Loss-of-function mutations in DNaseL13, the enzyme that restricts the amount of microparticle-associated DNA, cause SLE in humans and mice. In this issue of JEM, Hartl et al. uncover a reduction in plasma DNASE1L3 enzymatic activity due to the presence of autoantibodies in patients with non-familial SLE.
Abstract
Loss-of-function mutations in DNaseL13, the enzyme that restricts the amount of microparticle-associated DNA, cause SLE in humans and mice. In this issue of JEM, Hartl et al. (2021. J. Exp. Med. https://doi.org/10.1084/jem.20201138) uncover a reduction in plasma DNASE1L3 enzymatic activity due to the presence of autoantibodies in patients with nonfamilial SLE.
Systemic lupus erythematosus (SLE) is an autoimmune disease affecting predominantly women and presenting with a broad spectrum of clinical manifestations. Among a variety of immune alterations, autoantibodies (auto-Abs) to nuclear antigens (Ags), especially those recognizing double-stranded DNA (dsDNA), are a hallmark of this disease (Tsokos, 2011). While familial aggregation and monozygotic twin concordance are described, SLE normally presents as a sporadic disease resulting from the combined effect of multiple common genetic variants (Chen et al., 2017). Rare Mendelian or de novo mutations in a single gene also give rise to a lupus phenotype. To date, at least 36 of these have been characterized, including hypomorphic mutations in DNase1L3 (Demirkaya et al., 2020). First described 10 yr ago, DNASE1L3-deficient patients present with childhood onset disease characterized by a lack of gender bias and a high frequency of both anti-dsDNA Abs and lupus nephritis (LN; Al-Mayouf et al., 2011).

Insights from Zurong Wan and Virginia Pascual.
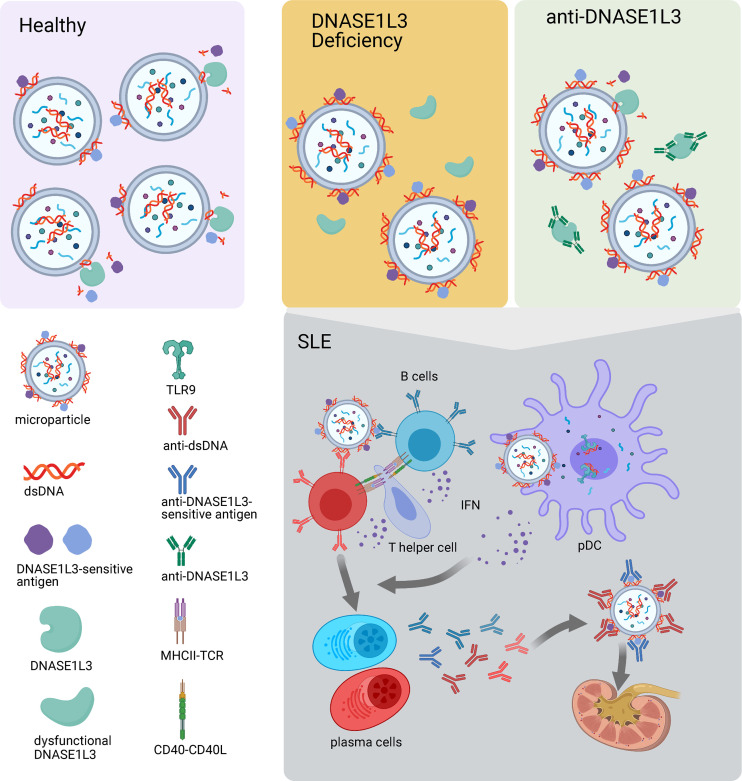
The work from Hartl et al. (2021) in this issue of JEM stems from the previous characterization by the same group of DNASE1L3 deficiency in mice. Consistent with the human phenotype, DNASE1L3-KO mice of different genetic backgrounds develop anti-chromatin and anti-dsDNA Abs in the first weeks of life irrespective of gender. SLE-like disease manifestations, including LN, appear, however, later in life and only in the context of a permissive background. The universal breakdown of tolerance to dsDNA in the absence of DNASE1L3 activity is remarkable and highlights the nonredundant role of this enzyme in chromatin-associated DNA fragmentation within apoptotic cell–derived microparticles (MPs). Thus, DNASE1L3 deficiency is enough to cause accumulation of and B cell responses to MP-associated DNA (Sisirak et al., 2016).
Using a novel functional assay, these authors now describe that DNASE1L3 enzymatic activity is reduced in plasma from more than 50% of patients with sporadic SLE and LN, and that this reduction correlates with the presence of neutralizing anti-DNASE1L3 auto-Abs (Hartl et al., 2021). As expected, cell-free DNA (cfDNA) associated with circulating MP, the physiological substrate of DNASE1L3, is increased in these patients while total cfDNA levels remain similar to those of controls. The authors go on to show that auto-Abs to DNASE1L3-sensitive Ags on MPs are also prevalent in patients with LN, especially those with evidence of active renal disease (see figure). Furthermore, the levels of these auto-Abs correlate with overall disease severity.
Auto-Ab–mediated inhibition of DNASE1L3 recapitulates its genetic deficiency and leads to increased levels of dsDNA and DNASE1L3-sensitive Ags on MPs. MPs may deliver these Ags to pDCs and induce IFN production, which, in combination with T cell costimulatory signals, promotes the differentiation of autoreactive B cells into plasma cells. Auto-Abs produced by these plasma cells form immune complexes, which deposit in kidney and contribute to tissue damage. Created with BioRender.com.
DNASE1L3 and DNASE1 are the main extracellular enzymes responsible for DNase activity in the circulation (Napirei et al., 2009). DNASE1L3 is unique in its capacity to digest membrane- and/or protein-associated DNA, including intact chromatin (Sisirak et al., 2016; Napirei et al., 2005; Wilber et al., 2002). In fact, Ab-mediated inhibition of plasma DNase activity had been described before in LN (Bruschi et al., 2020), but this paper is the first to demonstrate that these Abs mainly recognize DNASE1L3. Similarly, MPs have long been recognized as antigenic targets of Abs to DNA and nucleosomes in SLE (Ullal et al., 2011), but this group is the first to characterize “DNASE1L3-sensitive Ags” on MPs. Among them, they identify HMGB1, one of the major DNA-associated nonhistone proteins. Overall, this work highlights the relevance of the extracellular DNA degradation pathway as a safeguard to maintain tolerance to DNA and DNA-associated proteins beyond the rare genetic DNASE1L3 deficiency.
MPs are extracellular vesicles of 100–1,000 nm, derived from plasma membrane of different origins, including apoptotic cells (Piccin et al., 2007). Apoptosis-induced MPs not only enclose DNA as cargo but also present it on the surface, where it becomes accessible to anti-DNA Abs (Reich and Pisetsky, 2009). In addition to reporting that patients with LN display a significant increase in the MP cfDNA fraction, Hartl et al. (2021) show that this fraction is enriched in longer polynucleosomal fragments, which bind auto-Abs with higher affinity compared with soluble DNA and are more efficient at inducing IFN production by plasmacytoid dendritic cells (pDCs), a well-known pathogenic player in human SLE (Pascual et al., 2006). Compared with its soluble counterpart (Carrasco and Batista, 2006), engagement with MP membrane-bound DNA is also more effective at inducing Ag receptor signaling in B cells and contributes to autoreactive B cell survival and plasma cell differentiation. MP cfDNA may also engage in the formation of immune complexes, which, through their interactions with opsonic receptors such as the Fc and complement receptors, contribute to LN. Last but not least, through their capacity to convey their cargos to recipient cells by membrane fusion, micropinocytosis, or receptor-mediated endocytosis (Burbano et al., 2015), MPs may serve as a vehicle to internalize nucleic acids and their binding proteins (Mobarrez et al., 2019) and access TLRs and non-TLR pattern recognition receptors in multiple subcellular compartments.
LN, a major risk factor for overall morbidity and mortality in SLE, includes a heterogeneous group of conditions that can only be properly classified through invasive procedures such as a kidney biopsy (Almaani et al., 2017). The search for surrogate biomarkers to classify LN and monitor its progression is therefore an area of great interest. Hartl et al. (2021) bring a series of novel readouts that correlate with LN and proteinuria: (i) DNASE1L3 activity, (ii) Abs to DNASE1L3, (iii) MP-associated cfDNA, and (iv) Abs to DNASE1L3-sensitive Ags on MP. Among these, they propose the latter as a relatively simple and high-throughput assay with potential diagnostic value. While the preliminary data looks encouraging, validation of such an assay would require larger patient cohorts and well-annotated longitudinal clinical data to evaluate its sensitivity and specificity. Moreover, IgG binding to DNASE1L3-sensitive Ags on MPs and other biomarkers here described have not been correlated with histological LN classes, which carry different prognosis and respond differently to therapy.
Auto-Ab–mediated inhibition of DNASE1L3 is reminiscent of previously reported auto-Abs against disease-relevant targets that recapitulate their genetic deficiency. As noted by the authors, an example is C1q deficiency, one of the strongest genetic risk factors for early-onset SLE. Thus anti-C1q Abs, which reduce the levels of C1q and impair the clearance of dying cells, are associated with LN (Leffler et al., 2014). Interestingly, no correlation was found between the presence of anti-C1q and DNASE1L3 auto-Abs in the patients studied by Hartl et al. (2021) Beyond SLE, Bastard et al. (2020) recently reported that ∼10% of patients with life-threatening coronavirus disease 2019 pneumonia had neutralizing auto-Abs against type I IFNs, a B cell autoimmune phenocopy of inborn errors of type I IFN immunity that predispose to severe viral infections. Most SLE auto-Ab specificities, however, have not been identified as genetic risk factors. Ongoing technological progress, enabling high-throughput screening of thousands of auto-Ags in parallel, is paving the way to discovering novel auto-Abs at unprecedented speed (Ayoglu et al., 2016; Zandian et al., 2017). By 2014, for example, a total of 180 auto-Abs had been associated with SLE, among which 102 were found to correlate with severity of one or multiple clinical manifestations (Yaniv et al., 2015). Intriguinly, while a few auto-Ab specificities are known to play a pathogenic role in SLE (Ruiz-Irastorza et al., 2010), a significant fraction of auto-Abs in these patients recognize intracellular signaling proteins, making it unlikely that they play a direct role in pathogenesis and tissue damage. Clustering patients according to their auto-Ab specificities, however, classified them into groups with unique subphenotype characteristics and/or organ involvement, supporting their biomarker value (Lewis et al., 2018).
A number of questions arise from this work, including whether auto-Ab–mediated reduction in DNASE1L3 activity is upstream of the breakdown of tolerance to DNA in sporadic SLE patients. Chronologically, a number of SLE auto-Abs are detected up to a decade before clinical disease onset, with those against dsDNA appearing on average ∼2 yr before diagnosis (Arbuckle et al., 2003). Whether anti-DNASE1L3 Abs precede those to dsDNA remains to be determined, but in the patients studied by Hartl et al. (2021) their levels did not seem to correlate. As an alternative scenario, autoreactivity against MP-associated Ags may appear first, eventually facilitating the uptake of DNASE1L3 and its substrate–MP-associated DNA by B cells and other antigen-presenting cells. Interestingly, in the small cohort of anti-Ro/SSA–positive mothers of children born with heart block reported by Hartl et al. (2021), auto-Ab–mediated reduction in DNASE1L3 activity seemed a promising biomarker of progression to overt SLE and LN. Overall, as exemplified in the manuscript by Hartl et al. (2021), characterizing the Ab-accessible SLE autoantigen repertoire of extracellular and transmembrane proteins might be a low-hanging fruit in the search for pathogenic drivers of sporadic SLE.
References
- Al-Mayouf, S.M., et al. 2011. Nat. Genet. 10.1038/ng.975 [DOI] [Google Scholar]
- Almaani, S., et al. 2017. Clin. J. Am. Soc. Nephrol. 10.2215/CJN.05780616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbuckle, M.R., et al. 2003. N. Engl. J. Med. 10.1056/NEJMoa021933 [DOI] [Google Scholar]
- Ayoglu, B., et al. 2016. Bioanalysis. 10.4155/bio.16.31 [DOI] [PubMed] [Google Scholar]
- Bastard, P., et al. 2020. Science. 10.1126/science.abd4585 [DOI] [Google Scholar]
- Bruschi, M., et al. 2020. J. Rheumatol. 10.3899/jrheum.181232 [DOI] [Google Scholar]
- Burbano, C., et al. 2015. Mediators Inflamm. 10.1155/2015/267590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco, Y.R., and Batista F.D.. 2006. Curr. Opin. Immunol. 10.1016/j.coi.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Chen, L., et al. 2017. Curr. Opin. Rheumatol. 10.1097/BOR.0000000000000411 [DOI] [Google Scholar]
- Demirkaya, E., et al. 2020. J. Clin. Med. 10.3390/jcm9030712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, J., et al. 2021. J. Exp. Med. 10.1084/jem.20201138 [DOI] [Google Scholar]
- Leffler, J., et al. 2014. Ann. Rheum. Dis. 10.1136/annrheumdis-2014-205287 [DOI] [PubMed] [Google Scholar]
- Lewis, M.J., et al. 2018. J. Autoimmun. 10.1016/j.jaut.2018.02.009 [DOI] [Google Scholar]
- Mobarrez, F., et al. 2019. J. Autoimmun. 10.1016/j.jaut.2019.05.003 [DOI] [Google Scholar]
- Napirei, M., et al. 2005. Biochem. J. 10.1042/BJ20042124 [DOI] [Google Scholar]
- Napirei, M., et al. 2009. FEBS J. 10.1111/j.1742-4658.2008.06849.x [DOI] [PubMed] [Google Scholar]
- Pascual, V., et al. 2006. Curr. Opin. Immunol. 10.1016/j.coi.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Piccin, A., et al. 2007. Blood Rev. 10.1016/j.blre.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Reich, C.F. III, and Pisetsky D.S.. 2009. Exp. Cell Res. 10.1016/j.yexcr.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Ruiz-Irastorza, G., et al. 2010. Lancet. 10.1016/S0140-6736(10)60709-X [DOI] [Google Scholar]
- Sisirak, V., et al. 2016. Cell. 10.1016/j.cell.2016.05.034 [DOI] [Google Scholar]
- Tsokos, G.C. 2011. N. Engl. J. Med. 10.1056/NEJMra1100359 [DOI] [Google Scholar]
- Ullal, A.J., et al. 2011. J. Autoimmun. 10.1016/j.jaut.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Wilber, A., et al. 2002. Mol. Ther. 10.1006/mthe.2002.0625 [DOI] [PubMed] [Google Scholar]
- Yaniv, G., et al. 2015. Autoimmun. Rev. 10.1016/j.autrev.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Zandian, A., et al. 2017. J. Proteome Res. 10.1021/acs.jproteome.6b00916 [DOI] [PubMed] [Google Scholar]