general characterization of properties of the proteins in the data set. False discovery rate (FDR) <0.05 was set as the cutoff of significance. Pathway analysis was performed using Reactome (version 70, https://reactome.org/), an open-source software tool. p value <0.05 was set as the cutoff of significance.
Validation of DEPs by ELISA
First, human urine samples were centrifuged at 2000g for 20 min. VATB1 was detected by HUMAN ELISA kits (Quanzhou Ruixin Biotechnology Co., Ltd., Quanzhou, China) according to the manufacturer’s instructions. APOC3 and CFAD were detected by HUMAN ELISA kits (Abcam, Cambridge, U.K.) according to the manufacturer’s instructions. In brief, the samples and standards were added to appropriate wells and then the antibodies were added to all wells. The mixtures were incubated for 1 h; then, the wells were aspirated and washed with 350 μL of 1× wash buffer PT three (or five) times. The TMB development solution was added to each well and incubated for 10 (or 15) min. The stop solution was added to each well, and the absorbance at 450 nm was read on a Thermo Scientific Multiskan FC microplate reader (Thermo Scientific, Massachusetts). The concentration of urine creatinine was measured by Roche cobas 8000 (Roche, Basel, Switzerland). The final concentration of protein was corrected by the concentration of the creatinine for statistical analysis.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant nos. 82074376, 31971360, and 81774216), the Joint Innovation Project of National Center for Protein Sciences (Beijing) and Guangdong Provincial Hospital of Chinese Medicine (grant no. 2017KT1821), the Specific Research Fund for TCM Science and Technology of Guangdong Provincial Hospital of Chinese Medicine (grant no. YN2016XP01), the National Key Research and Development Program of China (grant no. 2017YFC0908403), the Specific Fund of State Key Laboratory of Dampness Syndrome of Chinese Medicine (grant nos. SZ2020ZZ04 and SZ2020ZZ05) and Guangdong Provincial Science and Technology Project (grant no. 2018B030322012).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c06229.
Detailed clinical data of all samples (Table S1), quantification information for 1102 proteins that were quantified in at least 50% of the samples in a certain group (Table S2), gene ontology analysis of DEPs in the HUA group (Table S3), pathway analysis of DEPs in the HUA group (Table S4), results of ELISA experiments (Table S5), and detailed clinical data of 15 additional samples used in the validation experiments (Table S6) (PDF)
(XLSX)
Author Contributions
¶ S.H. and H.W. contributed equally to this work.
The authors declare no competing financial interest.
Notes
Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository56 with data set identifier PXD016900.
Supplementary Material
References
- Stamp L.; Dalbeth N. Screening for hyperuricaemia and gout: a perspective and research agenda. Nat. Rev. Rheumatol. 2014, 10, 752–756. 10.1038/nrrheum.2014.139. [DOI] [PubMed] [Google Scholar]
- Zhu Y.; Pandya B. J.; Choi H. K. Prevalence of gout and hyperuricemia in the US general population: the National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum. 2011, 63, 3136–3141. 10.1002/art.30520. [DOI] [PubMed] [Google Scholar]
- Liu Rui.; Han Cheng.; Wu Di.; Xia Xinghai.; Gu Jianqiu.; Guan Haixia.; Shan Zhongyan.; Teng Weiping. Prevalence of hyperuricemia and gout in mainland China from 2000 to 2014: a systematic review and meta-analysis. BioMed Res. Int. 2015, 2015, 762820 10.1155/2015/762820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin T.; Richette P. Impact of comorbidities on gout and hyperuricaemia: an update on prevalence and treatment options. BMC Med. 2017, 15, 123 10.1186/s12916-017-0890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babio N.; Martínez-González M. A.; Estruch R.; Wärnberg J.; Recondo J.; Ortega-Calvo M.; Serra-Majem L.; Corella D.; Fitó M.; Ros E.; Becerra-Tomás N.; Basora J.; Salas-Salvadó J. Associations between serum uric acid concentrations and metabolic syndrome and its components in the PREDIMED study. Nutr., Metab. Cardiovasc. Dis. 2015, 25, 173–180. 10.1016/j.numecd.2014.10.006. [DOI] [PubMed] [Google Scholar]
- Stack A. G.; Hanley A.; Casserly L. F.; Cronin C. J.; Abdalla A. A.; Kiernan T. J.; Murthy B. V.; Hegarty A.; Hannigan A.; Nguyen H. T. Independent and conjoint associations of gout and hyperuricaemia with total and cardiovascular mortality[J]. QJM 2013, 106, 647–658. 10.1093/qjmed/hct083. [DOI] [PubMed] [Google Scholar]
- Kodama S.; Saito K.; Yachi Y.; Asumi M.; Sugawara A.; Totsuka K.; Saito A.; Sone H. Association between serum uric acid and development of type 2 diabetes. Diabetes care 2009, 32, 1737–1742. 10.2337/dc09-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartáková V.; Kuricová K.; Pácal L.; Nová Z.; Dvořáková V.; Švrčková M.; Malúšková D.; Svobodová I.; Řehořová J.; Svojanovský J.; Olšovský J.; Bělobrádková J.; Kaňková K. Hyperuricemia contributes to the faster progression of diabetic kidney disease in type 2 diabetes mellitus. J. Diabetes Complications 2016, 30, 1300–1307. 10.1016/j.jdiacomp.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Choi B. G.; Kim D. J.; Baek M. J.; Ryu Y. G.; Kim S. W.; Lee M. W.; Park J. Y.; Noh Y. K.; Choi S. Y.; Byun J. K.; Shim M. S.; Mashaly A.; Li H.; Park Y.; Jang W. Y.; Kim W.; Kang J. H.; Choi J. Y.; Park E. J.; Park S. H.; Lee S.; Na J. O.; Choi C. U.; Kim E. J.; Park C. G.; Seo H. S.; Oh D. J.; Rha S. W. Hyperuricaemia and development of type 2 diabetes mellitus in Asian population. Clin. Exp. Pharmacol. Physiol 2018, 45, 499–506. 10.1111/1440-1681.12911. [DOI] [PubMed] [Google Scholar]
- Johnson R. J.; Merriman T.; Lanaspa M. A. Causal or noncausal relationship of uric acid with diabetes. Diabetes 2015, 64, 2720–2722. 10.2337/db15-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T.; Hu H.; Zharikov S.; Tuttle K. R.; Short R. A.; Glushakova O.; Ouyang X.; Feig D. I.; Block E. R.; Herrera-Acosta J.; Patel J. M.; Johnson R. J. A causal role for uric acid in fructose-induced metabolic syndrome. Am. J. Physiol.: Renal Physiol. 2006, 290, F625–F631. 10.1152/ajprenal.00140.2005. [DOI] [PubMed] [Google Scholar]
- Baldwin W.; McRae S.; Marek G.; Wymer D.; Pannu V.; Baylis C.; Johnson R. J.; Sautin Y. Y. Hyperuricemia as a mediator of the proinflammatory endocrine imbalance in the adipose tissue in a murine model of the metabolic syndrome. Diabetes 2011, 60, 1258–1269. 10.2337/db10-0916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicerchi C.; Li N.; Kratzer J.; Garcia G.; Roncal-Jimenez C. A.; Tanabe K.; Hunter B.; Rivard C. J.; Sautin Y. Y.; Gaucher E. A.; Johnson R. J.; Lanaspa M. A. Uric acid-dependent inhibition of AMP kinase induces hepatic glucose production in diabetes and starvation: evolutionary implications of the uricase loss in hominids. FASEB J. 2014, 28, 3339–3350. 10.1096/fj.13-243634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M.; Borghi C.; Cicero A. F. G.; Hisatome I.; Niwa K.; Ohno M.; Johnson R. J.; Lanaspa M. A. Elevated serum uric acid increases risks for developing high LDL cholesterol and hypertriglyceridemia: A five-year cohort study in Japan. Int. J. Cardiol. 2018, 261, 183–188. 10.1016/j.ijcard.2018.03.045. [DOI] [PubMed] [Google Scholar]
- Tinahones F. J.; Collantes E.; C-Soriguer F. J.; González-Ruiz A.; Pineda M.; Añón J.; Sánchez Guijo P. Increased VLDL levels and diminished renal excretion of uric acid in hyperuricaemic–hypertriglyceridaemic patients. Rheumatology 1995, 34, 920–924. 10.1093/rheumatology/34.10.920. [DOI] [PubMed] [Google Scholar]
- Wang C. S.; McConathy W. J.; Kloer H. U.; Alaupovic P. Modulation of lipoprotein lipase activity by apolipoproteins. Effect of apolipoprotein C-III. J. Clin. Invest. 1985, 75, 384–390. 10.1172/JCI111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona F.; Tinahones F. J.; Collantes E.; Garcia-Fuentes E.; Escudero A.; Soriguer F. Response to a urate-lowering diet according to polymorphisms in the apolipoprotein AI-CIII-AIV cluster. J. Rheumatol. 2005, 32, 903–905. [PubMed] [Google Scholar]
- Major T. J.; Dalbeth N.; Stahl E. A.; Merriman T. R. An update on the genetics of hyperuricaemia and gout. Nat. Rev. Rheumatol. 2018, 14, 341–353. 10.1038/s41584-018-0004-x. [DOI] [PubMed] [Google Scholar]
- Woodward O. M.; Köttgen A.; Coresh J.; Boerwinkle E.; Guggino W. B.; Köttgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 10338–10342. 10.1073/pnas.0901249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo H.; Chiba T.; Nagamori S.; Nakayama A.; Domoto H.; Phetdee K.; Wiriyasermkul P.; Kikuchi Y.; Oda T.; Nishiyama J.; Nakamura T.; Morimoto Y.; Kamakura K.; Sakurai Y.; Nonoyama S.; Kanai Y.; Shinomiya N. Mutations in glucose transporter 9 gene SLC2A9 cause renal hypouricemia. Am. J. Hum. Genet. 2008, 83, 744–751. 10.1016/j.ajhg.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto A.; Kimura H.; Chairoungdua A.; Shigeta Y.; Jutabha P.; Cha S. H.; Hosoyamada M.; Takeda M.; Sekine T.; Igarashi T.; Matsuo H.; Kikuchi Y.; Oda T.; Ichida K.; Hosoya T.; Shimokata K.; Niwa T.; Kanai Y.; Endou H. Molecular identification of a renal urate–anion exchanger that regulates blood urate levels. Nature 2002, 417, 447–452. 10.1038/nature742. [DOI] [PubMed] [Google Scholar]
- Kawamura Y.; Nakaoka H.; Nakayama A.; Okada Y.; Yamamoto K.; Higashino T.; Sakiyama M.; Shimizu T.; Ooyama H.; Ooyama K.; Nagase M.; Hidaka Y.; Shirahama Y.; Hosomichi K.; Nishida Y.; Shimoshikiryo I.; Hishida A.; Katsuura-Kamano S.; Shimizu S.; Kawaguchi M.; Uemura H.; Ibusuki R.; Hara M.; Naito M.; Takao M.; Nakajima M.; Iwasawa S.; Nakashima H.; Ohnaka K.; Nakamura T.; Stiburkova B.; Merriman T. R.; Nakatochi M.; Ichihara S.; Yokota M.; Takada T.; Saitoh T.; Kamatani Y.; Takahashi A.; Arisawa K.; Takezaki T.; Tanaka K.; Wakai K.; Kubo M.; Hosoya T.; Ichida K.; Inoue I.; Shinomiya N.; Matsuo H. Genome-wide association study revealed novel loci which aggravate asymptomatic hyperuricaemia into gout. Ann. Rheum. Dis. 2019, 78, 1430–1437. 10.1136/annrheumdis-2019-215521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht E.; Waldenberger M.; Krumsiek J.; Evans A. M.; Jeratsch U.; Breier M.; Adamski J.; Koenig W.; Zeilinger S.; Fuchs C.; Klopp N.; Theis F. J.; Wichmann H. E.; Suhre K.; Illig T.; Strauch K.; Peters A.; Gieger C.; Kastenmüller G.; Doering A.; Meisinger C. Metabolite profiling reveals new insights into the regulation of serum urate in humans. Metabolomics 2014, 10, 141–151. 10.1007/s11306-013-0565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Sun L.; Zong G.; Gao X.; Zhang H.; Xiong Q.; Huo S.; Niu Z.; Sun Q.; Zeng R.; Lin X. Associations of amino acid and acylcarnitine profiles with incident hyperuricemia in middle-aged and older Chinese. Arthritis Care Res. 2019, 1305–1314. 10.1002/acr.24013. [DOI] [PubMed] [Google Scholar]
- Huang C. C.; Lou B. S.; Hsu F. L.; Hou C. C. Use of urinary metabolomics to evaluate the effect of hyperuricemia on the kidney. Food Chem. Toxicol. 2014, 74, 35–44. 10.1016/j.fct.2014.08.017. [DOI] [PubMed] [Google Scholar]
- Yu F.; Chen Z. Y.; Li W.; Zhang Z. X.; Yao H. Study of differentially expressed serun proteins in primary hyperuricemia in uyghur by 2-DE and MALDI-TOF-MS. Biotechnology 2012, 22, 52. [Google Scholar]
- O’Riordan E.; Goligorsky M. S. Emerging studies of the urinary proteome: the end of the beginning?[J]. Curr. Opin. Nephrol. Hypertens. 2005, 14, 579–585. 10.1097/01.mnh.0000168425.60729.36. [DOI] [PubMed] [Google Scholar]
- Zimmerli L. U.; Schiffer E.; Zürbig P.; Good D. M.; Kellmann M.; Mouls L.; Pitt A. R.; Coon J. J.; Schmieder R. E.; Peter K. H.; Mischak H.; Kolch W.; Delles C.; Dominiczak A. F. Urinary proteomic biomarkers in coronary artery disease[J]. Mol. Cell. Proteomics 2008, 7, 290–298. 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- Adachi J.; Kumar C.; Zhang Y.; Olsen J. V.; Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006, 7, R80 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husi H.; Stephens N.; Cronshaw A.; MacDonald A.; Gallagher I.; Greig C.; Fearon K. C.; Ross J. A. Proteomic analysis of urinary upper gastrointestinal cancer markers. Proteomics: Clin. Appl. 2011, 5, 289–299. 10.1002/prca.201000107. [DOI] [PubMed] [Google Scholar]
- Wasinger V. C.; Zeng M.; Yau Y. Current status and advances in quantitative proteomic mass spectrometry. Int. J. Proteomics 2013, 2013, 180605 10.1155/2013/180605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Ahn H. S.; Kim J. H.; Jeong H.; Yu J.; Yeom J.; Song S. H.; Kim S. S.; Kim I. J.; Kim K. Differential Urinary Proteome Analysis for Predicting Prognosis in Type 2 Diabetes Patients with and without Renal Dysfunction. Int. J. Mol. Sci. 2020, 21, 4236 10.3390/ijms21124236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Gómez E.; Bardají-de-Quixano B.; Ferrer S.; Brotons C.; Knepper M.A.; Carrascal M.; Abian J.; Mas J. M.; Calero F.; Ballarín J. A.; Fernández-Llama P. Urinary Proteome Analysis Identified Neprilysin and VCAM as Proteins Involved in Diabetic Nephropathy. J. Diabetes Res. 2018, 2018, 6165303 10.1155/2018/6165303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matafora V.; Lanzani C.; Zagato L.; Manunta P.; Zacchia M.; Trepiccione F.; Simonini M.; Capasso G.; Bachi A. Urinary proteomics reveals key markers of salt sensitivity in hypertensive patients during saline infusion. J. Nephrol. 2021, 10.1007/s40620-020-00877-z. [DOI] [PubMed] [Google Scholar]
- Leng W.; Ni X.; Sun C.; Lu T.; Malovannaya A.; Jung S. Y.; Huang Y.; Qiu Y.; Sun G.; Holt M. V.; Ding C.; Sun W.; Men X.; Shi T.; Zhu W.; Wang Y.; He F.; Zhen B.; Wang G.; Qin J. Proof-of-Concept Workflow for Establishing Reference Intervals of Human Urine Proteome for Monitoring Physiological and Pathological Changes. EBioMedicine. 2017, 18, 300–310. 10.1016/j.ebiom.2017.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T.; Fukatsu M.; Suzuki S.; Wada T.; Joh T. Elevated serum uric acid predicts impaired fasting glucose and type 2 diabetes only among Japanese women undergoing health checkups[J]. Diabetes Metab. 2011, 37, 252–258. 10.1016/j.diabet.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Krishnan E.; Pandya B. J.; Chung L.; Hariri A.; Dabbous O. Hyperuricemia in young adults and risk of insulin resistance, prediabetes, and diabetes: a 15-year follow-up study. Am. J. Epidemiol. 2012, 176, 108–116. 10.1093/aje/kws002. [DOI] [PubMed] [Google Scholar]
- White M. F.; Kahn C. R. The insulin signaling system. J. Biol. Chem. 1994, 269, 1–4. 10.1016/S0021-9258(17)42297-6. [DOI] [PubMed] [Google Scholar]
- Sprong T.; Roos D.; Weemaes C.; Neeleman C.; Geesing C. L.; Mollnes T. E.; van Deuren M. Deficient alternative complement pathway activation due to factor D deficiency by 2 novel mutations in the complement factor D gene in a family with meningococcal infections. Blood 2006, 107, 4865–4870. 10.1182/blood-2005-07-2820. [DOI] [PubMed] [Google Scholar]
- Lo J. C.; Ljubicic S.; Leibiger B.; Kern M.; Leibiger I. B.; Moede T.; Kelly M. E.; Chatterjee Bhowmick D.; Murano I.; Cohen P.; Banks A. S.; Khandekar M. J.; Dietrich A.; Flier J. S.; Cinti S.; Blüher M.; Danial N. N.; Berggren P. O.; Spiegelman B. M. Adipsin is an adipokine that improves β cell function in diabetes. Cell 2014, 158, 41–53. 10.1016/j.cell.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H.; Qiao Q.; Dong Y.; Gao W.; Tang B.; Qian R.; Tuomilehto J. The prevalence of hyperuricemia in a population of the coastal city of Qingdao, China. J. Rheumatol. 2006, 33, 1346–1350. [PubMed] [Google Scholar]
- Nestel P. J.; Fidge N. H. Apoprotein C metabolism in man. Adv. Lipid Res. 1982, 19, 55–83. 10.1016/B978-0-12-024919-0.50008-4. [DOI] [PubMed] [Google Scholar]
- Wang C. S.; McConathy W. J.; Kloer H. U.; Alaupovic P. Modulation of lipoprotein lipase activity by apolipoproteins. Effect of apolipoprotein C-III[J]. J. Clin. Invest. 1985, 75, 384–390. 10.1172/JCI111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson R. M.; O’Brien W. M. Dependence of serum-uric-acid on haemoglobin and other factors in the general population. Lancet 1966, 777–778. 10.1016/S0140-6736(66)90368-0. [DOI] [PubMed] [Google Scholar]
- Su P.; Hong L.; Zhao Y.; Sun H.; Li L. The Association Between Hyperuricemia and Hematological Indicators in a Chinese Adult Population. Medicine 2016, 95, e2822 10.1097/MD.0000000000002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmach M. J.; Szczerbinski L.; Wasilewska N.; Protas P.; Wasilewska A. Hematological parameters in adolescents with hyperuricemia. Indian Pediatr. 2014, 51, 1003–1005. 10.1007/s13312-014-0547-0. [DOI] [PubMed] [Google Scholar]
- Høieggen A.; Alderman M. H.; Kjeldsen S. E.; Julius S.; Devereux R. B.; De Faire U.; Fyhrquist F.; Ibsen H.; Kristianson K.; Lederballe-Pedersen O.; Lindholm L. H.; Nieminen M. S.; Omvik P.; Oparil S.; Wedel H.; Chen C.; Dahlöf B.; The impact of serum uric acid on cardiovascular outcomes in the LIFE study. Kidney Int. 2004, 65, 1041–1049. 10.1111/j.1523-1755.2004.00484.x. [DOI] [PubMed] [Google Scholar]
- Kırça M.; Oğuz N.; Çetin A.; Uzuner F.; Yeşilkaya A. Uric acid stimulates proliferative pathways in vascular smooth muscle cells through the activation of p38 MAPK, p44/42 MAPK and PDGFRβ. J. Recept. Signal Transduction 2017, 37, 167–173. 10.1080/10799893.2016.1203941. [DOI] [PubMed] [Google Scholar]
- Su H. Y.; Yang C.; Liang D.; Liu H.-F. “Research Advances in the Mechanisms of Hyperuricemia-Induced Renal Injury”. BioMed Res. Int. 2020, 2020, 5817348 10.1155/2020/5817348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzani R.; Salvi F.; Dessì-Fulgheri P.; Rappelli A. Renin-angiotensin system, natriuretic peptides, obesity, metabolic syndrome, and hypertension: an integrated view in humans. J. Hypertens. 2008, 26, 831–843. 10.1097/HJH.0b013e3282f624a0. [DOI] [PubMed] [Google Scholar]
- Pisitkun T.; Shen R. F.; Knepper M. A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 13368–13373. 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N.; Kulak N. A.; Cox J.; Neuhauser N.; Mayr K.; Hoerning O.; Vorm O.; Mann M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell. Proteomics 2012, 11, M111.013722 10.1074/mcp.M111.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.; Neuhauser N.; Michalski A.; Scheltema R. A.; Olsen J. V.; Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Cox J.; Hein M. Y.; Luber C. A.; Paron I.; Nagaraj N.; Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Chen T.; Wu S.; Yang C.; Bai M.; Shu K.; Li K.; Zhang G.; Jin Z.; He F.; Hermjakob H.; Zhu Y. iProX: an integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.