Abstract
We report aqueous, site-selective modification of proteins using a reactive peptide interface comprising a nine-residue sequence. This interface is the fastest (second-order rate constant of 152 M−1 s−1) catalyst-free, cysteine-based method for modifying proteins available to date, and enables near-quantitative labeling of antibodies in cell lysate.
Graphical Abstract
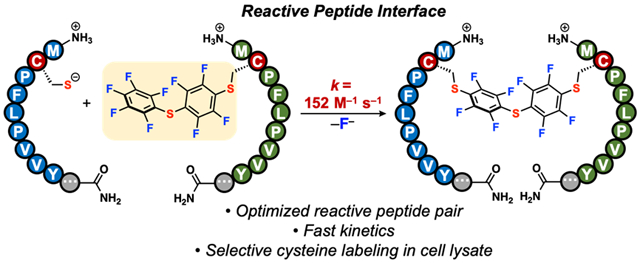
A nine-residue cysteine-based reactive peptide interface enables rapid site-selective modification of proteins in water.
Bioconjugation chemistry enables the development of new therapeutics,1,2 biomaterials,3,4 and probes for the exploration of complex biological processes.5 The site-selective modification of proteins, however, is challenging due to the chemical complexity of biomolecules and necessity for aqueous environments. Site-selective bioconjugation methods have been developed to address this challenge, some of which include unnatural amino acid incorporation into proteins,6-9 fusion of engineered, self-modifying enzymes,10,11 ligand-directed protein modification,12-14 and peptide tag-based modification.12-20
Peptide tags constitute a powerful approach to protein modification given that they are (1) small in size, making them less likely to perturb protein structure and function, and (2) usually composed of canonical amino acids, making them easier to genetically encode. The use of peptide sequences that accomplish otherwise sluggish reactions in water has been prevalent in the literature in this past decade.17-24 It is often hypothesized that the driving force for these reactions is the peptide’s unique sequence. This sequence could afford improved reaction rates through enhanced nucleophilicity of the reactive residue arising from side-chain interactions and/or interactions between the peptide and the electrophile. Synthetic peptide libraries can be used to rapidly develop sequences that facilitate site-selective bionconjugations.21-24 Screening such libraries has led to the discovery of peptides that react preferentially with a specific electrophile.
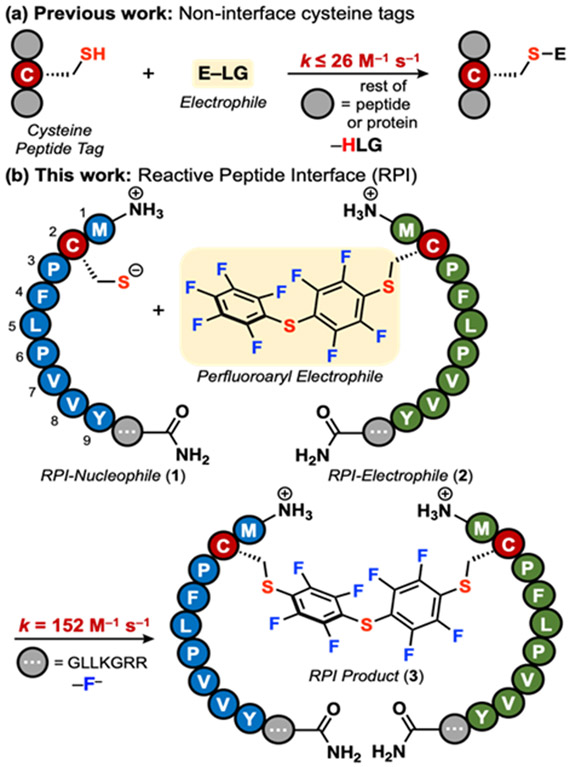
Cysteine is commonly leveraged as the reactive residue in peptide tags due to its inherent nucleophilicity. Various Cys-containing peptides have been developed that react with electrophiles such as p-(chloromethyl)benzamide,15 2-cyanobenzothiazole,16 aza-dibenzocyclooctynes,17 and perfluoro-aromatics (Fig. 1a).18-20 To date, the fastest of these catalyst-free Cys tags is a 29-residue peptide that exhibits a second-order rate constant (k) of 25.8 ± 1.8 M−1 s−1 when reacting with pentafluoro-phenyl sulfide (PS).20 Nonetheless, a rapid (k > 100 M−1 s−1) system capable of site-selective protein labeling that contains a short (≤10-mer) sequence, requires no catalyst and consists of exclusively canonical residues is not yet available.
Fig. 1.
The Reactive Peptide Interface (RPI) undergoes rapid Cys arylation with perfluoroaryl electrophiles. (a) Previous work has focused on modulating the reactivity of a Cys-containing peptide by screening approaches. (b) This work focuses on the RPI, composed of a Cys-containing, nucleophilic peptide (1) and a perfluoroaryl-containing, electrophilic peptide (2). Conditions in (b): 10 μM 1, 50 μM 2, 0.2 M sodium phosphate, 20 mM TCEP, pH 8, 37 °C.
To address these challenges, we developed a matched peptide-peptide pair termed the reactive peptide interface (RPI). This interface consists of two peptides: a nucleophilic, Cys-containing peptide (1) and an electrophilic, perfluoroarylated peptide (2, Fig. 1b). The unique sequences of these peptides enhance the reaction rate of a nucleophilic aromatic substitution reaction between 1 and 2. Our system contains an optimized electrophilic peptide that enhances the electrophilicity of its perfluoroarene and improves its solubility in aqueous buffers, in contrast to other water insoluble electrophilic reagents. In addition, potential favorable non-covalent interactions between these two interacting peptides could facilitate the rapid reaction rate and the site-selectivity of the system through molecular recognition. This peptide interface, discovered through peptide library screening, allows for rapid Cys arylation with a k = 152 ± 3 M−1 s−1. This RPI enabled the site-selective modification of a miniprotein and of an antibody in cell lysate.
The reactive peptide interface was discovered through iterative generations of synthetic peptide library screening. A one-bead-one-compound combinatorial peptide library was prepared via split-pool synthesis. This library was reacted on-bead with a biotinylated electrophilic peptide containing a decafluorobiphenyl (DB) moiety (S1, see Fig. 2b). Subsequent streptavidin-allophycocyanin staining allowed for isolation of hits on-bead via fluorescence-activated bead sorting (FABS). The hit peptides were cleaved from the beads, and analyzed via liquid chromatography-tandem mass spectrometry (LC-MS/MS) for de novo sequencing (Fig. S1a).18,25,26 The final, nucleophilic peptide library was designed so that Cys was fixed at the second position, with variable residues in positions 1 and 3 through 9. The variable residues had 16 possibilities: all 20 canonical amino acids except Cys, Ile, Asn, Arg and Glu (Fig. S1b). This was done to control the library size for effective screening, to minimize redundant side-chain groups, and to eliminate isobaric residues. An additional constant region (Gly-Leu-Leu-Lys-Gly-Arg-Arg) was incorporated at the C-termini of all library members to ensure chromatographic resolution of the peptides.
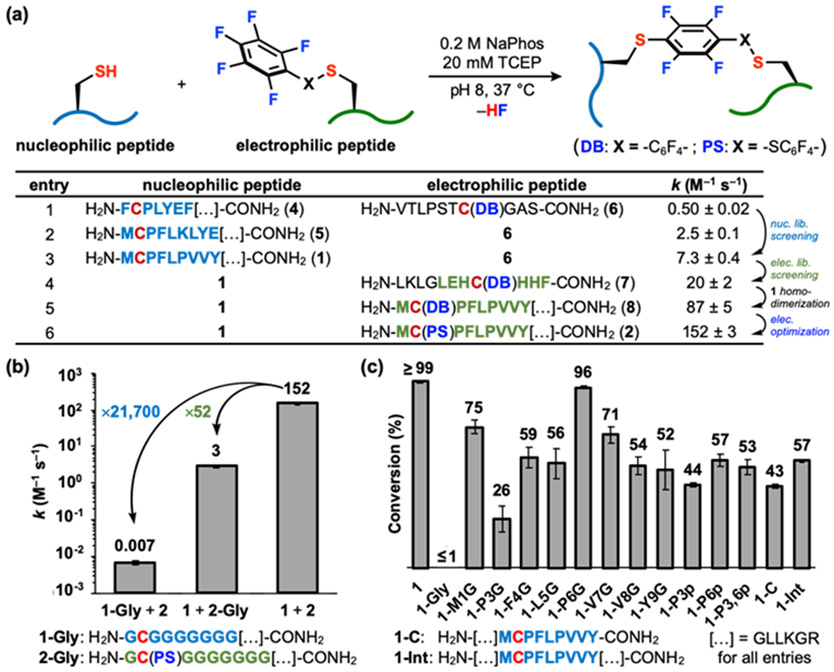
Fig. 2.
Sequence-reactivity studies give insight into the RPI mechanism of action. (a) Reaction scheme for nucleophilic and electrophilic peptide reactions in (c) and list of peptides for peptide library screening and reactions in (b) and (c). (b) Reaction scheme for nucleophilic and electrophilic peptide reactions in (d). (b) Relative contribution of 1 and 2 to the RPI’s rate constant. (c) Conversion (%) of 1 variants reacting with 2.
Nucleophilic peptide 1 was developed across three generations of library screening. In the first two generations of nucleophilic peptides, hits 4 and 5 were reacted with 6, with a k of 0.50 ± 0.02 and 2.5 ± 0.1 M−1 s−1, respectively (Figs. S1c, S5 and S6). Peptide 6 was chosen as the electrophilic partner, as this sequence was previously utilized in our group.18,27,28 Three generations of library screening led to 1 with RPI sequence “Met-Cys-Pro-Phe-Leu-Pro-Val-Val-Tyr”, or “MCPFLPVVY”. Peptide 1 reacted with 6 with a k = 7.3 ± 0.4 M−1 s−1 (Fig. S7). A glycine-variant analysis of 6 revealed sequence-dependent reactivity of 6 with the π-clamp peptide (Fig. S3), a previous Cys peptide tag discovered in our group.18 We hypothesized that the sequence in the electrophilic peptide could accelerate the reaction rate by (1) modulating the reactivity of the perfluoroaryl electrophile and/or (2) facilitating non-covalent interactions between the nucleophilic and electrophilic peptides. Optimization of the electrophilic peptide component of the RPI was also performed through library screening. A DB-installed electrophilic peptide library (Fig. S1b) was screened, resulting in the discovery of 7. Peptide 7 reacts with 1 with a k = 20 ± 2 M−1 s−1, a 2.7-fold improvement from the previous step (Fig. S8).
Peptide 1 serendipitously homodimerized. We hypothesized that 1 and non-perfluorarylated 7 (S2, free thiol) would dimerize in the presence of DB. The sole product of this transformation, however, was the homodimer of 1 (Fig. S9). Incorporating DB into this peptide (8), the reaction between 1 and 8 was found to display k = 87 ± 5 M−1 s−1, a 4.3-fold improvement from the previous step (Fig. S10).
The final stage of interface optimization consisted in optimizing the electrophile from DB to PS. The added sulfide functionality in PS is hypothesized to stabilize the Meisenheimer complex, thereby accelerating the reaction rate.29 Peptide 1 reacts with 2 with a k = 152 ± 3 M−1 s−1, a 1.7-fold improvement from the previous step, and a 20.8-fold increase over the reaction between 1 and 8 (Figs. S11 and S12). We refer to the reaction between 1 and 2 as the reactive peptide interface (RPI). For a complete list of the peptide sequences discovered from each generation of library screening and their corresponding kinetic analyses, please refer to Figs. 2a and S4-S40.
Sequence-reactivity studies were employed to elucidate the individual contributions of both components of the RPI to the reaction rate. To this end, we reacted Gly variants of 1 with 2 (1-Gly, 2-Gly) wherein the RPI residues were all substituted to glycine, except for Cys (Fig 2b). Nucleophilic peptide 1-Gly reacts with 2 with a k = 0.007 ± 0.001 M−1 s−1, a 21,700-fold decrease from the optimized reaction rate. The reaction between 1 and electrophilic peptide 2-Gly exhibited a k = 2.9 ± 0.2 M−1 s−1, constituting a 52-fold rate decrease (Figs. S13 and S14). Under similar conditions, no product formation was observed between 1-Gly and 2-Gly after 72 h (Fig. S15). Our results suggest that the peptide sequences of both 1 and 2 are critical for the RPI’s rapid reaction rate, with nucleophile 1 having a greater contribution than electrophile 2.
The importance of each residue in the RPI peptides was elucidated through Gly-variant studies. Glycine-variants of 1 were reacted with 2. Individual substitution of each RPI residue in 1 led to decreased reactivity with 2 compared to the optimized system (Fig. 2c). Substituting Pro-3 for Gly (1-P3G) results in the lowest conversion to product. A similar trend is seen when Pro-3 and/or Pro-6 are substituted for D-Pro. The RPI positional variants were also tested. The nucleophilic RPI sequence was incorporated at the N-terminus, C-terminus (1-C), and internal position (1-Int) of a model peptide. While the highest conversion was observed for reactions of the N-terminal RPI peptide (≥99%), tagging of the 1-C (43%) and 1-Int (57%) are also possible (Fig. 2c).
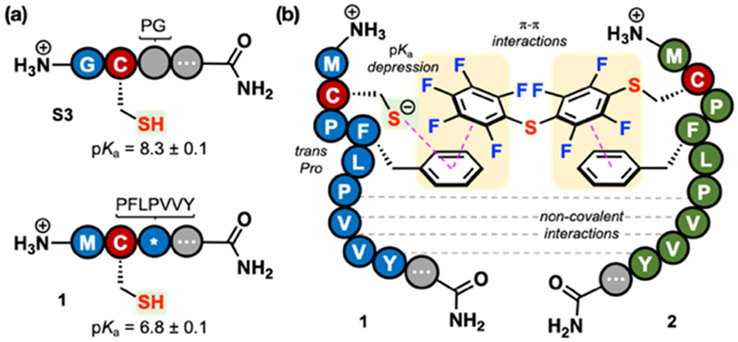
The unique structure of peptide 1 results in the pKa depression of its Cys thiol, presumably caused by the local microenvironment of this peptide. The pKa of the Cys thiol of a control peptide (S3) was determined to be 8.3 ± 0.1.18,27 The pKa of 1 was determined to be 6.8 ± 0.1 (Fig. 3a and Fig. S41), 1.5 units lower than control. The Cys thiolate of 1 is 32-fold more concentrated at pH 8 than that of the control peptide S3. Although pKa depression appears to be a large contributor to the increased reaction rate, we hypothesize that molecular recognition of 1 and 2 could also be playing a role. First, the RPI sequence appears to be prone to homodimerization (Fig. S9), and noncovalent interactions may explain this phenomenon. Secondly, both components of the RPI are essential to achieve the rapid rate, as demonstrated by the Gly-variant studies above.
Fig. 3.
Non-covalent interactions and pKa depression contribute to RPI rate enhancement. (a) Comparison of the pKa of the peptide 1’s Cys thiol with that of a control peptide (S3). (b) Proposed RPI mechanism of action.
The reactivity of 1 and 1-Gly toward a variety of electrophiles was surveyed (Fig. S44). Interestingly, 1 exhibits a clear preference for reacting with DB and PS, while 1-Gly does not react under the experimental conditions. The reactive electrophile c reacts indiscriminately with both peptides. We observed a slight preference of either peptide for electrophiles d, e and f. Despite having a lower pKa, 1 does not show a higher conversion for all electrophiles surveyed, pointing to possible molecular recognition events between 1 and the incoming perfluoroaryl electrophile.
A mechanistic model of the RPI is presented in Fig. 3b. Similar to reported precedent,27 we hypothesize that Pro-3 adopts a trans conformation that provides rigidity to the sequence. In particular, this conformation may stabilize the interactions of the Phe residue with (1) the Cys residue, depressing its pKa,18,27,28 and with (2) the perfluoroarene of 2.27,28,30-36 Additional non-covalent interactions between 1 and 2 are hypothesized, such as hydrogen bonding and hydrophobic interactions. The exact nature of these interactions is currently under further investigation.
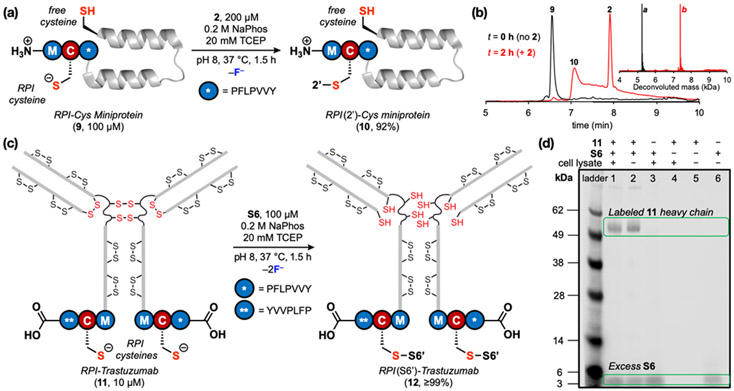
We applied the RPI to site-specifically modify protein biomolecules such as a miniprotein and an antibody. The RPI sequence and a free Cys were incorporated at the N- and C-termini, respectively, of a minimized, 33-residue Z domain of protein A (Z33), resulting in 9 (Fig. 3).37 Then, 9 was reacted with 2 to assess site-selectivity at the RPI versus the C-terminal free Cys. Only one modification was observed, with the RPI Cys site-selectively labeled with 2 to produce 10 in near quantitative yield after 1.5 h. No single-labeled C-terminal Cys (S4) or doubly-labeled side-products (S5) were observed (Fig. 3). We have yet to understand if the peak shape corresponding to 10 in Fig. 4b is due to the chromatography conditions used or due to molecular features of the product. The RPI sequence was incorporated at the C-terminal of the heavy chains of trastuzumab (RPI-trastuzumab, 11), a humanized recombinant monoclonal antibody that binds to the human epidermal growth factor receptor 2 (HER2) cell membrane receptor. Reaction of 11 with a biotinylated 2 (S6) resulted in conversion to the homogeneous 12 in near-quantitative yield after 1.5 h (Fig. 4a and Fig. S42a). 12 retained affinity to HER2 receptors (dissociation constant, Kd = 5.4 ± 0.2 nM) in an Octet binding assay (Fig. S42b). In addition, flow cytometry studies showed that 12 binds to HER2-positive BT474 cells (Fig. S42c). These results emphasize the practical utility of our new RPI in obtaining homogeneous bioconjugates rapidly and efficiently.
Fig. 4.
Site-selective bioconjugation was achieved in a miniprotein and an antibody using the RPI. (a) 9 reacts site-selectively with 2 at the RPI Cys to produce 10 in 92% yield after 90 min. (b) Total-ion count chromatograms and deconvoluted mass spectra show single modification of 9 to produce 10. a: obs. 5258.0., calc. 5258.6; b = obs. 7432.3., calc. 7432.6. (c) Site-selective bioconjugation at the RPI cysteine sites of a doubly C-terminal RPI-tagged trastuzumab antibody (11) with biotinylated 2 (S6) results in homogenous 12 in near-quantitative yield after 90 min. 11 contains multiple free cysteines due to reducing reaction conditions. (d) Streptavidin western blot shows selective labeling of 11 heavy chains with S6 in HeLa cell lysate (1 mg/mL). Reaction conditions were the same as in (a) plus the lysate.
Finally, we investigated the ability of the RPI to afford substrate selectivity in a complex cell environment with numerous competing functionalities and proteins. To this end, we employed the RPI to selectively label 11 with S6 directly in HeLa cell lysate. A streptavidin western blot analysis of the reaction mixture showed exclusive and quantitative labeling of the heavy chains of 11 after 90 min (Fig. 4b and Fig. S43). This result indicates that the RPI can be used as an efficient strategy to site-selectively label proteins in biological milieu, despite an abundance of Cys thiols and other nucleophiles available for reaction. This method could also allow for specific capture and subsequent pulldown of RPI sequence-tagged proteins in cell lysates.
A reactive peptide sequence was developed that undergoes catalyst-free Cys arylation at a rapid rate (k = 152 ± 3 M−1 s−1). The reactive peptide interface (RPI) was discovered through screening of synthetic combinatorial peptide libraries. Our results demonstrate that optimizing both the nucleophilic and electrophilic components of a bioconjugation interface can significantly accelerate the rate of reaction. RPI bioconjugation is advantageous in that it uses a short nine-residue sequence that is composed entirely of canonical amino acids. This feature allowed us to site-selectively label the cysteine thiol of RPI incorporated in two proteins containing additional free cysteines under mild conditions. In addition, RPI-tagged trastuzumab was selectively labeled in the complex environment of a cell lysate, which contains multiple proteins with free cysteines. Based on these results, we are confident the RPI has great potential to be the starting point in the development of the next-generation toolkit for ultra-rapid, site-selective, and quantitative bioconjugations. Current and future studies are aimed at deciphering how the molecular features of the RPI peptides affect the reaction parameters to further enhance the sequence reactivity, as well as implementing this platform for selective protein labeling at cell surfaces.
Supplementary Material
Acknowledgments
This research was funded by the National Institutes of Health (NIH, grant R01-GM110535 to B.L.P.), a Novartis Early Career Award (to B.L.P.) and a Bristol-Myers Squibb Unrestricted Grant in Synthetic Organic Chemistry (to B.L.P. and C. Z.). D.D.-M. and C.R.S. are grateful for the MIT Dean of Science fellowship and a NIH postdoctoral fellowship (F32-GM133073), respectively. C.Z. is a recipient of the George Büchi Summer Research Fellowship and the Koch Graduate Fellowship in Cancer Research from MIT. The authors acknowledge the Biophysical Instrumentation Facility at MIT for providing the Octet BioLayer Interferometry System (NIH S10 OD016326).
Footnotes
Conflicts of interest
B.L.P. is a co-founder of Amide Technologies and Resolute Bio. Both companies focus on developing protein and peptide therapeutics.
Electronic Supplementary Information (ESI) available: Supporting Information. See DOI: 10.1039/d1cc00095k
Notes and references
- 1.Chudasama V, Maruani A and Caddick S, Nat. Chem, 2016, 8, 114–119. [DOI] [PubMed] [Google Scholar]
- 2.Heinis C, Nat. Chem. Biol, 2014, 10, 696–698. [DOI] [PubMed] [Google Scholar]
- 3.Cobo I, Li M, Sumerlin BS and Perrier S, Nat. Mater, 2014, 14, 143–159. [DOI] [PubMed] [Google Scholar]
- 4.Lutz J-F and Zarafshani Z, Adv. Drug Deliv. Rev, 2008, 60, 958–970. [DOI] [PubMed] [Google Scholar]
- 5.Xue L, Karpenko IA, Hiblot J and Johnsson K, Nat. Chem. Biol, 2015, 11, 917–923. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Xie J and Schultz PG, Annu. Rev. Biophys. Biomol. Struct, 2006, 35, 225–249. [DOI] [PubMed] [Google Scholar]
- 7.Liu CC and Schultz PG, Annu. Rev. Biochem, 2010, 79, 413–444. [DOI] [PubMed] [Google Scholar]
- 8.Lang K and Chin JW, Chem. Rev, 2014, 114, 4764–4806. [DOI] [PubMed] [Google Scholar]
- 9.Sletten EM and Bertozzi CR, Angew. Chemie Int. Ed, 2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H and Johnsson K, Chem. Biol, 2003, 10, 313–317. [DOI] [PubMed] [Google Scholar]
- 11.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF and Wood KV, ACS Chem. Biol, 2008, 3, 373–382. [DOI] [PubMed] [Google Scholar]
- 12.Lotze J, Reinhardt U, Seitz O and Beck-Sickinger AG, Mol. Biosyst, 2016, 12, 1731–1745. [DOI] [PubMed] [Google Scholar]
- 13.Yamada K, Shikida N, Shimbo K, Ito Y, Khedri Z, Matsuda Y and Mendelsohn BA, Angew. Chemie Int. Ed, 2019, 58, 5592–5597. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T and Hamachi I, Acc. Chem. Res, 2012, 45, 1460–1469. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami T, Ogawa K, Goshima N and Natsume T, Chem. Biol, 2015, 22, 1671–1679. [DOI] [PubMed] [Google Scholar]
- 16.Ramil CP, An P, Yu Z and Lin Q, J. Am. Chem. Soc, 2016, 138, 5499–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang C, Dai P, Vinogradov AA, Gates ZP and Pentelute BL, Angew. Chemie Int. Ed, 2018, 57, 6459–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, Welborn M, Zhu T, Yang NJ, Santos MS, Van Voorhis T and Pentelute BL, Nat. Chem, 2015, 8, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans ED and Pentelute BL, ACS Chem. Biol, 2018, 13, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans ED, Gates ZP, Sun Z-YJ, Mijalis AJ and Pentelute BL, Biochemistry, 2019, 58, 1343–1353. [DOI] [PubMed] [Google Scholar]
- 21.Lam KS, Lebl M and Krchňák V, Chem. Rev, 1997, 97, 411–448. [DOI] [PubMed] [Google Scholar]
- 22.Lam KS, Lehman AL, Song A, Doan N, Enstrom AM, Maxwell J and T.-M. RB in Liu E, in Combinatorial Chemistry, Part B, Academic Press, 2003, vol. 369, pp. 298–322. [DOI] [PubMed] [Google Scholar]
- 23.Gray BP and Brown KC, Chem. Rev, 2014, 114, 1020–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauffman WB, Guha S and Wimley WC, Nat. Commun, 2018, 9, 2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Touti F, Gates ZP, Bandyopadhyay A, Lautrette G and Pentelute BL, Nat. Chem. Biol, 2019, 15, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semmler A, Weber R, Przybylski M and Wittmann V, J. Am. Soc. Mass Spectrom, 2010, 21, 215–219. [DOI] [PubMed] [Google Scholar]
- 27.Dai P, Williams JK, Zhang C, Welborn M, Shepherd JJ, Zhu T, Van Voorhis T, Hong M and Pentelute BL, Sci. Rep, 2017, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai P, Zhang C, Welborn M, Shepherd JJ, Zhu T, Van Voorhis T and Pentelute BL, ACS Cent. Sci, 2016, 2, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birchall JM, Green M, Haszeldine RN and Pitts AD, Chem. Commun, 1967, 338–339. [Google Scholar]
- 30.Hunter CA, Singh J and Thornton JM, J. Mol. Biol, 1991, 218, 837–846. [DOI] [PubMed] [Google Scholar]
- 31.Sinnokrot MO and Sherrill CD, J. Am. Chem. Soc, 2004, 126, 7690–7697. [DOI] [PubMed] [Google Scholar]
- 32.Arnstein SA and Sherrill CD, Phys. Chem. Chem. Phys, 2008, 10, 2646–2655. [DOI] [PubMed] [Google Scholar]
- 33.Pace CJ and Gao J, Acc. Chem. Res, 2013, 46, 907–915. [DOI] [PubMed] [Google Scholar]
- 34.Salonen LM, Ellermann M and Diederich F, 2011, 4808–4842. [DOI] [PubMed] [Google Scholar]
- 35.Ringer AL, Senenko A and Sherrill CD, Protein Sci., 2007, 16, 2216–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishio M, Umezawa Y, Fantini J, Weiss MS and Chakrabarti P, Phys. Chem. Chem. Phys, 2014, 16, 12648–12683. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson B, Moks T, Jansson B, Abrahmsén L, Elmblad A, Holmgren E, Henrichson C, Jones TA and Uhlén M, Protein Eng. Des. Sel, 1987, 1, 107–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.