Abstract
Kidney cancer is a malignant tumor of the urinary system. Although the 5-year survival rate of patients with kidney cancer has increased by ~30% in recent years due to the early detection of low-grade tumors using more accurate diagnostic methods, the global incidence of kidney cancer continues to increase every year. Therefore, identification of novel and efficient candidate genes for predicting the prognosis of patients with kidney cancer is important. The present study aimed to investigate the role of SEC61 translocon subunit-γ (SEC61G) in kidney cancer. The Cancer Genome Atlas database was screened to obtain the expression profile of SEC61G and identify its association with kidney cancer prognosis. Furthermore, the in vitro effect of SEC61G knockdown on kidney cancer cell proliferation, migration, invasion and apoptosis was investigated using a Cell Counting Kit-8 assay, wound healing assay, Transwell assay and flow cytometry. The results demonstrated that compared with healthy tissues, SEC61G was upregulated in human kidney tumor tissues, which was associated with poor prognosis. In addition, SEC61G knockdown significantly inhibited kidney cancer cell proliferation, migration and invasion compared with the negative control (NC) group. Furthermore, E-cadherin expression was significantly upregulated, and N-cadherin and β-catenin expression levels were significantly downregulated in SEC61G-knockdown kidney cancer cells compared with the NC group. In addition, compared with the NC group, SEC61G knockdown significantly promoted cell apoptosis in a caspase-dependent manner. The aforementioned results suggested that SEC61G might serve as a proto-oncogene to promote kidney tumor progression. Therefore, the present study provided a novel candidate gene for predicting the prognosis of patients with kidney cancer.
Keywords: kidney cancer, proto-oncogene, SEC61 translocon subunit-γ, tumor progression, The Cancer Genome Atlas database
Introduction
Kidney cancer is a malignant tumor of the urinary system, accounting for 2.2% of all adult malignancies worldwide (1). The most common type of kidney cancer is renal cell carcinoma (RCC), which accounts for 85–90% of all cases (2). According to the classification of International Society of Urological Pathology, RCC is primarily divided into three subtypes: i) Clear cell (ccRCC); ii) papillary; and iii) chromophobe (3), of which ccRCC is the most common subtype (4). In the past two decades, the incidence of kidney cancer has been increasing by 2% each year worldwide (5). Although progress has been made and the 5-year survival rate of patients with kidney cancer has increased by ~30% in recent years due to the early diagnosis of low-grade tumors, there were 403,262 new cases of kidney cancer and 175,098 related deaths worldwide in 2018 based on the GLOBOCAN 2018 estimates (1). Therefore, the identification of novel prognostic candidate genes for predicting the prognosis of patients with kidney cancer is important.
SEC61 translocon subunit-γ (SEC61G) is a member of the SEC61 translocon, which is composed of three subunits in mammals, namely Sec61α, Sec61β and Sec61γ (6). SEC61 translocon is the central component of the protein translocation apparatus of the endoplasmic reticulum (ER) membrane, which mediates the integration of ribosomally synthesized unfolded proteins into the ER membrane and the translocation of nascent polypeptides into the ER lumen (7). It has been also reported that SEC61 directly interacts with SEC62 and SEC63 to promote protein folding, modification and translocation (8).
Over the past few years, mutations in the SEC61A1 gene have been associated with autosomal dominant tubulointerstitial kidney disease in humans, diabetes and hepatosteatosis in mice, and professional secretory cell apoptosis (9–11). In addition, SEC63 gene mutations were associated with autosomal dominant polycystic liver disease (12,13). Furthermore, amplification and upregulation of SEC genes, primarily SEC62 and SEC63, was detected in several types of human cancer, including gastric, colorectal, cervical and non-small cell lung cancer, as well as small bowel cancer associated with hereditary non-polyposis colorectal cancer and hepatocellular carcinoma associated with Lynch syndrome (14–19). Regarding SEC61G, a previous study reported that SEC61G was remarkably overexpressed in 77% of glioblastoma multiforme cases, whereas SEC61G knockdown attenuated tumor cell proliferation and induced cell apoptosis (20). However, whether SEC61G is upregulated in kidney cancer tissues and associated with tumor progression is not completely understood.
In the present study, The Cancer Genome Atlas (TCGA) database was screened to obtain the expression profile of SEC61G and identify its association with kidney cancer prognosis. Furthermore, SEC61G was knocked down in kidney cancer cells and the in vitro effect of SEC61G knockdown on kidney cancer cell proliferation, migration, invasion and apoptosis was investigated. All these efforts aimed to investigate the role of SEC61G in kidney cancer, which will provide a novel target or biomarker for the treatment and prognosis of kidney cancer.
Materials and methods
Cell culture
A498 and 769-P cell lines were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. A498 cells were cultured in DMEM (Hyclone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin (Sigma-Aldrich; Merck KGaA) and 100 mg/µl streptomycin (Sigma). 769-P cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin. Cells were maintained at 37°C with 5% CO2. Cells were mycoplasma free and authenticated by STR assay. Cells were seeded into 6-well plates and allowed to grow to 80% confluence prior to use in subsequent experiments.
Bioinformatics analysis
The University of Alabama Cancer Database (UALCAN; ualcan.path.uab.edu/index.html) (21) was used to assess the expression of SEC61G in kidney cancer based on TCGA (https://cancergenome.nih.gov/) data. The Gene Expression Profiling Interactive Analysis (GEPIA; gepia.cancer-pku.cn/index.html) (22) was used to construct survival curves based on SEC61G gene expression data and clinical data in kidney cancer samples obtained from TCGA database, which were analyzed using the log-rank test. The cut-off value for overall survival (OS) was set to the median SEC61G gene expression value. The expression profile of SEC61G was clearly defined in human renal cancer using UALCAN and GEPIA.
Small interfering (si)RNA transfection
A498 and 769-P cells were seeded into 6-well plates at a density of 4–8×105 cell/well. At 2 h prior to transfection, culture medium was replaced with fresh complete medium (RPMI-1640 supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin). Subsequently, cells were transfected with 5 µl siRNAs against SEC61G (5′-CAUUGUUGGUGGCUGAAUATT-3′) or scrambled NC siRNA (5′-TTCTCCGAACGTGTCACGT-3′) at room temperature (~25°C) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following incubation for 6 h, cells were supplemented with fresh complete culture medium and cultured for an additional 48 h. Then, cells were used for subsequent experiments. siRNAs were purchased from Oligobio Biotechnology Co., Ltd. (http://www.oligobio.com.cn/).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from different transfected A498 and 769-P cells using an Ultrapure RNA kit (CoWin Biosciences) according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using HiFiScript cDNA synthesis kits (CoWin Biosciences) following the manufacturer's protocol. Subsequently, with the SYBR Master Mixture (CoWin Biosciences), qPCR was performed to assess the effect of SEC61G knockdown on SEC61G mRNA gene expression levels. The following primers were used for qPCR (synthesized by Genewiz, Inc.): GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′; and Sec61G forward, 5′-TGGATCAGGTAATGCAGTTT-3′ and reverse, 5′-TCAGCCACCAACAATGATG-3′. The PCR reaction was as follows: Pre-incubation at 95°C for 10 min and incubation at 95°C for 30 sec, followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min and a final dissociation stage (95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec). mRNA expression levels were quantified using the 2−ΔΔCq method (23) and normalized to the internal reference gene GAPDH.
Western blotting
Total protein was isolated from A498 and 769-P cells using RIPA lysis buffer. The concentration of total protein was detected using the BCA method. Proteins (20 µg per lane) were separated via 10% SDS-PAGE and transferred to PVDF membranes (EMD Millipore). After blocking with TBS containing 0.1% Tween-20 containing 5% skimmed milk at room temperature for 1 h, the membranes were incubated with primary antibodies targeted against: SEC61G (Rabbit; 1:500; cat. no. 11147-2-AP; ProteinTech Group, Inc.), E-cadherin (Rabbit; 1:1,000; cat. no. 20874-1-AP; ProteinTech Group, Inc.), N-cadherin (Rabbit; 1:2,000; cat. no. 22018-1-AP; ProteinTech Group, Inc.), Bax (Rabbit; 1:2,000; cat. no. 50599-2-lg; ProteinTech Group, Inc.), GAPDH (Mouse; 1:5,000; cat. no. 60004-1-Ig; ProteinTech Group, Inc.), β-catenin (Rabbit; 1:1,000; cat. no. AF6266; Affinity Biosciences), BCL-2 (Rabbit; 1:1,000; cat. no. AF6139; Affinity Biosciences), Cleaved Caspase-3 (Rabbit; 1:1,000; cat. no. AF7022; Affinity Biosciences), AKT (Rabbit; 1:1,000; cat. no. AF6261; Affinity Biosciences), and p-AKT (Rabbit; 1:1,000; cat. no. AF0016; Affinity Biosciences) overnight at 4°C. Following primary antibody incubation, the membranes were incubated with corresponding HRP-conjugated anti-rabbit IgG (Goat; 1:5,000; cat. no. S0001; Affinity Biosciences) or anti-mouse IgG (Goat; 1:5,000; cat. no. S0002; Affinity Biosciences) secondary antibodies at room temperature for 2 h. Protein bands were visualized using an ECL detection kit (Pierce; Thermo Fisher Scientific, Inc.). Protein expression levels were semi-quantified using Quantity One Software (version 4.2.2; Bio-Rad Laboratories, Inc.) with GAPDH as the loading control.
Cell proliferation assay
A498 and 769-P cell proliferation was evaluated by performing a Cell Counting Kit-8 (CCK-8) assay. Briefly, cells were harvested, resuspended in 100 µl medium (1.5×104 cells/ml), seeded into 96-well plates (2,000 cells/well) and incubated at 37°C. At 24 h intervals, 10 µl CCK-8 reagent (Beijing TransGen Biotech Co., Ltd.) was added to each well and incubated at 37°C for an additional 1.5 h. Absorbance was measured at a wavelength of 450 nm using a microplate reader.
Cell apoptosis assay
Flow cytometry was performed to determine A498 and 769-P cell apoptosis using the FITC-Annexin V Apoptosis Detection kit (BD Biosciences) according to the manufacturer's protocol. At 24 h post-transfection, cells (1–5×105 cells/ml) were supplemented in serum-free medium and cultured for an additional 24 h. Subsequently, cells were harvested, resuspended, incubated with 5 µl FITC-conjugated Annexin V at room temperature for 5 min and then incubated with 10 µl PI in the dark at room temperature for 5 min. Cell apoptosis was analyzed using FACSCalibur flow cytometer (BD Biosciences) and FlowJo software (version 7.6.1; FlowJo LLC). The percentage of total apoptotic events was defined as the sum of the apoptotic cells in the early stage (Annexin V positive/PI negative) and late stage (Annexin V positive/PI positive).
Cell invasion and migration assay
A498 and 769-P cell migration and invasion were assessed using 24-well Transwell chambers. To assess cell invasion, 100 µl Matrigel was added to the upper chamber and incubated for 2 h. Subsequently, 500 µl serum-free medium was plated into a 24-well Transwell chamber for 30 min. Then, 100 µl cell suspension (5×104 cells) was plated into the upper chamber, and 600 µl complete medium containing 10% FBS was plated into the lower chamber. Following incubation at 37°C for 24 h, cells on the upper surface of the filter membrane were removed using cotton swabs. Invading cells were fixed with 4% paraformaldehyde at room temperature for 15 min and stained with 0.1% Giemsa at room temperature for 5 min. Stained cells were quantified in five randomly selected fields of view per filter under a fluorescence microscope (CKX 51; Olympus Corporation). To assess cell migration, the aforementioned protocol was performed, but the upper chamber was not precoated with Matrigel.
Wound healing assay
Cells were seeded (5×105 cells/well) into 6-well plates containing RPMI-1640 with 10% FBS to form a confluent cell monolayer. The following day, an artificial wound was created using a sterile 200-µl pipette tip. Following washing with PBS, cells were cultured for 24 h. Wounds were observed at 0 and 24 h using an inverted fluorescence microscope (CKX51; Olympus Corporation). The wound closure area of the migrating monolayer of cells was quantified using ImageJ software (version 1.49; National Institutes of Health).
Statistical analysis
All experiments were repeated three times. Statistical analyses were performed using SPSS software (version 18.0; SPSS, Inc.). Comparisons between groups were analyzed using the unpaired Student's t-test. Data are presented as the mean ± SD. P<0.05 was considered to indicate a statistically significant difference.
Results
SEC61G expression and its association with survival in patients with kidney cancer
To investigate whether SEC61G served as a novel and efficient candidate gene for predicting kidney cancer, the expression profile of SEC61G was obtained using the UALCAN web-tool based on TCGA database. The results demonstrated that SEC61G expression was significantly increased in human kidney tumor tissues compared with healthy tissues (P<0.001; Fig. 1A). Subsequently, the association between increased expression levels of SEC61G and the survival of patients with kidney cancer was investigated. Kaplan-Meier curve analysis revealed that high SEC61G expression levels were significantly associated with poor OS in patients with kidney cancer compared with low SEC61G expression levels (Fig. 1B). The results indicated that SEC61G might serve as a biomarker for kidney cancer.
Figure 1.
SEC61G is a potential novel candidate gene for the prognosis of patients with kidney cancer. (A) SEC61G expression in human kidney tumor tissues (n=290) and healthy tissues (n=32). ***P<0.001 vs. healthy controls. (B) Overall survival was analyzed by Kaplan-Meier analysis based on the median expression of SEC61G in patients with kidney cancer. SEC61G, SEC61 translocon subunit-γ; TPM, transcripts per million; HR, hazard ratio.
SEC61G knockdown attenuates human kidney cancer cell proliferation
Tumor progression is a complex process, which is characterized by tumor cell proliferation, migration, invasion, metastasis, colony formation and adhesion (24,25). Therefore, the present study investigated the effects of SEC61G on cell proliferation, migration, invasion and survival.
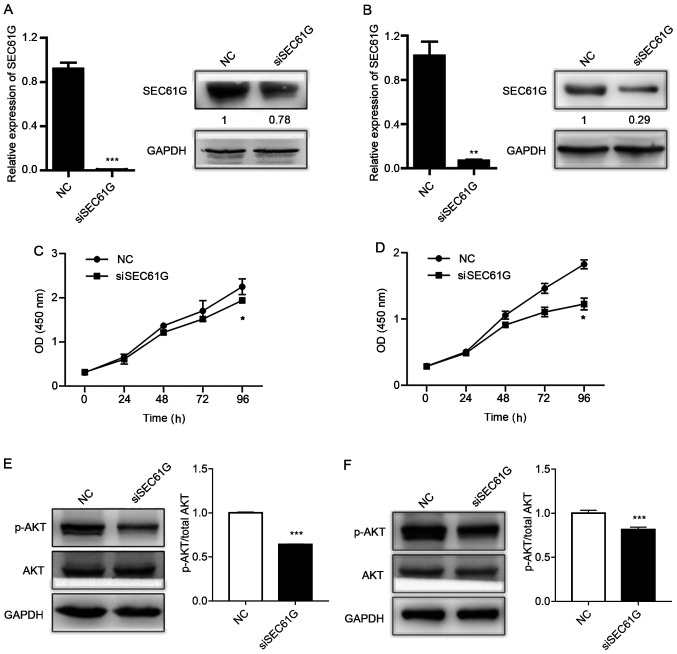
To determine the role of SEC61G in human kidney cancer cell proliferation, A498 and 769-P cells were transfected with SEC61G siRNA. Following transfection with SEC61G siRNA, SEC61G expression levels were dramatically decreased at the mRNA and protein levels in A498 (Fig. 2A) and 769-P (Fig. 2B) cells compared with the NC group. Subsequently, the present study examined whether SEC61G was essential for cell proliferation. The CCK-8 assay results demonstrated that SEC61G knockdown significantly inhibited A498 (Fig. 2C) and 769-P (Fig. 2D) cell proliferation compared with the NC group.
Figure 2.
SEC61G knockdown suppresses human kidney cancer cell proliferation. A498 and 769-P cells were transfected with siSEC61G or NC. Transfection efficiency of siSEC61G in (A) A498 cells and (B) 769-P cells. Effect of SEC16G knockdown on (C) A498 cell proliferation and (D) 769-P cell proliferation. Effect of SEC16G knockdown on AKT phosphorylation in (E) A498 cells and (F) 769-P cells. Data are presented as the mean ± SD from three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. NC. SEC16G, SEC61 translocon subunit-γ; si, small interfering RNA; NC, negative control; OD, optical density; p, phosphorylated.
The PI3K/AKT signaling pathway promotes cell proliferation in the majority of cell types, including kidney cancer cells (26,27). In addition, it has been reported that the PI3K/AKT signaling pathway is activated in ~50% of RCC cases (28–30). Therefore, the effect of SEC61G knockdown on the activation status of AKT in A498 and 769-P cells was assessed. The results revealed that AKT phosphorylation was significantly decreased by SEC61G knockdown in both kidney cancer cell lines compared with the NC group (Fig. 2E and F). The results indicated that SEC61G affected human kidney cancer cell proliferation via the PI3K/AKT signaling pathway.
SEC61G knockdown inhibits human kidney cancer cell migration
Subsequently, the effect of SEC61G on cell migration was investigated. Compared with the NC group, SEC61G knockdown significantly decreased A498 (Fig. 3A) and 769-P (Fig. 3B) cell migration. The effect of SEC61G on cell migration was further assessed by performing wound healing assays. Consistent with the previous findings, SEC61G knockdown significantly decreased the area of wound healing compared with the NC group (Fig. 3C and D). Therefore, the results suggested that SEC61G knockdown inhibited kidney cancer cell migration compared with the NC group.
Figure 3.
SEC61G knockdown inhibits human kidney cancer cell migration. A498 and 769-P cells were transfected with siSEC61G or NC. (A) Effect of SEC61G knockdown on A498 cell migration that was assessed by performing Transwell migration assay. (B) Effect of SEC61G knockdown on 769-P cell migration that was assessed by performing Transwell migration assay. (C) Effect of SEC61G knockdown on A498 cell migration that was assessed by performing wound healing assay. (D) Effect of SEC61G knockdown on 769-P cell migration that was assessed by performing wound healing assay. Data are presented as the mean ± SD of the number of cells in five randomly selected fields of view from three independent experiments (magnification, ×200). **P<0.01 and ***P<0.001 vs. NC. SEC61G, SEC61 translocon subunit-γ; si, small interfering RNA; NC, negative control.
SEC61G knockdown inhibits human kidney cancer cell invasion
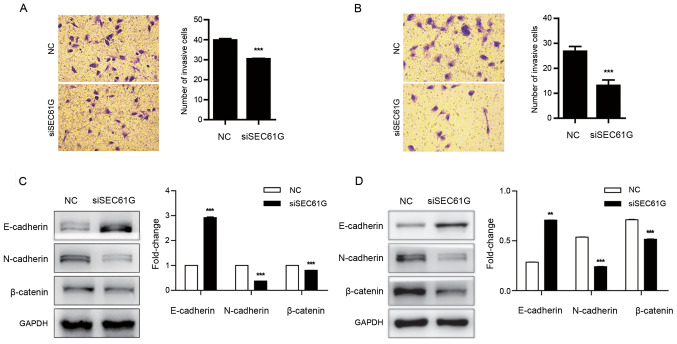
To evaluate the effect of SEC61G on kidney cancer cell invasion, a cell invasion assay was performed. The results demonstrated that SEC61G knockdown significantly inhibited A498 (Fig. 4A) and 769-P (Fig. 4B) cell invasion compared with the NC group.
Figure 4.
SEC61G knockdown suppresses human kidney cancer cell invasion. A498 and 769-P cells were transfected with siSEC61G or NC. Effect of SEC61G knockdown on (A) A498 cell invasion and (B) 769-P cell invasion. In the two panels, data are presented as the mean ± SD of the number of cells in five randomly selected fields of view from three independent experiments (magnification, ×200). (C) Effect of SEC61G knockdown on E-cadherin, N-cadherin and β-catenin protein expression levels in A498 cells. (D) Effect of SEC61G knockdown on E-cadherin, N-cadherin and β-catenin protein expression levels in 769-P cells. Data are presented as the mean ± SD of three independent experiments. **P<0.01 and ***P<0.001 vs. NC. SEC61G, SEC61 translocon subunit-γ; si, small interfering RNA; NC, negative control; E-cad, E-cadherin; N-cad, N-cadherin.
Several transcriptional factors are involved in cancer cell invasion and migration, including E-cadherin, N-cadherin and β-catenin (31,32). It has been reported that during cell invasion and migration, E-cadherin is downregulated, whereas N-cadherin and β-catenin are upregulated (33). Therefore, the expression levels of E-cadherin, N-cadherin and β-catenin were assessed following SEC61G knockdown in A498 and 769-P cells. Compared with the NC group, SEC61G knockdown significantly increased E-cadherin expression levels, and significantly decreased N-cadherin and β-catenin expression levels (Fig. 4C and D). The aforementioned results indicated that SEC61G enhanced kidney cancer cell invasion.
SEC61G knockdown promotes human kidney cancer cell apoptosis
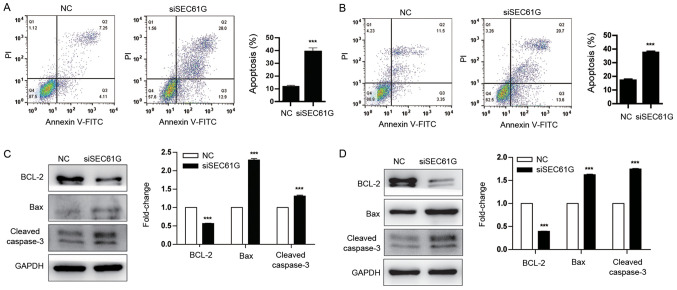
To further explore the role of SEC61G in kidney cancer cell apoptosis, Annexin V/PI staining was performed. Compared with the NC group, SEC61G knockdown significantly increased A498 and 769-P cell apoptosis (Fig. 5A and B). In addition, the expression levels of the apoptosis-related proteins BCL-2, Bax and Cleaved caspase-3 were also determined. The western blotting results demonstrated that compared with the NC group, SEC61G knockdown significantly downregulated the expression of the antiapoptotic protein BCL-2 and significantly upregulated the expression of the proapoptotic protein Bax (Fig. 5C and D). In addition, the protein expression levels of the apoptosis-related marker Cleaved caspase-3 were also significantly increased in SEC61G-knockdown A498 and 769-P cells compared with the NC group. Collectively, the results suggested that SEC61G promoted human kidney cancer cell survival.
Figure 5.
SEC61G knockdown promotes human kidney cancer cell apoptosis. A498 and 769-P cells were transfected with siSEC61G or NC. Effect of SEC61G knockdown on (A) A498 cell apoptosis and (B) 769-P cell apoptosis. (C) Effect of SEC61G knockdown on BCL-2, Bax, Cleaved caspase-3 protein expression levels in A498 cells. (D) Effect of SEC61G knockdown on BCL-2, Bax, Cleaved caspase-3 protein expression levels in 769-P cells. Data are presented as the mean ± SD of three independent experiments. ***P<0.001 vs. NC. SEC61G, SEC61 translocon subunit-γ; si, small interfering RNA; NC, negative control.
Discussion
By using TCGA database, it was found that SEC61G was upregulated in kidney cancer tissues compared with healthy tissues. Additionally, high SEC61G expression levels were associated with poor survival rates in patients with kidney cancer. Moreover, SEC61G knockdown significantly inhibited kidney cancer cell proliferation, migration and invasion, and enhanced cell apoptosis via a caspase-dependent signaling pathway. Taken together, the present study demonstrated that SEC61G might be a proto-oncogene in human kidney cancer. The results suggested that SEC61G might serve as a novel candidate gene for predicting the prognosis of patients with kidney cancer.
The SEC61 complex, composed of SEC61 α, β and γ, is the central component of the protein translocation apparatus for the translocation of nascent polypeptides into the ER lumen and the integration of transmembrane proteins into the ER bilayer (6,7). SEC61G is frequently amplified and overexpressed in several types of tumors, such as glioblastoma (20) and hepatocellular carcinoma (34). The present study demonstrated that SEC61G was upregulated in kidney cancer tissues compared with healthy tissues. High expression levels of SEC61G were associated with poor overall survival in patients with kidney cancer, suggesting that SEC61G might serve as a marker of poor prognosis for patients with kidney cancer.
The uncontrolled proliferation of cancer cells is considered as a hallmark of cancer (35). Previous studies have reported that SEC61β is involved in the trafficking of EGFR and EGFR-mediated activation of the PI3K/AKT signaling pathway (36,37). The PI3K/AKT signaling pathway promotes cell proliferation and inhibits apoptosis in the majority of cell types, including kidney cancer cells (26,27). In addition, AKT is a serine-threonine kinase downstream of the PTEN/PI3K signaling pathway (38). The PI3K/AKT signaling pathway was activated in ~50% of RCC cases (28–30). To reveal the roles and mechanism underlying SEC61G in kidney cancer cell proliferation, cell proliferation, apoptosis and AKT phosphorylation were assessed in SEC61G-knockdown kidney cancer cell lines. The results demonstrated that SEC61G knockdown significantly inhibited kidney cancer cell proliferation and AKT phosphorylation, and promoted cell apoptosis compared with the NC group. Therefore, the results suggested that SEC61G promoted kidney cancer cell proliferation via the PI3K/AKT signaling pathway.
Emerging evidence has indicated that increased tumor cell migration and invasion are hallmarks of cancer, resulting in tumor dissemination and aggressiveness (35,39,40). However, the effect of SEC61G on kidney tumor cell migration and invasion is not completely understood. In the present study, the Transwell and wound healing assay results demonstrated that SEC61G knockdown significantly suppressed kidney cancer cell migration and invasion compared with the NC group. In addition, the expression levels of E-cadherin were significantly upregulated, and the expression levels of N-cadherin and β-catenin were significantly decreased following SEC61G knockdown in human kidney cancer cells compared with the NC group. Overall, the results indicated that SEC61G knockdown inhibited kidney cancer cell migration and invasion compared with the NC group.
siRNA technology is widely used to inhibit the expression of cancer-specific genes for cancer treatment, and is considered a feasible strategy (41). The present study demonstrated that SEC61G knockdown significantly attenuated cell proliferation, migration and invasion, and induced cell apoptosis in human kidney cancer cells compared with the NC group. Therefore, the development of novel drugs containing SEC61G siRNA or inhibitor may be considered as a promising therapeutic strategy for patients with kidney cancer.
Tumor progression is a complex process, which is characterized by cell proliferation, migration, invasion, metastasis, colony formation and adhesion (24,25). A study reported that SEC61G was required for tumor cell survival in glioblastoma multiforme (20). In addition, the present study demonstrated that compared with healthy tissues, SEC61G was upregulated in kidney tumor tissues, which promoted tumor progression and predicted poor prognosis in patients with kidney cancer. However, the molecular signaling pathways underlying the effects of SEC61G are not completely understood. Furthermore, the expression of SEC61G in other types of cancer, including breast, lung and gastric cancer, as well as its role in cancer cell migration, invasion, metastasis and colony formation require further investigation. Therefore, further studies are required to reveal the effects of SEC61G in tumor progression.
Collectively, the present study demonstrated that compared with healthy tissues, SEC61G was upregulated in human renal tumor tissues, which was associated with poor overall survival in patients with kidney cancer based on data obtained from TCGA database. In addition, compared with the NC group, SEC61G knockdown significantly inhibited kidney cancer cell proliferation, migration and invasion, and induced cell apoptosis. Therefore, SEC61G may serve as a novel candidate gene for predicting the progression of patients with kidney cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HM and YW were responsible for the conception and design of the research, and drafting the manuscript or revising it critically for important intellectual content. LG and GY confirmed the authenticity of all the raw data, made substantial contributions to acquisition of data and performed the experiments. XJ and ZS performed the data analysis and interpretation. JW and YW participated in the design of the study and performed the statistical analysis. All authors have read and approved the manuscript and given final approval of the version to be published.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Choueiri TK, Motzer RJ. Systemic therapy for metastatic renal-cell carcinoma. N Engl J Med. 2017;376:354–366. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 3.Srigley JR, Delahunt B, Eble JN, Egevad L, Epstein JI, Grignon D, Hes O, Moch H, Montironi R, Tickoo SK, et al. The international society of urological pathology (ISUP) vancouver classification of renal neoplasia. Am J Surg Pathol. 2013;37:1469–1489. doi: 10.1097/PAS.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- 4.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko V, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ljungberg B, Bensalah K, Canfield S, Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L, Merseburger AS, et al. EAU guidelines on renal cell carcinoma: 2014 update. Eur Urol. 2015;67:913–924. doi: 10.1016/j.eururo.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Greenfield JJ, High S. The Sec61 complex is located in both the ER and the ER-Golgi intermediate compartment. J Cell Sci. 1999;112:1477–1486. doi: 10.1242/jcs.112.10.1477. [DOI] [PubMed] [Google Scholar]
- 7.Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann R, Muller L, Wullich B. Protein transport into the endoplasmic reticulum: Mechanisms and pathologies. Trends Mol Med. 2006;12:567–573. doi: 10.1016/j.molmed.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Schauble N, Lang S, Jung M, Cappel S, Schorr S, Ulucan O, Linxweiler J, Dudek J, Blum R, Helms V, et al. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 2012;31:3282–3296. doi: 10.1038/emboj.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolar NA, Golzio C, Zivna M, Hayot G, Van Hemelrijk C, Schepers D, Vandeweyer G, Hoischen A, Huyghe JR, Raes A, et al. Heterozygous loss-of-function SEC61A1 mutations cause autosomal-dominant tubulo-interstitial and glomerulocystic kidney disease with anemia. Am J Hum Genet. 2016;99:174–187. doi: 10.1016/j.ajhg.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd DJ, Wheeler MC, Gekakis N. A point mutation in Sec61alpha1 leads to diabetes and hepatosteatosis in mice. Diabetes. 2010;59:460–470. doi: 10.2337/db08-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davila S, Furu L, Gharavi AG, Tian X, Onoe T, Qian Q, Li A, Cai Y, Kamath PS, King BF, et al. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat Genet. 2004;36:575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- 13.Drenth JP, Martina JA, van de Kerkhof R, Bonifacino JS, Jansen JB. Polycystic liver disease is a disorder of cotranslational protein processing. Trends Mol Med. 2005;11:37–42. doi: 10.1016/j.molmed.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Mori Y, Sato F, Selaru FM, Olaru A, Perry K, Kimos MC, Tamura G, Matsubara N, Wang S, Xu Y, et al. Instabilotyping reveals unique mutational spectra in microsatellite-unstable gastric cancers. Cancer Res. 2002;62:3641–3645. [PubMed] [Google Scholar]
- 15.Schulmann K, Brasch FE, Kunstmann E, Engel C, Pagenstecher C, Vogelsang H, Krüger S, Vogel T, Knaebel HP, Rüschoff J, et al. HNPCC-associated small bowel cancer: Clinical and molecular characteristics. Gastroenterology. 2005;128:590–599. doi: 10.1053/j.gastro.2004.12.051. [DOI] [PubMed] [Google Scholar]
- 16.Casper M, Weber SN, Kloor M, Müllenbach R, Grobholz R, Lammert F, Zimmer V. Hepatocellular carcinoma as extracolonic manifestation of Lynch syndrome indicates SEC63 as potential target gene in hepatocarcinogenesis. Scand J Gastroenterol. 2013;48:344–351. doi: 10.3109/00365521.2012.752030. [DOI] [PubMed] [Google Scholar]
- 17.Heselmeyer K, Macville M, Schrock E, Blegen H, Hellström AC, Shah K, Auer G, Ried T. Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes Cancer. 1997;19:233–240. doi: 10.1002/(SICI)1098-2264(199708)19:4<233::AID-GCC5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 18.Allen DG, White DJ, Hutchins AM, Scurry JP, Tabrizi SN, Garland SM, Armes JE. Progressive genetic aberrations detected by comparative genomic hybridization in squamous cell cervical cancer. Br J Cancer. 2000;83:1659–1663. doi: 10.1054/bjoc.2000.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehan E, Ben-Dor A, Liao W, Lipson D, Frimer H, Rienstein S, Simansky D, Krupsky M, Yaron P, Friedman E, et al. Chromosomal aberrations and gene expression profiles in non-small cell lung cancer. Lung Cancer. 2007;56:175–184. doi: 10.1016/j.lungcan.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Lu Z, Zhou L, Killela P, Rasheed AB, Di C, Poe WE, McLendon RE, Bigner DD, Nicchitta C, Yan H. Glioblastoma proto-oncogene SEC61gamma is required for tumor cell survival and response to endoplasmic reticulum stress. Cancer Res. 2009;69:9105–9111. doi: 10.1158/0008-5472.CAN-09-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Steeg PS. Tumor metastasis: Mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 25.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 26.Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: An updated review. Ann Med. 2014;46:372–383. doi: 10.3109/07853890.2014.912836. [DOI] [PubMed] [Google Scholar]
- 27.Porta C, Figlin RA. Phosphatidylinositol-3-kinase/Akt signaling pathway and kidney cancer, and the therapeutic potential of phosphatidylinositol-3-kinase/Akt inhibitors. J Urol. 2009;182:2569–2577. doi: 10.1016/j.juro.2009.08.085. [DOI] [PubMed] [Google Scholar]
- 28.Hu H, Jiang C, Li G, Lu J. PKB/AKT and ERK regulation of caspase-mediated apoptosis by methylseleninic acid in LNCaP prostate cancer cells. Carcinogenesis. 2005;26:1374–1381. doi: 10.1093/carcin/bgi094. [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Jette N, Moussienko D, Bebb DG, Lees-Miller SP. ATM-deficient colorectal cancer cells are sensitive to the PARP inhibitor olaparib. Transl Oncol. 2017;10:190–196. doi: 10.1016/j.tranon.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross-Goupil M, Massard C, Ravaud A. Targeted therapies in metastatic renal cell carcinoma: Overview of the past year. Curr Urol Rep. 2012;13:16–23. doi: 10.1007/s11934-011-0232-y. [DOI] [PubMed] [Google Scholar]
- 31.Mrozik KM, Blaschuk OW, Cheong CM, Zannettino AC, Vandyke K. N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer. BMC Cancer. 2018;18:939. doi: 10.1186/s12885-018-4845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pal I, Rajesh Y, Banik P, Dey G, Dey KK, Bharti R, Naskar D, Chakraborty S, Ghosh SK, Das SK, et al. Prevention of epithelial to mesenchymal transition in colorectal carcinoma by regulation of the E-cadherin-β-catenin-vinculin axis. Cancer Lett. 2019;452:254–263. doi: 10.1016/j.canlet.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Tania M, Khan MA, Fu J. Epithelial to mesenchymal transition inducing transcription factors and metastatic cancer. Tumour Biol. 2014;35:7335–7342. doi: 10.1007/s13277-014-2163-y. [DOI] [PubMed] [Google Scholar]
- 34.Gao H, Niu W, He Z, Gao C, Peng C, Niu J. SEC61G plays an oncogenic role in hepatocellular carcinoma cells. Cell Cycle. 2020;19:3348–3361. doi: 10.1080/15384101.2020.1843816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007;18:1064–1072. doi: 10.1091/mbc.e06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorkin A. Internalization of the epidermal growth factor receptor: Role in signalling. Biochem Soc Trans. 2001;29:480–484. doi: 10.1042/bst029a063a. [DOI] [PubMed] [Google Scholar]
- 38.Hu M, Zhu S, Xiong S, Xue X, Zhou X. MicroRNAs and the PTEN/PI3K/Akt pathway in gastric cancer (Review) Oncol Rep. 2019;41:1439–1454. doi: 10.3892/or.2019.6962. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 40.Friedl P, Wolf K. Tumor-cell invasion and migration: Diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 41.Yagi N, Manabe I, Tottori T, Ishihara A, Ogata F, Kim JH, Nishimura S, Fujiu K, Oishi Y, Itaka K, et al. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer Res. 2009;69:6531–6538. doi: 10.1158/0008-5472.CAN-08-3945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.