SUMMARY
Advances in 3D imaging technology are transforming how radiologists search for cancer1,2 and how security officers scrutinize baggage for dangerous objects.3 These new 3D technologies often improve search over 2D images4,5 but vastly increase the image data. Here, we investigate 3D search for targets of various sizes in filtered noise and digital breast phantoms. For a Bayesian ideal observer optimally processing the filtered noise and a convolutional neural network processing the digital breast phantoms, search with 3D image stacks increases target information and improves accuracy over search with 2D images. In contrast, 3D search by humans leads to high miss rates for small targets easily detected in 2D search, but not for larger targets more visible in the visual periphery. Analyses of human eye movements, perceptual judgments, and a computational model with a foveated visual system suggest that human errors can be explained by interaction among a target’s peripheral visibility, eye movement under-exploration of the 3D images, and a perceived overestimation of the explored area. Instructing observers to extend the search reduces 75% of the small target misses without increasing false positives. Results with twelve radiologists confirm that even medical professionals reading realistic breast phantoms have high miss rates for small targets in 3D search. Thus, under-exploration represents a fundamental limitation to the efficacy with which humans search in 3D image stacks and miss targets with these prevalent image technologies.
In Brief
Will 3D imaging technologies always lead to improvements for the visual search targets? Lago et al. show that, when humans search 3D image stacks, they under-explore with eye movements, overestimate the area they have searched, and often miss small targets that are salient in 2D images.
Graphical Abstract
RESULTS AND DISCUSSION
Humans show a remarkable ability to find targets in cluttered scenes. A large body of literature has investigated the strategies and computations that enable successful search6–12 and the limitations that lead to errors.13–19 A majority of these investigations involve human observers searching for targets in 2D images. Today, technological advances in 3D imaging are changing how human experts search in important perceptual tasks ranging from radiology1,2,20–24 and security25 to science26,27 and natural disaster management. In many applications, human observers read the resulting 3D images as a stack of 2D slice images. Expert observers scroll through the volume to determine the presence or absence of a target23,28,29 that might only occupy a subset of the slices. The 3D image stacks provide additional depth information about the target objects of interest and better segmentation of the background structures and improve human perceptual decisions.4,30 But are there circumstances for which search with 3D images might negatively affect perceptual performance? We investigated human efficacy at searching in 3D image stacks relative to search in 2D images and assessed whether there are bottlenecks in the human visuo-cognitive capabilities for 3D search.
We used images created in the laboratory (power-law, filtered-noise backgrounds ~1/f2.8; Figure 1A) to evaluate 2D and 3D search effects. These synthetic images allow isolating the effect of visual search while controlling for image differences that arise from image-generation variations in 2D versus 3D real-world imaging systems. We evaluated search performance for a large target (a 3D Gaussian; 2 standard deviations = 0.44 degrees of visual angle, °) or a small target (sharp-edged sphere with diameter = 0.13°). The large target is analogous to a mass in breast X-ray images, although the small target mimics a calcification. A radiologist would look for both types of targets. The target-present images (50% of the trials) for the 2D search condition consisted of the slice in the 3D image stack containing the center slice of the 3D target. In all tasks, within a block of trials, observers were informed which target might be present.
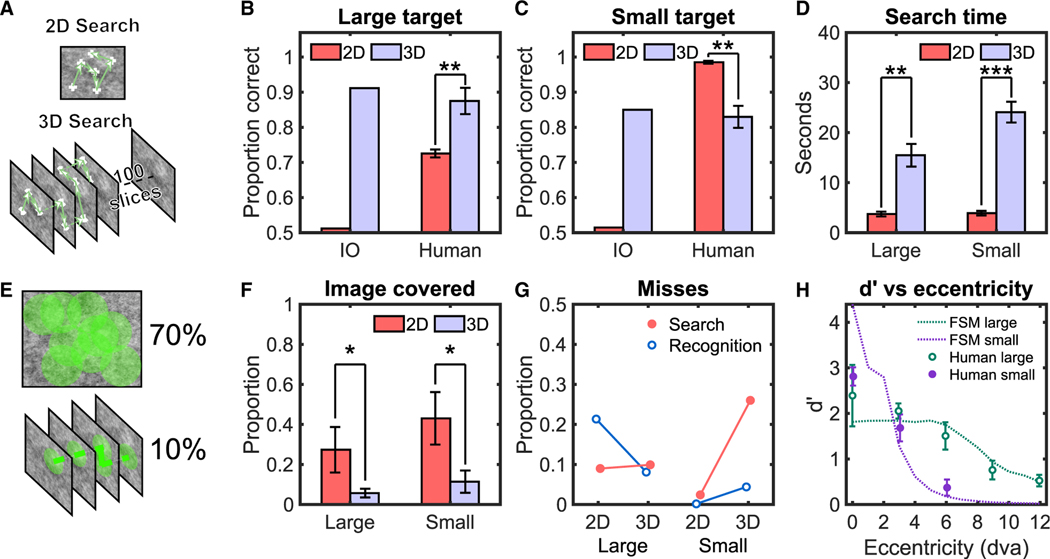
Figure 1. Human and Ideal Observer Results for 2D and 3D Search in Filtered Noise.
(A) A sample 2D image (top) and slices of a 3D volume (bottom) of filtered noise images with fixations and scrolls of a search by a human observer.
(B) Proportion correct for target detection for human observers and an ideal Bayesian observer (IO) for the large target. To avoid ceiling effects, IO performance was degraded with internal noise to approximately match human performance in 3D for each target type. The same value of internal noise was then used for the IO’s 2D search (STAR Methods).
(C) Proportion correct for target detection for human observers and IO for the small target.
(D) Average search time measured for human observers.
(E) A sample of the image covered for a 2D trial (top) and a 3D trial (bottom) with a useful field of view (UFOV) of 2.5° radius.
(F) Average proportion of image covered by human observers’ UFOV.
(G) Search and recognition errors for human observers.
(H) Symbols: average human target detectability (d’) versus retinal eccentricity for small and large targets during brief presentations of 2D images (500 ms).
Continuous lines: fit of the foveated search model (FSM) (STAR Methods).
Error bars are ±SEM. *p < 0.05; **p < 0.01; ***p < 0.001. See also Figure S1.
We first considered the performance trade-off between the benefits of additional target slices in 3D image stacks and the detriments of a vast increase in search space. In typical 3D imaging modalities, most targets are large enough to occupy multiple slices, adding visual information about the target relative to a single 2D image. However, the 3D image stacks also increase the possible locations at which the target might be, and this higher positional uncertainty is known to degrade perceptual performance.31–34 These trade-offs can be quantified for the lab-created images by the ideal Bayesian observer,35–38 which optimally uses the visual information to make inferences about the target’s presence. For an ideal observer in this search task, the benefits from additional target slices outweigh the detrimental effects of location uncertainty. Thus, 3D search improves ideal observer search accuracy for both target types over 2D search (Figures 1B and 1C).
For seven trained human observers, searching with no time limits, accuracy also increased significantly for the large target in 3D images relative to 2D images (Figures 1B and 1C; proportion correct: ΔPC = PC3D–PC2D = 0.17, t(6) = 4.815, p = 0.003; difference in indices of detectability, Δd’ = 1.01). But in contrast to the ideal Bayesian observer, human detection deteriorated significantly for the small target in 3D search (ΔPC = 0.16, p < 0.001, t(6) = 5.125 or Δd’ = 1.63, p = 0.002; 2-way interaction between target and search type, F[1, 4,732] = 153.34, p < 0.0001; Figures S1A and S1B for true-positive rate [TPR] and false-positive rate [FPR]). The observers’ decision confidence also significantly decreased for 3D search with small targets (t(6) = −4.24; p = 0.005), but not for large targets (Figure S1C). The human 3D deterioration detecting the small target generalized to search tasks with different target contrasts and scenarios in which observers do not know a priori which of the two targets might be present (Figures S1D and S1E).
Why do humans show such deficits in 3D search for small targets not present in an ideal observer? Response times by themselves cannot explain the human results. The response times for 3D search of small targets had the longest response times (t(6) = −10.24; p < 0.001; Figure 1D) and resulted in the lowest accuracy.
We evaluated the hypothesis that the deterioration of accuracy when searching for the small target in 3D images is related to the human observers’ greater reliance on visual processing in the visual periphery when compared to 2D search. Such a scenario would occur if observers explored 2D images more exhaustively with their fovea than the 3D image stacks. The under-exploration and greater utilization of peripheral processing in 3D search are posited as the limiting factors in detecting the small target.
To evaluate the role of peripheral processing in the searches, we estimated the percentage of the entire image scrutinized by an area around each measured fixation location during search (i.e., the useful field of view [UFOV] 2.5° radius circle;39 Figure 1E). Figure 1F shows that the percentage of the image covered by the observers’ UFOV for the 3D search is significantly less (~1/4) than for 2D (see Figure S1F for saccade frequency analysis).
We also partitioned human missed target trials into two categories: search and recognition errors.40,41 Recognition errors are related to foveal processing and refer to instances in which the target was fixated but was still missed in the final decision. Search error trials are related to peripheral processing and refer to cases where the target was not fixated and missed in the final decision. We found that the increase in human misses for the small target in 3D search is almost exclusively related to a rise in search errors (Figure 1G). However, the 3D search does not increase the search errors for the larger target (Figure 1G). We hypothesized that the dissociation in results across target types is related to differences in their visibility in the visual periphery. We conducted separate measurements of human observers’ ability to detect the presence of briefly presented (500 ms) large or small targets across a range of foveal eccentricities at cued locations while maintaining steady fixation. The results confirmed that the small target is highly detectable near the fovea, but its detectability drastically degrades with increasing retinal eccentricity (Figure 1H). In contrast, the detectability of the large target decreases less abruptly with retinal eccentricity. Thus, the detrimental effect of 3D images on search accuracy is not present on targets that are more detectable in the visual periphery. For these targets, the visual periphery guides eye movements42 to fixate the regions more likely to contain the target, and thus, even in 3D search, there are few search errors.
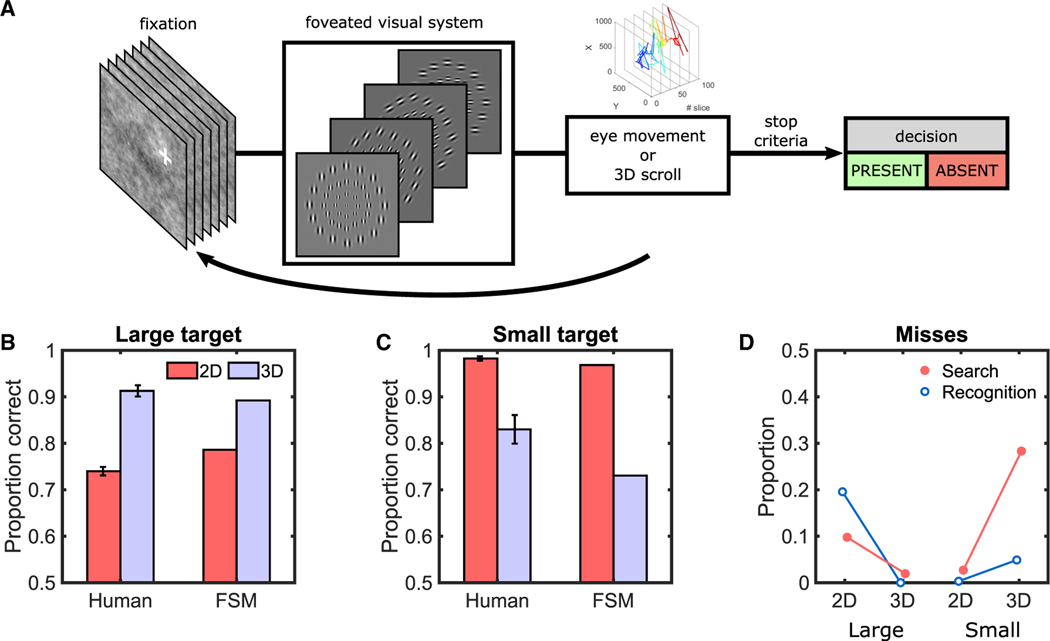
To provide further support that the human results can be explained by the peripheral visibility of a target and the under-exploration of eye movements, we implemented a computational model. The model processes the image in parallel43–45 but with varying spatial processing across the visual field. It uses an optimal linear combination of spatial frequency and orientation-tuned receptive fields at each eccentricity to detect the target.46,47 The model’s visual degradation with distance from fixation was adjusted (2 fitting parameters: scaling of Gabor receptive fields with eccentricity and internal noise; STAR Methods) to fit the average human detectability as a function of eccentricity for both targets (Figure 1H). For the search tasks, the model lines up its fovea with the actual fixations and scrolls of each individual on a given slice (~121,500 fixations across all observers’ trials; Figure 2A; Video S1). The model makes trial decisions by comparing the fixation/scroll with the strongest evidence for target presence against a decision threshold. The foveated search model (FSM) correctly predicts the human results: for 3D search (relative to 2D) reduced errors for the large target (Figure 2B) but increased the errors for the small target (Figure 2C). The FSM also shows a similar trend for error types (Figure 2D) as for humans. The modeling results provide further support that the human 3D misses are related to eye movement under-exploration and the targets’ peripheral visibility.
Figure 2. FSM Results in Synthetic Noise Images for 2D and 3D Search.
(A) FSM that incorporates receptive field sizes that scale with eccentricity and the eye movements and scrolling behavior measured in human observers. For illustration, the flowchart only shows the scaling with retinal eccentricity of a single spatial frequency channel and four orientations. See also Video S1 for the FSM’s eye movements.
(B) Proportion correct performance for human observers and the FSM for large target.
(C) Proportion correct performance for human observers and the FSM for the small target.
(D) Search and recognition errors during search for the FSM model for large and small targets.
Error bars are ±SEM.
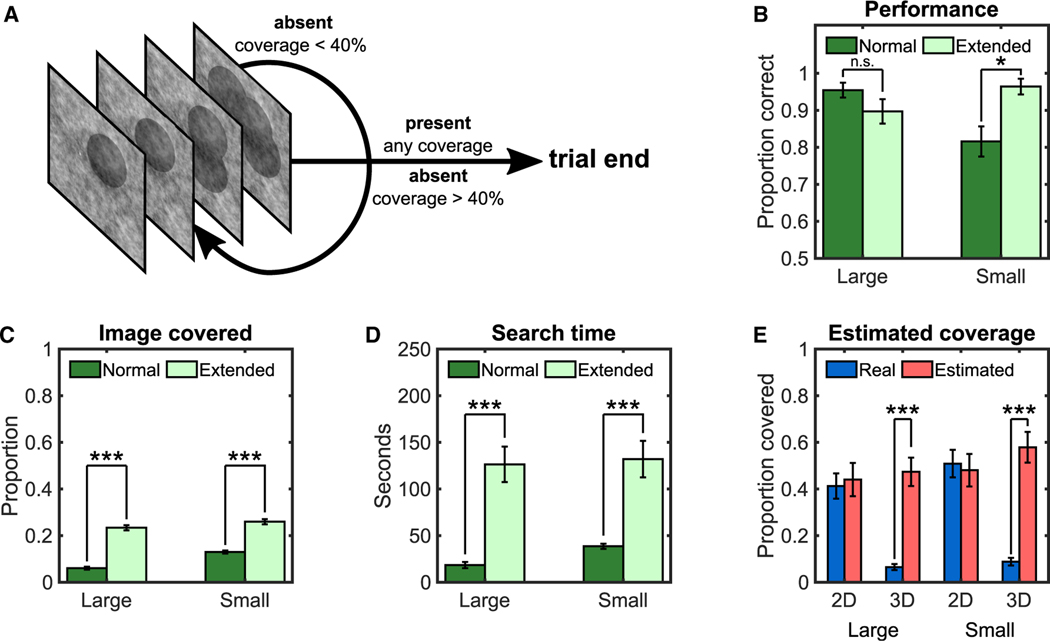
Given human observers’ tendency to under-explore 3D image stacks, can their eye movement strategy be influenced to reduce the search deficits? We conducted a study with four trained observers to test the premise that experimentally extending the eye movement exploration would reduce the 3D miss rates. Observers first completed a normal 3D search where they could terminate the search at any time. Observers then participated in a new condition in which they were not allowed to terminate their search until they either reported a target present or explored a percentage of the search area comparable to that in 2D search (40%; Figure 3A). When observers extended their visual search of 3D image stacks (Figures 3B and 3C), the search errors were vastly mitigated with the small targets (ΔPC = 0.15; p = 0.017), although introducing no difference for the large targets (p = 0.181). The extended search comes at the cost of an ~3.5-fold increase in search times (Figure 3D).
Figure 3. Human Results for Extended 3D Search and Estimated Coverage in 2D and 3D Search.
(A) Schematic of one trial in the extended 3D search experiment. Participants could not respond absent until they explored a minimum of 40% of the image calculated with a 2.5°-radius UFOV. To help observers keep track of the areas they searched, the computer shaded a 2.5° radius area around each observer’s fixations, which appeared after the observer scrolled to another slice.
(B) Proportion of correct target detection for the normal search and extended search.
(C) Proportion covered with a 2.5° UFOV.
(D) Average search times.
(E) Real and estimated coverage for participants using a 2.5° UFOV.
Error bars are ±SEM. *p < 0.05; ***p < 0.001.
But then, why do observers naturally terminate their search early if they could keep searching and greatly reduce their errors for the small target? There might be multiple reasons, including observers’ inability to estimate the performance benefits of extending the search, failure to account for the visibility of the targets in the visual periphery, or a secondary goal to minimize sensory-motor energetic costs.48,49 A follow-up experiment provided some clues as to why observers under-explore. Six new observers searched for the targets in 2D and 3D search and, after each trial, provided an estimate of the percentage of the total area they had explored. Results (Figure 3E) showed that observers accurately estimated the percentage of the 2D image areas covered but vastly overestimated the area covered in 3D image stacks. This may be one reason why subjects terminate their 3D search before more fully exploring the images.
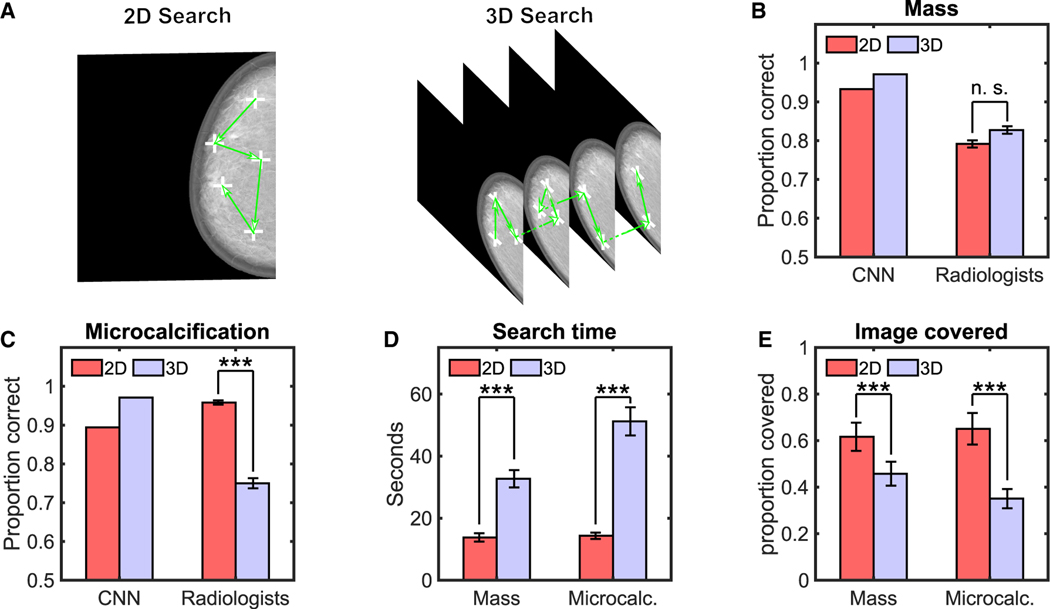
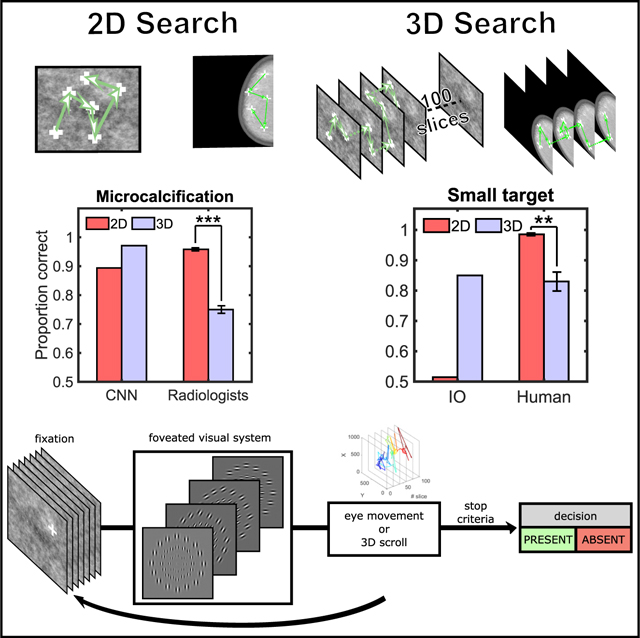
To assess the results’ generality to more real-world scenarios, we conducted a similar study with 12 radiologists. We used twenty-eight digital breast tomosynthesis (DBT) phantom images50,51 (Figure 4A) that included simulated targets (50% presence) that are typically spatially large (mass; size = 0.5°) and those that are small (microcalcification; size = 0.06°). We used the same images for 2D and 3D search to isolate the effects due to search from other variables related to the generation of the images by different imaging systems. For the 2D search, images consisted of the central slice of the 3D image stack.
Figure 4. Radiologists and CNN Results for 2D and 3D Search with Digital Breast Tomosynthesis Phantoms.
(A) Sample 2D and 3D of digital breast tomosynthesis (DBT) phantoms with sample fixations and scrolls of a radiologist.
(B) Proportion correct of convolutional neural network (CNN) and radiologists for detection of the simulated mass.
(C) Proportion correct of radiologists and CNN for detection of the simulated microcalcification.
(D) Average search time for radiologists.
(E) Proportion of image covered by radiologists (2.5° radius UFOV).
***p < 0.001. n.s., not statistically significant. Error bars are ±SEM. See also Figure S2.
The comparison of radiologists to an optimal Bayesian observer is not possible because the model requires knowledge of the images’ statistical properties, which are not available for the DBT breast phantoms. Thus, we computed performance for the best current convolutional neural network (CNN) to segment organs in medical images (nn 3D U-net)52 after training to detect our simulation targets. We found that the CNN’s detection for both mass and microcalcification improved for 3D relative to 2D search (Figures 4B and 4C). In contrast, radiologists’ performance for the microcalcification degraded significantly between the 3D and 2D search (ΔPC = 0.209, t(11) = − 4.5220, p < 0.001; Δd’ = −1.5, p < 0.001), but not for the larger mass target (ΔPC = 0.04, t(11) = 0.64; p = 0.54; Δd’ = −0.19; 2-way interaction for target type and search type; F[1, 668] = 19.91; p < 0.0001; Figures S2A and S2B for a breakdown in TPR and FPR). The confidence of radiologists decreased significantly for the 3D small target decisions relative to 2D (t(11) = 4.76; p < 0.001), but not the large target (Figure S2C). Radiologists spent more time in 3D search (Figure 4D) but covered a smaller fraction of all the images regions for the 3D search relative to the 2D search (Figure 4E; t(11) = −8.35; p < 0.001 for masses; t(11) = −9.21; p < 0.001 for microcalcifications; Figure S2D for eye movements).
One possible explanation for radiologists’ tendency to under-explore the 3D image stacks is their awareness that the readings were part of a laboratory study. An analysis of the data volume1 (64–128 slices) and reading times in clinical scenarios (2 to 3 min per case) suggests that radiologists typically under-explore 3D image stacks in clinical practice. And yet a survey we administered to 21 radiologists suggests a vast overestimation of the area they explore in clinical practice. They reported 94% (±3.5%) of the area for 2D mammograms and 90% (±4.1%) for 3D DBT images.
Together, our findings do not imply that 3D imaging techniques always lead to inferior human detection performance for small targets relative to 2D methods. Other differences arise between real-world 2D and 3D images related to the image acquisition and reconstruction processes. However, a recent study with radiologists found a degradation for small targets in real DBT images, suggesting that some of the visuo-cognitive bottlenecks revealed in the current paper are at play.4 Thus, our study motivates the need to find solutions to mitigate miss errors with small targets. With a prolonged practice focused only on the 3D search for small targets, observers might learn to search more exhaustively. Encouraging observers to extend their search might reduce errors. The prolonged search is impractical in terms of the associated added time cost. One practical solution is to present a 2D image along with the 3D image stack. This would allow radiologists to take advantage of the performance benefits of the 3D stacks for large targets and rely on the 2D image to find small targets easily missed in the 3D image stacks. A synthesized 2D image can be created from the 3D image stack and presented to the observer. Such a solution is typical in most clinics using DBT images.53,54 Alternatively, computer vision/artificial intelligence aids55–57 and multiple readers58–60 can also reduce search errors.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Miguel P. Eckstein (miguel.eckstein@psych.ucsb.edu).
Materials Availability
No materials are available for this study.
Data and Code Availability
All of the raw data from this article are accessible via Mendeley Data (https://doi.org/10.17632/tjy4h67z4j.1). Analysis scripts can be provided by requesting them to the Lead Contact. Other necessary software is listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Raw Data | This paper | https://doi.org/10.17632/tjy4h67z4j.1 |
| Software and Algorithms | ||
| MATLAB R2019a | The MathWorks | RRID: SCR_001622 |
| Python | https://www.python.org | RRID: SCR_008394 |
| Eyelink 1000 Eyetracker | SR Research, Mississauga, ON, Canada | RRID: SCR_009602 |
| Psychotoolbox | 61 | http://psychtoolbox.org/ |
| PyschoPy | 62 | https://www.psychopy.org/ |
| nn 3D U-net | 63 | https://github.com/MIC-DKFZ/nnUNet |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Seven undergraduate students and four graduate students from the University of California, Santa Barbara, participated in the 2D and 3D search study with filtered noise. All participants were verified to have normal or corrected-to-normal vision. Undergraduates received course credit in exchange for participation. A second group of twelve expert radiologists, naive to the hypothesis, were recruited from the Hospital of the University of Pennsylvania and Pennsylvania Hospital. Radiologists’ experience ranged from 1 month to 3 years of DBT reading with a volume of 50 to 500 cases read per year. The gender balance for the study performed at UC Santa Barbara was 28% male and 72% female, with ages ranging from 20 to 29 years. For the radiologist psychophysical study, it was 75% male and 25% female, with ages ranging from 27 to 34. All participants provided informed written consent and were recruited and treated according to approved human subject research protocols by the University of California, Santa Barbara (12-16-0806). Finally, the survey was conducted with 21 radiologists from the Sansum Clinic in Santa Barbara, Pennsylvania Hospital, and Hospital of the University of Pennsylvania.
METHOD DETAILS
Psychophysical Search Task with 1/f2.8 filtered noise
Seven observers searched for a target in 2D and 3D images with a 50% probability of the target being present on each trial. They were instructed to search for a known signal embedded within a correlated Gaussian noise field. The stimuli images were created using a 3D correlated Gaussian noise field (μ = 128, σ = 25) with a 3D power spectrum S(f) derived from X-ray breast mammograms, specifically NPS(f)= 1/f2:8 using frequency indices. The size of a 3D volumetric image was 1024 × 820 × 100 voxels. Subjects saw 800 trials in total. 2D and 3D search sessions were intermixed and counterbalanced. Each observer participated in 5 sessions of 80 trials per condition (2D and 3D) for a total of 160 trials per session. Two different targets were generated, designed to have high and low visibility in the periphery (large and small target, respectively). The searched target was randomized across trials. The larger target was a 3D Gaussian-shaped target (2σ = 20 voxels or 0.44 degrees of visual angle), and the smaller target was a sphere (diameter = 6 voxels or 0.13 degrees of visual angle), contrast for both targets was set to 65%; see Figure S3A for more information. While the correlated noise field and large target are mainly present at lower frequencies, the small target has a higher presence on higher frequencies due to its sharp edges. For each trial, before the presentation of the test image, a high contrast copy of the target that might be present in the trial (large or small) was shown to observers and remained visible during the trial. After pressing the space bar, the image was displayed.
In the 3D case, the volume was displayed as a stack of 2D images, and the first slice was initially displayed. The observers could scroll freely using the mouse with no time limit or other constraints. A non-overlapping scroll bar was presented on the right side of the screen showing the slice that was currently being displayed. In the 2D trials, only one slice was present (the one corresponding to the central slice of the target, if present), and the scrolling was disabled. At any time, to finalize the trial, observers had to press the space bar. Observers used an 8-scale confidence rating to indicate their decision about the presence (ratings 5–8) or absence (ratings 1–4) of the target. Subsequently, a feedback screen showing the correct response and the target location (in case of a target-present trial) was displayed. Before the experiment, we trained observers to navigate through volumetric images using 10 practice trials. We did not use the practice trials for the analysis.
The stimuli were displayed in a medical-grade monitor (Barco MDRC-1119 LCD) with a resolution of 1280 × 1024 pixels at a distance of ~75 cm (pixels per degree of visual angle was 45). The monitor was calibrated to have linear contrast with 0.05 cd/m2 at gray level 0 and 111 cd/m2 at gray level 255. The experiment took place in a darkened room. The screen area outside the image was set to a neutral gray level of 128. Figure S3B shows the outline of the search experiment. A real-time eye tracker (Eyelink 1000, SR Research) was used at all times, including a calibration screen at the beginning of each experimental session. Fixations were detected using the default parameters: eye velocity and acceleration thresholds of 30 deg/s and 9,500 deg/s2, respectively. The timing of the scrolling behavior was also recorded at a sampling rate of 60Hz. We used Psychtoolbox to develop this experiment.61
Detectability versus retinal eccentricity psychophysical task
This experiment measured the detectability of both targets with respect to the distance to the fixation point (eccentricity). The same seven observers that participated in the search experiment participated in this experiment. This forced-fixation experiment was designed as a location-known exactly and signal-known-exactly task in which the observers were instructed to fixate at a cross while fiduciary marks indicated the possible location of the target (Yes/No task; 50% probability of target presence). After pressing the space bar, the stimulus was shown for 500ms. Eye-position was monitored in real-time. If a change in eye position exceeded 1 degree of visual angle, the trial was interrupted. Observers responded using an 8-scale confidence rating about the absence or presence of the target. Small and large target trials were intermixed randomly, with 50% prevalence. The target contrasts were the same as the original search experiment. Figure S3C shows the outline of one trial of this experiment. We measured the detectability at eccentricities of 0, 3, and 6 degrees of visual angle for the small target and at 0, 3, 6, 9, and 12 degrees for the large target. Each observer participated in 1,000 and 800 trials, respectively, for large and small targets across eccentricities (total of 8,000 trials). We used the same eye tracker and monitor setup as for the search psychophysical experiment. We used Psychtoolbox to develop this experiment.61
Extended search psychophysical task
Four new observers participated in this experiment. In the control condition (normal search), observers freely searched small and large targets only in 3D image stacks. In a separate, extended search condition, participants had to explore at least 40% of the 3D image stack to terminate the trial, if their response was target absent. This restriction was not enforced when they responded that the target was present, allowing participants to terminate the trial at any time in these trials. Additionally, to allow observers to better track the regions that they have explored, circular shaded areas (2.5-degree radius) appeared around observer fixations when participants revisited a slice. At any time, participants could see their current explored percentage relative to the minimum explored percentage required to terminate the trial. By pressing a key, the percentage left to reach the threshold (from 0% to 100%) was shown on the screen. Participants were not informed that this threshold was set to 40%. Each observer participated in 100 trials for both large and small targets and was only performed for 3D search. We used the same eye tracker and monitor setup as for the search psychophysical experiment. We used PsychoPy to develop this experiment.62
Estimated search psychophysical task
Six participants participated (one out of seven from the 2D/3D search experiment and the same four from the extended search) in another variation of the 2D/3D search experiment, which included an additional question at the end of each trial. After responding to the absence or presence of the target, participants were asked to estimate the percentage of image/volume they had explored. They responded using a slider that ranged from 0% to 100%. Observers participated in 200 trials across all conditions. We used the same eye tracker and monitor setup as for the search psychophysical experiment. We used PsychoPy to develop this experiment.62
Image coverage evaluation
To calculate the percentage of the images explored during the search, we “painted” the image with 2.5-degree-radius circles centered on each fixation. In 3D images, we only “painted” these circles on the slice at the moment of each fixation. 3D scrolls counted as new fixations for the total computation. The final percentage is calculated by dividing the number of pixels (or voxels in 3D) covered by the circles by the total number of pixels (or voxels in 3D).
Psychophysical Search Task with radiologists
Twelve radiologists participated in the study. They sat ~75 cm away from a vertical medical-grade monitor placed in a darkened room. Images used in the radiologist study were generated by the OpenVCT virtual breast imaging tool from the University of Pennsylvania.50,64,65 This tool generates full phantom DBT images, including different tissues (skin, Cooper’s ligaments, adipose, and glandular) in a realistic manner. The phantom is projected using clinical acquisition geometry and clinical automatic exposure control settings (Selenia Dimensions, Hologic, Marlborough, MA). Reconstruction was performed at 100μm in-plane resolution and 1mm slice spacing (Briona Standard; Real Time Tomography, LLC, Villanova, PA). We used 700ml phantoms compressed in ML direction at 6.33mm thickness with glandular tissue prevalence of 15%–25%. The size of each 3D virtual DBT was 2048×1792×64 voxels. The stimuli were displayed in a 5Mpx grayscale DICOM calibrated monitor (2560×2048 pixels), keeping their aspect ratio. Two lesions were simulated and inserted in a random location on 50% of the trials. A small lesion, similar to a microcalcification, was simulated as a solid sphere of 0.3mm diameter (0.06 degrees of visual angle). A large lesion, similar to a mass, was simulated as a combination of several 3D ellipsoids with an average diameter of 7mm (0.5 degrees of visual angle). Density stepped from the center to the sides of the lesion to simulate blending with the background. For 2D stimuli, the central slice of the lesion was selected on signal-present trials, the central slice of the reconstruction was selected on signal-absent trials.
Radiologists were asked to search for a given signal (microcalcification or mass) in the phantom DBT. The experiment had 56 trials, 28 of them corresponding to the 2D single slice case, and 28 to the complete 3D volume. Each condition had 14 signal-absent trials and 14 signal-present trials. The prevalence between microcalcification and masses was also 50%. All four conditions were randomly intermixed. An eye tracker recorded the participant’s eye movements in real-time at a frequency of 500Hz (EyeLink Portable Duo, SR Research). We used Psychtoolbox to develop this experiment.61
Radiologist survey
An online survey was sent to a group of twenty-one radiologists. The survey asked radiologists to estimate the percentage of the image explored during the reading of a 2D Digital Mammogram and a 3D Digital Breast Tomosynthesis.
Calculating figures of merit for task performance
To compare performances, we calculated the True Positive Rate (TPR) and False Positive Rate (FPR). We calculated the proportion of correct trials as PC = (TPR + (1 – FPR))/2 for models and humans. The index of detectability d′ was calculated using the usual transformation for a yes/no task, where is the inverse of the cumulative normal distribution function.66
| (Equation 1) |
2D/3D Ideal Bayesian Observer
The ideal observer model was run on the same images with which the human observers were presented. The ideal observer calculates, for each location in the 2D or 3D image, the posterior probability of the target being present or absent given the image data to make optimal decisions.66–69 For search, the ideal observer’s first step reduces to correlating a 2D or 3D optimal template with the image at all locations that might contain the target.69,70 The ideal observer’ template w(x, y, z) is convolved (*) with the image71 g(x, y, z) and a decision variable λ was calculated for each pixel/voxel in the image as follows:
| (Equation 2) |
The optimal linear template, w(x, y, z) is calculated by taking into account the shape of the target and the spatio-temporal correlations in the noise. The template is calculated from the 3D image covariance matrix of the noise (Kg), which describes the noise variance and covariance between all pixels in the 3D image stack and a mathematical representation of each target. This optimal template calculation is typically expressed using 1D vectorized version of the 2D or 3D signal (s) and a 2D covariance matrix, Kg:
| (Equation 3) |
where wIO is a 1-D vectorized version of the optimal template.
The scalar template response (λ, Equation 2), at each location p, is used to calculate the likelihood of the template response given target presence and target absence using the expected mean and standard deviation of the ideal observer template responses under each hypothesis. A likelihood ratio can be calculated for each location. A yes/no decision is reached by summing the evidence for the target (likelihood ratios) across locations and comparing the resulting decision variable to a threshold.38,68–70 The likelihoods are given by Gaussian probability density functions with different means for target presence and target absence but the same variance. The 2D search ideal observer implementation can be derived similarly by considering single slices and 2-dimensions, and consistent with previous implementations.69,72–75 To compare the ideal observer and human performance across conditions, we added internal noise to the ideal observer to match human performance in 3D for each target type. Other options, such as matching 2D performance of human and ideal observer, led to ceiling performance for the ideal observer in 3D search of the small signal.
3D Foveated Search Model (FSM)
This model observer accounts for the target detectability as a function of the target’s distance from the observer’s fixation point (retinal eccentricity). The model processes the entire image in parallel with a foveated visual system for each fixation point. The model processes the visual input with spatial frequency and orientation tuned feature extraction channels (Gabor functions). To model the decreasing spatial resolution with increasing retinal eccentricity, we scaled the Gabor channels as a function of distance from the target to the point of fixation. At 0 degrees retinal eccentricity (the fovea), the center frequencies of the channels are the six spatial frequencies of a standard channel model (0.5, 1, 2, 4, 8, and 16 cycles per degree of visual angle) and the eight different orientations.76 The center frequencies for all the Gabor channels are non-linearly scaled as a function of the eccentricity, E, in degrees of visual angle.
| (Equation 4) |
Thus, the model loses access to high spatial frequencies as the retinal eccentricity increases (e.g., the highest spatial frequency channel ranged from 16 cycles per degree in the fovea to 0.33 cycles per degree at 9 degrees eccentricity). At each eccentricity, the model uses the best linear combination of channels to detect the target. To reduce computational complexity, we utilized ten sets of different channels to cover eccentricities from the fovea to 9 degrees of visual angle.
We calculated the proportion correct for a Yes/No task of the FSM model detecting each target (50% probability of target presence) at a single location at each eccentricity. The proportion correct was transformed into a d’ detectability index for the model at each eccentricity for each target. We fit the model to human measurements of d’ versus retinal eccentricity target detection data for both target types simultaneously with a single set of parameters. We used a global optimization algorithm based on the MATLAB implementation of the Mesh Adaptive Direct Search (MADS) to fit the two Gabor-channel scaling parameters α and β. To degrade the model’s overall performance to match that of humans, we included internal noise, which perturbed the model’s decision variable. The internal noise was a random scalar value sampled from a Gaussian distribution with a standard deviation proportional to the standard deviation of the model decision variable: as a fitting parameter fixed across target types. Optimized parameters were found to be α = 0.7063, β = 1.6953, and K = 2.7813. The different sets of channels per eccentricity result in ten templates that process the visual field, one per each specific eccentricity.
To apply the FSM to the visual search task, we assumed a fixation point and processed the entire image with the foveated model. For each location (p) in the image, the dot product between the corresponding template (wε) at the eccentricity (ε) and the image (gp) data at that location were used to generate a response for the model (λp). The result is modified by an additive the internal noise εint:
| (Equation 5) |
The 3D component of the model is constructed by building independent 2D foveated channelized templates, corresponding to each slice of the signal, and stacking them together to create a final 3D template. To model slice/depth integration, we measured the mean number of slices viewed during a fixation across observers and trials. Human observers scrolled, on average, across fives slices for each fixation. Thus, the model also utilized five slices: the signal’s central slice plus two slices above and below.
The scalar template response λp at each location p was used to calculate a likelihood of the response for that eccentricity given target presence or absence:
| (Equation 6) |
where μ and σ are the mean response and standard deviation of the template response, respectively. Finally, a likelihood ratio was then computed for each location by dividing the likelihood of the signal present by the likelihood of the signal absent.
| (Equation 7) |
Integration across fixations
We considered optimal integration of information, assuming statistical independence. The decision variable in every location of the image is the result of the product of likelihood ratios for all the fixations N in the current slice.
| (Equation 8) |
Final perceptual decisions after multiple fixations
In order to achieve a final decision, we combined the template responses in different ways. For the ideal observer, we used the optimal rule: the sum of the likelihood ratios across slices and then compared the resulting variable against a decision threshold. For the foveated model, we used the maximum across fixations and slices. Note that the distribution of varies with the number of fixations (N) executed by the model. Consequently, the optimal decision threshold also varies with N. We estimated the optimal thresholds for every N by training the model on a separate set of trials and images for N = [0…30].
Human Fixations
We utilized the measured human fixations as input for the fixations of the model for the same trials/images. The performance was calculated for each human observer separately by having the model fixate in the corresponding fixations of the human observer for each trial. There was an exclusion of fixations of less than 50 ms, which were fewer than 1% of all fixations
Convolutional Neural Network applied Digital Breast Tomosynthesis Phantoms
The medical segmentation decathlon77 is a general-purpose 3D segmentation challenge for the following organs: liver, brain, hippocampus, lung, prostate, cardiac, pancreas, colon, hepatic, and spleen tumor segmentations. Due to the wide variety of tasks, networks trained for this challenge are easily generalizable to new tasks. Fabian et al.63 implemented nn U-Net for this challenge and is currently the best performing network. It is based on a simple U-Net architecture and the training process is simplified by automating the selection of training parameters based on the properties of the dataset. The nn U-Net is optimized to work for 12 GB GPUs. This brings a restriction on the maximum patch size that can be processed by the network. Our DBT images are of size 2048×1664×64 and exceed the maximum size allowed by the network. Although nn U-Net provides a fallback option of using a cascade network, consisting of a low-resolution network followed by another network to refine the segmentation for images where the patch size in full resolution mode is much smaller than the image size, it is a much slower training process which would likely take ~20 days to train on a single GPU. In order to expedite the training process, we chose to crop the input image to make it conform to the requirements of the non-cascade network. The training input image was cropped to a size of 64×380×380. For the 2D cases, since the 3D network does not allow for a single slice, the central signal slice is replicated four times, resulting in input images of 4×380×380. The same network architecture is used for both 2D and 3D tasks. The network tries to minimize a combination of the Dice coefficient78 loss and cross-entropy loss. We used 524 images for training and 102 images for evaluation. No changes were made to the network or the hyperparameters, which were suggested by the automation part of the network. The evaluation was performed in the same full-resolution images (2048×1664×64) as those viewed by the radiologists.
Since the goal was to compare the segmentation model against radiologists’ performance when detecting the target, traditional evaluation metrics like Dice or Jaccard coefficients were not used for evaluation. In order to obtain a comparable metric with humans, we defined our own evaluation metric. If the input to the network is of size (2048×1664×64), the output of the network is (2×2048×1664×64), where the two values corresponding to each pixel/voxel correspond to background and foreground (i.e., target) probabilities for that pixel. We selected only those pixels/voxels with a foreground probability of 1.0. Since the goal is to detect the presence of a target, we computed all the 3D connected components (Q) with six connectivity in the binary output image. Then, we computed the volume of each of the connected components. Finally, we selected the number of pixels/voxels of the maximum out of those Q volumes to be the representative of a given image. This process resulted in N values corresponding to the N 2D images or 3D image sequences. A decision threshold was utilized to determine whether an image/image stack was target present or absent. We varied a decision threshold and selected the threshold that maximized the proportion correct.
QUANTIFICATION AND STATISTICAL ANALYSIS
The psychophysical data from readers was modeled as a dichotomous trial outcome coded as 0 (incorrect response) or 1 (correct response). The data were analyzed in MATLAB (version 2019a) using logistic regression (logit link function) and mixed-effects ANOVA in which cases and readers are modeled as random effects, and image modality (2D or 3D) and target size (large or small) are modeled as fixed effects. We modeled the main effects as well as 2-way interactions between effects. The primary endpoint of the statistical analysis was the interaction between modality and target size, which was consistent with the dissociation in performance due to the limitations of peripheral visual processing. Additionally, for the purpose of comparing 2D and 3D search parameters within a given signal size, we also calculated paired comparison t tests across subjects. The sample size and statistical details of the experiments are indicated in each study in the Results section. For all statistical tests, we used the significance criteria of p < 0.05. We additionally used standard t tests for pairwise comparisons.
Supplementary Material
Highlights.
Humans searching 3D image stacks miss small targets that are salient in 2D images
Optimal observers and deep neural networks do not show the deficits for 3D images
Human eye movement under-exploration in 3D search explains the misses
New 3D-imaging technologies should consider these human visuo-cognitive bottlenecks
ACKNOWLEDGMENTS
The research was funded by the National Institutes of Health grants R01 EB018958 and R01 EB026427 and the US Army Research Office under contract number W911NF-19-D-0001 for the Institute for Collaborative Biotechnologies. The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the official policies, either expressed or implied, of the US government. The US government is authorized to reproduce and distribute reprints for government purposes notwithstanding any copyright notation herein.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2020.12.029.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Rogalla P, Kloeters C, and Hein PA (2009). CT technology overview: 64-slice and beyond. Radiol. Clin. North Am 47, 1–11. [DOI] [PubMed] [Google Scholar]
- 2.Skaane P. (2017). Breast cancer screening with digital breast tomosynthesis. Breast Cancer 24, 32–41. [DOI] [PubMed] [Google Scholar]
- 3.Wetter OE (2013). Imaging in airport security: past, present, future, and the link to forensic and clinical radiology. J. Forens. Radiol. Imag 1, 152–160. [Google Scholar]
- 4.Georgian-Smith D, Obuchowski NA, Lo JY, Brem RF, Baker JA, Fisher PR, Rim A, Zhao W, Fajardo LL, and Mertelmeier T. (2019). Can digital breast tomosynthesis replace full-field digital mammography? A multireader, multicase study of wide-angle tomosynthesis. Am. J. Roentgenol 212, 1393–1399. [DOI] [PubMed] [Google Scholar]
- 5.Mayo JR, and Lam S. (2015). Computed tomography and the secrets of lung nodules. Can. Assoc. Radiol. J 66, 2–4. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe JM, and Horowitz TS (2017). Five factors that guide attention in visual search. Nat. Hum. Behav 1, 0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckstein MP, Koehler K, Welbourne LE, and Akbas E. (2017). Humans, but not deep neural networks, often miss giant targets in scenes. Curr. Biol 27, 2827–2832.e3. [DOI] [PubMed] [Google Scholar]
- 8.Malcolm GL, and Henderson JM (2009). The effects of target template specificity on visual search in real-world scenes: evidence from eye movements. J. Vis 9, 8. [DOI] [PubMed] [Google Scholar]
- 9.Eckstein MP (2017). Probabilistic computations for attention, eye movements, and search. Annu. Rev. Vis. Sci 3, 319–342. [DOI] [PubMed] [Google Scholar]
- 10.Peelen MV, and Kastner S. (2014). Attention in the real world: toward understanding its neural basis. Trends Cogn. Sci 18, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe JM, Võ ML-H, Evans KK, and Greene MR (2011). Visual search in scenes involves selective and nonselective pathways. Trends Cogn. Sci 15, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M, Feng J, Ma KT, Lim JH, Zhao Q, and Kreiman G. (2018). Finding any Waldo with zero-shot invariant and efficient visual search. Nat. Commun 9, 3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verghese P. (2012). Active search for multiple targets is inefficient. Vision Res. 74, 61–71. [DOI] [PubMed] [Google Scholar]
- 14.Wolfe JM, Horowitz TS, and Kenner NM (2005). Cognitive psychology: rare items often missed in visual searches. Nature 435, 439–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michel M, and Geisler WS (2011). Intrinsic position uncertainty explains detection and localization performance in peripheral vision. J. Vis 11, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulun VC, Schütz AC, Michel MM, Geisler WS, and Gegenfurtner KR (2015). Visual search under scotopic lighting conditions. Vision Res. 113 (Pt B), 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitroff SR, and Biggs AT (2014). The ultra-rare-item effect: visual search for exceedingly rare items is highly susceptible to error. Psychol. Sci 25, 284–289. [DOI] [PubMed] [Google Scholar]
- 18.Semizer Y, and Michel MM (2017). Intrinsic position uncertainty impairs overt search performance. J. Vis 17, 13. [DOI] [PubMed] [Google Scholar]
- 19.Ackermann JF, and Landy MS (2010). Suboptimal choice of saccade endpoint in search with unequal payoffs. J. Vis 10, 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Desolneux A, Muller S, de Carvalho PM, and Carton A-K(2018). Comparison of microcalcification detectability in FFDM and DBT using a virtual clinical trial. Proc. SPIE 10577, 105770D. [Google Scholar]
- 21.Gur D, Abrams GS, Chough DM, Ganott MA, Hakim CM, Perrin RL, Rathfon GY, Sumkin JH, Zuley ML, and Bandos AI (2009). Digital breast tomosynthesis: observer performance study. AJR Am. J. Roentgenol 193, 586–591. [DOI] [PubMed] [Google Scholar]
- 22.Badano A, Graff CG, Badal A, Sharma D, Zeng R, Samuelson FW, Glick SJ, and Myers KJ (2018). Evaluation of digital breast tomosynthesis as replacement of full-field digital mammography using an in silico imaging trial. JAMA Netw. Open 1, e185474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams LH, and Drew T. (2019). What do we know about volumetric medical image interpretation?: a review of the basic science and medical image perception literatures. Cogn. Res. Princ. Implic 4, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C-C, and Wolfe JM (2019). Eye movements in medical image perception: a selective review of past, present and future. Vision (Basel) 3, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karimi S, Jiang X, Cosman P, and Martz H. (2014). Flexible methods for segmentation evaluation: results from CT-based luggage screening. J. XRay Sci. Technol 22, 175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrand G, English J, and Irani P. (2016). 3D visualization of astronomy data cubes using immersive displays. arXiv, arXiv:1607.08874v1. https://arxiv.org/abs/1607.08874. [Google Scholar]
- 27.Goodman AA (2012). Principles of high-dimensional data visualization in astronomy. Astron. Nachr 333, 505–514. [Google Scholar]
- 28.Drew T, Vo ML, Olwal A, Jacobson F, Seltzer SE, and Wolfe JM(2013). Scanners and drillers: characterizing expert visual search through volumetric images. J. Vis 13, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aizenman A, Drew T, Ehinger KA, Georgian-Smith D, and Wolfe JM(2017). Comparing search patterns in digital breast tomosynthesis and full-field digital mammography: an eye tracking study. J. Med. Imaging (Bellingham) 4, 045501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noroozian M, Hadjiiski L, Rahnama-Moghadam S, Klein KA, Jeffries DO, Pinsky RW, Chan HP, Carson PL, Helvie MA, and Roubidoux MA (2012). Digital breast tomosynthesis is comparable to mammographic spot views for mass characterization. Radiology 262, 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohn TE, and Lasley DJ (1974). Detectability of a luminance increment: effect of spatial uncertainty. J. Opt. Soc. Am 64, 1715–1719. [DOI] [PubMed] [Google Scholar]
- 32.Palmer J. (1994). Set-size effects in visual search: the effect of attention is independent of the stimulus for simple tasks. Vision Res. 34, 1703–1721. [DOI] [PubMed] [Google Scholar]
- 33.Burgess AE, and Ghandeharian H. (1984). Visual signal detection. II. Signal-location identification. J. Opt. Soc. Am. A 1, 906–910. [DOI] [PubMed] [Google Scholar]
- 34.Bochud FO, Abbey CK, and Eckstein MP (2004). Search for lesions in mammograms: statistical characterization of observer responses. Med. Phys 31, 24–36. [DOI] [PubMed] [Google Scholar]
- 35.Burgess AE, Wagner RF, Jennings RJ, and Barlow HB (1981). Efficiency of human visual signal discrimination. Science 214, 93–94. [DOI] [PubMed] [Google Scholar]
- 36.Geisler WS (2011). Contributions of ideal observer theory to vision research. Vision Res. 51, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barlow HB (1980). The absolute efficiency of perceptual decisions. Philos. Trans. R. Soc. Lond. B Biol. Sci 290, 71–82. [DOI] [PubMed] [Google Scholar]
- 38.Eckstein MP, Whiting JS, and Thomas JP (1996). Role of knowledge in human visual temporal integration in spatiotemporal noise. J. Opt. Soc. Am. A Opt. Image Sci. Vis 13, 1960–1968. [DOI] [PubMed] [Google Scholar]
- 39.Kundel HL (1975). Peripheral vision, structured noise and film reader error. Radiology 114, 269–273. [DOI] [PubMed] [Google Scholar]
- 40.Kundel HL, Nodine CF, Thickman D, and Toto L. (1987). Searching for lung nodules. A comparison of human performance with random and systematic scanning models. Invest. Radiol 22, 417–422. [DOI] [PubMed] [Google Scholar]
- 41.Krupinski EA (2011). The role of perception in imaging: past and future. Semin. Nucl. Med 41, 392–400. [DOI] [PubMed] [Google Scholar]
- 42.Eckstein MP, Beutter BR, and Stone LS (2001). Quantifying the performance limits of human saccadic targeting during visual search. Perception 30, 1389–1401. [DOI] [PubMed] [Google Scholar]
- 43.Najemnik J, and Geisler WS (2008). Eye movement statistics in humans are consistent with an optimal search strategy. J. Vis 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig CJH, Davies JR, and Eckstein MP (2014). Foveal analysis and peripheral selection during active visual sampling. Proc. Natl. Acad. Sci. USA 111, E291–E299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akbas E, and Eckstein MP (2017). Object detection through search with a foveated visual system. PLoS Comput. Biol 13, e1005743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Myers KJ, and Barrett HH (1987). Addition of a channel mechanism to the ideal-observer model. J. Opt. Soc. Am. A 4, 2447–2457. [DOI] [PubMed] [Google Scholar]
- 47.Shimozaki SS, Eckstein MP, and Abbey CK (2003). An ideal observer with channels versus feature-independent processing of spatial frequency and orientation in visual search performance. J. Opt. Soc. Am. A Opt. Image Sci. Vis 20, 2197–2215. [DOI] [PubMed] [Google Scholar]
- 48.Araujo C, Kowler E, and Pavel M. (2001). Eye movements during visual search: the costs of choosing the optimal path. Vision Res. 41, 3613–3625. [DOI] [PubMed] [Google Scholar]
- 49.Kowler E. (2011). Eye movements: the past 25 years. Vision Res. 51, 1457–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pokrajac DD, Maidment AD, and Bakic PR (2012). Optimized generation of high resolution breast anthropomorphic software phantoms. Med. Phys 39, 2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakic PR, Pokrajac DD, De Caro R, and Maidment ADA (2014). Realistic simulation of breast tissue microstructure in software anthropomorphic phantoms. In Breast Imaging: 12th International Workshop, IWDM 2014, Gifu City, Japan, June 29–July 2, 2014. Proceedings, Fujita H, Hara T, and Muramatsu C, eds. (Springer International; ), pp. 348–355. [Google Scholar]
- 52.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, and Ronneberger O. (2016). 3D U-Net: learning dense volumetric segmentation from sparse annotation. arXiv, arXiv:1606.06650v1. https://arxiv.org/abs/1606.06650v1. [Google Scholar]
- 53.Zuckerman SP, Maidment ADA, Weinstein SP, McDonald ES, and Conant EF (2017). Imaging with synthesized 2D mammography: differences, advantages, and pitfalls compared with digital mammography. AJR Am. J. Roentgenol 209, 222–229. [DOI] [PubMed] [Google Scholar]
- 54.Zuckerman SP, Conant EF, Keller BM, Maidment ADA, Barufaldi B, Weinstein SP, Synnestvedt M, and McDonald ES (2016). Implementation of synthesized two-dimensional mammography in a population-based digital breast tomosynthesis screening program. Radiology 281, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenspan H, van Ginneken B, and Summers RM (2016). Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 35, 1153–1159. [Google Scholar]
- 56.Rodriguez-Ruiz A, Lång K, Gubern-Merida A, Teuwen J, Broeders M, Gennaro G, Clauser P, Helbich TH, Chevalier M, Mertelmeier T, et al. (2019). Can we reduce the workload of mammographic screening by automatic identification of normal exams with artificial intelligence? A feasibility study. Eur. Radiol 29, 4825–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N,Ashrafian H, Back T, Chesus M, Corrado GS, Darzi A, et al. (2020). International evaluation of an AI system for breast cancer screening. Nature 577, 89–94. [DOI] [PubMed] [Google Scholar]
- 58.Juni MZ, and Eckstein MP (2017). The wisdom of crowds for visual search. Proc. Natl. Acad. Sci. USA 114, E4306–E4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geijer H, and Geijer M. (2018). Added value of double reading in diagnostic radiology,a systematic review. Insights Imaging 9, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caumo F, Brunelli S, Zorzi M, Baglio I, Ciatto S, and Montemezzi S. (2011). Benefits of double reading of screening mammograms: retrospective study on a consecutive series. Radiol. Med. (Torino) 116, 575–583. [DOI] [PubMed] [Google Scholar]
- 61.Kleiner M, Brainard D, Pelli D, Ingling A, Murray R, and Broussard C(2007). What’s new in Psychtoolbox-3. Perception 36, 1–16. [Google Scholar]
- 62.Peirce JW (2007). PsychoPy–psychophysics software in Python. J. Neurosci. Methods 162, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Isensee F, Petersen J, Kohl SA, Jäger PF, and Maier-Hein KH (2019). nnu-net: breaking the spell on successful medical image segmentation. arXiv, arXiv:1904.08128v1. https://arxiv.org/abs/1904.08128v1. [Google Scholar]
- 64.Bakic PR, Pokrajac DD, and Maidment ADA (2017). Computer simulation of the breast subcutaneous and retromammary tissue for use in virtual clinical trials. Proc. SPIE 10132, 101325C. [Google Scholar]
- 65.Bakic PR, Barufaldi B, Higginbotham D, Weinstein SP, Avanaki AN, Espig KS, Xthona A, Kimpe TRL, and Maidment ADA (2018). Virtual clinical trial of lesion detection in digital mammography and digital breast tomosynthesis. Proc. SPIE 10573, 1057306. [Google Scholar]
- 66.Green DM, and Swets JA (1989). Signal Detection Theory and Psychophysics (Peninsula Pub; ). [Google Scholar]
- 67.Geisler WS (2003). Ideal Observer Analysis (MIT; ). [DOI] [PubMed] [Google Scholar]
- 68.Burgess A, and Ghandeharian H. (1984). Visual signal detection. I. Ability to use phase information. J. Opt. Soc. Am. A 1, 900–905. [DOI] [PubMed] [Google Scholar]
- 69.Abbey CK, and Eckstein MP (2014). Observer efficiency in free-localization tasks with correlated noise. Front. Psychol 5, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peterson W, Birdsall T, and Fox W. (1954). The theory of signal detectability. Trans. IRE Profess. Group Info. Theory 4, 171–212. [Google Scholar]
- 71.Yu L, Chen B, Kofler JM, Favazza CP, Leng S, Kupinski MA, and McCollough CH (2017). Correlation between a 2D channelized Hotelling observer and human observers in a low-contrast detection task with multislice reading in CT. Med. Phys 44, 3990–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barrett HH, and Myers KJ (2004). Foundation of Image Science (John Wiley and Sons; ). [Google Scholar]
- 73.Burgess AE (1994). Statistically defined backgrounds: performance of a modified nonprewhitening observer model. J. Opt. Soc. Am. A Opt. Image Sci. Vis 11, 1237–1242. [DOI] [PubMed] [Google Scholar]
- 74.Barrett HH, Yao J, Rolland JP, and Myers KJ (1993). Model observers for assessment of image quality. Proc. Natl. Acad. Sci. USA 90, 9758–9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eckstein MP, Abbey CK, and Bochud FO (2000). A practical guide to model observers for visual detection in synthetic and natural noisy images. In Handbook of Medical Imaging, Van Metter RL, Beutel J, and Kundel HL, eds. (SPIE; ). [Google Scholar]
- 76.Eckstein MP, and Whiting JS (1995). Lesion detection in structured noise. Acad. Radiol 2, 249–253. [DOI] [PubMed] [Google Scholar]
- 77.Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, Van Ginneken B, Kopp-Schneider A, Landman BA, Litjens G, Menze B, et al. (2019). A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv, arXiv:1902.09063v1. https://arxiv.org/abs/1902.09063. [Google Scholar]
- 78.Dice LR (1945). Measures of the amount of ecologic association between species. Ecology 26, 297–302. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the raw data from this article are accessible via Mendeley Data (https://doi.org/10.17632/tjy4h67z4j.1). Analysis scripts can be provided by requesting them to the Lead Contact. Other necessary software is listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| Raw Data | This paper | https://doi.org/10.17632/tjy4h67z4j.1 |
| Software and Algorithms | ||
| MATLAB R2019a | The MathWorks | RRID: SCR_001622 |
| Python | https://www.python.org | RRID: SCR_008394 |
| Eyelink 1000 Eyetracker | SR Research, Mississauga, ON, Canada | RRID: SCR_009602 |
| Psychotoolbox | 61 | http://psychtoolbox.org/ |
| PyschoPy | 62 | https://www.psychopy.org/ |
| nn 3D U-net | 63 | https://github.com/MIC-DKFZ/nnUNet |