SUMMARY
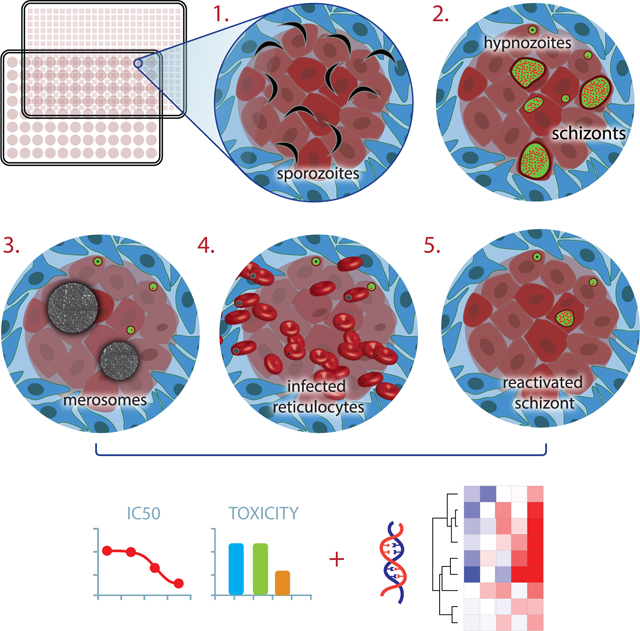
The unique relapsing nature of Plasmodium vivax infection is a major barrier to malaria eradication. Upon infection, dormant liver stage forms, hypnozoites, linger for weeks to months and then relapse to cause recurrent blood stage infection. Very little is known about hypnozoite biology; definitive biomarkers are lacking and in vitro platforms that support phenotypic studies are needed. Here, we recapitulate the entire liver stage of P. vivax in vitro, using a multiwell format that incorporates micropatterned primary human hepatocyte co-cultures (MPCCs). MPCCs feature key aspects of P. vivax biology, including establishment of persistent small forms and growing schizonts, merosome release, and subsequent infection of reticulocytes. We find that the small forms exhibit previously described hallmarks of hypnozoites, and we pilot MPCCs as a tool for testing candidate anti-hypnozoite drugs. Finally, we employ a hybrid capture strategy and RNA-seq to describe the hypnozoite transcriptome and gain insight into its biology.
eTOC Blurb
Plasmodium vivax hypnozoites are difficult to study due to lack of human liver platforms. Gural et al. recapitulated the entire liver stage of P. vivax in vitro, including formation and reactivation of hypnozoites and release of merosomes. Hybrid capture followed by RNAseq revealed a first look into the hypnozoite transcriptome.
Graphical Abstract
INTRODUCTION
Since it was first reported in 2700BC, the malaria parasite Plasmodium has successfully evaded all attempts at eradication. Combined, the two most prevalent human Plasmodium species put 3.2 billion people at risk of malaria infection (WHO, 2015). Plasmodium parasites enter the blood stream via the bite of an infected Anopheles mosquito, travel to the liver, and invade hepatocytes. In this obligate, yet clinically-silent stage, the invading sporozoites develop and replicate by schizogony, forming thousands of new haploid parasites called merozoites. Upon completion of the liver stage, merozoites are released into the blood to infect erythrocytes, initiating the cyclic and symptomatic blood stage. While Plasmodium falciparum is responsible for the majority of malaria-associated deaths, P. vivax presents a bigger barrier to eradication due to its propensity to cause chronic, relapsing disease weeks to years after the original infection. This species-specific aspect of P. vivax biology was discovered, only 3 decades ago, to be caused by a dormant liver stage form of the parasite, termed the hypnozoite. Originally identified in livers of rhesus monkeys infected with Plasmodium cynomolgi (Krotoski et al., 1980), the hypnozoite remains a relative biological mystery. In the absence of specific molecular or phenotypic markers, hypnozoites are generally described as small, uninucleate forms that persist for weeks to months after the initial infection (Krotoski et al., 1982), do not express late liver stage antigens, are sensitive to the only clinically-available hypnozoite-targeting drug (Dembele et al., 2011), and have the potential to relapse. Functionally, the cues that cause dormancy or promote reactivation are still poorly understood. This limited knowledge surrounding hypnozoite biology, due in large part to limited access to P. vivax sporozoites and the inability to establish primary human hepatocyte (PHH) cultures, has stymied drug development and represents a barrier to eradication. Today, the only clinically available hypnozoite-eliminating drug, primaquine, has an unknown mechanism of action and is contraindicated in a subset of the population in which a Glucose-6-dehydrogenase enzyme deficiency causes hemolysis upon administration of the drug (Wells et al., 2010). Moreover, increasing prevalence of drug resistance against blood stages (Price et al., 2014) underscores the urgent need for new liver stage-targeting agents, yet in order to develop new interventions, robust in vitro models that facilitate hypnozoite characterization, allow assessment of drug sensitivity, and help uncover cues that prompt both dormancy and reactivation are needed.
Historically, examples of successful in vitro culture of liver stage P. vivax are extremely limited. While P. vivax schizonts were first visualized in PHHs (Mazier et al., 1984), small forms were only reported in hepatoma cell lines, HepG2-A16 and HC04, after 9–14 days of culture (Chattopadhyay et al., 2010; Hollingdale et al., 1985)(Sattabongkot et al., 2006). Yet, the ongoing proliferation of infected hepatoma cells limited the utility of these platforms for the long-term assays necessary to interrogate P. vivax hypnozoite development. We have developed a microscale human liver platform that combines PHHs with supportive stromal cells in a multiwell micropatterned co-culture format (Khetani and Bhatia, 2008), where stable hepatocyte-specific function and metabolism is observed for 4–6 weeks. This platform supports infection with Hepatitis C and B viruses, P. falciparum and P. vivax (reviewed in (March et al., 2015)). In addition to providing a permissive host, hepatocytes cultured in MPCCs exhibit human-specific drug metabolism and long term stability, ideal traits for drug screening and studies of long-term dormancy and reactivation.
In this study, we progressed beyond our previous preliminary findings and confirmed the hypnozoite identity of small forms present in the MPCC system using clinical Thai P. vivax isolates. Specifically, we found that small forms exhibit the known hallmarks of hypnozoites in that they are small, uninucleate, persistent for weeks, negative for late stage liver antigens, sensitive to primaquine, and appear to have the capacity to reactivate. Furthermore, our multiwell in vitro platform enabled transcriptional characterization of both small and large P. vivax liver stage forms, by using a customized capture method prior to RNA sequencing. To demonstrate its potential as a drug screening platform, we compared the effectiveness of six clinical candidates in multi-point dosing. Finally, we have reengineered the MPCCs to support a 384-well format, paving the way for fully automated high throughput drug screening. Collectively, the data presented here highlight the potential of MPCCs to enhance our understanding of hypnozoite biology and advance drug development against this stage of the P. vivax life cycle.
RESULTS
Infection with clinical P. vivax isolates yields hypnozoites and schizonts in MPCCs
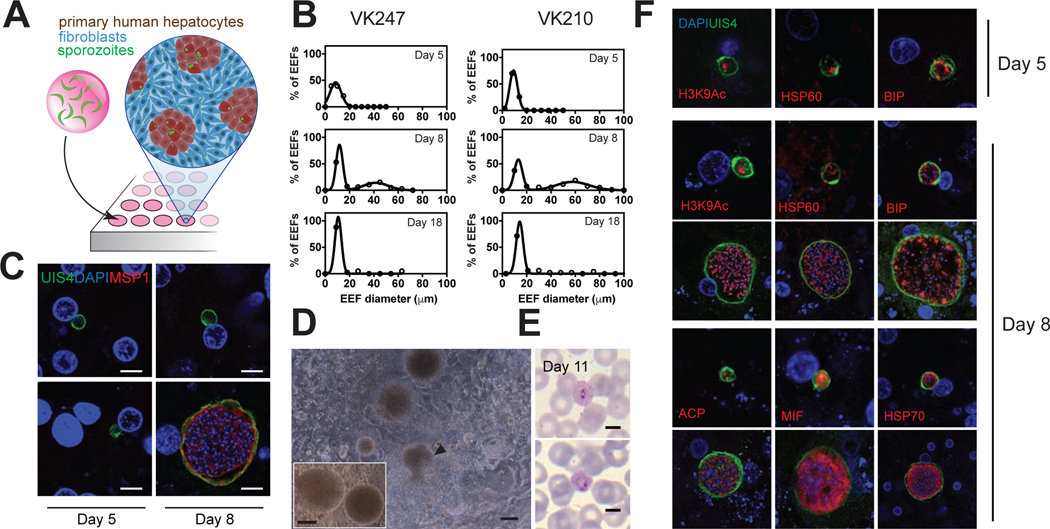
Previous work with P. vivax in the MPCCs has been performed with cryopreserved sporozoites passaged through monkeys. In order to establish liver stage hallmarks of clinical Thai P. vivax isolates, we infected MPCCs with sporozoites obtained from freshly dissected Anopheles dirus mosquitoes (Figure 1A). To assess growth kinetics, we fixed cultures over a series of time points, using two P. vivax subtypes observed in Thailand, VK210 and VK247, which differ in their central repeated region of the circumsporozoite protein (CSP) (Rosenberg et al., 1989). Both subtypes successfully infected PHHs, and gave rise to a subpopulation of exo-erythrocytic forms (EEFs) that grew in size, and a subpopulation of EEFs that remained small (Figure 1B). While day 5 forms were small, a bimodal separation of small and large forms became pronounced on day 8 for both subtypes tested. Consistent with clinical human data that reported P.vivax schizonts of up to 42 μm in diameter in a human liver biopsy (Shortt and Garnham, 1948), EEFs in day 8 MPCCs ranged from 7 to 80 μm in diameter. We further quantified day 10 forms and observed parasites as large as 130 μm in diameter (data not shown). To determine parasite development and maturation in vitro, we tested reactivity to an antibody against the merozoite surface protein 1 (MSP1) (Combe et al., 2009). At the earliest time point tested, 5 days post infection, no MSP1 expression was observed (Figure 1C), while on day 8 post infection, large parasites but not small forms were positive for MSP1. Remarkably, day 18 and 21 cultures of P. vivax sporozoites of both strains exhibited persistent small forms (Figures 1B and S1B) which, based on their size, morphology, lack of MSP1 expression, and lack of growth, were considered candidate hypnozoites.
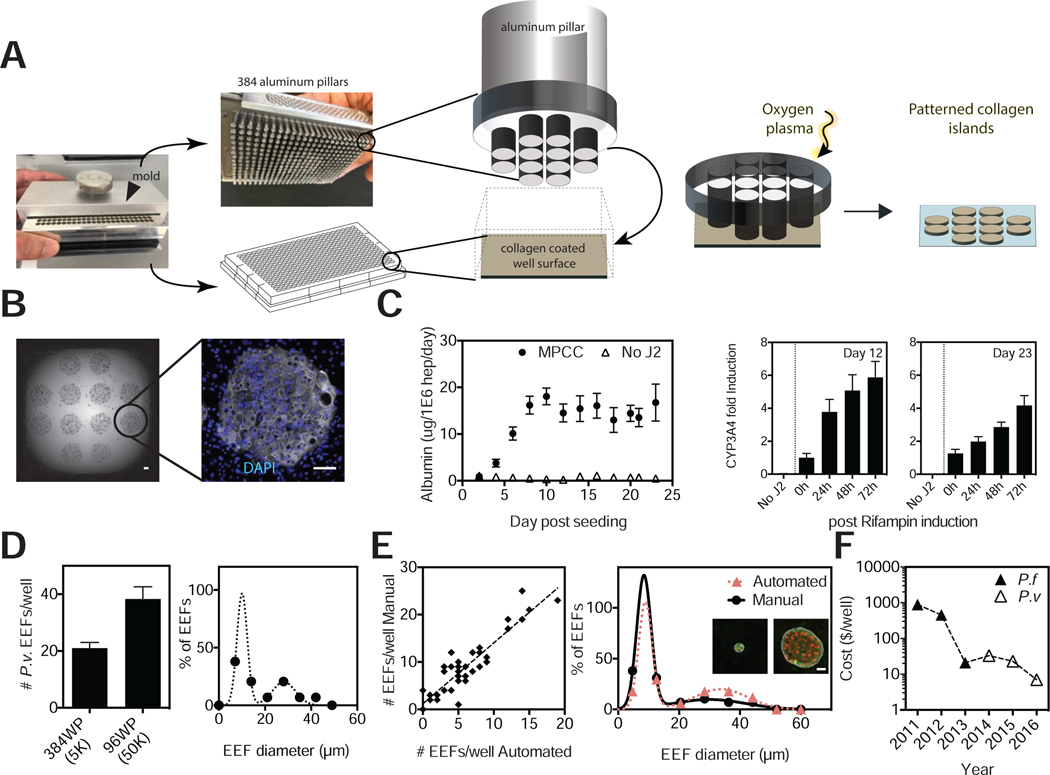
Figure 1: MPCCs Can Be infected With Clinical P. vivax Isolates.
(A) Schematic of the MPCC system where primary human hepatocyte (PHH) islands are patterned in 96-well plates. Sporozoites are overlaid onto cultures 1 day post seeding and incubated for 3 hours, followed by addition of mouse fibroblasts.
(B) Size distribution histograms of P. vivax EEFs in the MPCC system are shown after infection with 2 clinical isolates, performed in two separate experiments. VK247 experiment: day 5, 310 parasites; day 8, 199 parasites; day 18, 49 parasites. VK210 experiment: day 5, 207 parasites; day 8, 179 parasites; day 18, 41 parasites. At least 3 wells per time point pooled. See also Figure S1B.
(C) Representative images of small and large parasites on days 5 and 8, stained with an anti-MSP1 antibody. Scale bar, 5μm.
(D) Representative images of merozoite release in live P. vivax (VK210) cultures on day 12 (observed in five out of five wells). Released merosomes (white arrows) and merozoite-releasing merosome are shown (black arrow). Inset shows close up view of two merosomes on the same day. Scale bars, 50μm. See also Figure S1A and Supplementary Video 1–3.
(E) Representative images of P. vivax infected reticulocytes. Reticulocyte-enriched red blood cells were overlaid onto P. vivax-infected MPCCs, 10 days post-infection. Giemsa staining of collected blood cells revealed ring-stage parasites, one day later. Experiment performed 3 times by adding blood cells to at least 6 infected wells, all of which became positive for blood stage infection as assessed by blood smear. (6/1018, 10/1070 and 4/995 infected red blood cells counted) Scale bars, 5μm.
(F) P. vivax EEFs from MPCC cultures were fixed on days 5 and 8. Antibodies against UIS4, BIP, HSP60 and H3K9Ac were used to visualize parasite structures. Day 8 structures were further characterized with antibodies against HSP70, MIF and ACP. A representative small and large form is shown for all day 8 proteins. Scale bars, 5μm. See also Figures S1C and S2.
Upon completion of its liver stage development, Plasmodium parasites emerge from the host hepatocytes within membrane-bound vesicles; known as merosomes (Sturm et al., 2006). These structures contain the erythrocyte-invading merozoites. A hallmark of EEF maturation, merosome release, was observed in live cultures on days 11 and 12 (Figures 1D, S1A and Supplemental Videos 1–3). Furthermore, to confirm that MPCCs support the full liver stage life cycle of human malaria, MPCCs were overlaid with packed red blood cells, of which 33% were reticulocytes. Up to 1% of red blood cells (3% of reticulocytes) overlaid 10 days post infection became positive for ring stage P. vivax parasites, as assessed by analysis of Giemsa-stained smears on day 11 (Figure 1E). In two additional independent experiments, smears of overlaid reticulocytes were negative on day 8 post-infection but became positive on day 9.
To assay for longitudinal changes in cellular organelles in parasites, we used several antibodies targeted against P. vivax. Developing EEFs were visualized using an antibody that recognizes the parasitophorous vacuole membrane (PVM) (Mikolajczak et al., 2015). Over time, the PVM expanded, encapsulating the growing number of merozoites and isolating the parasite cytoplasm from that of the host (Figure 1F). Recently, in a humanized mouse model of P. vivax infection, a coalescence of UIS4, termed the ‘prominence’, has been proposed as a candidate hypnozoite marker, based on its exclusive observation in small forms found in sections of infected chimeric mouse livers (Mikolajczak et al., 2015). In contrast, in MPCC cultures, a UIS4 prominence is observed both in small forms and in a subpopulation of large forms (Figure S1C). The endoplasmic reticulum of the parasite was visualized using an anti-binding immunoglobulin protein (BIP) antibody (Noe et al., 2000) (Figure 1F). The structure appeared as a network that expanded in size and complexity as the parasite developed, and that included punctate bright spots of varying sizes. The mitochondria, visualized via the heat shock protein 60 (HSP60), formed a vast, scaffold-like network surrounding the individual nuclei of the EEFs on day 8 (Figure 1F). During the three-day growth period depicted, detection of the individual nuclei became difficult using the DAPI stain, especially in candidate hypnozoites, so we employed an additional nuclear antibody to track histone-acetylation marks (H3K9Ac), previously shown to stain P. vivax nuclei (Mikolajczak et al., 2015). While small forms maintained a single nuclear structure, growing schizonts contained numerous nuclei (Figure 1F). Day 8 forms were further visualized using antibodies against heat shock protein 70 (HSP70) and macrophage inhibitory factor (MIF) (Miller et al., 2012). The cytoplasm of the parasites expanded in growing schizonts compared to the small forms present at the same time point (Figure 1F). The apicoplast was visualized with an anti-acyl carrier protein (ACP) antibody and showed a complex, branched expression pattern in growing schizonts versus punctate spots in the small forms (Figure S2). Overall, cellular structures of the P. vivax EEFs became more complex over time.
Functional characterization of small forms fit pre-existing hypnozoite criteria
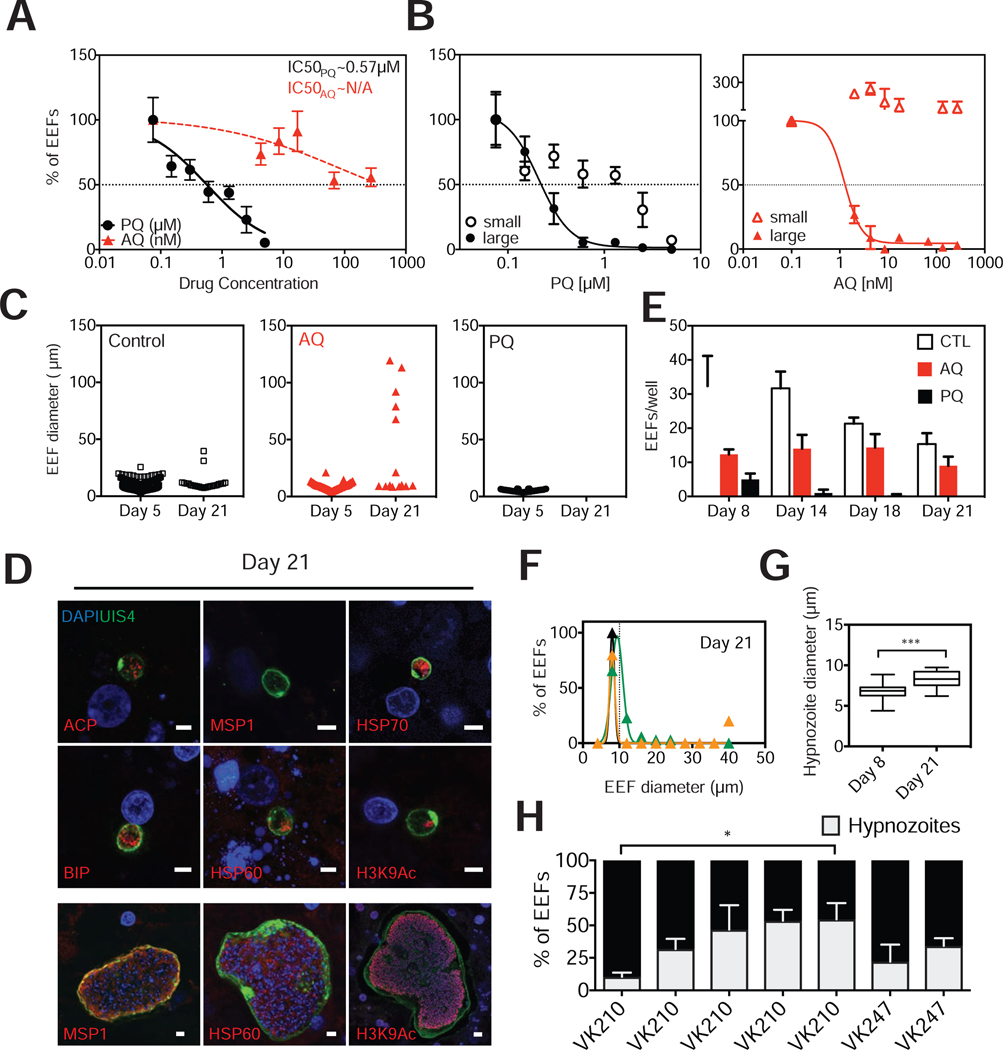
In addition to the size and kinetic characterization evidence that the small, persistent candidate hypnozoites identified in MPCCs are indeed bona fide dormant forms, we performed further functional characterization of the small forms. We treated P. vivax-infected MPCCs with two drugs that have been proposed to have differential hypnozoite-killing activity in clinical settings. We characterized the half-maximal inhibitory concentration (IC50) of primaquine and atovaquone on all forms, by assessing the number of parasites remaining in culture when exposed to a range of drug concentrations. Primaquine exhibited an IC50 of 0.32 μM (95% confidence interval; 0.26 to 0.4). In contrast, treatment with atovaquone, a drug that is clinically ineffective against hypnozoites, reduced the number of EEFs in culture, but not sufficiently to achieve an IC50, even at the highest concentrations tested (Figure 2A). Indeed, atovaquone eliminated the subpopulation of parasites that were larger than 10 μm in diameter, and left a residual subpopulation of only small forms. Primaquine, on the other hand, had killing activity against both small and large forms, consistent with its clinical use as the only available drug with activity against hypnozoites (Figure 2B).
Figure 2: Characterization of Drug Sensitivity, Size, Frequency, and Reactivation of Small Forms in MPCCs Match Hypnozoite Criteria.
(A) Primaquine (PQ) and atovaquone (AQ) were dosed at varying concentrations, starting 3 hours post infection (hpi) and replaced with daily media changes until day 5 when cultures were fixed (prophylactic mode). IC50 curves were produced by plotting fraction of EEFs remaining in culture at each concentration, as measured against untreated control wells. (mean±sem from triplicate wells, representative experiment shown, isolate VK210).
(B) Data from (a) was replotted, after segmenting the populations of parasites into ‘large’ and ‘small’ subpopulations, according to a 10μm diameter size threshold. Resulting curve fits show differential effect of PQ and AQ on small (<10 μm in diameter) and large (>10 μm in diameter) parasite populations.
(C) Cultures were dosed with PQ (5μM) or AQ (270nM) at 3 hpi until day 5. Cultures were maintained with daily media changes until day 21, when the sizes of any remaining parasites were assessed (n=3 wells per independent experiment. 3 experiments pooled for control, two experiments pooled for AQ, 1 experiment for PQ). See also Figures S3A and S4D.
(D) Representative hypnozoites and candidate reactivated schizonts in day 21 cultures, stained with a panel of antibodies, as indicated. Scale bar, 5μm. See also Figure S3C.
(E) Number of parasites remaining per well after treatment with PQ or AQ under the same treatment regimen as in (c) (mean±sem, n=3). See also Figure S4D.
(F) Size histograms of day 21 parasites to set a threshold for hypnozoite size. (3 independent experiments, triplicate wells per experiment).
(G) Hypnozoite diameters from day 8 and day 21 cultures (3 independent experiments, triplicate wells per experiment). *P=0.0004, two-tailed unpaired t-test with Welch’s correction.
(H) Hypnozoite frequency was evaluated in 6 separate experiments on day 8. CSP subtype of each experiment is indicated on the x-axis. (mean±sem shown from at least triplicate wells per experiment. Number of parasites interrogated in each experiment are as follows: 118,211,145,265,179,66,199) *Kruskall-Wallis test
Day 21 MPCC cultures revealed not only persistent hypnozoites, but also a collection of large schizonts (Figure 2C). The observation of large forms that appear beyond the first wave of merosome release is consistent with the interpretation that they originate from reactivated hypnozoites. These forms exhibited similar size ranges and morphologies as day 8 schizonts, based on their staining pattern with antibodies against MSP1, HSP60, and H3K9Ac (Figure 2D). To support the hypothesis that the large forms detected on day 21 represent reactivated small forms, we treated cultures prophylactically with atovaquone for five days in an attempt to deplete large forms. These pre-treated cultures also contained large forms on day 21, consistent with possible reactivation events, where large forms derive from previously dormant hypnozoites (Figure 2C). In an alternative approach, we treated cultures with a different schizonticide from days 5 to 8. While six days beyond the last drug treatment, on day 14, cultures solely exhibited small forms, on day 18, one of the cultures revealed re-emergence of large forms (Figure S3A). In contrast, prophylactic primaquine treatment of cultures completely cleared all parasites by day 21 (Figures 2C and 2E). Thus, in the MPCC system, persistent small forms that lack MSP1 expression also display characteristic functional traits of dormant forms: they are differentially drug sensitive to primaquine versus atovaquone, and appear to have the capacity to reactivate. We believe this set of phenotypic attributes supports their classification as bona fide hypnozoites.
In the course of our efforts to track the dynamic phenotype of P. vivax EEFs in MPCCs, our kinetic analysis has revealed that in addition to their capacity for reactivation, and despite their dormant appearance, hypnozoite sizes increase slightly over time (7 to 10 μm; Figures 2G and S3B), consistent with a previous description (Mikolajczak et al., 2015). Finally, the relative incidence of hypnozoite forms that arise after infection with a given P. vivax sporozoite pool was quantified. In our cultures, hypnozoite frequencies measured on day 8 ranged from 11 to 48%, and were independent of the CSP subtype of the parasite (Figure 2H).
Hypnozoites cultured in the MPCCs provide an antimalarial testing platform
An important clinical reality faced by providers is that primaquine sensitivity of patients is variable, in part due to differences in CYP2D6 metabolism, the enzyme complex thought to be primarily responsible for primaquine bioactivation, and thus not all patients respond similarly to primaquine (Bennett et al., 2013). The MPCC system has the potential to more closely predict not only clinical outcomes related to treatment with drugs such as primaquine that require adequate metabolic activity, but also liver toxicity in donor contexts which is a major problem in clinical drug development (Kaplowitz, 2005).
In order to query whether the MPCC platform can detect patient-specific variations in drug responsiveness, we interrogated the primaquine IC50 values obtained using different donors. MPCCs established with PHHs isolated from two different human donors were infected with the same P. vivax clinical isolate and showed a 6-fold difference in their responsiveness to primaquine (Figure 3A). This result is consistent with a 2-fold change in CYP2D6 activity between the two PHHs, in that the more sensitive cells exhibit 2-fold higher CYP2D6 activity (Figure S4A). Thus, infection of MPCCs can model complex patient-specific phenotypes that arise from a combination of host biology and drug metabolism. Notably, only relatively common genotypes are available using PHHs. For rare genotypes, induced pluripotent stem cells provide a complementary platform to study drug efficacy in a defined host (Berger et al., 2015; Ng et al., 2015).
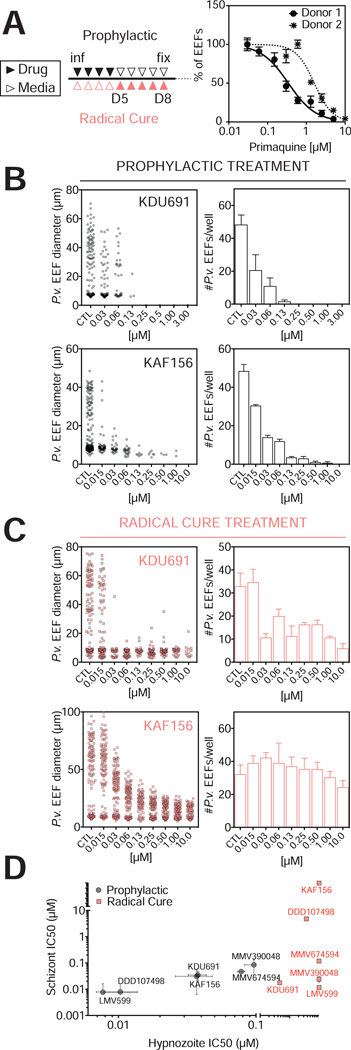
Figure 3: In vitro Hypnozoites Serve as an Antimalarial Testing Platform.
(A) Left: Representative dosing regimens for the drugs tested. Drug is added to cultures (filled triangles), or cultures are kept with daily drug-free media changes (open triangles). Prophylactic dosing begins 3hpi (black); radical cure dosing begins 5 days pi (red). Right: Prophylactic PQ dosing. PHHs with different CYP2D6 activities were used to create MPCCs that were infected with the same P. vivax clinical isolate (VK210). IC50 curves were produced by plotting fraction of EEFs remaining in culture at each concentration, as measured against untreated control wells (mean±sem, n=3, two donors were tested in three independent experiments). Donor 1 is used for all subsequent experiments.
(B) KDU691 and KAF156 were dosed following the prophylactic regimen in (A), and on day 8, the diameter and number of remaining EEFs were assayed (mean±sem, n=3).
(C) KDU691 and KAF156 were dosed following the radical cure regimen in (A), and on day 8, the diameter and number of remaining EEFs were assayed (mean±sem, n=3).
(D) IC50s of six compounds in prophylactic and radical cure modes. (mean±sem, n=3). Break in axes indicate highest concentration tested. See also Figure S4B.
Having established that MPCC infections can replicate clinical drug responsiveness outcomes using existing antimalarials, we applied our platform to assess the potential effectiveness of novel candidate compounds: 4 compounds (LMV599, KDU691, MMV390048 and MMV674594) that target the lipid kinase phosphatidylinositol-4-OH kinase (PI(4)K) (Ghidelli-Disse et al., 2014; McNamara et al., 2013; Younis et al., 2012), one compound (DDD107498) that targets translation elongation factor 2 (eEF2) in P. falciparum with activity against P. vivax blood stages (Baragaña et al., 2015), and one compound (KAF156) with an unknown mechanism of action (Kuhen et al., 2014) (Figures 3A–3D and S4B and S4C). All drugs were tested under two dosing regimens, termed ‘prophylactic’ (dosing day 0 to day 5) and ‘radical cure’ (dosing day 5 to day 8), and the effect of each drug on both parasite number and size were recorded on day 8 (Figure 3A). In prophylactic mode, the four PI(4)K inhibitors tested had varying activity on both small and large forms (Figure 3D). LMV599, the most potent of the four, cleared all parasites even at the lowest concentration tested. The other three PI(4)K inhibitors tested had similar IC50 values and achieved complete clearance. Notably, the PI(4)K inhibitors tested were more potent than primaquine, which was most effective at 5 μM. DDD107498 had similar potency to the most potent PI(4)K inhibitor tested, and KAF156, though less potent, had activity against both small and large forms. In radical cure mode, the compounds tested appeared less efficacious. PI(4)K inhibitors had activity against large forms but not all small forms were cleared from culture. KDU691 cleared a majority of large forms in radical cure mode, in contrast with the relatively small impact of KAF156 using this dosing regimen (Figure 3C). DDD107498 and KAF156 did not cause as significant a reduction in parasite number as the PI(4)K inhibitors, even at the highest concentrations tested, though the remaining parasites were small in size (Figure S4B).
Hybrid capture reveals P. vivax hypnozoite transcriptome in the MPCCs
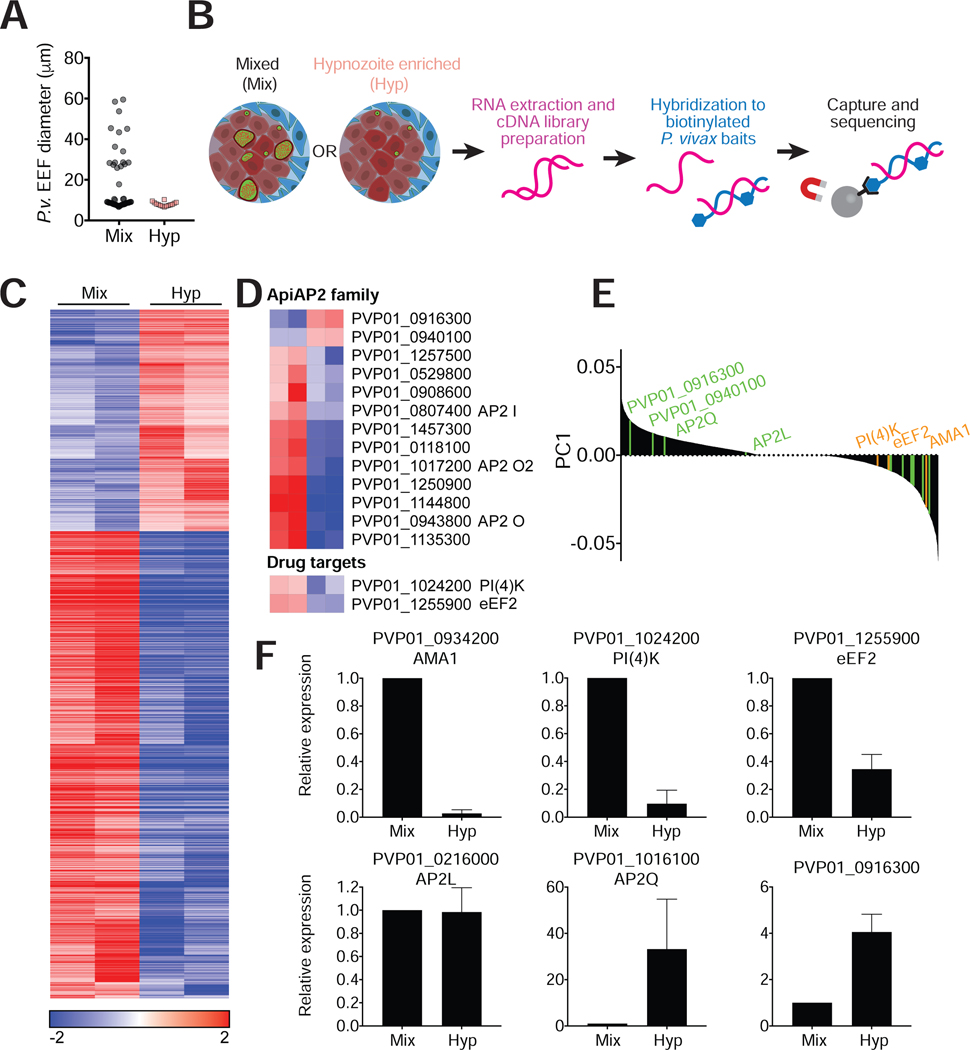
To further characterize the hypnozoite stage at a molecular level, we performed RNA-seq on hypnozoite-enriched MPCC cultures on day 9 post-infection. Hypnozoite enrichment was achieved by treatment with a PI(4)K inhibitor in ‘radical cure’ mode and processed in parallel with untreated cultures (referred to as mixed samples) (Figure 4A). Total RNA from infected MPCCs was prepared and enriched for P. vivax transcripts by magnetic pull-down using custom made baits tilling the recently assembled P. vivax P01 genome (Auburn et al., 2016). Baits were designed to include all P. vivax annotated genes and intergenic regions, and exclude ribosomal RNA transcripts and regions with homology to human or mouse. Selection by hybrid capture was followed by Illumina HiSeq2000 sequencing to generate over 100 million single end 40nt reads, with an average of 29 million reads per sample (Figure 4B). Alignment to P. vivax P01 transcriptome revealed robust enrichment, with approximately 70% and 20% of reads mapping uniquely to P. vivax in mixed and hypnozoite-enriched cultures respectively (Table S1; Figures S5 and S6). Notably, hypnozoite-enriched samples showed lower library complexity compared to mixed samples, suggestive of a preserved, albeit reduced transcriptional activity, consistent with a quiescent state (Figure 4C).
Figure 4: RNA-seq reveals differential expression patterns between hypnozoites and schizonts.
(A) Cultures were treated with a PI(4)K inhibitor in radical cure mode to enrich for hypnozoites. Drug was removed on day 8 pi and cultures were kept for one additional day in media before processing. Diameters of EEFs remaining in culture on day 9 were plotted for treated and untreated cultures.
(B) Schematic of sample processing. RNA extraction was performed on mixed and hypnozoite enriched cultures on day 9. cDNA libraries were hybridized to P. vivax specific baits for enrichment before sequencing.
(C) Heat map was generated for transcripts with adjusted P value (Padj)<0.01. Median log-transformed TPM values were calculated for each gene and the log2-fold-changes over the median was calculated for each sample. The resulting matrix was subjected to hierarchical clustering (two biological replicates per condition).
(D) Top heat map shows differential representation of ApiAP2 genes in mixed versus hypnozoite-enriched samples (Padj<0.01). Bottom heat map shows differential representation of drug targets PI(4)K and eEF2.
(E) Principal component analysis where positive values are biased towards hypnozoite-enriched samples. Genes in the ApiAP2 family are indicated in green and additional genes for which quantitative RT-PCR analysis was performed are indicated in orange.
(F) qRT-PCR analysis of 5 representative genes in non-captured RNA samples (mean±sem from at least 3 biological replicates of which one is an independent sample, not used for hybrid selection.)
Hypnozoite-enriched samples showed a significantly different transcriptional profile compared to mixed samples (Figure 4E). Gene Ontology (GO) enrichment analysis of the differentially expressed genes revealed suppression of functions related to maturity, merozoite invasion, and egress in the hypnozoite-enriched samples (Table S1). We also found that regulators of transcription, namely, members of the plant-derived Apicomplexan Apetala2 (ApiAP2) family of transcription factors (Balaji et al., 2005) were altered in the two populations (Figure 4D; Table S1). In one example, PVP01_1016100 (AP2-Q), which has been proposed as a quiescence marker in P. cynomolgi (Cubi et al., 2017), exhibited low representation in both mixed and hypnozoite-enriched samples (Transcripts Per Million (TPM)<25). Another AP2-encoding gene, PVP01_0916300, was observed at higher transcript abundance (TPM>150) and showed significantly higher representation in hypnozoite-enriched samples. Notably, the liver-specific AP2, PVP01_0216000 (Iwanaga et al., 2012) had equivalent representation in the two sample sets (TPM>160). Relative abundance of these transcripts, as well as transcripts of a subset of genes in the identified GO terms were confirmed by performing quantitative RT-PCR (qRT-PCR) on the same RNA samples prior to hybrid selection, as well as on independently collected samples (Figure 4F).
Finally, to evaluate the potential to apply insights from the transcriptome data towards drug discovery efforts, we mined the data sets for relative expression of several known drug targets. For the compounds tested against hypnozoites in MPCCs (Figure 3D), two targets have been identified: PI(4)K (McNamara et al., 2013) and eEF2 (Baragaña et al., 2015)). Hybrid capture and qRT-PCR analysis showed lower representation of genes coding for PI(4)K and eEF2 in hypnozoite-enriched samples relative to mixed samples (Figure 4D and 4E), which could explain the superior killing activity of the four PI(4)K inhibitors and DDD107498 on schizonts versus hypnozoites under radical cure treatment (Figure 3). However, we cannot exclude the possibility that the experimental framework selected for drug-insensitive hypnozoites, nor that the target pathway is downregulated in response to drug treatment.
MPCCs can be fabricated in 384 well plates
Anti-hypnozoite drug screening efforts can be improved by reducing biomass requirements, since access to sporozoites is a major logistical bottleneck. Towards this goal, we scaled down the MPCC platform to be compatible with industry standard 384-well plates. We have previously shown that precise ratios of homotypic and heterotypic interactions play essential roles in maintaining hepatocyte function in MPCCs (Khetani and Bhatia, 2008). In line with this observation, adapting the protocol for use in the smaller, 384-well system required that the island size and center-to-center distances were preserved. The monolithic (poly)dimethylsiloxane (PDMS) mold with elastomeric pillars and patterns was re-designed and precision-engineered to be compatible with the 384-well plate format. The 384-mold consists of individually spring loaded, composite metal-PDMS pillars with protruding patterns. The protruding patterns comprise 12 soft PDMS posts that, when in contact with the collagen-coated surface of each well, protect islands of collagen from ablation when exposed to oxygen plasma (Figure 5A). Spring-loading each pillar ensures uniform, conformal contact across an entire 384-well plate. After plasma treatment, seeded PHHs selectively attach to the remaining collagen pattern and are subsequently surrounded by supportive stromal cells.
Figure 5: P. vivax-infected MPCCs are Reengineered in 384-well Plates.
(A) Images of the 384 well plate MPCC mold and schematics describing the collagen ablation process.
(B) Representative image of PHHs seeded on collagen islands in a 384-well plate on day 1 before addition of fibroblasts (left panel). Representative image of CK18 expression on day 12 (right panel). Scale bars, 100μm.
(C) Albumin levels in PHHs with (MPCC) and without fibroblast (no J2) for at least 3 weeks in culture (left panel). CYP3A4 activity assay post rifampin treatment on days 12 and 23 (middle and right panels).
(D) Comparison of infection rates in 384 and 96 MPCCs. Sporozoite doses are indicated in parentheses. Histogram of day 8 forms in 384 MPCCs (right panel).
(E) Comparison of EEF numbers (n=36) and sizes (n=5) according to manual or automated image analysis. Inset shows a representative hypnozoite and schizont with white outline detected by the algorithm to measure size. Scale bar, 10μm.
(F) MPCC cost reduction, including PHHs, sporozoites, chemical screening, reagents, imaging, and labor. (2016: switch to 384 MPCCs).
Seeded PHHs, positive for host markers such as CK18 (Figure 5B), were functionally stable for 3 weeks, as depicted via stable albumin secretion levels. Furthermore, a primary drug metabolism enzyme, CYP3A4, remained both active and inducible for at least 23 days (Figure 5C). A pilot infection of the 384 MPCCs with clinical Thai P. vivax isolates revealed only a 2-fold reduction in infection rates per well compared to the 96-well format, despite the 10-fold reduction in initial parasite load and a 3-fold reduction in number of PHHs seeded. When expressed as a function of infection efficiency, this encouraging proof-of-concept translates to an enhanced outcome per biomass of 1.5-fold (hepatocytes), or 5-fold (parasites). On day 8, a bimodal population in parasite size became apparent, allowing distinction of hypnozoites and schizonts (Figure 5D). Furthermore, the platform is fully automatable. Cell seeding, media change, and drug dosing steps can be performed with liquid handlers; imaging with an automated microscope; and image analysis using Cell Profiler (Carpenter et al., 2006; Jones et al., 2008). In one example, automated imaging and subsequent Cell Profiler analysis revealed strong agreement between automated and manual parasite counts and sizes, suitable for high throughput drug screening (Figure 5E). Finally, we have achieved a 200X reduction in the cost per well of the MPCC platform since 2011 for anti-malarial screen purposes (Figure 5F). The decreased biomass needs achieved by the 384 MPCC represents the most significant contributor to the price reduction in 2016.
DISCUSSION
This study presents a rigorous in vitro characterization of hypnozoites, including assessment of their functional hallmarks and transcriptional profile. Hypnozoites detected in the MPCC system exhibit the known hallmarks of this liver stage: they are small, uninucleate, MSP1-negative, and differentially sensitive to primaquine versus atovoquone. Furthermore, in addition to persistent hypnozoites, we also observe large forms that reemerge on day 21, consistent with potential reactivation.
The demonstrated capacity to culture hypnozoites marks the MPCC system as a promising screening platform (in 96 and 384 well formats), and paves the way towards high throughput testing of existing and novel antimalarial candidates. Drug testing can be performed in both prophylactic and radical cure dosing strategies that target growing liver stage parasites or established hypnozoites, respectively (Campo et al., 2015). Here, we tested six compounds: KAF156, DDD107798 and four compounds that target PI(4)K. All six of the compounds cleared both hypnozoites and schizonts under prophylactic treatment. Under radical cure treatment, some of the drugs had killing activity against large forms, but were unable to eliminate remaining small forms, in line with the observed relapses reported in monkeys treated with the same compounds (Zeeman et al., 2016), demonstrating the predictive capacity of MPCCs in both prophylactic and radical cure dosing regimens. Notably, this screening platform offers the added benefit of using P. vivax as the test parasite, without the need for large animal studies. For more thorough characterization of the remaining small forms under drug pressure, long-term kinetic studies should be performed to measure their reactivation capacity.
Primaquine remains the only clinically approved drug with anti-hypnozoite activity. It should be noted that while, to the best of our knowledge, the MPCC system is the only in vitro system that has shown elimination of P. vivax parasites with prophylactic primaquine treatment, the radical cure treatment regimen used here has not achieved complete clearance of small forms as assessed by microscopy. The clinical standard of primaquine radical cure requires a 14-day regimen, usually co-administered with chloroquine, with blood stage breakthrough as readout. However, clinical studies show that even with this dosing regimen, relapses occur in individuals infected with P. vivax, likely due to inadequate dosing, geographical origin of the parasite, evolving primaquine resistance, or a combination of these factors (Collins and Jeffery, 1996; Goller et al., 2007). It is possible that radical cure in vitro might require a longer dosing regimen, in combination with a blood schizonticide. Furthermore, the observed lack of primaquine efficacy from day 5 cultures onwards raises questions regarding the biological changes that the hypnozoite might undergo. Given that the mechanism of hypnozoite clearance in the human liver has not yet been identified, to definitively assess whether the remaining small forms observed in culture post-treatment are viable parasites, their reactivation capacity should be interrogated with longer-term studies, combined with reticulocyte overlays to assess blood breakthrough.
Towards further molecular characterization of the elusive dormant parasites, we provide here the P. vivax hypnozoite transcriptome. This was achieved via a hybrid capture method whereby parasite transcripts were enriched using nucleic acid ‘baits’ designed specifically for the P. vivax genome. While a similar strategy has previously been used to enrich for pathogen DNA in clinical blood samples (Melnikov et al., 2011), here we apply hybrid selection for RNA enrichment of low input samples prior sequencing. The obtained results were successfully corroborated by an independent method (RT-PCR), for various targets in multiple samples, strengthening confidence in our findings. This new methodology can now be applied towards querying transcriptomes of not only Plasmodium liver-stages, but also other developmental stages of the parasite in mammalian and mosquito hosts, which have historically been challenging to perform due to major host contamination. We anticipate the utility of this tool towards elucidating the dynamic transcriptomes of the full parasite life-cycle, identifying stage-specific biomarkers, as well as elucidating novel drug and vaccine targets.
RNA capture and sequencing described here demonstrated that P. vivax hypnozoites exhibit reduced transcriptional activity, with low numbers of transcripts showing relatively high representation compared to the schizont stage. While 40% of the identified transcripts encode proteins of unknown function, the gene list contains RNA and DNA binding proteins, nucleases, proteases, and transferases, suggestive of preserved metabolic and catalytic activity, which could be linked to the observed increase in hypnozoite size in our long-term cultures. Consistent with a recent transcriptomic study of P. cycnomolgi hypnozoites, we also found members of the ApiAP2 family of transcription factors represented in hypnozoite-enriched samples. In addition to AP2Q, which was proposed as a transcriptional suppressor (but showed fairly low representation and high variation in our P. vivax samples), we identified a second putative AP2, PVP01_0916300 with more robust representation. PVP01_0916300 appears to also be highly expressed in P. falciparum gametocytes (López-Barragán et al., 2011), another quiescent form of the parasite. It would be interesting to investigate whether different quiescent parasite stages share similar regulatory mechanisms. ApiAP2 factors have been shown to regulate stage-specific transcription and parasite development (Iwanaga et al., 2012; Kafsack et al., 2014; Modrzynska et al., 2017; Sinha et al., 2014; Yuda et al., 2009, 2010), and thus are attractive candidates for further investigation and validation as regulators of the quiescent state. Notably, there are several putative AP2s that showed higher representation in mixed cultures and may be important as transcription activators, or even implicated in reactivation. One example that could be of interest to investigate is PVP01_0118100, which is one of the 25% of differentially detected genes that are exclusively found in P. vivax, P. cynomolgi, P. fragile, P. knowlesi and P. inui, with no orthologues in P. falciparum or rodent malaria parasites. Other unveiled candidates for which no function has been assigned could also be explored in the future as hypnozoite-specific biomarkers.
Finally, hypnozoite-specific properties of P. vivax infections, that have so far been observed largely in clinical studies, can be investigated in MPCCs. P. vivax strains that originate from different regions of the world are known to give rise to different relapse frequencies (Battle et al., 2014; Goller et al., 2007), which has been attributed to varying hypnozoite ratios (White, 2011). Recently, one study compared the two Thai isolate CSP subtypes using a humanized mouse system and found differences in hypnozoite frequencies (Mikolajczak et al., 2015). Our results however reveal hypnozoite frequencies ranging from 11 to 48%, regardless of the CSP subtype. It is not clear whether the VK210 and VK247 categorization of P. vivax is sufficient to capture hypnozoite frequency or other biological differences, if any, between the Thai strains. Future studies using parasites derived from other geographic locations should elucidate similarities and differences between strains, including drug sensitivity.
Overall, our data highlight the capacity of the MPCC system to facilitate interrogations of hypnozoite biology and testing of anti-hypnozoite compounds in the absence of dependence on human experimentation. MPCCs offer advantages over existing in vitro systems. The longevity of cultures allow monitoring of important P. vivax hallmarks such as merosome release and reactivation as well as testing of hypotheses regarding reactivation (Shanks and White, 2013); having a full repertoire of host functions allows for primaquine sensitivity and cross-screening for cellular toxicity; and its reproducibility allows drug sensitivity testing. MPCCs are compatible with robotic fluid handlers and high-content imaging readouts and are suitable for international dissemination, with training, as evidenced by the implementation of the platform at four sites in two countries. Compared to in vivo systems, MPCCs enable biomass enrichment due to the well-plate format, reduced biomass requirements, and dynamic monitoring of parasite biology via microscopy, such as time-lapse longitudinal studies of live cultures. Finally, the MPCCs allow querying of the liver stage transcriptome of P. vivax and will enable profiling of hypnozoite transcripts over time and in response to drug pressure. Taken together, the robust, high throughput-ready in vitro human liver system presented here offers the potential to gain new biological insights into P. vivax development in human hepatocytes, and represents a screening platform for candidate drugs directed against distinct stages of P. vivax, including the hypnozoite stage, a required asset in the push to achieve malaria eradication.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sangeeta N. Bhatia (sbhatia@mit.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
P. vivax parasites:
Anopheles dirus mosquitoes were fed on blood collected from symptomatic patients attending malaria clinics in Tak, Songkla, and Ubon-Ratchathani Provinces in Thailand, confirmed positive for only P. vivax via microscopy and RT-PCR.
Briefly, P. vivax infected blood was drawn into heparinized tubes and kept at 37C until processing. Infected blood was washed once with RPMI 1640 incomplete medium. Packed infected blood was resuspended in warm non-heat inactivated naïve human AB serum for a final hematocrit of 50%. Resuspended blood was fed to laboratory reared female Anopheles dirus mosquitoes for 30 minutes via an artificial membrane attached to a water-jacketed glass feeder kept at 37C. Engorged mosquitoes were kept on 10% sugar at 26C under 80% humidity at the designated insectary at the Mahidol Vivax Research Unit for 14 days. The confirmation of single species of P. vivax infection was performed by nested-PCR. Sporozoites were aseptically dissected from the salivary glands of infected mosquitoes 14–21 days after blood feeding and pooled in harvesting medium (DMEM media supplemented with 2% (v/v) penicillin-streptomycin).
Cells:
Cryopreserved primary human hepatocytes were purchased from vendors permitted to sell products derived from human organs procured in the United States by federally designated Organ Procurement Organizations. Vendors include Bioreclamation IVT and Thermo Fisher. Human hepatocytes (Donor 1: female age 35; donor 2: female age 77) were maintained in high glucose Dulbecco’s Modified Eagle’s Medium (DMEM with L-glutamine, Corning) with 10% (v/v) fetal bovine serum (FBS, Gibco), 1% (v/v) ITS+ (insulin/human ransferrin/selenous acid and linoleic acid) premix (BD Biosciences), 7 ng/ml glucagon (Sigma), 40 ng/ml dexamethasone (Sigma), 15 mM HEPES (Gibco), and 1% (v/v) penicillin-streptomycin (Corning).
J2–3T3 male murine embryonic fibroblasts (gift of Howard Green, Harvard Medical School) were cultured at <18 passages in fibroblast medium comprising of DMEM with high glucose, 10% (v/v) bovine serum (Thermo Fisher), and 1% (v/v) penicillin-streptomycin (Corning).
METHOD DETAILS
Micropatterned Co-cultures (MPCCs):
The technique has been previously explained in detail (March et al., 2015). Briefly; glass-bottomed 96 or 384-well plates were coated with rat-tail type I collagen (50 μg/ml) and subjected to soft lithographic techniques (Ploss et al., 2010) to pattern the collagen into microdomains (islands of 500 μm) that mediate selective hepatocyte adhesion. The mold for the 384MPCC was designed and constructed by Phenomyx (PHYX.384.MPCC, Cambridge, MA). Briefly, the system comprises composite aluminum-PDMS pillars with 12 circular protruding patterns arranged on an orthogonal grid. Each pillar is oriented in a polycarbonate plate and spring-loaded to ensure conformal contact with the plate bottom. A clamping plate with plasma access holes holds the 384-pillar assembly together. A sliding bracket with torque-control knob applies controlled pressure to the 384-pillar assembly during plasma ablation. To create MPCCs, cryopreserved primary human hepatocytes (Bioreclamation IVT) were pelleted by centrifugation and then seeded on collagen-micropatterned plates. 3T3-J2 murine embryonic fibroblasts were seeded 1 day after seeding, following infection with Plasmodium vivax sporozoites.
P. vivax infection of MPCCs:
30,000 to 60,000 sporozoites were overlaid onto MPCC cultures (5,000 for 384 MPCCs), seeded the day before, in hepatocyte medium and kept at 37°C and 5% CO2 for 3 hours for infection to occur. Post-infection, wells were washed twice and fresh media containing fibroblasts was added. Cultures were fixed on days 5, 8, 9, 10, 11, 18 and 21 for in paraformaldehyde (PFA) or ice-cold methanol for analysis by immunofluorescence.
Human Reticulocyte Overlay:
Adult human blood from the Thai Red Cross was passed through Pall filters (Pall Corporation) to remove leukocytes. Remaining red blood cells were washed with RPMI1640 and collected via centrifugation (100 g, 10 min). Packed cells were resuspended in OptiPrep (Axis Shield) and KCl buffer and centrifuged at 3,000 g for 30 minutes. Reticulocytes were collected from the interface and washed with RPMI1640 then resuspended to 50% hematocrit. Reticulocyte enrichment was calculated by methylene blue staining. 33×106 red blood cells (of which 33% were reticulocytes) were overlaid per well of the 96-well MPCC, diluted in hepatocyte medium. Fresh media containing red blood cells was added daily.
Drug Treatment of P. vivax EEFs in MPCCs:
Infected MPCCs were incubated with media containing the drug being tested (primaquine diphosphate (Sigma) ranging from 0.1–10 μM, atovaquone (Sigma) ranging from 0.1 to 270nM; MMV390048, MMV67494 KDU691, LMV599, KAF156, DDD107498 (Medicines for Malaria Venture) ranging from 0.03 μM to 10 μM. For prophylactic treatment, fresh drug-containing medium was added daily until day 5 with drug-free media changes until fixation on day 8. For radical cure treatment, fresh drug-containing medium was added daily from day 5 until day 8 when cultures were fixed. IC50 curves were generated by plotting the number of parasites left in culture, compared to control, under varying doses of drug.
Immunofluorescence Staining:
MPCCs were fixed with either ice-cold methanol for 10 minutes at 4°C or 4% paraformaldehyde (PFA) for 20 minutes at room temperature. PFA-fixed samples were permeabilized with 0.1% TritonX for 10 minutes at room temperature. Wells were washed twice with PBS, blocked with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), and incubated with primary antibodies for 1 hour at room temperature. Samples were washed with PBS then incubated with Alexa 546-conjugated secondary goat-anti-mouse (Invitrogen) and Alexa 647-conjugated goat anti-rabbit (Invitrogen) for 1 hour at room temperature. Samples were washed with PBS, counterstained with the DNA dye Hoechst 33258 (Invitrogen; 1:5,000), and kept in Aquamount (Lerner Laboratories). Images were captured on a Nikon Eclipse Ti fluorescence microscope or a Nikon 1AR Ultra-Fast confocal microscope. Areas of the developing liver stage parasites were measured using ImageJ and used to calculate the corresponding diameter. Liver stage P. vivax parasites were detected using rabbit polyclonal antibodies against UIS4, BIP, MIF, HSP60, HSP70, MSP1 and mouse monoclonal antibodies against UIS4 and ACP. The nuclei of the parasites were visualized using a rabbit polyclonal anti-acetyl-Histone H3 (Lys9) antibody (Millipore).
Hybrid capture, RNA-seq extraction and analysis:
SureSelectXT RNA Direct capture probes were designed for the P. vivax P01 genome using eArray with the assistance of Agilent Technologies. Preliminary RNA-seq experiments revealed significant transcription outside the annotated gene loci in P. vivax (data not shown). To accommodate novel transcripts, probes were designed to tile the P. vivax genome such that they included all known genes and intergenic regions while specifically excluding rRNA transcripts, known pseudogenes and regions with homology to human or mouse. Design is available as ELID: S3090564
Total RNA was extracted using TRIzol (Thermo Fisher) and purified using RNeasy Mini Kit (Qiagen) according to manufacturer’s instructions. Samples were DNase treated. RNA was quality controlled using an AATI Fragment Analyzer and 100ng was prepared following the SureSelectXT RNA Direct protocol version A0. Illumina libraries were quantitated using the Fragment Analyzer and by qPCR and sequenced as a single end 40nt read using a HiSeq2000.
Quantitative RT-PCR:
Total RNA was extracted with TRIzol® (Thermo Fisher), DNAse treated and purified using the RNeasy MinElute Cleanup Kit (Qiagen). cDNA synthesis was performed using SuperScript II (Thermo Fisher) and RT-PCR was carried out using PowerUp SYBR Green Master Mix (Applied Biosystems) in a Roche Light Cycler 480 Real-Time PCR Detection System according to the manufacturer’s instructions. The primers used are listed in Table S2. Relative gene expression was calculated with the ΔΔCt method, using PVP01_1213400 as housekeeping gene.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes and statistical analysis:
n represents the number wells from each plate as described in the figure legends. Data were analyzed using Prism 7.0 (GraphPad Software, San Diego, CA) and results represent means ± SEM. Methods used for computing statistical significance is indicated in figure legends. Statistically significant differences were defined as * when p values were < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001. To avoid plate position effects, setups of all conditions were randomly assigned in each experiment. Drugs tested in figures 3b - d were blinded and scored by independent researchers.
RNA-seq Data:
Illumina Offline BaseCaller1.9.3 software was used for basecalling. Reads were aligned against Plasmodium vivax PvP01 from PlasmoDB v. 34 (Sept 2017) using STAR v. 2.5.3a (Dobin et al., 2013) with flags -runThreadN 8 --runMode alignReads --outFilterType BySJout --outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --alignIntronMin 10 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --outSAMtype BAM SortedByCoordinate --quantMode TranscriptomeSAM with --genomeDir pointing to a 75nt-junction PvivaxP01 STAR suffix array.
Gene expression was quantitated using RSEM v. 1.3.0 (Li and Dewey, 2011) with the following flags for all libraries: rsem-calculate-expression --calc-pme --alignments -p 8 --forward-prob 0 against an annotation matching the STAR SA reference. Posterior mean estimates (pme) of counts were retrieved, and transcripts corresponding to rRNAs and tRNAs were removed.Resulting read counts were summarized by genes, then converted into RPKMs using RSEM effective gene length estimates, and finally to TPMs.
For principal components analysis (PCA), log-transformed, quantile-normalized TPM data were processed using a singular-value decomposition approach (SVD) as implemented in the prcomp function in the R statistical environment (v. 3.4.0). Gene loadings for each gene for component 1 were extracted and ranked. Differential-expression analysis was performed using DESeq2 on the rRNA-substracted raw counts (Anders and Huber, 2010; Love et al., 2014) Briefly, sequencing library size factors were estimated for each library, and differences in gene expression between conditions (expressed as log2-transformed fold-changes in expression levels) were estimated under a general linear model (GLM) framework fitted on the read counts. In this model, read counts of each gene in each sample were modeled under a negative binomial distribution, based on the fitted mean of the counts and aforementioned dispersion parameters. Differential expression significance was assessed using a Wald test on the fitted count data (all these steps were performed using the DESeq() function in DESeq2). Genes with at least a two-fold change between “Mixed” and “Hypnozoite” sample groups (with adjusted P values <0.01 using Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995) were selected for downstream analysis.
For heat map generation, median log-transformed TPM values were calculated for each gene and the log2-fold-changes over the median was calculated for each sample. The resulting matrix was subjected to hierarchical clustering using 1-Pearson correlation as a distance metrics and a [complete-single-sample average] linkage function.
DATA AND SOFTWARE AVAILABILITY
Raw Data:
Raw RNA-seq data has been deposited into the Gene Expression Omnibus (GEO) under the accession number GSE108016.
Supplementary Material
Supplemental Table 2 (Related to Figure 4): Primers used for quantitative RT-PCR
Supplemental Table 1 (Related to Figure 4): RNA-seq analysis of P. vivax in mixed and hypnozoite-enriched cultures on day 9.
Videos of merosomes in MPCC cultures on days 10–12. This video was captured on an iPhone 7S through the eyepiece of a light microscope.
Merosome release was captured for one of the merosomes. This video was captured on an iPhone 7S through the eyepiece of a light microscope.
Free merozoites in MPCC cultures on days 10–12 were captured. This video was captured on an iPhone 7S through the eyepiece of a light microscope.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa 546-conjugated secondary goat-anti-mouse | Invitrogen | Cat# A11030 |
| Alexa 647-conjugated goat anti-rabbit | Invitrogen | Cat# A21246 |
| Hoechst 33258 | Invitrogen | Cat# H3569 |
| rabbit polyclonal anti-acetyl-Histone H3 (Lys9) antibody | Millipore | Cat# 06-942 |
| rabbit polyclonal UIS4 antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| rabbit polyclonal BIP antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| rabbit polyclonal MIF antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| rabbit polyclonal HSP60 antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| rabbit polyclonal HSP70 antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| rabbit polyclonal MSP1 antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| mouse monoclonal UIS4 antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| mouse monoclonal ACP antibody | gift from Sebastian Mikolajcak at CIDR | N/A |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Human reticulocytes | Thai Red Cross | N/A |
| Primary Human Hepatocytes | Thermo Fisher, Bioreclamation IVT | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Aquamount | Lerner Laboratories | Cat# 41799-008 |
| Primaquine diphosphate | Sigma | Cat# 160393 |
| Atovaquone | Sigma | Cat# A7986 |
| MMV67494 KDU691, LMV599, KAF156, DDD107498 | Medicines for Malaria Venture | N/A |
| Critical Commercial Assays | ||
| TRIzol | Thermo Fisher | Cat# 15596026 |
| RNeasy MinElute Cleanup Kit | Qiagen | Cat# 74204 |
| Super Script II | Thermo Fisher | Cat# 18064014 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Cat# A25741 |
| SureSelectXT RNA | Agilent | Custom made |
| Deposited Data | ||
| Raw RNAseq data | This paper |
https://www.ncbi.nlm.nih.gov/geo/ accession number: GSE108016) |
| Experimental Models: Cell Lines | ||
| 3T3-J2 murine embryonic fibroblasts | gift of Howard Green, Harvard Medical School | N/A |
| Experimental Models: Organisms/Strains | ||
| Plasmodium vivax | Mahidol Vivax Research Unit (Bangkok, Thailand) | N/A |
| Oligonucleotides | ||
| Primers included in Table S2 | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Illumina Offline BaseCaller v1.9.3 | Illumina | N/A |
| STAR v. 2.5.3a | Github | https://github.com/alexdobin/STAR/releases |
| RSEM v. 1.3.0 | Dewey Lab | https://deweylab.github.io/RSEM/ |
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
Highlights.
The MPCC platform supports formation and reactivation of hypnozoites in vitro
P. vivax schizonts in the MPCCs mature, release merosomes, and infect reticulocytes
Hybrid capture and RNA-sequencing reveals the hypnozoite transcriptome in the MPCCs
MPCCs allow prophylactic and radical cure testing of anti-hypnozoite drugs
ACKNOWLEDGMENTS
We are grateful to MMV and Kelly Chibale for supplying the compounds; Wanlapa Roobsong for help with reticulocytes; Adam Falls for assistance with the 384 MPCC mold; Sabrina Hawthorne, Owen Hardy and the Koch Institute Swanson Biotechnology Center, specifically Jon Penterman in the Genomics Core Facility for technical support; Maria Mota, Stephen Hoffman, Brice Campo, Omar Vandal, Richard Elliott, Dan Neafsey, Bronwyn MacInnis, Dyann Wirth and James J. Collins for insightful discussions. This work was supported by the Bill & Melinda Gates Foundation and in part by the Koch Institute Support Grant P30-CA14051 from the National Cancer Institute. P. vivax sporozoite production was supported by MMV. NG is an HHMI International Student Research Fellow, SNB is an HHMI Investigator.
Footnotes
DECLARATION OF INTERESTS
S.N.B. is a co-founder of Ascendance.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auburn S, Böhme U, Steinbiss S, Trimarsanto H, Hostetler J, Sanders M, Gao Q, Nosten F, Newbold CI, Berriman M, et al. (2016). A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, and Aravind L (2005). Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baragaña B, Hallyburton I, Lee MCS, Norcross NR, Grimaldi R, Otto TD, Proto WR, Blagborough AM, Meister S, Wirjanata G, et al. (2015). A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle KE, Karhunen MS, Bhatt S, Gething PW, Howes RE, Golding N, Van Boeckel TP, Messina JP, Shanks GD, Smith DL, et al. (2014). Geographical variation in Plasmodium vivax relapse. Malar. J. 13, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 57, 289–300. [Google Scholar]
- Bennett JW, Pybus BS, Yadava A, Tosh D, Sousa JC, McCarthy WF, Deye G, Melendez V, and Ockenhouse CF (2013). Primaquine Failure and Cytochrome P-450 2D6 in Plasmodium vivax Malaria. N. Engl. J. Med. 369, 1381–1382. [DOI] [PubMed] [Google Scholar]
- Berger DR, Ware BR, Davidson MD, Allsup SR, and Khetani SR (2015). Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology 61, 1370–1381. [DOI] [PubMed] [Google Scholar]
- Campo B, Vandal O, Wesche DL, and Burrows JN (2015). Killing the hypnozoite - drug discovery approaches to prevent relapse in Plasmodium vivax. Pathog. Glob. Health 109, 107–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang I, Friman O, Guertin DA, Chang J, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay R, Velmurugan S, Chakiath C, Andrews Donkor L, Milhous W, Barnwell JW, Collins WE, and Hoffman SL (2010). Establishment of an in vitro assay for assessing the effects of drugs on the liver stages of Plasmodium vivax malaria. PLoS One 5, e14275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins WE, and Jeffery GM (1996). Primaquine resistance in Plasmodium vivax. Am J Trop Med Hyg 55, 243–249. [DOI] [PubMed] [Google Scholar]
- Combe A, Giovannini D, Carvalho TG, Spath S, Boisson B, Loussert C, Thiberge S, Lacroix C, Gueirard P, and Ménard R (2009). Clonal Conditional Mutagenesis in Malaria Parasites. Cell Host Microbe 5, 386–396. [DOI] [PubMed] [Google Scholar]
- Cubi R, Vembar SS, Biton A, Franetich J-F, Bordessoulles M, Sossau D, Zanghi G, Bosson-Vanga H, Benard M, Moreno A, et al. (2017). Laser capture microdissection enables transcriptomic analysis of dividing and quiescent liver stages of Plasmodium relapsing species. Cell. Microbiol. 19, e12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembele L, Gego A, Zeeman A-M, Franetich J-F, Silvie O, Rametti A, Le Grand R, Dereuddre-Bosquet N, Sauerwein R, van Gemert G-J, et al. (2011). Towards an in vitro model of Plasmodium hypnozoites suitable for drug discovery. PLoS One 6, e18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli-Disse S, Lafuente-Monasterio M, Waterson D, Witty M, Younis Y, Paquet T, Street LJ, Chibale K, Gamo-Benito F, Bantscheff M, et al. (2014). Identification of Plasmodium PI4 kinase as target of MMV390048 by chemoproteomics. Malar. J. 13, P38. [Google Scholar]
- Goller JL, Jolley D, Ringwald P, and Biggs B-A (2007). Regional differences in the response of Plasmodium vivax malaria to primaquine as anti-relapse therapy. Am. J. Trop. Med. Hyg. 76, 203–207. [PubMed] [Google Scholar]
- Hollingdale MR, Collins WE, Campbell C, and Schwartz AL (1985). In vitro culture of two populations (dividing and nondividing) of exoerythrocytic parasites of Plasmodium vivax. Am. J. Trop. Med. Hyg. 34, 216–222. [DOI] [PubMed] [Google Scholar]
- Iwanaga S, Kaneko I, Kato T, and Yuda M (2012). Identification of an AP2-family Protein That Is Critical for Malaria Liver Stage Development. PLoS One 7, e47557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TR, Kang I, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, and Carpenter AE (2008). CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafsack BFC, Rovira-Graells N, Clark TG, Bancells C, Crowley VM, Campino SG, Williams AE, Drought LG, Kwiatkowski DP, Baker DA, et al. (2014). A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplowitz N (2005). Idiosyncratic drug hepatotoxicity. Nat. Rev. Drug Discov. 4, 489–499. [DOI] [PubMed] [Google Scholar]
- Khetani SR, and Bhatia SN (2008). Microscale culture of human liver cells for drug development. Nat. Biotechnol. 26, 120–126. [DOI] [PubMed] [Google Scholar]
- Krotoski WA, Krotoski DM, Garnham PC, Bray RS, Killick-Kendrick R, Draper CC, Targett GA, and Guy MW (1980). Relapses in primate malaria: discovery of two populations of exoerythrocytic stages. Preliminary note. Br. Med. J. 280, 153–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krotoski WA, Collins WE, Bray RS, Garnham PC, Cogswell FB, Gwadz RW, Killick-Kendrick R, Wolf R, Sinden R, Koontz LC, et al. (1982). Demonstration of hypnozoites in sporozoite-transmitted Plasmodium vivax infection. Am. J. Trop. Med. Hyg. 31, 1291–1293. [DOI] [PubMed] [Google Scholar]
- Kuhen KL, Chatterjee AK, Rottmann M, Gagaring K, Borboa R, Buenviaje J, Chen Z, Francek C, Wu T, Nagle A, et al. (2014). KAF156 Is an Antimalarial Clinical Candidate with Potential for Use in Prophylaxis, Treatment, and Prevention of Disease Transmission. Antimicrob. Agents Chemother. 58, 5060–5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Barragán MJ, Lemieux J, Quiñones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, and Su X (2011). Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March S, Ramanan V, Trehan K, Ng S, Galstian A, Gural N, Scull MA, Shlomai A, Mota MM, Fleming HE, et al. (2015). Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens. Nat. Protoc. 10, 2027–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazier D, Landau I, Druilhe P, Miltgen F, Guguen-Guillouzo C, Baccam D, Baxter J, Chigot J-P, and Gentilini M (1984). Cultivation of the liver forms of Plasmodium vivax in human hepatocytes. Nature 307, 367–369. [DOI] [PubMed] [Google Scholar]
- McNamara CW, Lee MCS, Lim CS, Lim SH, Roland J, Nagle A, Simon O, Yeung BKS, Chatterjee AK, McCormack SL, et al. (2013). Targeting Plasmodium PI(4)K to eliminate malaria. Nature 504, 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Galinsky K, Rogov P, Fennell T, Van Tyne D, Russ C, Daniels R, Barnes KG, Bochicchio J, Ndiaye D, et al. (2011). Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 12, R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczak SA, Vaughan AM, Kangwanrangsan N, Roobsoong W, Fishbaugher M, Yimamnuaychok N, Rezakhani N, Lakshmanan V, Singh N, Kaushansky A, et al. (2015). Plasmodium vivax liver stage development and hypnozoite persistence in human liver-chimeric mice. Cell Host Microbe 17, 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JL, Harupa A, Kappe SHI, and Mikolajczak SA (2012). Plasmodium yoelii Macrophage Migration Inhibitory Factor Is Necessary for Efficient Liver-Stage Development. Infect. Immun. 80, 1399–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrzynska K, Pfander C, Chappell L, Yu L, Suarez C, Dundas K, Gomes AR, Goulding D, Rayner JC, Choudhary J, et al. (2017). A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe 21, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Schwartz RE, March S, Galstian A, Gural N, Shan J, Prabhu M, Mota MM, and Bhatia SN (2015). Human iPSC-Derived Hepatocyte-like Cells Support Plasmodium Liver-Stage Infection In Vitro. Stem Cell Reports 4, 348–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe AR, Fishkind DJ, and Adams JH (2000). Spatial and temporal dynamics of the secretory pathway during differentiation of the Plasmodium yoelii schizont. 108, 169–185. [DOI] [PubMed] [Google Scholar]
- Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, and White NJ (2014). Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect. Dis. 14, 982–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg R, Wirtz R. a, Lanar DE, Sattabongkot J, Hall T, Waters a P., and Prasittisuk C (1989). Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science 245, 973–976. [DOI] [PubMed] [Google Scholar]
- Sattabongkot JETSUMON, YIMAMNUAYCHOKE N, LEELAUDOMLIPI S, RASAMEESORAJ M, JENWITHISUK R, COLEMAN RE, UDOMSANGPETCH R, Cui L, and BREWER TG (2006). Establishment of a human hepatocyte line that supports in vitro development of the exo-erythrocytic stages of the malaria parasites Plasmodium falciparum and P.vivax. Am J Trop Med Hyg 74, 708–715. [PubMed] [Google Scholar]
- Shanks GD, and White NJ (2013). The activation of vivax malaria hypnozoites by infectious diseases. Lancet Infect. Dis. 13, 900–906. [DOI] [PubMed] [Google Scholar]
- Shortt HE, and Garnham PCC (1948). Pre-erythrocytic stage in mammalian malaria parasites. Nature 161, 126. [DOI] [PubMed] [Google Scholar]
- Sinha A, Hughes KR, Modrzynska KK, Otto TD, Pfander C, Dickens NJ, Religa AA, Bushell E, Graham AL, Cameron R, et al. (2014). A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 507, 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S, Rennenberg A, Krueger A, Pollok J-M, Menard R, and Heussler VT (2006). Manipulation of Host Hepatocytes by the Malaria Parasite for Delivery into Liver Sinusoids. Science (80-. ). 313, 1287–1290. [DOI] [PubMed] [Google Scholar]
- Wells TNC, Burrows JN, and Baird JK (2010). Targeting the hypnozoite reservoir of Plasmodium vivax: the hidden obstacle to malaria elimination. Trends Parasitol. 26, 145–151. [DOI] [PubMed] [Google Scholar]
- White NJ (2011). Determinants of relapse periodicity in Plasmodium vivax malaria. Malar. J. 10, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2015). World Malaria Report 2015 (World Health Organization; ). [Google Scholar]
- Younis Y, Douelle F, Feng TS, Cabrera DG, Manach C Le, Nchinda AT, Duffy S, White KL, Shackleford DM, Morizzi J, et al. (2012). 3,5-Diaryl-2-Aminopyridines As a Novel Class of Orally Active Antimalarials Demonstrating Single Dose Cure in Mice and Clinical Candidate Potential. J. Med. Chem. 55, 3479–3487. [DOI] [PubMed] [Google Scholar]
- Yuda M, Iwanaga S, Shigenobu S, Mair GR, Janse CJ, Waters AP, Kato T, and Kaneko I (2009). Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol. 71, 1402–1414. [DOI] [PubMed] [Google Scholar]
- Yuda M, Iwanaga S, Shigenobu S, Kato T, and Kaneko I (2010). Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol. 75, 854–863. [DOI] [PubMed] [Google Scholar]
- Zeeman A-M, Lakshminarayana SB, van der Werff N, Klooster EJ, Voorberg-van der Wel A, Kondreddi RR, Bodenreider C, Simon O, Sauerwein R, Yeung BKS, et al. (2016). PI4K is a prophylactic, but not radical curative target in Plasmodium vivax -type malaria parasites. Antimicrob. Agents Chemother. 60, AAC.03080–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 2 (Related to Figure 4): Primers used for quantitative RT-PCR
Supplemental Table 1 (Related to Figure 4): RNA-seq analysis of P. vivax in mixed and hypnozoite-enriched cultures on day 9.
Videos of merosomes in MPCC cultures on days 10–12. This video was captured on an iPhone 7S through the eyepiece of a light microscope.
Merosome release was captured for one of the merosomes. This video was captured on an iPhone 7S through the eyepiece of a light microscope.
Free merozoites in MPCC cultures on days 10–12 were captured. This video was captured on an iPhone 7S through the eyepiece of a light microscope.
Data Availability Statement
Raw Data:
Raw RNA-seq data has been deposited into the Gene Expression Omnibus (GEO) under the accession number GSE108016.