Abstract
Over the last decade, the use of proteomics in the setting of prematurity has increased and has enabled researchers to successfully identify biomarkers for an array of associated morbidities. The objective of this scoping review was to identify the existing literature, as well as any knowledge gaps related to proteomic biomarker discoveries in the setting of prematurity. A scoping review was conducted using PubMed, Embase and Medline databases following the Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) guidelines. The study selection process yielded a total of 700 records, of which 13 studies were included in this review. Most studies used a tandem Mass Spectrometry (MS/MS) proteomics approach to identify key biomarkers. The corresponding studies identified proteins associated with retinopathy of prematurity (ROP), bronchopulmonary dysplasia (BPD), necrotising enterocolitis (NEC), late onset sepsis (LOS) and gestational age. This scoping review demonstrates the limited use of proteomics to identify biomarkers associated with severe complications of prematurity. Further research is warranted to identify biomarkers of other important morbidities associated with prematurity, such as intraventricular haemorrhage (IVH) and cerebral palsy, and to investigate the mechanisms associated with these outcomes.
Keywords: Proteomics, Biomarkers, Premature Infants, Prematurity
Introduction
Proteomics is a methodological approach that allows for the analysis of many proteins simultaneously and has been successful in identifying many novel disease biomarkers [1]. Proteomic methodologies have been previously used in varying contexts, such as discovering biomarkers of diabetic nephropathy and identifying novel diagnostic markers of cancer [2, 3]. Plasma proteomics is advantageous as it only uses a small volume of blood to study hundreds and sometimes thousands of proteins, and can identify changes in protein expression that may occur with age and disease [4]. Proteomics is not limited to analysis of blood samples, and enables the use of biological fluids such as saliva and urine, and tissue samples (e.g. tumours) [5]. Due to the small volume required for analysis, plasma proteomics has become increasingly popular and has enabled investigations of plasma proteins in vulnerable populations such as in paediatrics, as well as in critically ill patients, where blood may be scarce and not readily available for research purposes [4].
Preterm birth is the leading cause of death among the paediatric population globally [6]. With major technological advances in neonatal care over the last few decades, there has been an increase in survival of infants born preterm (< 37 weeks’ gestation), in particular those born extremely preterm (< 28 weeks’ gestation) [7]. Despite the technological advances that have improved survival in these vulnerable populations, preterm birth is associated with significant morbidities including intraventricular haemorrhage (IVH), necrotising enterocolitis (NEC), bronchopulmonary dysplasia (BPD), and neurosensory impairments [8].
Within the last decade proteomics has enabled researchers to identify predictive biomarkers of NEC in preterm infants using buccal swabs [9]. More specifically, plasma proteomics has previously identified proteins that may play a role in the development of retinopathy of prematurity [10]. However, to date there has been limited research into plasma protein biomarkers in predicting other outcomes in preterm infants. Consequently, a scoping review was conducted to understand the current state of knowledge in this space, and to identify knowledge gaps that could be addressed by future studies. A preliminary search of MEDLINE, PubMed, JBI Evidence Synthesis and Embase was conducted and did not identify any current systematic reviews or scoping reviews on this topic. Thus, this review is novel and will make a significant contribution to the understanding and knowledge in the use of proteomics in preterm infants.
Review question
The following research question was formulated using the PCC (Population, Concept, Context) framework: What is the existing proteomic evidence of blood biomarker research in the setting of prematurity?
Methods
Study design
This scoping review was conducted based on the Preferred Reporting Items for Systematic reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) Checklist [11].
Search strategy
The following three electronic databases: Medline, Embase and PubMed were searched on the 24th September 2020 for all peer-reviewed studies. An additional search for grey literature was conducted using the OpenGrey and GreyLit databases. The specific search terms used for each database are detailed in Appendix A. In summary, studies included in this review were identified using the search terms [‘preterm’ OR ‘premature’) AND [‘proteome’ OR ‘protein-analysis’] AND [‘blood-protein’ OR [biomarker’], as well as including derivatives of these terms. Studies identified in this review were limited to those written in the English language and conducted in humans only. Studies retrieved using these search terms and parameters were screened by two authors (NL and TC), initially focusing on the eligibility of the studies’ titles and abstracts using the following inclusion and exclusion criteria.
Selection criteria
Inclusion criteria: (I) infants born preterm (< 37 weeks), (II) blood proteome assessed, (III) primary research, (IV) English language and (V) human study.
Exclusion criteria: (I) infants born at term or post-term (≥ 37 weeks), (II) proteome of other biological samples (e.g. saliva or urine) assessed, (III) case report, review, conference abstract or editorial correspondence and (IV) animal studies.
Data extraction and charting
Studies that were chosen for full-text assessment were assessed by NL and TC and with any discrepancies and uncertainties, a third reviewer (VI) was to assess the studies. Data extracted included publication year, disease/outcome assessed, aims, study population, comparative groups, proteomic methodology, protein-pathway analysis, key findings and study limitations. The detailed assessment for each critically reviewed study is presented in Table 1.
Table 1.
Summary of included studies in the scoping review of proteomics in setting of prematurity
| Author Year Country |
Outcome | Aim | Population | Comparative groups (n =) | Proteomic methods | Pathway analysis | Key findings | Limitations |
|---|---|---|---|---|---|---|---|---|
|
Byung et al. [23] 2004 Korea |
PDA | To investigate the usefulness of rapid BNP assay as a diagnostic marker of symptomatic PDA in preterm infants | Preterm infants aged 25–34 weeks’ gestation |
Symptomatic PDA (n = 23) Control (n = 43) |
Immunoassay kits | None |
Circulating BNP measurements correlated with clinical and echocardiographic assessments of PDA BNP concentration was significantly higher in the infants with symptomatic PDA 3 days after birth BMP concentration measurements were correlated with ductal shunts |
Not listed |
|
Ng et al. [16] 2010 Hong Kong |
LOS NEC |
To identify novel biomarkers for early and accurate diagnosis of NEC and/or septicaemia in premature infants Develop a novel clinical strategy of antibiotic treatment in different risk categories of infants |
Infants born < 31 weeks’ gestation and with a birth weight of < 1500 g |
Sepsis/NEC (n = 77) No sepsis infants (n = 77) |
MALDI-TOF MS Immunoassay kits Protein microarray 2D-Gel Electrophoresis |
None |
The ApoSAA score can potentially formulate antibiotic treatment strategies for suspected LOS and NEC patients The ApoSAA Score equation is practical and clinically useful for accurate identification of NEC and LOS in preterm infants Proteins that are useful biomarkers of NEC and LOS: Pro-apoC2 and a des-arginine variant of SAA |
Proteomic protocol may not differentially detect low-plasma concentration proteins |
|
Stewart et al. [12] 2015 UK |
LOS NEC |
To investigate serum and metabolome longitudinally in preterm infants with NEC and LOS | Infants born 23–30 weeks’ gestation |
NEC (n = 6) LOS (n = 4) Control (n = 9) |
LC–MS/MS | None |
All proteins and metabolites were comparable among all patient groups C-reactive protein increased in all NEC patients Upregulated proteins associated with NEC diagnosis: C-reactive protein (1–205), MIF and SAA-2 Proteins associated with LOS diagnosis: Haptoglobin, transthyretin and U5 small nuclear ribonucleoprotein |
Study was not sufficiently powered to determine biomarkers for clinical diagnosis Serum samples were salvaged post routine clinical tests |
|
Ruiz-Gonzalez et al. [17] 2015 Spain |
IUGR | To analyse and identify serum proteome changes in IUGR and AGA infants | Infants born 29- ≥ 37 weeks’ gestation |
Very preterm (29–32 weeks’ gestation) (n = 28) Moderate preterm (33–36 weeks’ gestation) (n = 30) Term (≥ 37 weeks’ gestation) (n = 30) |
MALDI-TOF MS 2D-Gel Electrophoresis Western blot |
None |
MBOAT7 was only detected in IUGR across all GA groups Lower levels of APOL1 and SUMO3 were detected in UGR compared to AGA FCN2 was downregulated in IUGR after one week in the very preterm group, whereas TF was upregulated in the very preterm and term groups |
Extremely preterm infants (< 29 weeks) were not included in the study |
|
Lynch et al. [18] 2016 USA |
ROP | Identify plasma proteins associated with ROP | Infants born < 31 weeks’ gestation or birth weight < 1500 g |
No ROP (n = 23) Clinically significant ROP (n = 12) Low-grade ROP (n = 27) |
SOMAscan proteomic assay | None |
Proteins associated with clinically significant ROP: MnSOD, CRDL1 and PCSK9 MnSOD could be used as a therapeutic intervention target Proteins associated with a high risk of ROP included: FGF-19, MST1R, LH, cystatin M and Plasminogen IGFBP-7 was linked to the signalling pathway for ROP |
Small sample size Proteomic analysis was conducted on one sample from neonatal period |
|
Suski et al. [13] 2018 Poland |
GA |
To compare plasma proteome compositions in preterm infants from varying gestational ages To identify signalling pathways that could be differentially regulated due to the duration of a pregnancy |
Infants born < 30 weeks’ gestation |
Preterm Group 1 (< 26 weeks’ gestation) (n = 19) Preterm Group 2 (27–28 weeks’ gestation) (n = 19) Preterm Group 3 (29–30 weeks’ gestation) (n = 19) |
iTRAQ LC–MS/MS |
None |
Protein changes between gestation ages across several pathways for inflammation, immunomodulation, complement activation and coagulation As gestational age increased there was an increase in plasma protease inhibitor (C1Inh) and fibrinogen isoforms As gestational age increased there was a decrease in Complement C3, Factor V and C4-A Concentration of LRG1 increased over time SAP correlated with gestation age Significant changes in plasma concentrations of Apolipoprotein compositions, specifically Apo-D |
Not listed |
|
Suski et al. [14] 2018 Poland |
Signalling Pathways | To analyse plasma proteome changes in preterm infants that are stratified by their gestational age in order to identify proteins of malfunctioning signalling pathways | Infants born < 30 weeks’ gestation |
Preterm Group 1 (< 26 weeks’ gestation) (n = 19) Preterm Group 2 (27–28 weeks’ gestation) (n = 19) Preterm Group 3 (29–30 weeks’ gestation) (n = 19) |
iTRAQ LC–MS/MS |
None |
Changes in plasma protein concentrations were associated with preterm delivery LRG1 was negatively correlated with gestation age Downregulation of ORM 1 and 2 isoforms ZAG and afamin downregulated in all groups Changes in the inflammatory, coagulation and complement pathways identified among infants born preterm |
Not listed |
|
Wagner et al.[21] 2018 USA |
PVD | Identify proteins associated with pathogenesis of PVD | Preterm infants aged 23–29 weeks’ gestation |
PVD (n = 44) Non-PVD group (n = 56) |
SOMAscan proteomic assay | None |
18 proteins associated with PVD at day 7 (PF-4, MST1R, APP and STK16) Proteins associated with novel pathways: Platelet degranulation, signalling by MST1 |
Single centre study Circulating proteins may not correctly represent target organ |
|
Zasada et al. [10] 2018 Poland |
ROP |
To identify biomarkers of ROP To validate the findings with a gene expression study |
Infants born < 30 weeks’ gestation |
Preterm infants with ROP (n = 28) Preterm infants without ROP (n = 29) |
iTRAQ Protein Microarray MS/MS |
None |
Significant difference in 33 proteins among those who developed ROP compared with infants who did not Concentrations of complement C3 and fibrinogen increased in infants who developed ROP Microarray results for fibrinogen did not validate the findings from the proteomic analysis |
Results may not be generalised due to differences across varying NICUs An additional validation method could have been used to strengthen the reported findings |
|
Zasada et al. [15] 2019 Poland |
BPD | To identify plasma biomarkers of BPD and provide a further molecular understanding of BPD | Infants born < 30 weeks’ gestation |
Preterm infants with BPD (n = 36) Preterm infants without BPD (n = 21) |
iTRAQ MS/MS |
None |
Infants with BPD had a decrease in the following protein concentrations: afamin, gelsolin, apolipoprotein A-1 and galectin-3 binding protein t 36 weeks’ postmenstrual (PMA) infants with BPD had increasing plasma concentrations of TF |
Sample size of infants with severe BPD is small An additional validation method could have been used to strengthen the reported findings |
|
Arjaans et al. [19] 2020 USA |
BPD PH |
Determine changes in circulating angiogenic peptides during the first week of life and their association with developing BPD and PH later in life Determine peptides and relevant signalling pathways associated with risk of BPD and PH |
Infants born < 34 weeks’ gestation and a birthweight between 500 and 1250 g |
No BPD (n = 20) Mild BPD (n = 34) Moderate BPD (n = 26) Severe BPD (n = 22) |
SOMAscan proteomic assay | Reactome |
Proteins associated with BPD severity include: FGF-19, PF-4, CTAP-III and PDGF-AA Proteins associated with BPD diagnosis: PF-4, VEGF121, ANG-1, ANG-2, BMP10 AND HGF Increasing BMP10 levels were associated with Preterm infants developing BPD and PH later in life |
Relatively small sample size Circulating proteins may not represent expression in lung tissue |
|
Tosson et al. [24] 2020 Egypt |
Sepsis | To investigate S100A12 and additional cytokines as biomarkers for neonatal sepsis | Infants born 24–36 weeks’ gestation |
Controls (n = 22) Not infected (n = 22) Infection probable (n = 37) Infected (n = 37) |
ELLSA Magnetic bead array assay |
None |
S100A12 demonstrated high specificity and sensitivity between infected and control groups IL-6 and IL-10 were significantly different between infected and control group S100A12 was also significantly different among control and infected groups |
Not listed |
|
Zhong et al. [25] 2020 Sweden |
Blood protein profiles | To investigate protein profiles in extremely preterm infants | Infants born < 28 weeks’ gestation | Extremely preterm infants (n = 14) | Multiplex PEA technology | None |
Proteins that increased after birth: C3dCR2, Factor VII, Factor XI, INHBC, SELL, IL2-RA and GP6 Proteins that decreased after birth: COLEC12, IGFBP-1, FSTL3, GDF15 and CGA Infants born extremely preterm have similar serum profiles directly at birth which changes dramatically during the first week of life |
Small sample size Some infants received blood products during the study period, which could have impacted the results |
ROP: retinopathy of prematurity; PVD: pulmonary vascular disease; PH: pulmonary hypertension; LOS: late onset sepsis; BPD: bronchopulmonary dysplasia; NEC: necrotising enterocolitis; GA: gestational age;Pro-apoC2: Proapolipoprotein CII; SAA: serum amyloid A; MALDI-TOF MS: matrix assisted laser desorption ionization-time of flight mass spectrometry; MnSOD: mitochondrial superoxide dismutase; CRDL1: chordin-like protein 1;PCSK9: proprotein convertase subtilisin/kexin type 9; FGF-19: Fibroblast growth factor 19; MSP: hepatocyte growth factor-like protein; LH: luteinizing hormone; IGFBP-7: insulin-like growth factor-binding protein-7; iTRAQ: isobaric tags for relative and absolute quantitation; LC–MS/MS: liquid chromatography and tandem mass spectrometry; C1Inh: C1-inhibitor; LRG1: leucine-rich alpha-2-gylcoprotein; SAP: serum amyloid P-complement; Apo-D: apolipoprotein D; ZAG: zinc-alpha-2-glycoprotein; ORM: Orosomucoid; MST1: macrophage stimulating 1; PF-4: platelet factor 4; MSP: macrophage-stimulating receptor protein; APP: amyloid precursor protein; STK16: serine/threonine-protein kinase 16; CTAP-III: connective tissue-activating peptide III; PDGF-AA: Platelet-derived growth factor AA; VEGF121: Vascular endothelial growth factor 121; ANG-1: Angiopoietin 1; ANG-2: Angiopoietin 2; BMP10: Bone morphogenetic protein 10; HGF: Hepatocyte growth factor; PEA: proximity extension assays; C3dCR2: complement C3d Receptor 2; COLEC12: collectin subfamily member 12; INHBC: inhibin beta C subunit; SELL: selectin L; IL2-RA: interleukin 2 Receptor alpha; GP6: glycoprotein 6 platelet; IGFBP-1: insulin-like growth factor-binding protein-1; FSTL3: follistatin like 3; GDF15: growth differentiation factor 15; CGA: glycoprotein hormone alpha polypeptide; ELLSA: enzyme-linked immunosorbent assay; MIF: macrophage migration inhibitory factor; IUGR: Intrauterine growth restriction; AGA: adequate gestational age; MBOAT7: lysophospholipid acyltransferase 7; SUMO3: small ubiquitin-related modifier 3; FCN2: ficolin-2; TF: serotransferrin; PDA: patent ductus arteriosus; BNP: B-type natriuretic peptide
Results
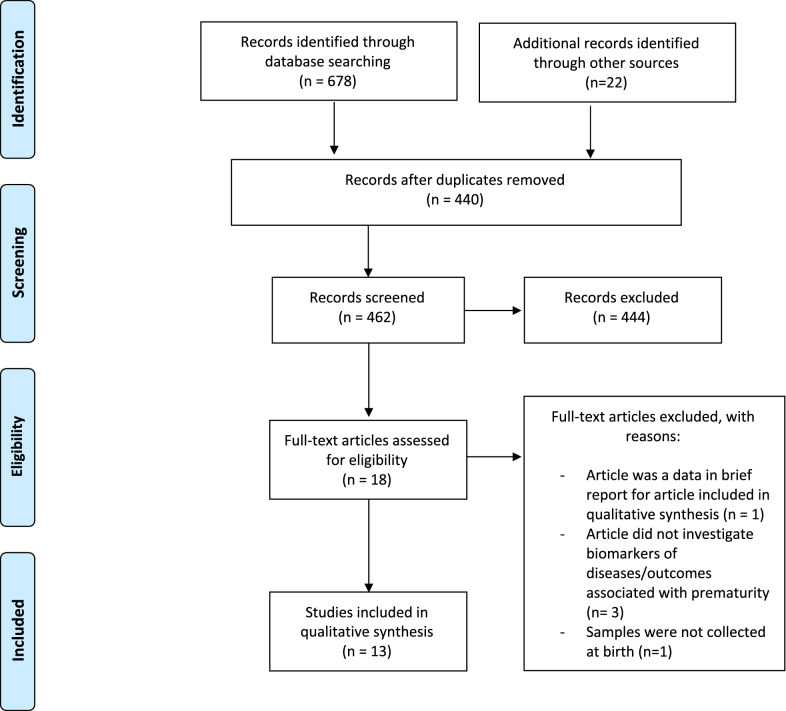
The initial search identified 678 studies using the scoping review search strategy, with an additional 22 studies identified using the grey literature search. After the removal of duplicates, 462 publications remained for title and abstract screening. A vast majority of studies (n = 444, 96%) were excluded due to not fulfilling the inclusion criteria or having no relevance to the topic of prematurity and blood biomarker discoveries. Eighteen studies underwent full-text review, with three studies excluded because they did not primarily investigate biomarkers of disease and outcomes. One study of children born preterm did not collect samples at birth and one study presented data in brief report, which did not include any proteomic data. Figure 1 illustrates the article screening and selection process, following the PRISMA guidelines (Fig. 2).
Fig. 1.
Summary of the study selection process for the scoping review
Fig. 2.
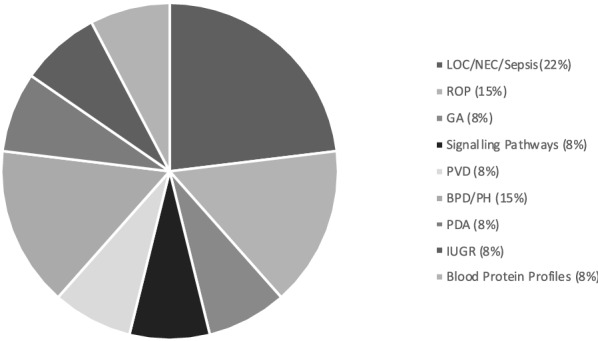
Blood proteomic studies identified were primarily conducted in the setting of LOC/NEC (23%, 3 studies) and ROP (15%, 2 studies)
Description of included studies
A total of thirteen studies met the inclusion criteria for this scoping review and are summarised in Table 1. Eleven of the thirteen included studies investigated proteins and their associations with known outcomes of prematurity. The participant gestational age at birth ranged from < 23 to 37 weeks, with sample sizes varying from 4 to 77 participants. Most studies used a tandem Mass Spectrometry method (MS/MS) to analyse the proteins of interest [10, 12–15]. Three of the fourteen studies also conducted protein validation and completed this task using protein microarray and immunoassay techniques [10, 16, 17]. Approximately half of the studies (n = 7, 47%) were completed using plasma samples (Fig. 3). The proteins identified as proteins of interest across the 13 studies included in this scoping review, with reference to the specific study/ies are summarised in Table 2.
Fig. 3.
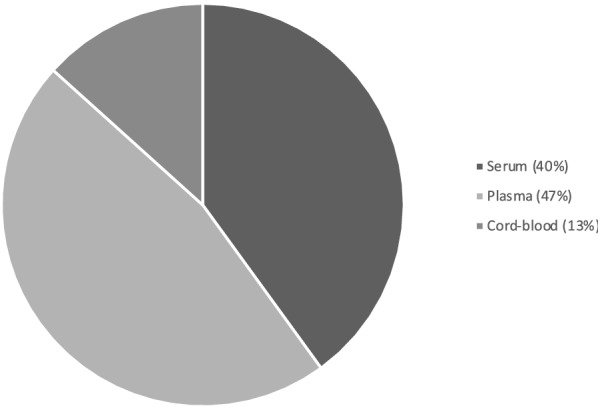
Sample types used in the identified studies were primarily conducted using plasma (47%, 7 studies) and serum (40%, 6 studies)
Table 2.
Proteins identified in the studies included in this scoping review
| Protein | Gene | UniProt accession number | Molecular function | Biological process | Study |
|---|---|---|---|---|---|
| Natriuretic peptides B | NPPB | P16860 | Diuretic hormone activity/ hormone receptor binding | Cell surface receptor signalling pathway/ body fluid secretion | [23] |
| Apolipoprotein C-II | APOC2 | P02655 | Lipoprotein lipase activator activity/ lipid binding | High-density lipoprotein particle remodelling/ retinoid metabolic process | [16] |
| Serum Amyloid A | SAA | P0DJI8 | G protein-coupled receptor/heparin binding | Activation of MAPK activity/ acute-phase response | [12, 16] |
| C-reactive protein (1–205) | CRP | P02741 | Calcium ion/ choline binding | Complement activation/ innate immune response | [12] |
| Macrophage migration inhibitory factor | MIF | P14174 | Cytokine activity/ receptor binding | Innate immune response/inflammatory response | [12] |
| Serum amyloid A-2 | SAA-2 | P0DJI9 | G protein-coupled receptor/heparin binding | Acute-phase response | [12] |
| Transthyretin | TTR | P02766 | Hormone activity | Cellular protein metabolic process/ extracellular matrix organization | [12] |
| Haptoglobin | HP | P00738 | Antioxidant activity/ haemoglobin binding | Acute inflammatory response | [12] |
| U5 small nuclear ribonucleoprotein | SNRNP40 | Q96DI7 | RNA binding | RNA splicing and processing | [12] |
| Lysophospholipid acyltransferase 7 | MBOAT7 | Q96N66 | Lysophospholipid acyltransferase activity | Lipid modification/regulation of triglyceride metabolic process | [17] |
| Apolipoprotein L1 | APOL1 | O14791 | Chloride channel activity/ lipid binding | Cellular Protein Metabolic Process/ cholesterol metabolic process | [15, 17] |
| Small ubiquitin-related modifier 3 | SUMO3 | P55854 | Protein tag/ ubiquitin-like protein ligase binding | Negative regulation of DNA binding | [17] |
| Ficolin-2 | FCN2 | Q15485 | Antigen/Calcium-dependant protein binding | Complement activation | [17] |
| Serotransferrin | TF | P02787 | ferric iron binding | Cellular iron ion homeostasis | [15, 17] |
| Mitochondrial superoxide2 | SOD2 | P04179 | DNA/enzyme binding | Cellular response to oxidative stress | [18] |
| Chordin-like protein 1 | CHRDL1 | Q9BU40 | Developmental protein | BMP signalling pathway/ post-translational protein modification | [18] |
| Proprotein convertase subtilisin/kexin type 9 | PCSK9 | Q8NBP7 | Apolipoprotein binding | Apoptotic process | [18] |
| Fibroblast growth factor 19 | FGF-19 | O95750 | Fibroblast Growth Factor Receptor Binding | MAPK cascade/ positive regulation of protein phosphorylation | [18, 19] |
| Macrophage-stimulating protein receptor | MST1R | Q04912 | ATP/ enzyme binding | Cell migration/ hepatocyte growth factor receptor signalling pathway | [18, 21] |
| Glycoprotein hormones alpha chain | CGA | P01215 | Follicle-stimulating hormone activity | Peptide hormone processing | [25] |
| Cystatin-M | CST6 | Q15828 | Cysteine-Type Endopeptidase Inhibitor Activity | Anatomical structure morphogenesis | [18] |
| Plasminogen | PLG | P00747 | Apolipoprotein Binding/ protein domain specific binding | Blood coagulation/ cellular protein metabolic process | [18] |
| Insulin-like growth factor-binding protein 7 | IGFBP-7 | Q16270 | Insulin-Like Growth Factor Binding | Cell Adhesion/ cellular protein metabolic process | [18] |
| Plasma protease C1 inhibitor | SERPING1 | P05155 | Serine-Type Endopeptidase Inhibitor Activity | Blood coagulation, intrinsic pathway/ complement activation, classical pathway | [13] |
| Complement C3 | C3 | P01024 | C5L2 anaphylatoxin chemotactic receptor binding | Cellular protein metabolic process | [10, 13] |
| Coagulation factor V | F5 | P12259 | Copper ion binding | Cellular protein metabolic process/ platelet degranulation | [13] |
| Complement C4-A | C4A | P0C0L4 | Endopeptidase inhibitor activity | Cellular protein metabolic process/ regulation of complement activation | [13] |
| Leucine-rich alpha-2-glycoprotein | LRG1 | P02750 | Transforming growth factor beta receptor binding | Neutrophil degranulation | [13, 14] |
| Serum amyloid P-component | APCS | P02743 | Calcium ion/ carbohydrate binding | cellular protein metabolic process/ complement activation | [13] |
| Apolipoprotein D | APOD | P05090 | Cholesterol binding/ lipid transporter activity | Angiogenesis/ lipid metabolic process | [13] |
| Alpha-1-acid glycoprotein 1 | ORM 1 | P02763 | Inflammatory response | Platelet/ neutrophil degranulation | [14] |
| Zinc-alpha-2-glycoprotein | AZGP1 | P25311 | Protein transmembrane transporter activity | Transmembrane transport/ retina homeostasis | [14] |
| Platelet factor 4 | PF4 | P02776 | Chemokine activity/ heparin binding | G protein-coupled receptor signalling pathway | [19, 21] |
| Amyloid-beta precursor protein | APP | P05067 | Acetylcholine receptor binding | Cellular protein metabolic process | [21] |
| Serine/threonine-protein kinase 16 | STK16 | O75716 | ATP binding/ protein serine/threonine kinase activity | Protein autophosphorylation | [21] |
| Afamin | AFM | P43652 | Fatty acid/ vitamin E binding | Vitamin transport/ protein stabilisation | [14, 15] |
| Gelsolin | GSN | P06396 | Actin/ calcium ion binding | Cellular protein metabolic process | [15] |
| Galectin-3 | LGALS3 | P17931 | Oligosaccharide/ RNA binding | Neutrophil degranulation/ innate immune response | [15] |
| Vascular endothelial growth factor A | VEGFA | P15692 | Vascular endothelial growth factor receptor binding | Activation of protein kinase activity/ angiogenesis | [19] |
| Angiopoietin-2 | ANGPT2 | O15123 | Metal ion binding/ receptor tyrosine kinase binding | Angiogenesis/ leukocyte migration | [19] |
| Angiopoietin-1 | ANGPT1 | Q15389 | Receptor tyrosine kinase binding | Angiogenesis/ leukocyte migration | [19] |
| Bone morphogenetic protein 10 | BMP10 | O95393 | Growth factor/ cytokine activity | Cell adhesion/ BMP signalling | [19] |
| Hepatocyte growth factor receptor | MET | P08581 | ATP binding/ protein tyrosine kinase activity | cell surface receptor signalling pathway/ cell migration | [19] |
| Protein S100-A12 | S100A12 | P80511 | Calcium/ion binding | Cytokine secretion/ inflammatory response | [24] |
| Interleukin-6 | IL6 | P05231 | Cytokine/ growth factor activity | Cellular protein metabolic process/ acute-phase response | [24] |
| Interleukin-10 | IL10 | P22301 | Cytokine/ growth factor activity | B cell differentiation/ cytokine-mediated signalling pathway | [24] |
| Complement receptor type 2 | CR2 | P20023 | Complement binding/ DNA binding | B cell differentiation/ immune response | [25] |
| Coagulation factor VII | F7 | P08709 | Calcium ion binding/ signalling receptor binding | Blood coagulation-extrinsic pathway | [25] |
| Coagulation factor XI | F11 | P03951 | Heparin binding | Blood coagulation-intrinsic pathway/ plasminogen activation | [25] |
| L-selectin | SELL | P14151 | Calcium ion binding | Leukocyte migration/ regulation of immune response | [25] |
| Interleukin-2 receptor subunit alpha | IL2RA | P01589 | Interleukin-2 binding/ receptor activity | cytokine-mediated signalling pathway | [25] |
| Platelet glycoprotein VI | GP6 | Q9HCN6 | Collagen binding/ signalling receptor activity | Blood coagulation/ platelet activation/ leukocyte migration | [25] |
| Collectin-12 | COLEC12 | Q5KU26 | Galactose binding/ low-density lipoprotein particle binding | Receptor-mediated endocytosis/ regulation of immune response | [25] |
| Follistatin-related protein 3 | FSTL3 | O95633 | Activin/ fibronectin binding | Cellular protein metabolic process/ cell differentiation | [25] |
| Growth/differentiation factor 15 | GDF15 | Q99988 | BMP receptor binding/ growth factor activity | Activation of MAPK activity/ BMP signalling | [25] |
| Insulin-like growth factor-binding protein 1 | IGFBP1 | P08833 | Insulin-like growth factor binding | Cellular protein metabolic process | [25] |
Retinopathy of prematurity (ROP)
Two studies investigated the outcome associated with prematurity, ROP [10, 18]. ROP is seen most commonly among infants born very preterm (< 32 weeks’ gestational age) or < 1250 g birth weight. Abnormal blood vessel development occurs in the retina in response to oxygen exposure, which can lead to retinal detachment and blindness in severe cases [18]. Currently there is no existing method to predict the occurrence of ROP in infants born preterm or born with a low birth weight and all high-risk infants are routinely screened. Hence, a proteomic approach was adopted to identify underlying biomarkers of the disease [10, 18]. Several biomarkers of the complement and inflammatory system were identified in infants who developed ROP [10]. Lynch et al. identified mitochondrial Superoxide dismutase (MnSOD), an antioxidant located in the mitochondria, as a potential therapeutic target for significant ROP [18].
Bronchopulmonary dysplasia (BPD) and pulmonary vascular disease (PVD)
Two of the thirteen included studies investigated plasma proteins and their association with BPD [15, 19]. BPD is a chronic lung disease that affects infants born preterm [20]. Arjaans et al. implemented the use of a SOMAscan proteomic assay, whereas Zasada et al. utilised MS/MS approach to identify key biomarkers of BPD. Both studies identified several proteins that may be used in future diagnosis of BPD as well associations between severity and disease prognosis [15, 19]. Wagner et al. investigated plasma proteins and their association with the pathogenesis of PVD, a term used to describe abnormal function and vascular growth of the lungs. They identified 18 proteins that were associated with PVD, including proteins associated with growth factors, angiogenesis and the extracellular matrix [21]. The protein analysis conducted by Wanger et al. also identified proteins of several different biological process pathways (e.g. Tissue Inhibitor of Metalloproteinases 3 (TIMP-3) used in platelet degradation and Bone proteoglycan II, involved in degradation of the extracellular matrix (ECM)) that may be associated with PVD.
Necrotising enterocolitis (NEC) and late onset sepsis (LOS)
Two of the thirteen studies examined biomarkers for NEC and LOS [12, 16]. Ng et al. investigated biomarkers for the early diagnosis of NEC among preterm infants. Ng et al. investigated their samples with a variety of proteomic methods, which included matrix-assisted laser desorption/ionisation (MALDI-ToF), 2D Gel-Electrophoresis (2DGE). The results of the discovery component of the study were validated using commercially available immunoassay kits and protein microarrays. Ng et al. identified a des-arginine variant of serum amyloid A (SAA) and Proapolipoprotein CII (Pro-apoC2) as very promising biomarkers of late-onset septicaemia and NEC [16]. Stewart et al. investigated the serum and metabolome of preterm infants with NEC and LOS longitudinally with a LC- MS/MS technique. Among all patient groups investigated the proteins and metabolite were comparable, with 12 proteins (e.g. Serum Amyloid A-2 and Haptoglobin) associated with NEC and LOS diagnosis [12]. Interestingly, the only protein common across the two studies was SAA [12, 16].
Gestational age and signalling pathways
Suski et al. completed several studies [13, 14] investigating plasma proteome changes in preterm infants comparing gestational ages [13] and malfunctioning proteins in various signalling pathways [14]. Utilising a tandem MS approach they were able to identify proteomic changes across varying gestational ages for several pathways which include; coagulation, inflammation, complement activations and immunomodulation [13, 14]. Suski et al. also observed Complement C3, Factor V and Complement C4-A were associated with gestational age [13]. LRG1 was the only common protein identified across the two studies [13, 14].
Discussion
In this scoping review we identified 13 primary studies that used proteomics to identify blood protein biomarkers in the setting of prematurity that used either plasma or serum as the sample which was analysed. It is important to note that studies conducted in serum cannot be directly compared to studies conducted in plasma as these are two entirely different samples. Unlike plasma which is prepared only via centrifugation, Preparation of serum entails formation and removal of a blood clot activating not only coagulation proteins but also changing the concentration of inflammatory proteins, a scenario that reflects the manipulation itself and not the physiological setting. Similarly, a cord-blood sample is different to the blood sample collected from babies at birth, due to differences in the vasculature of the umbilical cord and blood vessels within the newborn. Our findings indicate that the focus of research in the setting of blood protein biomarkers in the setting of prematurity focused on several diseases, such as ROP, BPD, LOS and NEC. However, there has been a lack of research focusing into other outcomes known to be associated with preterm birth such as cerebral palsy, intraventricular haemorrhage, or hypertension. To our best knowledge, none of the findings from the studies included in our scoping review have been translated into the clinical setting. Blood proteomic studies within this population may reflect a lack of collaboration between clinicians and proteomic experts, as well as difficulty in accessing samples from premature babies, factors that could be overcome, particularly in research institutes associated with tertiary hospitals [22].
Limitations of current published studies
The main limitation of the studies included in this review are the small sample sizes represented in those studies. Future studies should be adequately powered, and a shift of the primary focus from not only understanding mechanism of disease, but also on identifying proteins that are associated with outcomes or disease and which can be used in the clinical setting to improve outcomes for premature infants.
Conclusions
This scoping review identified a paucity of evidence around biomarker discoveries in the population of preterm infants. Several proteomic methods, including tandem mass spectrometry, immunoassays, and MALDI-TOF MS, have been used to identify biomarkers for various outcomes (e.g. ROP and BPD) associated with preterm birth. This review identifies the need for future research focusing on biomarkers to understand the possible mechanisms related to preterm birth, as well as to identify predictive protein biomarkers for complications or long-term sequelae associated with preterm birth, such as intraventricular haemorrhage and hypertension.
Acknowledgements
Not applicable.
Abbreviations
- ROP
Retinopathy of prematurity
- PVD
Pulmonary vascular disease
- PH
Pulmonary hypertension
- LOS
Late onset sepsis
- BPD
Bronchopulmonary dysplasia
- NEC
Necrotising enterocolitis
- GA
Gestational age
- Pro-apoC2
Proapolipoprotein CII
- SAA
Serum amyloid A
- MALDI-TOF MS
Matrix assisted laser desorption ionization-time of flight mass spectrometry
- MnSOD
Mitochondrial superoxide dismutase
- CRDL1
Chordin-like protein 1
- PCSK9
Proprotein convertase subtilisin/kexin type 9
- FGF-19
Fibroblast growth factor 19
- MSP
Hepatocyte growth factor-like protein
- LH
Luteinizing hormone
- IGFBP-7
Insulin-like growth factor-binding protein
- iTRAQ
Isobaric tags for relative and absolute quantitation
- LC–MS/MS
Liquid chromatography and tandem mass spectrometry
- C1Inh
C1-inhibitor
- SAP
Serum amyloid P
- Apo-D
Apolipoprotein D
- LRG1
Leucine-rich alpha-2-glycoprotein 1
- ZAG
Zinc-alpha-2-glycoprotein
- ORM
Orosomucoid
- MST1
Macrophage stimulating 1
- PF-4
Platelet factor 4
- MST1R
Macrophage-stimulating protein
- APP
Amyloid precursor protein
- STK16
Serine/threonine-protein kinase 16
- CTAP-III
Connective tissue-activating peptide III
- PDGF-AA
Platelet-derived growth factor AA
- VEGF121
Vascular endothelial growth factor 121
- ANG-1
Angiopoietin 1
- ANG-2
Angiopoietin 2
- BMP10
Bone morphogenetic protein 10
- HGF
Hepatocyte growth factor
- PEA
Proximity extension assays
- C3dCR2
Complement C3d Receptor 2
- COLEC12
Collectin subfamily member 12
- INHBC
Inhibin beta C subunit
- SELL
Selectin L
- IL2-RA
Interleukin 2 Receptor alpha
- GP6
Glycoprotein 6 platelet
- GFBP-1
Insulin-like growth factor-binding protein-1
- FSTL3
Follistatin like 3
- GDF15
Growth differentiation factor 15
- CGA
Glycoprotein hormone alpha polypeptide
- ELLSA
Enzyme-linked immunosorbent assay
- MIF
Macrophage migration inhibitory factor
- IUGR
Intrauterine growth restriction
- AGA
Adequate gestational age
- MBOAT7
Lysophospholipid acyltransferase 7
- SUMO3
Small ubiquitin-related modifier 3
- FCN2
Ficolin-2
- TF
Serotransferrin
- PDA
Patent ductus arteriosus
- BNP
B-type natriuretic peptide
Appendix A: Search strategies
A. 1. PubMed database
“Preterm” OR “pre-term” OR “prematur*”
“Proteom*” OR “protein-analysis”
“Blood-protein*” OR “serum-protein*” OR “plasma-protein*” OR “biomarker*” OR “marker*”
1 and 2 and 3
(“Animal” OR “animals” OR “rat” OR “rats” OR “mouse” OR “mice” OR “swine” OR “porcine” OR “murine” OR “sheep” OR “lamb” OR “lambs” OR “pig” OR “pigs” OR “piglet” OR “piglets” OR “rabbit” OR “rabbits” OR “cat” OR “cats” OR “dog” OR “dogs” OR “cattle” OR “bovine” OR “monkey” OR “monkeys” OR “trout” OR “marmoset” OR “marmosets”) NOT (“human” OR “humans” OR “patient” OR “patients” OR “newborn*” OR “baby” OR “babies” OR “neonat*” OR “infan*” OR “toddler*” OR “pre-schooler*” OR “preschooler*” OR “kindergarten” OR “boy” OR “boys” OR “girl” OR “girls” OR “child” OR “children” OR “childhood” OR “adolescen*” OR “pediatric*” OR “paediatric*” OR “youth*” OR “teen” OR “teens” OR “teenage*” OR “school-aged*” OR “school-child*” OR “school-girl*” OR “school-boy*” OR “schoolgirl*” OR “schoolboy*” OR “man” OR “men” OR “woman” OR “women” OR “adult” OR “adults” OR “middle-age*” OR “elderly”)
5 not 6
Limit to English language
A. 2. Embase database
Prematurity/
Exp low birth weight/
(Preterm or pre-term or prematur*).mp.
1 or 2 or 3
Exp proteomics/
Proteome/
Exp *protein analysis/
Proteom*.tw,kw,dq.
5 or 6 or 7 or 8
Exp plasma protein/
(Blood-protein* or serum-protein* or plasma-protein*).tw,kw,dq.
Biological marker/
(Biomarker* or marker*).tw,kw,dq.
10 or 11 or 12 or 13
4 and 9 and 14
(Rat or rats or mouse or mice or swine or porcine or murine or sheep or lamb or lambs or pig or pigs or piglet or piglets or rabbit or rabbits or cat or cats or dog or dogs or cattle or bovine or monkey or monkeys or trout or marmoset or marmosets).ti. and animal experiment
Animal experiment/ not (human experiment/ or human/)
Case report/
15 and 18
LIMIT 15 to (conference abstract or conference paper or "conference review" or editorial or letter)
15 not (16 or 17 or 19 or 20)
Limit 21 to English language
A. 3. Medline. database
Exp infant, low birth weight/ or infant, premature/
Exp infant, premature, diseases/
Premature Birth/
(Preterm or pre-term or prematur*).mp.
1 or 2 or 3 or 4
Exp Proteomics/
Proteome/
Proteom*.tw,kf.
6 or 7 or 8
Exp Blood Proteins/
(Blood-protein* or serum-protein* or plasma-protein*).tw,kf.
Exp biomarkers/
(Biomarker* or marker*).tw,kf.
10 or 11 or 12 or 13
5 and 9 and 14
(Exp animals/ or (rat or rats or mouse or mice or swine or porcine or murine or sheep or lamb or lambs or pig or pigs or piglet or piglets or rabbit or rabbits or cat or cats or dog or dogs or cattle or bovine or monkey or monkeys or trout or marmoset or marmosets).ti.) not human*.sh.
Limit 15 to (case reports or comment or editorial or guideline or letter or practice guideline)
15 not (16 or 17)
Limit 18 to English language
Authors' contributions
All authors listed have made a substantial, direct and intellectual contribution to the work. All authors read and approved the final manuscript.
Funding
There was no specific funding utilised for this review.
Availability of data and materials
Not applicable.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahram M, Petricoin EF. Proteomics discovery of disease biomarkers. Biomark Insights. 2008;6(3):325–333. doi: 10.4137/bmi.s689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanash S, Taguchi A. Application of Proteomics to Cancer Early Detection. Cancer J. 2011;17(6):423–428. doi: 10.1097/PPO.0b013e3182383cab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merchant ML, Klein JB. Proteomic discovery of diabetic nephropathy biomarkers. Adv Chronic Kidney Dis. 2010;17(6):480–486. doi: 10.1053/j.ackd.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ignjatovic V, Geyer PE, Palaniappan KK, Chaaban JE, Omenn GS, Baker MS, et al. Mass spectrometry-based plasma proteomics: considerations from sample collection to achieving translational data. J Proteome Res. 2019;18(12):4085–4097. doi: 10.1021/acs.jproteome.9b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics. 2006;6(23):6326–6353. doi: 10.1002/pmic.200600284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walani SR. Global burden of preterm birth. Int J Gynecol Obstet. 2020;150(1):31–33. doi: 10.1002/ijgo.13195. [DOI] [PubMed] [Google Scholar]
- 7.Cheong JLY, Spittle AJ, Burnett AC, Anderson PJ, Doyle LW. Have outcomes following extremely preterm birth improved over time? Semin Fetal Neonatal Med. 2020;25(3):101114. doi: 10.1016/j.siny.2020.101114. [DOI] [PubMed] [Google Scholar]
- 8.Glass HC, Costarino AT, Stayer SA, Brett CM, Cladis F, Davis PJ. Outcomes for extremely premature infants. Anesth Analg. 2015;120(6):1337–1351. doi: 10.1213/ANE.0000000000000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murgas Torrazza R, Li N, Young C, Kobeissy F, Chow M, Chen S, et al. Pilot study using proteomics to identify predictive biomarkers of necrotizing enterocolitis from buccal swabs in very low birth weight infants. Neonatology. 2013;104(3):234–242. doi: 10.1159/000353721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zasada M, Suski M, Bokiniec R, Szwarc-Duma M, Borszewska-Kornacka MK, Madej J, et al. An iTRAQ-based quantitative proteomic analysis of plasma proteins in preterm newborns with retinopathy of prematurity. Investig Opthalmology Vis Sci. 2018;59(13):5312. doi: 10.1167/iovs.18-24914. [DOI] [PubMed] [Google Scholar]
- 11.Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018;169(7):467. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 12.Stewart CJ, Nelson A, Treumann A, Skeath T, Cummings SP, Embleton ND, et al. Metabolomic and proteomic analysis of serum from preterm infants with necrotising entercolitis and late-onset sepsis. Pediatr Res. 2016;79(3):425–431. doi: 10.1038/pr.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suski M, Bokiniec R, Szwarc-Duma M, Madej J, Bujak-Gizycka B, Kwinta P, et al. Prospective plasma proteome changes in preterm infants with different gestational ages. Pediatr Res. 2018;84(1):104–111. doi: 10.1038/s41390-018-0003-2. [DOI] [PubMed] [Google Scholar]
- 14.Suski M, Bokiniec R, Szwarc-Duma M, Madej J, Bujak-Gizycka B, Borszewska-Kornacka MK, et al. Plasma proteome changes in cord blood samples from preterm infants. J Perinatol Off J Calif Perinat Assoc. 2018;38(9):1182–1189. doi: 10.1038/s41372-018-0150-7. [DOI] [PubMed] [Google Scholar]
- 15.Zasada M, Suski M, Bokiniec R, Szwarc-Duma M, Borszewska-Kornacka MK, Madej J, et al. Comparative two time-point proteome analysis of the plasma from preterm infants with and without bronchopulmonary dysplasia. Ital J Pediatr. 2019;45(1):112. doi: 10.1186/s13052-019-0676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng PC, Ang IL, Chiu RWK, Li K, Lam HS, Wong RPO, et al. Host-response biomarkers for diagnosis of late-onset septicemia and necrotizing enterocolitis in preterm infants. J Clin Invest. 2010;120(8):2989–3000. doi: 10.1172/JCI40196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruis-Gonzalez MD, Canete MD, Gomez-Chaparro JL, Abril N, Canete R, Lopez-Barea J. Alterations of protein expression in serum of infants with intrauterine growth restriction and different gestational ages. J Proteomics. 2015;119(101475056):169–182. doi: 10.1016/j.jprot.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Lynch AM, Wagner BD, Mandava N, Palestine AG, Mourani PM, McCourt EA, et al. The relationship of novel plasma proteins in the early neonatal period with retinopathy of prematurity. Invest Ophthalmol Vis Sci. 2016;57(11):5076–5082. doi: 10.1167/iovs.16-19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arjaans S, Wagner BD, Mourani PM, Mandell EW, Poindexter BB, Berger RMF, et al. Early angiogenic proteins associated with high risk for bronchopulmonary dysplasia and pulmonary hypertension in preterm infants. Am J Physiol Lung Cell Mol Physiol. 2020;318(4):L644–L654. doi: 10.1152/ajplung.00131.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramsay PL, DeMAYO FJ, Hegemier SE, Wearden ME, Smith CV, Welty SE. Clara cell secretory protein oxidation and expression in premature infants who develop bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2001;164(1):155–161. doi: 10.1164/ajrccm.164.1.2008022. [DOI] [PubMed] [Google Scholar]
- 21.Wagner BD, Babinec AE, Carpenter C, Gonzalez S, O’Brien G, Rollock K, et al. Proteomic profiles associated with early echocardiogram evidence of pulmonary vascular disease in preterm infants. Am J Respir Crit Care Med. 2018;197(3):394–397. doi: 10.1164/rccm.201703-0654LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017;13(9):942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byung MC, Kee HL, Baik LE, Kee HY, Young SH, Chang SS, et al. Utility of rapid B-type natriuretic peptide assay for diagnosis of symptomatic patent ductus arteriosus in preterm infants. Pediatrics. 2005;115(3):e255–e261. doi: 10.1542/peds.2004-1837. [DOI] [PubMed] [Google Scholar]
- 24.Tosson AMS, Glaser K, Weinhage T, Foell D, Aboualam MS, Edris AA, et al. Evaluation of the S100 protein A12 as a biomarker of neonatal sepsis. J Matern Fetal Neonatal Med. 2020;33(16):2768–2774. doi: 10.1080/14767058.2018.1560411. [DOI] [PubMed] [Google Scholar]
- 25.Zhong W, Danielsson H, Tebani A, Karlsson MJ, Elfvin A, Hellgren G, et al. Dramatic changes in blood protein levels during the first week of life in extremely preterm infants. Pediatr Res. 2020 doi: 10.1038/s41390-020-0912-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.