Abstract
Ribosomal protein genes are among the most highly expressed genes in most cell types. Their products are generally essential for ribosome synthesis, which is the cornerstone for cell growth and proliferation. Many cellular resources are dedicated to producing ribosomal proteins and thus this process needs to be regulated in ways that carefully balance the supply of nascent ribosomal proteins with the demand for new ribosomes. Ribosomal protein genes have classically been viewed as a uniform interconnected regulon regulated in eukaryotic cells by target of rapamycin and protein kinase A pathway in response to changes in growth conditions and/or cellular status. However, recent literature depicts a more complex picture in which the amount of ribosomal proteins produced varies between genes in response to two overlapping regulatory circuits. The first includes the classical general ribosome‐producing program and the second is a gene‐specific feature responsible for fine‐tuning the amount of ribosomal proteins produced from each individual ribosomal gene. Unlike the general pathway that is mainly controlled at the level of transcription and translation, this specific regulation of ribosomal protein genes is largely achieved through changes in pre‐mRNA splicing efficiency and mRNA stability. By combining general and specific regulation, the cell can coordinate ribosome production, while allowing functional specialization and diversity. Here we review the many ways ribosomal protein genes are regulated, with special focus on the emerging role of posttranscriptional regulatory events in fine‐tuning the expression of ribosomal protein genes and its role in controlling the potential variation in ribosome functions.
This article is categorized under:
Translation > Ribosome Biogenesis
Translation > Ribosome Structure/Function
Translation > Translation Regulation
Keywords: gene duplication, gene regulation, posttranscriptional regulation, ribosomal proteins, ribosome, TOR pathway
Global circuit of ribosome synthesis.
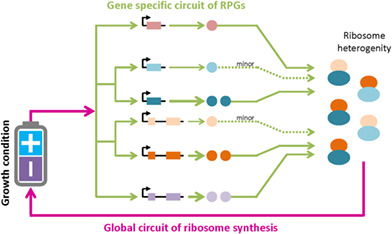
1. INTRODUCTION
Ribosomes are essential universally conserved ribonucleoprotein complexes composed of ribosomal RNA (rRNA) and ribosomal proteins (RPs) (Fox, 2010; Wilson & Doudna Cate, 2012; Yonath & Franceschi, 1998). These multimolecular nanomachines translate the genetic code in messenger RNA (mRNA) and catalyze protein synthesis in all cell types. Ribosomes function like machines forming a supply‐line of steady‐state levels of cellular proteins (Lafontaine & Tollervey, 2001; Wilson & Doudna Cate, 2012). The functional centers of the ribosome are highly conserved, but the overall molecular weight and number of components varies, especially between bacterial and eukaryotic cells. For example, while bacterial ribosomes boast a molecular weight of ~2.7 M Daltons (MDa), fungal and mammalian ribosome weigh in at ~3.5 and ~4.5 MDa, respectively (Ben‐Shem et al., 2011; Khatter, Myasnikov, Natchiar, & Klaholz, 2015; Nakao, Yoshihama, & Kenmochi, 2004; Pruitt, Tatusova, Brown, & Maglott, 2012; Schuwirth et al., 2005). Eukaryotic ribosomes are more complex in terms of both their rRNA and protein constituents. In bacteria the 30S small subunit contains a 16S rRNA measuring 1.5 kb, while the eukaryote equivalent (40S) RNA of over 1.9 kb (Chaker‐Margot, 2018; Garrett, Ungewickell, Newberry, Hunter, & Wagner, 1977; Melnikov et al., 2012; Moore, 1988; Rajendhran & Gunasekaran, 2011; Sogin & Gunderson, 1987). Likewise, the bacterial large 50S ribosomal subunit includes 23S rRNA, of approximately 2.9 kb, and the 120‐nucleotide 5S rRNA, while eukaryotic large subunit includes three different RNAs: the 25‐28S rRNA, at around 3.55–5.00 kb, the 5.8S rRNA of approximately 160 nucleotides and a 120‐nucleotide 5S rRNA (Biedka, Wu, LaPeruta, Gao, & Woolford Jr., 2017; Champney, 2001; Lee, Henry, & Yeh, 1983; Melnikov et al., 2012; Moore & Steitz, 2003). The number of RPs also increases substantially in eukaryotic ribosome with 12 or more new proteins added to each of the two subunits. Despite these variations in ribosomes in the kingdoms of life, their overall shape and core components, including 34 RPs, are universally conserved (Jansen et al., 1991; Yonath & Franceschi, 1998). This is consistent with the basic mechanism of translation being conserved in all living organisms, while the variations optimize ribosome function to the needs of different species. For example, the increased complexity of eukaryotic ribosomes may result from the increased complexity of the transcriptome and the higher demand for temporal and spatial regulation of translation. In higher eukaryotes, ribosomes must localize where proteins required and respond to the special needs of different tissues, including cues from both quiescent and proliferating cells (Genuth & Barna, 2018a; Komili, Farny, Roth, & Silver, 2007; Xue & Barna, 2012).
Regardless of the evolutionary rank of the organism or its ribosome complexity, all cells need an efficient, well‐coordinated and accurate ribosome production pipeline. To meet these demands, cells in general aim to co‐express related genes as interconnected units for a simple form of regulation. However, cells use different strategies to meet this goal of coordinated ribosome synthesis. The simplest and most compact of these strategies is found in the Escherichia coli genome where all the rRNA molecules are expressed from 7 operons, which also include a varying number of tRNAs (Kaczanowska & Ryden‐Aulin, 2007). Similarly, ribosomal protein genes (RPGs), translation elongation factors and the alpha subunit of RNA polymerase are arranged in 19 operons, where all genes of an operon are expressed from common promoters (Fu, Deiorio‐Haggar, Anthony, & Meyer, 2013; Lindahl & Zengel, 1986). Through this streamlined regulon‐type organization, the expression of rRNA and RP constituents are required, and thus of the resulting ribosomes themselves, are controlled through a unique transcriptional and translational machinery governed by strict feedback mechanisms (Aseev, Koledinskaya, & Boni, 2016). In eukaryotic cells, many features of the bacterial regulon are lost, while certain basic principles are maintained. For example, while most eukaryotic rRNA genes, other than the 5S rRNA gene, remained in clusters, and eukaryotic RPGs are produced from independent genes (Brown, Cole, & Erives, 2008; Chaker‐Margot, 2018; Eichler & Craig, 1994; Laferte et al., 2006). The relative complexity of the organization of eukaryotic RPGs is also increased by the physical separation between mature RPs accumulating in the cytoplasm and their genes and nascent mRNAs located in the nucleus. As a result of this different organization, the autoregulatory mechanism and the common response of RPGs to changes in growth conditions are much more complicated in eukaryotic cells (D. E. Martin, Soulard, & Hall, 2004; Mayer & Grummt, 2006; Pedruzzi et al., 2003). The production of several eukaryotic RPGs is controlled by the TOR and PKA pathways, which link ribosome production to nutrient availability (Mayer & Grummt, 2006; Warner, 1999). Indeed, common promoter and translation motifs were identified in certain RPGs (Bahler, 2005; Gingras, Raught, & Sonenberg, 2004; Liao, Majithia, Huang, & Kimmel, 2008; Mayer & Grummt, 2006). However, not all eukaryotic RPGs feature these regulatory elements (Guimaraes & Zavolan, 2016; Reja, Vinayachandran, Ghosh, & Pugh, 2015). In many cases, the promoter activity, transcription pattern, and RNA amount produced from these genes are very different (Ghulam, Catala, & Abou Elela, 2020). Specific RPGs may also respond individually to a myriad of growth conditions in the background of a relatively stable ribosome production (Xue & Barna, 2012).
In this review, we will summarize the different strategies used by eukaryotic cells to control the expression of RPGs with particular focus on yeast genes, which are the most extensively studied group of RPGs. The different features of eukaryotic RPGs will be described and their contribution to both the gene‐specific and the general regulatory mechanisms of ribosome synthesis will be highlighted. Differences between yeast and human organisms will be outlined and an overall model illustrating the basic principles guiding the regulation of eukaryotic RPGs described.
2. RPGS STRUCTURE, ORGANIZATION, AND DISTRIBUTION
2.1. Number and duplication of RP genes
The number of RPs is fairly constant between species. In bacteria the ribosome includes around 55 RPs while the eukaryotic counterpart includes about 79 RPs (Figure 1a). Despite the high conservation of RPs, the number of RPGs coding for these proteins varies greatly between different kingdoms of life and even between closely related species like Saccharomyces cerevisiae and Candida glabrata (Mullis et al., 2020). Most RPs in plants and fungi are encoded by more than one gene, while protists and animal genomes contain only a few verified duplicated RPGs (dRPGs; Dharia, Obla, Gajdosik, Simon, & Nelson, 2014; Nakao et al., 2004; Wapinski et al., 2010). These duplicated genes are believed to have risen from genome duplication, genome hybridization, retroduplication, or in some cases from polyploidy (Dharia et al., 2014; Nakao et al., 2004; Wapinski et al., 2010). The variation in RPGs number is not related to genome size but linked to the overall propensity of the organism to gene duplications (Dharia et al., 2014; Nakao et al., 2004; Wapinski et al., 2010). However, it is difficult to draw firm conclusions about the extent of dRPG in protists and mammalian genomes as only a few protist genomes are sequenced and many mammalian duplication events such as retroduplications are poorly documented and many pseudogenes are uncharacterized.
FIGURE 1.
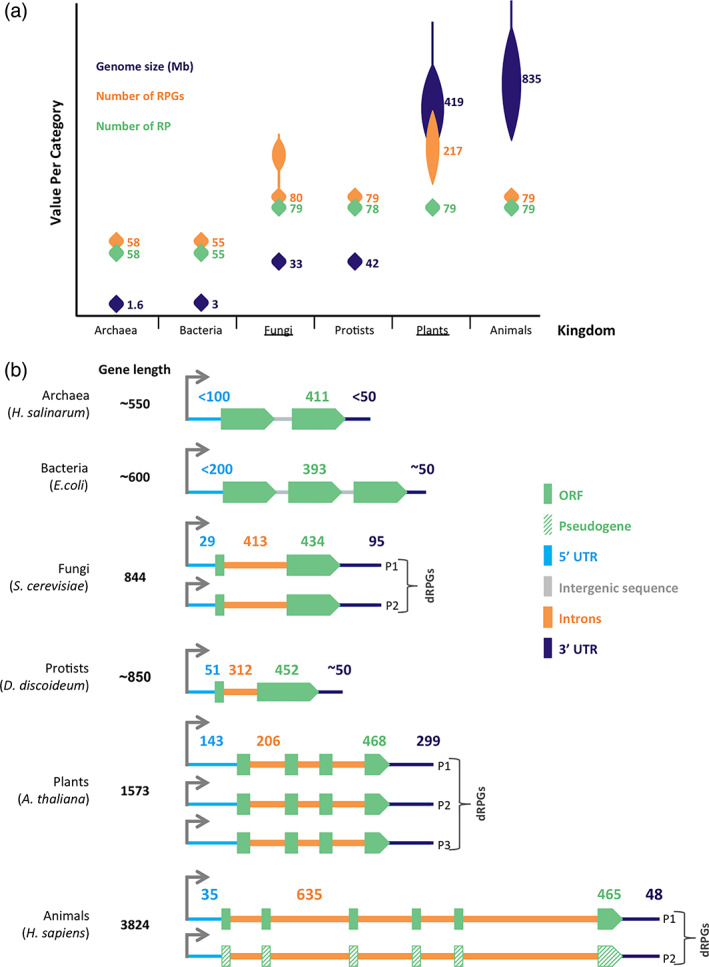
Ribosomal protein gene structure and distribution in the main kingdoms of life. (a) The number of ribosomal protein genes increases in duplicated genomes independent of the protein number and genome size. Genome sizes in mega bases (Mb) are illustrated based on data from NCBI genome browser overview tool for over 300 genomes per kingdom with the median size, the median number of ribosomal protein (RP) and RP genes (RPGs) calculated from 3 to 14 species per kingdom based on information taken from Ribosomal Protein Gene Database. Kingdoms with most genome duplication events are underlined. (b) General features of the different classes of RPGs. The most common structure of RPGs in a representative species from each of the six kingdoms of life are schematically illustrated to indicate the differences in gene size, regulatory region, and the gene organization in the genome. Green boxes, orange lines, gray lines, and blue lines represent the open reading frame (ORF), the introns, the intergenic sequences, 5′ and 3′ UTRs, respectively. The median size of the genes (bp) and ORFs are based on data from NCBI genome browser, SGD, Ribosomal Protein Gene Database, and ThaleMine (Yoshihama et al., 2002). The median number of introns per RPGs are illustrated on each schematic, while the median sizes (bp) of UTRs, ORF and introns are shown on top. Two or more copies of the same genes are shown for species with documented genome duplication events and pseudogenes are represented by green stripped boxes. In most cases the regulatory sequences (UTRs and introns) often vary between species, between genes, and even between duplicated genes. The UTR and gene sizes for archaea and bacteria are estimates based on limited number of genes that are likely to vary as more accurate mapping of the untranslated regions in these species becomes available. The transcription start sites are shown by the gray arrows on top
Studies of eight mammalian genomes including human, chimp, monkey, mouse, rat, dog, cow, and opossum identified 14,552 gene duplicates including dRPGs and suggest that 8% of these are expressed (Dharia et al., 2014). Although some dRPG are known, the best‐studied examples of metazoan dRPGs are those encoding RPL22/eL22. This eukaryote‐specific RP is encoded by two genes eL22 and eL22‐like, which are required for development in Drosophila (Gershman, Pritchard, Chaney, & Ware, 2020; Mageeney et al., 2018; Mageeney & Ware, 2019). Flies depleted of eL22 and rescued by eL22‐like overexpression have reduced fertility, confirming that eL22‐like cannot fully substitute for its paralog's function. Profiling ribosomes carrying one or the other copy of eL22 by RNA‐seq identified 12,051 transcripts (mRNAs/noncoding RNAs), of which half were enriched in the ribosome incorporating one or the other paralog. Analysis of the mRNAs with modified translation suggested that the specific paralogs indeed influenced the translation of a specific set of genes associated with development, especially spermatogenesis (Gershman et al., 2020; Mageeney et al., 2018; Mageeney & Ware, 2019). eL22 is also duplicated in mammals; work in mice suggests that unlike in Drosophila, the deletion of one or the other paralog affects the mRNAs bound but does not induce a major change in translation and both could be incorporated into active ribosomes (O'Leary et al., 2013). For example, specific effects of the paralog were observed in T and B cell development in mice and they exhibited antagonistic functions in regulating the emergence of hematopoietic stem cells in zebra fish (Anderson et al., 2007; Y. Zhang et al., 2013). Another suggestion of possible specialization comes in colorectal cancer where eL22‐like is upregulated and eL22 is not (Rao et al., 2019). Other RPGs duplication was also reported including S27/S27L and L39/L39L (Ruvinsky et al., 2009; Xiong, Zhao, He, & Sun, 2011). Together these data suggest functional specialization of dRPGs in animals but this needs more studied examples and mechanistic explanation. Meanwhile, information about the genome‐wide dynamics and functions of dRPGs are available from studies in S. cerevisiae. In this organism, 118 out of the 137 RPGs are duplicated (Cotney & Shadel, 2006; Dean, Davis, Davis, & Petrov, 2008; Mizuta, Hashimoto, & Otaka, 1995; Mullis et al., 2020; Wapinski et al., 2010). These dRPGs are often found in different chromosomes and are independently expressed from different promoters. Almost all of these duplicated genes are expressed and incorporated into ribosomes with the exception of L15B/eL15B, for which it is not yet clear if it is expressed under certain growth conditions or not at all (Ghulam et al., 2020; Simoff, Moradi, & Nygard, 2009). Since most of the dRPGs in yeast have quasi‐identical amino acids sequence it was suggested that dRPGs increases the regulatory spectrum and by consequence, the functions of RPGs (Parenteau et al., 2008, 2011, 2015). Later in this review, we will dissect in detail the regulatory and functional implication of dRPG in yeast and other organisms.
2.2. RPG architecture
Cross‐species comparison of RPG architecture remains a considerable challenge, despite the relatively high number of sequenced genomes. The annotations of the regulatory sequences are mostly absent or inaccurate. As a result, it is very difficult to draw conclusions about the overall architecture of RPGs in each kingdom. Instead, we will focus the discussion in this section on the few relatively well‐annotated genomes (Figure 1b). Comparison of RPGs from the halophile archaea Halobacterium salinarum, the bacteria Escherichia coli, the fungus Saccharomyces cerevisiae, the protist Dictyostelium discoideum, the plant Arabidopsis thaliana and human Homo sapiens indicates that while the length of the coding sequence and thus the amino acid sequence is very similar across distant species the gene size and organization varies greatly. This cross‐species variation in gene sizes stems mostly from the UTRs and introns. In H. salinarum and E. coli, genes are compact and most are organized in operons with highly variable and poorly characterized UTRs. Only a few RPG operons are characterized in these two species and the few studied operons feature 2–3 genes with each gene size ranging from 550 to 600 nucleotides (Coenye & Vandamme, 2005; Evfratov et al., 2017; Lindahl & Zengel, 1986; Schmitt, Coureux, Monestier, Dubiez, & Mechulam, 2019; Slinger, Newman, Lee, Pei, & Meyer, 2015; Turova, 2003; J. Wang, Dasgupta, & Fox, 2009). The 5′ and 3′ UTRs of the operon are usually around 50 nucleotides long, with few exceptions that are to up to 200 nucleotides (Coenye & Vandamme, 2005; Evfratov et al., 2017; Lindahl & Zengel, 1986; Schmitt et al., 2019; Slinger et al., 2015; Turova, 2003; J. Wang et al., 2009). For example, in H. Salinarum, ORF A encodes two large subunit proteins with typical 5′ and 3′ UTRs (leader and trailer sequence) of 63 and 44 nucleotides, respectively (Itoh, 1988). On the other hand, the E. coli S10–spc–alpha operon, which encodes eight large‐subunit and three small‐subunit proteins, exhibits relatively long 5′ and 3′ UTRs of 137 and 62 nucleotides, respectively (Freedman, Zengel, Archer, & Lindahl, 1987; Zurawski & Zurawski, 1985). The unusually long S10 5′ UTR, which is substantially longer than the average leader size of 25–35 nucleotides in E. coli, signifies the presence of a regulatory sequence controlling the transcription and translation rate of this operon (Dvir et al., 2013; Freedman et al., 1987). The number of RPGs with similar regulatory sequence in bacteria at large remains unclear. Based on these observations, one could safely conclude that, in general, the regulatory sequences have little impact on bacterial and archaeal RPG operon size, which mostly reflects the length of the coding sequence.
In stark contrast to archaea and bacteria, most eukaryotic RPGs, with a few exceptions like those of C. elegans, are individually expressed from genes dominated by long regulatory sequences that vary greatly in length (Figure 1b and (Allen, Hillier, Waterston, & Blumenthal, 2011). In general, the largest eukaryotic RPGs are found in humans and the smallest in fungi (Figure 1b). In general, eukaryotic RPG 5′ and 3′ UTRs are shorter than those of other genes. For example, while the median sizes of the RPGs 5′UTR and 3′ UTR are 29–35 and 48–95 nucleotides, respectively, and their averages are 42–74 and 56–114 nucleotides, respectively, the averages in the eukaryotic transcriptome are 100–200 and 100–800 nucleotides (David et al., 2006; Kotagama, Babb, Wolter, Murphy, & Mangone, 2015; Kreppel et al., 2004; Lin & Li, 2012; Pesole et al., 2002; Yoshihama, Nakao, Nguyen, & Kenmochi, 2006). The UTRs of plant RPGs, as represented here by A. thaliana, tend to be larger than those in other eukaryotes and their sizes do not deviate much from the average recorded for the entire A. thaliana transcriptome (Figure 1b and Krishnakumar et al., 2017; Yoshihama et al., 2006). The median sizes of RPGs 5′ and 3′ UTRs in this species are 143 and 206 with averages of 171 and 306, while the transcriptome averages are 155 and 242 nucleotides, respectively (Krishnakumar et al., 2017; Yoshihama et al., 2006). Like most eukaryotic genes, the 3′ UTRs of RPGs are longer and more heterogeneous in size than the 5′ UTRs. Alternative 3′ end formation and/or polyadenylation, which modify the position of the mRNA 3′ end, have been recorded for eukaryotic RPGs, including those in yeast and metazoan (Antione & Fried, 1995; Chakrabarti, Dinkins, & Hunt, 2018; Gudipati, Neil, Feuerbach, Malabat, & Jacquier, 2012; Parenteau et al., 2015; Stevens, Howe, & Hunt, 2018; Vallejos Baier, Picao‐Osorio, & Alonso, 2017). The relatively short size of RPGs 5′ UTRs may reflect the need for efficient translation, while the long and heterogeneous 3′ UTRs may provide opportunities for gene regulation.
Aside from the UTRs, the most visible regulatory element of eukaryotic RPGs is their introns as the vast majority of eukaryotic RPGs contain introns even in species where introns are less abundant like S. cerevisiae (Amaldi et al., 1995; Nakao et al., 2004; Parenteau et al., 2011; Yoshihama, Nguyen, & Kenmochi, 2007). The number of genes containing introns and the sizes of introns vary between species with the fewest and smallest introns being in the obligate parasite microsporidia and the most in human (Figure 1b and [Whelan, Lee, Lee, & Fast, 2019]). Introns have the most impact on eukaryotic RPG size especially in compact genomes like that of yeast. Indeed, while RPs are smaller than average proteins, RPGs are average‐sized genes (Grishkevich & Yanai, 2014; Hurowitz & Brown, 2003). The presence of introns dramatically increases the complexity of RPG expression and regulation as they variably slow the expression cycle (Chorev & Carmel, 2012; Mattick, 1994; Tycowski, Shu, & Steitz, 1996). In addition, many introns contain noncoding RNA requiring additional layers of coordination and regulation (Chorev & Carmel, 2012). In a later section of this review, we will discuss the details of how RPG structure can influence their expression and regulatory mechanisms.
3. TRANSCRIPTIONAL REGULATION OF RPGS
From the early days of ribosome biology, the obvious need for coordinating the expression of the different ribosome components prompted suggestions of common and uniform transcriptional control of all RPGs (Bollen, 1980; Davies & Nomura, 1972; Mager & Planta, 1991; Nomura & Morgan, 1977; Planta & Raue, 1988). In this simplistic model, RPs function as a single regulon transcribed in unison according to the availability of nutrients and rRNA. This vision is for the most‐part true in bacteria where RPGs form operons in which the expression of multiple genes is controlled by a single promoter and regulated through a strict feedback mechanism (An, Bendiak, Mamelak, & Friesen, 1981; Aseev et al., 2016; Cummings, Sands, Foreman, Fraser, & Hershey, 1991; Nomura & Morgan, 1977; Zurawski & Zurawski, 1985). In this bacterial model, free or excess RPGs will inhibit their own transcription and/or translation, ensuring an adequate supply of proteins while avoiding waste. In eukaryotes, the story is much more complicated and the link between the supply and demand is not as direct. Most eukaryotic RPGs are expressed from their own promoters producing different levels of mRNAs and do not always follow a uniform on and off state (e.g., certain genes might be less or more expressed than the others in different tissues or growth conditions; Genuth & Barna, 2018a; Ghulam et al., 2020; D. Li & Wang, 2020; Marcel, Catez, & Diaz, 2015; Nazar, Sitz, & Busch, 1974; Parenteau et al., 2015; Petibon, Parenteau, Catala, & Elela, 2016; Xue & Barna, 2012). Indeed, in some cases the RPGs do not share the same organization or promoter structure even within the same organisms (Figure 2, transcriptional panel).
FIGURE 2.
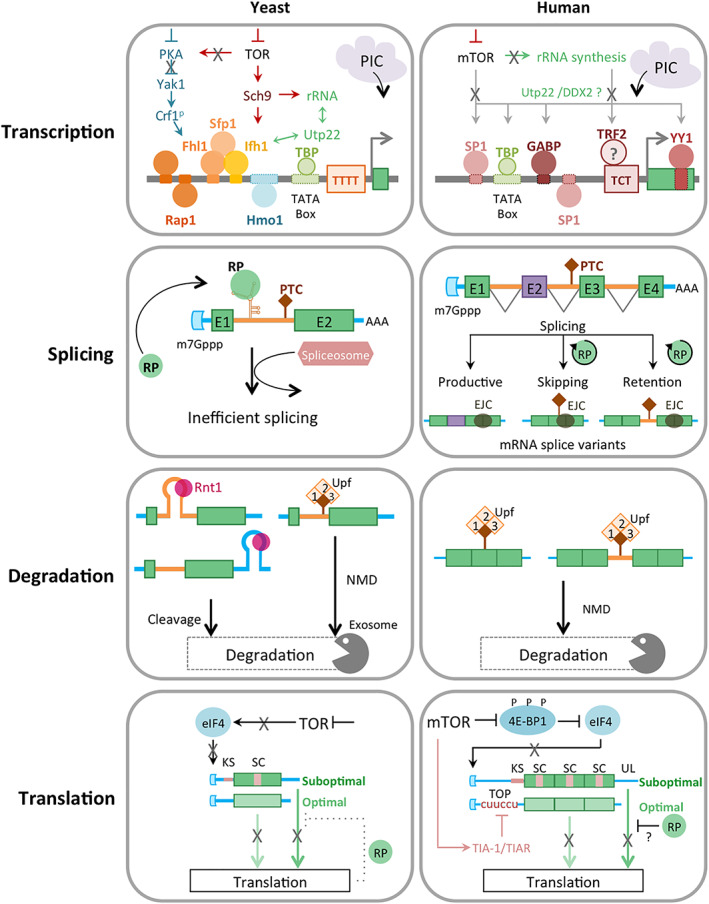
Mechanisms of regulation of ribosomal protein expression. The different levels of regulation modulating the expression of RPGs in yeast and human cells are outlined. Mechanisms lacking direct experimental evidence are indicated with question marks. The diagram summarizes factors found in different genes. Not all the factors are found at the same time and on the same genes. PTC, PIC, NMD, EJC, Upf, RP, KS, SC, and UL indicate premature termination codon, preinitiation complex, nonsense mediated decay, exon junction complex, UPF complex required for RNA degradation via NMD, ribosomal protein, suboptimal Kozak score, suboptimal codon, and long 3′ untranslated region, respectively
The extreme example of this variation between eukaryotic RPGs is found in C. elegans where certain RPGs are in operons while others are independent genes, and where there is no universally conserved motif (Sleumer et al., 2012). In other organisms, like yeast, there are similarities between the RPGs, and some of the motifs are conserved in other species (Figure 2, transcription panel and (Hu & Li, 2007)). However, despite the partial conservation of certain motifs, no universal promoter structure was identified, arguing against a comprehensive and universal transcription‐based regulatory system. The available literature does not permit cross‐eukaryote conclusions about the regulatory mechanisms controlling RPG transcription. Aside from the yeast, only partial studies of limited groups of genes are available in metazoans, including studies in flies, worm, mice, and human (X. Ma, Zhang, & Li, 2009; Maki, Rhoads, Stewart, Van Slyke, & Roufa, 1989; Perry, 2005; Saeboe‐Larssen, Urbanczyk Mohebi, & Lambertsson, 1997; Sleumer et al., 2012). Accordingly, in the next section we will focus on yeast RPGs, which is the most studied model, and compare yeast for the most part to human, with occasional mentions of select examples of divergent features from other metazoans, all to clarify the generality and specificity of RPG transcription.
3.1. General features of RPG promoters
Like most genes, RPGs have a promoter with a core section required for the general recruitment of RNA Pol II and a regulatory section containing motifs required for gene‐specific control of expression (Figure 2, Transcription panel and (Fermi, Bosio, & Dieci, 2017; B. Knight et al., 2014)). However, unlike many genes, most RPGs lack the conserved TATA box sequence, which is considered the central motif for the core promoter region that binds the general transcription factor TBP. In S. cerevisiae, few RPGs have TATA boxes. In humans, 7 RPGs have consensus TATA sequences (Yoshihama et al., 2002), while about 35% of the RPGs have TATA‐like sequence and 25% more have an A/T‐rich motif (Perry, 2005). However, it is not clear if the TATA‐like sequences in these genes are bound by TBP or other more specialized factors or if these are simply nonfunctional remnant motifs. Consistently, computational analysis of several mammalian and two amphibian RPGs found no canonical TATA‐box upstream of the transcription start site (TSS), but instead identified several TATA‐like sequences that may recruit TBP at least under special circumstances (Bosio, Negri, & Dieci, 2011; Genuario, Kelley, & Perry, 1993; Kirn‐Safran, Dayal, Martin‐DeLeon, & Carson, 2000; Marchioni, Morabito, Salvati, Beccari, & Carnevali, 1993). Therefore, it appears that while certain RPGs are transcriptionally activated by canonical TBP binding, most either use noncanonical TBP recruitment or find other ways to activate transcription.
Studies in yeast suggest that the activation of RPG transcription is achieved through partially specialized transcription factors that are not exclusive to RPG promoters (Bosio et al., 2011). Most yeast RPG promoters contain an upstream activating sequence (UAS) composed of either one or two repressor/activator binding sites for Rap1, which is an essential DNA‐binding transcription regulator (P. M. Goncalves et al., 1995). Rap1 often binds in duos, separated by 5–15 base pairs, and this constitutively recruits the Fork head‐like protein, Fhl1, keeping the region nucleosome‐free, in anticipation of transcription (B. Knight et al., 2014; Wade, Hall, & Struhl, 2004; Zhao et al., 2006). In growing cells, Sfp1 and Ifh1 will bind together with Fhl1 to form the FIS complex downstream of the Rap1 binding site (Jorgensen et al., 2004; Reja et al., 2015). In about half of the yeast RPGs, the FIS complex will recruit multiple Hmo1 proteins near the TSS. In the other half transcription proceeds without the binding of Hmo1 (Hall, Wade, & Struhl, 2006; Kasahara et al., 2007; B. Knight et al., 2014; Lavoie et al., 2010; Reja et al., 2015; Xiao, Kamau, Donze, & Grove, 2011). Overall, yeast RPG promoters can be grouped into three classes based on the factors implicated. Class I promoters bind Rap1, FIS, and Hmo1, Class II promoters bind Rap1 and FIS, while Class III includes eight promoters featuring Abf1 binding sites and none of the other RPG associated factors (B. Knight et al., 2014). Intracategory differences also involve variations in the binding pattern of all these transcription factors. Variation in the type and position of the binding factors was shown to impact transcription efficiency, suggesting that while certain common features are preserved between RPGs, their transcription is not fixed at one level or mechanism (Cusanovich, Pavlovic, Pritchard, & Gilad, 2014). Measurement of yeast RPG promoter activity in a reporter system revealed variations in RPGs promoter output. Promoters of single‐copy genes were about 1.5 fold more active than those found in dRPGs (Zeevi et al., 2011). In addition, a loose relation was observed between promoter output and the binding of the Fhl1 and Sfp1 factors or the presence of TATA‐boxes (Zeevi et al., 2011). Surprisingly, not all RPGs, even those with a given promoter architecture, have similar transcriptional activity and no linear relationship was identified between the promoter architecture, reporter‐based activity, and the amount of mRNA produced by the different RPGs (Ghulam et al., 2020; Zeevi et al., 2011).
The notion of a universal mechanism of RPG transcriptional regulation is most clearly struck down by considering yeast dRPGs (Ghulam et al., 2020; Mageeney et al., 2018; Mullis et al., 2020). Most dRPGs are expressed in a strict hierarchy where one of the two paralogs dominates the other to form the majority of the ribosomes (Ghulam et al., 2020; Parenteau et al., 2011; Parenteau et al., 2015; Petibon et al., 2016). As such, one would expect the major gene copies (i.e., the most highly expressed paralog at the protein level) to contain promoter features that foster their transcription, while the minor copies would share features that repress or attenuate transcription. At the very least, the major copies should be more transcribed than their minor counterpart. However, we found that protein levels of the two paralogs not necessary correlate, if any, with the transcriptional activity or mRNA levels (Ghulam et al., 2020). In fact, the promoters of many minor paralogs are more transcriptionally active than those of the major copies (Ghulam et al., 2020; Petibon et al., 2016). For example, although both paralogs of S. cerevisiae S9/uS4 share a similar type II promoter architecture, it is the promoter of the minor uS4A copy that is more transcribed (B. Knight et al., 2014; Zeevi et al., 2011). Clearly, and regardless of these irregularities, there must be some general coordination between the different RPGs. This is particularly evident from the rapid shutdown of RP production in response to stress and the paucity of common features in RPG promoters (Perry, 2005; Warner, 1999). More careful comparison between the temporal variations in RPGs promoters' activity in response to different growth condition is required to differentiate between concerted, coordinated and coupled regulation of transcription.
Mammals have a striking diversity of RPG promoters, but analysis of human genomes nevertheless identified several potential motifs that may form the basis of common RPG promoter structure (Yoshihama et al., 2002). Three transcription factors; GABP, SP1, and YY1, were predicted to bind in different combinations to the promoters of 90% of human RPGs. However, none of these factors is unique to RPGs and are often associated with other TATA, TATA‐like, or TATA‐less promoters (Perry, 2005). The recruitment of human RNA Pol II is proposed to take place through TBP‐related factor TRF2 binding to a TCT motif, which is conserved in metazoan RBP promoters. Experimental evidence for this type of recruitment is found in Drosophila where TCT and not TATA‐dependent transcription depends on TRF2 (Perry, 2005; Y. L. Wang et al., 2014). Motif searches in 13 eukaryotic species did not identify RPG promoter features that are conserved across all organisms, but instead found at least one motif to be conserved within species except in worm where no shared motif was found between RPGs (X. Li, Zhong, & Wong, 2005; Y. L. Wang et al., 2014). Partial cross‐species conservation was found between mammals and insects but some of these motifs might not be strictly linked to transcription. Indeed, some of the motifs were found in mammalian RPG introns, while others were found thousands of base pairs upstream of the TSS (X. Li et al., 2005; Y. L. Wang et al., 2014). Most RPG transcriptional motifs in nonyeast systems await validation with few exceptions like the most frequent motifs found in mouse RPL30/eL30, which affect transcription when mutated (Hariharan, Kelley, & Perry, 1989). More studies of metazoan RPGs are needed in order to understand how transcription of these RPGs is regulated. Nevertheless, given the complexity of metazoa it is likely that the transcription of RPGs will be even more gene‐specific than in yeast. Indeed, the emerging concepts of ribosome specialization, extra‐ribosomal functions, and the plasticity of RPGs are incompatible with the notion of uniformly expressed RPGs. The challenge now is to understand how cells reconcile RPG‐specific needs with the overall requirement of coordinated ribosome biogenesis in homeostasis and in response to stress.
3.2. RPGs transcriptional regulation pathways
Despite their wide variations in promoter sequences and transcriptional activities, many RPGs respond to common signals linking ribosome biogenesis to changes in the cell state and growth conditions. The most conserved and well‐studied of these signaling networks is the nutrient sensing target of rapamycin protein kinase (TOR) pathway. TOR controls ribosome synthesis in response to several cellular cues including changes in nutrient availability, growth factors, and cell size (Loewith & Hall, 2011; Powers, Dilova, Chen, & Wedaman, 2004; Wullschleger, Loewith, & Hall, 2006; X. Yang et al., 2008). The TOR pathway has many effects on cell functions, but RPGs are the predominant target of TOR in yeast. TOR may regulate the transcription of RPGs either directly by modulating the availability or activity of transcription factors controlling RPG expression, or indirectly by altering rRNA synthesis that in turn affects the transcription of RPGs.
In yeast, the TOR pathway includes two multi‐protein complexes named TORC1 and TORC2 (Loewith & Hall, 2011). The two complexes are biochemically and functionally distinct. Only TORC1 responds to rapamycin, and it is TORC1 which is responsible for most regulation of RPG expression through the modulation of transcription factor Fhl1 activity (M. S. Kim & Hahn, 2016; D. E. Martin et al., 2004). When cells are growing and TORC1 is active, Ifh1 is phosphorylated and bound to Fhl1 to stimulate RPG transcription. Conversely, inhibition of TORC1 in the absence of nutrients results in the phosphorylation of Crf1, which displaces Ifh1 to repress RPG transcription. The exact signaling events upstream of Ifh1 are not clear, but a cross talk between TORC1 and the PKA pathway that controls Crf1 through Yak1 has been suggested (D. E. Martin et al., 2004). The causal relationship between PKA and TORC1 in this case is not clear, but it was proposed that TORC1 functions upstream of PKA for a subset of PKA targets, putting TORC1 both upstream and parallel to PKA (Soulard et al., 2010). Fhl1 may not be the only conduit through which TORC1 regulates transcription; a study from the Warner laboratory suggests that TOR inhibition could act independent of Ifh1, at least in the context of a reporter promoter (Zhao et al., 2006). TORC1 may also regulate the transcription of RPGs through interaction between rRNA processing factors Utp22 and Ifh1 (Albert et al., 2016). In this case, repression of transcription of RPGs via the TOR or other pathways will lead to release of Utp22 from the pre‐rRNA in the nucleolus, leaving it free to interact with Ifh1, CK2, and Rrp7 forming the CURI complex. The interaction between Utp22 and Ifh1 invokes a sustained release of Ifh1 from the promoter and thus repression of RPG expression (Albert et al., 2016). This mechanism directly links the transcription of RPGs to the availability of rRNA and shows that TORC1 effects on transcription likely follow multiple paths toward global repression of RPG expression. Despite these and other exciting advances in understanding the mechanism of transcription repression by TOR n yeast, many questions remain open. For example, it is not clear if all RPGs respond to TORC1 in the same way or to the same extent, and the possibility of transcription‐independent regulation of RPG expression remains open. Indeed, recent studies have shown that splicing may play an important role in repressing the expression of certain RPGs in response to TORC1 (Parenteau et al., 2019).
The mammalian TOR pathway (mTOR) signaling cascade is well studied. Maps are available showing hundreds of reactions and targets connecting different cell activities including ribosome biogenesis (Caron et al., 2010). Unlike the yeast, there is little knowledge about whether and how mTOR controls RPG transcription. Most of what we know about mTOR's transcriptional activity concerns the repression of rDNA transcription through the modulation of both RNA Pol I and III (Iadevaia, Huo, Zhang, Foster, & Proud, 2012; Iadevaia, Liu, & Proud, 2014). rRNA synthesis is controlled by the S6K1 kinase, affecting TIF‐IA activity (Iadevaia et al., 2014). One would expect the change in rRNA availability to feed back on the transcription of RPGs perhaps through a mechanism analogous to that found in yeast via Utp22 (Albert et al., 2016; Rudra, Mallick, Zhao, & Warner, 2007), but this remains to be validated. Alternatively, mTOR could perhaps regulate RPG transcription. It was shown that transcription factors involved in RPG transcription, like SP1 or YY1, are also implicated in the mTOR pathway (Astrinidis et al., 2010; Cunningham et al., 2007; Jablonska & Polouliakh, 2014). In Figure 2, we illustrate a possible mechanism by which mTOR might regulate global RPG transcription based on the scarce information available from mammalian cells and by analogy to the yeast. Clearly, more in‐depth studies are needed to clarify the contribution of transcription to the titration of RPG expression in response to changes in growth conditions and particularly to the TOR pathway.
4. RNA DEPENDENT REGULATION OF RPGS
Traditionally, transcription is viewed as the principal step for controlling RPG expression, followed by a streamlined mRNA maturation process meeting the enormous demands for ribosome synthesis (Warner, 1999). According to this transcription‐centric model, polyadenylation, splicing, and RNA stability are mostly constitutive and uniform processes (Warner, 1999; Warner, Vilardell, & Sohn, 2001). Indeed, even alternative splicing, which is the hallmark of protein diversity in mammalian, is thought to be absent from human RPGs, supporting the view of RNA‐independent regulation of RPG expression (Gupta & Warner, 2014). However, RNAseq and RT‐qPCR quantification indicated variation in the amount mRNA produced by individual RPGs ranging from 3‐ to 10‐fold (Gupta & Warner, 2014; Lipson et al., 2009). Notably, it was recently shown that these variations in mRNA quantity exist even between genes coding for the same RPs like the near‐identical dRPG pairs in yeast (Ghulam et al., 2020). Indeed, recent studies start to paint a new picture of ribosome biogenesis where post‐transcriptional regulation plays a central role in determining the amount of proteins produced from each gene and permits gene‐specific regulation (Figures 2 and 3 and [Ahamad, Ojha, Srivastava, Bhattacharya, & Bhattacharya, 2015; de la Cruz et al., 2018; Parenteau et al., 2011; Parenteau et al., 2015; Petibon et al., 2016]). In this section, we will review the recent evidence supporting the role of RNA as major regulator of ribosome biogenesis with emphasis on the role of transcription termination, splicing, and RNA stability in adjusting the dose of proteins produced from each RPG.
FIGURE 3.
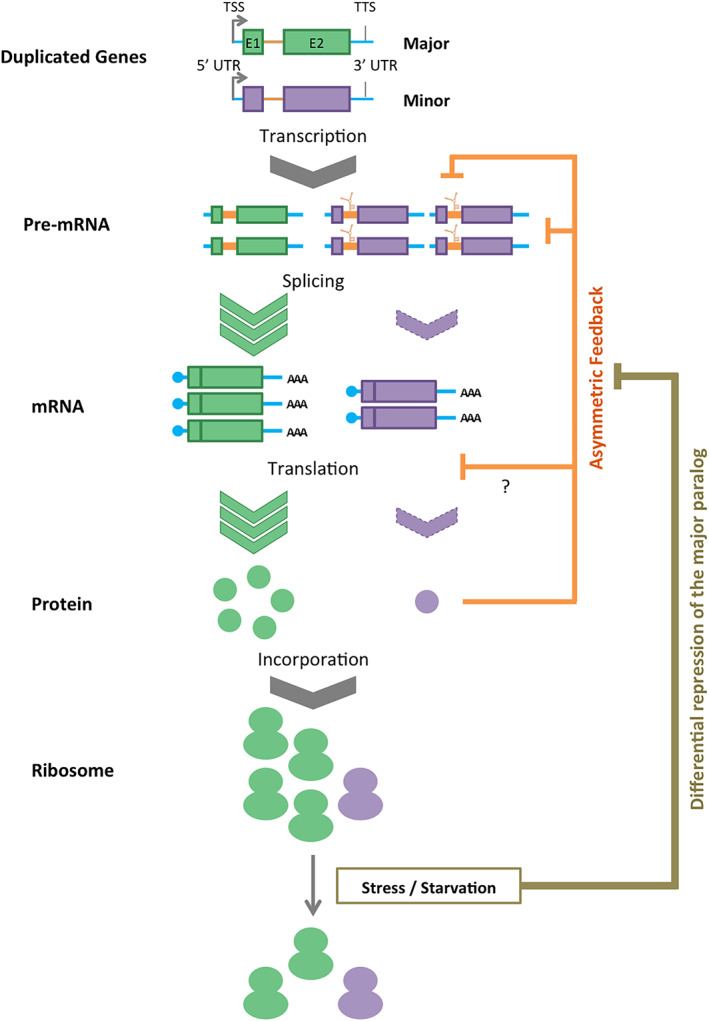
Strategies of interparalog regulation in yeast. Schematic representation of different regulatory elements controlling the expression of dRPGs and their relative contribution to the hierarchy of RPGs in Saccharomyces cerevisiae. In this model, the transcription of duplicated genes does not reflect their final expression outcome. Instead, it is the difference in splicing efficiency of the paralog's pre‐mRNA enforced by asymmetric feedback mechanism differentially inhibiting the splicing of the minor paralog that mostly determine the hierarchy of paralog expression. Differential translation of the major paralog further widens the gap between the two paralogs by favoring the synthesis of the major paralog mRNA. In some isolated cases, differential integration of the major paralog protein into ribosomes also occurs. When cells are exposed to stress or run out of nutrients, the major paralogs are differentially repressed at the level of splicing and translation, tipping the balance toward the production of ribosomes with the minor paralogs, which are in general less affected by stress and starvation. In the cases of duplicated genes lacking introns, most of the regulation takes place through the 5′ and 3′ UTRs that are often different in sequence and length leading to alternative transcription termination, stability and translation efficiency. TSS, TTS, E1, I, E2, 5′ UTR, 3′ UTR indicate transcription starts site, TTS, Exon 1, Intron, Exon 2, 5′ untranslated region, 3′ untranslated region
4.1. RNA maturation and 3′ end formation
The contribution of RNA to the regulation of RPGs is most evident in the form of alternative or differential splicing and alternative transcription termination or polyadenylation. In S. cerevisiae, these modes of gene regulation play a particularly important role in determining the output of dRPGs (Parenteau et al., 2011; Parenteau et al., 2015). In the case of the genes coding for the large subunit protein RPL9/eL9, the transcription of the minor copy eL9B has two alternative transcription termination sites (TTS). One leads to polyadenylation and hence expression of the gene, while the other form gives nonadenylated RNA that is rapidly degraded (Gudipati et al., 2012). The balance of these two termination pathways is determined by the binding of eL9 to a stem‐loop structure at the 3′ end of the genes. When eL9 is in excess, it will preferentially bind and inhibit the minor copy. As such, this mechanism functions as an asymmetric feedback mechanism favoring the expression of eL9A at the cost of eL9B as described in Figure 3. Consistently, comparison of the 3′ UTR length indicated that in most dRPGs, the minor paralog features two alternative termination sites and/or a longer 3′UTR (Parenteau et al., 2015). In the case of RPL8A/eL8A it was shown that the choice of polyadenylation site depends on growth conditions and exposure to stress (Parenteau et al., 2015). Therefore, at least in yeast, it seems that alternative 3′ end formation plays a role in determining the amount of RNA produced from RPGs and shapes the expression hierarchy of dRPGs. Alternative polyadenylation sites were also observed in metazoan cells, but their contribution to RPG regulation remains unclear (Antione & Fried, 1995; Chakrabarti et al., 2018). Interestingly, the 3′ UTR was also shown to influence the cellular distribution of RPs in human cells through the association with actin microfilaments leading to localized translation (Corral‐Debrinski, 2007; Russo, Russo, Cuccurese, Garbi, & Pietropaolo, 2006).
4.2. RPG regulation by splicing
The most versatile and multifaceted regulatory step in the eukaryotic gene expression pathway is splicing, since it can modify not only the quantity, but also the type of proteins produced from each gene. Splicing increases protein diversity through (i) alternative splicing, (ii) titrating mRNA amount by varying the rate of pre‐mRNA maturation, and (iii) triggering mRNA degradation by the inclusion of degradation signals (Black, 2000; Blencowe, 2006; Chew, 1997; Cooper, 2005; Fedor, 2008; Jo & Choi, 2015). In the case of eukaryotic RPGs, splicing is indeed a quasi‐ubiquitous feature found even in the smallest of genomes (Gill et al., 2010; Grisdale, Bowers, Didier, & Fast, 2013; Peyretaillade et al., 2009; Slamovits & Keeling, 2009; Whelan et al., 2019). Notably, introns are over‐represented in S. cerevisiae RPGs where only 5% of the genes contain introns in genome and 89% of all spliced RNA in this organism is generated from RPGs (Hooks, Delneri, & Griffiths‐Jones, 2014; Parenteau et al., 2011; Warner, 1999). Unlike most eukaryotic genes, RPGs rarely produce more than one protein from the same gene through the process of alternative splicing (Gupta & Warner, 2014). As such, it appears for the moment that splicing contributes indirectly to ribosome heterogeneity by modulating the pool of RP to be incorporated into ribosomes, but not in terms of modifying the structure of the proteins. As such, a more detailed, focused and comprehensive analysis of alternative splicing isoforms at mRNA and protein levels and their incorporation in the ribosome in higher eukaryotes is required. It is clear that splicing does play an important role in regulating RPG expression and functions as a tool for feedback regulation (Figure 2, splicing panel). Systematic intron deletion from RPGs in yeast indicated that most RPG mRNAs are regulated through splicing (Parenteau et al., 2011); 84% of intron deletions altered the amount of mRNA produced from RPGs (Parenteau et al., 2011). Surprisingly, introns do not always repress the expression of RPGs as would be expected from the delay generated by the splicing kinetics. Instead, near half of the intron deletions resulted in reduced expression of RPGs, suggesting that introns may promote the production of RPGs. The extent to which introns affected the expression varied between genes. This supports a model whereby gene‐specific splicing efficiency may determine the level of RPG expression not only as negative regulator, but also as enhancer. (Parenteau et al., 2011). Consistently, assaying RPG splicing efficiency using a heterologous reporter system also indicated significant variations in splicing efficiency between RPGs (Yofe et al., 2014).
Variation in splicing efficiency of RPGs plays an important role in adjusting the expression level of each RPG and defines the expression ratio of dRPGs mostly through an asymmetric feedback mechanism (Ghulam et al., 2020; Parenteau et al., 2011; Petibon et al., 2016). As described in Figures 2 and 3, variation in RPG splicing may stem from innate differences in intron sequences that determine the steady‐state splicing efficiency, or through proteins that bind a specific intronic structure and inhibit splicing in specific conditions (Hooks et al., 2014; Neuveglise, Marck, & Gaillardin, 2011; Yoshihama et al., 2007). The intron sequences of many RPG pairs differ by 40%, leaving ample room for gene‐specific splicing repressors and enhancers. However, the specific mechanisms by which splicing efficiency varies between RPGs remain to be fully characterized. The situation is complex, not least because of a known feedback mechanism by which free RPs act themselves as splicing factors to affect their own splicing and hence their own production. Many RPs are RNA‐binding proteins and they can repress splicing by binding specific RNA structures near the splice site (Macias, Bragulat, Tardiff, & Vilardell, 2008; Petibon et al., 2016; Vilardell & Warner, 1997). In the case of L30/eL30, free protein (not incorporated into ribosome) binds to a structure near the 5′ splice site and inhibits U2 binding, leading to reduction in splicing and the amount of mature eL30 RNA (Macias et al., 2008; Vilardell & Warner, 1997). In the dRPGs coding for S9/uS4, the binding of uS4 protein to an intronic structure inhibited splicing of the minor paralog uS4A, favoring the expression the major paralog uS4B (Petibon et al., 2016). A similar mechanism was reported for the duplicated genes coding for L22/eL22, where the minor copy mRNA featured a structure that binds to eL22 protein, leading to the inhibition of splicing (Gabunilas & Chanfreau, 2016). The role of introns in defining the hierarchy of dRPGs appears to be a general mode of autoregulation in yeast, as at least 60% of all gene pairs in this organism require the presence of at least one intron for interparalog regulation (Parenteau et al., 2011). More recent studies have shown that these changes in splicing in turn change protein levels and the number of ribosomes containing specific RP paralogs (Ghulam et al., 2020; Petibon et al., 2016). As well as its likely homeostatic role, splicing‐dependent regulation also contributes to adjusting RPG expression ratios in response to stress and nutrient depletion (Ghulam et al., 2020; Parenteau et al., 2011; Petibon et al., 2016). This clearly indicates the regulation of dRPGs through splicing is not static but rather a dynamic regulatory element that may adjust the ratio of the RP produced from each paralog according to growth conditions.
Splicing‐dependent regulation of RPGs is much less understood in metazoa, but the few available studies point to a role analogous to that found in yeast. For example, the small subunit protein S9/uS4 in D. melanogaster was shown to regulate the expression of its own gene (Plocik & Guthrie, 2012). In at least five eukaryotes including human, alternative splicing represses the expression of uS4 by directing the RNA to nonsense mediated decay (NMD; Plocik & Guthrie, 2012). It is also likely that this type of regulation is not restricted to uS4, but also found in many, if not the majority, of metazoan and especially human RPGs. In human cells, alternatively spliced transcripts often contain premature termination codons permitting splicing based regulation of gene expression (Mitrovich & Anderson, 2000). Notably, L3/uL3, L7A/eL8A, L10A/uL1, and L12/uL11 genes were shown to be auto‐regulated through alternative splicing induced NMD (Cuccurese, Russo, Russo, & Pietropaolo, 2005). Alternative splicing was also implicated in inducing the expression of several RP isoforms in hypoxia (Brumwell, Fell, Obress, & Uniacke, 2020). Cell type specific exon‐skipping event was also reported for S24/eS24. In this case, splicing introduces a stop codon and removes three amino acids from the protein C‐terminal (Song et al., 2017). Additional and more mechanistic information was deduced from human S26/eS26 where it was shown, in vitro and in vivo, that indeed the RP binds directly to its intron and influences splicing (Ivanov, Malygin, & Karpova, 2005). Experimentally, unproductive mRNA isoforms for four RPs (L3, eL8A, uL1, and uL11) were shown to accumulate in C. elegans when NMD was inhibited (Mitrovich & Anderson, 2000). Overexpressing uL11 reprogrammed the splicing of its pre‐mRNA leading to degradation of its own transcript (Mitrovich & Anderson, 2000). Therefore, like in yeast, splicing may function as a tool for RPG autoregulation in metazoan cells. However, it is also clear that the scope of RPG splicing regulation is largely unexplored as only a few metazoan RPGs were tested in a very limited number of tissues and conditions.
4.3. Stability and degradation of RPGs mRNA
The half‐life of cytoplasmic RPG mRNA is within the average range of mRNA half‐lives, suggesting that once exported to the cytoplasm, most RPGs are translated with little inter‐RPG variation or specialized regulation (Y. Wang et al., 2002). In S. cerevisiae, the half‐life of RPG mRNAs is 22 ± 6 min, while those of other cellular mRNAs average 23 min with a median of 20 min, so RPGs as a group are indistinguishable from other yeast RNAs in this respect. However, five RPG mRNAs (S4A/eS4A, S4B/eS4B, L3/uL3, S27A/eS31A, and S28A/eS28A) do have shorter half‐lives of under 10 min (Y. Wang et al., 2002). It is not yet clear if this peculiarly rapid turnover rate reflects a special regulatory requirement or random variation in the degradation process mitigated by upregulation of one or many levels of the gene expression pipeline. In any case, it remains clear that cytoplasmic mRNA degradation does not play a major role in varying the amount nor the timing of RP expression.
In contrast, nuclear RNA degradation clearly has an important role in determining RPG RNA levels, as it acts as a sweeper for miss‐spliced and terminated RPGs. As discussed in above sections, reduced splicing, alternative splicing or transcription read‐through and alternative polyadenylation of RPGs can introduce degradation signals leading to RNA degradation. Currently, it is thought that splicing and/or transcription termination acts as the decisional step defining the fate of the mRNA while RNA degradation functions as the constitutive executioner (Figure 2). However, it is clear that not all RPGs mRNA are degraded by the same mechanism in the nucleus and as such, it is possible that the specific pathway used influences the rate of RPG degradation downstream of transcription and splicing. For example, while inhibiting the splicing of certain RPGs can lead to degradation by the double‐stranded RNA‐specific ribonuclease Rnt1p as is the case of RPS22B/uS8B and RPL18A/eL18A, others like RPS9/uS4 are degraded by NMD (Danin‐Kreiselman, Lee, & Chanfreau, 2003; Peccarelli & Kebaara, 2014; Plocik & Guthrie, 2012). Similarly, while modifying the site of transcription termination led to the degradation of both RPL8A/eL8A and RPL9/uL6, eL8A termination failure leads to degradation by Rnt1p whereas uL6 degradation is carried out by the exosome (Gudipati et al., 2012; Parenteau et al., 2015). There are also some examples of splicing and transcription‐independent RPG mRNA degradation. For example, direct binding of the de‐capping protein Edc3 to a stem‐loop structure at the 3′ end of RPS28B/eS28B mRNA leads to its de‐capping and thus to degradation by 5′‐3′ exoribonucleases (He, Li, Roy, & Jacobson, 2014; Kolesnikova, Back, Graille, & Seraphin, 2013). This mode of degradation helps regulate eS28B levels since free eS28B binds and inhibits the activity of Edc3 in a gene‐specific pattern (He et al., 2014; Kolesnikova et al., 2013). Accordingly, all steps of mRNA maturation might be targeted for regulation through variation in RNA stability and/or sensitivity to ribonucleases. The notion of RPG‐specific nuclear degradation pattern was also supported by the great variation in RPG transcripts' susceptibility to nuclear ribonucleases. Indeed, all the RPG mRNAs are affected by different sets of ribonucleases and most minor dRPG paralogs are more sensitive to ribonucleases than the major ones (Parenteau et al., 2015). It will take a more systematic analysis of metazoan mRNA degradation patterns to accurately assess the contribution of RNA degradation to the control of RPG expression and ribosome biogenesis in general.
5. TRANSLATION REGULATION OF RPGS
RPGs are among the most efficiently translated genes in all living organisms and as such they are built for speed (Warner, 1999; Warner et al., 2001). However, like all genes, RPGs do obey the general signals regulating translation in response to changes in growth conditions or cell status, and variations in the translation efficiency between RPGs are starting to be reported, even in yeast (Ghulam et al., 2020; Gingras et al., 2004; W. Zhang, Du, Zhou, & Chen, 2018). In general, regulation of RPG translation can be classified into two types; the first being global translation regulation and the second specialized gene‐specific regulation (Figure 2, translation panel). Global regulation usually takes place in the form of general repression of RPG translation as a group along with other genes in response to nutrient sensing regulators, like the TOR pathway (Gingras et al., 2004; W. Zhang et al., 2018). On the other hand, gene‐specific regulation takes the form of either innate variations in translation efficiency between genes or either a fine‐tuning mechanism that varies the response of certain RPGs to stress (Ghulam et al., 2020). Global regulation is conserved in several eukaryotes, while the gene‐specific mechanisms have mostly been reported in yeast. In this section, we will review the literature supporting these two types of translation regulation and their utility.
5.1. General regulation of RPGs translation
RPGs are subject and particularly sensitive to most global mechanisms of translation regulation, including the nutrient‐sensing TOR pathways and other developmental or cell differentiation regulator mechanisms (Gingras et al., 2004; Xiao & Grove, 2009; X. Yang et al., 2008). Nutrients and growth factors are the most prominent signals for ribosome biogenesis. They often regulate the translation of RPG mRNAs as part of the collective induction of ribosome synthesis that also includes stimulation of rRNA and RPG transcription (Abe, Yamashita, & Isono, 1994; Gingras et al., 2004; Hamilton, Stoneley, Spriggs, & Bushell, 2006; Masvidal, Hulea, Furic, Topisirovic, & Larsson, 2017; X. Yang et al., 2008). Translation of RPGs is particularly crucial for boosting the synthesis of new ribosomes in newly divided cells and for jump‐starting the growth cycle (Arribere, Doudna, & Gilbert, 2011; Johnstone & Lasko, 2001; Richter, 1991). Notably, translation activation of the mRNAs might be the most important regulatory target for situations requiring a rapid response. Rapid translation activation allows cells to produce new proteins faster than starting from de novo transcriptional activation of the same genes. Translation acts as an amplifier of the effect of transcriptional gene control by enhancing the protein output from transcriptionally induced genes or else as a damper reducing the protein output from already‐transcriptionally repressed genes (Melamed, Pnueli, & Arava, 2008; Preiss, Baron‐Benhamou, Ansorge, & Hentze, 2003). The most studied mechanism of translation regulation is that controlled by the TOR pathway, which links translation to nutrients availability, cell size, and cell division (Caron et al., 2010; Gingras et al., 2004; Iadevaia et al., 2014; Masvidal et al., 2017; Mayer & Grummt, 2006). When nutrients are limited or another off signal is received, the main regulator of the TOR pathway, TORC1, is inactivated leading to the dephosphorylation of the translation initiation factor 4E‐BP, which disrupts translation initiation complexes (Figure 2, Translation panel and (X. M. Ma & Blenis, 2009)). In mammalian cells, phosphorylation of mTORC1 by PI3K and AKT kinases leads to the phosphorylation of the S6K protein and the repression of a specific class of mRNAs containing 5′ terminal oligopyrimidine (TOP) tracts (Thoreen et al., 2012). This group of mRNAs, which include many RPGs, are the primary translation targets of mTOR (Fenton & Gout, 2011). The presence of pyrimidine‐rich translational elements (PRTE) within their 5′ UTRs was proposed to mediate at least in part the mTOR effect on translation (Hsieh et al., 2012). Other triggers of translation repression in mammalian cells include p53, which may act either in parallel or through the mTOR pathway to repress translation through the titration of the amount of free eIF4E through the binding 4E‐BP1 association (Constantinou & Clemens, 2007; Horton et al., 2002; Jessica et al., 2012). The role of p53 in the regulation of translation is currently a subject of extensive study and its effect on RPGs expression underlines yet another link between RPGs expression and the cell cycle and proliferation.
5.2. RPGs specific regulation of translation
For decades, RP mRNAs were thought to be equally translated as a single regulon fluctuating in unison between on and off states in response to cellular cues. However, recent work in yeast has started to project a different image where RPGs and especially dRPGs are translated at different levels and in response to different stimuli (Ghulam et al., 2020; Komili et al., 2007). Surprisingly, in silico comparison of RPG mRNAs indicated a great variation in the translation regulatory elements, including the number of suboptimal codons, Kozak sequence, UTR lengths, and GC content. Consistently, comparison of the level of RPG association with ribosomes found notable variation between RPGs and even between duplicated RPGs. In 56% of the cases, the most efficiently translated RPGs exhibited more optimal Kozak score. On the other hand, in 35% of the cases the most translated genes were also those with the most optimal Kozak access, having lower GC content in the 5′ UTR or exhibiting shorter 3′ UTR. Interestingly, while no strict correlation was found between translation rate and any single translation regulatory element, the more translated genes always had a more favorable overall regulatory element composition. This indicates that RPG translation efficiency is the result of a combination of factors that may take various forms from enhanced initiation to improved elongation. In general, RPGs producing most RNA were also those having the most efficiently translated mRNA, indicating that at least in these cases translation works as an amplifier of RP production (Ghulam et al., 2020).
The 3′ UTR of the minor paralog of RPL8/eL8 contains a specific translation regulatory element; substitution of the 3′ UTR of the minor paralog eL8A, which is under‐translated, with a heterologous UTR increased translation, while the same substitution in the major and more translated paralog did not (Parenteau et al., 2015). Strikingly, this type of gene‐specific regulation is not static but may also change depending on growth conditions. For example, it was shown that exposure to stress differentially represses genes that are normally over‐translated under normal conditions (Parenteau et al., 2015). Aside from the above‐mentioned translation regulatory elements, it was recently discovered that the amino acid charge of RPGs might play an important role in their rate of translation by slowing the elongation rates. In this new model, RPG translation is determined by the balance between a highly efficient initiation rate and a relatively inefficient elongation rate (Riba et al., 2019). It is not clear, however, if variation between RPGs' amino acids charge could cause differences in translation or whether these changes are responsive to changes in cell growth. Overall, it is now clear that translation may play an important role in adjusting the output of RPGs on a gene‐by‐gene basis, the exact formula of this type of regulation remains unclear.
6. POST‐TRANSLATION REGULATION OF RPGS
Once translated, RPs may undergo a number of post‐translational modifications including phosphorylation, ubiquitination, methylation, acetylation, and SUMOylation (Dalla Venezia, Vincent, Marcel, Catez, & Diaz, 2019; Genuth & Barna, 2018a). Unlike other regulatory steps in RPG production, most post‐translational modifications appear to be protein‐specific (D. Simsek & Barna, 2017). Indeed, the dependence of these modifications on the amino acid sequence makes them highly suitable for gene‐specific regulation. In general, RP modifications can be divided into two groups; the first includes modifications regulating protein stability like the proteasome inducing ubiquitination. The second includes modifications that alter the function or the behavior of the proteins, like phosphorylation, which may alter subcellular localization or protein interactions of RPs. Phosphorylation can also alter ribosome biogenesis or even lead to extra ribosomal functions. These protein modifications may occur either co‐translationally or post‐translationally, and they are considered the best source for a potential ribosome specialization (Genuth & Barna, 2018b; Ruiz‐Canada, Kelleher, & Gilmore, 2009; Sauert, Temmel, & Moll, 2015; C. I. Yang, Hsieh, & Shan, 2019). In the following section, we will discuss the impact of specific modifications on RP regulation, functions, and stability.
The very first RP modification was discovered in RPS6/eS6 more than 4 decades ago by the Wool lab, and it remains highly studied and functionally relevant (Gressner & Wool, 1974; Khalaileh et al., 2013; Ruvinsky et al., 2009). The phosphorylation of eS6 is implicated in cell size control, pancreatic cancer, glucose homeostasis, activation of neurons, integration of light signals, and circadian clock signals (Enganti et al., 2017; Khalaileh et al., 2013; Z. A. Knight et al., 2012; Ruvinsky et al., 2005). The small subunit protein eS6 is highly conserved from yeast to human and several of the phosphorylated residues are also conserved. In mammals, the two serine residues close to the carboxy‐terminus of eS6 are directly phosphorylated by S6K in response to mTORC1 (Nakashima & Tamanoi, 2010). In yeast, the conserved serine residues (Ser232 and Ser233) are also phosphorylated, likely by the functional S6K homolog Sch9 (Gonzalez et al., 2015). However, mutation of this residue in yeast did not have major effect on growth in rich media (Johnson & Warner, 1987). In contrast, The role of phosphorylation in mammalian cells remains unclear since mutation in these residues had little phenotypic effect (Ruvinsky et al., 2009).
Phosphorylation is not restricted to eS6 but was also detected in a large number of RPs, and several of these modifications altered the function of their respective proteins. For example, in the case of RPL12/uL11, phosphorylation altered translation during mitosis while phosphorylation of RPS15/uS19 resulted in LRRK2 neurodegeneration in Parkinson's disease (Imami et al., 2018; I. Martin et al., 2014). In yeast, phosphorylation of Ser223 of RPS7/eS7 is implicated in the biogenesis of the small ribosomal subunit, translation, and cellular proliferation (Tomioka et al., 2018). Eventually, phosphorylation was found in almost all RPs tested, increasing the spectrum of RP regulation (Dinman, 2016; Ferretti & Karbstein, 2019). Notably, phosphorylation may also expand the function of RPs beyond their function in the ribosome. For example, stoichiometric phosphorylation of RPL13A/uL13 at a single serine triggers its release from assembled ribosomes to function as an essential component of the interferon‐ϒ activated inhibitor of translation complex (Mazumder et al., 2003; Mukhopadhyay et al., 2008). In time, it is likely that more extra‐ribosomal functions will be discovered. In addition to protein phosphorylation, other types of modifications also alter the function of RPs. For example, methylation of RPL42/eL42 modulates translation and cellular proliferation in S. pombe, while methylation of RPL3/uL3 promotes translational elongation fidelity, and histidine methylation of uL3 contributes to the assembly of the 60S ribosomal subunit in S. cerevisiae (Al‐Hadid et al., 2014; Shirai, Sadaie, Shinmyozu, & Nakayama, 2010). SUMOylation is another common RP modification that alters the subcellular localization of RPL22/eL22 in Drosophila, and contributes to RPL11/uL5‐mediated activation of p53 in mice (El Motiam et al., 2019; Kearse, Ireland, Prem, Chen, & Ware, 2013).
One of the most recent developments in the study of post‐translational modification of RPs is the discovery of the association of the UFMylation enzyme (UFL1) with the assembled 60S LSU and 80S ribosome and its involvement in metazoan‐specific post‐translational modifications (PTM) of RPs including RPL26/uL24 (Deniz Simsek et al., 2017; Xu & Barna, 2020). UFMylation of uL24 plays a direct role in co‐translational protein translocation through the degradation of a translocation‐arrested endoplasmic reticulum protein by targeting it to the lysosomes (Walczak et al., 2019; L. Wang et al., 2020). Finally, glycosylation, hydroxylation, and acetylation can modulate RP function by altering their secretion, RNA binding, and protein synthesis properties (Kamita et al., 2011; Y. Kim, Lee, Kim, & Kim, 2016; Yanshina, Bulygin, Malygin, & Karpova, 2015). Despite these numerous examples of RP modifications, the mechanistic details and impact on protein synthesis are mostly unknown leading to a debate. Indeed, the capacity of a single or a cluster of protein modifications to alter translation is contested given that the peptidyl transferase center of the ribosome is made mostly of rRNA (Dinman, 2016; Ferretti & Karbstein, 2019). The challenge now is not to find more RP modifications but rather to better understand their contribution to the functions of RPs and ribosomes.
In addition to altering protein functions, modifications were also shown to change the stability of RPs by increasing resistance to proteases or by tagging them for degradation. For example, while SUMOylation of human protein RPS3/uS3 makes it resistant to proteolysis, the phosphorylation of the yeast protein P1/P1 triggers its degradation (Jang et al., 2011; Nusspaumer, Remacha, & Ballesta, 2000). Under certain circumstances, modification and specifically ubiquitination may lead to RP degradation at least in the experimental systems tested. However, it is unclear if this degradation is a natural mechanism for controlling RP doses within the normal cycle of ribosome biogenesis or functions as a special feature triggered by artificial overexpression of RPGs. It is also not clear how, when, and why a specific RPG may escape, under natural conditions, the layers of transcriptional and post‐transcriptional regulation to produce excess proteins out of sync with the other components. An emerging paradigm of RP function is the post translationally regulated sub‐stoichiometric association of RPs to actively translating ribosomes that results in the production of functional heterogeneous ribosomal population. Indeed, it was shown that among others, RPL38/eL38 and RPL10A/uL1 are under associated with the ribosome under certain conditions. The association of these proteins was implicated in the translation of mRNAs with internal ribosome entry site (IRES) (Zhen Shi et al., 2017; Xue et al., 2015).
7. REGULATION OF RIBOSOME BIOGENESIS A TUG OF WAR BETWEEN DIVERSITY AND HOMOGENEITY
The textbook view of regulation of RPGs and ribosome biogenesis is one of conformity, streamlining, and tight regulation (Warner, 1989; Warner et al., 2001; Warner, Kumar, Udem, & Wu, 1973). Key research articles have purveyed concepts like a uniform regulatory program controlled by the TOR pathway at the transcription level in yeast and at the translational level in mammalian cells (Berger et al., 2007; Iadevaia et al., 2012; Thoreen et al., 2012; Wullschleger et al., 2006; Xiao & Grove, 2009). However, these views are being constantly challenged by recent experiments highlighting the variation in RPG levels of expression and regulatory mechanisms leading to the heterogeneity of ribosome composition (Ferretti & Karbstein, 2019; Genuth & Barna, 2018a; Kondrashov et al., 2011; Mageeney & Ware, 2019; Sauert et al., 2015). Indeed, while the concept of specialized ribosomes may continue to be debated, it is hard to challenge the heterogeneity of ribosome composition and regulatory programs. As evident from the examples discussed earlier, most eukaryotes produce variable amounts of mRNA from different RPGs, and mass‐spectrometry analyses continue to detect variations in the protein contents of ribosome populations extracted from different tissues and cells grown under different conditions (Genuth & Barna, 2018b; Giavalisco et al., 2005; D. Li & Wang, 2020; Marcel et al., 2015; Nazar et al., 1974; Sauert et al., 2015; Xue & Barna, 2012). The challenge now is to reconcile these different views in a single model that permits heterogeneity while maintaining a level of conformity allowing the coordination of ribosome biogenesis. In the next section, we will discuss examples of RPG regulatory heterogeneity and discuss possible models for combining the needs for gene‐specific and ribosome‐wide regulatory programs.
7.1. Heterogeneity of RPG expression profiles and control mechanisms
Most RPG studies compare their responses to different stimuli as a group in relation to the rest of the genome. This type of comparison over‐emphasizes the homogeneity of response within the group and underestimates intergene variations and the existence of outlier genes (Ishii et al., 2006; X. Li, Zheng, Hu, & Li, 2016; Reja et al., 2015). However, suggestions of gene‐specific regulation in response to changes in growth conditions were advanced more than two decades ago. Comparison of RPG expression in D. discoideum spores and exponentially growing cells identified 12 developmentally regulated RPs that deviate from the general RPG expression profile (Ramagopal, 1990; Ramagopal & Ennis, 1981, 1982). Tissue‐specific expression of certain RPGs was also observed in plants as the expression of a reporter gene driven by the Rpl16A promoter was differentially detected in proliferating tissues (Williams & Sussex, 1995). Another example in plants is S5A/uS7A, which is differentially expressed in dividing cells while its paralog is expressed in differentiated cells (Weijers et al., 2001). In several multicellular organisms like fruit flies, zebra fish, and mice, mutations in RPGs resulted in tissue‐specific phenotypes supporting the notion of tissue‐specific expression, or at least function of RPGs (Kongsuwan et al., 1985; Z. Shi & Barna, 2015). In human, tissue‐specific expression of a specific set of RPGs was observed in different organisms and mutation in certain RPGs led to defects in one tissue and not the others, as the case in Diamond‐Blackfan Anemia. In this case, mutations in any one of 12 RPGs (RPL11/uL5, RPL15/eL15, RPL26/uL24, RPL31/eL31, RPL36A/eL42, RPL5/uL18, RPS7/eS7, RPS10/eS10, RPS17/eS17, RPS19/eS19, RPS24/eS24, or RPS26/eS26) specifically impaired bone marrow function (Cmejla & Cmejlova, 2003; Gazda & Sieff, 2006; P. Goncalves, Pereira, Kidd, & Ribeiro, 2004; Mirabello et al., 2017; R. Wang et al., 2015).
Other RPG mutations can lead to tissue‐specific predisposition to cancer (Sulima, Hofman, De Keersmaecker, & Dinman, 2017). Variation in expression of RPGs can also stem from the utilization of certain RPGs for nonribosomal function. In yeast, at least 10 genes were assigned nonribosomal functions including rRNA processing and assembly, transcription regulation, translation, DNA repair, and replication (Lu, Zhu, Xiong, Wang, & Jia, 2015). Similar extra‐ribosomal functions were also detected in mice and human (Briggs & Dinman, 2017; Molavi, Samadi, & Hosseingholi, 2019). To perform these dual functions, the RPGs in question need to be independently regulated to meet not only the need for new ribosomes, but also to fulfill their specialized function. However, this notion awaits experimental validation to robustly link RPG specialization to defined patterns of gene expression. Meanwhile, the well‐documented variation in the expression and in some cases the virtual absence of one or more RPs from certain tissues (Genuth & Barna, 2018a, 2018b; D. Simsek & Barna, 2017; Xue & Barna, 2012) prove the ribosome tolerance to variations in RP amounts and as such, negates any necessity for regulatory conformity of RPGs.
7.2. Duplicated RPGs: An example of controlled regulatory diversity
The strongest argument for individualistic regulation of RPs is the great difference in the expression of dRPG. Almost all yeast dRPGs and the few dRPGs studied in higher eukaryotes are expressed at differing levels and in many cases, they respond to different stimuli. The Silver lab demonstrated that paralog‐specific expression of RPL7/uL30, RPL12/uL11, RPS18B/uS13B, and RPL22/eL22 genes is required for yeast bud site selection and localized translation of Ash1 mRNA (Komili et al., 2007), while our lab subsequently demonstrated that most yeast dRPG pairs in yeast are differentially regulated to favor the expression of one gene copy (Ghulam et al., 2020; Parenteau et al., 2011; Parenteau et al., 2015; Petibon et al., 2016). Surprisingly, this expression hierarchy is not established by different transcription levels but mostly through differential splicing (Parenteau et al., 2011). In most cases, one of the two copies is more spliced than the other and often the predominant protein plays an important role in repressing the splicing of its paralog (Petibon et al., 2016). This gene copy‐specific repression is not static as in many cases, it can be overturned by cellular stress. For example, stress affects the expression of RPS9/uS4, RPS17/eS17, RPS29/uS14, and RPL33/eL33 in an intron‐dependent manner (Parenteau et al., 2011). Indeed, splicing rate is the major determinant of the expression hierarchy of yeast dRPGs. In general, genes that are more spliced are more incorporated into ribosomes confirming the role of splicing as the major determinant of intergene regulation in yeast (Ghulam et al., 2020). Curiously, in certain cases the most transcribed paralog was the least spliced, producing the least amount of mature mRNA (Parenteau et al., 2011; Petibon et al., 2016).
In intron‐less genes the situation is different of course. Here, expression control in response to stress is mediated by differential transcription termination and 3′ end formation (Gudipati et al., 2012). In these cases, the minor paralog features alternative TTS and a longer 3′ UTR (Parenteau et al., 2015). The available data on dRPG regulation in yeast suggests that efficient splicing or strong transcription termination dictates which copy is most expressed at the protein level (Figure 3). Under normal growth conditions, expression of dRPGs is auto‐regulated through an asymmetrical feedback mechanism whereby the free RPs binds to the intron or the 3′ UTR to repress the minor paralog. In most cases, this differential repression of the minor paralog is cemented by higher translation efficiency of the major paralog. As a result, under normal growth conditions the cell will always contain more ribosomes featuring the major RP, even when the minor paralog is the more transcribed. When cells are exposed to stress, the abundance of the major paralog is reduced, at least in part, through the repression of is translation and/or transcription. This reduction in the major paralog expression leads consequently to the de‐repression of the minor paralog splicing or transcription termination, which are otherwise repressed by the proteins produced from the major paralog. As a result, the number of ribosomes containing the minor RP forms increases, while those featuring the major RP form decreases. Duplication of RPGs in yeast likely serves as a specialized stress‐response mechanism replacing the highly expressed RPGs, which are suitable for growth under optimal conditions, with a layer of poorly expressed genes better suited for quiescent, stressed, or starved cells.
7.3. Combined model for regulation and diversity of RPGs
Careful examination of both classical and recent studies of RPG expression suggests two partially overlapping regulatory circuits (Figure 4). The first is a global ribosome synthesis regulatory network that controls the overall expression of RPGs, while the second is a gene specific tuning system that adjusts the amount of proteins produced from each gene individually. In yeast, TOR primarily controls the transcription of RPGs in response to signals emanating from the nutrient and cell status sensing pathways (TOR and PKA pathways), while in mammalian cells these same pathways mostly control the translation initiation of RPGs. On the other hand, the gene specific circuitry adjusts the mRNA amount produced from each RPG in relation to the need of this particular RP for either extra‐ribosomal function, response to stress or paralog‐specific function. The global circuitry provides a rough coordination of the different ribosome components and network with the CURI (Ck2/Utp22/Rrp7/Ifh1) transcriptional regulatory complex controlling both rRNA and RPGs expression. On the other hand, the stochastic variations that are inevitably created by GST as it tunes the expression of individual genes would function as a source for new RP functions or as a toolbox for stress response. As illustrated in the model presented in Figure 4, the integration of these two regulatory circuits provides cells with the mean to diversify its RPGs functions while keeping control of the overall process of ribosome production.
FIGURE 4.
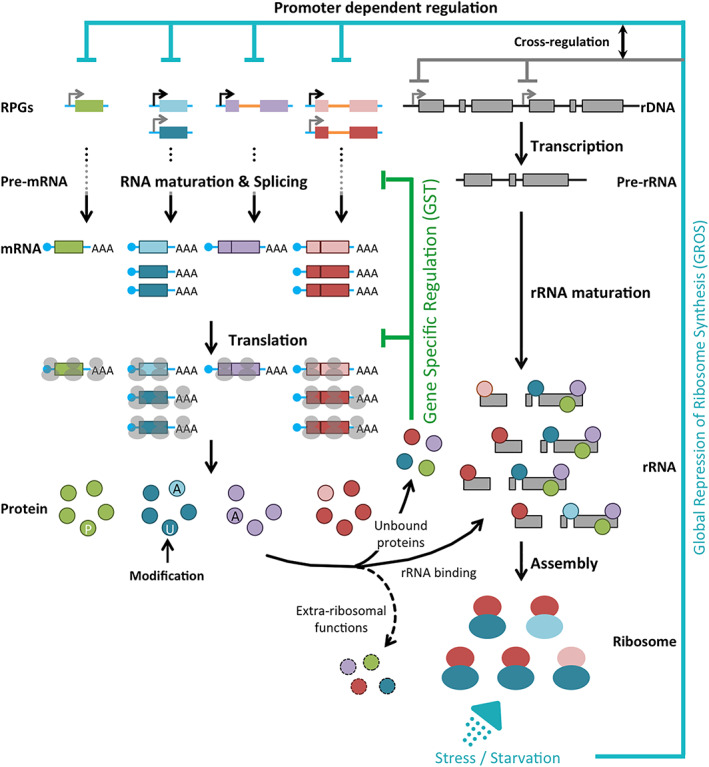
General mechanisms for controlling and coordinating the expression of RPGs. The illustrated schematics depict the basic principles guiding the control of RPG expression. In this hypothetical model, the RPGs will respond collectively to changes in the amount of mature rRNA and signals for global repression of ribosome synthesis transcription repression guided by common promoter elements. On the other hand, gene specific regulation required for adjusting the dose of duplicated RPGs and genes playing extra‐ribosomal roles may take place through regulation of splicing, RNA stability, transcription termination, and translation. A, P, and U indicates examples of modifications of RPs through acetylation, phosphorylation, and ubiquitination
8. CONCLUSION
Ribosome biogenesis is the most conserved cellular process predating life as we know it. Traditionally, this synthesis was seen as a streamlined and uniform process regulated as one regulon through transcription and/or translation. In this review, we presented examples from the literature that dispute this elementary view of ribosome biogenesis, highlight the differences in the expression and focus on regulatory mechanisms of the different RPGs. Overall, the current state of knowledge indicates that while certain levels of a common regulatory network do exist, as illustrated by common promoter and translation regulatory elements, they are far from universal. In no species do RPGs feature the exact same promoter structure; no universal elements are detected cross‐species. Indeed, while the coding sequence of RPGs is highly conserved even in distant species like bacteria and mammals, their regulatory sequences vary even between duplicated genes coding for the same RP subunit. Recent studies comparing the amount of RNA and proteins produced from different RPGs also show great diversity in gene output, negating the notion of conformity. Compensation for imbalance through differential protein degradation is possible but was not established a naturally occurring regulatory program. Instead, in this review, we propose a different vision of ribosome biogenesis where the expression of RPGs is controlled mostly by gene‐specific regulatory elements that define the dose of each protein under the umbrella of global regulatory system overseeing ribosome production as a whole.
This new paradigm of ribosome synthesis permits RPGs to be expressed at different levels, in some cases at different times or in different tissues. These variations are adjusted based on each gene's function inside and outside the ribosome. At the same time, the process is controlled by common elements or interconnected networks linking many but not all RPGs' transcription and/or translation to the expression of rRNA. This model would now benefit from experimental scrutiny to understand the interplay between the general and gene‐specific regulatory sequences and their mechanisms of action. It is not clear how maverick RPGs that do not have common regulatory elements respond to general regulatory pathways like the TOR pathway. Do these genes use different regulatory elements or continue to be produced at steady levels and their protein products are degraded when the transcription of rRNA and certain RPGs are shut down? With more systematic studies of RPG expression in organisms other than yeast, we should be able to understand the essence of RPG regulatory processes in the coming years.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
AUTHOR CONTRIBUTIONS
Cyrielle Petibon: Data curation; visualization; writing‐original draft; writing‐review and editing. Mustafa Malik Ghulam: Conceptualization; data curation; writing‐original draft; writing‐review and editing. Mathieu Catala: Conceptualization; data curation; writing‐original draft. Sherif Abou Elela: Conceptualization; resources; supervision; validation; visualization; writing‐original draft; writing‐review and editing.
RELATED WIREs ARTICLES
ACKNOWLEDGMENTS
We thank Julie Parenteau and John Woolford for critical reading of the manuscript. Work in Abou Elela Lab is supported by grants from Canadian Institutes of Health Research (CIHR) and Natural Sciences and Engineering Research Council, NSERC. Sherif Abou Elela holds Canada Research Chair in RNA Biology and Cancer Genomics.
Petibon C, Malik Ghulam M, Catala M, Abou Elela S. Regulation of ribosomal protein genes: An ordered anarchy. WIREs RNA. 2021;12:e1632. 10.1002/wrna.1632
Cyrielle Petibon and Mustafa Malik Ghulam contributed equally to this work.
Funding information Natural Sciences and Engineering Research Council of Canada
REFERENCES
- Abe, R. , Yamashita, A. , & Isono, K. (1994). Cloning and characterization of the ribosomal protein genes in the spc operon of a prokaryotic endosymbiont of the pea aphid, Acyrthosiphon kondoi . DNA Research, 1(3), 103–114. [DOI] [PubMed] [Google Scholar]
- Ahamad, J. , Ojha, S. , Srivastava, A. , Bhattacharya, A. , & Bhattacharya, S. (2015). Post‐transcriptional regulation of ribosomal protein genes during serum starvation in Entamoeba histolytica . Molecular and Biochemical Parasitology, 201(2), 146–152. 10.1016/j.molbiopara.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Albert, B. , Knight, B. , Merwin, J. , Martin, V. , Ottoz, D. , Gloor, Y. , … Shore, D. (2016). A molecular titration system coordinates ribosomal protein gene transcription with ribosomal RNA synthesis. Molecular Cell, 64(4), 720–733. 10.1016/j.molcel.2016.10.003 [DOI] [PubMed] [Google Scholar]
- Al‐Hadid, Q. , Roy, K. , Munroe, W. , Dzialo, M. C. , Chanfreau, G. F. , & Clarke, S. G. (2014). Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly. Molecular and Cellular Biology, 34(15), 2903–2916. 10.1128/MCB.01634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M. A. , Hillier, L. W. , Waterston, R. H. , & Blumenthal, T. (2011). A global analysis of C. elegans trans‐splicing. Genome Research, 21(2), 255–264. 10.1101/gr.113811.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaldi, F. , Camacho‐Vanegas, O. , Cardinall, B. , Cecconi, F. , Crosio, C. , Loreni, F. , … Pierandrei‐Amaldi, P. (1995). Structure and expression of ribosomal protein genes in Xenopus laevis . Biochemistry and Cell Biology, 73(11–12), 969–977. [DOI] [PubMed] [Google Scholar]
- An, G. , Bendiak, D. S. , Mamelak, L. A. , & Friesen, J. D. (1981). Organization and nucleotide sequence of a new ribosomal operon in Escherichia coli containing the genes for ribosomal protein S2 and elongation factor Ts. Nucleic Acids Research, 9(16), 4163–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, S. J. , Lauritsen, J. P. , Hartman, M. G. , Foushee, A. M. , Lefebvre, J. M. , Shinton, S. A. , … Wiest, D. L. (2007). Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53‐dependent checkpoint. Immunity, 26(6), 759–772. 10.1016/j.immuni.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Antione, M. , & Fried, M. (1995). Use of alternative polyadenylation sites by the chicken S6 ribosomal protein gene. Biochemical and Biophysical Research Communications, 210(2), 384–391. 10.1006/bbrc.1995.1673 [DOI] [PubMed] [Google Scholar]
- Arribere, J. A. , Doudna, J. A. , & Gilbert, W. V. (2011). Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Molecular Cell, 44(5), 745–758. 10.1016/j.molcel.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aseev, L. V. , Koledinskaya, L. S. , & Boni, I. V. (2016). Regulation of ribosomal protein operons rplM‐rpsI, rpmB‐rpmG, and rplU‐rpmA at the transcriptional and translational levels. Journal of Bacteriology, 198(18), 2494–2502. 10.1128/JB.00187-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrinidis, A. , Kim, J. , Kelly, C. M. , Olofsson, B. A. , Torabi, B. , Sorokina, E. M. , & Azizkhan‐Clifford, J. (2010). The transcription factor SP1 regulates centriole function and chromosomal stability through a functional interaction with the mammalian target of rapamycin/raptor complex. Genes, Chromosomes and Cancer, 49(3), 282–297. 10.1002/gcc.20739 [DOI] [PubMed] [Google Scholar]
- Bahler, J. (2005). A transcriptional pathway for cell separation in fission yeast. Cell Cycle, 4(1), 39–41. [DOI] [PubMed] [Google Scholar]
- Ben‐Shem, A. , Garreau de Loubresse, N. , Melnikov, S. , Jenner, L. , Yusupova, G. , & Yusupov, M. (2011). The structure of the eukaryotic ribosome at 3.0 a resolution. Science, 334(6062), 1524–1529. 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- Berger, A. B. , Decourty, L. , Badis, G. , Nehrbass, U. , Jacquier, A. , & Gadal, O. (2007). Hmo1 is required for TOR‐dependent regulation of ribosomal protein gene transcription. Molecular and Cellular Biology, 27(22), 8015–8026. 10.1128/MCB.01102-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedka, S. , Wu, S. , LaPeruta, A. J. , Gao, N. , & Woolford, J. L., Jr. (2017). Insights into remodeling events during eukaryotic large ribosomal subunit assembly provided by high resolution cryo‐EM structures. Catalysis, 14(10), 1306–1313. 10.1080/15476286.2017.1297914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D. L. (2000). Protein diversity from alternative splicing: A challenge for bioinformatics and post‐genome biology. Cell, 103(3), 367–370. [DOI] [PubMed] [Google Scholar]
- Blencowe, B. J. (2006). Alternative splicing: New insights from global analyses. Cell, 126(1), 37–47. 10.1016/j.cell.2006.06.023 [DOI] [PubMed] [Google Scholar]
- Bollen, A. (1980). Genetics, structure and function of bacterial ribosomes. Bulletin et Memoires de l Academie Royale de Medecine de Belgique, 135(6), 382–430. [PubMed] [Google Scholar]
- Bosio, M. C. , Negri, R. , & Dieci, G. (2011). Promoter architectures in the yeast ribosomal expression program. Transcription, 2(2), 71–77. 10.4161/trns.2.2.14486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, J. W. , & Dinman, J. D. (2017). Subtractional heterogeneity: A crucial step toward defining specialized ribosomes. Molecular Cell, 67(1), 3–4. 10.1016/j.molcel.2017.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S. J. , Cole, M. D. , & Erives, A. J. (2008). Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics, 9, 442. 10.1186/1471-2164-9-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumwell, A. , Fell, L. , Obress, L. , & Uniacke, J. (2020). Hypoxia influences polysome distribution of human ribosomal protein S12 and alternative splicing of ribosomal protein mRNAs. RNA, 26, 361–371. 10.1261/rna.070318.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, E. , Ghosh, S. , Matsuoka, Y. , Ashton‐Beaucage, D. , Therrien, M. , Lemieux, S. , … Kitano, H. (2010). A comprehensive map of the mTOR signaling network. Antineoplastic Agents/Pharmacology/Therapeutic Use, 6, 453. 10.1038/msb.2010.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker‐Margot, M. (2018). Assembly of the small ribosomal subunit in yeast: Mechanism and regulation. RNA, 24(7), 881–891. 10.1261/rna.066985.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti, M. , Dinkins, R. D. , & Hunt, A. G. (2018). Genome‐wide atlas of alternative polyadenylation in the forage legume red clover. Gene Expression Regulation, Plant, 8(1), 11379. 10.1038/s41598-018-29699-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champney, W. S. (2001). Bacterial ribosomal subunit synthesis: A novel antibiotic target. Anti‐Bacterial Agents/Pharmacology, 1(1), 19–36. [DOI] [PubMed] [Google Scholar]
- Chew, S. L. (1997). Alternative splicing of mRNA as a mode of endocrine regulation. Trends in Endocrinology and Metabolism, 8(10), 405–413 doi:S1043‐2760(97)00167‐7 [pii]. [DOI] [PubMed] [Google Scholar]
- Chorev, M. , & Carmel, L. (2012). The function of introns. Exon‐Junction Complex, 3, 55. 10.3389/fgene.2012.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmejla, R. , & Cmejlova, J. (2003). Molecular basis of diamond‐Blackfan anaemia: What have we learnt so far? Sbornik Lekarsky, 104(2), 171–181. [PubMed] [Google Scholar]
- Coenye, T. , & Vandamme, P. (2005). Organisation of the S10, spc and alpha ribosomal protein gene clusters in prokaryotic genomes. FEMS Microbiology Letters, 242(1), 117–126. 10.1016/j.femsle.2004.10.050 [DOI] [PubMed] [Google Scholar]
- Constantinou, C. , & Clemens, M. J. (2007). Regulation of translation factors eIF4GI and 4E‐BP1 during recovery of protein synthesis from inhibition by p53. Cell Death and Differentiation, 14(3), 576–585. [DOI] [PubMed] [Google Scholar]
- Cooper, T. A. (2005). Alternative splicing regulation impacts heart development. Cell, 120(1), 1–2. 10.1016/j.cell.2004.12.030 [DOI] [PubMed] [Google Scholar]
- Corral‐Debrinski, M. (2007). mRNA specific subcellular localization represents a crucial step for fine‐tuning of gene expression in mammalian cells. Biochimica et Biophysica Acta, 1773(4), 473–475. 10.1016/j.bbamcr.2006.06.008 [DOI] [PubMed] [Google Scholar]
- Cotney, J. , & Shadel, G. S. (2006). Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. Journal of Molecular Evolution, 63(5), 707–717. 10.1007/s00239-006-0075-1 [DOI] [PubMed] [Google Scholar]
- de la Cruz, J. , Gomez‐Herreros, F. , Rodriguez‐Galan, O. , Begley, V. , de la Cruz Munoz‐Centeno, M. , & Chavez, S. (2018). Feedback regulation of ribosome assembly. Current Genetics, 64(2), 393–404. doi: 10.1007/s00294-017-0764-x [DOI] [PubMed] [Google Scholar]
- Cuccurese, M. , Russo, G. , Russo, A. , & Pietropaolo, C. (2005). Alternative splicing and nonsense‐mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Research, 33(18), 5965–5977. 10.1093/nar/gki905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, H. S. , Sands, J. F. , Foreman, P. C. , Fraser, J. , & Hershey, J. W. (1991). Structure and expression of the infA operon encoding translational initiation factor IF1. Transcriptional control by growth rate. Journal of Biological Chemistry, 266(25), 16491–16498. [PubMed] [Google Scholar]
- Cunningham, J. T. , Rodgers, J. T. , Arlow, D. H. , Vazquez, F. , Mootha, V. K. , & Puigserver, P. (2007). mTOR controls mitochondrial oxidative function through a YY1‐PGC‐1alpha transcriptional complex. Nature, 450(7170), 736–740. 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]
- Cusanovich, D. A. , Pavlovic, B. , Pritchard, J. K. , & Gilad, Y. (2014). The functional consequences of variation in transcription factor binding. Evolution, Molecular, 10(3), e1004226. 10.1371/journal.pgen.1004226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Venezia, N. , Vincent, A. , Marcel, V. , Catez, F. , & Diaz, J. J. (2019). Emerging role of eukaryote ribosomes in translational control. International Journal of Molecular Sciences, 20(5), 1226. 10.3390/ijms20051226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin‐Kreiselman, M. , Lee, C. Y. , & Chanfreau, G. (2003). RNAse III‐mediated degradation of unspliced pre‐mRNAs and lariat introns. Molecular Cell, 11(5), 1279–1289. [DOI] [PubMed] [Google Scholar]
- David, L. , Huber, W. , Granovskaia, M. , Toedling, J. , Palm, C. J. , Bofkin, L. , … Steinmetz, L. M. (2006). A high‐resolution map of transcription in the yeast genome. Proceedings of the National Academy of Sciences of the United States of America, 103(14), 5320–5325. 10.1073/pnas.0601091103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, J. , & Nomura, M. (1972). The genetics of bacterial ribosomes. Annual Review of Genetics, 6, 203–234. 10.1146/annurev.ge.06.120172.001223 [DOI] [PubMed] [Google Scholar]
- Dean, E. J. , Davis, J. C. , Davis, R. W. , & Petrov, D. A. (2008). Pervasive and persistent redundancy among duplicated genes in yeast. Evolution, Molecular, 4(7), e1000113. 10.1371/journal.pgen.1000113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharia, A. P. , Obla, A. , Gajdosik, M. D. , Simon, A. , & Nelson, C. E. (2014). Tempo and mode of gene duplication in mammalian ribosomal protein evolution. Base Sequence, 9(11), e111721. 10.1371/journal.pone.0111721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman, J. D. (2016). Pathways to specialized ribosomes: The Brussels lecture. Journal of Molecular Biology, 428(10 Pt B), 2186–2194. 10.1016/j.jmb.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvir, S. , Velten, L. , Sharon, E. , Zeevi, D. , Carey, L. B. , Weinberger, A. , & Segal, E. (2013). Deciphering the rules by which 5'‐UTR sequences affect protein expression in yeast. Proceedings of the National Academy of Sciences of the United States of America, 110(30), E2792–E2801. 10.1073/pnas.1222534110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler, D. C. , & Craig, N. (1994). Processing of eukaryotic ribosomal RNA. Progress in Nucleic Acid Research and Molecular Biology, 49, 197–239. [DOI] [PubMed] [Google Scholar]
- El Motiam, A. , Vidal, S. , de la Cruz‐Herrera, C. F. , Da Silva‐Alvarez, S. , Baz‐Martinez, M. , Seoane, R. , … Rivas, C. (2019). Interplay between SUMOylation and NEDDylation regulates RPL11 localization and function. FASEB Journal, 33(1), 643–651. 10.1096/fj.201800341RR [DOI] [PubMed] [Google Scholar]
- Enganti, R. , Cho, S. K. , Toperzer, J. D. , Urquidi‐Camacho, R. A. , Cakir, O. S. , Ray, A. P. , … von Arnim, A. G. (2017). Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Arabidopsis, 8, 2210. 10.3389/fpls.2017.02210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evfratov, S. A. , Osterman, I. A. , Komarova, E. S. , Pogorelskaya, A. M. , Rubtsova, M. P. , Zatsepin, T. S. , … Dontsova, O. A. (2017). Application of sorting and next generation sequencing to study 5‐UTR influence on translation efficiency in Escherichia coli . Nucleic Acids Research, 45(6), 3487–3502. 10.1093/nar/gkw1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor, M. J. (2008). Alternative splicing minireview series: Combinatorial control facilitates splicing regulation of gene expression and enhances genome diversity. Journal of Biological Chemistry, 283(3), 1209–1210. 10.1074/jbc.R700046200 [DOI] [PubMed] [Google Scholar]
- Fenton, T. R. , & Gout, I. T. (2011). Functions and regulation of the 70kDa ribosomal S6 kinases. International Journal of Biochemistry and Cell Biology, 43(1), 47–59. 10.1016/j.biocel.2010.09.018 [DOI] [PubMed] [Google Scholar]
- Fermi, B. , Bosio, M. C. , & Dieci, G. (2017). Multiple roles of the general regulatory factor Abf1 in yeast ribosome biogenesis. Current Genetics, 63(1), 65–68. 10.1007/s00294-016-0621-3 [DOI] [PubMed] [Google Scholar]
- Ferretti, M. B. , & Karbstein, K. (2019). Does functional specialization of ribosomes really exist? RNA, 25(5), 521–538. 10.1261/rna.069823.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, G. E. (2010). Origin and evolution of the ribosome. Binding Sites/Genetics, 2(9), a003483. 10.1101/cshperspect.a003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman, L. P. , Zengel, J. M. , Archer, R. H. , & Lindahl, L. (1987). Autogenous control of the S10 ribosomal protein operon of Escherichia coli: Genetic dissection of transcriptional and posttranscriptional regulation. Proceedings of the National Academy of Sciences of the United States of America, 84(18), 6516–6520. 10.1073/pnas.84.18.6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. , Deiorio‐Haggar, K. , Anthony, J. , & Meyer, M. M. (2013). Most RNAs regulating ribosomal protein biosynthesis in Escherichia coli are narrowly distributed to Gammaproteobacteria. Nucleic Acids Research, 41(6), 3491–3503. 10.1093/nar/gkt055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabunilas, J. , & Chanfreau, G. (2016). Splicing‐mediated autoregulation modulates Rpl22p expression in Saccharomyces cerevisiae . Evolution, Molecular, 12(4), e1005999. 10.1371/journal.pgen.1005999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett, R. A. , Ungewickell, E. , Newberry, V. , Hunter, J. , & Wagner, R. (1977). An RNA core in the 30S ribosomal subunit of Escherichia coli and its structural and functional significance. Cell Biology International Reports, 1(6), 487–502. [DOI] [PubMed] [Google Scholar]
- Gazda, H. T. , & Sieff, C. A. (2006). Recent insights into the pathogenesis of Diamond‐Blackfan anaemia. British Journal of Haematology, 135(2), 149–157. 10.1111/j.1365-2141.2006.06268.x [DOI] [PubMed] [Google Scholar]
- Genuario, R. R. , Kelley, D. E. , & Perry, R. P. (1993). Comparative utilization of transcription factor GABP by the promoters of ribosomal protein genes rpL30 and rpL32. Gene Expression, 3(3), 279–288. [PMC free article] [PubMed] [Google Scholar]
- Genuth, N. R. , & Barna, M. (2018a). The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Molecular Cell, 71(3), 364–374. 10.1016/j.molcel.2018.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth, N. R. , & Barna, M. (2018b). Heterogeneity and specialized functions of translation machinery: From genes to organisms. Nature Reviews Genetics, 19(7), 431–452. 10.1038/s41576-018-0008-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman, B. W. , Pritchard, C. E. , Chaney, K. P. , & Ware, V. C. (2020). Tissue‐specific expression of ribosomal protein Paralogue eRpL22‐like in Drosophila melanogaster eye development. Developmental Dynamics, 249, 1147–1165. 10.1002/dvdy.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghulam, M. M. , Catala, M. , & Abou Elela, S. (2020). Differential expression of duplicated ribosomal protein genes modifies ribosome composition in response to stress. Nucleic Acids Research, 48(4), 1954–1968. 10.1093/nar/gkz1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco, P. , Wilson, D. , Kreitler, T. , Lehrach, H. , Klose, J. , Gobom, J. , & Fucini, P. (2005). High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Molecular Biology, 57(4), 577–591. 10.1007/s11103-005-0699-3 [DOI] [PubMed] [Google Scholar]
- Gill, E. E. , Lee, R. C. , Corradi, N. , Grisdale, C. J. , Limpright, V. O. , Keeling, P. J. , & Fast, N. M. (2010). Splicing and transcription differ between spore and intracellular life stages in the parasitic microsporidia. Molecular Biology and Evolution, 27(7), 1579–1584. 10.1093/molbev/msq050 [DOI] [PubMed] [Google Scholar]
- Gingras, A. C. , Raught, B. , & Sonenberg, N. (2004). mTOR signaling to translation. Current Topics in Microbiology and Immunology, 279, 169–197. [DOI] [PubMed] [Google Scholar]
- Goncalves, P. , Pereira, J. C. , Kidd, A. M. , & Ribeiro, M. L. (2004). Gene symbol: RPS19. Disease: Diamond‐Blackfan anaemia. Human Genetics, 115(6), 534. [PubMed] [Google Scholar]
- Goncalves, P. M. , Griffioen, G. , Minnee, R. , Bosma, M. , Kraakman, L. S. , Mager, W. H. , & Planta, R. J. (1995). Transcription activation of yeast ribosomal protein genes requires additional elements apart from binding sites for Abf1p or Rap1p. Nucleic Acids Research, 23(9), 1475–1480 doi:5c0027 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, A. , Shimobayashi, M. , Eisenberg, T. , Merle, D. A. , Pendl, T. , Hall, M. N. , & Moustafa, T. (2015). TORC1 promotes phosphorylation of ribosomal protein S6 via the AGC kinase Ypk3 in Saccharomyces cerevisiae . Base Sequence, 10(3), e0120250. 10.1371/journal.pone.0120250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressner, A. M. , & Wool, I. G. (1974). The phosphorylation of liver ribosomal proteins in vivo. Evidence that only a single small subunit protein (S6) is phosphorylated. Journal of Biological Chemistry, 249(21), 6917–6925. [PubMed] [Google Scholar]
- Grisdale, C. J. , Bowers, L. C. , Didier, E. S. , & Fast, N. M. (2013). Transcriptome analysis of the parasite Encephalitozoon cuniculi: An in‐depth examination of pre‐mRNA splicing in a reduced eukaryote. Algorithms, 14, 207. 10.1186/1471-2164-14-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishkevich, V. , & Yanai, I. (2014). Gene length and expression level shape genomic novelties. Genome Research, 24(9), 1497–1503. 10.1101/gr.169722.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati, R. K. , Neil, H. , Feuerbach, F. , Malabat, C. , & Jacquier, A. (2012). The yeast RPL9B gene is regulated by modulation between two modes of transcription termination. EMBO Journal, 31(10), 2427–2437. 10.1038/emboj.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes, J. C. , & Zavolan, M. (2016). Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biology, 17(1), 236. 10.1186/s13059-016-1104-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, V. , & Warner, J. R. (2014). Ribosome‐omics of the human ribosome. RNA, 20(7), 1004–1013. 10.1261/rna.043653.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D. B. , Wade, J. T. , & Struhl, K. (2006). An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae . Molecular and Cellular Biology, 26(9), 3672–3679. 10.1128/MCB.26.9.3672-3679.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, T. L. , Stoneley, M. , Spriggs, K. A. , & Bushell, M. (2006). TOPs and their regulation. Biochemical Society Transactions, 34(Pt 1, 12–16. 10.1042/BST20060012 [DOI] [PubMed] [Google Scholar]
- Hariharan, N. , Kelley, D. E. , & Perry, R. P. (1989). Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes and Development, 3(11), 1789–1800. 10.1101/gad.3.11.1789 [DOI] [PubMed] [Google Scholar]
- He, F. , Li, C. , Roy, B. , & Jacobson, A. (2014). Yeast Edc3 targets RPS28B mRNA for decapping by binding to a 3′ untranslated region decay‐inducing regulatory element. Molecular and Cellular Biology, 34(8), 1438–1451. 10.1128/MCB.01584-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks, K. B. , Delneri, D. , & Griffiths‐Jones, S. (2014). Intron evolution in Saccharomycetaceae. Genome Biology and Evolution, 6(9), 2543–2556. 10.1093/gbe/evu196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, L. E. , Bushell, M. , Barth‐Baus, D. , Tilleray, V. J. , Clemens, M. J. , & Hensold, J. O. (2002). p53 activation results in rapid dephosphorylation of the eIF4E‐binding protein 4E‐BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation. Oncogene, 21(34), 5325–5334. 10.1038/sj.onc.1205662 [DOI] [PubMed] [Google Scholar]
- Hsieh, A. C. , Liu, Y. , Edlind, M. P. , Ingolia, N. T. , Janes, M. R. , Sher, A. , … Ruggero, D. (2012). The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature, 485(7396), 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. , & Li, X. (2007). Transcriptional regulation in eukaryotic ribosomal protein genes. Genomics, 90(4), 421–423. 10.1016/j.ygeno.2007.07.003 [DOI] [PubMed] [Google Scholar]
- Hurowitz, E. H. , & Brown, P. O. (2003). Genome‐wide analysis of mRNA lengths in Saccharomyces cerevisiae . Genome Biology, 5(1), R2. 10.1186/gb-2003-5-1-r2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia, V. , Huo, Y. , Zhang, Z. , Foster, L. J. , & Proud, C. G. (2012). Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochemical Society Transactions, 40(1), 168–172. 10.1042/BST20110682 [DOI] [PubMed] [Google Scholar]
- Iadevaia, V. , Liu, R. , & Proud, C. G. (2014). mTORC1 signaling controls multiple steps in ribosome biogenesis. Seminars in Cell and Developmental Biology, 36, 113–120. 10.1016/j.semcdb.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Imami, K. , Milek, M. , Bogdanow, B. , Yasuda, T. , Kastelic, N. , Zauber, H. , … Selbach, M. (2018). Phosphorylation of the ribosomal protein RPL12/uL11 affects translation during mitosis. Molecular Cell, 72(1), 84–98 e89. 10.1016/j.molcel.2018.08.019 [DOI] [PubMed] [Google Scholar]
- Ishii, K. , Washio, T. , Uechi, T. , Yoshihama, M. , Kenmochi, N. , & Tomita, M. (2006). Characteristics and clustering of human ribosomal protein genes. Algorithms, 7, 37. 10.1186/1471-2164-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, T. (1988). Complete nucleotide sequence of the ribosomal 'A' protein operon from the archaebacterium, Halobacterium halobium. European Journal of Biochemistry, 176(2), 297–303. 10.1111/j.1432-1033.1988.tb14281.x [DOI] [PubMed] [Google Scholar]
- Ivanov, A. V. , Malygin, A. A. , & Karpova, G. G. (2005). Human ribosomal protein S26 suppresses the splicing of its pre‐mRNA. Biochimica et Biophysica Acta, 1727(2), 134–140. 10.1016/j.bbaexp.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Jablonska, A. , & Polouliakh, N. (2014). In silico discovery of novel transcription factors regulated by mTOR‐pathway activities. Bioinformatic Tools and Databases, 2, 23. 10.3389/fcell.2014.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, C. Y. , Shin, H. S. , Kim, H. D. , Kim, J. W. , Choi, S. Y. , & Kim, J. (2011). Ribosomal protein S3 is stabilized by sumoylation. Biochemical and Biophysical Research Communications, 414(3), 523–527. 10.1016/j.bbrc.2011.09.099 [DOI] [PubMed] [Google Scholar]
- Jansen, R. P. , Hurt, E. C. , Kern, H. , Lehtonen, H. , Carmo‐Fonseca, M. , Lapeyre, B. , & Tollervey, D. (1991). Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. Journal of Cell Biology, 113(4), 715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessica, P. , Malin, A. , Carl, S. , Tor, O. , Urban, G. , & Kristina, D. (2012). The p53 target gene TRIM22 directly or indirectly interacts with the translation initiation factor eIF4E and inhibits the binding of eIF4E to eIF4G. Biology of the Cell, 104(8), 462–475. 10.1111/boc.201100099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, B. S. , & Choi, S. S. (2015). Introns: The functional benefits of introns in genomes. Genomics & Informatics, 13(4), 112–118. 10.5808/GI.2015.13.4.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S. P. , & Warner, J. R. (1987). Phosphorylation of the Saccharomyces cerevisiae equivalent of ribosomal protein S6 has no detectable effect on growth. Molecular and Cellular Biology, 7(4), 1338–1345. 10.1128/mcb.7.4.1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone, O. , & Lasko, P. (2001). Translational regulation and RNA localization in Drosophila oocytes and embryos. Annual Review of Genetics, 35, 365–406. 10.1146/annurev.genet.35.102401.090756 [DOI] [PubMed] [Google Scholar]
- Jorgensen, P. , Rupes, I. , Sharom, J. R. , Schneper, L. , Broach, J. R. , & Tyers, M. (2004). A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes and Development, 18(20), 2491–2505. 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska, M. , & Ryden‐Aulin, M. (2007). Ribosome biogenesis and the translation process in Escherichia coli . Microbiology and Molecular Biology Reviews, 71(3), 477–494. 10.1128/MMBR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamita, M. , Kimura, Y. , Ino, Y. , Kamp, R. M. , Polevoda, B. , Sherman, F. , & Hirano, H. (2011). N(alpha)‐acetylation of yeast ribosomal proteins and its effect on protein synthesis. Journal of Proteomics, 74(4), 431–441. 10.1016/j.jprot.2010.12.007 [DOI] [PubMed] [Google Scholar]
- Kasahara, K. , Ohtsuki, K. , Ki, S. , Aoyama, K. , Takahashi, H. , Kobayashi, T. , … Kokubo, T. (2007). Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae . Molecular and Cellular Biology, 27(19), 6686–6705. 10.1128/MCB.00876-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. G. , Ireland, J. A. , Prem, S. M. , Chen, A. S. , & Ware, V. C. (2013). RpL22e, but not RpL22e‐like‐PA, is SUMOylated and localizes to the nucleoplasm of Drosophila meiotic spermatocytes. Nucleus, 4(3), 241–258. 10.4161/nucl.25261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaileh, A. , Dreazen, A. , Khatib, A. , Apel, R. , Swisa, A. , Kidess‐Bassir, N. , … Zamir, G. (2013). Phosphorylation of ribosomal protein S6 attenuates DNA damage and tumor suppression during development of pancreatic cancer. Cancer Research, 73(6), 1811–1820. 10.1158/0008-5472.CAN-12-2014 [DOI] [PubMed] [Google Scholar]
- Khatter, H. , Myasnikov, A. G. , Natchiar, S. K. , & Klaholz, B. P. (2015). Structure of the human 80S ribosome. Nature, 520(7549), 640–645. 10.1038/nature14427 [DOI] [PubMed] [Google Scholar]
- Kim, M. S. , & Hahn, J. S. (2016). Role of CK2‐dependent phosphorylation of Ifh1 and Crf1 in transcriptional regulation of ribosomal protein genes in Saccharomyces cerevisiae . Biochimica et Biophysica Acta, 1859(8), 1004–1013. 10.1016/j.bbagrm.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Kim, Y. , Lee, M. S. , Kim, H. D. , & Kim, J. (2016). Ribosomal protein S3 (rpS3) secreted from various cancer cells is N‐linked glycosylated. Oncotarget, 7(49), 80350–80362. 10.18632/oncotarget.10180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn‐Safran, C. B. , Dayal, S. , Martin‐DeLeon, P. A. , & Carson, D. D. (2000). Cloning, expression, and chromosome mapping of the murine hip/Rpl29 gene. Genomics, 68(2), 210–219. 10.1006/geno.2000.6283 [DOI] [PubMed] [Google Scholar]
- Knight, B. , Kubik, S. , Ghosh, B. , Bruzzone, M. J. , Geertz, M. , Martin, V. , … Shore, D. (2014). Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes and Development, 28(15), 1695–1709. 10.1101/gad.244434.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, Z. A. , Tan, K. , Birsoy, K. , Schmidt, S. , Garrison, J. L. , Wysocki, R. W. , … Friedman, J. M. (2012). Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell, 151(5), 1126–1137. 10.1016/j.cell.2012.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova, O. , Back, R. , Graille, M. , & Seraphin, B. (2013). Identification of the Rps28 binding motif from yeast Edc3 involved in the autoregulatory feedback loop controlling RPS28B mRNA decay. Nucleic Acids Research, 41(20), 9514–9523. 10.1093/nar/gkt607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komili, S. , Farny, N. G. , Roth, F. P. , & Silver, P. A. (2007). Functional specificity among ribosomal proteins regulates gene expression. Cell, 131(3), 557–571. 10.1016/j.cell.2007.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, N. , Pusic, A. , Stumpf, C. R. , Shimizu, K. , Hsieh, A. C. , Ishijima, J. , … Barna, M. (2011). Ribosome‐mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell, 145(3), 383–397. 10.1016/j.cell.2011.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsuwan, K. , Yu, Q. , Vincent, A. , Frisardi, M. C. , Rosbash, M. , Lengyel, J. A. , & Merriam, J. (1985). A Drosophila minute gene encodes a ribosomal protein. Nature, 317(6037), 555–558. 10.1038/317555a0 [DOI] [PubMed] [Google Scholar]
- Kotagama, K. , Babb, C. S. , Wolter, J. M. , Murphy, R. P. , & Mangone, M. (2015). A human 3'UTR clone collection to study post‐transcriptional gene regulation. BMC Genomics, 16, 1036. 10.1186/s12864-015-2238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreppel, L. , Fey, P. , Gaudet, P. , Just, E. , Kibbe, W. A. , Chisholm, R. L. , & Kimmel, A. R. (2004). dictyBase: A new Dictyostelium discoideum genome database. Nucleic Acids Research, 32(Database issue), D332–D333. 10.1093/nar/gkh138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar, V. , Contrino, S. , Cheng, C. Y. , Belyaeva, I. , Ferlanti, E. S. , Miller, J. R. , … Chan, A. P. (2017). ThaleMine: A warehouse for Arabidopsis data integration and discovery. Plant and Cell Physiology, 58(1), e4. 10.1093/pcp/pcw200 [DOI] [PubMed] [Google Scholar]
- Laferte, A. , Favry, E. , Sentenac, A. , Riva, M. , Carles, C. , & Chedin, S. (2006). The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes and Development, 20(15), 2030–2040. 10.1101/gad.386106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D. L. , & Tollervey, D. (2001). The function and synthesis of ribosomes. Nature Reviews Molecular Cell Biology, 2(7), 514–520. 10.1038/35080045 [DOI] [PubMed] [Google Scholar]
- Lavoie, H. , Hogues, H. , Mallick, J. , Sellam, A. , Nantel, A. , & Whiteway, M. (2010). Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biology, 8(3), e1000329. 10.1371/journal.pbio.1000329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. C. , Henry, B. , & Yeh, Y. C. (1983). Binding of proteins from the large ribosomal subunits to 5.8 S rRNA of Saccharomyces cerevisiae . Journal of Biological Chemistry, 258(2), 854–858. [PubMed] [Google Scholar]
- Li, D. , & Wang, J. (2020). Ribosome heterogeneity in stem cells and development. Journal of Cell Biology, 219(6), e202001108. 10.1083/jcb.202001108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zheng, Y. , Hu, H. , & Li, X. (2016). Integrative analyses shed new light on human ribosomal protein gene regulation. Gene Expression Regulation, Plant, 6, 28619. 10.1038/srep28619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zhong, S. , & Wong, W. H. (2005). Reliable prediction of transcription factor binding sites by phylogenetic verification. Proceedings of the National Academy of Sciences of the United States of America, 102(47), 16945–16950. 10.1073/pnas.0504201102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, X. H. , Majithia, A. , Huang, X. , & Kimmel, A. R. (2008). Growth control via TOR kinase signaling, an intracellular sensor of amino acid and energy availability, with crosstalk potential to proline metabolism. Amino Acids, 35(4), 761–770. 10.1007/s00726-008-0100-3 [DOI] [PubMed] [Google Scholar]
- Lin, Z. , & Li, W. H. (2012). Evolution of 5′ untranslated region length and gene expression reprogramming in yeasts. Molecular Biology and Evolution, 29(1), 81–89. 10.1093/molbev/msr143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, L. , & Zengel, J. M. (1986). Ribosomal genes in Escherichia coli . Annual Review of Genetics, 20, 297–326. 10.1146/annurev.ge.20.120186.001501 [DOI] [PubMed] [Google Scholar]
- Lipson, D. , Raz, T. , Kieu, A. , Jones, D. R. , Giladi, E. , Thayer, E. , … Causey, M. (2009). Quantification of the yeast transcriptome by single‐molecule sequencing. Nature Biotechnology, 27(7), 652–658. 10.1038/nbt.1551 [DOI] [PubMed] [Google Scholar]
- Loewith, R. , & Hall, M. N. (2011). Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics, 189(4), 1177–1201. 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , Zhu, Y. F. , Xiong, J. , Wang, R. , & Jia, Z. (2015). Potential extra‐ribosomal functions of ribosomal proteins in Saccharomyces cerevisiae . Microbiological Research, 177, 28–33. 10.1016/j.micres.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Ma, X. , Zhang, K. , & Li, X. (2009). Evolution of Drosophila ribosomal protein gene core promoters. Gene, 432(1–2), 54–59. 10.1016/j.gene.2008.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. M. , & Blenis, J. (2009). Molecular mechanisms of mTOR‐mediated translational control. Nature Reviews Molecular Cell Biology, 10(5), 307–318. 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- Macias, S. , Bragulat, M. , Tardiff, D. F. , & Vilardell, J. (2008). L30 binds the nascent RPL30 transcript to repress U2 snRNP recruitment. Molecular Cell, 30(6), 732–742. 10.1016/j.molcel.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Mageeney, C. M. , Kearse, M. G. , Gershman, B. W. , Pritchard, C. E. , Colquhoun, J. M. , & Ware, V. C. (2018). Functional interplay between ribosomal protein paralogues in the eRpL22 family in Drosophila melanogaster . Fly (Austin), 12(3–4), 143–163. 10.1080/19336934.2018.1549419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mageeney, C. M. , & Ware, V. C. (2019). Specialized eRpL22 paralogue‐specific ribosomes regulate specific mRNA translation in spermatogenesis in Drosophila melanogaster . Molecular Biology of the Cell, 30(17), 2240–2253. 10.1091/mbc.E19-02-0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager, W. H. , & Planta, R. J. (1991). Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Molecular and Cellular Biochemistry, 104(1–2), 181–187. 10.1007/BF00229818 [DOI] [PubMed] [Google Scholar]
- Maki, C. , Rhoads, D. D. , Stewart, M. J. , Van Slyke, B. , & Roufa, D. J. (1989). The Drosophila melanogaster RPS17 gene encoding ribosomal protein S17. Gene, 79(2), 289–298. 10.1016/0378-1119(89)90211-4 [DOI] [PubMed] [Google Scholar]
- Marcel, V. , Catez, F. , & Diaz, J. J. (2015). Ribosome heterogeneity in tumorigenesis: The rRNA point of view. Molecular & Cellular Oncology, 2(3), e983755. 10.4161/23723556.2014.983755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchioni, M. , Morabito, S. , Salvati, A. L. , Beccari, E. , & Carnevali, F. (1993). XrpFI, an amphibian transcription factor composed of multiple polypeptides immunologically related to the GA‐binding protein alpha and beta subunits, is differentially expressed during Xenopus laevis development. Molecular and Cellular Biology, 13(10), 6479–6489. 10.1128/mcb.13.10.6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D. E. , Soulard, A. , & Hall, M. N. (2004). TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell, 119(7), 969–979. 10.1016/j.cell.2004.11.047 [DOI] [PubMed] [Google Scholar]
- Martin, I. , Kim, J. W. , Lee, B. D. , Kang, H. C. , Xu, J. C. , Jia, H. , … Dawson, V. L. (2014). Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson's disease. Cell, 157(2), 472–485. 10.1016/j.cell.2014.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masvidal, L. , Hulea, L. , Furic, L. , Topisirovic, I. , & Larsson, O. (2017). mTOR‐sensitive translation: Cleared fog reveals more trees. RNA Biology, 14(10), 1299–1305. 10.1080/15476286.2017.1290041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J. S. (1994). Introns: Evolution and function. Current Opinion in Genetics and Development, 4(6), 823–831. [DOI] [PubMed] [Google Scholar]
- Mayer, C. , & Grummt, I. (2006). Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene, 25(48), 6384–6391. 10.1038/sj.onc.1209883 [DOI] [PubMed] [Google Scholar]
- Mazumder, B. , Sampath, P. , Seshadri, V. , Maitra, R. K. , DiCorleto, P. E. , & Fox, P. L. (2003). Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript‐specific translational control. Cell, 115(2), 187–198. 10.1016/s0092-8674(03)00773-6 [DOI] [PubMed] [Google Scholar]
- Melamed, D. , Pnueli, L. , & Arava, Y. (2008). Yeast translational response to high salinity: Global analysis reveals regulation at multiple levels. RNA, 14(7), 1337–1351. 10.1261/rna.864908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov, S. , Ben‐Shem, A. , Garreau de Loubresse, N. , Jenner, L. , Yusupova, G. , & Yusupov, M. (2012). One core, two shells: Bacterial and eukaryotic ribosomes. Nature Structural & Molecular Biology, 19(6), 560–567. 10.1038/nsmb.2313 [DOI] [PubMed] [Google Scholar]
- Mirabello, L. , Khincha, P. P. , Ellis, S. R. , Giri, N. , Brodie, S. , Chandrasekharappa, S. C. , … Savage, S. A. (2017). Novel and known ribosomal causes of diamond‐Blackfan anaemia identified through comprehensive genomic characterisation. Journal of Medical Genetics, 54(6), 417–425. 10.1136/jmedgenet-2016-104346 [DOI] [PubMed] [Google Scholar]
- Mitrovich, Q. M. , & Anderson, P. (2000). Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans . Genes and Development, 14(17), 2173–2184. 10.1101/gad.819900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta, K. , Hashimoto, T. , & Otaka, E. (1995). The evolutionary relationships between homologs of ribosomal YL8 protein and YL8‐like proteins. Current Genetics, 28(1), 19–25. [DOI] [PubMed] [Google Scholar]
- Molavi, G. , Samadi, N. , & Hosseingholi, E. Z. (2019). The roles of moonlight ribosomal proteins in the development of human cancers. Journal of Cellular Physiology, 234(6), 8327–8341. 10.1002/jcp.27722 [DOI] [PubMed] [Google Scholar]
- Moore, P. B. (1988). Structure‐function correlations in the small ribosomal subunit from Escherichia coli . Annual Review of Biophysics and Biophysical Chemistry, 17, 349–367. 10.1146/annurev.bb.17.060188.002025 [DOI] [PubMed] [Google Scholar]
- Moore, P. B. , & Steitz, T. A. (2003). The structural basis of large ribosomal subunit function. Annual Review of Biochemistry, 72, 813–850. 10.1146/annurev.biochem.72.110601.135450 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, R. , Ray, P. S. , Arif, A. , Brady, A. K. , Kinter, M. , & Fox, P. L. (2008). DAPK‐ZIPK‐L13a axis constitutes a negative‐feedback module regulating inflammatory gene expression. Molecular Cell, 32(3), 371–382. 10.1016/j.molcel.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis, A. , Lu, Z. , Zhan, Y. , Wang, T. Y. , Rodriguez, J. , Rajeh, A. , … Lin, Z. (2020). Parallel concerted evolution of ribosomal protein genes in Fungi and its adaptive significance. Molecular Biology and Evolution, 37(2), 455–468. 10.1093/molbev/msz229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao, A. , Yoshihama, M. , & Kenmochi, N. (2004). RPG: The ribosomal protein gene database. Nucleic Acids Research, 32(Database issue), D168–D170. 10.1093/nar/gkh004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, A. , & Tamanoi, F. (2010). Conservation of the Tsc/Rheb/TORC1/S6K/S6 signaling in fission yeast. Enzyme, 28, 167–187. 10.1016/S1874-6047(10)28008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar, R. N. , Sitz, T. O. , & Busch, H. (1974). Heterogeneity in the methylation and 5′ termini of Novikoff ascites hepatoma 5.8 S ribosomal RNA. FEBS Letters, 45(1), 206–212. [DOI] [PubMed] [Google Scholar]
- Neuveglise, C. , Marck, C. , & Gaillardin, C. (2011). The intronome of budding yeasts. Comptes Rendus Biologies, 334(8–9), 662–670. 10.1016/j.crvi.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Nomura, M. , & Morgan, E. A. (1977). Genetics of bacterial ribosomes. Annual Review of Genetics, 11, 297–347. 10.1146/annurev.ge.11.120177.001501 [DOI] [PubMed] [Google Scholar]
- Nusspaumer, G. , Remacha, M. , & Ballesta, J. P. (2000). Phosphorylation and N‐terminal region of yeast ribosomal protein P1 mediate its degradation, which is prevented by protein P2. EMBO Journal, 19(22), 6075–6084. 10.1093/emboj/19.22.6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary, M. N. , Schreiber, K. H. , Zhang, Y. , Duc, A. C. , Rao, S. , Hale, J. S. , … Kennedy, B. K. (2013). The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genetics, 9(8), e1003708. 10.1371/journal.pgen.1003708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau, J. , Durand, M. , Morin, G. , Gagnon, J. , Lucier, J. F. , Wellinger, R. J. , … Elela, S. A. (2011). Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell, 147(2), 320–331. 10.1016/j.cell.2011.08.044 [DOI] [PubMed] [Google Scholar]
- Parenteau, J. , Durand, M. , Veronneau, S. , Lacombe, A. A. , Morin, G. , Guerin, V. , … Abou Elela, S. (2008). Deletion of many yeast introns reveals a minority of genes that require splicing for function. Molecular Biology of the Cell, 19(5), 1932–1941. 10.1091/mbc.E07-12-1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau, J. , Lavoie, M. , Catala, M. , Malik‐Ghulam, M. , Gagnon, J. , & Abou Elela, S. (2015). Preservation of gene duplication increases the regulatory Spectrum of ribosomal protein genes and enhances growth under stress. Cell Reports, 13(11), 2516–2526. 10.1016/j.celrep.2015.11.033 [DOI] [PubMed] [Google Scholar]
- Parenteau, J. , Maignon, L. , Berthoumieux, M. , Catala, M. , Gagnon, V. , & Abou Elela, S. (2019). Introns are mediators of cell response to starvation. Nature, 565(7741), 612–617. 10.1038/s41586-018-0859-7 [DOI] [PubMed] [Google Scholar]
- Peccarelli, M. , & Kebaara, B. W. (2014). Regulation of natural mRNAs by the nonsense‐mediated mRNA decay pathway. Eukaryotic Cell, 13(9), 1126–1135. 10.1128/EC.00090-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi, I. , Dubouloz, F. , Cameroni, E. , Wanke, V. , Roosen, J. , Winderickx, J. , & De Virgilio, C. (2003). TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Molecular Cell, 12(6), 1607–1613. [DOI] [PubMed] [Google Scholar]
- Perry, R. P. (2005). The architecture of mammalian ribosomal protein promoters. BMC Evolutionary Biology, 5, 15. 10.1186/1471-2148-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole, G. , Liuni, S. , Grillo, G. , Licciulli, F. , Mignone, F. , Gissi, C. , & Saccone, C. (2002). UTRdb and UTRsite: Specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Research, 30(1), 335–340. 10.1093/nar/30.1.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petibon, C. , Parenteau, J. , Catala, M. , & Elela, S. A. (2016). Introns regulate the production of ribosomal proteins by modulating splicing of duplicated ribosomal protein genes. Nucleic Acids Research, 44(8), 3878–3891. 10.1093/nar/gkw140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyretaillade, E. , Goncalves, O. , Terrat, S. , Dugat‐Bony, E. , Wincker, P. , Cornman, R. S. , … Peyret, P. (2009). Identification of transcriptional signals in Encephalitozoon cuniculi widespread among Microsporidia phylum: Support for accurate structural genome annotation. Algorithms, 10, 607. 10.1186/1471-2164-10-607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta, R. J. , & Raue, H. A. (1988). Control of ribosome biogenesis in yeast. Trends in Genetics, 4(3), 64–68. [DOI] [PubMed] [Google Scholar]
- Plocik, A. M. , & Guthrie, C. (2012). Diverse forms of RPS9 splicing are part of an evolving autoregulatory circuit. PLoS Genetics, 8(3), e1002620. 10.1371/journal.pgen.1002620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, T. , Dilova, I. , Chen, C. Y. , & Wedaman, K. (2004). Yeast TOR signaling: A mechanism for metabolic regulation. Current Topics in Microbiology and Immunology, 279, 39–51. [DOI] [PubMed] [Google Scholar]
- Preiss, T. , Baron‐Benhamou, J. , Ansorge, W. , & Hentze, M. W. (2003). Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nature Structural Biology, 10(12), 1039–1047. 10.1038/nsb1015 [DOI] [PubMed] [Google Scholar]
- Pruitt, K. D. , Tatusova, T. , Brown, G. R. , & Maglott, D. R. (2012). NCBI reference sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Research, 40(Database issue, D130–D135. 10.1093/nar/gkr1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendhran, J. , & Gunasekaran, P. (2011). Microbial phylogeny and diversity: Small subunit ribosomal RNA sequence analysis and beyond. Microbiological Research, 166(2), 99–110. 10.1016/j.micres.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Ramagopal, S. (1990). Induction of cell‐specific ribosomal proteins in aggregation‐competent nonmorphogenetic Dictyostelium discoideum. Biochemistry and Cell Biology, 68(11), 1281–1287. 10.1139/o90-190 [DOI] [PubMed] [Google Scholar]
- Ramagopal, S. , & Ennis, H. L. (1981). Regulation of synthesis of cell‐specific ribosomal proteins during differentiation of Dictyostelium discoideum. Proceedings of the National Academy of Sciences of the United States of America, 78(5), 3083–3087. 10.1073/pnas.78.5.3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopal, S. , & Ennis, H. L. (1982). Ribosomal protein synthesis during spore germination and vegetative growth in Dictyostelium discoideum. Journal of Biological Chemistry, 257(2), 1025–1031. [PubMed] [Google Scholar]
- Rao, S. , Peri, S. , Hoffmann, J. , Cai, K. Q. , Harris, B. , Rhodes, M. , … Wiest, D. L. (2019). RPL22L1 induction in colorectal cancer is associated with poor prognosis and 5‐FU resistance. PLoS One, 14(10), e0222392. 10.1371/journal.pone.0222392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reja, R. , Vinayachandran, V. , Ghosh, S. , & Pugh, B. F. (2015). Molecular mechanisms of ribosomal protein gene coregulation. Genes and Development, 29(18), 1942–1954. 10.1101/gad.268896.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riba, A. , Di Nanni, N. , Mittal, N. , Arhne, E. , Schmidt, A. , & Zavolan, M. (2019). Protein synthesis rates and ribosome occupancies reveal determinants of translation elongation rates. Proceedings of the National Academy of Sciences of the United States of America, 116(30), 15023–15032. 10.1073/pnas.1817299116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, J. D. (1991). Translational control during early development. BioEssays, 13(4), 179–183. 10.1002/bies.950130406 [DOI] [PubMed] [Google Scholar]
- Rudra, D. , Mallick, J. , Zhao, Y. , & Warner, J. R. (2007). Potential Interface between ribosomal protein production and pre‐rRNA processing. Molecular and Cellular Biology, 27(13), 4815–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Canada, C. , Kelleher, D. J. , & Gilmore, R. (2009). Cotranslational and posttranslational N‐glycosylation of polypeptides by distinct mammalian OST isoforms. Cell, 136(2), 272–283. 10.1016/j.cell.2008.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, A. , Russo, G. , Cuccurese, M. , Garbi, C. , & Pietropaolo, C. (2006). The 3′‐untranslated region directs ribosomal protein‐encoding mRNAs to specific cytoplasmic regions. Biochimica et Biophysica Acta, 1763(8), 833–843. 10.1016/j.bbamcr.2006.05.010 [DOI] [PubMed] [Google Scholar]
- Ruvinsky, I. , Katz, M. , Dreazen, A. , Gielchinsky, Y. , Saada, A. , Freedman, N. , … Meyuhas, O. (2009). Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One, 4, e5618. 10.1371/journal.pone.0005618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky, I. , Sharon, N. , Lerer, T. , Cohen, H. , Stolovich‐Rain, M. , Nir, T. , … Meyuhas, O. (2005). Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes and Development, 19(18), 2199–2211. 10.1101/gad.351605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeboe‐Larssen, S. , Urbanczyk Mohebi, B. , & Lambertsson, A. (1997). The Drosophila ribosomal protein L14‐encoding gene, identified by a novel minute mutation in a dense cluster of previously undescribed genes in cytogenetic region 66D. Molecular and General Genetics, 255(2), 141–151. 10.1007/s004380050482 [DOI] [PubMed] [Google Scholar]
- Sauert, M. , Temmel, H. , & Moll, I. (2015). Heterogeneity of the translational machinery: Variations on a common theme. Biochimie, 114, 39–47. 10.1016/j.biochi.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, E. , Coureux, P. D. , Monestier, A. , Dubiez, E. , & Mechulam, Y. (2019). Start codon recognition in eukaryotic and Archaeal translation initiation: A common structural core. International Journal of Molecular Sciences, 20(4), 939. 10.3390/ijms20040939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth, B. S. , Borovinskaya, M. A. , Hau, C. W. , Zhang, W. , Vila‐Sanjurjo, A. , Holton, J. M. , & Cate, J. H. (2005). Structures of the bacterial ribosome at 3.5 a resolution. Science, 310(5749), 827–834. 10.1126/science.1117230 [DOI] [PubMed] [Google Scholar]
- Shi, Z. , & Barna, M. (2015). Translating the genome in time and space: Specialized ribosomes, RNA regulons, and RNA‐binding proteins. Annual Review of Cell and Developmental Biology, 31, 31–54. 10.1146/annurev-cellbio-100814-125346 [DOI] [PubMed] [Google Scholar]
- Shi, Z. , Fujii, K. , Kovary, K. M. , Genuth, N. R. , Röst, H. L. , Teruel, M. N. , & Barna, M. (2017). Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome‐wide. Molecular Cell, 67(1), 71–83. 10.1016/j.molcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai, A. , Sadaie, M. , Shinmyozu, K. , & Nakayama, J. (2010). Methylation of ribosomal protein L42 regulates ribosomal function and stress‐adapted cell growth. Journal of Biological Chemistry, 285(29), 22448–22460. 10.1074/jbc.M110.132274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoff, I. , Moradi, H. , & Nygard, O. (2009). Functional characterization of ribosomal protein L15 from Saccharomyces cerevisiae . Current Genetics, 55(2), 111–125. 10.1007/s00294-009-0228-z [DOI] [PubMed] [Google Scholar]
- Simsek, D. , & Barna, M. (2017). An emerging role for the ribosome as a nexus for post‐translational modifications. Current Opinion in Cell Biology, 45, 92–101. 10.1016/j.ceb.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek, D. , Tiu, G. C. , Flynn, R. A. , Byeon, G. W. , Leppek, K. , Xu, A. F. , … Barna, M. (2017). The mammalian Ribo‐interactome reveals ribosome functional diversity and heterogeneity. Cell, 169(6), 1051–1065. 10.1016/j.cell.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamovits, C. H. , & Keeling, P. J. (2009). Evolution of ultrasmall spliceosomal introns in highly reduced nuclear genomes. Molecular Biology and Evolution, 26(8), 1699–1705. 10.1093/molbev/msp081 [DOI] [PubMed] [Google Scholar]
- Sleumer, M. C. , Wei, G. , Wang, Y. , Chang, H. , Xu, T. , Chen, R. , & Zhang, M. Q. (2012). Regulatory elements of Caenorhabditis elegans ribosomal protein genes. BMC Genomics, 13, 433. 10.1186/1471-2164-13-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinger, B. L. , Newman, H. , Lee, Y. , Pei, S. , & Meyer, M. M. (2015). Co‐evolution of bacterial ribosomal protein S15 with diverse mRNA regulatory structures. PLoS Genetics, 11(12), e1005720. 10.1371/journal.pgen.1005720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin, M. L. , & Gunderson, J. H. (1987). Structural diversity of eukaryotic small subunit ribosomal RNAs. Evolutionary Implications Annals of the New York Academy of Sciences, 503, 125–139. 10.1111/j.1749-6632.1987.tb40603.x [DOI] [PubMed] [Google Scholar]
- Song, Y. , Botvinnik, O. B. , Lovci, M. T. , Kakaradov, B. , Liu, P. , Xu, J. L. , & Yeo, G. W. (2017). Single‐cell alternative splicing analysis with expedition reveals splicing dynamics during neuron differentiation. Molecular Cell, 67(1), 148–161. 10.1016/j.molcel.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard, A. , Cremonesi, A. , Moes, S. , Schutz, F. , Jeno, P. , & Hall, M. N. (2010). The rapamycin‐sensitive phosphoproteome reveals that TOR controls protein kinase a toward some but not all substrates. Molecular Biology of the Cell, 21(19), 3475–3486. 10.1091/mbc.E10-03-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, A. T. , Howe, D. K. , & Hunt, A. G. (2018). Characterization of mRNA polyadenylation in the apicomplexa. Base Sequence, 13(8), e0203317. 10.1371/journal.pone.0203317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima, S. O. , Hofman, I. J. F. , De Keersmaecker, K. , & Dinman, J. D. (2017). How ribosomes translate Cancer. Cancer Discovery, 7(10), 1069–1087. 10.1158/2159-8290.CD-17-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen, C. C. , Chantranupong, L. , Keys, H. R. , Wang, T. , Gray, N. S. , & Sabatini, D. M. (2012). A unifying model for mTORC1‐mediated regulation of mRNA translation. Nature, 485(7396), 109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka, M. , Shimobayashi, M. , Kitabatake, M. , Ohno, M. , Kozutsumi, Y. , Oka, S. , & Takematsu, H. (2018). Ribosomal protein uS7/Rps5 serine‐223 in protein kinase‐mediated phosphorylation and ribosomal small subunit maturation. Scientific Reports, 8(1), 1244. 10.1038/s41598-018-19652-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turova, T. P. (2003). Copy number of ribosomal operons in prokaryotes and its effect on phylogenic analyses. Mikrobiologiia, 72(4), 437–452. [PubMed] [Google Scholar]
- Tycowski, K. T. , Shu, M. D. , & Steitz, J. A. (1996). A mammalian gene with introns instead of exons generating stable RNA products [see comments]. Nature, 379(6564), 464–466. [DOI] [PubMed] [Google Scholar]
- Vallejos Baier, R. , Picao‐Osorio, J. , & Alonso, C. R. (2017). Molecular regulation of alternative Polyadenylation (APA) within the Drosophila nervous system. Journal of Molecular Biology, 429(21), 3290–3300. 10.1016/j.jmb.2017.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardell, J. , & Warner, J. R. (1997). Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Molecular and Cellular Biology, 17(4), 1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, J. T. , Hall, D. B. , & Struhl, K. (2004). The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature, 432(7020), 1054–1058. 10.1038/nature03175 [DOI] [PubMed] [Google Scholar]
- Walczak, C. P. , Leto, D. E. , Zhang, L. , Riepe, C. , Muller, R. Y. , DaRosa, P. A. , … Kopito, R. R. (2019). Ribosomal protein RPL26 is the principal target of UFMylation. Proceedings of the National Academy of Sciences of the United States of America, 116(4), 1299–1308. 10.1073/pnas.1816202116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Dasgupta, I. , & Fox, G. E. (2009). Many nonuniversal archaeal ribosomal proteins are found in conserved gene clusters. Archaea, 2(4), 241–251. 10.1155/2009/971494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Xu, Y. , Rogers, H. , Saidi, L. , Noguchi, C. T. , Li, H. , … Ye, Y. (2020). UFMylation of RPL26 links translocation‐associated quality control to endoplasmic reticulum protein homeostasis. Cell Research, 30(1), 5–20. 10.1038/s41422-019-0236-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Yoshida, K. , Toki, T. , Sawada, T. , Uechi, T. , Okuno, Y. , … Ito, E. (2015). Loss of function mutations in RPL27 and RPS27 identified by whole‐exome sequencing in diamond‐Blackfan anaemia. British Journal of Haematology, 168(6), 854–864. 10.1111/bjh.13229 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Liu, C. L. , Storey, J. D. , Tibshirani, R. J. , Herschlag, D. , & Brown, P. O. (2002). Precision and functional specificity in mRNA decay. Proceedings of the National Academy of Sciences of the United States of America, 99(9), 5860–5865. 10.1073/pnas.092538799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. L. , Duttke, S. H. , Chen, K. , Johnston, J. , Kassavetis, G. A. , Zeitlinger, J. , & Kadonaga, J. T. (2014). TRF2, but not TBP, mediates the transcription of ribosomal protein genes. Genes and Development, 28(14), 1550–1555. 10.1101/gad.245662.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski, I. , Pfiffner, J. , French, C. , Socha, A. , Thompson, D. A. , & Regev, A. (2010). Gene duplication and the evolution of ribosomal protein gene regulation in yeast. Proceedings of the National Academy of Sciences of the United States of America, 107(12), 5505–5510. 10.1073/pnas.0911905107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J. R. (1989). Synthesis of ribosomes in Saccharomyces cerevisiae . Microbiological Reviews, 53(2), 256–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J. R. (1999). The economics of ribosome biosynthesis in yeast. Trends in Biochemical Sciences, 24(11), 437–440. [DOI] [PubMed] [Google Scholar]
- Warner, J. R. , Kumar, A. , Udem, S. A. , & Wu, R. S. (1973). Ribosomal proteins and the assembly of ribosomes in eukaryotes. Biochemical Society Symposia, 37(0), 3–22. [PubMed] [Google Scholar]
- Warner, J. R. , Vilardell, J. , & Sohn, J. H. (2001). Economics of ribosome biosynthesis. Cold Spring Harbor Symposia on Quantitative Biology, 66, 567–574. 10.1101/sqb.2001.66.567 [DOI] [PubMed] [Google Scholar]
- Weijers, D. , Franke‐van Dijk, M. , Vencken, R. J. , Quint, A. , Hooykaas, P. , & Offringa, R. (2001). An Arabidopsis minute‐like phenotype caused by a semi‐dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development, 128(21), 4289–4299. [DOI] [PubMed] [Google Scholar]
- Whelan, T. A. , Lee, N. T. , Lee, R. C. H. , & Fast, N. M. (2019). Microsporidian introns retained against a background of genome reduction: Characterization of an unusual set of introns. Genome Biology and Evolution, 11(1), 263–269. 10.1093/gbe/evy260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M. E. , & Sussex, I. M. (1995). Developmental regulation of ribosomal protein L16 genes in Arabidopsis thaliana . Plant Journal, 8(1), 65–76. 10.1046/j.1365-313x.1995.08010065.x [DOI] [PubMed] [Google Scholar]
- Wilson, D. N. , & Doudna Cate, J. H. (2012). The structure and function of the eukaryotic ribosome. Cold Spring Harbor Perspectives in Biology, 4(5), a011536. 10.1101/cshperspect.a011536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger, S. , Loewith, R. , & Hall, M. N. (2006). TOR signaling in growth and metabolism. Cell, 124(3), 471–484. 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Xiao, L. , & Grove, A. (2009). Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Current Genomics, 10(3), 198–205. 10.2174/138920209788185261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L. , Kamau, E. , Donze, D. , & Grove, A. (2011). Expression of yeast high mobility group protein HMO1 is regulated by TOR signaling. Gene, 489(1), 55–62. 10.1016/j.gene.2011.08.017 [DOI] [PubMed] [Google Scholar]
- Xiong, X. , Zhao, Y. , He, H. , & Sun, Y. (2011). Ribosomal protein S27‐like and S27 interplay with p53‐MDM2 axis s a target, a substrate, and a regulator. Oncogene, 30(15), 1798–1811. 10.1038/onc.2010.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, A. , & Barna, M. (2020). Cleaning up stalled ribosome‐translocon complexes with ufmylation. Cell Research, 30(1), 1–2. 10.1038/s41422-019-0249-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, S. , & Barna, M. (2012). Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nature Reviews Molecular Cell Biology, 13(6), 355–369. 10.1038/nrm3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, S. , Tian, T. , Fujii, K. , Kladwang, W. , Das, B. , & Barna, M. (2015). RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature, 517(7532), 33–38. 10.1038/nature14010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. I. , Hsieh, H. H. , & Shan, S. O. (2019). Timing and specificity of cotranslational nascent protein modification in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 116(46), 23050–23060. 10.1073/pnas.1912264116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Yang, C. , Farberman, A. , Rideout, T. C. , de Lange, C. F. , France, J. , & Fan, M. Z. (2008). The mammalian target of rapamycin‐signaling pathway in regulating metabolism and growth. Journal of Animal Science, 86(14 Suppl), E36–E50. 10.2527/jas.2007-0567 [DOI] [PubMed] [Google Scholar]
- Yanshina, D. D. , Bulygin, K. N. , Malygin, A. A. , & Karpova, G. G. (2015). Hydroxylated histidine of human ribosomal protein uL2 is involved in maintaining the local structure of 28S rRNA in the ribosomal peptidyl transferase center. The FEBS Journal, 282(8), 1554–1566. 10.1111/febs.13241 [DOI] [PubMed] [Google Scholar]
- Yofe, I. , Zafrir, Z. , Blau, R. , Schuldiner, M. , Tuller, T. , Shapiro, E. , & Ben‐Yehezkel, T. (2014). Accurate, model‐based tuning of synthetic gene expression using introns in S. cerevisiae . PLoS Genetics, 10(6), e1004407. 10.1371/journal.pgen.1004407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath, A. , & Franceschi, F. (1998). Functional universality and evolutionary diversity: Insights from the structure of the ribosome. Structure, 6(6), 679–684. 10.1016/s0969-2126(98)00069-0 [DOI] [PubMed] [Google Scholar]
- Yoshihama, M. , Nakao, A. , Nguyen, H. D. , & Kenmochi, N. (2006). Analysis of ribosomal protein gene structures: Implications for intron evolution. Evolution . PLoS Genetics, 2(3), e25. 10.1371/journal.pgen.0020025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama, M. , Nguyen, H. D. , & Kenmochi, N. (2007). Intron dynamics in ribosomal protein genes. PLoS One, 2(1), e141. 10.1371/journal.pone.0000141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama, M. , Uechi, T. , Asakawa, S. , Kawasaki, K. , Kato, S. , Higa, S. , … Kenmochi, N. (2002). The human ribosomal protein genes: Sequencing and comparative analysis of 73 genes. Genome Research, 12(3), 379–390. 10.1101/gr.214202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevi, D. , Sharon, E. , Lotan‐Pompan, M. , Lubling, Y. , Shipony, Z. , Raveh‐Sadka, T. , … Segal, E. (2011). Compensation for differences in gene copy number among yeast ribosomal proteins is encoded within their promoters. Genome Research, 21(12), 2114–2128. 10.1101/gr.119669.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Du, G. , Zhou, J. , & Chen, J. (2018). Regulation of sensing, transportation, and catabolism of nitrogen sources in Saccharomyces cerevisiae . Microbiology and Molecular Biology Reviews, 82(1), e00040–17. 10.1128/MMBR.00040-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Duc, A. C. , Rao, S. , Sun, X. L. , Bilbee, A. N. , Rhodes, M. , … Wiest, D. L. (2013). Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Developmental Cell, 24(4), 411–425. 10.1016/j.devcel.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , McIntosh, K. B. , Rudra, D. , Schawalder, S. , Shore, D. , & Warner, J. R. (2006). Fine‐structure analysis of ribosomal protein gene transcription. Molecular and Cellular Biology, 26(13), 4853–4862. 10.1128/MCB.02367-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski, G. , & Zurawski, S. M. (1985). Structure of the Escherichia coli S10 ribosomal protein operon. Nucleic Acids Research, 13(12), 4521–4526. 10.1093/nar/13.12.4521 [DOI] [PMC free article] [PubMed] [Google Scholar]


