Abstract
Three‐dimensional quantitative phase imaging is an emerging method, which provides the 3D distribution of the refractive index (RI) and the dry mass in live and fixed cells as well as in tissues. However, an insufficiently answered question is the influence of chemical cell fixation procedures on the results of RI reconstructions. Therefore, this work is devoted to systematic investigations on the RI in cellular organelles of live and fixed cells including nucleus, nucleolus, nucleoplasm, and cytoplasm. The research was carried out on four different cell lines using a common paraformaldehyde (PFA)‐based fixation protocol. The selected cell types represent the diversity of mammalian cells and therefore the results presented provide a picture of fixation caused RI changes in a broader context. A commercial Tomocube HT‐1S device was used for 3D RI acquisition. The changes in the RI values after the fixation process are detected in the reconstructed phase distributions and amount to the order of 10−3. The RI values decrease and the observed RI changes are found to be different between various cell lines; however, all of them show the most significant loss in the nucleolus. In conclusion, our study demonstrates the evident need for standardized preparation procedures in phase tomographic measurements. © 2020 The Authors. Cytometry Part A published by Wiley Periodicals LLC. on behalf of International Society for Advancement of Cytometry.
Keywords: holography, holographic tomography, refractive index, cell analysis, paraformaldehyde fixation, quantitative phase imaging, tomographic phase microscopy
Introductions
Label‐free three‐dimensional quantitative phase imaging (3D QPI) is developing to an important tool for visualization and measuring physical and biological properties of cells and tissues (1, 2). Minimally invasive measurements of optical properties of biological objects without chemical markers provide new information about the investigated samples. The refractive index (RI), which is the basic optical parameters in 3D QPI, is closely related to the mass concentration in biological samples (3, 4). Measuring and visualization of the 2D and 3D distribution of the RI quantifies in detail morphological and biochemical internal structures of cells (5, 6) and tissues (7, 8). In order to introduce this quantitative parameter to life science applications and medical diagnostics, it is necessary to develop standard sample preparation and data processing procedures for usage in laboratories. Standardized protocols for the preparation of biological samples, for example, drying methods used for scanning electron microscopy (SEM), paraformaldehyde fixation protocols for QPI and fluorescence microscopy or tissue clearing as used for confocal microscopy, optical diffraction tomography (ODT), optical coherence tomography (OCT) or Raman microscopy, are critical for interlaboratory comparison of the results virtual staining (9), for remote diagnostics (7) as well as for novel methods of enhanced 3D RI distribution determination and analysis by use of artificial intelligence (AI) or machine learning (10, 11, 12).
Among a variety of 3D QPI methods, tomographic phase microscopy (TPM) (13, 14, 15) represents a wide range of label‐free methods that aim to reconstruct volumetric distribution of refractive index (RI). Within the TPM methods, the coherent linear ODT represents the oldest and most‐established technique, which has recently created and demonstrated a wealth of possibilities for 3D imaging of live and fixed cells and tissues (16). This is already reflected in the presence of commercial devices, namely the products of Nanolive Ltd (17) and Tomocube, Inc. (18), which become more and more popular among biomedical community. Both systems represent the laser‐driven limited‐angle variant of ODT (LAODT) with holographic projections (latter referred as LA holographic tomography, LAHT) and the scanning illumination configuration. It is relatively easy to implement and appropriate for characterization of adhesive cells and cell cultures directly on a Petri dish and tissues at sample slides. Unfortunately, despite indisputable advantages, the principle of operation of LAODT is a source of its biggest drawback known as the “missing cone” problem, namely there is a limited angular range of illumination directions, within which the holographic projections can be acquired. This inherent property of LAHT results in highly distorted tomographic reconstructions of analyzed samples along the optical axis of an optical system when simple reconstruction procedures, like direct inversion (19), are used. This problem may be solved through hardware modifications either by using hybrid approach in which the sample is rotated for consecutive LAHT reconstructions and their spectra are combined for full object representation (20) or by acquiring projections of a freely rotated sample in a fluidic system (21). This, however, does not allow to avoid any mechanical perturbation of the measured biological object and it is difficult to implement for investigations of samples at a Petri dish or a sample slide. In many LAHT systems, the “missing cone” problem is attempted to be solved by using a software approach with the most popular Gerchberg–Papoulis (GP) method with various regularization schemes (22). The currently best algorithmic solution, which offers increased reconstruction quality with highly minimized LAHT artifacts, is the total variation iterative constraint method (TVIC) combined with GP algorithm (23). Another, recently introduced, approach, which shows ability to reconstruct RI correctly in whole measurement volume, is utilizing a deep neural network (DNN) (24). However, the method is object specific and presently it is difficult for efficient usage in the commercial systems.
The RI distribution at cellular scale provided by 3D QPI can be obtained by means of two typical approaches for analysis, such as measuring live cells or chemically fixed ones. Chemical fixation of biological samples is common and often necessary, especially when cryofixation is not applicable or correlative imaging with other modalities like staining with dyes or fluorescence markers is required. The chemicals used to fix cells allow to preserve their shape, but they also introduce artifacts that affect sample structures or may interfere with the measurement (25). The mechanisms of fixation depend on the reagent used. Alcohol‐based fixations dehydrate cells, can cause proteins to denature and precipitate in situ (26, 27). Paraformaldehyde (PFA) induces covalent cross links between molecules, effectively gluing them together into an insoluble meshwork (28). Since chemical fixation strongly can change the internal structure of cells, it is reasonable to assume that it influences physical properties such as the RI values as well.
In order to get reliable and repeatable QPI results, there is a need for standardized preparation protocols of live and fixed cells. However, currently, there is lack of information and data about these procedures and their effect on RI values across various cell lines. In previous studies, the RI of organelles from HeLa cells (15, 29), HUH‐7 cells (6), and Jurkat cells (30) was analyzed. The data published show that the RI values for specific organelles are within similar limits but depend on the cell type. In order to use QPI tools and RI as a biomedical marker, thus it is necessary to collect data for a library of RI values for a wide range of cell lines and their internal organelles. Previous studies have also reported that there were changes in cell and tissue volumes after fixation (31). Also, mechanical properties of samples after PFA fixation were investigated (32) and the effects of chemical fixation on the cellular nanostructure were analyzed (25). However, a systematic approach for the determination of the PFA fixation process effect on the internal structures of cells, their RI distribution and dry mass changes after fixation have not been fully elucidated. Up to now, there are just a few published studies that carry out 3D RI measurements and such analyses have been conducted only for simple internal structures such as lipid droplets (6) and only for slightly internally differentiated cells (32).
This article aims to increase our knowledge on the effect of cell fixation on RI distribution and cell mass density. For this reason, the measurements by means of holographic tomography on RAW 264.7 mouse macrophages, NIH‐3T3 mouse embryonic fibroblasts, RLE‐6TN rat lung epithelial cells and NRK‐52E rat kidney epithelial cells cell lines were carried out.
Materials and Methods
Cell Lines and Cell Culture
In our study, we used four different cell lines that were chosen in order to consider features and cellular diversity of different organs of origin (33): NIH‐3T3 mouse fibroblasts (connective tissue), NRK‐52E rat epithelial cells (kidney), RLE‐6TN alveolar epithelial cells (lung), and RAW 264.7 mouse macrophages (immune system). Moreover, the cell lines were selected as they were found highly feasible for usage in earlier in vitro studies on the analysis and mechanisms of various different cytotoxic nanomaterials (33, 34, 35) and the successful application in the exploration of novel approaches for the label‐free quantification of nano‐cytotoxic effects and to probe the uptake of nanoparticles (36).
We performed all experiments with commercially available cell lines. All cell lines were cultured according to standard cell culture procedures without antibiotics. RAW 264.7 mouse macrophages, obtained by ATCC (No. ATCC® TIB‐71TM), were cultured in Dulbecco's modified eagle medium (DMEM) supplemented with 10% focal calf serum (FCS), 1 mM pyruvate, and 2 mM glutamine, passaged twice a week. NIH‐3T3 mouse embryonic fibroblasts, obtained by ATCC (No. ATCC® CRL‐1658TM), were cultured in DMEM supplemented with 10% FCS, 1 mM pyruvate, and 2 mM glutamine, passaged three times a week. NRK‐2E rat kidney epithelial cells, obtained by ATCC (No ATCC® CRL‐1571TM), were cultured in DMEM supplemented with 10% FCS, 1 mM pyruvate, and 2 mM glutamine, passaged twice times a week. RLE‐6TN rat lung epithelial cells, obtained by ATCC (No. ATCC® CRL‐2300TM), cultured in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% FCS, 1 mM pyruvate, and 2 mM glutamine, passaged twice times a week, passages 5–25 used for holotomography experiments.
For the experiments with fixation, cells were harvested and seeded into Tomodishes (Tomocube, Inc., Daejeon, Republic of Korea) from the culture and cultivated overnight (for RAW 264.7‐30,000 cells per Petri dish, for NIH‐3T3, NRK‐52E, RLE‐6TN – 10,000 cells per Petri dish in 800 μl of medium). Then the medium was removed and dishes were washed with 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES) buffered medium (with 850 mg/l NaHCO3; Lonza HEPES buffer 1 M, 17‐737E). 3D‐QPI measurements were performed with special medium prepared to preserve stable pH value, without CO2 atmosphere, with 10x DMEM (Biochrom, order F 0455), 4 mM l‐glutamine (200 mM, sample Biochrom, order K0283), 20 mM HESPES (1 mM, Biochrom, order L1613), 1 mM sodium pyruvate (100 mM, Biochrom, order L0473), 10% FBS Good (PAN Biotech), 850 mg/l NaHCO3 (7.5% sodium bicarbonate, Biochrom, order L1713). In the next step, the dish was prepared for the 3D‐QPI measurements by mounting in the above‐mentioned medium and covered with a cover glass or provided to further fixation procedures.
Fixation Protocol
Paraformaldehyde fixation is one of the most widely used fixation methods in cell biology and histopathology. Most frequently used concentrations of the fixative solution are 4% or 2% PFA for standard fixation protocols. An advantage of PFA fixation is the preservation of cell structures and antigens. In this study, 2% PFA solution was used. For fixation, overnight cultivated cells were washed with warm phosphate buffered saline (PBS, Sigma, Dulbecco's Phosphate Buffered Saline, order D8537, with MgCl, CaCl), with the same density as for experiment with live cells. In the next step, 2% PFA (electron microscopy sciences, 16% paraformaldehyde, methanol free) in PBS (without MgCl, CaCl) as a fixation solution were added and left for 2 h at the room temperature. Then dishes were washed three times with PBS (without MgCl, CaCl), and a glass cover slip as placed on the top of the dish.
Data Acquisition and 3D Refractive Index Reconstruction
The 3D refractive index distribution in cells was obtained by means of a limited‐angle holographic tomography (LAHT) approach, which is the one of the most popular version of optical diffraction tomography in biomedical applications (2, 14, 22). In LAHT, the projections required for the 3D RI reconstruction are captured as individual holographic images of a sample that had been illuminated from different angles by scanning object illuminating beam while the camera and the sample are stationary. The hologram acquisition was performed using a commercial off‐axis Mach–Zehnder interferometric setup with a digital micromirror device, DMD (HT‐1S; Tomocube Inc., Daejeon, Republic of Korea) (Fig. 1a). The incident angles of an object illumination beam were rapidly scanned by projecting a series of binary holograms on the installed DMD (37, 38). The light scattered by the sample was transmitted to the camera by means of an imaging system consisting of an objective lens (60×, numerical aperture NA = 0.8) and a tube lens. The object beam interfered with a plane reference beam on the camera plane and in‐plane off‐axis holograms were captured. 3D‐QPI measurements were performed in the laboratory with ambient temperature of (25°C). Additional effort has been made to properly calibrate the instrument and to follow the same measurement protocol during the whole study (one calibration procedure per five measurements).
Figure 1.
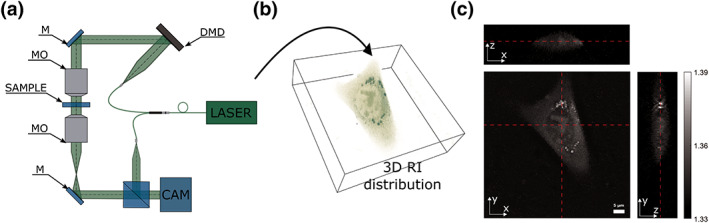
(a) Optical set‐up of the applied holotomographic microscope (HT‐1S, Tomocube Inc., Daejeon, Republic of Korea), MO – Microscope objectives, M – Mirrors, DMD ‐ digital micromirror device (adapted from reference (25)); (b) 3D RI distribution visualization; (c) refractive index distribution on x‐y, x‐z, and y‐z cross sections in live cell from NIH3T3 line. Cross sections location is marked with red, dashed lines. [Color figure can be viewed at wileyonlinelibrary.com]
The phase and amplitude (the complex amplitude) of the scattered light were retrieved from the recorded holograms and then processed for reconstruction of the 3D RI map (Fig. 1b). The tomographic reconstruction algorithm is based on the Fourier diffraction theorem (19) under the first‐order Rytov approximation (39). In short, during this procedure, the 2D Fourier spectrum of the measured complex amplitude of a sample illuminated at a certain angle is mapped to the surface of a hemisphere called the Ewald sphere in the 3D Fourier space. The 3D Fourier space filled with the spectra is than 3D inverse Fourier transformed providing a 3D RI distribution. However, due to the limited angle of numerical aperture of the imaging system (NA = 0.8), there is missing some information in the 3D Fourier spectrum of the sample. To fill this information, the iterative constraint algorithm is executed to the tomograms reconstructed from the diffraction algorithm. The detailed descriptions of the reconstruction algorithm and its constrains implemented in the commercial software (TomoStudio 2.6.24, Tomocube, Inc., Daejeon, Republic of Korea), which was used for 3D refractive index calculations can be found elsewhere (40). However, the applied tomographic reconstruction procedure does not fully overcome the “missing cone” problem and anisotropic resolution along z‐axis (41). This is illustrated for the example of representative RI cross sections in the (x‐z) and (y‐z) planes that are depicted in Figure 1c. On the other hand, it was shown that the RI distribution in the best focused (x‐y) plane provides the retrieval of very stable and highly accurate values (41, 42). Therefore, in this work, it was decided to perform the systematic analysis of refractive index in live and fixed cells based on the 2D RI values distribution reconstructed at the best focused plane of the analyzed samples.
Refractive Index Images Processing
The full scheme of the evaluation process for the retrieval of refractive index images is illustrated in Figure 2. At the first stage, the holograms acquired through the 3D‐QPI microscope were used for determination of the three‐dimensional distribution of the refractive index of the sample by utilizing the TomoStudio software. All 3D visualizations were rendered using the open‐source tomography platform tomviz (43). Next, the 2D RI distributions (x‐y plane) were selected manually from 3D RI data, starting from default middle position in the reconstruction and introducing manual correction for the position based on the best contrast of the substructures. Each selected 2D RI image underwent global (whole cells) and local (internal cell organelles) segmentation by using a MATLAB R2018b (MathWorks Inc., Natick, MA) script.
Figure 2.
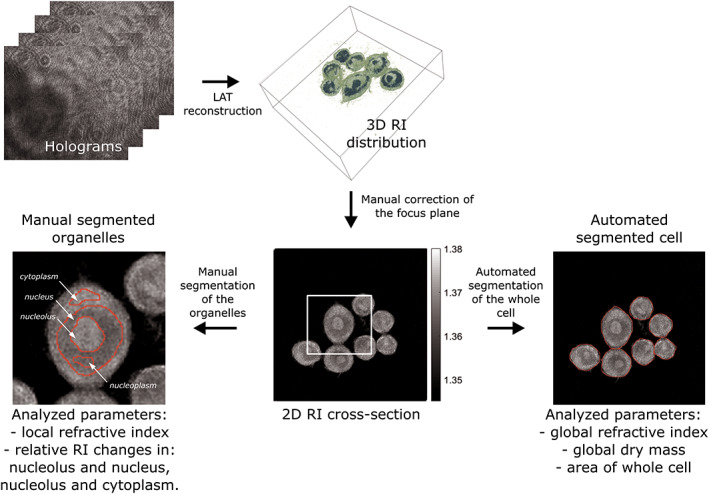
Evaluation pipeline for analyzing global and local biophysical parameters of cells based on holotomography measurements. [Color figure can be viewed at wileyonlinelibrary.com]
The global segmentation was performed automatically by an algorithm based on the cluster method (44). From a segmented image, the refractive index of whole live and fixed cells and cell surfaces were calculated. The average values of RI and surface were calculated for both live and fixed cells for each cell line and the differences in the average values were determined and analyzed. The refractive index of cells is closely related to the cellular dry mass, which is an important biophysical parameter and thus also was analyzed in our study. As described with details in reference (6) the mass concentration Δm(x, y, z) of biomolecules highly correlates with the spatial distribution of the refractive index difference Δn(x,y,z) = n s(x,y,z)‐n medium between the sample refractive index n s (in this study, the spatial cellular refractive index) and the homogeneous refractive index n medium of the of the surrounding medium:
| (1) |
The parameter α in Eq. (1) is denoted as the specific refractive increment and has been determined for human proteins to α = 0.190 ml/g with a standard deviation of 0.003 ml/g (3). In our work, the cellular dry mass was calculated for each cell by integration of the spatial distribution of the mass concentration Δm(x,y,z) as obtained by Eq. (1) from the corresponding cellular refractive index distribution. The dry mass was calculated only for live cells, as the relationship between mass concentration and refractive index change is unknown for fixed cells.
The local segmentation of internal organelles (nucleus, nucleolus, nucleoplasm, and cytoplasm) in the RI images was performed manually by using a MATLAB R2018b script. Based on the segmented images, the average, local RI of these organelles were determined. The average values of the local refractive index between live cells and PFA fixed cells were compared. The RI difference between nucleolus and nucleus, nucleolus, and cytoplasm were determined and the changes of this difference before and after PFA fixation were analyzed.
Statistical Analysis
The statistical analysis of global and local refractive index, surface and global dry mass was carried out using MATLAB R2018b scripts. Descriptive parameters were presented in Figure 4 as mean ± the errors—95% confidence intervals of the average and in Figure 5 as median, minimum, maximum of a data with all outliers included. Relative changes of the RI difference between nucleolus and nucleus, nucleolus and cytoplasm were determined and the changes of this difference before and after PFA fixation were calculated and the significance of the differences was assessed by the Student's t test, or by the Mann–Whitney U test, if the assumption of normality was not met (which was evaluated by Shapiro–Wilk test). Statistical significance level was P < 0.05.
Results
Global Refractive Index of Cells before and after PFA Fixation
Figure 3 shows the three‐dimensional reconstructions, the two‐dimensional refractive index distributions within the corrected focused plane and corresponding bright field images of representative cells from the investigated NIH‐3T3, RAW 264.7, NRK‐52E, RLE‐6TN cell lines. Bright field imaging was performed complementary utilizing an inverted microscope (iMIC, TilPhotonics, Gräfelfing, Germany) with a 20x microscope objective (Zeiss LD Acroplan, 20x/0.4 Korr). Live cells were measured in earlier mentioned physiological buffer medium (n medium = 1.3379, 334 mOsmol/kg) and fixed cells were analyzed in PBS (Sigma, order D8537; n PBS = 1.3332, 287 mOsmol/kg). Due to differences in molecular density and the significantly higher molecular density of nucleolus, it is easy to distinguish between different cellular organelles in NIH‐3T3, NRK‐52E, RLE‐6TN cells. The spherical, more condensed structure and higher mass density of RAW 264.7 macrophages made finding the best focus plane in the reconstruction of these cells more difficult. In NIH‐3T3, NRK‐52E and RLE‐6TN cells, internal structures can be easily identified based on the 3D‐QPI measurements. In RAW 264.7 cells, the nucleus and nucleolus were better visible than other organelles and their refractive index can be measured much easier than other structures.
Figure 3.
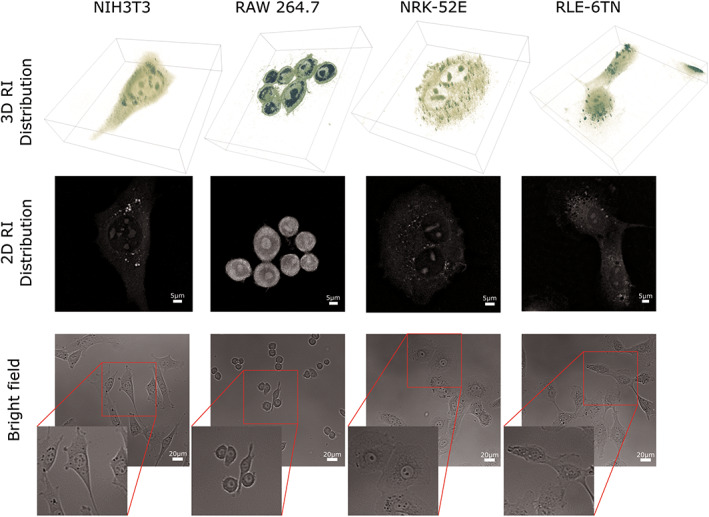
Representative 3D rendered isosurface images of an entire 3D RI distribution, 2D cross section of the RI distribution within the best focal plane and complementary captured bright field images of living NIH3T3, RAW 264.7, NRK‐52E, RLE‐6TN cells. [Color figure can be viewed at wileyonlinelibrary.com]
For statistical analysis, measurements on individual cells were performed. To investigate the effect of PFA fixation on cells, for each cell type live and fixed cells from lines NIH‐3T3 (n ive cells = 88, n fixed cells = 59), RAW 264.7 (n live cells = 126, n fixed cells = 154), NRK‐52E (n live cells = 68, n fixed cells = 87), RLE‐6TN (n live cells = 60, n fixed cells = 54) were measured in N = 2 independently conducted experiments. Statistical analysis was carried out on the global and local RI of the cells, cell surface area and dry mass. Measurements were performed in a room temperature (T = 25°C) and the time delay between two subsequent measurements was approximately 1 min. Comparison of the average value of refractive index in whole cells before and after fixation (mean ± errors as 95% confidence intervals of the average) is shown in Figure 4. The global mean value of RI in live RAW 264.7 cells is the highest from all measured cell lines (n RAW264.7 = 1.3666 ± 0.0004). Mean values of cells from three other lines are similar in the range of thousandths (n NIH3T3 = 1.3448 ± 0.0006, n NRK − 52E = 1.3426 ± 0.0003, n RLE−6TN = 1.3438 ± 0.0006). After fixation, the cells indicate a significant decrease in refractive index. The largest change occurs in the macrophages (Δn RAW264.7 = 0.0131), accompanied with a slight increase in the surface area on the selected cross section (ΔSRAW264.7 = 21.7 μm2). The cells from the other lines also tend to a decreased refractive index after the fixation process, the RI change is found in range of thousandth (Δn NIH3T3 = 0.0025, Δn NRK − 52E = 0.0045, Δn RLE−6TN = 0.0035), but in the case of these cell lines, the surface area on the selected cross section, contrary to macrophages, decreases significantly (ΔSNIH3T3 = 196.8 μm2, ΔSNRK − 52E = 412.7 m2, ΔSRLE−6TN = 196, 7 μm2).
Figure 4.
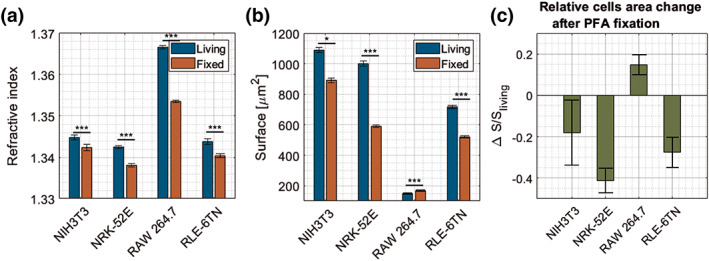
Comparison between live and fixed cells (a) global refractive index, (b) surface area of cells in the optimum focus plane (c) relative change of the surface area after fixation. [Color figure can be viewed at wileyonlinelibrary.com]
As the specific refractive increment relates the change in refractive index with an increase in biomolecular mass density (α ~ n), the results from average RI obtained by systematic measurements with the 3D‐QPI system of four cell lines were converted to the average density of the cellular dry mass. Fibroblast cells (line NIH3T3) and epithelial cells (lines NRK‐52E and RLE‐6TN) show a similar average global mass density (ρ NIH3T3 = 7.471 pg/μm3 ± 0.003, ρ NRK − 52E = 7.459 pg/μm3 ± 0.002, ρ RLE−6TN = 7.466 pg/μm3 ± 0.004), while the average density of macrophages from line RAW 264.7 is much higher (ρRAW264.7 = 7.592 pg/μm3 ± 0.002).
Decrease of Local Refractive Index in Organelles of Cells after PFA Fixation across Four Cell Lines
Figure 5 shows representative local average refractive index data from live and fixed cells in 2% PFA solution. RAW 264.7, NIH‐3T3, RLE‐6TN, and NRK‐52E cells were grown under standard protocols and then measured with 3D‐QPI as described above. For all data sets minimum, maximum and outliers are included. The red line shows the median of the data. Literature data on the refractive index of individual cellular organelles in measured cell lines are insufficient as a basis to reject outliers. In each cell line, four cellular organelles were analyzed: nucleus, nucleolus, nucleoplasm, and cytoplasm. The region of interest in each measurement was chosen manually (see example in Fig. 2). The results retrieved from the local refractive index analysis show that the nucleolus has the highest value of refractive index from the analyzed organelles across all four cell lines (n RAW264.7 = 1.3720 ± 0.0006, n NIH3T3 = 1.3497 ± 0.0008, n NRK‐52E = 1.3507 ± 0.0006, n RLE−6TN = 1.3494 ± 0.0008). In RAW 264.7 and NRK‐52E cells, the nucleus has lower refractive index than the cytoplasm (45). In cells from the NIH‐3T3, NRK‐52E, RLE‐6TN cell lines, RI values in nucleus, nucleoplasm and cytoplasm are similar to each other within the range of 0.002. However, organelles in RAW 264.7 cells do not represent this tendency. There are significant differences between refractive index values before and after fixation in the analyzed cell organelles in all cell lines. In all cell lines, in all organelles, there is decrease of refractive index after fixation. In cells from NIH‐3T3, NRK‐52E, RLE‐6TN lines, the largest decrease in RI is observed in the nucleolus. In the case of RAW 264.7 line, the major decrease in RI value after fixation occurs in the cytoplasm (Δn = 0.016). The findings for the average values in Figure 5 are also reflected by the data from each of the two independently performed experiments that are plotted as bar diagrams in Figure S1.1 of the Supporting Information file S1. The results from both measurement series show for all four investigated cell lines highly similar refractive index values and tendencies, with respect to the intracellular compartments.
Figure 5.
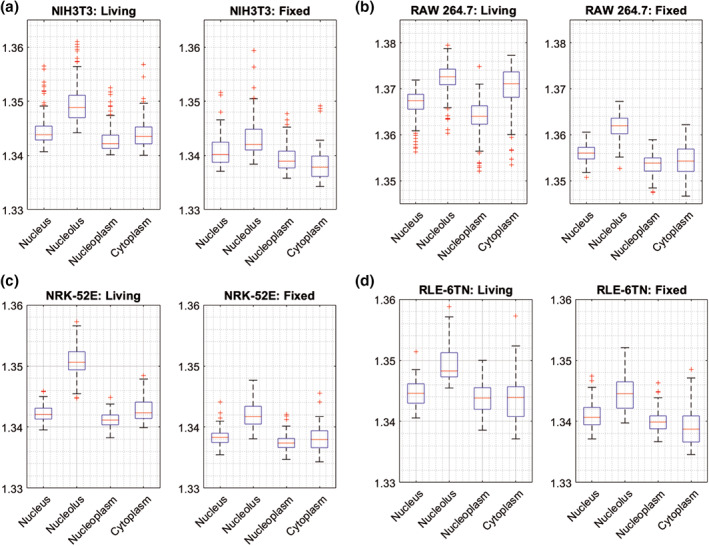
Comparison of the average refractive index values between live and fixed cells (median, minimum, maximum of a data and all outliers) in four cell lines: (a) NIH‐3T3 mouse embryonic fibroblasts, (b) RAW 264.7 mouse macrophages (c) NRK‐52E rat kidney epithelial cells, (d) RLE‐6TN rat lung epithelial cells. In each cell line four cellular organelles were analyzed: Nucleus, nucleolus, nucleoplasm and cytoplasm. [Color figure can be viewed at wileyonlinelibrary.com]
Effect of Paraformaldehyde Cell Fixation on Image Contrast in 3D‐QPI Measurements
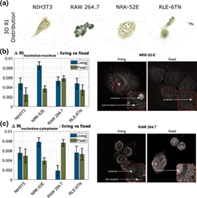
The refractive index values inside the cells, in the internal organelles, were compared before and after fixation. The analysis showed that in three out of four cell lines (NIH‐3T3, NRK‐52E, RLE‐6TN) the refractive index difference between nucleus and nucleolus and between nucleus and cytoplasm decreases, thus reducing the contrast in the RI image. Signal‐to‐noise ratio for fixed cells decreased and lower values of Δn after the fixation process indicate a lower quality and lower contrast of an image. This is most visible in cells from the NRK‐52E line (for illustration see upper right panel in Fig. 6), where the difference between nucleus and nucleolus, and between nucleus and cytoplasm decreases the most (by 0.0048, 0.0041, respectively). In the case of cells from the RAW264.7 macrophages, the differences between nucleolus‐nucleus and nucleolus‐cytoplasm increase after the fixation process and thus increase the contrast of the image (see Fig. 6a,b). The increase in the refractive index difference between the nucleus and the cytoplasm before and after fixation is significantly greater than the increase in the difference between the nucleolus and the nucleus.
Figure 6.
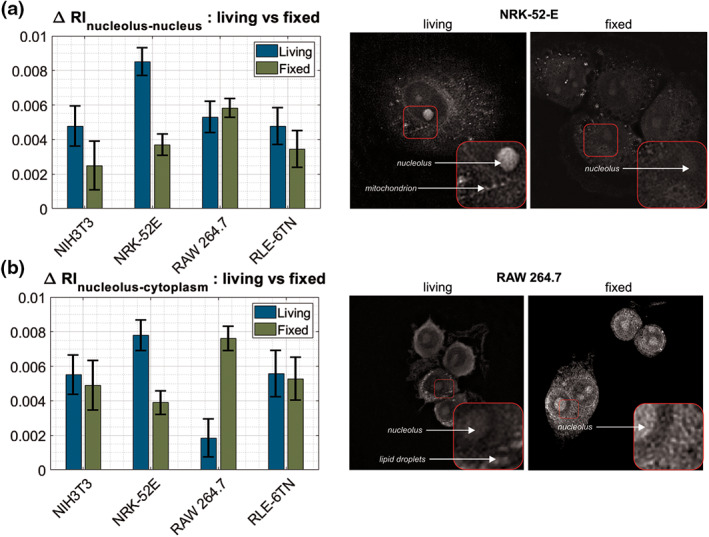
Differences in RI between (a) nucleolus‐nucleus and (b) nucleolus‐cytoplasm in all the cell lines with image comparison of contrast change of live and fixed cells on the example of two the most affected cell lines: NRK‐52E and RAW 264.7. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
The refractive index values of cells are directly connected to the dry mass concentration of the specimen and thus is an important biophysical feature, with prospects to be applied in digital phase histopathology or remote diagnostics (7) supported by deep learning procedures. The results in Figures 2, 3, 4, 5, 6 show that 3D‐QPI allows reliable high‐quality measurements of the refractive index already for quite low cell numbers compared to other techniques, like flow cytometry (46), but for measuring larger cell populations, as done in this study, an automated segmentation tool for the cellular compartments is essential to make this process more feasible for routine usage in laboratories. Here, for statistical analysis, measurements of individual cells were performed; hence, the acquired data are limited (in agreement with the findings in reference (29)). In order to introduce an 3D‐QPI‐based tool for further extensive medical analysis, the tools for analysis of 3D refractive index reconstructions should be further automated.
RI values of live cells and cell organelles are still not standardized and appear to be cell type dependent as indicated by the results in Figures 4 and 5. There thus is a need to standardized preparation protocols of live and fixed cells measurements to have reliable quantitative data that is comparable across different laboratories and users. The retrieved refractive index values of RAW 264.7 cells (Fig. 4) are comparable to those published earlier (47, 48). The refractive indices of the other cell lines that were investigated in this study, to the best of our knowledge, have not been published before. The comparison with data from other cell types published show that the refractive index values are within similar limits but dependent on the cell type (48). In order to use QPI tools and refractive index data as a biomedical marker, it is necessary to create a library of refractive indices for wide number of cell lines and their internal organelles.
In Figure 5, the median values of the cytoplasm RI in living cells, in two cell lines RLE‐6TN and NIH3T3 are found to be lower than the median value of the RI of nucleus (RLE‐9TN: ΔRI = −0.0007, NIH3T3: ΔRI = −0.0003), but, in contrast, for RAW 264.7 and NRK‐52E cells the median RI of cytoplasm in living cells is higher than that of nucleus (RAW 264.7: ΔRI = +0.0037, NRK‐52E: ΔRI = +0.0002). This is contrary to the information published in the literature (45, 49), where it was shown that the refractive index of the nucleus is lower than RI of cytoplasm. However, isolated cellular nuclei or cells in suspension were studied there, and the isolation process or state of the cell could have affected the value of the refractive index of the nucleus. It should also be taken into account that in the spatial resolved measurements made for this study, the refractive index values of the nucleoplasm is lower than RI of cytoplasm across all four lines and that the RI of nucleolus is much higher than RI of cytoplasm, which was also reported in Kim's paper (29).
Our experimental results show that PFA fixation of cells causes a refractive index decrease across all four investigated cell lines (Fig. 4a). Moreover, the average dry mass and surface area retrieved from in focus cross sections were found reduced after PFA fixation compared to those of the living cells (Fig. 4b). These effects may be explained by dissolution of cell membrane lipids, dissolved intracellular solutes and fixation induced shrinking due to PFA induced protein cross‐link effects. In early published data (31), a RI increase after PFA fixation was reported. However, in (31), the research was conducted on HeLa cells and different cell fixation procedures were applied, which may be an explanation for these refractive index differences. In our study, it was found that changes in refractive index before and after fixation process are different across all four cell lines (Fig. 4). The highest decrease of RI after fixation occurs in RAW 264.7 cells, the lowest in the NIH‐3T3 cell line.
Analysis of the cells cross‐sectional areas (Figs. 4b, c) has revealed that cells from three PFA‐treated cell lines (NIH‐3T3, NRK‐52E, RLE‐6TN) are shrunken, which is in good agreement with earlier observations (27, 31, 32). The average area change is different for each cell line. The highest shrinkage is found for NRK‐52E cells. In cells from the RAW 264.7 cell line, the average area slightly increases after fixation (25, 50). The data acquired in this article support the results from previous studies (26, 31, 32, 45). PFA fixation also caused forming of micron scale clusters in nucleus, the nanostructure of cells is changed (25). Furthermore, the cells surface roughness is larger in PFA‐treated cells (32). Even though, it was proven that it is still possible to extract clinically relevant nanoscopic information after fixation.
The influence of chemical substances on the contrast in phase measurements was previously studied and a chemical preparation was searched for which the contrast would improve (51, 52, 53). Fixation allows to stop the vital processes and degradation of the cells; they serve to stabilize the fine structural details of cells and tissues prior the examination. In our study, a nonuniform decrease in the refractive index of individual organelles after the fixation across all four cell lines caused changes in the difference between the refractive index of the nucleus and the nucleolus, and the nucleolus and cytoplasm (Figs. 5, 6) and thus resulted a change of the contrast. Higher ΔRI results in a higher visibility of nucleolus (for illustration see upper right panel in Fig. 6). The largest decrease in the difference between the nucleus and cytoplasm occurs in NRK‐52E and is 3.884∙10−3, the smallest changes in contrast were observed in the RLE‐6TN and NIH‐3T3, where the difference between the nucleus and cytoplasm after fixation decreases within the range of 3∙10−4 to 5∙10−4. In these three cell lines, fixation with PFA caused lower contrast. The situation is different when measuring the RAW 264.7 macrophages. In these cells, fixation causes an increase in the refractive index difference between the nucleolus and the nucleus (lower panel of Fig. 6) and between the nucleolus and the cytoplasm, where increase is significant and equals to 5.773∙10−3. The contrast improvement for fixed RAW 264.7 cells was already clearly observable during the conduction of the measurements. Moreover, in a complementary performed documentation, living and fixed cells were observed with bright field microscopy under white light illumination (see Fig. S1.2 in the Supporting Information). For NIH‐3T3 fibroblasts, NRK‐52E kidney cells and RLE‐6TN macrophages, the bright field image contrast for fixed cells was found lower than for living cells. Instead, RAW 264.7 macrophages appeared with increased contrast of the intracellular structures. These observed bright field image contrast changes correlate with the determined refractive index differences between the internal cellular organelles after fixation and the images that are plotted in Figure 6. The reduced bright field image contrast can be explained by the decreased refractive index difference between internal cell compartments as detected for NIH‐3T3, NRK‐52E, RLE‐6TN cells that leads to reduced sample induced diffraction patterns. Contrary, a higher contrast of the intracellular organelles can be explained by an increase of the refractive index differences, as measured for the RAW 264.7 cells, that causes increased diffraction effects in the bright field images, respectively. Any future experimental study applied in medical or basic research, that focuses on the RI of cells and their compartments, should take these findings into account when fixed and live cells are analyzed.
Conclusions and Future Prospects
In our study, we have presented in detail the significant impact of PFA fixation on the global refractive index of cells and the local spatial RI distribution that is related to their intracellular organelles. We found that fixation causes in most of the investigated cell types a decrease in global and local refractive index and also in the related dry cell mass density. We have also shown that depending on the cell line these changes are different. In addition, we have noticed that in immunological cells (such as RAW 264.7 macrophages) PFA fixation caused an increase in refractive index and dry mass density. For the future implementation of quantitative phase imaging techniques, for example, toward remote diagnostics supported by machine learning procedures, it is necessary to take into account these differences between live and fixed preparations (54). In order to obtain reliable data for fixed cells and tissues using QPI techniques, it is also necessary to use standardized preparation processes of cell cultures for the measurements.
All measurements in this study have been performed using 3D‐QPI techniques. This allowed us to perform 3D measurement and visualization of the cells and to retrieve the local refractive indices of cellular organelles. Due to the anisotropic distribution of the RI along the z‐axis and the challenges of three‐dimensional segmentation of internal structures, the analysis was conducted based on the best focused 2D cross sections. For future developments, it is crucial to extend the analysis to volumetric refractive index values by improvement of the tomographic reconstruction algorithms (23, 55) and the enhancement and automation of 3D segmentation processes based on machine learning and neural networks.
Finally, the results of our research show that it is difficult to determine the rules of for conversion factors to describe the RI changes between fixed and live samples in case of machine learning procedures which can be considered, for example, in the interpretation or inter‐laboratory diagnostics. Therefore, procedures need to be standardized to allow a reliable interpretation of QPI images acquired with different equipment at different locations and for an efficient application of machine learning algorithms. For this reason, it is also important to build in the future detailed libraries of refractive index values for biological samples. Round robin tests between different laboratories can also contribute as an important step to standardize cell culture preparation and measurement procedures to speed up the development of QPI technologies and AI‐based diagnostics.
In conclusion, the PFA fixation process significantly affects the global and local refractive index of cells with significant impact in quantitative phase imaging. This effect is cell line dependent and has to be taken into account when using QPI data for reliable cell studies in life sciences, as well as for diagnostics and for machine learning procedures design.
AUTHOR CONTRIBUTIONS
Maria Baczewska: Conceptualization; data curation; formal analysis; investigation; methodology; visualization; writing‐original draft; writing‐review and editing. Kai Eder: Data curation; formal analysis; investigation; writing‐original draft; writing‐review and editing. Steffi Ketelhut: Data curation; resources. Bjoern Kemper: Conceptualization; formal analysis; investigation; methodology; resources; supervision; validation; writing‐original draft; writing‐review and editing. Małgorzata Kujawińska: Conceptualization; formal analysis; funding acquisition; investigation; methodology; project administration; supervision; validation; writing‐original draft; writing‐review and editing.
Supporting information
Figure S1.1 Average refractive index (RI) data of intracellular structures retrieved from two independently performed series of measurements on living and fixed cells of the 4 investigated cell lines (a): RLE‐6TN lung cells, (b): RAW 264.7 macrophages, (c): NRK‐52E kidney cells, (d): NIH‐3 T3 fibroblasts. Notice that the RI values retrieved from of both series show highly similar values and likewise tendencies which illustrates the reproducibility of experimental data.
Figure S1.2 Representative white light images of living (upper row) and fixed (lower row) cells captured with identical adjustments of an utilized bright field microscope (iMIC, Till Photonics, Germany). Note that for NIH‐3 T3 fibroblasts, NRK‐52E kidney cells and RLE‐6TN macrophages, the observed image contrast for fixed cells is lower than for living cells, while RAW 264.7 macrophages appear with increased contrast, in particular, of the intracellular structures. The observed bright field image contrast changes for living and fixed cells corelate with the determined refractive index differences between the internal cellular organelles after fixation that are plotted in Figure 6. A decreased refractive index difference between internal cell compartment as observed for NIH‐3 T3, NRK‐52E, RLE‐6TN cells lead to reduced diffraction patters and thus to a reduced bright field image contrast while an increase of the refractive index differences as observed for RAW 264.7 macrophages results in an increased contrast of the intracellular organelles caused increased diffraction effects, respectively.
ACKNOWLEDGMENTS
The research leading to the described results was carried out within the program TEAM TECH/2016‐1/4 of Fundacja na rzecz Nauki Polskiej, co‐financed by the European Union under the European Regional Development Fund and PROM program (POWR.03.03.00‐00‐PN13/18), financed by European Commission, European Regional Development, non‐competitive project entitled International Scholarship Exchange for PhD students and academic staff. Moreover, the authors gratefully acknowledge support from Tomocube Inc., Korea.
Literature Cited
- 1. Majeed H, Sridharan S, Mir M, Ma L, Min E, Jung W, Popescu G. Quantitative phase imaging for medical diagnosis. J Biophotonics 2017;10:177–205. [DOI] [PubMed] [Google Scholar]
- 2. Park Y, Depeursinge C, Popescu G. Quantitative phase imaging in biomedicine. Nat Photonics 2018;12:578–589. [Google Scholar]
- 3. Barer R. Determination of dry mass, thickness, solid and water concentration in living cells. Nature 1953;172:1097–1098. [DOI] [PubMed] [Google Scholar]
- 4. Popescu G, Park Y, Lue N, Best‐Popescu C, Deflores L, Dasari R, Feld M, Badizadegan K. Optical imaging of cell mass and growth dynamics. Am J Physiol Cell Physiol 2008;2:345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zangle T, Teitell M. Live‐cell mass profiling: An emerging approach in quantitative biophysics. Nat Methods 2014;11:1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim K, Lee S, Yoon J, Heo J, Choi C, Park Y. Three‐dimensional label‐free imaging and quantification of lipid droplets in live hepatocytes. Sci Rep 2016;6:36815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vodovnik A, Aghdam M. Complete routine remote digital pathology services. J Pathol Inform 2018;1:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pesapane F, Codari M, Sardanelli F. Artificial intelligence in medical imaging: Threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur Radiol Exp 2018;2:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rivenson Y, Wang H, Wei Z, de Haan K, Zhang Y, Wu Y, Gunaydin H, Zuckerman J, Chong T, Sisk A, et al. Virtual histological staining of unlabelled tissue autofluorescence images via deep learning. Nat Biomed Eng 2019;3:466–477. [DOI] [PubMed] [Google Scholar]
- 10. Yoon J, Jo Y, Kim Y, Kim K, Lee S, Park Y. Identification of non‐activated lymphocytes using three dimensional refractive index tomography and machine learning. Sci Rep 2017;7:6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nygate Y, Levi M, Mirsky S, Turko N, Rubin M, Barnea I, Dardikman‐Yoffe G, Haifler M, Shalev A, Shaked N. Holographic virtual staining of individual biological cells. Proc Natl Acad Sci USA 2020;117(17):9223–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J, Kim H, Cho H, Jo Y, Song Y, Ahn D, Lee K, Park Y, Ye S. Deep‐learning‐based label‐free segmentation of cell nuclei in time‐lapse refractive index tomograms. IEEE Access 2019;7:83449–83460. [Google Scholar]
- 13. Kuś A, Krauze W, Kujawińska M. Active limited‐angle tomographic phase microscope. J Biomed Opt 2015;20:111216. [DOI] [PubMed] [Google Scholar]
- 14. Sung Y, Choi W, Fang‐Yen C, Badizadegan K, Dasari R, Feld M. Optical diffraction tomography for high resolution live cell imaging. Opt Express 2009;17:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi W, Fang‐Yen K, Badizadegan K, Oh S, Lue N, Dasari R, Feld M. Tomographic phase microscopy. Nat Methods 2007;4:717–719. [DOI] [PubMed] [Google Scholar]
- 16. Kim K, Yoon J, Shin S, Lee S, Yang S, Park Y. Optical diffraction tomography techniques for the study of cell pathophysiology. J Biomed Photonics Eng 2016;020201:2. [Google Scholar]
- 17. Nanolive Cell Explorer hardware description. http://nanolive.ch/hardware/.
- 18. Tomocube, Inc., Republic of Korea. Tomocube Technology. http://www.tomocube.com/product/technology/.
- 19. Wolf E. Three‐dimensional structure determination of semi‐transparent objects from holographic data. Opt Commun 1969;1:153–156. [Google Scholar]
- 20. Simon B, Debailleul M, Houkal M, Ecoffet C, Bailleul J, Lambert J, Spangenberg A, Liu H, Soppera O, Haeberlé O. Tomographic diffractive microscopy with isotropic resolution. Optica 2017;4:460–463. [Google Scholar]
- 21. Merola F, Memmolo P, Miccio L, Savoia R, Mugnano M, Fontana A, D'Ippolito G, Sardo A, Iolascon A, Gambale A, et al. Tomographic flow cytometry by digital holography. Light‐Sci Appl 2017;6:e16241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuś A, Krauze W, Makowski P, Kujawińska M. Holographic tomography: Hardware and software solution for 3D quantitative biomedical imaging. ETRI J 2019;41:61–71. [Google Scholar]
- 23. Krauze W, Makowski P, Kujawińska M, Kuś A. Generalized total variation iterative constraint strategy in limited angle optical diffraction tomography. Opt Express 2016;24:4924–4936. [DOI] [PubMed] [Google Scholar]
- 24. Lim J, Ayoub A, Psaltis D. Three‐dimensional tomography of red blood cells using deep learning. ASC Photon 2020;2(2):026001. [Google Scholar]
- 25. Li Y, Almassalha L, Chandler J, Zhou X, Stypula‐Cyrus Y, Hajsak K, Roth E, Bleher R, Subramanian H, Szleifer I, et al. The effects of chemical fixation on the cellular nanostructure. Exp Cell Res 2017;358:253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Perry C, Chung J, Ylaya K, Choi C, Simpson A, Matsumoto K, Smith W, Hewitt S. A buffered alcohol‐based fixative for histomorphologic and molecular applications. J Histochem Cytochem 2016;64:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kiernan J. Formaldehyde, formalin, paraformaldehyde and glutaraldehyde: What they are and what they do. Micros Today 2000;00‐1:8–12. [Google Scholar]
- 28. Thavarajah R, Mudimbaimannar V, Elizabeth J, Rao U, Ranganathan K. Chemical and physical basics of routine formaldehyde fixation. J Oral Maxillofac Pathol 2012;16:400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim T, Lee B, Fujii J, Kim J, Pack C. Physicochemical properties of nucleoli in live cells analyzed by label‐free optical diffraction tomography. Cell 2019;8:699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Q, Zhong L, Tang P, Yuan Y, Liu S, Tian J, Lu X. Quantitative refractive index distribution of single cell by combining phase‐shifting interferometry and AFM imaging. Sci Rep 2017;7:2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Su J, Hsu W, Tjiu J, Chiang C, Huang C, Sung K. Investigation of influences of the paraformaldehyde fixation and paraffin embedding removal process on refractive indices and scattering properties of epithelial cells. J Biomed Opt 2019;19:075007. [DOI] [PubMed] [Google Scholar]
- 32. Kim S, Kim J, Okajima T, Cho N. Mechanical properties of paraformaldehyde treated individual cells investigated by atomic force microscopy and scanning ion conductance microscopy. Nano Converg 2017;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Farcal L, Lernando Torres Andón F, Di Cristo L, Rotoli BM, Bussolati O, Bergamaschi E, Mech A, Hartmann NB, Rasmussen K, Riego‐Sintes J, et al. Comprehensive in vitro toxicity testing of a panel of representative oxide nanomaterials: First steps toward an intelligent testing strategy. PLoS ONE 2015;10(5):e0127174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kroll A, Dierker C, Rommel C, Hahn D, Wohlleben W, Schulze‐Isfort C, Göbbert C, Voetz M, Hardinghaus F, Schnekenburger J. Cytotoxicity screening of 23 engineered nanomaterials using a test matrix of ten cell lines and three different assays. Part Fibre Toxicol 2011;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Driessen MD, Mues S, Vennemann A, Hellack B, Bannuscher A, Vimalakanthan V, Riebeling C, Ossig R, Wiemann M, Schnekenburger J, et al. Proteomic analysis of protein carbonylation: A useful tool to unravel nanoparticle toxicity mechanisms. Part Fibre Toxicol 2015;12:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mues S, Antunovic J, Ketelhut S, Kemper B, Schnekenburger J. Novel optical approaches for label‐free quantification of nano‐cytotoxic effects proc. SPIE 2016;9719. 10.1117/12.2213269 [DOI] [Google Scholar]
- 37. Shin S, Kim K, Yoon J, Park Y. Active illumination using a digital micromirror device for quantitative phase imaging. Opt Lett 2015;40:5407–5410. [DOI] [PubMed] [Google Scholar]
- 38. Lee K, Kim K, Kim G, Shin S, Park Y. Time‐multiplexed structured illumination using a DMD for optical diffraction tomography. Opt Lett 2017;42:999–2017. [DOI] [PubMed] [Google Scholar]
- 39. Devaney A. Inverse‐scattering theory within the Rytov approximation. Opt Lett 1981;6:374–376. [DOI] [PubMed] [Google Scholar]
- 40. Kim K, Yoon H, Diez‐Silva M, Dao M, Dasari R, Park Y. High‐resolution three‐dimensional imaging of red blood cells parasitized by plasmodium falciparum and in situ hemozoin crystals using optical diffraction tomography. J Biomed Opt 2014;9:011005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kostencka J, Kozacki T, Kuś A, Kemper B, Kujawińska M. Holographic tomography with scanning of illumination: Space‐domain reconstruction for spatially invariant accuracy. Biomed Optics Express 2016;7:4086–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kujawińska M, Krauze W, Baczewska M, Kuś A, Ziemczonok M. Comparative study of laboratory and commercial limited‐angle holographic tomography setups. Proc SPIE 2019;10887:1088708. [Google Scholar]
- 43. Hanwell M, Ayachit U, Muller D, Hovden R. 2020: https://tomviz.org/
- 44. Hartigan J, Wong M, Algorithm AS. 136: A K‐means clustering algorithm. J Royal Stat Soc 1979;28:100–108. [Google Scholar]
- 45. Schurmann M, Scholze J, Muller P, Guck J, Chan C. Cell nuclei have lower refractive index and mass density than cytoplasm. J Biophotonics 2016;9:1068–1076. [DOI] [PubMed] [Google Scholar]
- 46. Picot J, Guerin C, Kim L, Boulanger C. Flow cytometry: Retrospective fundamentals and recent instrumentation. Cytotechnology 2012;64:109–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mues S, Ketelhut S, Kemper B, Schnekenburger J. Digital holographic microscopy as multimodal read out for in vitro nanomaterial cytotoxicity testing. Proc SPIE‐OSA 2017;10414. 10.1117/12.2287269 [DOI] [Google Scholar]
- 48. Kemper B, Bauwens A, Bettenworth D, Gotte M, Greve B, Kastl L, Ketelhut S, Lenz P, Mues S, Schnekenburger J, et al. Label‐free monitoring of cells in vitro: Label‐free quantitative in vitro live cell imaging with digital holographic microscopy. Bioanal Rev 2019;2:219–272. [Google Scholar]
- 49. Habaza M, Kirchbaum M, Guernth‐Marschner C, Dardikman G, Barnea I, Korenstein R, Duschl C, Shaked N. Rapid 3D refractive‐index imaging of live cells in suspension without labeling using dielectrophoretic cell rotation. Adv Sci 2017;4:1600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tran T, Sundaram C, Bahler C, Eble J, Grignon D, Monn M, Simper N, Cheng L. Correcting the shrinkage effects of formalin fixation and tissue processing for renal tumors: Toward standardization of pathological reporting of tumor size. J Cancer 2015;6:759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stewart J, Villasmil M, Frampton M. Changes in fluorescence intensity of selected leukocyte surface markers following fixation. Cytom Part A 2017;71A:379–385. [DOI] [PubMed] [Google Scholar]
- 52. Kemper B, Przibilla S, Rommel C, Vollmer A, Ketelhut S, Schnekenburger J, von Bally G. Simplified setup for imaging with digital holographic microscopy and enhanced quantitative phase contrast by osmotic stimulation of living cells. Proc SPIE 2011;7904:790404. [Google Scholar]
- 53. Rommel C, Dierker C, Schmidt L, Przibilla S, von Bally G, Kemper B, Schnekenburger J. Manipulating intracellular refractive index for contrast enhanced digital holographic imaging of subcellular structures. Proc. SPIE 2011;7902:790203. [DOI] [PubMed] [Google Scholar]
- 54. Jo Y, Cho H, Lee S, Choi G, Kim G, Min H‐S, Park Y. Quantitative phase imaging and artificial intelligence: A review. IEEE J Sel Top Quantum Electron 2018;25:1. [Google Scholar]
- 55. Krauze W. Optical diffraction tomography with finite object support for the minimization of missing cone artifacts. Biomed Opt Express 2020;11:1919–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.1 Average refractive index (RI) data of intracellular structures retrieved from two independently performed series of measurements on living and fixed cells of the 4 investigated cell lines (a): RLE‐6TN lung cells, (b): RAW 264.7 macrophages, (c): NRK‐52E kidney cells, (d): NIH‐3 T3 fibroblasts. Notice that the RI values retrieved from of both series show highly similar values and likewise tendencies which illustrates the reproducibility of experimental data.
Figure S1.2 Representative white light images of living (upper row) and fixed (lower row) cells captured with identical adjustments of an utilized bright field microscope (iMIC, Till Photonics, Germany). Note that for NIH‐3 T3 fibroblasts, NRK‐52E kidney cells and RLE‐6TN macrophages, the observed image contrast for fixed cells is lower than for living cells, while RAW 264.7 macrophages appear with increased contrast, in particular, of the intracellular structures. The observed bright field image contrast changes for living and fixed cells corelate with the determined refractive index differences between the internal cellular organelles after fixation that are plotted in Figure 6. A decreased refractive index difference between internal cell compartment as observed for NIH‐3 T3, NRK‐52E, RLE‐6TN cells lead to reduced diffraction patters and thus to a reduced bright field image contrast while an increase of the refractive index differences as observed for RAW 264.7 macrophages results in an increased contrast of the intracellular organelles caused increased diffraction effects, respectively.


