Abstract
Many cellular processes involve the participation of large macromolecular assemblies. Understanding their function requires methods allowing to study their dynamic and mechanistic properties. Here we present a method for quantitative analysis of native protein or ribonucleoprotein complexes by mass spectrometry following their separation by density – qDGMS. Mass spectrometric quantitation is enabled through stable isotope labelling with amino acids in cell culture (SILAC). We provide a complete guide, from experimental design to preparation of publication-ready figures, using a purposely-developed R package – ComPrAn. As specific examples, we present the use of sucrose density gradients to inspect the assembly and dynamics of the human mitochondrial ribosome (mitoribosome), its interacting proteins, the small subunit of the cytoplasmic ribosome, cytoplasmic aminoacyl-tRNA synthetase complex and the mitochondrial PDH complex. ComPrAn provides tools for analysis of peptide-level data as well as normalization and clustering tools for protein-level data, dedicated visualization functions and graphical user interface. Although, it has been developed for the analysis of qDGMS samples, it can also be used for other proteomics experiments that involve 2-state labelled samples separated into fractions. We show that qDGMS and ComPrAn can be used to study macromolecular complexes in their native state, accounting for the dynamics inherent to biological systems and benefiting from its proteome-wide quantitative and qualitative capability.
Keywords: Proteomics, Density gradient ultracentrifugation, Complexome profiling, Mitochondrial ribosome, SILAC, R package
Graphical abstract
Highlights
-
•
qDGMS is a novel method to study macromolecular complex composition and assembly.
-
•
Complexes are separated in near-native form by density gradient ultracentrifugation.
-
•
SILAC enables simultaneous quantitative proteomic analysis of two biological samples.
-
•
R package ComPrAn allows analysis of SILAC complexome profiling and qDGMS data sets.
1. Introduction
Many cellular processes involve the participation of large proteinaceous macromolecular assemblies. Biochemical functions of many of these complexes have been extensively investigated. More recently, their structures have started to be unveiled at a greater speed, especially due to technological advances in techniques such as cryo-electron microscopy. Nonetheless, in many cases more dynamic and mechanistic aspects of their roles and functions remain elusive.
Complexome profiling, or complexomics, is a recently developed mass spectrometry-based method used to study macromolecular complexes close to their native state. It allows investigation of their protein subunit composition and transient interactions with other proteins such as accessory and assembly factors. This approach typically involves the separation of protein complexes by blue-native polyacrylamide gel electrophoresis (BN-PAGE) and has been successfully used in the study of several mitochondrial oxidative phosphorylation complexes [[1], [2], [3], [4], [5], [6], [7]]. However, separation by BN-PAGE may not be suitable for all macromolecular complexes (e.g. subunits of mammalian ribosomes). Sucrose density gradient ultracentrifugation in combination with proteomics has previously been used to study interactors of yeast mitochondrial ribosomes [8,9]. However, these studies focus on analysis of only selected regions of the gradient [8] or label free mass spectrometry analysis of protein migration across the density gradient within a single sample [9]. To address the above limitations, we developed and optimized quantitative Density Gradient analysis by Mass Spectrometry (qDGMS) which combines stable isotope labelling with amino acids in cell culture (SILAC) and density gradient ultracentrifugation followed by fractionation and analysis by liquid chromatography – mass spectrometry (LC–MS2). To illustrate the potential of this method, we applied qDGMS to the study of the human mitochondrial ribosome [10].
Data analysis and visualization of complexomics results is challenging, since protein identification by mass spectrometry requires proteins to be digested into peptides, and because meaningful relative protein abundance estimates can only be made when comparing the observed signals of identical peptides between samples. When a protein mixture is separated into many fractions then for certain proteins present at low abundances, different subsets of characteristic peptides can be obtained in different fractions, reflecting detection limit of mass spectrometry. Labelling and multiplexing samples reduces further the complement of peptides detected in all labelling states, limiting reliable comparisons between the samples. When estimating protein abundance on the basis of peptide abundance, to make results comparable between fractions and across samples, a strategy needs to be used that accounts for the acquisition of different peptide subsets.
In recent years, software tools have been developed for the analysis of complexomics experiments [4,11,12]. However, the functionality of these tools is limited to a single sample [11] or to a specific task [12]. Visualization of complexomics results is often done manually in spreadsheets [1,2,6], which is time consuming, offers limited functionality, and is prone to errors.
Here we describe the details of the qDGMS method and present the ComPrAn (Complexome Profiling Analysis) R package, a dedicated software for the data analysis of qDGMS experiments. ComPrAn uses peptide data identified and quantified by widely used proteomics software (e.g. the peptide search engine Mascot and the mass spectrometry pipeline application Proteome Discoverer, respectively) and includes tools for estimation of protein abundance from peptide chromatographic peak area, the normalization of such protein abundance estimates, cluster analysis of protein abundance profiles and visualization tools. The package is accompanied by a user-friendly graphical interface. Although, ComPrAn has been developed for analysis of qDGMS experiments, it can also be used for any other type of protein complex chromatography methods that fractionate 2-state labelled samples. ComPrAn was developed in R and is available as an open source package which allows easy integration with other R packages.
Therefore, the methodology presented in this report provides a complete guide, from experimental design to publication-ready figures, rendering the study of native protein complexes accessible to a broader audience.
2. Method description
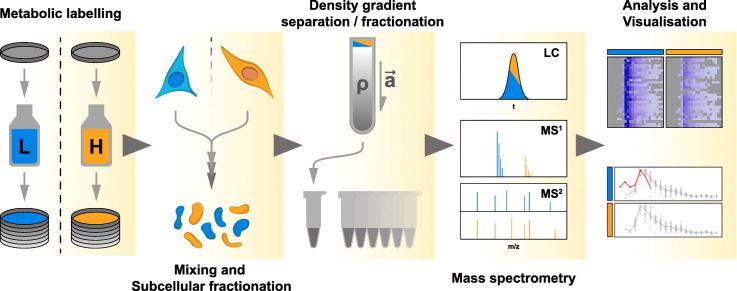
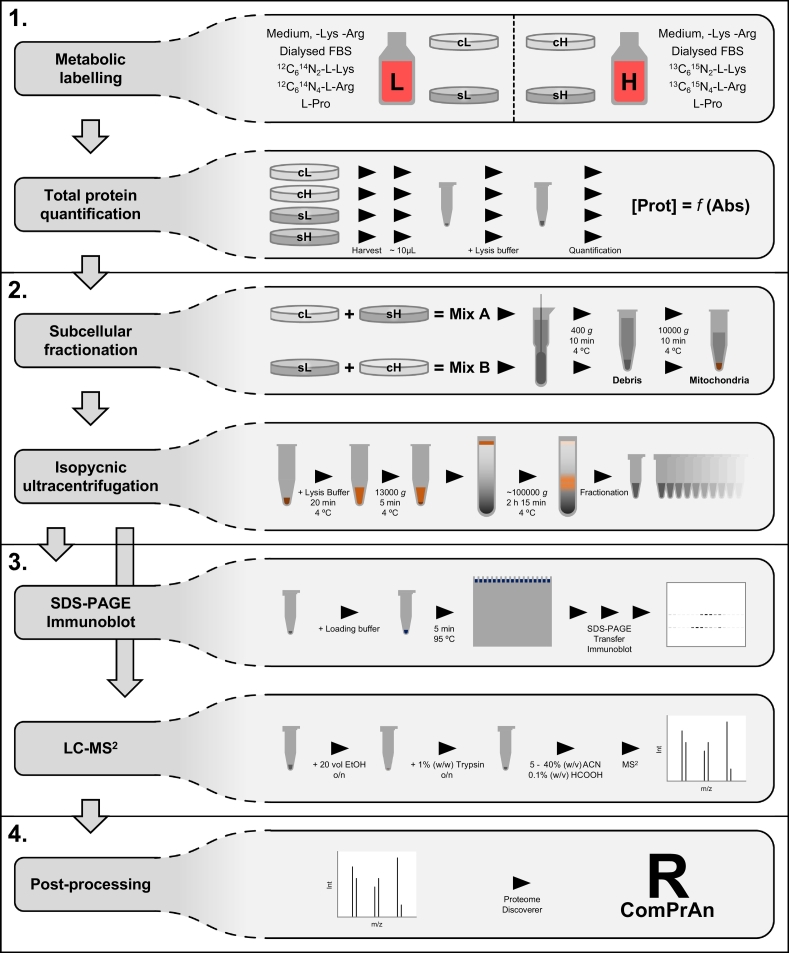
There are four main steps in the qDGMS procedure (Fig. 1): 1. Reciprocal labelling of cells using SILAC, 2. Cellular compartment enrichment and separation of macromolecular complexes, 3. Analysis of fractions by mass spectrometry, 4. Data analysis. We provide a detailed, step-by-step protocol as a supplementary file (Supplementary File S1).
Fig. 1.
Schematic overview of the experimental procedure. Experimental stages of the qDGMS method: Step1 – metabolic labelling of cells, harvesting and total protein quantification; Step 2 – optional subcellular fractionation to enrich the samples for the organelle of choice (e.g. mitochondria), separation of sample components by density (i.e. density gradient ultracentrifugation); Step 3 – verification of gradient integrity and resolution (e.g. SDS-PAGE followed by immunoblotting), mass spectrometric (MS2) analysis; Step 4 – data processing using the ComPrAn R package.
2.1. Reciprocal labelling of cells using SILAC
The aim of qDGMS experiments is to compare the composition and amount of protein complexes between biological samples. In the first stage of qDGMS, the two samples to be compared are grown in SILAC media. In this metabolic labelling procedure, one set of cells is cultured with amino acids containing carbon and nitrogen isotopes at natural abundance ratios (predominantly 12C and 14N), while the other set is cultured with amino acids highly enriched in less naturally abundant isotopes (13C and 15N). Since the latter have a higher mass number, they are often termed “heavy”, and the former “light”. Commonly used amino acids in SILAC are heavy and light arginine and lysine. When arginine is used, SILAC growth medium is typically supplemented with excess proline to prevent conversion of arginine to proline, which would reduce the availability of labelled arginine to be incorporated into proteins. To ensure that virtually all cellular proteins are labelled, cells are cultured in the presence of light and heavy amino acids for a minimum of 7 doubling times (see Supplementary File S1, section A). At this point, cells are harvested, and total protein mass quantified (see Supplementary File S1, sections C and D). This measure is used as a proxy for the amount of material in each sample, and equal amounts of heavy- and light-labelled sample are mixed for subsequent processing stages. Importantly, biological duplicates should be performed, with reciprocal labelling of cell lines; this not only provides opportunity to inspect the reproducibility of the results, but also serves to mitigate any variations isotopic effects and metabolic labelling may introduce.
2.2. Cellular compartment enrichment and separation of macromolecular complexes
In case the complex of interest is localized in a sub-cellular compartment, enrichment of this compartment may increase resolution and promote detection of the constituents. In the example presented below, mitochondria are isolated from differentially labelled cells (see Supplementary File S1, sections E and F). After obtaining purified mitochondria (e.g. by differential centrifugation), they are lysed and layered on a 10%–30% (w/v) sucrose gradient (see Supplementary File S1, section G). By ultracentrifugation, the constituents of the lysate migrate through the gradient and reach equilibrium in the region where the density of the surrounding solution matches their own. This step can be adjusted to accommodate different procedures required to prepare samples for density gradient ultracentrifugation. Likewise, the composition and range of the density gradient can also be optimized to suit the complexes under study. The main aspect to take into consideration regarding this step is that the composition of the gradient buffer should be compatible with the subsequent mass spectrometric analysis (see Section 2.3).
2.3. Analysis of fractions by mass spectrometry
Upon completion of density gradient ultracentrifugation, the gradient is fractionated into portions of equal volume. This can be performed either manually or automatically, using a fractionator. In either case, fractions can be collected from the top or the bottom of the gradient, depending on the convenience and/or available equipment. Automatic fractionation typically leads to a more uniform and reproducible collection. Measurement of UV absorption during automatic fractionation allows the profiling of protein content and of complexes that contain, for instance, nucleic acids.
To verify that separation on the density gradient was successful, a portion of the collected fractions can be analyzed by immunoblotting. This profiling not only informs on the integrity and resolution of the gradient, but also identifies fractions where the complexes of interest are enriched. Furthermore, the results from this experiment can later be matched with mass spectrometry findings to confirm the profile of proteins of interest.
Subsequently, the components of collected fractions are analyzed by mass spectrometry (see Supplementary File S1, section H). To prepare fractions for mass spectrometry, proteins can be directly precipitated from solution. Alternatively, fractions can be briefly resolved by polyacrylamide gel electrophoresis in cases where buffer components (such as detergents, sucrose or polymers of glycols or glycerol) need to be removed owing to potential interferences with subsequent procedures; in this case, gel slabs are excised, diced into small pieces and then subjected to in-gel proteolysis.
Since proteins from the two samples undergo the same procedures prior to the mass spectrometric analysis, and are ionized in parallel, the results are endowed with a quantitative power, in addition to be used to evaluate qualitative changes.
2.4. Data analysis
Analysis of data produced by qDGMS experiments is complex and involves multiple steps, some of which are unique for complexome profiling-type experiments. In order to facilitate this, we developed an R package – Complexome Profiling Analysis (ComPrAn) – which is freely available and can be installed from https://github.com/Scavetta/ComPrAn.
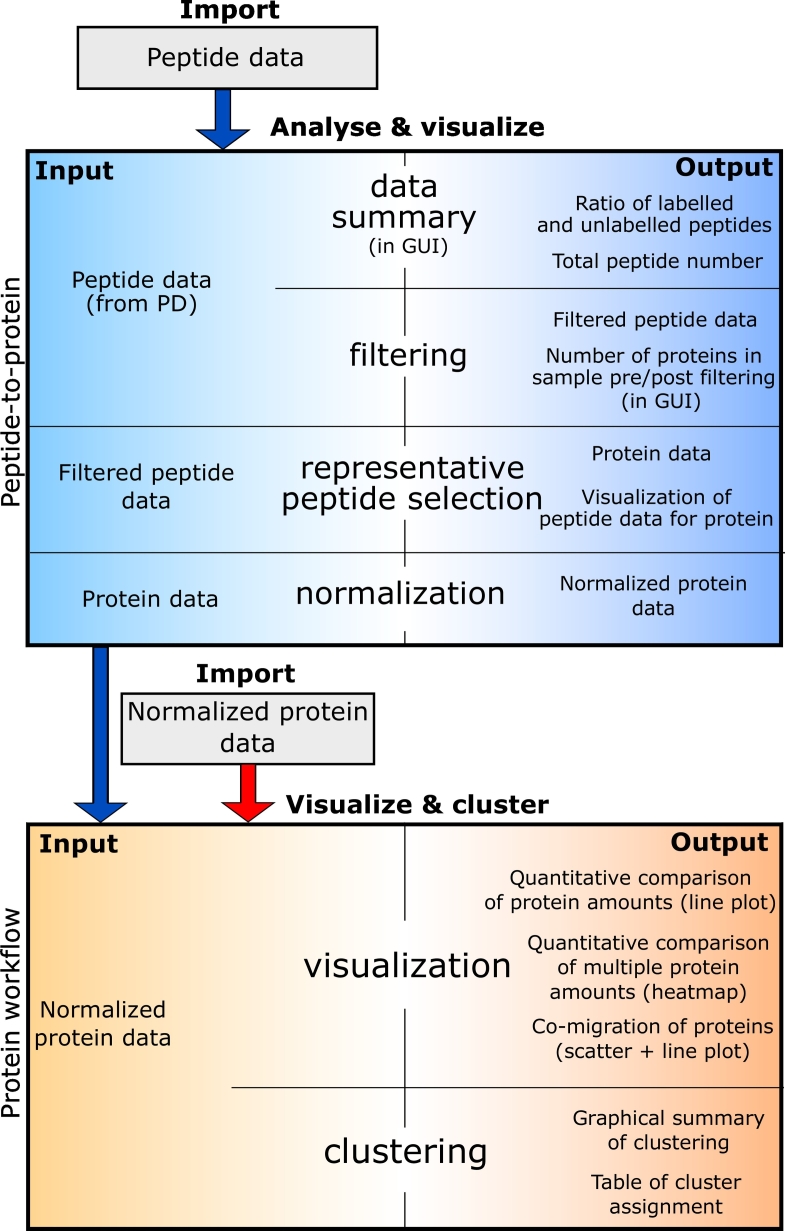
The main application of ComPrAn is to compare changes in protein complex composition and/or quantity between 2-state labelled samples. The workflow is divided into two parts: (i) “peptide-to-protein” (Fig. 2, blue box), which uses peptide-level data (for example generated by Proteome Discoverer) to estimate protein abundance and to normalize those data by scaling values between 0 and 1 for each identified protein (ii) “protein workflow” (Fig. 2, orange box), which allows for clustering and visualizing normalized data. The approach to protein abundance estimation used in ComPrAn is based on the principles used for the analysis of published SILAC complexome profiling experiments [3,6]. For step-by-step data analysis procedures see Supplementary File S1, section I.
Fig. 2.
Schematic overview of the data analysis using ComPrAn. The data flow (middle), with input (left) and output (right) of each step performed for peptide and normalized protein data inputs. Peptide-to-protein part of the workflow does not need to be performed each time a new visualization/clustering is done; normalized protein data can be used as a direct input in the “protein workflow” (red arrow). GUI = graphical user interface, PD = Proteome Discoverer™.
2.4.1. Estimation of protein abundance from peptide abundance
Before using ComPrAn, raw mass spectrometry data need to be interpreted by identification and relative quantification of the peptides in each fraction, for both labelling states (heavy and light). The input data format corresponds to the peptide data produced by Proteome Discoverer (Thermo Scientific, version 1.4) (see Supplementary File S1, Materials, section “ComPrAn input files description”). However, as this is a tab-delimited text file, peptide data produced in other software should be convertible into a format compatible with ComPrAn.
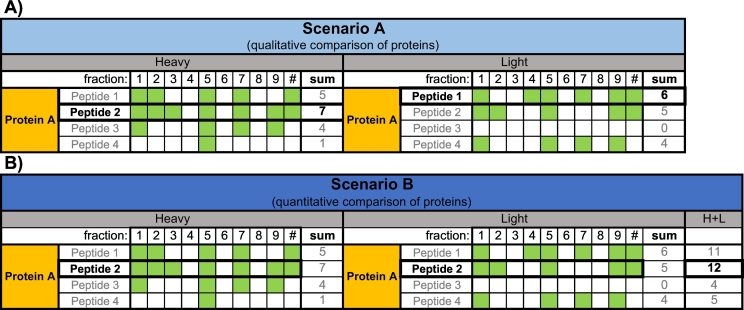
qDGMS enables comparison of migration profiles of proteins along the density gradient and between the two studied samples, both from a qualitative and quantitative perspective. The first step of the ComPrAn analysis is to estimate protein abundance from peptide abundance. For each protein, multiple peptides are usually detected in each fraction; however, the sets of detected peptides often differ across fractions and between the two studied biological samples. The detection and quantification of peptides can be affected by multiple factors including peptide sequence, ionizability, charge state, modifications and fragmentation efficiency. Using peptides that differ in these parameters for estimation of protein abundance may lead to false conclusions, as they do not constitute a real quantitative identity across fractions and samples. Therefore, to enable reliable relative protein quantification between fractions and samples, a single representative peptide of the protein is selected in the pipeline as a proxy of protein abundance (rather than estimating protein quantity from the median or average values of peptide abundances). During the analysis, peptides are divided into “heavy” and “light” batches, based on their isotopic labelling status. Each peptide is identified by its sequence, charge state and modifications. The representative peptide can be selected in two ways: one aimed to optimally estimate protein abundances separately for each sample (i.e. within each labelling state) and is intended for qualitative comparisons (henceforth Scenario A), or to compare the protein abundance quantitatively between labelling states (Scenario B).
When the objective is to evaluate qualitative differences between samples, such as protein co-migration, it is advantageous to select a representative peptide independently for heavy and light samples (Scenario A). By treating the two labelling states independently, migration profiles can be defined for all identified proteins, even if some of their peptides were not detected in both labelling states (Fig. 3A). By comparing protein migration profiles, it is possible to assess changes in the composition of complexes and identify their potential interactors.
Fig. 3.
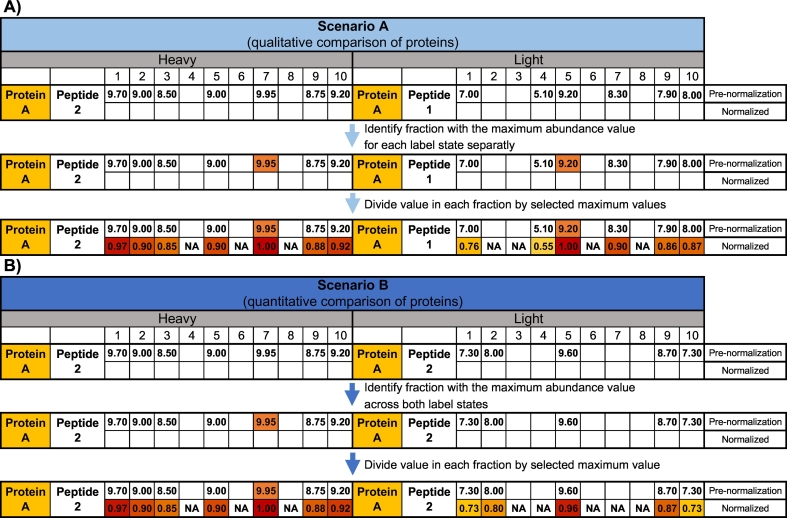
Schematic example of the process of selecting peptides to represent protein abundance, for heavy and light labelling states (peptides are split into two groups according to their attributed labelling states). Green boxes represent that a given peptide was detected in a given fraction. A) Scenario A. The peptide present in the highest number of fractions within each labelling state is considered the representative peptide (in this example Peptide 2 for heavy and Peptide 1 for light). Thus, for the same protein, the peptide picked for one labelling state may be different from the peptide selected for the other. This scenario picks a representative peptide for all proteins, even if they are present only in one labelling state. B) Scenario B. The selected representative peptide is the one that is present in the highest number of fractions, for both labelling states considered together. Thus, for the same protein, the representative peptide selected is the same for both labelling states. In this scenario a representative peptide cannot be selected for proteins that do not have shared peptides or are not present in both labelling states. The peptide present in both samples and in the highest number of fractions (column H + L) is selected as representative (Peptide 2).
When the objective is to evaluate the difference in protein abundance between labelling states, for any given protein, the selected representative peptide is the same for both labelling states (Scenario B). By comparing peptides that match in their sequence, charge and modifications between heavy and light samples, changes in the protein amounts can be quantified between these states. Although only proteins for which at least one matched peptide was identified in both samples will be analyzed, potentially restricting the number of assessed proteins (particularly those present at low abundance), this procedure ensures high reliability of quantitation (Fig. 3B).
2.4.2. Normalization of protein intensities
The range of abundance of different proteins in a cell spans several orders of magnitude [13]. This variability associated to protein abundance may hinder an effective visualization of the migration profiles. To ease identification of co-migrating proteins (Scenario A) or comparison of relative quantities (Scenario B), protein abundance is scaled to values between 0 and 1. However, due to the differences in representative peptide selection between Scenarios A and B, also the methods of normalization differ between these scenarios: protein abundance values are scaled by the maximum value either within each labelling state (Fig. 4A), or across both labelling states (Fig. 4B).
Fig. 4.
Schematic example of the process protein abundance normalization. A) Schematic of a simplified case of normalization according to scenario A (see also Fig. 3A). First, the fraction with the highest value is identified, separately for heavy and light samples. Next, the value of each fraction is divided by this maximum value within each labelling state. This means that, for Protein A, there will be a value of 1 present at least in one fraction for the heavy sample and at least in one fraction for light sample. B) Schematic of a simplified case of normalization according to scenario B (see also Fig. 3B). First, the fraction with the highest value across both labelling states is identified. Next, the value of each fraction is divided by this maximum value. This means that, for Protein A, there will be a value of 1 present at least in one fraction in at least one of the heavy or light samples.
2.4.3. Hierarchical clustering
Hierarchical clustering of proteins can be performed as part of the ComPrAn workflow. Changes in the migration profile of proteins of interest can be verified manually; however, methods such as hierarchical clustering allow an assessment of overall changes in protein migration patterns across all detected proteins. In agglomerative hierarchical clustering, each protein is first assumed to be its own cluster and is progressively linked to others to form larger clusters. Proteins are assigned to clusters based on their “distance” from each other given a metric of similarity between their migration profiles. Several metrices can be used as a distance measure, such as Pearson's correlation or the Euclidian distance in multidimensional space. In each step, clusters that are closest to each other are joined. The distance between clusters that are formed by more than one protein is determined by the choice of linkage method. In single linkage (also known as nearest neighbor), distance between clusters is the minimal distance between any two components of clusters. Conversely, complete linkage is defined as the maximum distance between any two components of clusters. Average linkage defines distance between clusters as the mean distance between all components of clusters (for further information on clustering techniques used in proteomics see [14,15]).
Parameters available for cluster analysis in ComPrAn are based upon centered or uncentered Pearson's correlation as a distance measure with options of complete, average or single linkage. The output is provided in a tabular format with cluster numbers associated with each protein, separately for each labelling state.
2.4.4. Visualization
ComPrAn provides dedicated functions for visualization of qDGMS results. There are several different types of plots available, examples of which are described in detail in the Results and discussion section. In proteome-wide experiments, such as qDGMS, datasets can be incomplete owing to insufficient peptide coverage for some proteins. Ideally, each protein will have peptides detected in a continuous range of fractions, generating a continuous migration profile. Methods of representing missing data vary between the software packages chosen for visualization. Interpreting missing data as numeric zero, especially when data are scaled between 0 and 1, might lead to difficulties in distinguishing low signal from no detection in the visualizations. To overcome this problem, ComPrAn allows the presentation of missing data either by omitting the connection between data points or by using a different color for the profile markers (Supplementary Fig. S1).
2.4.5. Graphical user interface
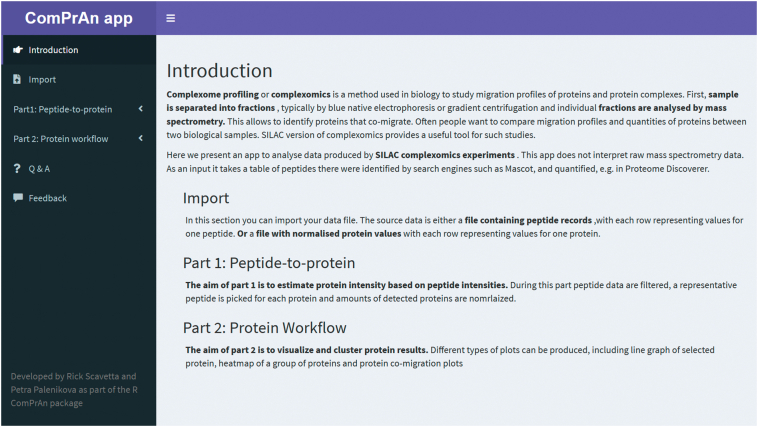
In addition to command line tools, ComPrAn also incorporates a webpage-like graphical user interface (“ComPrAn app”, Fig. 5) created using the Shiny R package, which enables building of interactive applications straight from the R code [16]. Availability of the ComPrAn app makes the package easy to use without any prior programming experience. Each section of the user interface contains instructions for use. By displaying intermediate steps in the workflow, the user is able to perform exploratory data analysis and to verify technical aspects of the experiment.
Fig. 5.
Introduction page of the ComPrAn graphical user interface (app). Different sections of the analysis are displayed in the panel on the left.
Analysis steps of the ComPrAn app are grouped into two parts. In Part 1, once peptide data are imported, information is provided in the app section Summary (Supplementary Fig. S2A) pertaining to the total number of peptides in the file and how many of them passed the initial quality control. In the Filter and Select app section peptides are filtered based on the parameters defined by the user. At this point, plots summarizing numbers of proteins in the two biological samples are shown (Supplementary Fig. S2B). In the Rep Peptides app section, it is possible to visualize how many peptides were detected for each protein and which peptide was selected as representative. This is particularly helpful when proteins appear to be absent in the later stages of the analysis. The last section of Part 1 allows to normalize protein abundance and export an intermediate file containing this information. Procedures in Part 2 consist of various visualizations that are described in the Results and discussion section, and hierarchical clustering. For detailed descriptions of the analysis steps using the interactive app see Supplementary File S1, section I-1.
3. Results and discussion
In order to illustrate the capability of qDGMS in combination with ComPrAn, we applied these tools to study mitochondrial complexes in cells depleted of METTL15. This protein has been recently identified as the mitochondrial methyltransferase responsible for the N4-methylcytidine modification in position 839 (m4C839) of the mitochondrial 12S rRNA [10,17,18] In particular, we used qDGMS to characterize mitochondrial ribosome subunits in METTL15 knock-out cells using data from Van Haute et al. [10] to showcase the potential and highlight points for considerations of this method. We re-analyzed the data from this study with ComPrAn and provide the results of this analysis as an illustration of expected outcomes of the integration of qDGMS with ComPrAn.
3.1. qDGMS allows early sample multiplexing
Complexome profiling is a powerful method to study protein-containing macromolecular complexes. However, as this experimental procedure includes several preparation steps (e.g. isolation, purification, fraction or gel section preparation), it can introduce technical biases during sample processing, making it more difficult to evaluate true biological differences. To reduce technical variability, biological samples can be labelled by means detectable by mass spectrometry such as SILAC and processed/analyzed simultaneously. In qDGMS, thanks to SILAC metabolic labelling samples can be combined at a very early stage of sample processing. This is critical for the mitigation of putative methodologic artefacts, which could arise if samples were processed separately, while enabling the tracing of the source of peptides at later stages.
An important consideration with metabolic labelling strategies like SILAC is the verification of full incorporation of heavy isotopes into peptides in labelled cells. This involves the quantification of peptides containing the heavy amino acids relative to the respective light counterparts. For that, either a sample of the relevant cell populations is collected and analyzed during the labelling period, which is prolonged until a satisfactory heavy isotope incorporation is attained, or a sample is retained before samples labelled with different isotopes are mixed. It should be noted that reliable relative quantification between mixed heavy and light samples can only be achieved with complete incorporation of the heavy isotopes.
3.2. Optional steps of qDGMS
SILAC labelling, separation of complexes and other cellular components by density gradient ultracentrifugation and mass spectrometric analysis comprise the core steps of qDGMS. Depending on the target of study, biological samples may require more or less extensive preparation steps before separation of their components. Factors to consider include the context and abundance of the complexes of interest within the cellular environment. Another aspect is the complexity of the final product, as more heterogeneous samples may present more hurdles to the mass spectrometric identification of all components. In that sense, it may be advisable to perform some form of purification, at least to remove contaminant species and/or very abundant cellular components. However, this may come as a trade-off of more purification steps, which may contribute to a decrease in the amount of biological material, and the loss of the native ambient of the complexes, for example by exposure to harsher conditions or substances. Nevertheless, since both samples intended to be compared have been combined prior to the application of any experimental procedure, they will experience the same conditions, and any interference or bias will most likely equally affect both.
Since METTL15 modifies the 12S rRNA, we focused our analysis on the mitochondrial ribosome. While mitoribosome is relatively abundant within mitochondria, there are other complexes that share similar properties (e.g. cytosolic ribosomes) which considerable abundance may impinge on the complete identification of components of the former. To circumvent this, in the experiment presented here, mitochondria were enriched from the homogenized mix of differently labelled cell populations by differential centrifugation. Although density gradient ultracentrifugation is the core step of the qDGMS, the properties of the density gradient can be adjusted according to the protein complex of interest (Section 3.5).
3.3. Quantitative assessment of protein complexes using ComPrAn
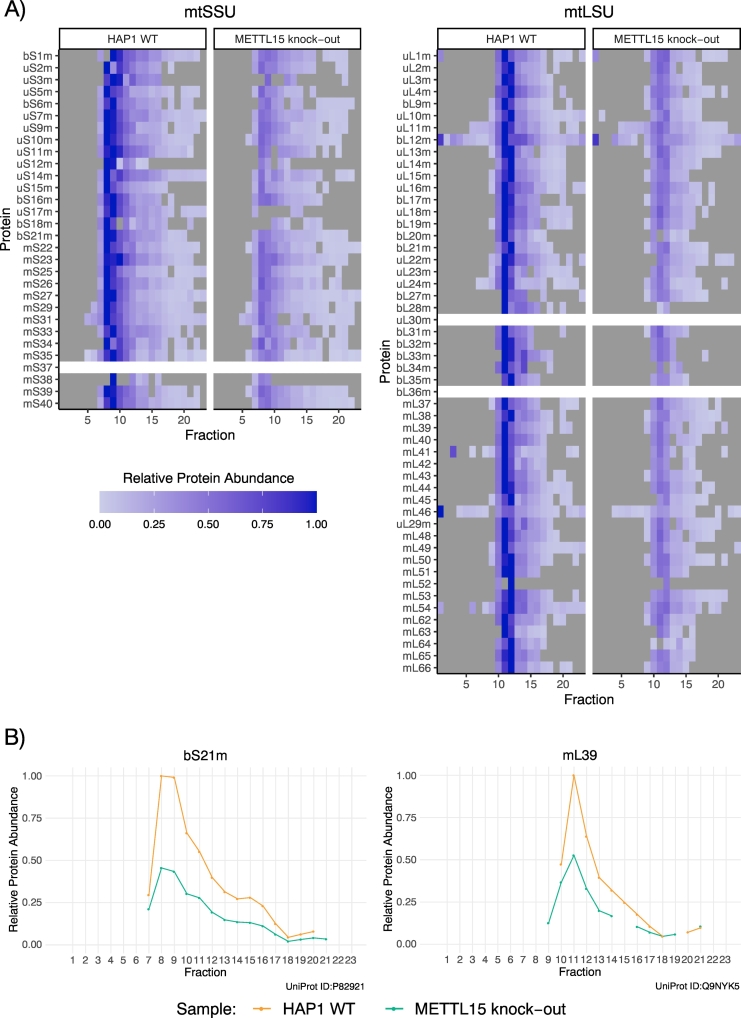
ComPrAn offers several visualization functions for the profiling and quantitative comparison of components of protein-containing complexes across samples and along density gradients. A heatmap representation can be used for a simultaneous visualization of a large number of proteins (Fig. 6A). When a component of a complex is not present in the protein level data, an empty white row is shown in the heatmap (Fig. 6A, uL30m). For any given protein, this absence in quantitative protein data can be caused by lack of detection of any corresponding peptides, lack of detection of one of the SILAC labelling states, or absence of peptides shared between labelling states. The package includes a functionality to display profiles of all peptides detected for proteins of interest, which allows identification of the cause of protein absence from the heatmap (Supplementary Fig. S3). While heatmaps provide a good overview across a number of proteins of interest, it is often advantageous to be able to draw attention to individual proteins and to compare their profiles with line-plots, which is also included as a visualization alternative in ComPrAn (Fig. 6B).
Fig. 6.
A) Quantitative comparison of proteins of small (mtSSU) and large (mtLSU) subunits of the mitoribosome in wild-type and METTL15 knock-out cells. Grey squares represent the absence of detection of a given protein in a given fraction. When a protein was not detected in any fraction a white line is shown. B) Quantitative comparison of selected proteins of the small (bS21m) and large (mL39) mitoribosomal subunits.
3.4. Comparing protein co-migration using ComPrAn
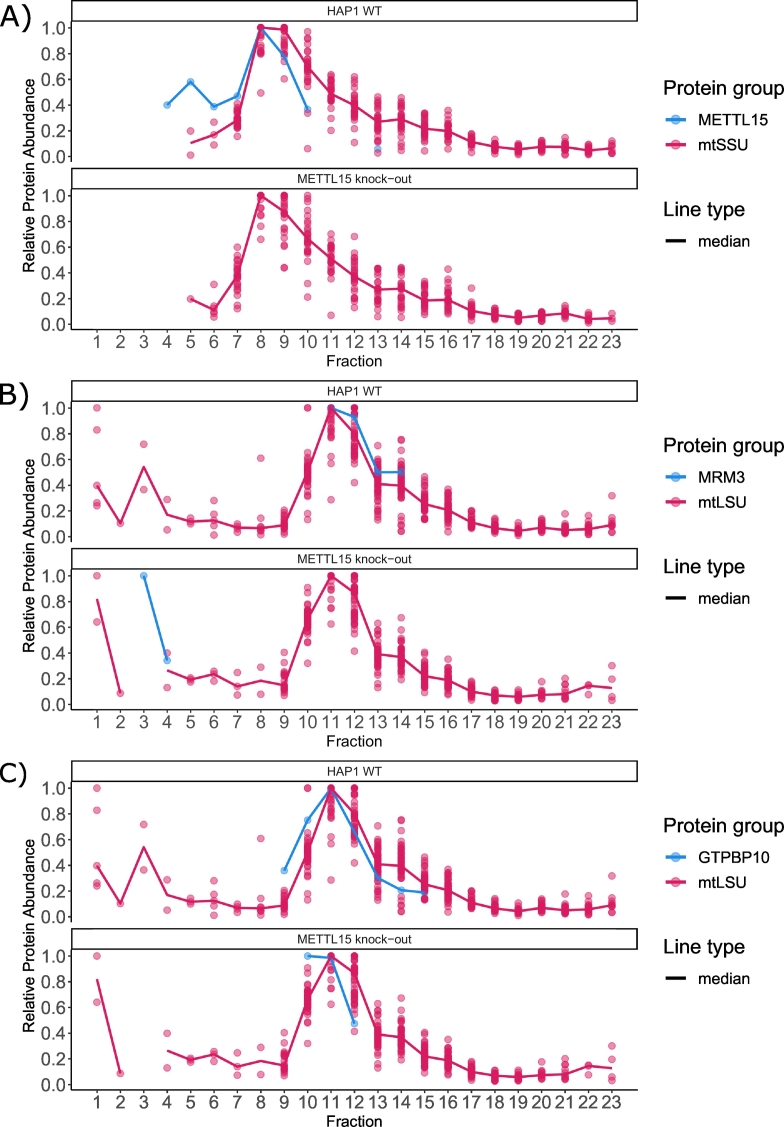
Changes in protein complex composition can be detected as shifts in the migration profile of its constituents. Furthermore, co-migration can be used as a prediction for putative protein interactions. In the qDGMS dataset from Van Haute et al. [10], METTL15 was found to co-migrate with the small mitoribosomal subunit (mtSSU) in the wild-type cell line (Fig. 7A). Other known mitoribosome biogenesis factors, namely MRM3 and GTPBP10, co-migrated with the large mitoribosomal subunit (mtLSU), consistent with previously published data [[19], [20], [21]], Fig. 7B, C). When hierarchical clustering was applied to data from wild-type and METTL15 knock-out cell lines, those biogenesis factors were contained in the clusters where mtLSU components were present (Supplementary Table S1).
Fig. 7.
Qualitative profiling of the human mitoribosome subunit and interacting proteins. ComPrAn-generated qDGMS migration profiles for wild-type and METTL15 knock-out cells are shown. A) METTL15 co-migrates with the mitoribosome small subunit (mtSSU) in the wild-type cells and is not detected in knock-out cells. B) Co-migration of the MRM3 protein with the large mitoribosomal subunit (mtLSU) is observed only in the wild-type cell line. C) The ribosome biogenesis factor GTPBP10 has a similar migration profile to mtLSU in both cell lines.
3.5. Expanded data analysis using ComPrAn
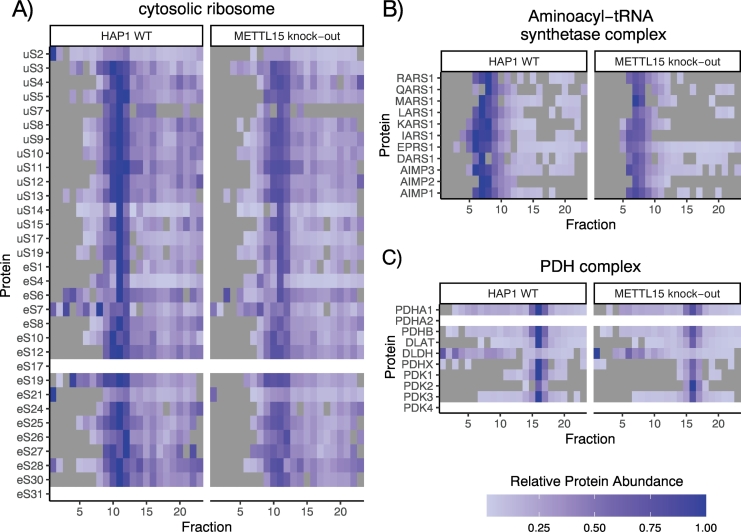
Mass spectrometric analysis of qDGMS and other complexome profiling techniques produces proteome-wide data. These experiments are usually performed by the investigators to address specific research questions, such as characterizing the assembly of mitoribosome and its perturbation. However, ComPrAn as a dedicated data analysis and visualization tool, allows data exploration beyond the original study purpose. To further illustrate the proteome-wide analysis capability of the qDGMS method, we analyze here three other macromolecular complexes, namely the small subunit of the cytoplasmic ribosome, the cytoplasmic aminoacyl-tRNA synthetase complex and the mitochondrial pyruvate dehydrogenase (PDH) complex (Fig. 8).
Fig. 8.
Quantitative analysis of additional protein complexes. Migration profile of cytoplasmic complexes: A) the small subunit of cytosolic ribosome (SSU) and B) the aminoacyl-tRNA synthetase complex. Due to the overall abundance of these complexes, they can still be detected in mitochondria-enriched samples. C) The PDH complex is presented as an example of another mitochondrial protein complex.
Therefore, even without changes in experimental setup and gradient buffer composition, it was possible to easily detect components of other complexes, and evaluate their composition, confirming the applicability of qDGMS in studying of a wide variety of macromolecular complexes.
4. Conclusions
Here we presented qDGMS, a method for the study of protein-containing macromolecular complexes. Sample processing in qDGMS preserves complexes close to their native state, enabling the study of their composition and assembly. Compared to classic complexome profiling experiments where complexes are separated by blue-native electrophoresis (BN-PAGE), qDGMS provides higher flexibility in adjusting and optimizing the composition of the gradient to better match the characteristics of the studied complex.
Furthermore, we introduce ComPrAn, a dedicated data analysis tool with easy to use graphical user interface that helps in making qDGMS more accessible. With a growing interest and use of complexomics-type experiments, including qDGMS, and complexome profiling data sharing platforms, such as CEDAR [22, this issue], the availability of user-friendly data analysis and visualization tools will be crucial for achieving the full potential of this method. While the current version of ComPrAn has been developed for 2-state labelled data, analysis of multiplexed mass spectrometry data could be implemented into future versions, as the outreach and demand for the method grows.
In summary, the qDGMS method in combination with the ComPrAn analysis software add to the portfolio of available techniques for the study of protein complexes in their native form, including complexes involved in mitochondrial oxidative phosphorylation.
The following are the supplementary data related to this article.
Cluster analysis of normalized protein data. ComPrAn's Scenario A was used for peptide selection and normalization. The parameters of clustering were uncentered Pearson's correlation, complete linkage and 0.65 threshold for cluster assignment.
Supplementary figures.
Steb-by-step description of the qDGMS protocol and ComPrAn pipeline.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We are grateful to the members of the MRC MBU Mitochondrial Genetics Group for their input during the development of this method. This work was supported by core funding from Medical Research Council, UK (MC_UU_00015/4). PP and MM were supported by the Marie Sklodowska-Curie ITN-REMIX grant (Grant 721757). PRG was supported by Fundação para a Ciência e a Tecnologia, Portugal (PD/BD/105750/2014).
Contributor Information
Michal Minczuk, Email: michal.minczuk@mrc-mbu.cam.ac.uk.
Pedro Rebelo-Guiomar, Email: pfrg2@mrc-mbu.cam.ac.uk.
References
- 1.Guerrero-Castillo S., Baertling F., Kownatzki D., Wessels H.J., Arnold S., Brandt U., Nijtmans L. The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 2017;25:128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Heide H., Bleier L., Steger M., Ackermann J., Dröse S., Schwamb B., Zörnig M., Reichert A.S., Koch I., Wittig I., Brandt U. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16:538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 3.M. Protasoni, R. Pérez-Pérez, T. Lobo-Jarne, M.E. Harbour, S. Ding, A. Peñas, F. Diaz, C.T. Moraes, I.M. Fearnley, M. Zeviani, C. Ugalde, E. Fernández-Vizarra, Respiratory supercomplexes act as a platform for complex III-mediated maturation of human mitochondrial complexes I and IV, The EMBO Journal. 39 (2020) e102817. doi:10.15252/embj.2019102817. [DOI] [PMC free article] [PubMed]
- 4.Senkler J., Senkler M., Eubel H., Hildebrandt T., Lengwenus C., Schertl P., Schwarzländer M., Wagner S., Wittig I., Braun H.-P. The mitochondrial complexome of Arabidopsis thaliana. Plant J. 2017;89:1079–1092. doi: 10.1111/tpj.13448. [DOI] [PubMed] [Google Scholar]
- 5.J. Senkler, N. Rugen, H. Eubel, J. Hegermann, H.-P. Braun, Absence of complex I implicates rearrangement of the respiratory chain in european mistletoe, Current Biology. 28 (2018) 1606–1613.e4. doi: 10.1016/j.cub.2018.03.050. [DOI] [PubMed]
- 6.Vidoni S., Harbour M.E., Guerrero-Castillo S., Signes A., Ding S., Fearnley I.M., Taylor R.W., Tiranti V., Arnold S., Fernandez-Vizarra E., Zeviani M. MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Rep. 2017;18:1727–1738. doi: 10.1016/j.celrep.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 7.Wessels H.J.C.T., Vogel R.O., Lightowlers R.N., Spelbrink J.N., Rodenburg R.J., van den Heuvel L.P., van Gool A.J., Gloerich J., Smeitink J.A.M., Nijtmans L.G. Analysis of 953 human proteins from a mitochondrial HEK293 fraction by complexome profiling. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woellhaf M.W., Sommer F., Schroda M., Herrmann J.M. Proteomic profiling of the mitochondrial ribosome identifies Atp25 as a composite mitochondrial precursor protein. MBoC. 2016;27:3031–3039. doi: 10.1091/mbc.e16-07-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Möller-Hergt B.V., Carlström A., Stephan K., Imhof A., Ott M. The ribosome receptors Mrx15 and Mba1 jointly organize cotranslational insertion and protein biogenesis in mitochondria. MBoC. 2018;29:2386–2396. doi: 10.1091/mbc.E18-04-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Haute L., Hendrick A.G., D’Souza A.R., Powell C.A., Rebelo-Guiomar P., Harbour M.E., Ding S., Fearnley I.M., Andrews B., Minczuk M. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 2019;47:10267–10281. doi: 10.1093/nar/gkz735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giese H., Ackermann J., Heide H., Bleier L., Dröse S., Wittig I., Brandt U., Koch I. NOVA: a software to analyze complexome profiling data. Bioinformatics. 2015;31:440–441. doi: 10.1093/bioinformatics/btu623. [DOI] [PubMed] [Google Scholar]
- 12.Van Strien J., Guerrero-Castillo S., Chatzispyrou I.A., Houtkooper R.H., Brandt U., Huynen M.A. COmplexome profiling ALignment (COPAL) reveals remodeling of mitochondrial protein complexes in Barth syndrome. Bioinformatics. 2019;35:3083–3091. doi: 10.1093/bioinformatics/btz025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albaum S.P., Hahne H., Otto A., Haußmann U., Becher D., Poetsch A., Goesmann A., Nattkemper T.W. A guide through the computational analysis of isotope-labeled mass spectrometry-based quantitative proteomics data: an application study. Proteome Sci. 2011;9:30. doi: 10.1186/1477-5956-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meunier B., Dumas E., Piec I., Béchet D., Hébraud M., Hocquette J.-F. Assessment of hierarchical clustering methodologies for proteomic data mining. J. Proteome Res. 2007;6:358–366. doi: 10.1021/pr060343h. [DOI] [PubMed] [Google Scholar]
- 16.W. Chang, J. Cheng, J.J. Allaire, Y. Xie, J. McPherson, shiny: Web Application Framework for R. R Package Version 1.5.0. https://CRAN.R-project.org/package=shiny, 2020. https://CRAN.R-project.org/package=shiny (accessed October 17, 2020).
- 17.Chen H., Shi Z., Guo J., Chang K., Chen Q., Yao C.-H., Haigis M.C., Shi Y. The human mitochondrial 12S rRNA m4C methyltransferase METTL15 is required for mitochondrial function. J. Biol. Chem. 2020;295:8505–8513. doi: 10.1074/jbc.RA119.012127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laptev I., Shvetsova E., Levitskii S., Serebryakova M., Rubtsova M., Zgoda V., Bogdanov A., Kamenski P., Sergiev P., Dontsova O. METTL15 interacts with the assembly intermediate of murine mitochondrial small ribosomal subunit to form m4C840 12S rRNA residue. Nucleic Acids Res. 2020;48:8022–8034. doi: 10.1093/nar/gkaa522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavdovskaia E., Kolander E., Steube E., Mai M.M.-Q., Urlaub H., Richter-Dennerlein R. The human Obg protein GTPBP10 is involved in mitoribosomal biogenesis. Nucleic Acids Res. 2018;46:8471–8482. doi: 10.1093/nar/gky701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maiti P., Kim H.-J., Tu Y.-T., Barrientos A. Human GTPBP10 is required for mitoribosome maturation. Nucleic Acids Res. 2018;46:11423–11437. doi: 10.1093/nar/gky938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rorbach J., Boesch P., Gammage P.A., Nicholls T.J.J., Pearce S.F., Patel D., Hauser A., Perocchi F., Minczuk M. MRM2 and MRM3 are involved in biogenesis of the large subunit of the mitochondrial ribosome. Mol. Biol. Cell. 2014;25:2542–2555. doi: 10.1091/mbc.E14-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. Van Strien, A. Haupt, S. Arnold, H.-P. Braun, A. Cabrera-Orefice, J.S. Choudhary, F. Evers, E. Fernandez-Vizarra, S. Guerrero-Castillo, T. Kooij, P. Páleníková, M. Pardo, C. Ugalde, I. Wittig, L. Wöhlbrand, U. Brandt, U. Schulte, M.A. Huynen 2021, CEDAR, an online resource for the reporting and exploration of complexome profiling data, Biochimica et Biophysica Acta (BBA) - Bioenergetics. (this issue). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cluster analysis of normalized protein data. ComPrAn's Scenario A was used for peptide selection and normalization. The parameters of clustering were uncentered Pearson's correlation, complete linkage and 0.65 threshold for cluster assignment.
Supplementary figures.
Steb-by-step description of the qDGMS protocol and ComPrAn pipeline.