Abstract
Species distribution data are fundamental to the understanding of biodiversity patterns and processes. Yet, such data are strongly affected by sampling biases, mostly related to site accessibility. The understanding of these biases is therefore crucial in systematics, biogeography, and conservation. Here we present a novel approach for quantifying sampling effort and its impact on biodiversity knowledge, focusing on Africa. In contrast to previous studies assessing sampling completeness (percentage of species recorded in relation to predicted), we investigate whether the lack of knowledge of a site attracts scientists to visit these areas and collect samples of species. We then estimate the time required to sample 90% of the continent under a Weibull distributed biodiversity sampling rate and the number of sampling events required to record 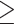 50% of the species. Using linear and spatial regression models, we show that previous sampling has been strongly influencing the resampling of areas, attracting repeated visits. This bias has existed for over two centuries, has increased in recent decades, and is most pronounced among mammals. It may take between 172 and 274 years, depending on the group, to achieve at least one sampling event per grid cell in the entire continent. Just one visit will, however, not be enough: in order to record
50% of the species. Using linear and spatial regression models, we show that previous sampling has been strongly influencing the resampling of areas, attracting repeated visits. This bias has existed for over two centuries, has increased in recent decades, and is most pronounced among mammals. It may take between 172 and 274 years, depending on the group, to achieve at least one sampling event per grid cell in the entire continent. Just one visit will, however, not be enough: in order to record 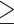 50% of the current diversity, it will require at least 12 sampling events for amphibians, 13 for mammals, and 27 for birds. Our results demonstrate the importance of sampling areas that lack primary biodiversity data and the urgency with which this needs to be done. Current practice is insufficient to adequately classify and map African biodiversity; it can lead to incorrect conclusions being drawn from biogeographic analyses and can result in misleading and self-reinforcing conservation priorities. [Amphibians; birds; mammals; sampling bias; sampling gaps; Wallacean shortfall.]
50% of the current diversity, it will require at least 12 sampling events for amphibians, 13 for mammals, and 27 for birds. Our results demonstrate the importance of sampling areas that lack primary biodiversity data and the urgency with which this needs to be done. Current practice is insufficient to adequately classify and map African biodiversity; it can lead to incorrect conclusions being drawn from biogeographic analyses and can result in misleading and self-reinforcing conservation priorities. [Amphibians; birds; mammals; sampling bias; sampling gaps; Wallacean shortfall.]
Although the number of scientists, scientific organizations, and publications are increasing worldwide (Stork and Astrin 2014), our knowledge on the distribution of biodiversity—a cornerstone in our understanding of life on Earth—may not be expanding to the same extent (Bini et al. 2006; Boakes et al. 2010; Feeley and Silman 2011; Stropp et al. 2016).
Georeferenced specimens in natural history collections and observations are fundamental for the classification and understanding of biodiversity patterns. Besides the increasing availability of observational data derived from citizen science initiatives, museum specimens are still the main source of information for taxonomic, systematic, and ecological studies (Shaffer et al. 1998; Graham et al. 2004). However, there remain important gaps of knowledge in the distribution of organisms—the “Wallacean shortfall” (Lomolino 2004). Species distribution data sets are often strongly affected by temporal, spatial, and taxonomic biases (Meyer et al. 2016). Temporal biases can be influenced by intensive collecting periods or by seasonality (Ward 2012). Spatial biases often relate to accessibility (Reddy and Dávalos 2003), protected areas and particular habitats (Sánchez-Fernández et al. 2008), or climatic zones (Loiselle et al. 2008). Sampling biases are also known to be strongly affected by variables such as body size and taxonomic group (Schmidt-Lebuhn et al. 2013; Troudet et al. 2017). As an additional concern, there are differences between whether how well-studied groups are and how represented they are in taxonomic collections. One example is the reluctance of taking vouchers of supposedly “well-studied” groups such as birds (Bates et al. 2004; Schmitt et al. 2019).
Although it has been widely documented that certain parts of continents are visited and sampled more frequently than others (Meyer et al. 2015), the underlying causes for this unevenness may be attributed to several factors. These include language barriers (Harford 2015), lack of basic resources, and poor infrastructure (Walker et al. 2006; Foster and Briceño-Garmendia 2009; Beegle et al. 2016). Additional factors such as political regimes (Rydén et al. 2019), corruption (Mbaku 2010; Bello-Schünemann and Moyer 2018), dangerous tropical diseases (Hotez and Kamath 2009; Amarasinghe et al. 2011; Bhatt et al. 2015), and expensive or burdensome permit requirements (Engel et al. 2015) may further discourage work in particular countries and regions. Although many of these factors have been previously described in the literature and are well known to the systematic community, the influence of existing previous knowledge of the biodiversity of a site in attracting scientists remains unknown. Scientists may either preferentially visit areas that are more accessible or, alternatively, prefer to sample in well-known areas. Quantifying (Zizka et al. 2020) and creating awareness around sampling biases are crucial to the efficient implementation of conservation policies and are essential to both scientists and decision-makers.
Here we test whether the degree of previous knowledge of biodiversity within an area increases the likelihood that additional sampling will be done in the same cell. We then estimate the time and effort necessary to sample Africa’s biodiversity under current practices. We perform our analyses on data from amphibians, mammals, and birds, given their relatively high level of baseline knowledge derived from digitally available natural history collections, as compared with many other organisms.
We outline two possible scenarios. Under the first scenario, researchers may actively seek data-deficient areas because they offer an opportunity to find new species and fill gaps in biodiversity knowledge. Under this scenario, researchers may be more likely to sample accessible areas but the extent of knowledge of an area should reduce the desire to revisit it for additional sampling—a phenomenon previously documented for the collection of individual species (Steege et al. 2011). This is, for example, the case for certain biological surveys in the 16th and 17th centuries, when collected specimens were often treated as art pieces and hidden from competitors until their economic value was estimated (Ritterbush 1969; Impey and MacGregor 1985).
Under the second scenario, sampling planning may not be primarily driven by the attempt to fill in gaps, but rather by the likelihood of retrieving data, often under time constraints. Therefore, researchers may return to visited areas because finding a focal species to obtain appropriate tissue for molecular phylogenetic analyses may be easier, more certain, and more cost-efficient. One example of this scenario is the Mount Namuli in Mozambique, surveyed repeatedly in 1931–1932, 1998, 2007, 2011, 2014, and 2016, which rendered it considerably better sampled than any surrounding areas (Vincent 1933; Ryan et al. 1999; Timberlake et al. 2009; Portik et al. 2013; Farooq and Conradie 2015; Conradie et al. 2016). This repeated sampling resulted in new range expansions (Farooq and Conradie 2015), new species (Conradie et al. 2018), and a better understanding of the biogeography of the region through phylogenetic studies (Branch et al. 2014; Bittencourt-Silva et al. 2016).
Materials and Methods
Using a grid-cell size equivalent to 100 by 100 km (more specifically the cells were 100 by 100 km at 30 degrees North or South; cells at lower latitudes were wider and lower, whereas cells are higher latitude were thinner and higher), we tested whether knowledge of biodiversity within a cell changes the likelihood of additional sampling within it. One advantage of this approach is that it shows whether sampling is spatially restricted because some sites are easier to reach, or if the very existence of knowledge is causing scientists to revisit well-known areas.
For all analyses, we worked on a cylindrical equal-area Berhmann projection. We estimated the time it would take to sample at least once in 90% of the land area of Africa, and the number of sampling events required to record at least 50% of the species of an area of 10,000 km . Our estimation of the time to sample 90% of Africa was based on the assumption that the rate of biodiversity sampling since the 1800s can be adequately described by a Weibull distribution, whereas in the sampling effort analysis, we removed the temporal aspect by randomizing the years 100 times.
. Our estimation of the time to sample 90% of Africa was based on the assumption that the rate of biodiversity sampling since the 1800s can be adequately described by a Weibull distribution, whereas in the sampling effort analysis, we removed the temporal aspect by randomizing the years 100 times.
All analyses were conducted using three groups: amphibians, mammals, and birds. Our species occurrence data set consisted of records retrieved from the Global Biodiversity Information Facility (GBIF) for amphibians (https://doi.org/10.15468/dl.hyyea9), mammals (https://doi.org/10.15468/dl.gms3up) excluding bats, and birds (https://doi.org/10.15468/dl.unxn5u) recorded in Africa from 1801 until the end of 2019 (31 December 2019). Bats and marine mammals were excluded from the analyses of mammals because they are generally sampled by different methods and researchers than for non-flying terrestrial mammals.
We focused on species occurrence records contributed by scientists, because these provide the primary source of information and material for the community of professional systematists. Citizen science observations, although important for popular engagement and data gathering, were therefore excluded due to their mixed systematic value (e.g., Troudet et al. 2018). Although not all countries in Africa are formal participants of the GBIF Network, which could potentially lead to underestimation of the completeness of each grid cell, most species collections in Africa are housed by members of the GBIF network, such as in South Africa, western European countries, and the United States. Other intrinsic limitations of GBIF are that it does not provide access to records collected and stored in nonparticipant countries or in scientific articles or reports where no voucher was collected, such as in photography-based inventories characteristic of Environmental impact assessments. This might also contribute to the underestimation of the true completeness rate.
To update and synonymize the taxonomy from GBIF, we used the R package RangeBuilder (version 1.4) (Rabosky et al. 2016) and removed the records of species not present in the IUCN’s polygon list. We applied the R package CoordinateCleaner (Zizka et al. 2019) to exclude duplicates and records outside the IUCN range for each species.
We define a sampling event in each cell as sampling within a given calendar year.
To estimate a simple measure of sampling completeness, we followed a similar approach to Meyer et al. (2015). We assume that the range polygons created by IUCN using version 2019-3 (IUCN 2019) are accurate. We consider as unsampled those cells overlapped by a range polygon but without the respective species record, whereas any records outside the range polygons are assumed to be errors. Although we acknowledge that none of these assumptions fully capture the complexity of species distributions and occurrences, we consider them sufficiently close to reality for the purpose of our analysis and unlikely to result in major systematic biases.
Effect of Previous Knowledge on Sampling Probability
We calculated the probability of visiting any grid cell according to its ratio of completeness using a Logistic model of the sampling events by completeness plus year. Although few cells in nondesert parts of Africa have communities of amphibians, mammals, and birds with fewer than 10 species, the same did not hold true for the threshold of at least five sampling events, resulting in the exclusion of a large portion of grid cells from our analysis
Cells with fewer sampling events are more likely to have more extreme effect sizes, resulting from the size of the difference between few data points. We therefore restricted our analyses to test only whether the effect was positive or negative. To distinguish between primarily colonial sampling and recent sampling, we conducted separate analyses for two time periods: 1801–1940, 1980–2019, and then for the total period: 1801–2019. Although most countries attained independence between 1950 and 1980, separating colonial times from independence dates can be misleading. We assumed the end of World War II as an igniter of the emancipation of African countries following Cooper (2019) and considered the period of 1940–1980 uncertain and impossible to assign across diverse African countries. The lower boundary of 1800 was arbitrarily chosen to encapsulate all subsequent colonial periods and to avoid any dubious earlier records. In summary, we considered pre-1940 colonial, post-1980 noncolonial, and 1940–1980 unknown and therefore excludable from our analyses. The term colonial is used here as a shorthand for the time period prior to the emergence of the independent African nations we see now, and that, depending on the part of Africa, this definition includes a period before the formal establishment of the colonial structure.
Spatial patterns might affect the degree at which sampling completeness increases the probability of researchers visiting a cell. To investigate this possibility, we carried out a multiple correlation analysis with variables that cover both environmental and social aspects. These included annual precipitation (Worldclim v 2: Fick and Hijmans 2017), human influence (WCS and CIESIN 2005), net primary production (Imhoff et al. 2004), protected areas coverage (UNEP-WCMC 2019), elevation (Jarvis et al. 2008), Human Development Index (HDI), and Gross Domestic Product (GDP) per capita (Kummu et al. 2018).
To account for spatial autocorrelation, regressions were conducted through simultaneous autoregressive models with spatial error (i.e. SAR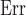 models; Haining and Haining 2003). We tested 40 different neighborhoods (10 with a fixed number of neighbors between 1 and 10, and 30 containing all combinations of cells up to 250, 500, 750, 1000, 1250, or 1500 km away with either of the five default weighting schemes). The best neighborhood was chosen as the one minimizing the corrected Akaike Information Criterion (AICc) by using the R package wiqid version 0.2.2 (Meredith 2017). To obtain an estimate of how close our data are to the fitted regression line, we calculated the Nagelkerke pseudo
models; Haining and Haining 2003). We tested 40 different neighborhoods (10 with a fixed number of neighbors between 1 and 10, and 30 containing all combinations of cells up to 250, 500, 750, 1000, 1250, or 1500 km away with either of the five default weighting schemes). The best neighborhood was chosen as the one minimizing the corrected Akaike Information Criterion (AICc) by using the R package wiqid version 0.2.2 (Meredith 2017). To obtain an estimate of how close our data are to the fitted regression line, we calculated the Nagelkerke pseudo 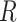 -squared using the function summary.sarlm from the R package spdep version 1.1-2 (Bivand et al. 2005).
-squared using the function summary.sarlm from the R package spdep version 1.1-2 (Bivand et al. 2005).
To predict the required time to sample 90% of cells in Africa, we used survival analyses (Demetrius 1978). We treated unsampled cells as “alive” and sampled cells as “dead,” and treated all cells still not visited in 2019 as “censored.” This treatment is usually done in survival analysis to describe unknown survival time. We then fitted our data with a Weibull distribution, which is able to accommodate the three basic survival curves with constant, monotonically increasing or monotonically decreasing mortality (Pinder et al. 1978). Mathematically, this assumes that the sampling rate 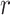 at time
at time  can be modeled as:
can be modeled as:
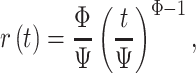 |
which for 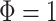 means a constant sampling rate, whereas
means a constant sampling rate, whereas 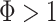 means that sampling rates are increasing over time in a consistent manner.
means that sampling rates are increasing over time in a consistent manner.
Our assumption that the sampling rate of collection in Africa can be modeled by a single Weibull distribution constitutes a technical oversimplification, given the idiosyncrasies of each country and region. This rate is likely to have been affected by socio-political events (Rydén et al. 2019), and the implementation of regulations such as CITES, the Nagoya Protocol, and national and regional legislations, potentially leading to reduced fieldwork. The implementation of this assumption is, however, required by our modeling approach, which is limited by the requirement of a single rate for the temporal predictions.
To predict the number of sampling events necessary to find at least half of all the predicted species in a particular cell, we did a spatial regression analysis using the formula:
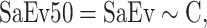 |
where SaEv50 corresponds to the number of sampling events to record at least 50% of completeness, SaEv is the number of sampling events, and C is the completeness value. To remove the effect of a particular year on our model, we randomized the year of visit in our data set 100 times and used the median value.
Both the SARerr regressions and the linear regression were conducted in R 3.5.1 (Team 2018) using glm (Team 2018), errorsarlm from the package spdep (Bivand et al. 2005), and the R package survival ver 2.44-1.1 (Therneau and Lumley 2015).
Additional R packages used for data manipulation and visualization were: BBmisc version 1.11 (Bischl et al. 2017), Rphylopic version 0.2.0 (Chamberlain 2018), Tidyverse version 1.2.1 (Hadley Wickham and Wickham 2017), gridExtra version 2.3 (Auguie et al. 2017), raster version 2.9-5 (Hijmans et al. 2015), pbapply version 1.4-0 (Solymos 2016), ggplot2 version 3.1.0 (Wickham 2016), sf version 0.7-1 (Pebesma 2018), and package rnaturalearth version 0.1.0 (South 2017).
Results
By overlapping the map of Africa obtained from the R package rnaturalearth version 0.1.0 (South 2017) with our grid, we obtained a total of 3212 cells that include any portion of continental Africa. Among the groups surveyed, birds were the most well sampled in our study area, for which we retrieved 775,131 records distributed across 1798 cells. Mammals had 63,521 records across 1578 cells and amphibians only had 15,991 records distributed across 936 cells (Fig. 1). The lower number of cells for amphibians is due to the substantial areas of dryland habitats in Africa.
Figure 1.
Top: Number of sampling events per grid cell for each taxon group, as estimated from the Global Biodiversity Information Facility. White areas represent areas without any records. Sampling events can be interpreted as the number of different years in which a particular cell has been visited. Bottom: Number of sampling events by country. We divided the number of sampling events by country area and transformed the data by dividing each value by the maximum value of each taxon for visualization purposes. Much of Africa has rarely been visited. South Africa and Tanzania are in the top three with the greatest number of cells—more than five sampling events for all three taxa. Regarding the least-sampled countries—those lacking any cells with more than 5 sampling events—we identified 25 countries for amphibians, 16 for mammals, and 5 for birds. Comoros, Libya, and Djibouti have no cells with more than 5 sampling events for any of these groups. The full list of sampling events by country per taxa is provided in the Supplementary Tables S1–S3 available on dryad.
Effect of Previous Knowledge on the Probability of Visiting a Cell
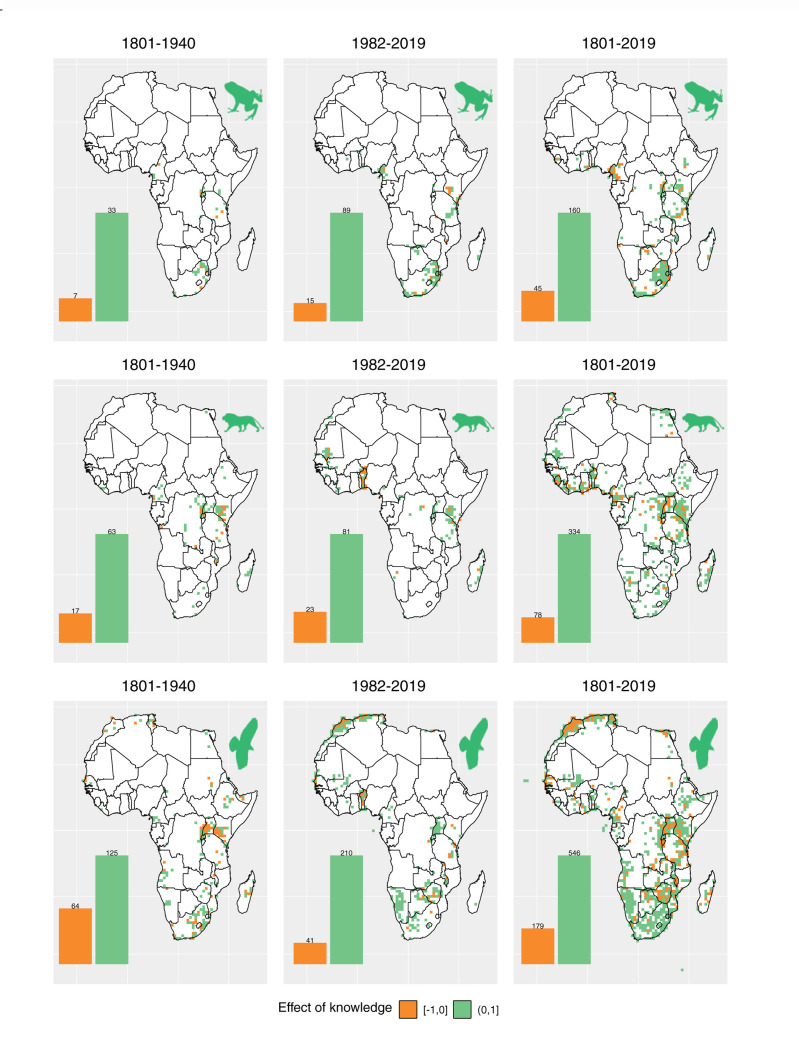
We assessed the effect of biodiversity knowledge available for a particular cell on the probability of sampling the cell, by using a logistic model between sampling completeness, sampling events, and year. Negative values correspond to cells in which the amount of existing knowledge decreases the probability of resampling a cell. In contrast, a positive value corresponds to a positive effect of existing knowledge on the probability of sampling.
By analyzing data that are readily available to the scientific community, we found that amphibians were seldom recorded between 1801 and 1940, when compared with mammals and birds (Fig. 2, left panel). In our analyses, we explicitly accounted for overall temporal changes in sampling. Our results therefore investigate any patterns in sampling probability happening in addition to overall temporal changes. Across most of Africa, previous sampling in amphibians has strongly influenced the resampling of areas: between 1801 and 1940, 83% of the previously sampled cells had a positive effect. This is similar to the period between 1982 and 2019, where 78% of the previously sampled cells were positive (Fig. 2). Mammologists also show the strongest preference for revisiting areas previously sampled, with similar values of 79% before 1940 and 81% after 1982 (Fig. 2). The bird sampling shows that before 1940, there was an increased preference of discovery inferred by a higher sampling frequency in unsampled areas among researchers, when compared with after 1982. In both time periods, previous sampling decreased the likelihood of visits in the majority of cells, and the pattern became more pronounced in the later time period. This is demonstrated by an increase from 66% to 75% in the proportion of positive cells (Fig. 2).
Figure 2.
Effect of previous knowledge on sampling of amphibians, mammals, and birds. The spatial patterns were calculated through logistic regressions between completeness and sampling events in amphibians, mammals, and birds, and the sign of the effect mapped per cell. White grid cells were not used in this analysis because they either did not contain any records, had less than five visits, or had an expected richness lower than 10. The left panel corresponds to the filtered data available between 1801 and 1940; the middle panel corresponds to the period of 1982 to 2019; and the right panel includes data from 1801 to 2019. Only cells with five or more sampling events were included to reduce the effects of overfitting. Bar values represent the number of cells corresponding to negative/positive values. For all three taxa analyzed, there is strong and pervasive evidence that previous knowledge leads to increased sampling. All taxa also show a temporal increase in this trend. Maps with only cells with P value 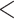 0.05 can be found in the Supplementary Figs. S1–S3 available on dryad.
0.05 can be found in the Supplementary Figs. S1–S3 available on dryad.
Sampling Events Required to Record 50% of Species
Our analysis revealed small effects between the tested predictors and their effect on the attractiveness of an area based on pre-existing knowledge. The general pattern is that the existence of increased knowledge increases the likelihood of increased sampling (Table 1). We only found three instances of small significant effects. In amphibians, increased knowledge was slightly less likely to lead to increased sampling in areas with higher net primary productivity (NPP) (Table 1). In birds, increased knowledge was slightly less likely to lead to increased sampling in areas with more roads and slightly more likely to lead to increased sampling in areas with higher precipitation (Table 1).
Table 1.
Predictors for the effect of previous sampling on the probability of sampling for amphibians, mammals, and birds in Africa.
| Amphibians | Mammals | Birds | |
|---|---|---|---|
| Estimate (SE) | Estimate (SE) | Estimate (SE) | |
| (Intercept) | 0.777(0.023)*** | 0.806(0.011)*** | 0.758(0.031)*** |
| IUCN richness | -0.089(0.038)* | -0.081(0.018) | -0.025(0.032) |
| Protected areas | -0.017(0.03) | 0.009(0.019) | 0.011(0.017) |
| Human influence | 0.101(0.032)** | -0.036(0.021) | -0.069(0.023)** |
| HDI | -0.048(0.034) | -0.019(0.02) | -0.004(0.027) |
| GDP | -0.012(0.03) | 0.02(0.021) | -0.03(0.018) |
| NPP | -0.036(0.04) | 0.006(0.026) | -0.059(0.031) |
| Precipitation | -0.052(0.042) | -0.043(0.022) | 0.053(0.03) |
| Elevation | -0.003(0.028) | 0.02(0.017) | -0.019(0.019) |
| Road density | -0.002(0.028) | 0.005(0.017) | -0.031(0.016) |
| AICc | 214 | 390.4 | 774.8 |
Nagelkerke pseudo 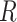 -squared -squared |
0.141 | 0.073 | 0.104 |
Notes: Significance of the following predictors for the effect of previous sampling on the probability of sampling: IUCN predicted richness, protected area coverage, human influence, HDI, GDP per capita, NPP, precipitation, elevation, and road density. Human influence is a significant positive predictor for amphibians and negative for birds. Predicted richness is a significant negative predictor for amphibians.
*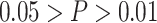 ; **
; **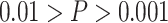 ; ***
; ***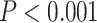 .
.
Time to Sample at Least Once in 90% of Africa
The model predicted the time it would take, assuming a Weibull distributed biodiversity sampling rate, to sample at least once in 90% of Africa. For amphibians, we predicted the sampling coverage of Africa to be achieved between 2192 and 2233, for mammals between 2222 and 2257, and for birds between 2253 and 2294 (Fig. 3).
Figure 3.
Proportion of grids (10,000 km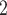 ) in Africa sampled at least once and containing at least 10 species. We used only the cells where the groups are expected to occur, meaning that amphibians, birds, and mammals had a different number of cells where they could be recorded. Ribbons indicate 95% confidence intervals. These analyses predict that, for birds, species occurrence data for 90% of Africa will only be achieved somewhere between years 2253 and 2294; for mammals between 2222 and 2257; and for amphibians between 2192 and 2233.
) in Africa sampled at least once and containing at least 10 species. We used only the cells where the groups are expected to occur, meaning that amphibians, birds, and mammals had a different number of cells where they could be recorded. Ribbons indicate 95% confidence intervals. These analyses predict that, for birds, species occurrence data for 90% of Africa will only be achieved somewhere between years 2253 and 2294; for mammals between 2222 and 2257; and for amphibians between 2192 and 2233.
Sampling Events Required to Achieve 50% Inventory
Our spatial regression analysis showed that the number of sampling events required to record 50% of the species was mainly positively associated with HDI and elevation of the grid cells for amphibians. For mammals, HDI was positively associated with the number of sampling events. For birds, richness was positively associated with the number of sampling events, unlike NPP, which displayed a negative association. We found that it would take on average 11.5 visits for amphibians, 12.7 visits for mammals, and 27.0 visits for birds to recover 50% of all species within a cell (Supplementary Table S4 available on dryad at https://doi.org/10.5061/dryad.ngf1vhhsg).
Discussion
In this study, we unveil, quantify, and map a new bias in biodiversity data: that knowledge in itself is leading to an increase in sampling bias. For all three examined vertebrate groups, researchers tend to return to areas based on the existence of previous knowledge rather than visiting and sampling new areas.
The lack of biological specimens and tissues for most of the African continent is likely to impact the comprehensiveness of phylogenetic reconstructions, our understanding of taxonomic relationships, and the estimation of divergence times and biogeographic history (Hortal et al. 2015). These are just some of the expected effects caused by the exclusion of critical species, populations, and genetic diversity in phylogenetic analyses. Since there are few biological barriers across the African continent (Fjeldså 1994), and political borders in Africa seldom follow natural features, it is essential that the systematics community acknowledges the need for wide spatial sampling in their research. In addition, conservation practitioners must also be aware of this bias. The need to reliably assess global biodiversity in an era where conservation relies increasingly on “big data” (Arts et al. 2015) demands the use of estimation and extrapolation (Colwell 2009). However, such procedures can be heavily compromised by uneven sampling across large land extensions (Reddy and Dávalos 2003). The effect of sampling bias on diversity estimates may also obstruct solid inference on underlying drivers of biodiversity build-up as well as loss (Loiselle et al. 2008; Engemann et al. 2015). The reluctance to visit new areas may also affect the diversity pattern for microendemics, because such species may only be known from studied areas. For instance, species endemic to individual mountains or inselbergs in eastern Africa (Branch et al. 2014; Bittencourt-Silva et al. 2016) would only be known from sites visited by a specialist.
Since more than a third of Africa lacks digitally accessible information (Meyer et al. 2015), we expected that researchers conducting inventories would be attracted to data-deficient areas (Scenario 1 above). However, our results show the contrary: sampling events tend to occur where knowledge already exists (Scenario 2). We find a temporal increase in this trend, when comparing the sampling events from before 1940 with the sampling events after 1982. This pattern might be explained as a result of the spread of the reputation of a particular area for harboring high diversity, as mentioned by Reddy and Dávalos (2003) when they recorded a disproportionate amount of sampling effort toward areas rich in biodiversity. It may also be that previous sampling indicates the area is accessible and so can be surveyed with fewer resources than a site that has not been sampled. There is also a higher risk of a nonsurveyed site not containing the targeted species.
In order to exclude the influences of other predictors on the effect of previous knowledge in attracting more sampling events, we used a spatial regression analysis of completeness in relation to visits. We demonstrated that these effects cannot be attributed to any of the traditionally tested predictors (the predictors were not significant or had minimal effects). For amphibians, we observed a negative significant effect for predicted richness and a significant positive effect of human influence; however, in both cases, those effects were quantitatively minimal and much smaller than the effect of previous knowledge (Table 1).
Completing the Biodiversity Inventory of Africa
Our analyses indicated that it could take between 172 and 274 years for the research community to carry out at least one sampling effort in 90% of all cells across Africa. These estimates are based on current and historic rates of biological exploration, and encompass only some of the most well-studied organism groups of all: birds, mammals, and amphibians. In addition, a single sampling effort is far from enough to correctly characterize the diversity of any site: in our estimates, between 12 and 27 events are required to record at least 50% of the existing species.
Our models showed significant positive effects of elevation and HDI on the number of sampling events for amphibians; significant positive effects of HDI on the number of sampling events for mammals; significant positive effects of IUCN predicted richness; and significant negative effects of protected areas and net primary production on the sampling of birds. Even though not all protected areas used in this analysis were officially established since the 19th century, many of them that eventually became protected were likely to have been under some form of protection earlier on, under classifications such as reserves or hunting concessions. The significant positive effect of elevation on the number of sampling events on amphibians might be a consequence of the high endemism within the group in mountains such as in the Eastern Arc (Burgess et al. 2007).
Analyzing data collected through long periods of time at a continental scale poses numerous challenges, one of them being the continuous process of human expansion and consequent habitat transformation. Some areas may experience the emergence of mining or even become urban centers. We expect this to be a problem at fine scales, but due to our grid-cell area of  10 000 km
10 000 km , we expect the complete transformation of entire cells to be relatively rare. We further note that we only focus on species that, according to IUCN, currently occur in the cell. This means that species that are locally extirpated from much of Africa, such as lions, are not included even if there are records from the species prior to its local extirpation from any given area.
, we expect the complete transformation of entire cells to be relatively rare. We further note that we only focus on species that, according to IUCN, currently occur in the cell. This means that species that are locally extirpated from much of Africa, such as lions, are not included even if there are records from the species prior to its local extirpation from any given area.
Conclusions and Recommendations
This study conveys an urgent and crucial message: unless a radical and widespread change in research practice takes place, Africa’s rich biodiversity will remain largely unknown. We cannot protect or understand what we do not know about, yet the data available for most of Africa to adequately identify and delimit species boundaries, understand spatial biodiversity patterns, or to effectively promote species conservation are insufficient.
Waiting more than a century to complete the biodiversity inventory of Africa is not a viable option. Africa is experiencing the highest population growth of any continent (Gerland et al. 2014), with an expected 209% increase between 2000 and 2050 (United Nations 2017). Between 2015 and 2050, an additional 2.4 billion people are expected, which in combination with a rapidly changing climate will exert a tremendous pressure on natural ecosystems and their biodiversity. The recent Living Planet Report 2020 (WWF 2020) shows that wild populations of African vertebrates have declined an alarming 65% over the last 50 years alone.
It is also important to note that our estimates are based only on birds, mammals, and amphibians—three well-studied groups. The knowledge bias and spatial patterns we report are likely to be considerably worse for other groups such as plants, fungi, and insects (Stropp et al. 2016; Willis 2017, 2018). Such diversity may hold important solutions to help achieve the Sustainable Development Goals (Antonelli et al. 2019), but will be largely lost if not effectively mapped and conserved.
To tackle the challenges outlined in this study, we make the following four recommendations:
Funding providers (agencies, companies, and philanthropists) should actively promote projects aiming to sample areas that lack baseline biodiversity data. We recognize the difficulties in weighing up the costs and benefits of allocating limited time and resources toward additional data collection (Grand et al. 2007). Although additional sampling is also needed in well-sampled areas (such as already recognized biodiversity or endemism hotspots), for instance, to increase our full understanding of biodiversity and biotic interactions in those areas, it is essential to increase the focus on poorly sampled areas (Reddy and Dávalos 2003).
Researchers should, whenever possible, increase the taxonomic and methodological scope of their collection efforts. Biodiversity inventories usually involve producing point-locality biological data and conducting basic taxonomy (da Fonseca et al. 2020). Given the logistic and legislative challenges of carrying out fieldwork across most of Africa, we urge scientists to collaborate with specialists in different institutions and with varied taxonomic expertise to responsibly sample the maximum possible number of taxa (in full or as tissue samples, especially for endangered or large species). Expeditions with multitaxa foci may be especially valuable for creating baseline biodiversity data in numerous data-deficient areas, and should be encouraged by funding agencies and biodiversity institutions. It is also imperative to concentrate on physical specimens (whole specimens, tissue samples for DNA analyses, seeds for cultivation in the case of plants, among others), rather than only photographic evidence (Troudet et al. 2018), while preventing negative effects on the survival of threatened populations and species. Whenever possible, duplicates of samples should be deposited in multiple organizations, to increase their long-term safety and accessibility. The collection of rich metadata will increase the value of collections for many and as yet unforeseen uses (Bakker et al. 2019; Fernández et al. 2019).
Engage globally and locally. Biological sampling in Africa has to a large extent been carried out by European and North American institutions, with limited benefits returning to the countries of origin. Under the Access and Benefit-Sharing agreements of the Convention of Biological Diversity, it is crucial that future sampling activities are always done in close partnership with African institutions and researchers, for mutual benefits (Pearce et al. 2020).
Clarity on processing research permits. There is certain evidence that excessive in-country legislation that regulates research and collection permits can sometimes hinder research (Rydén et al. 2019; Williams et al. 2020). If possible, clarification on the process for sampling permits should be made transparent and available online for every country in the continent, to encourage and streamline biodiversity research.
Acknowledgments
H.F. and A.S. are supported thanks to financial support of CESAM (UIDP/50017/2020+UIDB/50017/2020) from FCT/MCTES through national funds, and the co-funding by the FEDER, within the PT2020 Partnership Agreement and Compete 2020, H.F. is also supported by WCS Christensen Conservation Leaders Scholarship, World Wildlife Foundation—Education for Nature Scholarship, and FCT/MCTES reference PD/BD/128123/2016. J.A.R.A. is supported by CAPES (#001292/2015-03) and the Instituto Serrapilheira. S.F. is supported by the Swedish Research Council (#2017-03862). A.A. is supported by grants from the Swedish Research Council, the Swedish Foundation for Strategic Research, Knut and Alice Wallenberg Foundation (through a Wallenberg Academy Fellowship), and the Royal Botanic Gardens, Kew. We thank our colleague Rhian Smith for proofreading this manuscript and also the three anonymous reviewers and editors whose suggestions greatly helped improve this study.
Supplementary Material
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.ngf1vhhsg
References
- Amarasinghe A., Kuritsky J.N., Letson G.W., Margolis, H.S.. 2011. Dengue virus infection in Africa. Emerg. Infect. Dis. 17(8):1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli A., Smith R.J., Simmonds M.S.. 2019. Unlocking the properties of plants and fungi for sustainable development. Nat. Plants. 5(11):1100–1102. [DOI] [PubMed] [Google Scholar]
- Arts K., van der Wal R., Adams W.M.. 2015. Digital technology and the conservation of nature. Ambio. 44(4):661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguie B., Antonov A., & Auguie M.B.. 2017. Package ‘gridExtra’. Miscellaneous Functions for “Grid” Graphics. [Google Scholar]
- Bakker F.T., Antonelli A., Clarke J.A., Cook J.A., Edwards S. V., Ericson P. G.,... & Irestedt, M.. 2020. The Global Museum: natural history collections and the future of evolutionary science and public education. PeerJ, 8, e8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates J.M., Bowie R.C.K., Willard D.E.. et al. 2004. A need for continued collecting of avian voucher specimens in Africa: why blood is not enough. Ostrich 74:187–191 [Google Scholar]
- Beegle, K., Christiaensen, L., Dabalen, A., & Gaddis, I.. 2016. Poverty in a rising Africa. The World Bank. [Google Scholar]
- Bello-Schünemann J., Moyer J.D.. 2018. Structural pressures and political instability-trajectories for sub-Saharan Africa. ISS Africa Report, 2018(9):1–32. [Google Scholar]
- Bhatt S., Weiss D., Cameron E., Bisanzio D., Mappin B., Dalrymple U., Battle K., Moyes C., Henry A., Eckhoff P.. 2015. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 526(7572):207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischl B., Lang M., Bossek J., Horn D., Richter J., Surmann D.. 2017. BBmisc: Miscellaneous Helper Functions for B. Bischl. R package version, 1. https://rdrr.io/cran/BBmisc/. [Google Scholar]
- Bischl B., Lang M., Bossek J., Horn D., Richter J., Surmann D.. 2017. BBmisc: Miscellaneous Helper Functions for B. Bischl. R package version, 1. [Google Scholar]
- Bittencourt-Silva G.B., Conradie W., Siu-Ting K., Tolley K.A., Channing A., Cunningham M., Farooq H.M., Menegon M., Loader S.P.. 2016. The phylogenetic position and diversity of the enigmatic mongrel frog Nothophryne Poynton, 1963 (Amphibia, Anura). Mol. Phylogenet. Evol. 99:89–102. [DOI] [PubMed] [Google Scholar]
- Bivand R., Bernat A., Carvalho M., Chun Y., Dormann C., Dray S., Halbersma R., Lewin-Koh N., Ma J., Millo G.. 2005. The spdep package. Comprehensive R Archive Network, Version, 05-83. [Google Scholar]
- Boakes E.H., McGowan P.J., Fuller R.A., Chang-Qing D., Clark N.E., O’Connor K., Mace, G.M.. 2010. Distorted views of biodiversity: spatial and temporal bias in species occurrence data. PLoS Biol. 8(6):e1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch W.R., Bayliss J., Tolley K.A.. 2014. Pygmy chameleons of the Rhampholeon platyceps complex (Squamata: Chamaeleonidae): Description of four new species from isolated ‘sky islands’ of northern Mozambique. Zootaxa. 3814(1):1–36. [DOI] [PubMed] [Google Scholar]
- Burgess N., Butynski T., Cordeiro N., Doggart N., Fjeldså J., Howell K., Kilahama F., Loader S., Lovett J.C., Mbilinyi B.. 2007. The biological importance of the Eastern Arc Mountains of Tanzania and Kenya. Biol. Conserv. 134(2):209–231. [Google Scholar]
- Chamberlain S. 2018. rphylopic: Get ’Silhouettes’ of ’Organisms’ from ’Phylopic’. R package version 0.2.0. Available from: https://CRAN.R-project.org/package=rphylopic. [Google Scholar]
- Colwell R.K. 2009. Biodiversity: concepts, patterns, and measurement. In: The Princeton Guide to Ecology (ed Levin SA), pp. 257–263. Princeton Univ. Press, Princeton, New Jersey. [Google Scholar]
- Conradie W., Bittencourt-Silva G., Farooq H., Loader S.P., Menegon M. & Tolley K.A.. 2018. New species of Mongrel Frogs (Pyxicephalidae: Nothophryne) for northern Mozambique inselbergs. African Journal of Herpetology, 67:61–85. 10.1080/21564574.2017.1376714. [DOI] [Google Scholar]
- Conradie W., Bittencourt-Silva G., Engelbrecht H.M., Loader S. P., Menegon M., Nanvonamuquitxo C., Scott M. & Tolley K.A.. 2016. Exploration into the hidden world of Mozambique’s sky island forests: new discoveries of reptiles and amphibians. Zoosystematics and Evolution, 92, 163–180. 10.3897/zse.92.9948. [DOI] [Google Scholar]
- Cooper F. 2019. Africa since 1940: the past of the present. Vol. 13. Cambridge, Cambridge University Press. [Google Scholar]
- da Fonseca G.A., Balmford A., Bibby C., Boitani L., Corsi F., Brooks T., Gascon C., Olivieri S., Mittermeier R.A., Burgess N.. 2000. … following Africa’s lead in setting priorities. Nature. 405(6785):393. [DOI] [PubMed] [Google Scholar]
- Demetrius L. 1978. Adaptive value, entropy and survivorship curves. Nature. 275(5677):213–214. [DOI] [PubMed] [Google Scholar]
- Engel U., Gebauer C. & Hüncke, A.. 2015. Notes from Within and Without: Research Permits between Requirements and’Realities’. SPP 1448 Working Paper Series; 16. [Google Scholar]
- Engemann K., Enquist B.J., Sandel B., Boyle B., Jørgensen P.M., Morueta-Holme N., Peet R.K., Violle C., Svenning J.C.. 2015. Limited sampling hampers “big data” estimation of species richness in a tropical biodiversity hotspot. Ecol. Evol. 5(3):807–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq H.O.M., Conradie, W.. 2015. Second record of a Scolecomorphus kirkii Boulenger, 1883 (Gymnophiona: Scolecomorphidae) for Mozambique. Herpetol. Notes. 8:59–62. [Google Scholar]
- Feeley K.J., Silman M.R.. 2011. The data void in modeling current and future distributions of tropical species. Global Change Biol. 17(1):626–630. [Google Scholar]
- Fernández N., Guralnick R., Kissling W.D.. 2019. A minimum set of Information Standards for Essential Biodiversity Variables. Biodiversity Information Science and Standards 3:e35212. 10.3897/biss.3.35212. [DOI] [Google Scholar]
- Fick S.E., Hijmans R.J. (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37(12):4302–4315. [Google Scholar]
- Fjeldså J. 1994. Geographical patterns for relict and young species of birds in Africa and South America and implications for conservation priorities. Biodivers. Conserv. 3(3):207–226. [Google Scholar]
- Foster V. and Briceno-Garmendia C.. 2010. Africa’s infrastructure: a time for transformation. World Bank, Washington, DC. [Google Scholar]
- Gerland P., Raftery A.E., Ševèíková H., Li N., Gu D., Spoorenberg T., Alkema L., Fosdick B.K., Chunn J., Lalic N.. 2014. World population stabilization unlikely this century. Science. 346(6206): 234–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham C.H., Ferrier S., Huettman F., Moritz C., Peterson A.T.. 2004. New developments in museum-based informatics and applications in biodiversity analysis. Trends Ecol. Evol. 19(9):497–503. [DOI] [PubMed] [Google Scholar]
- Grand J., Cummings M.P., Rebelo T.G., Ricketts T.H., Neel M.C.. 2007. Biased data reduce efficiency and effectiveness of conservation reserve networks. Ecol. Lett. 10(5):364–374. [DOI] [PubMed] [Google Scholar]
- Haining R. 2003. Spatial Data Analysis: Theory and Practice. Cambridge University Press: Cambridge, UK. [Google Scholar]
- Harford J.B. 2015. Barriers to overcome for effective cancer control in Africa. Lancet Oncol. 16(8):e385–e393. [DOI] [PubMed] [Google Scholar]
- Hijmans R. 2010. Package raster. R package version 2.9-5. http://CRAN.R-project.org/package=raster. [Google Scholar]
- Hortal J., de Bello F., Diniz-Filho J.A.F., Lewinsohn T.M., Lobo J.M., Ladle R.J.. 2015. Seven shortfalls that beset large-scale knowledge of biodiversity. Ann. Rev. Ecol. Evol. Syst. 46:523–549. [Google Scholar]
- Hotez P.J., Kamath A.. 2009. Neglected tropical diseases in sub-Saharan Africa: review of their prevalence, distribution and disease burden. PLoS Negl Trop Dis 3:e412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhoff M.L., Bounoua L., Ricketts T., Loucks C., Harriss R., Lawrence W.T.. 2004. Global patterns in net primary productivity. Data distributed by the Socioeconomic Data and Applications Center (SEDAC). Available at: http://sedac.ciesin.columbia.edu/es/hanpp.html. [Google Scholar]
- Impey O., MacGregor A.. 1985. The origins of museums: the cabinet of curiosities in sixteenth-and seventeenth-century Europe. Oxford: Clarendon Press. [Google Scholar]
- IUCN. 2019. The IUCN Red List of Threatened Species. Version 2019-3. Available from: http://www.iucnredlist.org. [Google Scholar]
- Jarvis A., Reuter H.I., Nelson A., Guevara E.. 2008. Hole-Filled SRTM for the Globe Version 4 (Int Center for Trop Agric, Cali, Colombia). [Google Scholar]
- Kummu M., Taka M., Guillaume J.H.. 2018. Gridded global datasets for gross domestic product and Human Development Index over 1990–2015. Sci. Data. 5:180004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiselle B.A., Jørgensen P.M., Consiglio T., Jiménez I., Blake J.G., Lohmann L.G., Montiel O.M.. 2008. Predicting species distributions from herbarium collections: does climate bias in collection sampling influence model outcomes? J. Biogeogr. 35(1):105–116. [Google Scholar]
- Lomolino M.V. 2004. Conservation biogeography. Frontiers of Biogeography: new directions in the geography of nature (ed. by Lomolino M.V. and Heaney L.R.), pp. 293–296. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Mbaku M.J. 2010. Corruption in Africa: Causes, Consequences and Cleanup (Lexington Books, USA: ). [Google Scholar]
- Meredith M. Quick and Dirty Estimates for Wildlife Populations. In: http://www.r-project.org/, editor. 2018. p. 81. [Google Scholar]
- Meyer C., Kreft H., Guralnick R., Jetz W.. 2015. Global priorities for an effective information basis of biodiversity distributions. Nat. Commun. 6:8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C., Weigelt P., Kreft H.. 2016. Multidimensional biases, gaps and uncertainties in global plant occurrence information. Ecol. Lett. 19(8):992–1006. [DOI] [PubMed] [Google Scholar]
- Pearce T.R., Antonelli A., Brearley F.Q., Couch C., Campostrini Forzza R., Gonçalves S.C., Magassouba S., Morim M.P., Mueller G.M., Nic Lughadha E., Obreza M., Sharrock S., Simmonds M.S.J., Tambam B.B., Utteridge T.M.A. & Breman E.. 2020. International collaboration between collections-based institutes for halting biodiversity loss and unlocking the useful properties of plants and fungi. Â Plants, People, Planet, 2(5), 515–534. 10.1002/ppp3.10149. [DOI] [Google Scholar]
- Pebesma E. 2018. Simple features for R: standardized support for spatial vector data. R J., 10(1):439–446. [Google Scholar]
- Pinder J.E., Wiener J.G., Smith M.H.. 1978. The Weibull distribution: a new method of summarizing survivorship data. Ecology. 59(1):175–179. [Google Scholar]
- Portik D.M., Mulungu E., Sequeira D., McEntee J.. 2013. Herpetological surveys of the Serra Jeci and Namuli massifs, Mozambique, and an annotated checklist of the southern Afromontane archipelago. Herpetol. Rev. 44(3):394–406. [Google Scholar]
- Rabosky A.R.D., Cox C.L., Rabosky D.L., Title P.O., Holmes I.A., Feldman A., McGuire J.A.. 2016. Coral snakes predict the evolution of mimicry across New World snakes. Nat. Commun. 7:11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S., Dávalos L.M.. 2003. Geographical sampling bias and its implications for conservation priorities in Africa. J. Biogeogr. 30(11):1719–1727. [Google Scholar]
- Ritterbush P.C. 1969. Science teaching and the future. Sci. Teacher. 36(6):32–39. [Google Scholar]
- Ryan P.G., Bento C, Cohen C, Graham J, Parker V, Spottiswoode C.. 1999. The avifauna and conservation status of the Namuli Massif, northern Mozambique. Bird Conserv. Int. 9(4):315–331. [Google Scholar]
- Rydén O., Zizka A., Jagers S.C., Lindberg S.I., Antonelli A.. Linking democracy and biodiversity conservation: empirical evidence and research gaps Ambio, 49 (2019), pp. 419-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Fernández D., Lobo J.M., Abellán P., Ribera I., Millán A.. 2008. Bias in freshwater biodiversity sampling: the case of Iberian water beetles. Divers. Distrib 14(5):754–762. [Google Scholar]
- Schmidt-Lebuhn A.N., Knerr N.J., Kessler M.. 2013. Non-geographic collecting biases in herbarium specimens of Australian daisies (Asteraceae). Biodivers. Conserv. 22(4):905–919. [Google Scholar]
- Schmitt C.J., Cook J.A., Zamudio K.R., Edwards S.V.. 2019. Museum specimens of terrestrial vertebrates are sensitive indicators of environmental change in the Anthropocene. Philos. Trans. R. Soc. B. 374(1763):20170387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer H.B., Fisher R.N., Davidson C.. 1998. The role of natural history collections in documenting species declines. Trends Ecol. Evol. 13(1):27–30. [DOI] [PubMed] [Google Scholar]
-
Solymos P.
2016. Zawadzki Z. pbapply: Adding Progress Bar to
 apply’Functions. R package version 1.2-1. http://CRAN.R-project.org/package=pbapply. [Google Scholar]
apply’Functions. R package version 1.2-1. http://CRAN.R-project.org/package=pbapply. [Google Scholar] - South, A. 2017. R natural earth: world map data from natural earth. In: R package version 0.1. Avaialble from: https://CRAN.R-project.org/package.... [Google Scholar]
- Steege H.T., Haripersaud P.P., Bánki O.S., Schieving F.. 2011. A model of botanical collectors’ behavior in the field: never the same species twice. Am. J. Bot. 98(1):31–37. [DOI] [PubMed] [Google Scholar]
- Stork H., Astrin J.J.. 2014. Trends in biodiversity research—a bibliometric assessment. Open J. Ecol. 4(7):354. [Google Scholar]
- Stropp J., Ladle R. J., Malhado A.C.M., Hortal J., Gaffuri J., Temperley W.H., Olav Skøien J., Mayaux P.. 2016. Mapping ignorance: 300 years of collecting flowering plants in Africa. Global Ecol. Biogeogr. 25(9):1085–1096. [Google Scholar]
- Team R.C. 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing Vienna Austria. 2018; version 3.5.1. [Google Scholar]
- Therneau T.M. & Lumley T.. 2015. Package ‘survival’. R Top Doc, 128, 112. https://cran.r-project.org/package=survival. [Google Scholar]
- Timberlake J., Dowsett-Lemaire F., Bayliss J., Alves T., Baena S., Bento C., Cook K., Francisco J., Harris T., Smith P.. 2009. Mt Namuli, Mozambique: biodiversity and conservation. Rep. Darwin Initiative Award. 15:36. [Google Scholar]
- Troudet J., Grandcolas P., Blin A., Vignes-Lebbe R., Legendre F.. 2017. Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 7(1):9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troudet J., Vignes-Lebbe R., Grandcolas P., Legendre F.. 2018. The increasing disconnection of primary biodiversity data from specimens: how does it happen and how to handle it? Syst. Biol. 67(6):1110–1119. [DOI] [PubMed] [Google Scholar]
- UNEP-WCMC I. 2019. Protected Planet: the World Database on Protected Areas (WDPA). August28. [Google Scholar]
- United Nations. 2017. World population prospects 2017. Retrieved from https://population.un.org/wpp/Download/Standard/Population/. [Google Scholar]
- Vincent J. 1933. The Namuli Mountains, Portuguese East Africa. Geogr. J., 81(4):314–327. [Google Scholar]
- Walker A., Bush T. and Oduro G.K.T.. 2006. “New principals in Africa: preparation, induction and practice”, Journal of Educational Administration, 44(4):359–375. 10.1108/09578230610676587. [DOI] [Google Scholar]
- Ward D.F. 2012. More than just records: analysing natural history collections for biodiversity planning. PLoS One. 7(11):e50346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag. [Google Scholar]
- Wickham H., Wickham M.H.. 2017. Package tidyverse. Easily Install and Load the ‘Tidyverse. [Google Scholar]
- Wildlife Conservation Society - WCS, Center for International Earth Science Information Network - CIESIN - Columbia University. (2005). Last of the Wild Project, Version 2, 2005 (LWP-2): Global Human Influence Index (HII) Dataset (Geographic). Available from: 10.7927/H4BP00QC. [DOI] [Google Scholar]
- Williams C., Walsh A., Vaglica V., et al. Conservation Policy: Helping or hindering science to unlock properties of plants and fungi. Plants, People, Planet. 2020; 2:535–545. 10.1002/ppp3.10139. [DOI] [Google Scholar]
- Willis, K. 2017. State of the World’s Plants 2017. Report. Royal Botanic Gardens, Kew. [Google Scholar]
- Willis, K. 2018. State of the World’s fungi 2018. Royal Botanic Gardens, Kew. [Google Scholar]
- WWF. 2020. Living Planet Report 2020 – Bending the Curve of Biodiversity Loss. Almond R. E. A., Grooten M., and Petersen T. Eds. Gland, Switzerland: WWF. [Google Scholar]
- Zizka A., Antonelli A. and Silvestro D.. 2020. sampbias, a method for quantifying geographic sampling biases in species distribution data. Ecography. 10.1111/ecog.05102. [DOI] [Google Scholar]
- Zizka A., Silvestro D., Andermann T., Azevedo J., Duarte Ritter C., Edler D., Farooq H., Herdean A., Ariza M., Scharn R.. 2019. CoordinateCleaner: Standardized cleaning of occurrence records from biological collection databases. Methods Ecol. Evol. 10(5):744–751. [Google Scholar]