Summary
Small-cell neuroendocrine cancers (SCNCs) are recalcitrant cancers arising from diverse primary sites that lack effective treatments. Using chemical genetic screens, we identified inhibition of ataxia telangiectasia and Rad3-related (ATR), primary activator of the replication stress response, and topoisomerase I (TOP1), nuclear enzyme that suppresses genomic instability, as synergistically cytotoxic in small cell lung cancer (SCLC). In a proof-of-concept study, we combined M6620 (berzosertib), first-in-class ATR inhibitor, and TOP1 inhibitor topotecan in patients with relapsed SCNCs. Objective response rate among patients with SCLC was 36% (9/25), achieving the primary efficacy endpoint. Durable tumor regressions were observed in patients with platinum-resistant SCNCs, typically fatal within weeks of recurrence. SCNCs with high neuroendocrine differentiation, characterized by enhanced replication stress, were more likely to respond. These findings highlight replication stress as a potentially transformative therapeutic SCNC vulnerability, paving the way for rational patient selection in these cancers, now treated as a single disease.
eTOC Blurb
Thomas et al identify DNA replication stress as a therapeutic vulnerability of small cell neuroendocrine cancers (SCNCs). SCNCs with high neuroendocrine and enhanced replication stress are more likely to respond to ATR and topoisomerase 1 inhibition, which yielded durable tumor regressions in patients with platinum-resistant tumors.
Graphical Abstract
Introduction
Small cell neuroendocrine cancers (SCNCs) are an exceptionally lethal cancer subtype arising in multiple tissues, most commonly the lung (small cell lung cancer; SCLC). SCNCs arise de novo or as a result of cellular plasticity when major growth programs and lineage-directing factors are effectively suppressed (Quintanal-Villalonga et al., 2020). Regardless of their tissue of origin, SCNCs share morphological features such as scant cytoplasm, lack of prominent nucleoli, and finely granular chromatin (Klimstra et al., 2015). They also share molecular profiles including expression of common neuroendocrine (NE) markers and frequent loss of TP53 and RB1 (Balanis et al., 2019; Beltran et al., 2016; Chang et al., 2018; George et al., 2015).
SCLCs are generally detected after they are already widely metastasized (Thomas et al., 2018a). There are few actionable alterations, and genomic subgroups that inform therapy are yet to be defined. Anti-programmed death-ligand 1 (PD-L1) antibody combined with platinum and etoposide is the current standard treatment for SCLC, but responses to this regimen are typically brief (Horn et al., 2018). Standard treatments for extra-pulmonary small cell neuroendocrine cancers (EP-SCNCs) – SCNCs arising at non-lung primary sites – are undefined, and due in part to their rarity and heterogeneity, clinical investigation remains sparse. There are no effective treatments for SCNCs after failure of first-line therapies. Life expectancy is typically less than one year. Outcomes are particularly poor for patients with platinum-resistant disease – defined as disease progression within 90 days of completing first-line chemotherapy – which is generally fatal within weeks of relapse (O’Brien et al., 2006). A major barrier to progress in SCLCs is the relative paucity of high-quality tumor material for genetic and molecular analyses. Biopsies at relapse are not standard and are challenging due to rapid cancer progression and patient comorbidities. As a result, SCLC genomic studies performed to date have been far smaller in scale relative to those of the tumor types represented in sequencing initiatives, such as The Cancer Genome Atlas.
In cancers that lack actionable oncogenes, targeting the well-defined phenotypic hallmarks could be of therapeutic benefit (Hanahan and Weinberg, 2011). One such hallmark is genomic instability, which promotes genetic diversity and thereby drives the acquisition of multiple hallmark capabilities. DNA damage resulting from unabated replication – referred to as DNA replication stress – is a major source of genomic instability in cancers and precancerous lesions (Bartkova et al., 2005; Bartkova et al., 2006; Gorgoulis et al., 2005; Zeman and Cimprich, 2014). Replication stress is a common feature of cancers with activated oncogenes or absent tumor suppressors that accelerate the rate of S phase entry and disrupt the DNA replication schedule (Halazonetis et al., 2008; Hills and Diffley, 2014). SCLCs are characterized by high degree of genomic instability. They exhibit ubiquitous loss of tumor suppressors, frequently amplify and overexpress oncogenes such as MYC, and have high expression of lineage transcription factors (Beltran et al., 2016; Chang et al., 2018; George et al., 2015; Semenova et al., 2015), all of which contribute to replication stress (Halazonetis et al., 2008; Kotsantis et al., 2016). We reasoned that replication stress is an SCLC hallmark, and as a result of this dependence SCLCs could be vulnerable to targeting the replication stress response (Thomas and Pommier, 2016).
Here we performed cell-based chemical genetic screens seeking out clinically actionable drug combinations targeting replication stress. Pharmacological inhibition of ataxia-telangiectasia–mutated and rad3-related (ATR), the primary responder to replication stress (Blackford and Jackson, 2017), exacerbated cell death and enhanced anti-tumor activity of clinical inhibitors of DNA topoisomerase I (TOP1), a nuclear enzyme that suppresses genomic instability (Tuduri et al., 2009). Extending this preclinical work, we investigated the anti-tumor activity of M6620 (berzosertib, VX-970), the first-in-class ATR inhibitor in combination with topotecan, a highly selective TOP1 inhibitor in patients with relapsed SCNCs (Clinicaltrials.gov identifier: NCT02487095). Tumor exomes and transcriptomes were investigated to dissect responses and rationalize patient stratification for future studies.
Results
High-throughput screening identifies active drugs and combinations in SCLC cells
First, we evaluated the activity of 2480 oncology-focused, approved and investigational drugs in a library termed MIPE 5.0 (Lin et al., 2019) across a panel of seven SCLC cell lines (DMS114, DMS79, H187, H196, H446, H524 and H889) (Table S1). The library exploits redundancy by including multiple inhibitors of well-explored targets while encompassing mechanistic diversity, targeting more than 860 distinct mechanisms of action.
Similar single-drug responses were observed across all seven cell lines. This coherent database of SCLC chemical vulnerabilities was used to prioritize drugs for further analyses. Drugs suppressing cell growth with an average Z-transformed area under the curve [Z-AUC] <−1 and active in at least five of the seven cell lines tested were classified as “hits” (Table S1, Fig. 1A). A total of 244 drugs met this threshold. Drug classes with more than 30% hit-rate included agents targeting nicotinamide phosphoribosyltransferase (NAMPT, 3/4 agents tested), tubulins (12/37), the proteasome (PSMD1, 5/9), histone deacetylases (HDACs, 9/18), TOP1 (5/10), ATR (2/5), and checkpoint kinase 1 (CHK1, 4/13) (Table S1). The enrichment of TOP1, ATR, and CHK1 inhibitors likely reflects SCLC vulnerability to drugs that exacerbate genomic instability and replication stress. Consistently, SCLC cells exhibit high expression of CHEK1, a checkpoint kinase activated in response to genotoxic stress, and CLSPN and TOPBP1, key adaptors that facilitate ATR-dependent phosphorylation of CHK1 (Kumagai and Dunphy, 2000; Zeman and Cimprich, 2014), compared with other solid tumor types in the Cancer Cell Line Encyclopedia (Fig. S1A) (Rajapakse et al., 2018; Tlemsani et al., 2020).
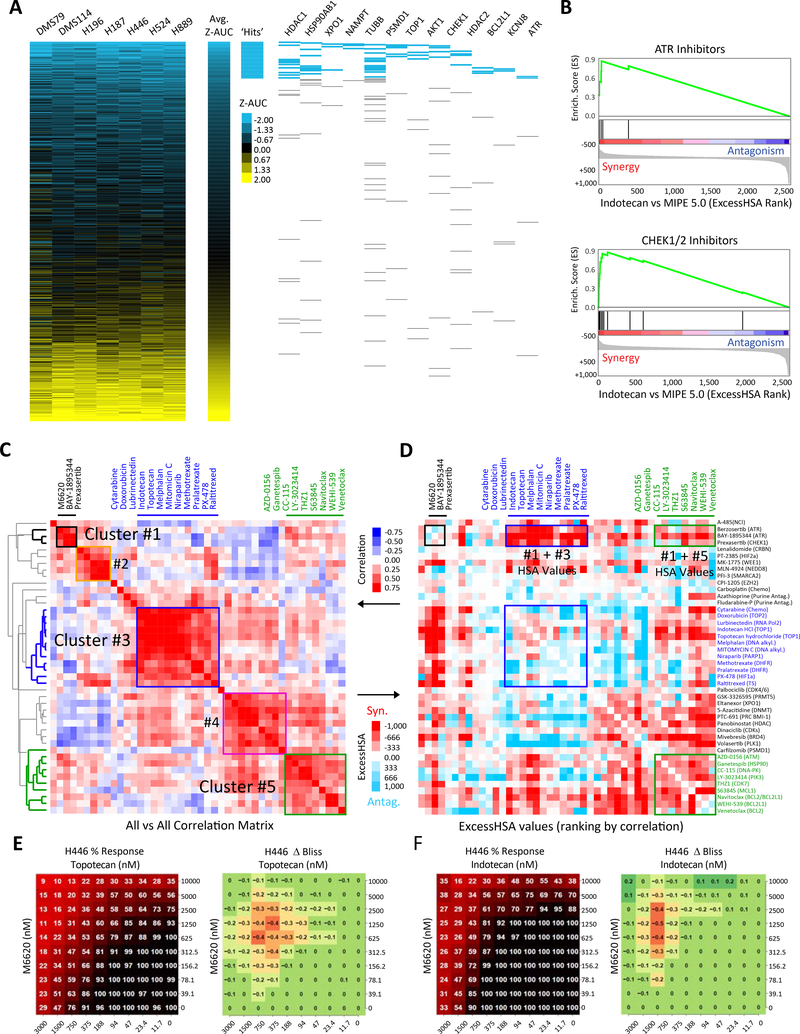
Figure 1: In vitro drug response profiles of SCLC reveal DNA damage response-related vulnerabilities.
A) Heatmap of ranked drug activities for seven SCLC cell lines (DMS114, DMS79, H187, H196, H446, H524 and H889) screened using the MIPE 5.0 library of approved and investigational drugs (n=2480). Activity scores based on Z-transformed area under the curve values. Mechanistic classes enriched among highly active agents are shown on the right panel. B) Drug-target enrichment analysis plots highlighting synergy of TOP1 inhibitors with CHK1 and ATR inhibitors. TOP1-inhibitor combinations examined in combination with MIPE 5.0 library using indotecan (LMP400), a next-generation clinical TOP1 inhibitor. TOP1 inhibitor-drug pairs ranked using the ExcessHSA metric. C) Unsupervised hierarchical clustering based on the correlation of the ExcessHSA showing drug-clusters with similar combination profiles. In this screen, 44 drugs selected based on single-drug activities, synergies from indotecan-combinations, and mechanistic interest were tested in combination (n=946 combinations). D) Correlation heatmap of the 44-drug all-versus-all combination screen highlighting synergistic clusters. 10 × 10 % response and ΔBliss heat maps for the combination of E) topotecan and M6620, and F) indotecan and M6620 across defined concentration ranges in H446 SCLC cell line. SCLC: small cell lung cancer; TOP1: topoisomerase 1; ExcessHSA: Excess over the Highest Single Agent. See also Table S1 and Fig. S1.
Next, we sought to identify synergistic drug combinations. Given the high cytotoxicity of TOP1 inhibitors topotecan and irinotecan, both active agents in SCLC patients (Hanna et al., 2006; von Pawel et al., 1999), we explored TOP1-inhibitor combinations with the entire MIPE 5.0 library (Table S1) using indotecan (LMP400), a next-generation clinical TOP1 inhibitor (Coussy et al., 2020). TOP1 inhibitor-drug pairs were ranked using the Excess over the Highest Single Agent (ExcessHSA) metric to quantitatively assess synergism and antagonism (Fig. S1B). We then exploited the mechanistic redundancy built into MIPE 5.0 to generate a pre-ranked drug-target enrichment analysis (DTEA) of the TOP1 inhibitor-drug interaction landscape. Notably, indotecan displayed a selective combination profile with only five drug targets enriched (false discovery rate [FDR] <10%). These included ATR (e.g. BAY-1895344: rank #1, M6620: # 32), HSP90 (e.g. ganetespib: #5), and CHK1 (e.g. prexasertib: #9) (Fig. 1B, Fig. S1C, D, Table S1).
Finally, we selected 44 drugs based on single-drug activities, synergies from indotecan-combinations, and mechanistic interest, and tested all possible pair-wise combinations (n=946) (Table S1). Unsupervised hierarchical clustering based on the correlation of the ExcessHSA values revealed drug-clusters with highly similar combination profiles (Fig. 1C). Importantly, ATR and CHK1 inhibitors grouped in a well-resolved cluster (#1). Other clusters of interest included DNA synthesis/replication-targeted chemotherapies (#3) and pro-apoptotic Bcl-2 family inhibitors (#5). Notably, the ATR/CHK1 cluster possessed strong synergy with both clusters #3 and #5 (Fig. 1D). This was confirmed for ATR inhibitors using different agents and multiple assay formats (Fig. 1E, F, Fig. S1E). This phenotype was specific for ATR/CHK1, as inhibitors of DNA-PK catalytic subunit (DNA-PKcs) and ataxia telangiectasia mutated (ATM), two other phosphoinositol 3-kinase-related kinases with principal roles in activating the DNA damage response (Blackford and Jackson, 2017), clustered separately.
Together, these studies highlighted several combinations of DNA damage response inhibitors and chemotherapies that could be effective treatments for SCLC. We centered our subsequent analyses on ATR inhibitors as they appeared better suitable for combinatorial approaches than CHK1 inhibitors (Sakurikar and Eastman, 2015), and investigated the mechanistic basis of their synergy with topotecan and irinotecan, standard-of-care chemotherapeutics for relapsed SCLC, with the goal of clinical translation.
ATR inhibition by M6620 enhances TOP1-mediated DNA damage by topotecan
As TOP1 inhibitors trap TOP1 on chromatin during its catalytic cycle, resulting in TOP1 DNA-protein crosslinks (TOP1-DPCs), we sought to examine the levels of topotecan-induced TOP1 DPCs in M6620-treated cells using the in vivo complex of enzyme (ICE) bioassay (Anand et al., 2018). M6620 pre-treatment in DMS114 cells led to a 1.4-fold increase in TOP1-DPCs after 2-hour treatment with topotecan (Fig. S2A), suggesting a role of ATR in the clearance of TOP1 DPCs and repair of TOP1-mediated DNA damage. Importantly, neither an ATM inhibitor (KU55933) nor a DNA-PKcs inhibitor (VS984) potentiated topotecan-generated TOP1-DPCs. Single-strand breaks are associated with the formation of TOP1-DPCs, and their repair determines the viability of topotecan-exposed cells (Pommier et al., 2016). We evaluated the extent of DNA breaks using alkaline comet assay and found elevated average comet tail moment in cells co-treated with M6620 and topotecan compared to cells treated with topotecan or M6620 alone (Fig. 2A; Fig. S2B). Replication combing assay further demonstrated that topotecan treatment led to hindered replication fork progression that was suppressed upon the addition of M6620 in two SCLC cell models (DMS114 and H209) (Fig. 2B; Fig S2C). In both models, we observed significantly reduced replication velocity within 30 minutes of topotecan treatment, consistent with TOP1 trapping by topotecan activating the replication checkpoint (Josse et al., 2014; Seiler et al., 2007). ATR inhibition by M6620 abrogated the replication block, consistent with ATR being critical for the topotecan-induced DNA replication checkpoint, and M6620 inhibiting the checkpoint to ensure continued replication (Table S2). ATR inhibition by M6620 also abrogated the S-phase arrest induced by TOP1 inhibition and forced progression to G2/M phase (Fig. S2D). Western blot analysis showed rapid phosphorylation of replication protein A (RPA) at S33, an established ATR phosphorylation site (Shao et al., 1999; Shiotani et al., 2013), following exposure to topotecan in DMS1114 cells. M6620 treatment reduced the topotecan-induced RPA32 phosphorylation (Fig. S2E). Together, these data confirm the critical role of ATR in the resolution of topotecan-induced DNA damage in SCLC cells.
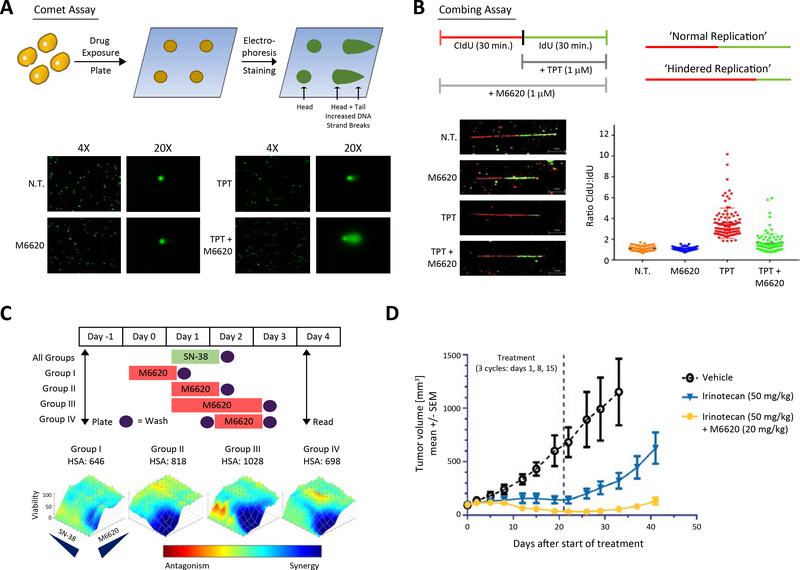
Figure 2: Concurrent inhibition of ATR and TOP1 exacerbates replication stress and DNA damage.
A) Schematic illustrating the comet assay (single cell gel electrophoresis) for the detection of DNA damage and specified imaging outcomes, highlighting increased DNA strand breaks caused by the combination of M6620 and topotecan. B) Schematic illustrating the combing assay (spatiotemporal analysis of DNA replication) for the detection of replication stress and specified imaging outcomes (mid line is mean+/− standard deviation, n = 100), highlighting the re-establishment of normal replication when M6620 is added to topotecan in the SCLC cell model DMS114. C) Cell viability with SN-38 and M6620 across different treatment schedules in the SCLC cell line H446. M6620 followed by SN-38 does not yield a synergistic effect on cell viability, SN-38 followed by M6620 and co-administration of both drugs creates a synergistic effect. Group I: M6620 for 24 hours followed by 24 hours of SN-38; Group II: concurrent M6620 and SN-38 (24 hours); Group III: concurrent M6620 and SN-38 (24 hours) followed by an additional 24 hours of M6620; Group IV: SN-38 for 24 hours followed by 24 hours of M6620. D) In vivo assessment (mean tumor volume +/− standard deviation, n = 5) of irinotecan (50 mg/kg, IP) alone and combination of irinotecan (50 mg/kg, IP) and M6620 (20 mg/kg, IV) in SCLC PDX LXFS573 model using a 3-cycle schedule with treatments on days 1, 8 and 15. Vertical dashed line marks end of third treatment cycle after 21-days. See also Table S2, Fig. S2.
Next, given the critical importance of drug scheduling in combination therapies, we performed in vitro and in vivo studies to determine the optimal dosing strategy for this combination. H446 SCLC cells were treated with M6620 and SN-38, the active metabolite of irinotecan, in four different schedules: M6620 administered a day prior to and a day after SN-38, and in two concurrent schedules. Drug synergism was noted to be schedule-dependent, with concurrent treatment producing maximal cytotoxicity (Fig. 2C, Table S2). Similar results were obtained with the combination of topotecan and M6620 (Fig. S2F, Table S2). We also evaluated the combination of irinotecan and M6620 in a SCLC patient derived xenograft model (Fig. 2D, Table S2) wherein concurrent administration of M6620 and irinotecan significantly limited tumor growth.
In order to establish the relationship between drug exposure, pharmacodynamic effects, and anti-tumor activity, we utilized previously reported colorectal adenocarcinoma models. After establishing that concurrent treatment produced a maximal cytotoxic response in a cell line model HT-29 (Fig. S2G, Table S2), we utilized the Colo 205 xenograft model to further test the in vivo schedule dependency, pharmacokinetics, and pharmacodynamics (Fig. S5H–J, Table S2). Maximal tumor growth inhibition was observed with concurrent administration of M6620 and irinotecan (Fig. S5B, Table S2). Pharmacokinetic analysis at three relevant doses of M6620 (10, 20 and 30 mg/kg) coupled with a pharmacodynamic examination of pCHK1 (Ser345) (Fig. S5I, J, Table S2) demonstrated that M6620 blocks irinotecan induced pCHK1 in a dose- and time-dependent manner. Together, these data show that concurrent inhibition of ATR and TOP1 yields robust synergy in multiple models, with evidence of pharmacodynamic modulation of ATR activity at clinically achievable drug concentrations (Terranova et al., 2020).
Clinical trial combining ATR and TOP1 inhibitors in SCLC patients
The demonstration of enhanced efficacy of ATR inhibitor with topotecan, the SCLC second-line standard-of-care chemotherapeutic, provided compelling rationale to investigate this combination in the clinic. In a proof-of-concept phase 2 trial, we combined M6620, the first-in-class, ATP-competitive inhibitor of ATR, and topotecan in SCLC patients who had relapsed after at least one prior chemotherapy. The primary endpoint was objective response rate (ORR) measured by Response Evaluation Criteria in Solid Tumors (RECIST) (Eisenhauer et al., 2009). Response had to be confirmed by the investigator at least four weeks later. The trial was conducted using Simon two-stage Minimax design to rule out an unacceptably low 10% response rate (p0=0.10) in favor of targeted response rate of 30% (p1=0.30), considered clinically meaningful based on outcomes with topotecan alone in relapsed SCLC (von Pawel et al., 1999).
M6620 (210 mg/m2 intravenously on days 2 and 5) was administered concurrently with topotecan (1.25 mg/m2 intravenously on days 1 through 5), in 21-day cycles (Fig. 3A) at doses previously identified as safe and associated with pharmacodynamic activity in patients with advanced solid tumors (Terranova et al., 2020; Thomas et al., 2018b). Dose reductions and interruptions were permitted for toxicities not relieved by supportive care. Treatment continued until disease progression. Radiographic evaluation was performed at baseline and every two cycles. Peripheral blood was collected at several time points to assess pharmacokinetics and circulating tumor cells (CTCs). Tumor biopsies were performed at baseline.
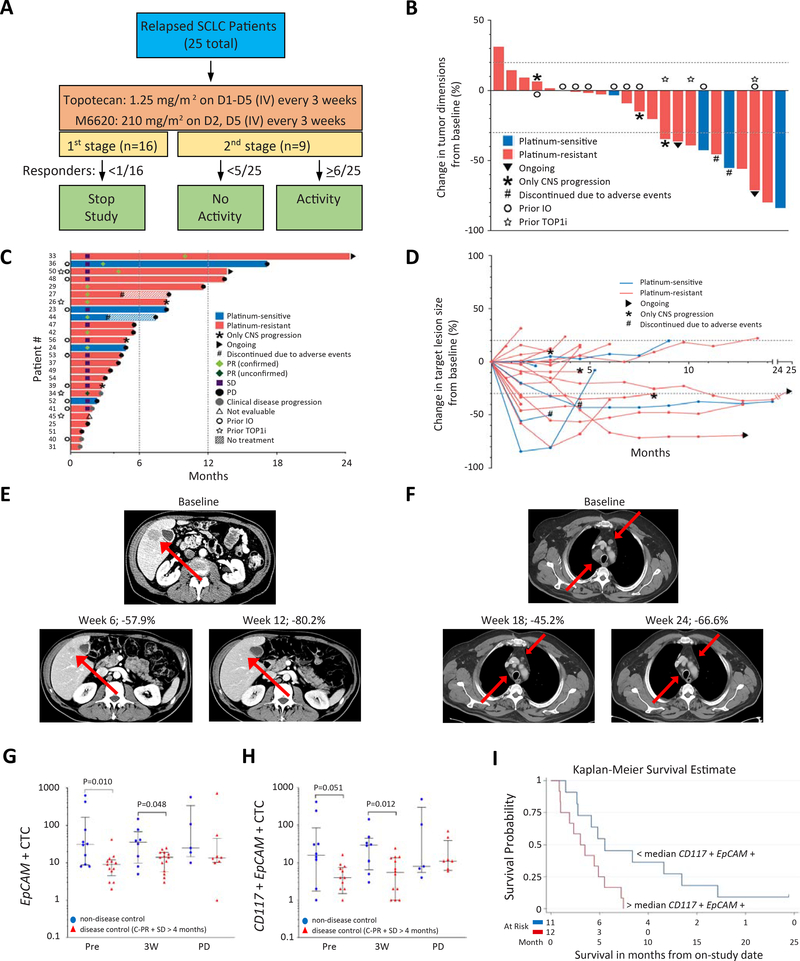
Figure 3: Trial design and efficacy of topotecan and M6620 in relapsed SCLC.
A) Study schema. B) Tumor responses to the combination of M6620 and topotecan in SCLC patients based on maximum change in tumor dimensions from baseline. Each bar represents a patient’s tumor response. C) Efficacy of the combination based on duration of response, and D) change in target lesion size from baseline. E) CT abdomen showing tumor regression in a patient (patient # 42) with platinum-resistant SCLC and liver metastasis (red arrows). F) CT chest showing tumor regression in a patient (patient #50) with platinum-resistant SCLC and hilar lymph node metastases (red arrows). G) Differences of EpCAM+ CTCs in patients with disease control (PR + SD ≥4 months, red triangles) vs. those without (blue circles). The number of samples (non-disease control vs. disease control): 10 vs. 13, 8 vs. 14, 5 vs. 8 pre-treatment, 3 weeks after treatment, and at disease progression, respectively. Median and interquartile ranges are shown. Statistical differences are evaluated by Mann-Whitney U test. H) Differences of CD117+ EpCAM+ CTCs in patients with disease control (PR + SD ≥4 months, red triangles) vs. those without (blue circles). Median and interquartile ranges are shown. I) Kaplan-Meier curve of PFS in patients with < median level of CD117+ EpCAM+ CTCs at pretreatment vs. those with ≥median CD117+ EpCAM+ CTCs. CTCs: circulating tumor cells; PR: partial response; SD: stable disease; PFS: progression-free survival. See also Table S3, Fig. S3.
A total of 26 patients were enrolled, all of whom had evidence of disease progression before study participation. Patients were self-referred or were referred by treating physicians from across the United States, and were unselected for sex, stage of disease, platinum-sensitivity, or other prognostic determinants. Tumor samples underwent central histopathologic review confirming SCLC diagnosis (Table S3). All patients completed at least one cycle (median: 5.5 cycles; range: 1–33) of treatment. Twenty-five patients were evaluable for the primary endpoint. Median age was 63 years (range: 51–78), and 17 patients (65.4%) were women (Table 1). Most patients had extensive-stage at diagnosis (18/26, 69.2%), and platinum-resistant disease (21/26, 80.8%).
Table 1.
Patient Characteristics
| Patient characteristic | N = 26 |
|---|---|
| Median (range) age in years | 63 (51–78) |
| Gender (male/female) | 17/9 (65%/35%) |
| Race (White/Black) | 24/2 (92%/8%) |
| ECOG performance status (0/1) | 1/25 (4%/96%) |
| Smoking status (never/former/current) | 2/6/18 (8%/23%/69%) |
| Disease stage at diagnosis (limited/extensive) | 8/18 (31%/69%) |
| Liver metastasis | 10 (38%) |
| History of brain metastasis | 9 (35%) |
| Type of treatment for brain metastasis (WBRT/SRS/Surgery) | 6/2/1 (23%/8%/4%) |
| Prophylactic whole brain radiation | 9 (35%) |
| Type of prior therapy (chemotherapy/radiation/immunotherapy/investigational agents) | 26/11/10/2 (100%/42%/38%/8%) |
| Number of prior systemic treatments (1/2/3/4) | 17/6/2/1 (65%/23%/8%/4%) |
| Prior chemotherapy type (Cisplatin/carboplatin + etoposide, Cisplatin/carboplatin + irinotecan1, Gemcitabine + nab-paclitaxel2) | 26/3/1 (100%/11%/4%) |
| Sensitivity to first-line chemotherapy (sensitive/resistant) | 5/21 (19.2%/80.8%) |
| Median (range) time from diagnosis to treatment (months) | 8.2 (3.5–32.3) |
Data are presented as median (range) or percentage (%)
Two patients were treated with cisplatin + irinotecan temporarily in first line due to etoposide shortage.
One patient was treated with gemcitabine + nab-paclitaxel as second line under clinical trial.
ECOG: Eastern Cooperative Oncology Group; WBRT: whole brain radiation therapy; SRS: stereotactic radiosurgery.
Plasma pharmacokinetics of topotecan and M6620 (Fig. S3A–C, Table S3) appeared consistent with the individual agents [M6620 Investigator’s Brochure, Hycamtin® Prescribing Information], suggesting absence of drug-drug interactions. Peak plasma concentrations of M6620 exceeded the doses that induced pharmacodynamic modulation of ATR activity in vivo (Fig. S2J). The most common treatment-related adverse events (n, %) of any grade were anemia (25, 96.2%), lymphopenia (25, 96.2%), thrombocytopenia (24, 92.3%), neutropenia (13, 50.0%) followed by gastrointestinal symptoms nausea (13, 50.0%) and vomiting (11, 42.3%) (Table S3). The most common grade 3 or 4 adverse events were lymphopenia (18, 69.2%), thrombocytopenia (15, 57,7%), anemia (14, 53.8%), and neutropenia (4, 15.4%). Complications such as febrile neutropenia were rare (1 patient; 3.8%), and no treatment-related deaths occurred.
Fifteen of twenty-six (57.7%) patients required at least one dose reduction, most commonly for thrombocytopenia (n=11, 42.3%) (Table S3). Two (7.7%) patients discontinued treatment due to treatment adverse events (both grade 4 thrombocytopenia despite two dose reductions) after four and five cycles of treatment, respectively. In general, the toxicity profile of the combination mirrored that of the phase I population of patients with treatment-refractory solid tumors (Thomas et al., 2018b). Most adverse events were attributable to topotecan, which as monotherapy is associated with a high frequency of myelosuppression (von Pawel et al., 1999). The differentiating features of rapidly dividing cancer cells, including higher replication stress that render them more susceptible to ATR inhibition may explain the absence of major overlapping toxicities with this combination.
ATR inhibition enhances the efficacy of TOP1 inhibition in chemotherapy-resistant SCLC patients
Seven of 16 (43.8%) patients had confirmed partial responses (PRs) in the first stage, allowing continued enrollment onto the second stage. In the overall study, nine of 25 patients [36.0%, 95% confidence interval (CI), CI: 18.0–57.5] achieved a confirmed PR, meeting the primary endpoint for response (Fig. 3B, C, D, Table S3). One patient had an unconfirmed response at first imaging (39.7% tumor reduction) but disease progression soon after. Most patients (17/25 patients; 68.0%) experienced tumor regressions (Fig. 3B, D). After a median potential follow up of 20.7 months, the median progression-free survival (PFS) was 4.8 months (95% CI: 2.8–7.4) (Fig. 3C, Fig. S3D). The PFS at 4 months and 6 months were 60.0% (38.4–76.1) and 36.0% (18.2–54.2), respectively. The median overall survival (OS) was 8.5 months (5.6–13.6). OS at 6 months and 12 months were 68.0% (46.1–82.5) and 32.0% (15.2–50.2) respectively (Fig. S3E).
Responses were observed in patients with both platinum-sensitive and platinum-resistant disease (ORR 60.0% [3/5 patients; 14.7–94.7] and 30.0% [6/20; 11.9–54.3] respectively; p = 0.31 by Fisher’s exact test) (Fig. 3E, F, Fig. S3F, Table S3). The median duration of response (DOR) was 6.4 months (1.1–14.3) (Fig. S3G, Table S3). Durable responses among patients with platinum-resistant disease were notable given that these tumors rarely respond to topotecan monotherapy, and are typically fatal within weeks of relapse (Horita et al., 2015; O’Brien et al., 2006). Four of six (66.7%) responders with platinum-resistant SCLC had DOR more than 6 months. Two of the platinum-resistant responders (33.3%; patient # 33 and 50) remain without disease progression and on treatment at data cut-off, 24 and 13 months respectively after starting treatment. The DOR, PFS and OS were not significantly different between patients with platinum-sensitive and resistant disease (Fig. S3H–J, Table S3), and patients who were previously treated with PD-1/PD-L1 inhibitors and those treated without (Fig. S3K, S3L, Table S3). In general, responses were not affected by dose reductions, with some of the responses maintained long-term despite dose modifications early on in the treatment course (Fig. S3M).
In an exploratory analysis, circulating tumor cells (CTCs) expressing EpCAM or CD117 were assessed pre-treatment, 3 weeks after treatment, and at disease progression. Patients with disease control [PR + stable disease (SD) ≥4 months] had significantly fewer CTCs pre-treatment and 3 weeks after treatment (Fig. 3G, H, Table S3). Consistently, patients with lower pretreatment CD117+ EpCAM+ CTCs than the median had better PFS than those with higher CD117+ EpCAM+ CTCs (median PFS: 5.5 vs. 3.0 months, p = 0.019 by log-rank test) (Fig. 3I).
Efficacy of ATR and TOP1 inhibition in SCNC patients independent of tissue of origin
Together, the trial in SCLC patients clinically validated the strategy of combined ATR and TOP1 inhibition and suggested that this approach might generally be effective at enhancing efficacy of topotecan. Given that SCLCs share a common molecular phenotype with EP-SCNCs (Balanis et al., 2019), we reasoned that EP-SCNCs – independent of their tissue of origin – may also be responsive to combined ATR and TOP1 inhibition.
To address this hypothesis, we amended the clinical trial and enrolled 10 patients with EP-SCNCs and tumor progression after at least one previous systemic chemotherapy (Tables S3). This expansion included patients with EP-SCNCs from distinct primary sites including bladder and cervix (two patients each), among others. Based on emerging preclinical data suggesting ATR inhibition overcoming DNA damaging therapy-resistance (Murai et al., 2018; Yazinski et al., 2017), patients whose tumor had progressed on TOP1 inhibitors were included in this cohort. Most patients had platinum-resistant disease (6/10, 60%) and had progressed on multiple systemic therapies, including four patients who progressed previously on topotecan.
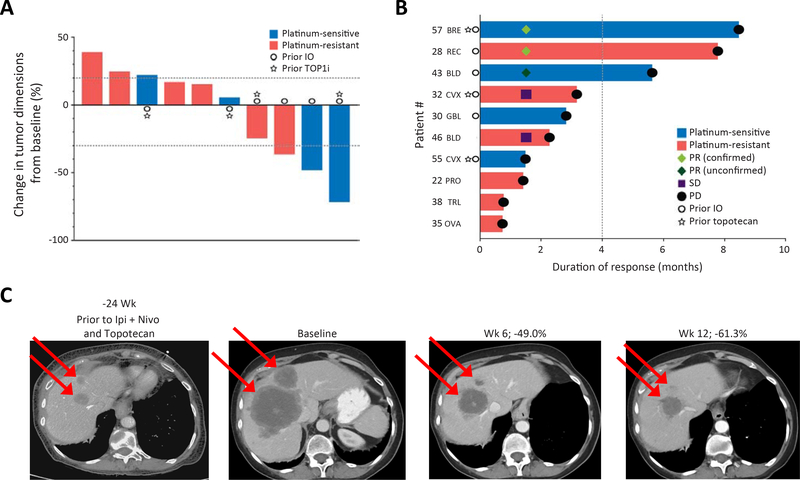
All ten patients were treated with at least one cycle (range: 1–10) and were evaluable for treatment efficacy. Treatment with M6620 and topotecan resulted in two confirmed PRs and an unconfirmed PR, yielding ORR of 20.0% (Fig. 4A, B). Notably, deep and durable responses were observed in tumors refractory to prior TOP1 inhibitor treatments. Two of five patients who achieved PR or SD had tumor progression previously on TOP1 inhibitors (Fig. 4B), including a patient with treatment-refractory breast SCNC (patient #57), who had major tumor regression lasting 8.5 months (Fig. 4C).
Figure 4: Efficacy of M6620 and topotecan in SCNCs regardless of tissue of origin.
A) Responses to the combination in EP-SCNCs based on change in tumor dimensions from baseline. Each bar represents a patient’s tumor response. The top and bottom dotted lines indicate the cutoffs of disease progression and partial response per RECIST criteria, respectively. B) Efficacy in EP-SCNCs based on duration of response. The dotted line indicates the 4-month timepoint after treatment. C) CT abdomen showing tumor regression in a patient with breast SCNC and liver metastases (patient #57). The patient had undergone extensive previous treatments including topotecan. REC, rectal; BLD, bladder; TRL, transformed lung cancer; BRE: breast; CVX, cervix; GBL, gall bladder; PRO, prostate; OVA, ovarian. See also Table S3.
Genomic alterations associated with response to ATR and TOP1 inhibition
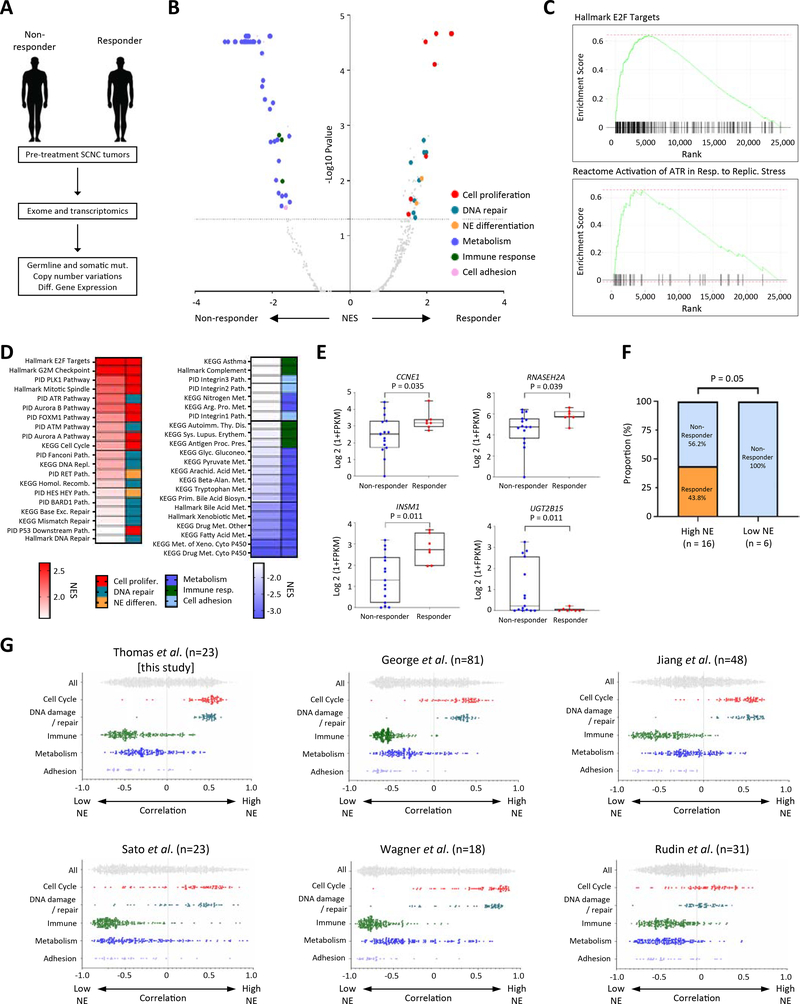
Although the combination yielded durable tumor regressions, there was significant heterogeneity in responses. Therefore, in an exploratory analysis, we sought to identify genomic correlates of response using whole exome sequencing (WES) of pre-treatment tumor (13 SCLCs and 6 EP-SCNCs) and matched germline blood. Responding tumors (n=6) were derived from patients who went on to have confirmed or unconfirmed PR whereas non-responding tumors (n=13) were from patients with stable or progressive disease. Tumor biopsies were obtained immediately prior to M6620 and topotecan (n=9) or before an earlier course of therapy (n=10) (Table S4).
Tumor mutational burden and somatic mutations in SCNC-related genes were not significantly associated with tumor responses (Fig. S4A, B). Mutations in putative genes associated with replication stress (e.g. CCNE1and MYC) (Murga et al., 2011; Toledo et al., 2011) or DNA repair (Williamson et al., 2016) (Table S4) were not enriched among responders. However, one responder (#36 SCLC, −37.3% tumor response, lasting 15 months) notably harbored germline splice site mutation of RAD54L, which encodes a recombinational protein critical for maintenance of genome stability (Heyer et al., 2006).
Next, we investigated somatic copy number alterations (SCNAs) in specific genes (Fig. S4A). Three of the six responding tumors harbored SCNAs in genes driving replication stress, including CCNE1 gain (Toledo et al., 2011) (#34 SCLC, #43 bladder SCNC) and ARID1A loss (Williamson et al., 2016) (#28 rectal SCNC). Among the three responding tumors with no SCNAs in the canonical replication stress-related genes, two harbored focal gains in SOX2 and SOX4, with gain of both genes observed in one case. In both instances, the patients had platinum-resistant SCLCs, achieved PRs (#50, #33), and remain on treatment over 13 and 24 months respectively, at the time of data cut-off. SOX2 is a neural stem maintenance gene, and transcriptional target of ASCL1 (Borromeo et al., 2016), a driver of NE differentiation across multiple tissue types. SOX genes are recurrently amplified in SCLC (Rudin et al., 2012), and have been implicated in the phenotypic switch from epithelial to NE differentiation under selection pressure from targeted therapy (Mu et al., 2017). Thus, focal SCNAs in genes related to replication-stress and NE differentiation may in part explain the variance in patient responses to ATR and TOP1 inhibition.
Association of tumor neuroendocrine differentiation and clinical benefit
To gain further insight into potential unifying mechanisms underlying tumor responses, we analyzed the transcriptomes through RNA-seq of responding (n=6) and non-responding (n=17) tumors with available high-quality RNA (Fig. 5A, Table S4; 13 tumors pre-treatment and 10 before an earlier therapy). Using a previously described molecular classifier that predicts the SCNC phenotype (Balanis et al., 2019), samples in our cohort (16 SCLCs and 7 EP-SCNCs) were more similar to each other and to NE prostate cancer than to adenocarcinoma or normal tissue (Fig. S4C).
Figure 5: Association of high replication stress and high NE differentiation with responses to M6620 plus topotecan; co-enrichment of high NE differentiation and replication stress among SCLCs.
A) Schema illustrating exomic and transcriptomic profiling of pre-treatment tumors among responding and non-responding SCNCs. B) Volcano plot showing gene set enrichments among SCNC responders and non-responders. Gene sets in the upper left and right quadrants are significantly enriched in non-responders and responders, respectively. Selected gene sets related to cell proliferation, DNA repair, NE differentiation, metabolism, immune response and adhesion are highlighted. C) Relative RNA expression-based enrichment of hallmark E2F targets and Reactome activation of ATR in response to replication stress pathways in responders compared with non-responders. The y-axis represents enrichment score and the x-axis indicates differential expression rank for all genes, with gene set-specific genes marked by vertical black lines. Rank positions toward the left indicate increased expression in responders. D) Hallmark, KEGG and PID gene sets differentially upregulated and downregulated among responders. Color gradation is based on GSEA normalized enrichment score. E) Box plot expression summaries for selected genes upregulated or downregulated among 7 responders (P <0.05) and 16 non-responders. The boxes and error bars indicate median with interquartile range and maximum/minimum ranges, respectively. Statistical differences are evaluated by Mann-Whitney U test. F) Response proportions among SCNCs with high and low NE differentiation. The statistical difference is evaluated by chi-square test. G) Scatter plots showing the distribution of correlations between NE signature scores (ranging from low NE to high NE correlation on the x-axis) and cell cycle, DNA damage, immune, metabolism, and adhesion signature scores (assessed by ssGSEA) in this study and five published, independent SCLC datasets. Within each study, the category-associated correlation distributions were significantly different based on pairwise Kolmogorov-Smirnov tests (P < 0.0001). See also Table S4, Figs. S4–6.
First, we examined differentially expressed genes (DEGs) between responding and non-responding tumors, and key biological processes (FDR <5%) represented by the DEGs, using Gene Set Enrichment Analysis (GSEA). Across PID, Hallmark and KEGG gene sets, genes upregulated in responding tumors displayed marked enrichment for multiple pathways associated with cell-cycle progression and DNA repair, including E2F target genes, the G2-M checkpoint, and ATM, ATR and Fanconi DNA repair pathways, consistent with responding tumors harboring a replication stress phenotype (Fig. 5B–D, Table S4) (Zhang et al., 2016). The enriched pathways represented essential components of the checkpoint response, including fork stalling, stabilization, and resolution, which allows cancer cells to cope with the increased rates of replication, and prevent replication stress-induced DNA damage (Bertoli et al., 2016).
Accordingly, responding tumors also exhibited collectively increased expression of genes upregulated in response to activation of the ATR pathway triggered by replication stress (Fig. 5C, D, Fig. S4D, E, Table S4). In line with these findings, genes upregulated in responding tumors displayed enrichment for pathways associated with genomic instability (normalized enrichment score [NES]=2.3, adjusted P<0.0001), replicative immortality and sustained proliferative signaling (NES=2.0, adjusted P=0.0001) (Fig. S5A) (Alcala et al., 2019; Hanahan and Weinberg, 2011). Genes downregulated in responding tumors were enriched for immune response, metabolism and adhesion. These patterns of gene set enrichment were maintained even after omitting EP-SCNCs from the analysis (Fig. S5B, C, Table S4). DEGs with higher expression in the responding tumors included those related to replication stress (CCNE1), maintenance of genomic stability (RNASEH2) (Reijns et al., 2012) and NE differentiation (ASCL1, BEX1, INSM1) (Fig. 5E, Fig. S5C, Table S4) (Borromeo et al., 2016; Kidd et al., 2014; Rooper et al., 2018).
Although SCNCs are NE tumors, inter-tumor heterogeneity in NE differentiation is documented (Rudin et al., 2019; Zhang et al., 2018). Given the striking upregulation of genes determining neuronal and NE fate among responders, we sought to determine whether the NE phenotype might stratify tumors based on responses. We characterized NE differentiation of each tumor using a 50-gene signature and single-sample gene set enrichment analysis (ssGSEA) (Zhang et al., 2018). Notably, seven of 16 (43.8%) tumors with high NE phenotype responded whereas none of the low NE tumors responded to ATR and TOP1 inhibition (P=0.05 by chi-square test) (Fig. 5F). Applying the recently proposed SCLC subtyping based on differential expression of transcription regulators (Rudin et al., 2019), all the responding tumors were of high NE ASCL1 or NEUROD1 subtypes (Fig. S6A–C), whereas none of the non-NE POU2F3 or YAP1 subtypes responded to treatment.
The observation, based on correlation of respective ssGSEA scores, that high NE phenotype was co-enriched with cell proliferation and DNA repair signatures implied co-regulated tumor phenotypes that together define a transcriptomic subset. To evaluate whether co-enrichment of the signatures was an exclusive feature of our cohort, we queried five additional SCLC cohorts with available RNA-seq or microarray data (George et al., 2015; Jiang et al., 2016; Rudin et al., 2012; Sato et al., 2013; Wagner et al., 2018). Across all five cohorts, we found NE signature co-enriched with cell proliferation and DNA repair pathways (Fig. 5G), and inversely associated with enrichment of metabolism, adhesion and immune response genesets. NE tumors in general have enhanced proliferation relative to non-NE tumors such as adenocarcinomas (Balanis et al., 2019; Wang et al., 2020). We extend these observations to define a distinct transcriptomic subset within SCNCs, characterized by high NE differentiation and enhanced replication stress, and upregulation of proliferation and DNA-damage repair pathways.
Importantly, patients with high NE SCNCs had significantly improved PFS [median: 4.5 (1.3–7.2) vs. 2.1 months (0.7–not reached), P = 0.038 by log-rank test] and a trend towards better OS [8.1 (2.5–13.6) vs. 2.8 months (1.5–not reached), P = 0.11] (Fig. S6D, E) compared with patients with low NE SCNCs. Further, patients with high NE SCNCs had significantly prolonged DOR compared to those with low NE SCNCs (3.8 months [1.2–6.9] vs. 0.7 months [0.03–not reached], P = 0.0019) (Fig. S6F). Although confirmation in independent cohorts is required, our findings indicate that SCNCs with high replication stress and high NE differentiation are more likely to respond to ATR and TOP1 inhibition.
Discussion
SCNCs are recalcitrant cancers with extremely poor prognosis and no effective therapeutic options at relapse. With no targetable alterations or molecular subtypes informing treatment, they represent a major clinical challenge. Using high throughput drug screens, we uncovered and validated a potent combination of ATR and TOP1 inhibitors, with marked antiproliferative synergy in SCLC cells. These observations were translated to the clinic in a study combining ATR inhibitor M6620 with standard-of-care TOP1 inhibitor topotecan. The study met its primary objective, with 36.0% confirmed ORR. Remarkably, the combination resulted in durable tumor regressions in platinum-resistant SCLC patients. Comprehensive exomic and transcriptomic profiling of pretreatment tumors demonstrated striking enrichment of high NE differentiation and enhanced replication stress among responding tumors.
The antitumor effects of the M6620 and topotecan combination appear to exceed those of current standard regimens and agents in development (Farago et al., 2019; Horita et al., 2015; Pietanza et al., 2018), particularly among platinum-resistant patients, although cross-trial comparisons should be interpreted with caution. Among platinum-resistant SCLC patients in the present study, the confirmed ORR was 30.0%, median DOR 6.9 months, and 66.7% of patients with confirmed responses had DOR>6 months. In comparison, topotecan, the approved second-line agent yields responses in approximately 5% of patients with platinum-resistant SCLC (Horita et al., 2015). The confirmed ORR was 22.2%, median DOR 4.7 months, and 11.7% of patients with confirmed responses had DOR>6 months with second-line lurbinectedin (Trigo et al., 2020). In contrast to previous studies, which for the most part, included patients with limited number of prior therapies (Farago et al., 2019; Pietanza et al., 2018; Trigo et al., 2020), more than a third of the patients in the present study had undergone extensive previous therapies, yet responses of notable depth and duration were observed.
SCLC is treated as a homogeneous disease with a one-size-fits-all approach. Yet, it has been known for many years that SCLC tumors and cell lines exhibit distinct inter-tumor heterogeneity with respect to expression of NE features (Gazdar et al., 1985). Recently, a consensus nomenclature proposed four SCLC molecular subtypes defined by differential expression of key transcription regulators ASCL1, NEUROD1, YAP1 and POU2F3 (Rudin et al., 2019). A 50-gene NE score distinguishes SCLC-A and most SCLC-N from the non-neuroendocrine subtypes (SCLC-Y and SCLC-P) (Zhang et al., 2018). Whether these molecular subtypes – defined using human cell lines and surgically resected tumors – are relevant to prognosis and treatment outcomes remains unknown. Here we demonstrate that SCNCs with high NE differentiation are characterized by enhanced replication stress, as evidenced by heightened requirement for cell proliferation and DNA-damage repair pathways. Our observations on subtype-specific vulnerabilities could prove useful for rational patient selection in future studies.
EP-SCNCs have extremely poor prognoses, and given their rarity and heterogeneity, advancing care through clinical trials is challenging. The striking responses in patients who had undergone extensive previous therapies provide compelling rationale to rigorously investigate the effect of this combination in EP-SCNC patients, and serve as a template for other signal-finding combination studies in EP-SCNCs, regardless of tissue of origin.
Owing to its phase 2 single-arm design, the trial is a proof-of-concept study, able to provide signals of activity and to generate preliminary evidence, supported by a sound translational background, to be further confirmed in a larger clinical trial. Due the small sample size of the clinical trial, the characteristics of patients included may not reflect the typical relapsed SCLC population, including a higher proportion of platinum resistant patients and females, affecting the generalizability of these findings. These limitations notwithstanding, our findings warrant further investigation. A randomized phase II study of the combination is underway in SCLC patients (NCT03896503), with a separate cohort for patients with EP-SCNC, regardless of primary site of origin.
In summary, we discovered the therapeutic vulnerability of SCLC cells to multiple drugs targeting the replication stress response. Combination of M6620 and topotecan led to substantial clinical benefit in a subset of relapsed SCNC patients, with higher than expected response rates and durable responses in patients with platinum-resistant disease. Comprehensive molecular assessment of pre-treatment tumors in this study provides a framework to understand the therapeutic responses and paves the way for development of personalized treatments in SCLC.
Star Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Anish Thomas (anish.thomas@nih.gov).
Materials Availability
This study did not generate new, unique reagents.
Data and Code Availability
The accession number for the sequencing data reported in this paper is dbGaP: phs2327.v1. Gene sets used for ssGSEA are available in Molecular Signatures Database https://www.gsea-msigdb.org/gsea/msigdb/. Gene expression data (RNA-seq or microarray) from independent SCLC cohorts were retrieved from: European Genome-phenome Archive EGAS00001000925, Gene Expression Omnibus (GEO): GSE60052,GSE43346, dbGAP: phs001049.v1.p1, and EGAS00001000334. Processed RNA-seq data (1+TMM_FPKM) and code for GSEA and ssGSEA are available at: https://github.com/alwaysblack777/Therapeutic-targeting-of-ATR-yields-durable-regressions-in-high-replication-stress-tumors.
Experimental Model and Subject Details
Cell lines
SCLC cell lines H446, H196, H524, DMS114, DMS79, H187, H889, and HT-29, and colorectal adenocarcinoma cell line Colo205 were purchased from ATCC and maintained in culture. Cell lines were authenticated using short tandem repeat DNA profiling and tested for mycoplasma contamination before experiments. Cells were cultured at 37°C and 5% CO2 in RPMI-1640 supplemented with 10% FBS, except HT-29 and DMS-114, which were grown in McCoy’s 5a Medium Modified supplemented with 10% FBS and Waymouth’s MB 75/21 medium supplemented with 10% FBS respectively. Colo205 cells were cultured in DMEM with 10% fetal bovine serum at 37°C and 10% CO2.
Mice
All animal experiment protocols were approved by the regional council Committee on the Ethics of Animal Experiments (Regierungspräsidium Darmstadt, Hessen, Germany for Colo 205, and Regierungspräsidium Freiburg, Baden-Wuerttemberg, Germany for LXFS573). Seven- to nine-week-old female athymic Nude-Foxn1nu (for Colo205 xenografts), or five- to seven-week old female NMRI-Foxn1 nu nude (for LXFS573 PDX) mice were used (Envigo, France, or Charles River, Germany).
Patient-derived xenograft model
Patient derived SCLC LXFS573 xenograft mouse models were obtained from Charles River Discovery Research Services Freiburg, Germany.
Clinical trial design and patients
Patients were enrolled on an investigator-initiated, single-arm, phase 2 trial (NCT02487095) conducted at the Center for Cancer Research, National Cancer Institute (NCI) in Bethesda, MD. This trial was approved by the NCI institutional review board and conducted per Good Clinical Practice guidelines, defined by the International Conference on Harmonization. All patients provided written informed consent per Declaration of Helsinki principles.
Patient characteristics are indicated in Table 1. Patients were eligible if they were aged 18 years or older and had histologically confirmed SCLC; measurable, progressing disease as defined by Response Evaluation Criteria in Solid Tumors (RECIST; version 1.1) (Eisenhauer et al., 2009) and an Eastern Cooperative Oncology Group (ECOG) performance status score of 2 or less. Eligible patients had at least one line of chemotherapy. Patients also had to have adequate renal, liver, and bone marrow function. Patients with symptomatic brain metastases were excluded from trial although patients who have had treatment for their brain metastasis and whose brain disease was stable without steroid therapy for 1 week were eligible. Before initiating therapy, we required patients to have at least 2 weeks without previous chemotherapy or major surgery and 24 hours without radiation. Platinum-sensitive SCLC was defined as relapse or disease progression ≥90 days after completion of first-line platinum-based chemotherapy, whereas platinum-resistant was defined as relapse or disease progression <90 days or during first-line chemotherapy.
M6620 was supplied under a Collaborative Research and Development Agreement (CRADA) among the National Cancer Institute (NCI) and initially Vertex Pharmaceuticals, and currently Merck KGaA, Darmstadt, Germany. Topotecan was obtained from commercial sources. The trial was conducted under an NCI Center for Cancer Research (CCR)–sponsored investigational new drug application (IND#118173) with institutional review board approval. All participants provided written, informed consent. Data were analyzed through 18th March 2020.
Method Details
Quantitively high-throughput and matrix screening
H446, H196, H524, DMS114, DMS79, H187, H889 Cells were dissociated with TrypLE as needed to remove attached cells. Cells were seeded in 5 uL of growth media using a Multidrop Combi dispenser into 1536 well white polystyrene tissue culture treated Corning plates at a density of (500–1000) cells/well, depending on the specific cell line, to allow for compounds to be present during exponential growth phase. After cell addition, 23 nL of MIPE 5.0 compounds were added to individual wells (11 dosing tested for all compounds in separate well) via a 1536 pin-tool. Bortezomib (final concentration 2.3 μM) was used as a positive control for cell cytotoxicity. For drug combination screening, 10 nL of compounds were acoustically dispensed into 1536 well white TC treated plates. Cells were then added to compound-containing plates at the same density used for single agent screening in 5 uL of media. A 5-point custom concentration, with constant 1:4 dilution between points was used for the primary 6×6 matrix screening, and a 9-point concentration range with 1:2 dilution between points was used for secondary 10×10 matrix screening. Plates were incubated for 48 hours at standard incubator conditions, covered by a stainless steel gasketed lid to prevent evaporation. 3 uL of Cell Titer Glo (Promega, Madison, WI, USA) were added to each well and plates were incubated at room temperature for 15 minutes with a stainless-steel lid in place. Luminescence readings were taken using a Viewlux (PerkinElmer, Waltham, MA, USA) with a 2 second exposure time per plate. Compound dose response curves were normalized to DMSO and empty well controls on each plate. All single agent screening data is publicly available within Table S1. All combination screening data is publicly available at https://matrix.ncats.nih.gov/. Each unique screen is independently searchable, and a ‘help’ tab provides an overview of methods and a tutorial to aid users as they search this database.
In vitro combination cell viability assay
Briefly, 2,000 HT-29 cells were seeded into the internal wells of 96 well plates and incubated in a 37 °C / 5% CO2 incubator. HT-29 cells were grown in McCoy’s 5A medium + 10% fetal bovine serum (FBS). Outer wells contained PBS only without cells. SN-38 was added to wells to achieve 9 final concentrations of (1.0, 0.333, 0.111, 0.037, 0.0123, 0.0041, 0.0014, 0.0005, 0.0002 μM). M6620 was added to wells to achieve 5 final concentrations of (0.8, 0.4, 0.2, 0.1, 0.05 μM). Drug concentrations were chosen to cover the linear range of the dose-response curves for each individual agent. Both drugs were resuspended in DMSO (10 mM M6620 and 50 mM SN-38) and dilutions were made in medium + 10% FBS. Cells were washed 2X with 200 μL fresh medium at each indicated wash step. The experiments were conducted once. Cell viability was measured using CellTiter Glo reagent (Promega) and the Envision luminescence plate reader (Perkin Elmer) on Day 4 for all scheduling groups. Synergy plots and Bliss Sum scores were calculated using MacSynergy II software(Prichard and Shipman, 1990), available free online https://www.uab.edu/medicine/peds/macsynergy and as described previously. All synergy plots and associated log volumes above the plane were calculated with a 95% confidence interval.
For combination analysis using H446 cells, cells were treated similarly except for the following: 1,000 cells were seeded into the internal wells of 384 well plates and H446 cells were grown in RPMI 1640 medium + 10% fetal bovine serum (FBS) and 2mM L-Glut. Outer wells contained PBS only without cells. Drugs (topotecan, irinotecan and M6620) were diluted in the H446 media and added to wells to achieve 9 final concentrations with a 1:3 dilution factor. Highest single agent (HSA) synergy was calculated by calculating the difference between the most effective single agent, and the results observed for any combination of agents.
In vivo of complex (ICE) assay
TOP1-DPCs were measured with in vivo complex of enzyme (ICE) assay as previously described (Anand et al., 2018). DMS114 cells were pretreated with ATR inhibitor M6620 (1 μM), ATM inhibitor KU-55933 (1 μM) or DNA-PKcs inhibitor VX984 (1 μM) for 4 h before co-treatment with topotecan (1 μM) for 2 h. Cells were lysed in sarkosyl solution (1% w/v) and then sheared through a 25g 5/8 needle (10 strokes). The lysates were layered onto CsCl solution (150% w/v), followed by centrifugation in NVT 65.2 rotor (Beckman coulter) at 42,000 RPM for 20 hours at 25 °C. The The resulting DNA samples were quantitated with NanoDrop spectrophotometer and subjected to slot-blot for immunoblotting with anti-TOP1 antibody (Cat #556597, Biosciences). TOP1-DPC was quantitated by densitometric analysis using ImageJ.
Alkaline comet assay
DMS114 cells were treated 1 μM topotecan or 1 μM topotecan + 1 μM M6620 for 8 h. Cells were then collected for alkaline comet assays using the CometAssay Kit (R&D Systems®, Catalog # 4250–050-K) following manufacturer’s instructions. Images were captured using BioSpa Live Cell Analysis System (Biotek) and tail moment was calculated using OpenComet(Gyori et al., 2014), a plugin for the image processing program ImageJ.
DNA combing assay
DNA combing assay was performed as described(Josse et al., 2014). After drug treatments and CldU-IdU pulses, DMS114 cells were embedded in low-melting agarose for digestion with β-agarase (New England Biolabs). DNA was then combed on silanized surfaces (Microsurfaces, Inc.) and replicons were detected with anti-CIdU and anti-IdU antibodies. Signal was captured and analyzed with FiberVision automated scanner (Genomic Vision).
Flow cytometry analysis
Analyses of EdU incorporation and cell cycle were performed as described(Josse et al., 2014). DMS114 cells were incubated with 50 μM 5-ethynyl-2’-deoxyuridine (EdU) for 30 min before adding 1 μM topotecan or 1 μM topotecan + 1 μM M6620 for 1h. Topotecan was then removed and M6620 was kept in medium for another 18 h before cells were harvested. EdU was detected by flow cytometry (Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay, Invitrogen), and DNA using DAPI. Data were acquired using BD LSRFortessa Flow Cytometer and further analyzed using FlowJo.
Xenograft studies
In vivo efficacy data were generated in the human colorectal adenocarcinoma Colo 205 (ATCC®, CCL-222™) xenograft model in mice, and in the patient derived SCLC LXFS573. The study designs and animal usage were conducted according to all applicable international, national and local laws and followed the national guidelines for the Care and Use of Laboratory Animals of the Society of Laboratory Animal Science (GV-SOLAS). Colo205 experiment was conducted by Merck Healthcare KGaA in Darmstadt, Germany and LXFS573 by Charles River Discovery Research Services Freiburg, Germany. Colo205 were grafted by subcutaneous injections of 4 million Colo205 cells in suspension, LXFS573 tumor pieces of 3–4 mm edge length. When tumor xenografts reached a mean volume of 60 mm3 (Colo205), or 110 mm3 (LXFE597), mice (n=10 per treatment arm, randomized from 15 mice per arm, LXFS597: n=5 per treatment arm, randomized from 8 mice to obtain a similar mean and median, within the treatment groups) received M6620 (clinical formulation, diluted in 5% dextrose/water) irinotecan (cell pharm Stada Pharma GmbH, infusion solution, 20 mg/ml stock solution, diluted in 0.9% NaCL) or the combination thereof in different doses and schedules. Tumor length (L) and width (W) were measured with calipers and tumor volumes were calculated using L×Ŵ2/2.
Pharmacodynamic and pharmacokinetic studies in xenograft models
Pharmacodynamic (PD) data were generated in Colo 205 human xenograft models in mice. The study was conducted by Merck Healthcare KGaA, Darmstadt, Germany. The animal experiment protocols wereas approved by the regional council Committee on the Ethics of Animal Experiments and conducted according to all applicable international, national and local laws and following the national guidelines for the Care and Use of Laboratory Animals of the GV-SOLAS. M6620 was administered i.v. once (10, 20, and 30 mg/kg) in combination with 50 mg/kg irinotecan given i.p.. Animals (4 to 5 animals per group) were sacrificed at different time points (1, 3, 6, 8, or 24 hours) after giving both drugs at the same time. For the vehicle control animals were sacrificed 12 h after vehicle administration. Tumors were removed, frozen in liquid nitrogen, lysed with HGNT buffer, and homogenized using a Precellys-24 homogenizer.
For analysis of pCHK1 and tCHK1 level, Luminex technology was used. 25 μg total protein were analyzed in the Luminex assay. After adding the diluted bead suspension (3000 beads) with coupled antibodies against total CHK1 (CHK1 (EP691Y); rabbit monoclonal ab, Abcam ab210964) the plate was incubated over night at 4°C. The beads were washed 3 times with TBST (Pierce/Thermo Scientific 28358 + 0,05% Tween) and 25 μl blocking reagent (Roche 11112589001 + 1% Tween) as well as 25 μl antibody dilution of the biotinylated detection antibody were added (tCHK1: CHK1 (2G1D5) mouse monoclonal ab; Cell Signaling #2360BF, pCHK1: pCHK1 (133D3) rabbit mAb; Cell Signaling #2348BF, final concentration 1.5 μg/ml). After 1 h incubation at RT, the beads were washed again with TBST and 25 μl blocking reagent and the SAPE dilution (25 μl, Moss #001NB) were added for 45 min at RT. After washing and adding of blocking reagent, the probes were analyzed in the Magpix instrument following the instructor’s manual. The instrument settings were as follows: sample volume: 35 μl, count: 50, unit: MFI. To control the Luminex assay, different concentrations of HT29 control lysate which were used as standard were analyzed in parallel. After background correction the signals for the phospho proteins were normalized to signals of the total protein. Samples were measured in singlicate. The mean value of the vehicle control was set to 100%. The values were analyzed in GraphPad and mean +/− SEM was calculated. The quantitative determination of M6620 in mouse plasma was done by HPLC-MS/MS. Data were analyzed in GraphPad and mean +/− SEM was calculated.
Clinical trial procedures
Study therapy consisted of M6620 administered at 210 mg/m2 intravenously over 60 minutes on day 2 and day 5, topotecan 1.25 mg/m2 intravenously over 30 minutes every 23 hours on days 1 through 5, and pegfilgrastim 6mg subcutaneously on day 6 in 21-day cycles. Dose reductions, interruptions, or both were permitted for toxicities not relieved by supportive care. Treatment was continued until disease progression, death, unacceptable toxicity, or withdrawal of consent.
A history and physical examination were performed at baseline and before each cycle. Complete blood counts and serum chemistries were performed on days 1, 8 and 15 of the first 2 cycles and on day 1 only thereafter. Adverse events were graded according to Common Terminology Criteria for Adverse Events version 4.0. Doses of M6620 and topotecan were not modified in the first cycle. Based on tolerability of the previous cycle, the dose of M6620 were modified in subsequent cycles as follows: for grade 4 hematologic toxicity, the dose was reduced by 25%; for grade 3 non-hematologic toxicity (except disease-related hyponatremia), the dose was reduced by 25%; for grade 4 non-hematologic toxicity, the dose was reduced by 50%. Based on the criteria above, a maximum of 2 dose reductions of M6620 were permitted. Topotecan dose was modified for renal impairment and myelosuppression. In addition, incase of grade 3 or higher toxicity during any cycle beyond cycle 1, treatment was interrupted and resumed when all toxicities had returned to grade 2 or less, at the discretion of the investigator.
Radiographic evaluation was performed at baseline and every two cycles for tumor response on the basis of RECIST version 1.1 (1). Peripheral blood was collected on day 1 and day 5 of each cycle, and at the time of progression. Tumor biopsies were performed at baseline. Patients were followed up for adverse events for 30 days from last dose of study therapy, and survival after discontinuation of study treatment until death or withdrawal of consent.
Circulating tumor cells
CTCs were detected from 10 mL of peripheral blood drawn into EDTA tubes pre-treatment, 3 weeks after treatment, and at the timepoint of disease progression. Epithelial cell adhesion molecule (EpCAM)-positive CTCs were isolated using magnetic pre-enrichment and quantified using multiparameter flow cytometry. CTCs were identified as viable, nucleated, EpCAM+ cells that did not express the common leukocyte antigen CD45, as described previously(Kauffman et al., 2016; Thomas et al., 2015). After enumeration of viable nucleated, CD45−, EpCAM+ cells, CTCs were further characterized for expression of CD117, a tyrosine kinase receptor associated with cancer progression and normal stem cell maintenance.
Pharmacokinetics
Blood samples for pharmacokinetic (PK) analyses of topotecan were collected before, during, and up to 24 hr following the end of a 30-min IV infusion on cycle 1 day 1. Plasma concentrations were measured using a validated LC-MS/MS assay with a lower limit of quantification (LLOQ) of 0.5 ng/mL. M6620 PK sampling was performed before, during, and up to 48 hr after the end of the 1-hr IV infusion given on cycle 1 day 2 (when the first dose was given). Plasma concentrations were measured using a validated LC-MS/MS assay with an LLOQ of 3 ng/mL. A noncompartmental analysis was used to estimate patient-specific PK parameters for both total topotecan and M6620. The maximum plasma concentration (Cmax) and time to Cmax (Tmax) were recorded as observed values. The area under the curve extrapolated to time infinity (AUCINF) was calculated using the log-linear trapezoidal rule and the elimination rate (lambda z). The terminal half-life was calculated as ln2/lambda z. The clearance rate was calculated as dose/AUCINF and the volume of distribution as clearance/lambda z.
DNA and RNA sequencing
Tumor samples were obtained immediately prior to M6620 and topotecan treatment or before an earlier course of therapy. Formalin-fixed, paraffin-embedded (FFPE) tumor tissue samples were prepared for whole-exome sequencing (WES) and RNA sequencing (RNA-Seq). One hundred nanograms of DNA was sheared to approximately 200 base pairs (bp) by sonication (Covaris, Woburn, MA). Exome enrichment was performed using SureSelect Clinical Research Exome Kits according to the manufacturer’s instructions (Agilent, Santa Clara, CA) and RNA enrichment was performed using TruSeq RNA Exome Library Prep according to manufacturer’s instructions (Illumina, San Diego). Paired-end sequencing (2 × 75 bp) was performed on an Illumina NextSeq500 instrument. The sequences were compared to the human reference genome hg19 using internally developed ClinOmics somatic Bioinformatic Pipeline v3.1. Peripheral blood DNA extracted from individual patients was used for germline exome sequencing. In brief, raw sequencing data in FASTQ format were aligned against the reference human genome (hg19) with BWA. The Genome Analysis Toolkit (GATK) and HaplotypeCaller (HAPLOC) were used for germline SNV and indel calling; whereas Strelka was used for somatic single nucleotide variant (SNV) and small indel calling. ANNOVAR was used to functionally annotate genetic variants. Variants with variant allele frequency (VAF) > 0.10, tumor sequencing depth > 50, and matched germline sequencing depth > 50 were considered. FACETS algorithm was used to determine total and allele-specific DNA copy number from WES. RNA was extracted from FFPE tumor cores using RNeasy FFPE kits according to the manufacturer’s protocol (QIAGEN, Germantown, MD). RNA-seq libraries were generated using TruSeq RNA Access Library Prep Kits (TruSeq RNA Exome kits; Illumina) and sequenced on NextSeq500 sequencers using 75bp paired-end sequencing method (Illumina, San Diego, CA).
Clinical trial outcomes
The primary endpoint was objective response rate (ORR; complete response plus PR as documented by RECIST version 1.1). Secondary endpoints were PFS, OS, and duration of response. Exploratory objectives were to characterize tumor gene expression and mutations as well as circulating tumor cell (CTC) changes that predict response.
Quantification and Statistical Analysis
All quantification and generation of figures and graphs in the pre-clinical part were done using software described in methods above.
Clinical and demographic characteristics of study participants were summarized using descriptive statistics. For response as the primary outcome, the trial was conducted using a Simon two-stage Minimax design in order to rule out an unacceptably low 10% response rate (p0=0.10) in favor of a targeted response rate of 30% (p1=0.30) based on historical controls(von Pawel et al., 1999). With alpha=0.10 and beta = 0.10, 16 evaluable patients were initially enrolled, and we considered accrual would continue to a total of 25 subjects if 2 or more of the first 16 subjects had a response. If there were 5 or more subjects of the 25 (20.0%) who have a response, we considered the combination treatment as sufficiently interesting.
All patients were evaluable for toxicity if they received at least one dose of the study drug. Only patients who had measurable disease present at baseline, had received at least one cycle of therapy, and had their disease re-evaluated were considered evaluable for response. Duration of response was measured from the time these criteria were met for response until progressive disease. We summarized adverse events as the proportion of the total number of patients treated. Time to event analyses were done using the Kaplan-Meier method. Progression free survival (PFS) was determined from on-study date until date of progression or date of death without progression, overall survival (OS) was calculated until death from any cause or date last known alive. Duration of response was calculated from date response was noted until date of progression or last follow-up without progression. The significance of the difference of Kaplan-Meier curves was determined by a log-rank test.
For PK data, Microsoft® Excel® was used to calculate statistics and GraphPad Prism® was used to generate figures. We did all statistical tests for clinical outcomes using STATA software version 16.0. All tests were two-sided. The study is registered with ClinicalTrials.gov, number NCT02487095. Hierarchical clustering for SCLC transcriptome subtypes was done using Qlucore Omics Explorer ver. 3.6.
For transcriptomic analyses, raw RNA-Seq count data were normalized for inter-gene/sample comparison using TMM-FPKM, followed by log2(x+1) transformation, as implemented in the edgeR R/Bioconductor package.(Robinson et al., 2010) Gene set enrichment analyses were performed using the fgsea R/Bioconductor package and a gene ranking based on sign(Median(xR(i)) – Median(xNR(i)) * [-log10(p(i))], where xR(i) and xNR(i) are vectors of expression values for the ith gene in responder and non-responder samples, respectively, while p(i) is the t-test p-value relative to the latter sample sets. This measure ranks genes from those most upregulated in responders to those most upregulated in non-responders. ssGSEA enrichment scores were computed using the GSVA R/Bioconductor package(Hanzelmann et al., 2013) and gene sets obtained from MSigDB.(Liberzon et al., 2011)
KEY RESOURCE TABLE
Supplementary Material
Table S1. High-throughput single-agent and combination drug screens in SCLC cells, related to Figure 1.
Table S2. In vitro and in vivo characterization of ATRi plus TOP1i, related to Figure 2.
Table S3. Patient characteristics, pharmacokinetics, adverse events, dose modifications, and CTCs, related to Figures 3 and 4.
Table S4. Biopsy characteristics, and exomic and transcriptomic assessment of responders and non-responders, related to Figure 5.
KEY RESOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CelltiterGlo | Promega | Cat# G7570 |
| CaspaseGlo | Promega | Cat# G881C |
| Anti-TOP1 antibody | Biosciences | Cat #556597 |
| CHK1 (EP691Y); rabbit monoclonal antibody | Abcam | ab210964 |
| tCHK1: CHK1 (2G1D5) mouse monoclonal antibody | Cell Signaling | #2360BF |
| pCHK1: pCHK1 (133D3) rabbit monoclonal antibody | Cell Signaling | #2348BF |
| Blocking reagent BR | Roche | 11112589001 |
| R-Phycoerythrin-conjugated streptavidin (SAPE) | Moss | 001NB |
| MagPlex Microsphere region 72 | Luminex | MC10072–01 |
| TBS | Pierce / Thermo Scientific | 28358 |
| Sulfo-NHS-LC-LC-Biotin | Thermo Scientific | 21338 |
| Sulfo-NHS | Thermo Scientific | 24510 |
| 1-Ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) | Thermo Scientific | 22980 |
| Precellys lysing kit, hard tissue homogenizing CK28-R | Bertin-instruments | POOO916-LYSKO-A |
| EpCAM (HEA-125) mouse monoclonal antibody | Miltenyi | 130–113–264 |
| CD117 (104D2) mouse monoclonal antibody | BioLegend | 313212 |
| CD45 (HI30) mouse monoclonal antibody | BioLegend | 304014 |
| FcR blocking reagent | Miltenyi | 130–059–901 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Dye | ThermoFisher | L34957 |
| Biological Samples | ||
| Pre-treatment SCNC tumors | This paper | https://clinicaltrials.gov/ct2/show/NCT02487095 |
| Peripheral blood samples to detect CTCs | This paper | https://clinicaltrials.gov/ct2/show/NCT02487095 |
| Peripheral blood mononuclear cells for germline analysis | This paper | https://clinicaltrials.gov/ct2/show/NCT02487095 |
| LXFS573 (patient-derived lung cancer model passaged as subcutaneous xenografts in female NMRI nude mice | Charles River Discovery Research Services Germany GmbH | Freiburg, Germany |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MIPE library compounds | Various Vendors | See table S2 from https://stm.sciencemag.org/content/11/519/eaaw0064 |
| Berzosertib (M6620) | ActiveBioChem | A-1014 |
| KU-55933 | MedChem Express | HY-12016 |
| VX984 | AstaTech | 41568 |
| Topotecan | Prestwick | Prestw-1196 |
| Irinotecan | Selleck | S1198 |
| Critical Commercial Assays | ||
| SureSelect Clinical Research Exome Kits | Agilent | https://www.agilent.com/ |
| TruSeq RNA Exome Library Prep | Illumina | https://emea.illumina.com/ |
| RNeasy FFPE kits | QIAGEN | https://www.qiagen.com/us/ |
| NextSeq500 sequencers | Illumina | https://emea.illumina.com/ |
| Deposited Data | ||
| MSigDB | Liberzon et al., 2011 | https://www.gsea-msigdb.org/gsea/msigdb/ |
| RNA-seq file | This paper | dbGaP: phs2327.v1 |
| Whole exome sequencing file | This paper | dbGaP: phs2327.v1 |
| Independent SCLC RNA-seq data by George et al. | George et al., 2015 | EGAS00001000925 |
| Independent SCLC RNA-seq data by Jiang et al. | Jiang et al., 2016 | GEO: GSE60052 |
| Independent SCLC microarray data by Sato et al. | Sato et al., 2013 | GEO: GSE43346 |
| Independent SCLC RNA-seq data by Wagner et al. | Wagner et al., 2018 | dbGAP: phs001049.v1.p1 |
| Independent SCLC RNA-seq data by Rudin et al. | Rudin et al., 2012 | EGA: EGAS00001000334 |
| Experimental Models: Cell Lines | ||
| H446 | ATCC | HTB-171 |
| H196 | ATCC | CRL-5823 |
| H524 | ATCC | CRL-5831 |
| DMS114 | ATCC | CRL-2066 |
| DMS79 | ATCC | CRL-2049 |
| H187 | ATCC | CRL-5804 |
| H889 | ATCC | CRL-5817 |
| HT-29 | ATCC | HTB-38 |
| Colo 205 | ATCC | CCL-222 |
| Experimental Models: Organisms/Strains | ||
| Female NMRI nu/nu mice (NMRI-Foxn1nu), 5–7 weeks of age | Charles River | Sulzfeld, Germany |
| Female Hsd:Athymic Nude-Foxn1nu, 6–8 weeks of age | Envigo | France |
| Software and Algorithms | ||
| NCATS matrix | National Center for Advancing Translational Sciences | https://matrix.ncats.nih.gov/ |
| MacSynergy II software | Prichard et al. 1990 PMID: 2088205 | https://www.uab.edu/medicine/peds/macsynergy |
| BWA | Li et al., 2009; PMID: 20080505 | http://bio-bwa.sourceforge.net/ |
| The Genome Analysis Toolkit (GATK) | Broad Institute | https://gatk.broadinstitute.org/hc/en-us |
| HaplotypeCaller (HAPLOC) | Broad Institute | https://gatk.broadinstitute.org/hc/en-us |
| Strelka | Illumina | https://www.illumina.com/ |
| ANNOVAR | Wang K et al., 2010; PMID: 20601685 | http://annovar.openbioinformatics.org/en/latest/#annovar-documentation |
| FACETS | Shen et al. 2016; PMID: 27270079 | https://bioinformaticshome.com/tools/cnv/descriptions/FACETS.html |
| EdgeR | Robinson et al., 2010. PMID: 19910308 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| fgsea R | Korotkevich G et al. 2019. bioRxiv. doi: 10.1101/060012, http://biorxiv.org/content/early/2016/06/20/060012. | http://www.bioconductor.org/packages/release/bioc/html/fgsea.html |
| GSVA | Hanzelmann et al., 2013. PMID: 23323831 | http://www.bioconductor.org/packages/release/bioc/html/GSVA.html |
| GraphPad Prism version 8.4.3 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| STATA software version 16.0 | StataCorp | https://www.stata.com/ |
| Qlucore Omics Explorer ver. 3.6 | Qlucore AB | https://qlucore.com/ |
| FlowJo v10.6.1 | Biosciences | https://flow.com |
Highlights.
Concurrent inhibition of ATR and TOP1 exacerbates replication stress in SCLC cells
M6620 plus topotecan yields durable tumor responses in platinum-resistant SCNC patients
SCNCs with high neuroendocrine differentiation are more likely to respond
DNA replication stress is a hallmark of high neuroendocrine SCNCs
Acknowledgements
The authors gratefully acknowledge the contributions of the patients who participated in the study. This work was supported by the intramural programs of the Center for Cancer Research, National Cancer Institute (ZIA BC 011793) and the Division of Preclinical Innovation, National Center for Advancing Translational Sciences of the National Institutes of Health. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Declaration of Interests
H.D., A.Z., F.T.Z. are employees of Merck KGaA, Darmstadt, Germany. B.E. is an employee of the EMD Serono Research & Development Institute Inc., Billerica, MA USA; a business of Merck KGaA, Darmstadt, Germany. JP was an employee of the EMD Serono Research & Development Institute Inc., Billerica, MA, USA at the time of study. A.T., and Y.P. report research funding to the institution from the following entities: EMD Serono Research & Development Institute Inc., Billerica, MA USA, AstraZeneca, Tarveda Therapeutics, and Prolynx Inc.
Footnotes
Additional Resources
The ClinicalTrials.gov identifier for this study is NCT02487095.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcala N, Leblay N, Gabriel AAG, Mangiante L, Hervas D, Giffon T, Sertier AS, Ferrari A, Derks J, Ghantous A, et al. (2019). Integrative and comparative genomic analyses identify clinically relevant pulmonary carcinoid groups and unveil the supra-carcinoids. Nat Commun 10, 3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand J, Sun Y, Zhao Y, Nitiss KC, and Nitiss JL (2018). Detection of Topoisomerase Covalent Complexes in Eukaryotic Cells. Methods Mol Biol 1703, 283–299. [DOI] [PubMed] [Google Scholar]
- Balanis NG, Sheu KM, Esedebe FN, Patel SJ, Smith BA, Park JW, Alhani S, Gomperts BN, Huang J, Witte ON, and Graeber TG (2019). Pan-cancer Convergence to a Small-Cell Neuroendocrine Phenotype that Shares Susceptibilities with Hematological Malignancies. Cancer Cell 36, 17–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. (2005). DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LVF, Kolettas E, Niforou K, Zoumpourlis VC, et al. (2006). Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637. [DOI] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. (2016). Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22, 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C, Herlihy AE, Pennycook BR, Kriston-Vizi J, and de Bruin RAM (2016). Sustained E2F-Dependent Transcription Is a Key Mechanism to Prevent Replication-Stress-Induced DNA Damage. Cell Rep 15, 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, and Jackson SP (2017). ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Molecular Cell 66, 801–817. [DOI] [PubMed] [Google Scholar]
- Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH, and Johnson JE (2016). ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep 16, 1259–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MT, Penson A, Desai NB, Socci ND, Shen R, Seshan VE, Kundra R, Abeshouse A, Viale A, Cha EK, et al. (2018). Small-Cell Carcinomas of the Bladder and Lung Are Characterized by a Convergent but Distinct Pathogenesis. Clin Cancer Res 24, 1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussy F, El-Botty R, Chateau-Joubert S, Dahmani A, Montaudon E, Leboucher S, Morisset L, Painsec P, Sourd L, Huguet L, et al. (2020). BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci Transl Med 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. (2009). New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45, 228–247. [DOI] [PubMed] [Google Scholar]
- Farago AF, Yeap BY, Stanzione M, Hung YP, Heist RS, Marcoux JP, Zhong J, Rangachari D, Barbie DA, Phat S, et al. (2019). Combination Olaparib and Temozolomide in Relapsed Small-Cell Lung Cancer. Cancer Discov 9, 1372–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Carney DN, Nau MM, and Minna JD (1985). Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 45, 2924–2930. [PubMed] [Google Scholar]
- George J, Lim JS, Peifer M, Sage J, and Thomas R (2015). Comprehensive genomic characterization of small cell lung cancer. Cancer Res 75. [Google Scholar]
- Gorgoulis VG, Vassiliou LVF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, DiTullio RA, Kastrinakis NG, Levy B, et al. (2005). Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913. [DOI] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, and Clement MV (2014). OpenComet: an automated tool for comet assay image analysis. Redox Biol 2, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, and Bartek J (2008). An oncogene-induced DNA damage model for cancer development. Science 319, 1352–1355. [DOI] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Hanna N, Bunn PA, Langer C, Einhorn L, Guthrie T, Beck T, Ansar R, Ellis P, Byrne M, Morrison M, et al. (2006). Randomized phase III trial comparing irinotecan/cisplatin with etoposide/cisplatin in patients with previously untreated extensive-stage disease small-cell lung cancer. J Clin Oncol 24, 2038–2043. [DOI] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, and Guinney J (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W-D, Li X, Rolfsmeier M, and Zhang X-P (2006). Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res 34, 4115–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills SA, and Diffley JF (2014). DNA replication and oncogene-induced replicative stress. Curr Biol 24, R435–444. [DOI] [PubMed] [Google Scholar]
- Horita N, Yamamoto M, Sato T, Tsukahara T, Nagakura H, Tashiro K, Shibata Y, Watanabe H, Nagai K, Inoue M, et al. (2015). Topotecan for Relapsed Small-cell Lung Cancer: Systematic Review and Meta-Analysis of 1347 Patients. Sci Rep 5, 15437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, Huemer F, Losonczy G, Johnson ML, Nishio M, et al. (2018). First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 379, 2220–2229. [DOI] [PubMed] [Google Scholar]
- Jiang L, Shen HB, and Yao YH (2016). Genomic Landscape Survey Identifies SRSF1 as a Key Oncodriver in Small Cell Lung Cancer. Chest 149, 328a–328a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse R, Martin SE, Guha R, Ormanoglu P, Pfister TD, Reaper PM, Barnes CS, Jones J, Charlton P, Pollard JR, et al. (2014). ATR Inhibitors VE-821 and VX-970 Sensitize Cancer Cells to Topoisomerase I Inhibitors by Disabling DNA Replication Initiation and Fork Elongation Responses. Cancer Res 74, 6968–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EC, Lee MJ, Alarcon SV, Lee S, Hoang AN, Walton Diaz A, Chelluri R, Vourganti S, Trepel JB, and Pinto PA (2016). Lack of Impact of Robotic Assisted Laparoscopic Radical Prostatectomy on Intraoperative Levels of Prostate Cancer Circulating Tumor Cells. J Urol 195, 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M, Modlin IM, and Drozdov I (2014). Gene network-based analysis identifies two potential subtypes of small intestinal neuroendocrine tumors. Bmc Genomics 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimstra DS, Beltran H, Lilenbaum R, and Bergsland E (2015). The spectrum of neuroendocrine tumors: histologic classification, unique features and areas of overlap. Am Soc Clin Oncol Educ Book, 92–103. [DOI] [PubMed] [Google Scholar]
- Kotsantis P, Silva LM, Irmscher S, Jones RM, Folkes L, Gromak N, and Petermann E (2016). Increased global transcription activity as a mechanism of replication stress in cancer. Nat Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, and Dunphy WG (2000). Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Molecular Cell 6, 839–849. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GL, Wilson KM, Ceribelli M, Stanton BZ, Woo PJ, Kreimer S, Qin EY, Zhang X, Lennon J, Nagaraja S, et al. (2019). Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen C-C, Wongvipat J, Ku S-Y, Gao D, Cao Z, et al. (2017). SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Tang SW, Leo E, Baechler SA, Redon CE, Zhang H, Al Abo M, Rajapakse VN, Nakamura E, Jenkins LMM, et al. (2018). SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol Cell 69, 371–384 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, Artista L, Schleker T, Guerra C, Garcia E, et al. (2011). Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol 18, 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien MER, Ciuleanu TE, Tsekov H, Shparyk Y, Cucevia B, Juhasz G, Thatcher N, Ross GA, Dane GC, and Crofts T (2006). Phase III trial comparing supportive care alone with supportive care with oral topotecan in patients with relapsed small-cell lung cancer. J Clin Oncol 24, 5441–5447. [DOI] [PubMed] [Google Scholar]
- Pietanza MC, Waqar SN, Krug LM, Dowlati A, Hann CL, Chiappori A, Owonikoko TK, Woo KM, Cardnell RJ, Fujimoto J, et al. (2018). Randomized, Double-Blind, Phase II Study of Temozolomide in Combination With Either Veliparib or Placebo in Patients With Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 36, 2386–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, and Nitiss JL (2016). Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, and Shipman C Jr. (1990). A three-dimensional model to analyze drug-drug interactions. Antiviral Res 14, 181–205. [DOI] [PubMed] [Google Scholar]
- Quintanal-Villalonga A, Chan JM, Yu HA, Pe’er D, Sawyers CL, Triparna S, and Rudin CM (2020). Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse VN, Luna A, Yamade M, Loman L, Varma S, Sunshine M, Iorio F, Sousa FG, Elloumi F, Aladjem MI, et al. (2018). CellMinerCDB for Integrative Cross-Database Genomics and Pharmacogenomics Analyses of Cancer Cell Lines. iScience 10, 247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MAM, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. (2012). Enzymatic Removal of Ribonucleotides from DNA Is Essential for Mammalian Genome Integrity and Development. Cell 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooper LM, Bishop JA, and Westra WH (2018). INSM1 is a Sensitive and Specific Marker of Neuroendocrine Differentiation in Head and Neck Tumors. Am J Surg Pathol 42, 665–671. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. (2012). Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44, 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, Johnson JE, Lehman JM, MacPherson D, et al. (2019). Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 19, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurikar N, and Eastman A (2015). Will Targeting Chk1 Have a Role in the Future of Cancer Therapy? J Clin Oncol 33, 1075–+. [DOI] [PubMed] [Google Scholar]
- Sato T, Kaneda A, Tsuji S, Isagawa T, Yamamoto S, Fujita T, Yamanaka R, Tanaka Y, Nukiwa T, Marquez VE, et al. (2013). PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep-Uk 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler JA, Conti C, Syed A, Aladjem MI, and Pommier Y (2007). The intra-S-phase checkpoint affects both DNA replication initiation and elongation: Single-cell and - DNA fiber analyses. Molecular and Cellular Biology 27, 5806–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova EA, Nagel R, and Berns A (2015). Origins, genetic landscape, and emerging therapies of small cell lung cancer. Gene Dev 29, 1447–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao RG, Cao CX, Zhang HL, Kohn KW, Wold MS, and Pommier Y (1999). Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA : DNA-PK complexes. Embo Journal 18, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Nguyen HD, Hakansson P, Marechal A, Tse A, Tahara H, and Zou L (2013). Two Distinct Modes of ATR Activation Orchestrated by Rad17 and Nbs1. Cell Rep 3, 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova N, Jansen M, Falk M, and Hendriks BS (2020). Population pharmacokinetics of ATR inhibitor berzosertib in phase I studies for different cancer types. Cancer Chemother Pharmacol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Pattanayak P, Szabo E, and Pinsky P (2018a). Characteristics and Outcomes of Small Cell Lung Cancer Detected by CT Screening. Chest 154, 1284–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, and Pommier Y (2016). Small cell lung cancer: Time to revisit DNA-damaging chemotherapy. Sci Transl Med 8, 346fs312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Rajan A, Berman A, Tomita Y, Brzezniak C, Lee MJ, Lee S, Ling A, Spittler AJ, Carter CA, et al. (2015). Sunitinib in patients with chemotherapy-refractory thymoma and thymic carcinoma: an open-label phase 2 trial. Lancet Oncol 16, 177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Redon CE, Sciuto L, Padiernos E, Ji J, Lee MJ, Yuno A, Lee S, Zhang Y, Tran L, et al. (2018b). Phase I Study of ATR Inhibitor M6620 in Combination With Topotecan in Patients With Advanced Solid Tumors. J Clin Oncol 36, 1594–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlemsani C, Pongor L, Elloumi F, Girard L, Huffman KE, Roper N, Varma S, Luna A, Rajapakse VN, Sebastian R, et al. (2020). SCLC-CellMiner: A Resource for Small Cell Lung Cancer Cell Line Genomics and Pharmacology Based on Genomic Signatures. Cell Rep 33, 108296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, Oyarzabal J, Pastor J, Bischoff JR, and Fernandez-Capetillo O (2011). A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 18, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigo J, Subbiah V, Besse B, Moreno V, Lopez R, Sala MA, Peters S, Ponce S, Fernandez C, Alfaro V, et al. (2020). Lurbinectedin as second-line treatment for patients with small-cell lung cancer: a single-arm, open-label, phase 2 basket trial. Lancet Oncol 21, 645–654. [DOI] [PubMed] [Google Scholar]
- Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, et al. (2009). Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11, 1315–U1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Pawel J, Schiller JH, Shepherd FA, Fields SZ, Kleisbauer JP, Chrysson NG, Stewart DJ, Clark PI, Palmer MC, Depierre A, et al. (1999). Topotecan versus cyclophosphamide, doxorubicin, and vincristine for the treatment of recurrent small-cell lung cancer. J Clin Oncol 17, 658–667. [DOI] [PubMed] [Google Scholar]
- Wagner AH, Devarakonda S, Skidmore ZL, Krysiak K, Ramu A, Trani L, Kunisaki J, Masood A, Waqar SN, Spies NC, et al. (2018). Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Smith BA, Balanis NG, Tsai BL, Nguyen K, Cheng MW, Obusan MB, Esedebe FN, Patel SJ, Zhang H, et al. (2020). A genetically defined disease model reveals that urothelial cells can initiate divergent bladder cancer phenotypes. Proc Natl Acad Sci U S A 117, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CT, Miller R, Pemberton HN, Jones SE, Campbell J, Konde A, Badham N, Rafiq R, Brough R, Gulati A, et al. (2016). ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun 7, 13837–13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, Ho CK, Todorova Kwan T, Morris R, Lauffer S, Nussenzweig A, et al. (2017). ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev 31, 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, and Cimprich KA (2014). Causes and consequences of replication stress. Nat Cell Biol 16, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dai Q, Park D, and Deng X (2016). Targeting DNA Replication Stress for Cancer Therapy. Genes (Basel) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Girard L, Zhang YA, Haruki T, Papari-Zareei M, Stastny V, Ghayee HK, Pacak K, Oliver TG, Minna JD, and Gazdar AF (2018). Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res 7, 32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. High-throughput single-agent and combination drug screens in SCLC cells, related to Figure 1.
Table S2. In vitro and in vivo characterization of ATRi plus TOP1i, related to Figure 2.
Table S3. Patient characteristics, pharmacokinetics, adverse events, dose modifications, and CTCs, related to Figures 3 and 4.
Table S4. Biopsy characteristics, and exomic and transcriptomic assessment of responders and non-responders, related to Figure 5.
Data Availability Statement
The accession number for the sequencing data reported in this paper is dbGaP: phs2327.v1. Gene sets used for ssGSEA are available in Molecular Signatures Database https://www.gsea-msigdb.org/gsea/msigdb/. Gene expression data (RNA-seq or microarray) from independent SCLC cohorts were retrieved from: European Genome-phenome Archive EGAS00001000925, Gene Expression Omnibus (GEO): GSE60052,GSE43346, dbGAP: phs001049.v1.p1, and EGAS00001000334. Processed RNA-seq data (1+TMM_FPKM) and code for GSEA and ssGSEA are available at: https://github.com/alwaysblack777/Therapeutic-targeting-of-ATR-yields-durable-regressions-in-high-replication-stress-tumors.