Abstract
Background
Many studies have investigated the role of the microbiome in inflammatory bowel disease (IBD), but few have focused on surgery specifically or its consequences on the metabolome that may differ by surgery type and require longitudinal sampling. Our objective was to characterize and contrast microbiome and metabolome changes after different surgeries for IBD, including ileocolonic resection and colectomy.
Methods
The UC San Diego IBD Biobank was used to prospectively collect 332 stool samples from 129 subjects (50 ulcerative colitis; 79 Crohn’s disease). Of these, 21 with Crohn’s disease had ileocolonic resections, and 17 had colectomies. We used shotgun metagenomics and untargeted liquid chromatography followed by tandem mass spectrometry metabolomics to characterize the microbiomes and metabolomes of these patients up to 24 months after the initial sampling.
Results
The species diversity and metabolite diversity both differed significantly among groups (species diversity: Mann-Whitney U test P value = 7.8e-17; metabolomics, P-value = 0.0043). Escherichia coli in particular expanded dramatically in relative abundance in subjects undergoing surgery. The species profile was better able to classify subjects according to surgery status than the metabolite profile (average precision 0.80 vs 0.68).
Conclusions
Intestinal surgeries seem to reduce the diversity of the gut microbiome and metabolome in IBD patients, and these changes may persist. Surgery also further destabilizes the microbiome (but not the metabolome) over time, even relative to the previously established instability in the microbiome of IBD patients. These long-term effects and their consequences for health outcomes need to be studied in prospective longitudinal trials linked to microbiome-involved phenotypes.
Keywords: inflammatory bowel disease, gut microbiome, intestinal surgery, metagenomics, metabolomics
INTRODUCTION
The role of the microbiome1–3 and metabolome4, 5 in inflammatory bowel disease (IBD) has been well established, and many studies have reported specific biomarkers of ulcerative colitis (UC) and Crohn’s disease (CD) at the microbiome6 or metabolome level.7 Prior work on microbiome dynamics in IBD includes the intriguing observation that ileal Crohn’s patients with resection have especially unstable stool microbiome dynamics; 8, 9 however, prior studies have not investigated the impact on the microbiome associated with different types of intestinal surgery in detail. Notably, a prospective study of 20 patients with ileal CD undergoing ileocolonic resection showed that there was no change in microbial diversity 6 months after surgery; however, the microbial community structure was altered in the setting of endoscopic recurrence.10
Investigating the effect of surgery is important for several reasons. First, an increasing body of evidence on dysbiosis suggests that occasional excursions into deleterious regions of microbiome space are important for triggering adverse events.8 Second, surgery is seen from a clinical perspective as a way to manage IBD but is irreversible, and the long-term adverse effects on the microbiome that may worsen disease have not been studied extensively.8, 10 Surgery may have meaningful, durable effects on the microbiome and metabolome and as a consequence should be a key variable to incorporate at a minimum into all future studies of the microbiome, particularly in the setting of IBD. Third, in general it is not known which therapies for IBD have large vs small effects on the microbiome, and surgery has been especially poorly studied in this respect. Studies of the microbiome in other disease areas, most notably diabetes, have shown significant effects of treatment that can be confounded by consequences of a disease, particularly when the treatment effects are unknown.11 Furthermore, durable effects on the microbiome from intestinal surgery may have potential implications for guiding future therapies.
Although 16S rRNA amplicon analysis has been a very useful tool for revealing microbiome differences and dynamics in IBD,3, 8 there are several important limitations in terms of taxonomic resolution and insight into function.12 To overcome these limitations in this study, we perform deep-coverage shotgun metagenomics, allowing species- and strain-level profiling and functional analysis of the microbiome and untargeted metabolomics with LC/MS/MS (liquid chromatography followed by tandem mass spectrometry), giving a direct chemical readout of the metabolome profile. This combination of techniques allows us to assess the value of these different data types as biomarkers for clinical states, including microbiome volatility and clinical status.
An important clinical consideration is the role and timing of surgery in the treatment of IBD. Although surgery is typically reserved as a last resort for medically refractory disease, a randomized trial compared medical therapy with surgery early in the treatment of ileal Crohn’s and showed similar outcomes, suggesting that ileocolonic resection could be considered as an alternative to medical therapy early in the course of Crohn’s disease.13 However, the potential long-term adverse effects of surgery, especially in terms of impact on the microbiome and/or metabolome, have not yet been fully elucidated.
To address these questions, we investigate the effect of surgical resection in a cohort of 129 subjects with IBD, stratified by disease subtype and type of surgery.
MATERIALS AND METHODS
Recruitment
Patients with a diagnosis of Crohn’s disease or ulcerative colitis who were seen at the University of California, San Diego (UCSD) at the Inflammatory Bowel Disease Center were prospectively recruited and consented into the UCSD IBD Biobank. Diagnosis was confirmed by an IBD specialist. The study participants provided written informed consent, and the study was approved by the institutional review board at the University of California, San Diego.
Specimen Collection
Participants collected samples at home in Covidien 2450SA stool specimen containers, then refrigerated samples for transport in a cooler. Samples were returned within 72 hours, aliquoted, and frozen at −80°C until DNA isolation.
UCSD Inflammatory Bowel Disease Biobank
Each patient’s clinical phenotype was assessed by an IBD specialist to define disease subtype (UC or CD), location, and phenotype based on Montreal disease classification for UC and CD (Table 1).14 Clinical and endoscopic data were collected prospectively, and disease phenotypes were confirmed by 2 IBD specialist physicians. Stool samples for each subject were collected approximately every 6 months.
TABLE 1.
Inflammatory Bowel Disease Patient Demographics (N = 129)
| Ulcerative Colitis (N = 50) | Crohn’s Disease (N = 79) | |
|---|---|---|
| Age (years) | ||
| Median (IQR) | 44 (30–59) | 36 (27–44) |
| Sex | ||
| Male, N (%) | 25 (50%) | 36 (46%) |
| Female, N (%) | 25 (50%) | 43 (54%) |
| Body Mass Index (kg/m2) | ||
| Median (IQR) | 23.4 (21.2–28.0) | 24.1 (21.3–27.9) |
| Disease duration (years) | ||
| Median (IQR) | 7 (2–12) | 9 (4–18) |
| UC Montreal Classification, N (%) | ||
| Proctitis (E1) | 9 (18%) | |
| Left sided colitis (E2) | 10 (20%) | |
| Extensive colitis (E3) | 27 (54%) | |
| J poucha | 4 (8%) | |
| Crohn’s Disease Location, N (%) | ||
| Ileal (L1) | 19 (24%) | |
| Colonic (L2) | 19 (24%) | |
| Ileocolonic (L3) | 36 (46%) | |
| Crohn’s disease of J poucha | 5 (6%) | |
| Crohn’s Disease Behavior, N (%) | ||
| Inflammatory (B1) | 8 (10%) | |
| Stricturing (B2) | 16 (20%) | |
| Fistulizing (B3) | 55 (70%) | |
| Surgical Resection, N (%) | ||
| None | 45 (90%) | 46 (58%) |
| Ileocolonic | 0 | 21 (27%) |
| Subtotal colectomy with ileorectal | ||
| anastomosis | 0 | 3 (4%) |
| Colectomy with end ileostomy | 0 | 4 (5%) |
| Colectomy with J pouch | 5 (10%) | 5 (6%) |
| Smoking, N (%) | ||
| Never | 36 (72%) | 55 (70%) |
| Prior | 13 (26%) | 18 (23%) |
| Current | 1 (2%) | 6 (7%) |
| TNF-inhibitor Exposure, N (%) | 30 (60%) | 65 (82%) |
| Biologic use at baseline, N (%) | ||
| TNF-inhibitor use | 15 (30%) | 42 (53%) |
| Integrin-inhibitor use | 5 (10%) | 8 (10%) |
| p40-inhibitor use | 0 (0%) | 4 (5%) |
aNot part of the Montreal Classification system but included for clarification
Shotgun Metagenomic Data Collection and Profiling
DNA was extracted with the Qiagen MagAttract PowerSoil DNA kit as previously described15 and constructed the shotgun metagenomics libraries using 100 ng of DNA from each sample. DNA was sheared to fragment sizes of 300 bp and input to the TruSeq Nano library prep kit. Amplified libraries were then pooled and sequenced using HiSeq 4000 platform.
For the sequenced reads, we trimmed the adaptors and performed quality filtering using atropos1.1.2116 (default parameters) and filtered out host reads using Bowtie2 2.3.0.17 After filtering out low-quality samples, we worked with 300 metagenomics samples. The average number of reads per sample after quality filtering is 32,103,916 reads. The taxonomic profiles were generated using MetaPhlan2 2.7.718 (default parameters), and the functional profiles were generated with HUMAnN2 0.11.219 (default parameters).
We investigated the profiles of dominant Escherichia coli (E. coli) strains using PanPhlan20 for 147 samples with E. coli abundance >1% using the “ecoli16” database downloaded from the PanPhlan website. We then constructed a panreactome matrix that describes the metabolic capability of the dominating E. coli strain in each sample following the method outlined in a previous study.21 We also performed multiple correspondence analysis on the panreactome matrix using python package mca22 with Benzecri correction, with the parameter of TOL set to 1e-9.
Untargeted Metabolomics Profiling + Data Processing
Sample extraction
Chemically cleaned stainless sterile beads were added to 100 mg to 50 mg of human fecal samples, along with 50% methanol (spiked with 2 uM of sulfamethazine) at a volume ratio of 1 mg per sample to 10 uL extraction solvent followed by a tissue homogenization using a Qiagen TissueLyzer II for 5 minutes at 25 Hz. Samples were centrifuged for 15 minutes at 14,000 rpm, and 400 uL of the resulting supernatant were transferred to a 96-well deep-well plate and dried via a centrifugal low-pressure system (SpeedVac Plus, Savant) and stored at −80°C until mass spectrometry analysis. Samples were resuspended with 130 uL of 50% methanol (spiked with 1 uM of sulfadimethoxine) and sonicated for 5 minutes. After centrifugation at 14,000 RPM for 15 minutes, 100 uL of supernatant were transferred to a new shallow-well 96-well plate. The 96-well plate was then diluted 20-fold.
Data acquisition
The fecal samples were analyzed using an ultra-high performance liquid chromatography (Ultimate 3000, Thermo) coupled to a quadrupole time-of-flight mass spectrometer (maXis Impact, Bruker). Chromatographic separation was accomplished using a Kinetex C18 1.7 uM, 100 Å, 2.1 mm by 50 mm column (Phenomenex) maintained at 40°C during separation. Five uL of extract was injected per sample. Mobile phase composition was A, LC-MS grade water with 0.1% formic acid (v/v), and B, LC-MS grade acetonitrile with 0.1% formic acid (v/v). The chromatographic elution gradient parameters were the following: 0.0 to 1.0 minutes, 5% B; 1.0 to 9.0 minutes, 100% B; and 9.0 to 10 minutes, 100% B. An MS1 scan from 50 to 1500 at 3 Hz was followed by MS2 scans. The heated electrospray ionization parameters were the following: drying gas, 9.0 L min-1; dry gas temperature 200°C; capillary voltage, 3.5 kV; end plate offset, −0.5 kV; and nebulizer, 2.0 bar. Hexakis (2,2-difluoroexthoxy) phosphazene, lock mass standard, was added to the ionization source.
Data processing
The acquired qTOF files (.d) were exported using DataAnalysis (Bruker) as.mzXML files after lock mass correction. Feature finding was performed on MS1 data in MZmine2,23 producing a data matrix of MS1 features (ie, m/z and retention time) and associated peak area. The MS2 data were analyzed using GNPS (estimated false discovery rate used is 0.005 at the settings).24
Microbiome-metabolite vectors (MMVEC) analysis to integrate multi-omics data
We performed MMVEC analysis25 to interrogate the relationship between metabolites and microbes using the MMVEC software (github.com/biocore/mmvec). The input of this workflow are 2 matrices—metabolite abundance and species abundance. We first performed MMVEC analysis on the entire data set to investigate for an association between the microbes and metabolites in the entire data sets (without stratifying for disease and surgery subtypes). We also ran MMVEC on subpopulations based on disease subtypes and surgery status. Specifically, we compared 4 subpopulations: UC without prior intestinal surgery, CD without prior intestinal surgery, CD with colectomy, and CD with ileocolonic resection.
Genomic structural variants analysis
We performed analysis to characterize the In accordance with a previous study,26 we defined structural variation as segments of varying lengths (potentially containing multiple genes) that are absent from certain bacteria in some people or present in a variable number of copies in others. We identified 2 categories of structural variants (SVs) following the workflow in the previously mentioned study using iterative coverage-based read assignment (ICRA) and structural gene variant (SGV)-finder with default settings. The first category is deletion SVs that are deleted in 25% to 75% of our samples, and the other category is variable SVs that have highly variable coverage across samples. We examined the association between the SVs and the surgery status using statistical tests (Mann-Whitney U test for deletion SVs and Spearman correlation for variable SVs as done in 2) and reported SVs statistically associated with surgery status after adjusting P values using Bonferroni correction. We used the SVexplorer (https://genie.weizmann.ac.il/SV/) to identify genes and annotations.
Statistical Analyses
The following description summarizes the main software packages used in this analysis. For reading and writing data, we used scikit-bio 0.5.5, the BIOM format 2.1.727 and QIIME 2 version 2019.1.28 Data visualization was done using Seaborn 0.9.0,29 Matplotlib 3.0.3,30 and QIIME2. The machine learning and linear algebra tasks were performed using scikit-learn 0.20.2,31 SciPy 1.2.1,32 Pandas 0.24.2,33 and NumPy 1.16.2.34 A detailed description of the individual steps has been published as a collection of Jupyter notebooks (https://github.com/knightlab-analyses/ibd-surgery).
This survey was represented by 3 contingency matrices: one for the taxonomic profile, one for the functional profiles, and one for the untargeted metabolomics. Alpha diversity calculations for the metabolomics and metagenomics matrices were performed using scikit-bio. For metabolomics we used the Shannon index. For the taxonomic profiles, we used the Faith phylogenetic diversity (PD)35 based on the NCBI taxonomy of the represented bacteria. In both cases, low-quality samples were removed from analyses. Similarly for beta diversity calculations, we used the Bray-Curtis distance for the metabolomics matrix and the unweighted UniFrac36 matrix for the taxonomic profiles as implemented in SciPy and scikit-bio. The differentially abundant features were estimated using analysis of composition of microbiomes (ANCOM)37 as implemented in scikit-bio and QIIME2.
For the evaluation of a model to classify samples according to the surgery status, we used a Random Forests classifier38 and a Precision-Recall curve for each data matrix. The performance of the classifier was evaluated using the average precision over 100 independent iterations. At each iteration, the subjects were exclusively split in a training and a test set. The Precision-Recall curve was selected to account for the class imbalance (70% of the subjects did not undergo surgery).
Data Availability
The metagenomic sequencing data have been deposited to the European Bioinformatics Institute (EBI accession is ERP121770), and the untargeted metabolomics have been deposited to the MASSIVE repository (MSV000082221). In addition, the full data set including sample metadata has been made public on Qiita39 (https://qiita.ucsd.edu/study/description/11546).
ETHICAL CONSIDERATIONS
The study was approved by the institutional review board at the University of California, San Diego, and the participants each underwent informed consent.
RESULTS
Overall demographics for the IBD patient population are shown in Table 1. Of 129 patients with IBD, 50 patients have ulcerative colitis, and 79 have Crohn’s disease. A total of 332 stool samples were collected (single sample from 18 patients; 2 samples from 36 patients; 3 samples from 40 patients; 4 samples from 23 patients; 5 samples from 6 patients). There is a median disease duration of 8 years, and 95 (73.6%) patients have current or prior TNF inhibitor exposure. In total, 91 (70.5%) patients have no history of intestinal surgery, 21 with Crohn’s disease underwent ileocolonic resection, and 17 including patients with UC and CD have had different types of colectomies. Of the patients who underwent colectomy, 10 with diagnosis of UC underwent subtotal colectomy with ileoanal pouch, and 5 out of 10 progressed to develop CD of the pouch. Three patients with CD had a subtotal colectomy with ileorectal anastomosis, and 4 with CD underwent total proctocolectomy with end ileostomy. These surgeries occurred a median of 3 years (interquartile range [IQR], 1–5.5 years) before the baseline stool sample collection.
Surgery Lowered Alpha Diversity in Both Microbiome and Metabolome
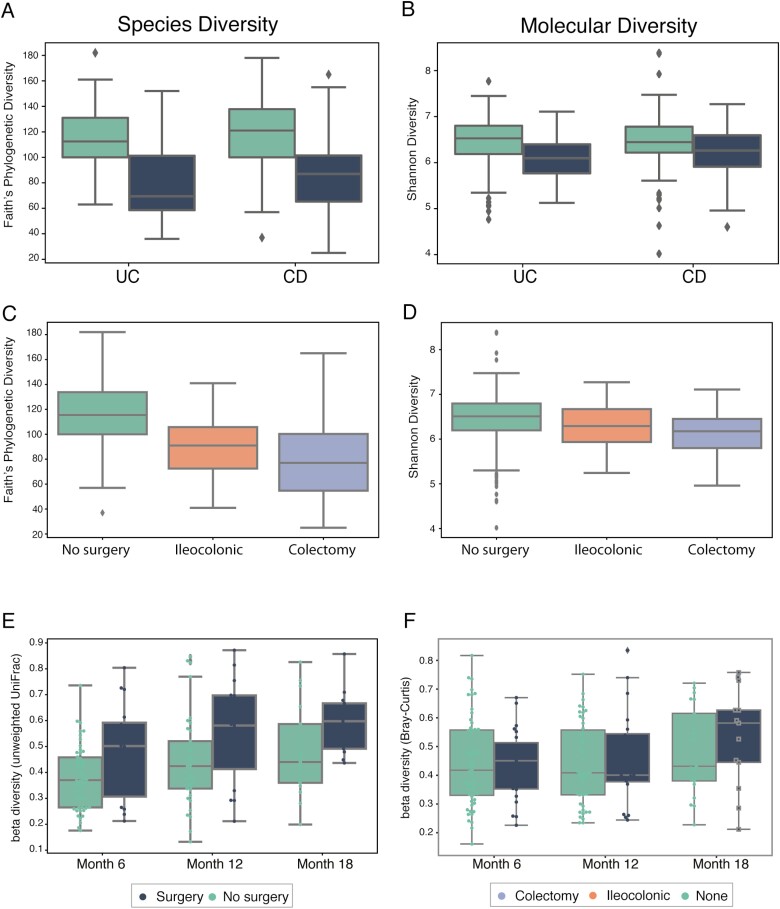
Prior intestinal surgery similarly decreases alpha diversity in both UC and CD patients (Fig. 1). In patients with UC, alpha diversity is not significantly different from those with a normal pouch, pouchitis as compared with CD of the pouch (Fig. S1). In patients with CD, different types of intestinal surgeries reduce alpha diversity with a trend toward the greatest reduction in samples after a total colectomy with end ileostomy (Kruskal-Wallis test, P = 4.51e-3), and this trend is consistent when repeating the analysis using only one sample per patient (Kruskal-Wallis test, P = 2.01e-3). Small sample sizes, however, limit the comparisons among UC and CD surgical subtypes.
FIGURE 1.
Comparison of alpha diversity and stability between surgery and non surgery groups. A, Phylogenetic diversity (metric: Faith) of species abundance for UC and CD samples. B, Molecular diversity (metric: Shannon) of molecular intensity evenness for UC and CD samples. C, Phylogenetic diversity (metric: Faith) of species abundance for samples with different types of surgery. D, Molecular diversity (metric: Shannon) of molecular intensity evenness for samples with different types of surgery. E, Phylogenetic volatility (metric: unweighted UniFrac) comparing each follow-up sample to the baseline time point. F, Molecular volatility (metric: Bray-Curtis) comparing each follow-up sample to the baseline time point.
Since UC and CD represent a disease spectrum, we combined their analysis and found that ileocolonic resection and colectomy significantly decrease phylogenetic diversity, and in particular, colectomy has a large impact on both microbial species diversity (Fig. 1C) and molecular diversity (Fig. 1D). Alpha diversity for species, as measured by Faith phylogenetic diversity, is lower in individuals with prior intestinal surgery, and samples from patients who have had a colectomy have the lowest alpha diversity (Kruskal test, P = 7.09e-16). Permutational multivariate analysis of variance (PERMANOVA) analysis of taxonomic profiles shows that specific surgery type (ileocolonic vs colectomy) explains 9.84% of the variation in the microbiome, followed by disease subtype (7.63%), then antibiotic use (4.69%), then disease activity (3.1%; Table S1). Other variables such as sex and age had much smaller effect sizes (Table S1). Notably, the number of years since surgery did not affect the overall reduction in alpha diversity (Spearman correlation, P > 0.05).
Both disease activity and antibiotic use are important potential confounding factors. To account for disease activity, we separated samples into those from patients with active endoscopic disease vs inactive endoscopic disease activity in those with an endoscopic assessment within 3 months of the stool specimen. We find that there were no significant differences in alpha diversity between patients with active vs inactive disease activity. This result suggests that the differences in alpha diversity cannot entirely be explained by disease activity. Furthermore, antibiotic use has been shown to reduce diversity in the short term and long term40 and may represent another potential confounding factor. Although there is significant variation among surgical protocols, at a minimum, one dose of multiple intravenous antibiotics is routinely given at the time of surgery. It is difficult, however, to unravel the precise effect of antibiotics during surgery, as it is an integral part of the procedure. To examine the effect of antibiotics, we investigated whether current or recent (defined as within 90 days) antibiotic use affected alpha diversity. By stratifying the samples based on both surgery status and current/recent antibiotic use, we found that regardless of surgery status, current/recent antibiotic use consistently decreased alpha diversity (Mann Whitney U test, P = 0.004 [surgery] and P = 0.04 [no surgery]), suggesting that antibiotic administration during surgery may contribute to the reduction in diversity observed in surgery samples (Fig. S2).
To assess volatility of the microbiome and metabolome and the possible effects of surgery on this variability, we performed longitudinal analyses to compare the differences in species and metabolite abundance for samples from patients with and without surgery. We find that the microbiomes of subjects who had prior surgery are much more variable in terms of their overall composition. The boxplots in Figures 1E and 1F show the differences between baseline, 6, 12, and 18 months for these groups, demonstrating that surgery increases microbiome—but interestingly not metabolome—volatility.
Surgery Affected Overall Taxonomic, Functional, and Metabolite Profiles
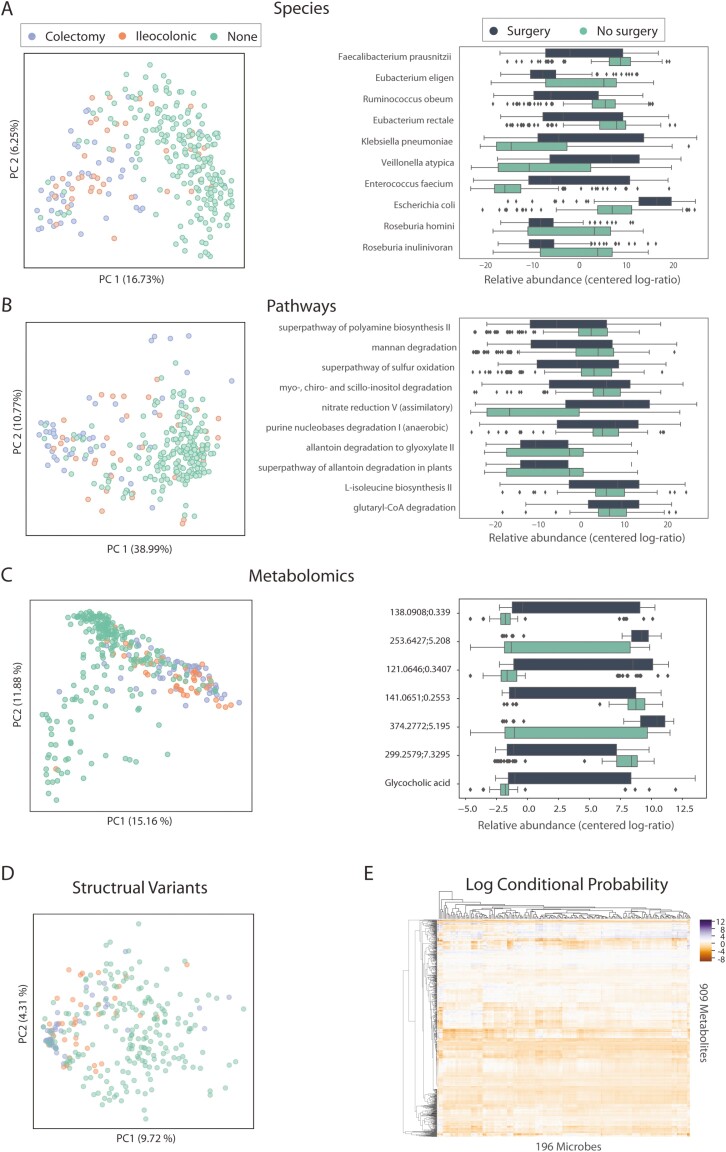
The beta diversity plots (Fig. 2A–C, left panels) show dissimilarity among samples, reduced to 2 dimensions for visualization purposes. These plots show that prior surgery affects overall taxonomic (PERMANOVA, P = 1.0e-3) and functional profiles and metabolite abundances. Although samples from IBD patients who had prior surgery are not identically distributed to samples from patients who did not undergo surgery in these Principal Coordinates Analysis (PCoA) plots, the distributions overlap, so we cannot differentiate between different types of surgery in all PCoA plots.
FIGURE 2.
Comparison of surgery types vs nonsurgery for taxonomic, functional, and metabolomics profiles. A, PCoA plot of species abundance (metric: unweighted unifrac) labeled by surgery subtypes, and top 10 differentiating species between surgery and nonsurgery samples. B, PCoA plot of pathway abundance (metric: Bray-Curtis) labeled by surgery subtypes, and top 10 differentiating pathways between surgery and nonsurgery groups. C, PCoA plot of metabolomics abundance (metric: Bray-Curtis) labeled by surgery subtypes, and top 7 differentiating metabolomics between surgery and nonsurgery groups. D, The first 2 components in the PCA analysis of deletion structural variants labeled by surgery subtype. E, Heatmap of log conditional probability matrix generated from MMVEC analysis for 196 microbes and 909 metabolites. The conditional probability is a log scale: purple represents high probability of co-occurrence of metabolites and microbes, whereas orange represents low probability.
To identify the specific taxa that contribute to these overall microbiome differences, we used the compositionally aware method ANCOM37 to identify the top 10 species that differentiate between surgery and nonsurgery samples. Of all species identified as differentially abundant, potential pathogens such as Klebsiella pneumoniae, Enterococcus faecium, and E. coli, in addition to Veillonella atypica, a known oral bacterium, have higher relative abundance in the surgery group compared with the nonsurgery group. In contrast, butyrate producers such as Eubacterium rectale, Faecalibacterium prausnitzii, Eubacterium Eligens, and Roseburia inulinivorans and gut symbionts including Roseburia hominis and Ruminococcus obeum have lower relative abundance in surgery samples than nonsurgery samples. Repeating these analyses at the pathway level, nitrate reduction is significantly elevated in surgery samples, consistent with previous studies that have shown that nitrate respiration occurs in the inflamed gut and promotes the growth of pathogens such as E. coli.41
We also performed an in-depth analysis of the metagenomics data to investigate the genomic SVs of the gut microbiome and their association with surgery status following methodology described by Zeevi et al.26 Based on this study, we defined structural variation as segments of varying lengths, potentially containing multiple genes that are deleted from certain bacteria in some individuals or present in a variable number of copies in others. We investigated 2 categories of SVs, deletion SVs (deleted and not covered in 25%–75% of samples), and variable SVs (with highly variable coverage across samples). We used the 2 tools developed by Zeevi et al 40: ICRA, which assigns reads to the representative microbial genomes, and SGV-finder, which characterizes SVs based on coverage depth. We identified a total of 7397 deletion SVs and 4015 variable SVs from 114 species. We first performed principal component analysis (PCA) on the deletion SVs and labeled samples stratified by surgery status. Surgery samples cluster together more than the nonsurgery samples (Fig. 2D); however, there is no distinct separation between these 2 groups of samples. Principal component analysis of the variable SVs provided little additional information and showed no strong associations (data not shown). To test for associations between surgery status and SVs, we performed Mann-Whitney U test (on deletion SVs) and Spearman correlation (variable SVs) following the methods from Zeevi et al.26 After correction for multiple testing, we identified 7 statistically significant deletion SVs and 26 variable SVs (see Supplementary Tables S3 and S4). The 7 identified deletion SVs are from various organisms, and 4 out of 7 deletion SVs are identified in the differentially abundant organisms shown in Figure 2A including E. coli, Faecalibacterium prausnitzii, and Eubacterium rectale, suggesting that these organisms differed not only in their abundances but also in their genomic content between postsurgery and nonsurgery samples. Many of these deletions SVs are involved in metabolism, likely induced by the change of environment and nutrient availability after surgery. For variable SVs, 14 out of 26 SVs were identified in Ruminococcus torques, and 9 SVs were identified in Blautia wexlerae. All variable SVs identified have higher coverage in groups without prior surgery compared with postsurgery samples. Interestingly, SVs in Ruminococcus torques are mostly involved in transporter systems that contribute to drug and antimicrobial transportation, in addition to site-specific recombinases, which suggest these variable SVs could be involved in plasmids carrying antimicrobial resistant genes.
We used an unbiased approach to examine differences in the fecal metabolome. Combining all samples with prior surgery shows a significant decrease of Shannon diversity in UC (Mann-Whitney U test, P = 0.027) but not in CD (Mann-Whitney U test, P = 0.100; Fig. S5A, B). Specifically, the metabolomics from the CD samples with prior surgery cluster together with the lowest relative evenness in those who underwent total colectomy with ileostomy. Several metabolites are differentially abundant in individuals with prior surgery; most of these were bile acids. Only cholic acid and amino-2-ethoxybenzene are more abundant in the setting of prior surgery. Both tyrosine and glutamic acid are less abundant in subjects with surgery.
In order to integrate metabolomics and metagenomics data to elucidate interaction between microbes and metabolites, we performed a co-occurrence analysis using a neural-network approach MMVEC25 on all samples and subgroups of the samples. However, the analysis did not reveal strong associations between particular microbes and metabolites. First, we performed this MMVEC analysis on the entire data set to investigate if there is any overall association between the microbes and metabolites in the entire data sets (without stratifying for disease and surgery subtypes). We identified conditional probability between 909 metabolites and 196 microbes from all metagenomics samples, yet most metabolites are associated with many microbes or no microbes at all (Fig. 2E), which may be a consequence of significant sample heterogeneity. Therefore, we then ran MMVEC on subpopulations based on disease subtypes and surgery status. Specifically, we compared 4 subpopulations: UC without prior intestinal surgery, CD without prior intestinal surgery, CD with colectomy, and CD with ileocolonic resection. However, this analysis was limited by the smaller sample size within each of the subgroups. Specifically, fewer individual microbes were identified in the colectomy and ileocolonic groups, making it difficult to compare across the different groups. Based on our interest in bile acids, we also examined bile acids and found that many microbes contribute to bile acids in every subgroup without a dominant contributor. Using MMVEC, we interrogated the relationships between microbes and metabolites but were not able to identify strong associations that were driven by disease subtype or prior surgery. This methodology and analysis may have been limited by the small sample size in each subgroup and the overall heterogeneity of the entire population, including multiple disease subtypes, different current and prior treatments, and disease activity. Though the MMVEC analysis did not provide significant additional insights, our data set will serve as an excellent resource for future analyses with methodologies that may evolve and improve in the future.
Higher Abundance of Primary Bile Acids Detected in Surgery Samples
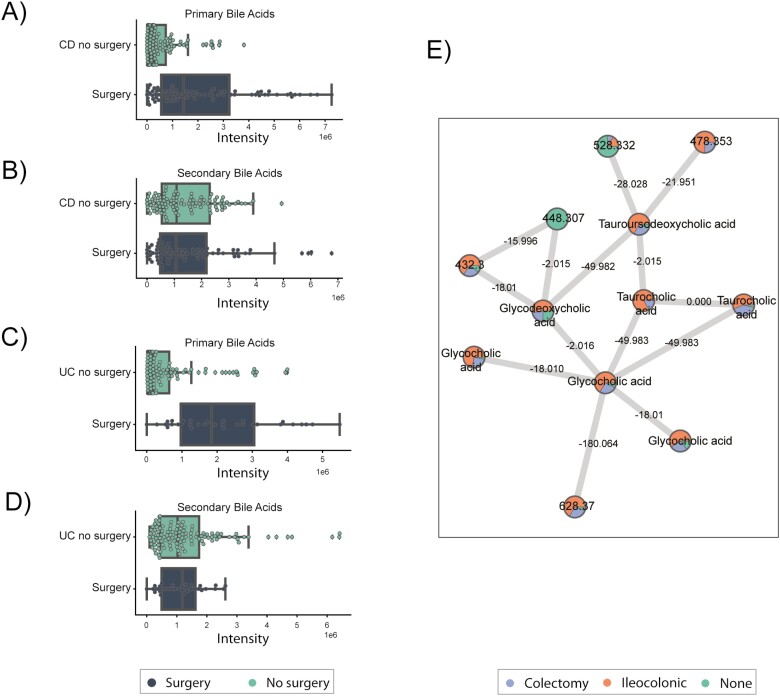
Overall, primary bile acids are increased for subjects that underwent surgery (in both UC and CD), whereas secondary bile acids are not significantly different (Fig. 3A–D). Primary biliary acids (BAs), including cholic acid and chenodeoxycholic acid, are produced by the liver, and then gut microbiota are responsible for deconjugation of BA to generate secondary BA. Fecal primary BAs have been shown to be enriched in IBD, with relative depletion of secondary BA in Crohn’s disease.5, 42, 43 Resection of the terminal ileum, as is the case of ileocolonic resections and most colectomies, may reduce reabsorption of primary BA and increase the concentration of BA in the colon. We detected 21 distinct BAs, including 14 primary BAs, predominantly cholic acid and chenodeoxycolic acid, and 7 secondary BAs, but few conjugated BAs. Notably in CD, primary BAs are increased in patients with prior ileocolonic resection (Fig. 3A, Fig. S6). There is a nonsignificant trend toward lower secondary bile acids in surgery samples without any specific signal based on surgery subtype. In UC, there is a similar trend toward increased primary bile acids in those with colectomy and J pouch; however, there are no significant changes in secondary bile acids stratified by prior surgery, though these analyses are limited by small sample sizes in subgroups (Fig. S7).
FIGURE 3.
Metabolomic analysis of the primary and secondary bile acids identified in the cohort. A, Primary bile acids for subjects with Crohn’s disease. B, Secondary bile acids for subjects with Crohn’s disease. C, Primary bile acids for subjects with Ulcerative Colitis. D, Secondary bile acids for subjects with ulcerative colitis. E, Molecular network of the metabolomics data, each node represents a metabolite and the edges represent the cosine similarity between the metabolite pairs.
Elevated E. coli Relative Abundance Observed in Surgery Samples
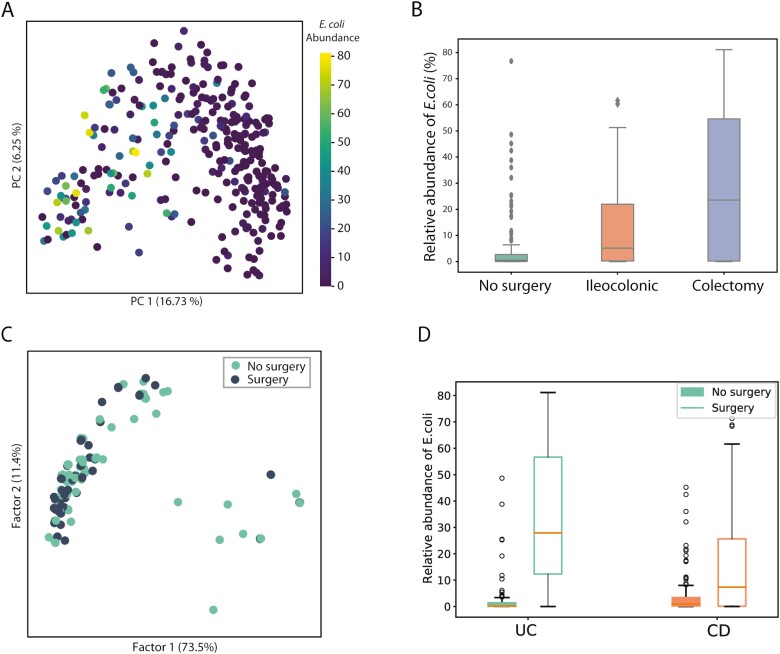
Given the observed importance of E. coli in IBD44 and its dominance of the overall community patterns, we performed a pangenome analysis of this species specifically. By replotting the PCoA plot and labeling by E. coli abundance, we examined the association between prior intestinal surgery and E. coli abundance. Examining UC and CD, we clearly observed that nonsurgery samples have markedly lower E. coli abundance as compared with surgery samples (Fig. 4A). Specifically, samples from all IBD patients who underwent colectomy have the highest abundance of E. coli, and samples from patients without prior surgery have the lowest (Fig. 4B). In CD, relative E. coli abundance is higher in patients with prior surgery, with the highest abundance occurring in patients with a colectomy with an end ileostomy (Fig. S4, Kruskal-Wallis test, P = 2.39 e-2). In UC, similar nonsignificant trends exist when samples were analyzed by UC postsurgery subtype (Fig. S4). The E. coli level also has a negative association with alpha diversity (Spearman correlation −0.26; P = 6.47e-6). Multiple factors may contribute to the expansion of E. coli after surgery, including loss of resistance to potential pathogens of the gut microbiome due to changes in the habitat and antibiotic use,45 increased epithelia oxygenation that allows expansion of E. coli through aerobic respiration,46 and the capability of E. Coli to obtain nutrients from the mucus that provides an advantage in colonization and persistence in the gut.47
FIGURE 4.
Comparison of E. coli abundance and characteristics of dominant E. coli strains between surgery and nonsurgery samples. A, PCoA plot of taxonomic profile labeled by E. coli abundance. B, Relative abundance of E. coli in no surgery, ileocolonic, and colectomy samples. C, MCA plot that describes the similarity of dominant E. coli strains from surgery and nonsurgery samples in terms of metabolic and virulent functions. D, Relative abundance of E. coli stratified by UC and CD.
We then ran PanPhlan on samples with >1% E. coli abundance and obtained genomic contents of dominant E. coli strains in 147 samples. We then constructed metabolic networks and identified the presence/absence of adherent-invasive E. coli–associated virulence genes. Multiple correspondence analysis (MCA) on the matrix describing metabolic reaction and virulence factor content in the 147 strains (at 1 strain per sample, Fig. 4C) suggests that E. coli strains from surgery as compared with no prior surgery samples have similar metabolic and virulence functions.
Taxonomic Profiles Differentiate Surgery Status Better Than Metabolic or Functional Profiles
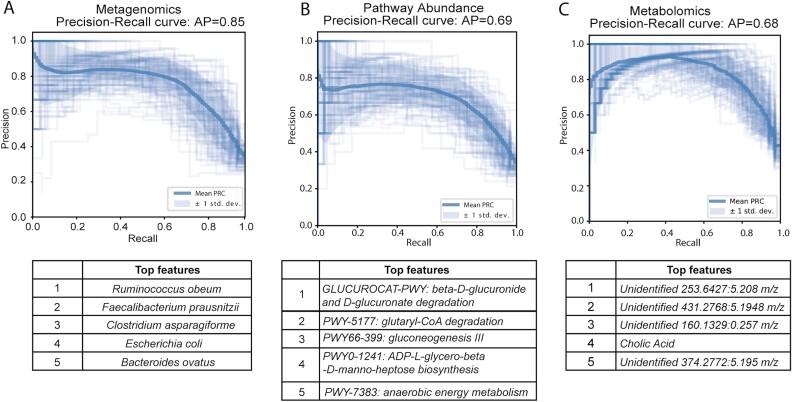
Finally, we tested the ability of these different data layers to discriminate between patients who had or had not previously undergone surgery. Specifically, we trained Random Forest classifiers to classify each sample according to whether it came from a patient who had undergone surgery, using the relative abundance tables for bacterial species, pathways, and metabolites described previously. We split the samples by subject to ensure that subjects in the training samples do not overlap with the test data set. We randomly split the data (70% train, 30% split) and tested and trained the model 100 times, yielding an average precision of 0.80 (Fig. 5A). Species adding the greatest weight to the classifier included Ruminococcus obeum, Clostridium asparagiforme, Faecalibacterium prausnitzii, Escherichia coli, and Bacteroides ovatus. Repeating this analysis at the pathway and the metabolite level, we found that pathway abundance and metabolomics data yielded substantially worse classifiers (average precision of 0.69 and 0.68, Fig. 5B, C). The top 5 contributing pathway features involve carbon and energy metabolism, whereas the top five contributing metabolite features are all unidentified, except cholic acid. It is worth noting that the microbiome, host factors, diet, medication use, and additional factors influence the metabolome, so sample heterogeneity may have a greater confounding effect on the metabolome.
FIGURE 5.
Random forest classifier to differentiate surgery from nonsurgery samples using species, pathway, and metabolite abundance. A, Precision-recall curve of species classifier and the top 5 features. B, Precision-recall curve of pathway classifier and the top 5 features. C, Precision-recall curve of metabolite classifier and the top 5 features.
We did not identify differences based on response to TNF-inhibitors with and without stratifying based on prior surgery (Mann-Whitney U test, P > 0.05). We did not have sufficient statistical power to stratify by response to vedolizumab or by subsequent need for surgery after the time points sampled in this study; however, these topics are of intense clinical interest and would be valuable to explore in an adequately powered prospective longitudinal study.
DISCUSSION
Our study has shown that surgery has a persistent effect in decreasing the alpha diversity (ie, the diversity within each sample) both of the gut microbiome and gut metabolome. Additionally, we show that the instability of the microbiome appears to increase following surgery, but the instability of the metabolome seems to be unchanged. Not all surgeries show the same magnitude of an effect in our study. Colectomy has a larger effect compared with ileocolonic resection. Intuitively, this finding is not entirely unanticipated given the amount of intestine typically removed in each of these surgeries. This observation is interesting though because in previous work, it has been shown that ileal CD with resection led to unstable microbiome dynamics,8 but it seems that colectomy may have an even larger effect. In our study, the type of surgery explained more variation in the microbiome data than by any other variable (9.84%), followed by disease subtype (7.63%), then antibiotic use (4.69%), and then disease activity (3.1%). Although diet also has a large known effect, particularly in studies in IBD, it was unfortunately not assessed in this cohort to date. Integrating dietary assessment into future studies will also be of considerable importance. The large effect size of surgery on the microbiome and metabolome indicates that surgery is a significant variable that must be measured and controlled for in studies of the microbiome and IBD, particularly in studies that seek to assess smaller effect-size variables, such as sex-specific or age-specific factors.
This study also reinforces the potential value and challenges in collecting multi-omics data, as the metagenomic and metabolomic data provide different views into IBD, providing a concordant view of alpha and beta diversity changes and a discordant view of instability with surgery. With ongoing advances in our ability to integrate large data sets, additional data layers such as the metatranscriptome and the metaproteome, as were collected in iHMP,48 may be useful for further untangling these relationships, as will more extensive host immune phenotyping and other host profiling. Intriguingly, the metagenomic data and the pathway information generated from it were better able to detect changes associated with surgery than the metabolome data, suggesting that integration of capabilities over a longer period rather than immediate readout of current state may be most important for explaining phenomena associated with surgery in IBD. These findings, however, may also suggest that surgery predominantly alters the composition of the microbiome, whereas the metabolome, which is influenced by other host factors, such as the immune system and diet, remains more stable.
Integrating metagenomics and metabolomic data remains a significant analytic challenge, and subsequent advances in the computational approaches may facilitate additional revealing analyses within this data set. A significant obstacle in data-driven clinical investigations is coping with the often limited numbers of samples, as the methods frequently rely on large sample sizes to produce statistically robust results. Our analyses using MMVEC were likely affected by this issue. Despite all these challenges, the differences between surgery- and nonsurgery-associated samples in this study can potentially be useful as a way to establish the biological correctness of future computational methods.
As an observational study with a cohort of IBD patients, there were significant confounding factors that were difficult to control for in our analyses and warrant further discussion. First, any intestinal surgery, whether it is ileocolonic resection or colectomy, is a significant invasive procedure with numerous potential downstream consequences. By evaluating surgery as a single exposure or variable, we are simplifying and likely combining the effects of multiple interventions that could be affecting the microbiome. Second, surgery is typically reserved for individuals who have severe disease and have exhausted medical therapies. Individuals undergoing surgical resection may have more biologically aggressive disease, which is challenging to measure. We tried to control for disease activity using endoscopic disease activity as a surrogate; however, this is an imperfect measure for disease severity and biological aggressiveness. In addition, matched objective disease activity assessment is not available for each stool sample because a significant proportion of the stool samples were collected longitudinally between endoscopic assessments. Alternative biomarkers, such as fecal calprotectin, would improve our ability to correct for disease activity, but these were not consistently obtained and measured. There remains the possibility that patients with more severe disease and corresponding microbiome changes are more likely to have received surgery and have more pronounced reductions in alpha diversity. Third, antibiotics are known to have a significant and potentially lasting effect on the microbiome,40, 49 and they are routinely given during colorectal surgery with extended courses of antibiotics administered in some cases. This is likely a confounding factor contributing to and potentially exaggerating the reduction in microbial diversity, in addition to effects from surgery; however, this effect is difficult to separate from surgery, though we examined the effect of current and recent antibiotic use. In addition, we did not have samples both pre- and postsurgery from most individuals, limiting some of our ability to draw definitive conclusions. This would ideally be addressed in future longitudinal studies with presurgery and rigorous postsurgery stool collection from the same individual to help define the timing and persistence of fecal microbial changes after surgery. The more important question, however, is whether these microbial changes have meaningful functional effects and may inform future therapeutic interventions.
Although there were a large number of patients included in our study, we included a heterogenous group of patients with IBD who were on a variety of medical therapies with multiple potential confounding variables. As a result, the sample sizes for specific disease subtypes or specific surgeries were relatively small. Our relatively small sample size may have limited the ability to detect significant differences in the microbiome and metabolome due to lack of statistical power. Ideally, this point would to be addressed in future work with prospective longitudinal study design within targeted populations. Such studies can be guided by our recent work on determining the appropriate sampling interval and number of samples required to characterize IBD dynamics.9
The results of this study expand upon what is known about the microbiome and its central role in the pathogenesis of disease recurrence in CD patients after ileocolonic resection50 and in UC patients after colectomy with pouchitis, showing reduction in the microbial diversity that persists for years. The durable effect on reduction of diversity with surgery, especially in the longer term, has not previously been well characterized in many studies where it represents a significant potential confounder. Moreover, current research strategies are working to harness the microbiome into both diagnostic and treatment strategies, and postsurgical patients represent a potential target population that may particularly benefit from approaches such as fecal microbial transplantation or other targeted means of modifying the microbiome. One such recent study demonstrated the short-term alterations in the microbiome of patients with ileal Crohn’s disease who underwent ileocolonic resection, identifying bacterial species that may aid with diagnosis and prediction of recurrence.10 Further mechanistic and longitudinal studies and more detailed targeted biomarker discovery efforts are required to understand the functional and clinical effects of the reduction in microbiome and metabolome diversity or the increase in microbiome instability and to develop inexpensive assays that allow clinicians to predict or explain relapse.
CONCLUSION
In this study, the collection and analysis of metagenomics and metabolomics data of an IBD cohort suggest that intestinal surgeries may have long-term effect on the gut microbiome, including reduced diversity of the microbes and metabolites, and further increased the instability in the gut microbiome of IBD patients. These long-term consequences of intestinal surgery may need to be taken into consideration carefully in future IBD microbiome studies.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Tara Schwartz and Andre Matti for assistance with sample processing and Daniel Freed for his contributions to the project.
Author Contribution: BSB, JN, AM, PD, SS, MT, DB, and WJS contritubed to patient recruitment and sample collection. XF, YVB, GA, GH, RL, JGS, and CC contributed to metagenomics data generation/analysis. YVB, EE, AKJ, KCW, FV, and MP contributed to metabolomics data generation/analysis. BSB, WJS, RK, PCD, ADS, LS, BOP, and JTC contributed to design and mentor study. XF, YVB, and RKB drafted the manuscript. All authors read, revised, and approved the final manuscript.
Supported by: This work was supported by Crohn’s and Colitis Foundation Career Development Award and UCSD 1KL2TR001444, NIDDK 1K23DK123406 and 1K23DK123406 (BSB), Microbial Science Initiative Graduate Research Fellowship and Seed Grant by UC San Diego Center for Microbiome Innovation, Novo Nordisk Foundation Center for Biosustainability and the Technical University of Denmark (NNF10CC1016517), Janssen Human Microbiome Initiative, NIDDK-funded San Diego Digestive Diseases Research Center (P30 DK120515), NIDDK T32DK007202 (MST), and Clinical and Translational Science Award grant (UL1- TR-001442).
Conflicts of Interest: WS reports the following involvement in an organization/entity with a financial or nonfinancial interest in the subject matter discussed in this manuscript; research grants from Atlantic Healthcare Limited, Amgen, Genentech, Gilead Sciences, Abbvie, Janssen, Takeda, Lilly, Celgene/Receptos,Pfizer, Prometheus Laboratories (now Prometheus Biosciences); consulting fees from Abbvie, Allergan, Amgen, Arena Pharmaceuticals, Avexegen Therapeutics, BeiGene, Boehringer Ingelheim, Celgene, Celltrion, Conatus, Cosmo, Escalier Biosciences, Ferring, Forbion, Genentech, Gilead Sciences, Gossamer Bio, Incyte, Janssen, Kyowa Kirin Pharmaceutical Research, Landos Biopharma, Lilly, Oppilan Pharma, Otsuka, Pfizer, Progenity, Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories), Reistone, Ritter Pharmaceuticals, Robarts Clinical Trials (owned by Health Academic Research Trust, HART), Series Therapeutics, Shire, Sienna Biopharmaceuticals, Sigmoid Biotechnologies, Sterna Biologicals, Sublimity Therapeutics, Takeda, Theravance Biopharma, Tigenix, Tillotts Pharma, UCB Pharma, Ventyx Biosciences, Vimalan Biosciences, Vivelix Pharmaceuticals; and stock or stock options from BeiGene, Escalier Biosciences, Gossamer Bio, Oppilan Pharma, Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories), Progenity, Ritter Pharmaceuticals, Ventyx Biosciences, Vimalan Biosciences. Spouse: consultant for and stock options in Opthotech, consultant for and stock options in Progenity, employee of and stock options in Oppilan Pharma, employee of and stock options in Escalier Biosciences, employee of and stock options in Prometheus Biosciences (merger of Precision IBD and Prometheus Laboratories), employee of and stock options in Ventyx Biosciences, employee of and stock options in Vimalan Biosciences. BB reports research grant from Prometheus Biosciences and consulting for Pfizer.The other authors have no competing interests.
REFERENCES
- 1. Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morgan XC, Tickle TL, Sokol H, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. ; IBDMDB Investigators . Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut. 2017;66:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One. 2009;4:e6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vázquez-Baeza Y, Gonzalez A, Xu ZZ, et al. Guiding longitudinal sampling in IBD cohorts. Gut. 2018;67:1743–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mondot S, Lepage P, Seksik P, et al. ; GETAID . Structural robustness of the gut mucosal microbiota is associated with Crohn’s disease remission after surgery. Gut. 2016;65:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baxter NT, Lesniak NA, Sinani H, et al. The glucoamylase inhibitor acarbose has a diet-dependent and reversible effect on the murine gut microbiome. mSphere. 2019;4. doi: 10.1128/mSphere.00528-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Quince C, Walker AW, Simpson JT, et al. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–844. [DOI] [PubMed] [Google Scholar]
- 13. Ponsioen CY, de Groof EJ, Eshuis EJ, et al. ; LIR!C study group . Laparoscopic ileocaecal resection versus infliximab for terminal ileitis in Crohn’s disease: a randomised controlled, open-label, multicentre trial. Lancet Gastroenterol Hepatol. 2017;2:785–792. [DOI] [PubMed] [Google Scholar]
- 14. Satsangi J, Silverberg MS, Vermeire S, et al. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marotz C, Amir A, Humphrey G, et al. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques. 2017;62:290–293. [DOI] [PubMed] [Google Scholar]
- 16. Didion JP, Martin M, Collins FS. Atropos: specific, sensitive, and speedy trimming of sequencing reads. Peerj. 2017;5:e3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. [DOI] [PubMed] [Google Scholar]
- 19. Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scholz M, Ward DV, Pasolli E, et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13:435–438. [DOI] [PubMed] [Google Scholar]
- 21. Fang X, Monk JM, Nurk S, et al. Metagenomics-based, strain-level analysis of escherichia coli from a time-series of microbiome samples from a Crohn’s disease patient. Front Microbiol. 2018;9:2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anon. mca. PyPI. Accessed October 8, 2019.https://pypi.org/project/mca/.
- 23. Pluskal T, Castillo S, Villar-Briones A, et al. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang M, Carver JJ, Phelan VV, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morton JT, Aksenov AA, Nothias LF, et al. Learning representations of microbe-metabolite interactions. Nat Methods. 2019;16:1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeevi D, Korem T, Godneva A, et al. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–48. [DOI] [PubMed] [Google Scholar]
- 27. McDonald D, Clemente JC, Kuczynski J, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 2012;1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Waskom M, Botvinnik O, O’Kane D, et al. mwaskom/seaborn: v0.9.0 2018. (July 2018).https://zenodo.org/record/1313201.
- 30. Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9:90–95. [Google Scholar]
- 31. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 32. Jones E, Oliphant T, Peterson P, et al. SciPy: Open source Scientific Tools for Python. 2001. [Google Scholar]
- 33. McKinney W, et al. Data structures for statistical computing in python. In: Proceedings of the 9th Python in Science Conference.Vol 445. Austin, TX, 2010:51–56. [Google Scholar]
- 34. Oliphant TE A guide to NumPy. USA: Trelgol Publishing; ; 2006. [Google Scholar]
- 35. Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 36. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mandal S, Van Treuren W, White RA, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 39. Gonzalez A, Navas-Molina JA, Kosciolek T, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15:796–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hughes ER, Winter MG, Duerkop BA, et al. Microbial respiration and formate oxidation as metabolic signatures of inflammation-associated dysbiosis. Cell Host Microbe. 2017;21:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hakala K, Vuoristo M, Luukkonen P, et al. Impaired absorption of cholesterol and bile acids in patients with an ileoanal anastomosis. Gut. 1997;41:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miettinen TA. The role of bile salts in diarrhoea of patients with ulcerative colitis. Gut. 1971;12:632–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Palmela C, Chevarin C, Xu Z, et al. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut. 2018;67:574–587. [DOI] [PubMed] [Google Scholar]
- 45. Kim S, Covington A, Pamer EG. The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol Rev. 2017;279:90–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cevallos SA, Lee J-Y, Tiffany CR, et al. Increased epithelial oxygenation links colitis to an expansion of tumorigenic bacteria. MBio. 2019;10. doi: 10.1128/mBio.02244-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Conway T, Cohen PS. Commensal and Pathogenic Escherichia coli Metabolism in the Gut. Microbiol Spectr. 2015;3. doi: 10.1128/microbiolspec.MBP-0006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Integrative HMP (iHMP) Research Network Consortium. The integrative human microbiome project. Nature. 2019;569:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108:4554–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rutgeerts P, Goboes K, Peeters M, et al. Effect of faecal stream diversion on recurrence of Crohn’s disease in the neoterminal ileum. Lancet. 1991;338:771–774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metagenomic sequencing data have been deposited to the European Bioinformatics Institute (EBI accession is ERP121770), and the untargeted metabolomics have been deposited to the MASSIVE repository (MSV000082221). In addition, the full data set including sample metadata has been made public on Qiita39 (https://qiita.ucsd.edu/study/description/11546).