Summary
Background
Acute infections can cause an individual to have an elevated resting heart rate (RHR) and change their routine daily activities due to the physiological response to the inflammatory insult. Consequently, we aimed to evaluate if population trends of seasonal respiratory infections, such as influenza, could be identified through wearable sensors that collect RHR and sleep data.
Methods
We obtained de-identified sensor data from 200 000 individuals who used a Fitbit wearable device from March 1, 2016, to March 1, 2018, in the USA. We included users who wore a Fitbit for at least 60 days and used the same wearable throughout the entire period, and focused on the top five states with the most Fitbit users in the dataset: California, Texas, New York, Illinois, and Pennsylvania. Inclusion criteria included having a self-reported birth year between 1930 and 2004, height greater than 1 m, and weight greater than 20 kg. We excluded daily measurements with missing RHR, missing wear time, and wear time less than 1000 min per day. We compared sensor data with weekly estimates of influenza-like illness (ILI) rates at the state level, as reported by the US Centers for Disease Control and Prevention (CDC), by identifying weeks in which Fitbit users displayed elevated RHRs and increased sleep levels. For each state, we modelled ILI case counts with a negative binomial model that included 3-week lagged CDC ILI rate data (null model) and the proportion of weekly Fitbit users with elevated RHR and increased sleep duration above a specified threshold (full model). We also evaluated weekly change in ILI rate by linear regression using change in proportion of elevated Fitbit data. Pearson correlation was used to compare predicted versus CDC reported ILI rates.
Findings
We identified 47 249 users in the top five states who wore a Fitbit consistently during the study period, including more than 13·3 million total RHR and sleep measures. We found the Fitbit data significantly improved ILI predictions in all five states, with an average increase in Pearson correlation of 0·12 (SD 0·07) over baseline models, corresponding to an improvement of 6·3–32·9%. Correlations of the final models with the CDC ILI rates ranged from 0·84 to 0·97. Week-to-week changes in the proportion of Fitbit users with abnormal data were associated with week-to-week changes in ILI rates in most cases.
Interpretation
Activity and physiological trackers are increasingly used in the USA and globally to monitor individual health. By accessing these data, it could be possible to improve real-time and geographically refined influenza surveillance. This information could be vital to enact timely outbreak response measures to prevent further transmission of influenza cases during outbreaks.
Introduction
In the USA, approximately 7% of working adults and 20% of children younger than 5 years of age get influenza annually.1 Traditional influenza surveillance relies largely on a combination of virologic and syndromic influenza-like illness (ILI) surveillance to estimate influenza trends.2 However, ILI surveillance has a 1–3 week reporting lag and is often revised weeks later by the US Centers for Disease Control and Prevention (CDC).3
Several groups have attempted to use rapid influenza tests,4 data on internet search terms (eg, Google Flu Trends),5 and social media outlets such as Twitter6 to provide real-time influenza surveillance. However, despite some success, Google Flu Trends was found to miss early waves of the 2009 H1N1 pandemic influenza7 and overestimate activity during outbreaks.7,8 Although Twitter could improve traditional ILI surveillance, it had variable success on its own.6,9 The challenge with using these methods is distinguishing between activity related to an individual’s own illness and those related to media or heightened awareness and interest about influenza during the influenza season. Consequently, there is a great need to enhance traditional ILI surveillance with new objective data streams that can provide real-time information on influenza activity.
A 2016 study estimated that 12% of US consumers owned a fitness band or smartwatch10 and this number continues to grow. Wearable sensors that continuously track an individual’s physiological measurements, such as resting heart rate (RHR), activity, and sleep, might be able to identify abnormal fluctuations indicting perturbations in one’s health, such as an acute infection. It is a normal physiological response to have an elevated RHR as a result of infection, especially when it is accompanied by a fever.11 Sleep and activity are also likely to differ from the norm when someone does not feel well. The purpose of our study was to evaluate whether wearable sensor data could improve influenza surveillance at the state level—so-called nowcasting. Enhanced ILI surveillance would improve the ability to enact quick outbreak response measures to prevent further spread of new influenza strains.
Methods
Data collection
Through a research collaboration between Scripps Research Translational Institute and Fitbit, we obtained de-identified data from a convenience sample of 200 000 consistent users who wore a Fitbit device from March 1, 2016, until March 1, 2018. These users wore their Fitbit for at least 60 days during this study time and had only one Fitbit tracker for the whole period. Inclusion criteria included having a self-reported birth year between 1930 and 2004, height greater than 1 m, and weight greater than 20 kg. User location (ie, state) was only collected for measurements after Dec 1, 2016, and was inferred for the previous period on the basis of the most frequent state reported. To sufficiently measure changes at a population level, we only evaluated users from the top five states with the most Fitbit users in our dataset: California, Texas, New York, Illinois, and Pennsylvania. De-identified Fitbit data were used for this study, which was determined by the Scripps institutional review board to be exempt from institutional review board review. All Fitbit users, including those whose data are used in this study, are notified that their de-identified data could potentially be used for research in the Fitbit Privacy Policy.
The dataset included daily measurements of RHR, sleep minutes from main sleep (ie, the longest sleep of the day), and wear time. Daily measurements with missing RHR, missing wear time, and wear time less than 1000 min per day were excluded from the study dataset. We also excluded data obtained in the first 2 weeks of March, 2016, because Fitbit implemented a change in their RHR algorithm at that time. Daily activity data were not available.
We obtained final end-of-season unweighted ILI rates from the CDC’s FluView database.12 CDC ILI rates are calculated as the weekly percentage of outpatient office visits for ILI, which is defined as fever (temperature >37·8°C) and a cough or sore throat without a known cause other than influenza, and are collected from sentinel surveillance clinics.2
Calculation of the RHR
According to Fitbit, RHR is calculated as follows: periods of still activity during the day are identified by looking at the accelerometer signal provided by the device. If inactivity is observed for a sufficiently long time (eg, 5 min), then it is assumed that the person is in a resting state, and their heart rate at that time is used to estimate their RHR. If the user wears the device to sleep at night, their sleeping heart rate is also used to improve this estimate. Note that the lowest heart rate during sleep can be lower than the RHR since the RHR is intended to capture the heart rate when a user is awake and at rest.13
The manufacturer has found that the estimated RHR based on this algorithm closely matches the value reported by the Fitbit device when measured by users in a supine position immediately after waking.13 The manufacturer has also verified the accuracy of the device in measuring heart rate during still periods by direct comparison with an electrocardiogram (ECG) reference and found a mean average error of less than 1 beat per min (bpm).13 Fitbit devices have shown good agreement with polysomnography and ECGs in measuring sleep and heart rate during sleep, with average heart rate less than 1 bpm lower than that recorded by ECG.14,15
Data analysis
For each user, overall mean (SD) of RHR and sleep duration during the entire study period were calculated. Any users with fewer than 100 RHR measures were excluded. Each user’s weekly RHR and sleep averages were also calculated to align with CDC ILI surveillance data reported on a weekly basis. Users with fewer than four RHR measures during a given week were omitted from downstream analyses pertaining to that week.
We hypothesised that elevated RHR and increased sleep duration compared with an individual’s average might be indicative of ILI. During each week, a user’s data were identified as abnormal if their weekly average exceeded a given threshold: a sleep time that was longer than 0·5 SD below their overall average and an RHR that was either 0·5 SD (model 1) or 1·0 SD (model 2) above their overall average. Additionally, thresholds that included a constant value higher than average were also evaluated. Users were stratified by state, and the proportion of users meeting these thresholds each week was calculated. Thus, for a given state k, the proportion of users with abnormal data for week j is defined as xj,k,l where l represents the 0·5 SD (model 1) or 1·0 SD (model 2) thresholds above average.
The number of CDC-reported ILI cases yj,k among the number of outpatient office visits nj,k during each week over the observation period across each state k was likewise collected. To simplify analytic issues dealing with 0 case counts in a given week, 1 was added to both measures. The proportion of cases in each state (ie, yj,k/nj,k) is defined as pj,k.
Various state-stratified models were considered to evaluate the relationships between ILI rates and Fitbit data. The first naive model, mnaive, simply modelled the CDC ILI case count as a function of the proportion xj,k,l of Fitbit users with abnormal data in a given week using a negative binomial model with offset nj,k. Because CDC ILI data are often delayed by several weeks and later revised, a 3-week lagged autoregressive term pj −3,k was added to the mnaive model to create the mabs model. This model was similar, but more conservative, to the autoregressive AR(3) model used by Yang and colleagues3 to evaluate the predictive power of Google Flu Trends using CDC ILI rates from up to 3 weeks before, and models the absolute ILI count yj,k in each week j. Formally:
where mabs is a negative binomial model with offset term log(nj,k). The H1 model shown assumes the ILI case count yj,k is affected by the proportion of users with abnormal data, whereas the baseline model mabs,H0 omits xj,k,l such that the null hypothesis is H0 : βx = 0 for each state k. Decisions to stratify by state were based on modifications of the mabs model; the modified model combined data across states and included a state main effect and stateby-xj,k,l interaction term:
where 1(k) represents an indicator variable for state k, βk is the coefficient for the main effect, and βx*k is the coefficient for the interaction term. The presence of significant interactions indicated that the effect of the Fitbit variable might differ by state, and thus we opted for a stratified approach.
Finally, we created a linear regression model to predict change in ILI rate from week to week. For each state k, change in ILI rate is given by and change in the proportion of users with abnormal data is given by , and the resulting mchange model more appropriately accounts for autocorrelation that remains present in mabs:
This change is evaluated by linear regression for each state k with elevated sleep and RHR thresholds l. In the first instance, parameters corresponding to the change in proportion xj,k,l of elevated RHR and sleep were of main interest, and compared with models omitting this term. Cross-correlation was used to evaluate 1-week lead (xj − 1,k,l) and 1-week lag (xj + 1,k,l) of the Fitbit data—ie, whether changes in Fitbit data occurred before or after corresponding changes in ILI rates. Pearson correlation (r) was used to compare predicted rates with CDC-reported ILI rates for time-matched, 1-week-lag, and 1-week-lead time periods. Additionally, we assessed correlation using only influenza-season data (week 40 up to week 20 the following year).
Model validation
We did a validation analysis, in which we used data from the first season (season 1: 2016 [week 11]–2017 [week 10]) for model training and data from the second season (season 2: 2017 [week 11]–2018 [week 9]) for model validation. Our validation analysis showed the addition of the Fitbit variable improved the correlations in all states except New York when using just one season of data. When season 2 data were used to predict ILI rates using the model fit with season 1 data, the Fitbit variable also improved correlations in all states except New York (appendix p 10).
We were limited to 2 years of Fitbit data, and therefore only had one season each for training and validation. Consequently, we found that New York, for which ILI cases were not reported during summer weeks in 2017 (season 1), had the lowest correlations for the mabs,H0 and mabs,H1 models compared with the other states (appendix p 10). Additionally, since influenza can peak at different times from season to season, and it had much higher activity in the second season, especially in California and Illinois, the mabs,H0 model had lower Pearson correlations in season 2 than in season 1 in those states. However, overall correlations showed improvements with the addition of the Fitbit variable, and reduced error terms (root mean squared error and mean absolute percentage error), indicating a better overall fit when the Fitbit variable was added to the models (appendix p 9).
Role of the funding source
The funder did not play any role in data collection, analysis, interpretation, writing of the manuscript, or decision to submit. JMR had access to all the data and was responsible for the decision to submit the manuscript. The US National Institutes of Health National Center for Advancing Translational Sciences grant UL1TR002550 supported part of the salary for SRS and EJT. Fitbit pulled the data with input from Scripps Research Translational Institute.
Results
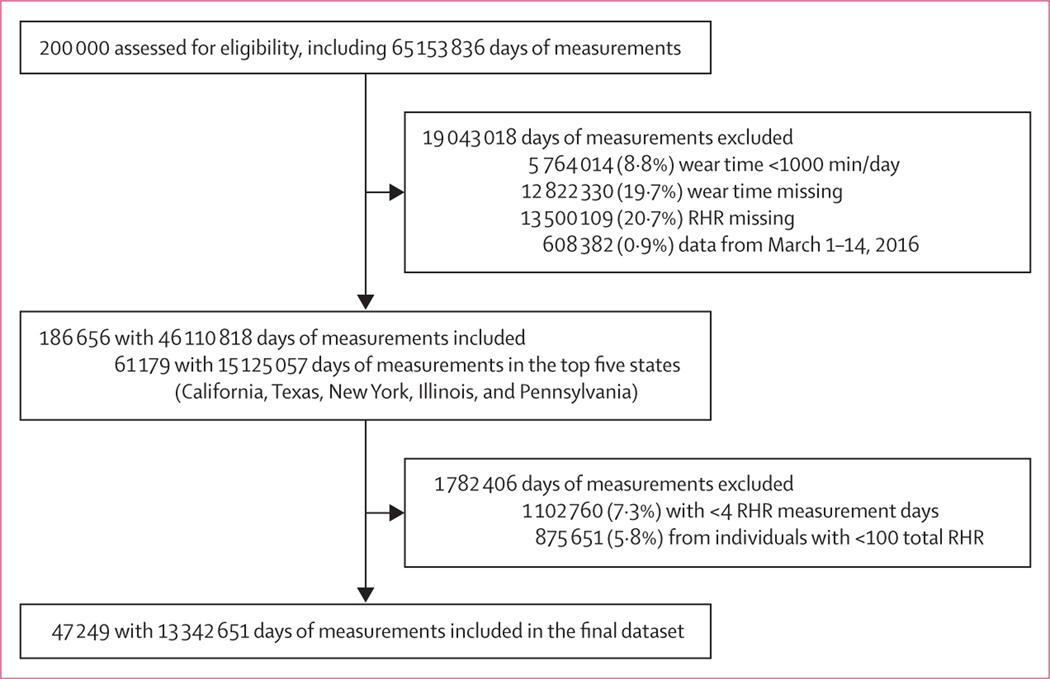
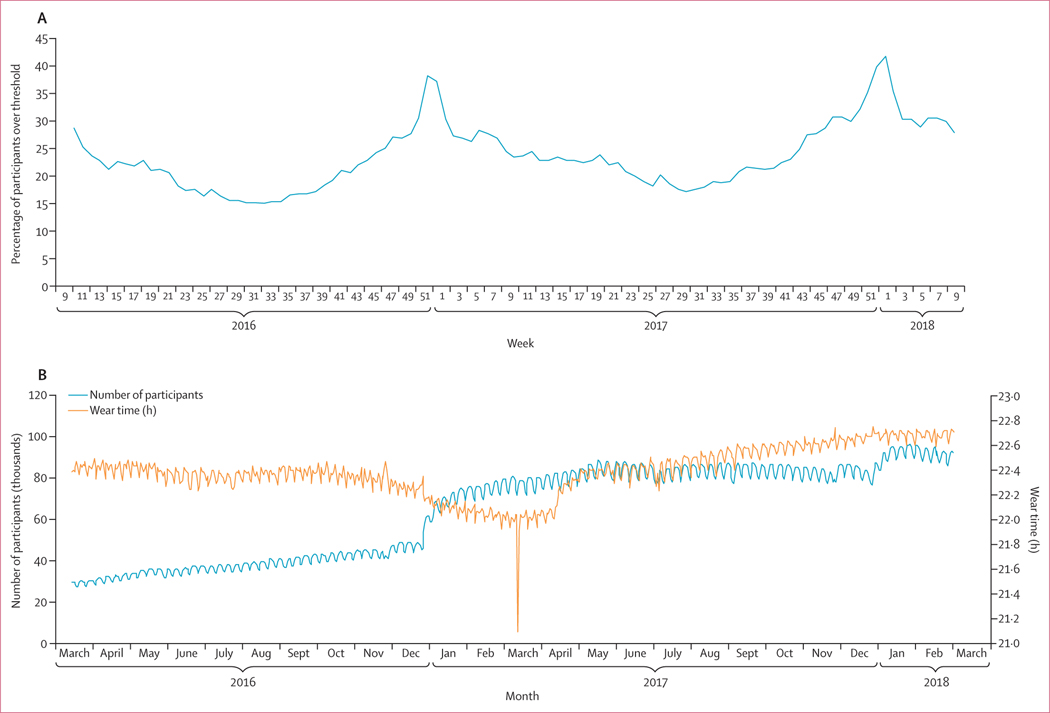
We originally obtained more than 65 million measurements from 200 000 Fitbit users (figure 1). Among those, 47 249 users totalling 13 342 651 daily measurements from five of the most populous states met inclusion criteria (figure 1). The mean age of included individuals was 42·7 years (SD 14·6) and 28 465 (60·2%) were female (table 1). The number of Fitbit users grew during the study period, especially around January, 2017 (figure 2).
Figure 1: Study profile.
RHR=resting heart rate.
Table 1:
Frequency of self-reported participant characteristics by state from March 15, 2016, to March 1, 2018 (n=47 249)
| Top five states (n=47 249) | California (n=13 632) | Texas (n=12 399) | New York (n=7872) | Illinois (n=7132) | Pennsylvania (n=6214) | |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Female | 28 465 (60·2%) | 8126 (60·0%) | 7139 (57·6%) | 4860 (61·7%) | 4444 (62·3%) | 3896 (62·7%) |
| Male | 18 594 (39·4%) | 5457 (40·0%) | 5205 (42·0%) | 3977 (37·8%) | 2658 (37·3%) | 2297 (37·0%) |
| Unknown | 190 (0·4%) | 49 (0·4%) | 55 (0·4%) | 35 (0·4%) | 30 (0·4%) | 21 (0·3%) |
| Age (years) | 42·7 (14·6) | 43·5 (14·9) | 41·9 (14·1) | 42·6 (14·8) | 42·6 (14·6) | 42·7 (14·8) |
| BMI | ||||||
| Underweight (<18·5 kg/m²) | 585 (1·2%) | 175 (1·3%) | 144 (1·2%) | 90 (1·1%) | 98 (1·4%) | 78 (1·3%) |
| Normal (18·5–24·9 kg/m²) | 12 751 (27·0%) | 4034 (29·6%) | 3158 (25·5%) | 2137 (27·2%) | 1822 (25·6%) | 1600 (25·8%) |
| Overweight (25·0–29·9 kg/m²) | 17 064 (36·1%) | 5010 (36·8%) | 4500 (36·3%) | 2890 (36·7%) | 2481 (34·8%) | 2183 (35·1%) |
| Obese (≥30·0 kg/m²) | 16 849 (35·7%) | 4413 (32·4%) | 4597 (36·3%) | 2755 (35·0%) | 2731 (38·3%) | 2353 (37·9%) |
Data are n (%) or mean (SD). BMI=body-mass index.
Figure 2: Percentage of participants with weekly data above threshold of the mnaive model (A) and average daily wear time against number of users (B).
Data are from March 15, 2016, to March 1, 2018. (A) Measurements from 144 360 users from all states were included. Measurements with missing wear time, wear time less than 1000 min/day or missing RHR were excluded, as well as weeks with fewer than four RHR measurements and users with less than 100 total RHR measurements. Model 1 thresholds were used: participants were over the threshold for any given week if they had a sleep time that was greater than 0·5 SD below their overall average and an RHR that was 0·5 SD above their overall average. (B) Measurements from 186 656 users from all states were included. Measurements with missing wear time, wear time less than 1000 min/day, and missing RHR were excluded for this analysis. The sharp downwards spike in wear time in March, 2017, is the result of daylight saving time. RHR=resting heart rate.
On average, users in the full dataset had an RHR of 65·6 bpm (SD 8·4), slept 6·6 h (SD 1·9) per night, and wore their device for 22·5 h (1·6) daily (table 2). RHR and sleep and wear time among users in the final dataset did not vary substantially by state (table 2). SDs for RHR (range 0·2–18·3 bpm) and sleep time (24–336 min) varied considerably from individual to individual.
Table 2:
Number of measurements and average resting heart rate, sleep time, and wear time for full dataset and top five states
| Users | Total measurements | Mean resting heart rate, bpm | Mean sleep time, h | Mean wear time, h | |
|---|---|---|---|---|---|
| USA* | 200 000 | 46 110 818 | 65·6 (8·4) | 6·6 (1·9) | 22·5 (1·6) |
| California | 13 632 | 616 646 | 65·3 (7·6) | 6·5 (0·9) | 22·4 (0·7) |
| Texas | 12 399 | 591 431 | 65·9 (7·8) | 6·6 (0·8) | 22·4 (0·6) |
| New York | 7872 | 351 768 | 65·5 (7·7) | 6·6 (0·9) | 22·4 (0·7) |
| Illinois | 7132 | 340 347 | 66·1 (7·8) | 6·6 (0·9) | 22·5 (0·6) |
| Pennsylvania | 6214 | 286 257 | 66·0 (7·9) | 6·6 (0·9) | 22·4 (0·7) |
Data are n or mean (SD). State data show population averages of individuals’ mean resting heart rate, sleep time, and wear time during entire study period, using data from the final dataset. bpm=beats per min.
Full dataset (before exclusions). Measurements taken from March 15, 2016, to March 1, 2018.
We tested varying levels of data abnormality depend ing on different RHR and sleep measurements. Our model 1 threshold definitions classified 531 648 (24·3%) of 2 186 559 weekly measure ments as abnormal, whereas our model 2 definitions classified 245 060 (11·2%) measurements as abnormal. We found the highest correlation with CDC-reported ILI rates when using the model 1 thresholds—ie, defining abnormal Fitbit data as 0·5 SD above a user’s average RHR combined with sleep more than 0·5 SD below the user’s average—and made it our final model (table 3). We also found that the addition of the sleep threshold improved our models slightly over ones that only incorporated RHR. We found that using an individual’s RHR SD from the entire study period, rather than using a constant value higher than their average, resulted in higher correlations. We also found that the proportion of participants with Fitbit data above the threshold was higher during the 2017–18 influenza season compared with the 2016–17 influenza season (figure 2).
Table 3:
Pearson correlations comparing CDC ILI rates with predicted rates in naive, null, and full negative binomial models and comparing change in CDC ILI rates with change in Fitbit data with a 1-week lag and a 1-week lead
| Negative binomial model predicting ILI case counts |
Linear regression model predicting weekly change in ILI rates |
|||||||
|---|---|---|---|---|---|---|---|---|
| mnaive | mabs,H0 | mabs,H1 | p value* | mchange | mchange(1-week lag) | mchange(1-week lead) | ||
| Model 1 (lower RHR threshold) | ||||||||
| California | 0·92 | 0·73 | 0·97 | <0·0001 | 0·62† | 0·31† | 0·32† | |
| Texas | 0·77 | 0·84 | 0·92 | <0·0001 | 0·24† | 0·22† | 0·10 | |
| New York | 0·33 | 0·79 | 0·84 | <0·0001 | 0·15 | 0·20† | −0·05 | |
| Illinois | 0·72 | 0·80 | 0·92 | <0·0001 | 0·35† | 0·34† | 0·16 | |
| Pennsylvania | 0·48 | 0·78 | 0·89 | <0·0001 | 0·27† | 0·16 | −0·11 | |
| Model 2 (higher RHR threshold) | ||||||||
| California | 0·90 | 0·73 | 0·96 | <0·0001 | 0·66† | 0·36† | 0·28† | |
| Texas | 0·73 | 0·84 | 0·90 | <0·0001 | 0·19 | 0·24† | 0·04 | |
| New York | 0·30 | 0·79 | 0·82 | <0·0001 | 0·11 | 0·19† | −0·06 | |
| Illinois | 0·70 | 0·80 | 0·90 | <0·0001 | 0·35† | 0·42† | 0·08 | |
| Pennsylvania | 0·42 | 0·78 | 0·88 | <0·0001 | 0·23† | 0·24† | −0·14 | |
Individuals were classified as having a week with abnormal Fitbit data if their weekly average exceeded a given threshold: a sleep time that was longer than 0·5 SD below their overall average and an RHR that was either 0·5 SD (model 1) or 1·0 SD (model 2) above their overall average. Naive models included just Fitbit data. H0 models assumed the ILI case count was not affected by the proportion of users with abnormal Fitbit data, whereas H1 models assumed that it was. CDC=US Centers for Disease Control and Prevention. ILI=influenza-like illness.RHR=resting heart rate.
p value comparing H0 to H1 models.
Pearson correlations were significant (p<0·05).
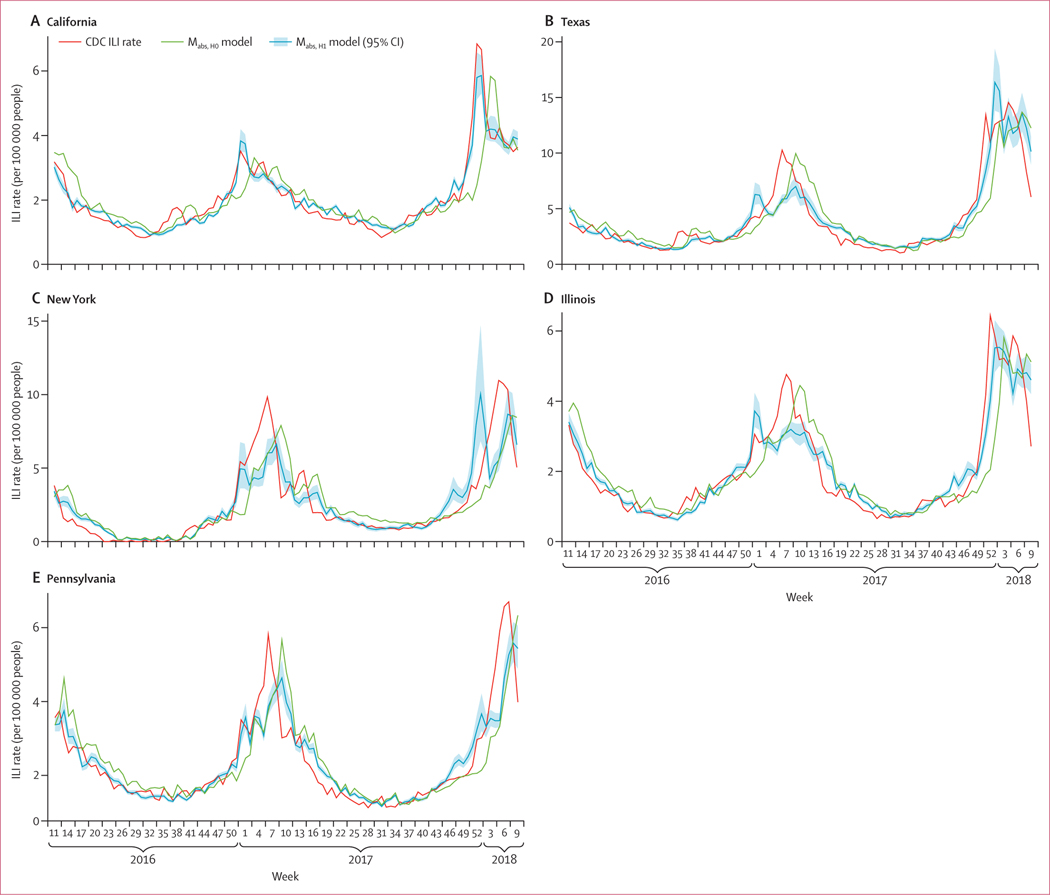
In all states, the mabs,H1 models had significantly higher correlations with ILI rates than the baseline mabs,H0 models, with improvements in Pearson correlations ranging from 6·3% (New York, model 1) to 32·9% (California, model 1), indicating that the Fitbit variable was a significant predictor of ILI (table 3, figure 3). The average increase in Pearson correlation was 0·12 (SD 0·07) over baseline. In general, prediction levels from the full mabs,H1 model were high, although more consistently for model 1, with California having the highest correlation (r = 0·97; p<0·0001) and New York the lowest (r = 0·89; p<0·0001; table 2). We found a significant interaction between the state variable and the Fitbit variable (p<0·0001) in our modified mabs model, indicating that the role of the Fitbit variable varied by state. We also tested the correlation for the same model but restricted to data from the influenza seasons only (ie, week 40 up to week 20 in the following year) and found similar correlations (table 4).
Figure 3: Weekly CDC ILI rates, predicted ILI rates from the baseline mabs,H0 model, and predicted rates and 95% CIs for the mabs,H1 model, by state.
Model 1 is used, with the lower heart rate cutoff. Data are from March 16, 2016, to March 1, 2018. CDC=Centers for Disease Control and Prevention. ILI=influenza-like illness.
Table 4:
Pearson correlations from model 1 restricted to influenza season only comparing CDC ILI rates with predicted rates in naive, null, and full negative binomial models and comparing change in CDC ILI rates with change in Fitbit data with a 1-week lag and a 1-week lead
| Negative binomial model predicting ILI case counts |
Linear regression model predicting weekly change in ILI rates |
|||||||
|---|---|---|---|---|---|---|---|---|
| mnaive | mabs,H0 | mabs,H1 | p value* | mchange | mchange (1-week lag) | mchange(1-week lead) | ||
| California | 0·91 | 0·61 | 0·97 | <0·0001 | 0·71† | 0·32† | 0·33† | |
| Texas | 0·72 | 0·79 | 0·89 | <0·0001 | 0·27† | 0·20 | 0·11 | |
| New York | 0·31 | 0·71 | 0·79 | <0·0001 | 0·15 | 0·21 | −0·07 | |
| Illinois | 0·61 | 0·71 | 0·88 | <0·0001 | 0·42† | 0·37† | 0·13 | |
| Pennsylvania | 0·34 | 0·71 | 0·85 | <0·0001 | 0·29† | 0·16 | −0·11 | |
Influenza season is defined as week 40 to week 20 in the following year. Individuals were classified as having a week with abnormal Fitbit data if their weekly average exceeded a given threshold: a sleep time that was longer than 0·5 SD below their overall average and an RHR that was 0·5 SD (model 1) above their overall average. Naive models included just Fitbit data. H0 models assumed the ILI case count was not affected by the proportion of users with abnormal Fitbit data, whereas H1 models assumed that it was. CDC=US Centers for Disease Control and Prevention. ILI=influenza-like illness. RHR=resting heart rate.
p value comparing H0 to H1 models.
Pearson correlations were significant (p<0·05).
When modelling the change in ILI rates from one week to the next, the mchange models mostly showed statistical association with proportions of elevated weekly RHR and abnormal sleep across all states, for either the time-matched or lagged data, at either RHR threshold (table 3). Inspection of the cross-correlation between fitted and observed models showed the Fitbit data generally did not lead the ILI rate data—that is, changes in Fitbit data were not observed before changes in ILI rate data. Instead, it was more common that Fitbit changes occurred in the week of changes in ILI rates (time matched) or in the following week (1-week lag). This implies that the changes in Fitbit data occur during or after the changes in ILI rates, and are therefore less predictive at forecasting future ILI events.
Discussion
Improved characterisation of an individual’s average values through wearable sensors will allow us to better identify deviations that could indicate the incidence of acute disease states, such as cold and influenza infections. To our knowledge, this is the first study to evaluate the use of RHR and sleep data in a large population to predict real-time ILI rates at the state level. We saw significant improvements in our ability to predict influenza when incorporating the proportion of users with abnormal sleep and RHR values in our full mabs,H1 model and in our mchange model, as well as reduced prediction errors (appendix p 9). Currently, CDC ILI data are typically reported 1–3 weeks late and reported numbers are often revised months later. The ability to harness wearable device data at a large scale might help to improve objective, real-time estimates of ILI rates at a more local level, giving public health responders the ability to act quickly and precisely on suspected outbreaks.
When someone is unwell, their RHR increases, their total sleep is likely to increase, and their activity is likely to decline. However, an elevated amount of sleep or elevated RHR for one person might be a normal level for someone else. Consequently, tracking an individual’s physiological changes over time and comparing their values over time to their individual norm or average could be a means of identifying assaults to their health. Our findings also supported the benefit of using individual health metrics: in our models, we found higher correlations from our predicted values with CDC ILI rates when we used an individual’s SD above normal to identify abnormal values instead of using the same value above average across the entire population.
The impact of infections on an individual’s RHR has been documented in several studies. One study found ill participants had RHRs that were elevated by 2·02–4·66 SD above their normal measurements.16 A study that examined 27 young men with acute febrile infections found that heart rates increased by 8·5 bpm per every 1°C increase in temperature.11 Similarly, a study among children with acute infections found that heart rate rose by 9·9–14·1 bpm for every 1°C increase in temperature, with higher increases in younger children.17 These studies indicate that infections can increase heart rate, probably due to increased body temperature and inflammatory responses as the body fights off an infection.
Our mchange models were better at predicting change with a 1-week lag compared with a 1-week lead. It is possible that an ILI infection results in an elevated RHR for several weeks after initial infection. Previous studies have also indicated that an elevated heart rate can occur before symptom onset.16 Since influenza has an incubation period of 1–4 days, there is only a short opportunity to identify infections before symptom onset. However, since individuals with febrile respiratory illness typically seek care 3–8 days after symptom onset,18 it is conceivable that ILI cases could be identified via sensor data earlier than through traditional, clinic-based ILI surveillance. Early identification via our method might be more likely if rates were predicted at a daily, rather than weekly, rate.
Lack of sleep can be a marker of stress, which can also raise RHR. In our study, users were considered to have normal sleep values if their weekly sleep average was less than 0·5 SD below their overall sleep average, as nights of short sleep duration have been shown to result in elevated heart rate the following day.19,20 We found that our correlations improved slightly when we classified people as displaying normal values when they had low sleep. In the future, improved measurements of stress by wearable devices, either by detection of voice changes or galvanic skin response, could further improve our ability to identify other non-infectious causes of elevated RHR.
Previous models to predict ILI rates have mainly used International Classification of Diseases codes,21 ILInet (CDC’s influenza database), Twitter, Google Flu Trends, Wikipedia, weather, crowd-sourced data, and school vacation schedule data.22 However, Twitter, Google Flu Trends, Wikipedia, and self-reported crowd-sourced data—and even ILInet—are all affected by outside factors such as media coverage of the influenza, with more of the so-called worried well seeking care or searching for information about influenza during epidemic periods. Use of sensor-based data would offer the first objective and real-time measurement of illness in a population that could potentially reduce the effect of overestimation during epidemics
By incorporating Fitbit data, we were able to improve ILI predictions at the state level. The predicted values from our mnaive model that just used the Fitbit variable with no lag indicate that this sensor-based method could potentially be useful on its own in local regions where ILI surveillance data might not be available. With greater volumes of data to analyse, this sensor-based surveillance method could be applied to more geo graphically refined areas in the future, such as county-level or city-level data.
Variation in individual characteristics can affect illness risk and physiological response to illness. In general, owners of wearable devices are usually wealthier than the general population, potentially making them less likely to have comorbidities that could make them more susceptible to severe infections. Additionally, these users might be more likely to get influenza vaccines or receive antivirals or other medications if they do get sick, which could reduce disease severity. A study that administered intravenous acetaminophen to critically ill febrile patients found that it significantly reduced their heart rate after 2 h.23 Individuals with comorbidities, as well as young children and people older than 65 years, typically have more severe responses to influenza infections24,25 and could have higher heart rate responses. In the future, understanding the role of individual characteristics such as age, comorbidities, obesity, and sex on abnormal values will be important for improving ILI prediction using this method.
It is likely that non-influenza or even non-respiratory infections are also captured by our Fitbit variable, which predominately relies on elevated RHR. It is possible that different infections, or even different influenza strains, could result in different physiological responses, with varying changes of heart rate or length of elevation. For example, H3N2 typically causes more severe illness24,25 than other strains. Like the CDC, which identified higher rates of ILI for 2017–18, we also saw higher peaks of the proportion of users with elevated Fitbit data during this influenza season compared with the previous year. It is also possible that our algorithm could pick up less severe infections that would not necessarily be captured by traditional ILI surveillance, which requires a visit to a health-care provider. Future work to better understand typical heart rate responses to specific viral or bacterial infections or even different influenza subtypes could improve our ability to track infections.
Additionally, there are external factors, other than illness, that could influence a person’s RHR and sleep. It is possible that our model is capturing some seasonal trends in RHR from changes in activity, holidays, or weather, rather than changes that result from just influenza or cold infections. Winter holidays have been associated with changes in weight gain,26 social mixing, increases in health-care seeking, differences in surveillance reporting,27 and potentially changes in alcohol consumption and stress. These factors could increase susceptibility to infection and can also affect ILI surveillance. A study found that RHR is higher at very cold or hot temperatures28 and heart rate can also be elevated when someone is dehydrated, which could be more likely to happen during certain seasons. Additionally, people might be less active during colder, winter months, resulting in deconditioning and increased heart rate. Future prospective studies should attempt to measure and adjust for these external variables and link individual Fitbit data to reported symptoms or laboratory influenza confirmation.
Our data had several limitations, including no activity data, which is typically collected by Fitbit devices. An activity variable could have improved the predictive ability of our models by allowing us to control for seasonal fitness changes or more short-term activity changes that could result from an illness. Another limitation is that our weekly RHR averages might include both days when an individual is sick and days when they are not sick, and therefore might be calculated using both normal and abnormal RHR and sleep measurements. Consequently, this could result in underestimation of illness by lowering the weekly averages. Additionally, sleep measuring devices have been found to have low accuracy.29 However, accuracy of devices will continue to improve as technology evolves.
Every year, up to 650 000 people die from influenza, globally.30 Quick detection of increases in ILI, indicating potential influenza epidemics, is key to early initiation of important non-pharmaceutical (eg, staying home when sick or handwashing) and pharmaceutical interventions (deploying antivirals and vaccines) that can help to prevent further spread and infection in the most susceptible populations. This study shows that using RHR and other metrics from wearables has the potential to improve real-time ILI surveillance. New wearables that include continuous sensors for temperature, blood pressure, pulse oximetry, ECG, or even cough recognition 31,32 are likely to further improve our ability to identify population and even individual-level influenza activity. In the future, with access to real-time data from these devices, it might be possible to identify ILI rates on a daily, instead of weekly, basis, providing even more timely surveillance. As these devices become more ubiquitous, this sensor-based surveillance technique could even be applied at a more global level where surveillance sites and laboratories are not always available.
Supplementary Material
Research in context.
Evidence before this study
Influenza results in up to 650 000 deaths worldwide each year. Traditional influenza surveillance reporting in the USA and globally is often delayed by 1–3 weeks, if not more, and revised months later. This delay can allow outbreaks to go unnoticed, quickly spreading to new susceptible populations and geographical regions. We searched PubMed from Jan 1, 1990, to July 20, 2019, using combinations of words or terms that included “influenza” OR “influenza-like illness” AND “predictions” OR “modeling” OR “nowcasting”. Previous studies have attempted to use crowd-sourced data, such as Google Flu Trends and Twitter, to provide real-time influenza surveillance information—a method known as nowcasting. However, these methods typically overestimate rates during epidemic periods and have variable success on their own, especially at the state level.
Added value of this study
To our knowledge, this is the first study to evaluate and show that objective data collected from wearables significantly improved nowcasting of influenza-like illness. This result held in all five states that we examined, with an average increase in Pearson correlation of 0·12 over baseline, resulting in correlations ranging from 0·84 to 0·97 in the final models. These associations remained consistent when correcting for first-order autocorrelation in time-matched or 1-week-lagged models.
Implications of all the available evidence
In the future, wearables could include additional sensors to prospectively track blood pressure, temperature, electrocardiogram, and cough analysis, which could be used to further characterise an individual’s baseline and identify abnormalities. Future prospective studies will help to differentiate deviations from an individual’s normal levels resulting from infectious versus non-infectious causes, and might even be able to identify infections before symptom onset. Capturing physiological and behavioural data from a growing number of wearable device users globally could greatly improve timeliness and precision of public health responses and even inform individual clinical care. It could also fill major gaps in regions where influenza surveillance data are not available.
Acknowledgments
This study was supported in part by the US National Institutes of Health National Center for Advancing Translational Sciences grant UL1TR001114.
Funding Partly supported by the US National Institutes of Health National Center for Advancing Translational Sciences.
Footnotes
Declaration of interests
We declare no competing interests.
Data sharing
Programming code can be requested by contacting the corresponding author. Data were made available by Fitbit according to appropriate security control and research agreements. For more on Fitbit’s work with the research community, please see the Fitbit Research Pledge.
References
- 1.Molinari NA, Ortega-Sanchez IR, Messonnier ML, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 2007; 25: 5086–96. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Influenza (flu): overview of influenza surveillance in the United States. Oct 15, 2019. https://www.cdc.gov/flu/weekly/overview.htm (accessed Jan 7, 2020).
- 3.Yang S, Santillana M, Kou SC. Accurate estimation of influenza epidemics using Google search data via ARGO. Proc Natl Acad Sci USA 2015; 112: 14473–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gren LH, Porucznik CA, Joy EA, Lyon JL, Staes CJ, Alder SC. Point-of-care testing as an influenza surveillance tool: methodology and lessons learned from implementation. Influenza Res Treat 2013; 2013: 242970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature 2009; 457: 1012–14. [DOI] [PubMed] [Google Scholar]
- 6.Paul MJ, Dredze M, Broniatowski D. Twitter improves influenza forecasting. PLoS Curr 2014; 6: ecurrents.outbreaks.90b9ed0f59bae4ccaa683a39865d9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olson DR, Konty KJ, Paladini M, Viboud C, Simonsen L. Reassessing Google Flu Trends data for detection of seasonal and pandemic influenza: a comparative epidemiological study at three geographic scales. PLoS Comput Biol 2013; 9: e1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler D. When Google got flu wrong. Nature 2013; 494: 155–56. [DOI] [PubMed] [Google Scholar]
- 9.Nagel AC, Tsou MH, Spitzberg BH, et al. The complex relationship of realspace events and messages in cyberspace: case study of influenza and pertussis using tweets. J Med Internet Res 2013; 15: e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai A. Study: 12 percent of US consumers own a fitness band or smartwatch. mobihealthnews. May 5, 2016. http://www.mobihealthnews.com/content/study-12-percent-us-consumers-ownfitness-band-or-smartwatch (accessed Jan 7, 2020). [Google Scholar]
- 11.Karjalainen J, Viitasalo M. Fever and cardiac rhythm. Arch Intern Med 1986; 146: 1169–71. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. FluView: national, regional, and state level outpatient illness and virus surveillance. December 15, 2017. https://gis.cdc.gov/grasp/fluview/fluportaldashboard.html (accessed Jan 7, 2020).
- 13.Heneghan C, Venkatraman S, Russell A. Investigation of an estimate of daily resting heart rate using a consumer wearable device. medRxiv 2019; published online Oct 18. DOI: 10.1101/19008771 (preprint). [DOI] [Google Scholar]
- 14.de Zambotti M, Baker FC, Willoughby AR, et al. Measures of sleep and cardiac functioning during sleep using a multi-sensory commercially-available wristband in adolescents. Physiol Behav 2016; 158: 143–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haghayegh S, Khoshnevis S, Smolensky MH, Diller KR. Accuracy of PurePulse photoplethysmography technology of Fitbit Charge 2 for assessment of heart rate during sleep. Chronobiol Int 2019; 36: 927–33. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Dunn J, Salins D, et al. Digital health: tracking physiomes and activity using wearable biosensors reveals useful health-related information. PLoS Biol 2017; 15: e2001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson M, Harnden A, Perera R, et al. Deriving temperature and age appropriate heart rate centiles for children with acute infections. Arch Dis Child 2009; 94: 361–65. [DOI] [PubMed] [Google Scholar]
- 18.Radin JM, Hawksworth AW, Kammerer PE, et al. Epidemiology of pathogen-specific respiratory infections among three US populations. PLoS One 2014; 9: e114871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antunes BM, Campos EZ, Parmezzani SS, Santos RV, Franchini E, Lira FS. Sleep quality and duration are associated with performance in maximal incremental test. Physiol Behav 2017; 177: 252–56. [DOI] [PubMed] [Google Scholar]
- 20.Kuetting DLR, Feisst A, Sprinkart AM, et al. Effects of a 24-hr-shiftrelated short-term sleep deprivation on cardiac function: a cardiac magnetic resonance-based study. J Sleep Res 2019; 28: e12665. [DOI] [PubMed] [Google Scholar]
- 21.Gog JR, Ballesteros S, Viboud C, et al. Spatial transmission of 2009 pandemic influenza in the US. PLoS Comput Biol 2014; 10: e1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGowan CJ, Biggerstaff M, Johansson M, et al. Collaborative efforts to forecast seasonal influenza in the United States, 2015–2016. Sci Rep 2019; 9: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schell-Chaple HM, Liu KD, Matthay MA, Sessler DI, Puntillo KA. Effects of IV acetaminophen on core body temperature and hemodynamic responses in febrile critically ill adults: a randomized controlled trial. Crit Care Med 2017; 45: 1199–207. [DOI] [PubMed] [Google Scholar]
- 24.Thompson WW, Shay DK, Weintraub E, et al. Influenza-associated hospitalizations in the United States. JAMA 2004; 292: 1333–40. [DOI] [PubMed] [Google Scholar]
- 25.Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003; 289: 179–86. [DOI] [PubMed] [Google Scholar]
- 26.Helander EE, Wansink B, Chieh A. Weight gain over the holidays in three countries. N Engl J Med 2016; 375: 1200–02. [DOI] [PubMed] [Google Scholar]
- 27.Gao H, Wong KK, Zheteyeva Y, Shi J, Uzicanin A, Rainey JJ. Comparing observed with predicted weekly influenza-like illness rates during the winter holiday break, United States, 2004–2013. PLoS One 2015; 10: e0143791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madaniyazi L, Zhou Y, Li S, et al. Outdoor temperature, heart rate and blood pressure in Chinese adults: effect modification by individual characteristics. Sci Rep 2016; 6: 21003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J, Finkelstein J. Consumer sleep tracking devices: a critical review. Stud Health Technol Inform 2015; 210: 458–60. [PubMed] [Google Scholar]
- 30.WHO. Up to 650 000 people die of respiratory diseases linked to seasonal flu each year. December 14, 2017. https://www.who.int/en/newsroom/detail/14-12-2017-up-to-650-000-people-die-of-respiratorydiseases-linked-to-seasonal-flu-each-year (accessed Jan 7, 2020).
- 31.Abeyratne UR, Swarnkar V, Setyati A, Triasih R. Cough sound analysis can rapidly diagnose childhood pneumonia. Ann Biomed Eng 2013; 41: 2448–62. [DOI] [PubMed] [Google Scholar]
- 32.Amrulloh YA, Abeyratne UR, Swarnkar V, Triasih R, Setyati A. Automatic cough segmentation from non-contact sound recordings in pediatric wards. Biomed Signal Proces 2015; 21: 126–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.