Abstract
Actin is essential for key processes in all eukaryotic cells. Cellpermeable optojasps provide spatiotemporal control of the actin cytoskeleton, confining toxicity and potentially rendering F‐actin druggable by photopharmacology. Here, we report cryo electron microscopy (cryo‐EM) structures of both isomeric states of one optojasp bound to actin filaments. The high‐resolution structures reveal for the first time the pronounced effects of photoswitching a functionalized azobenzene. By characterizing the optojasp binding site and identifying conformational changes within F‐actin that depend on the optojasp isomeric state, we refine determinants for the design of functional F‐actin photoswitches.
Keywords: actin filaments, drug design, electron microscopy, photoswitch, protein structures
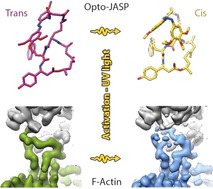
Targeting actin requires spatiotemporal control of drug activity. Optojasps are photo‐switchable small molecules, providing direct optical spatiotemporal control of the actin cytoskeleton. We present high‐resolution cryo‐EM structures of both isomeric states of an optojasp bound to F‐actin and describe in detail the binding pocket and conformational changes associated with switching of the azobenzene.
Actin is a key player in eukaryotic cell biology and involved in cellular motility, cytokinesis, intra‐cellular cargo transport and endocytosis. [1] The 42 kDa globular protein (G‐actin) assembles into polar filaments (F‐actin) that form complex and dynamic networks. To fulfill its diverse functions, the conformation and dynamics of actin filaments are clocked by ATP hydrolysis and the subsequent release of phosphate.[ 2 , 3 ] Furthermore, actin polymerization and the assembly of higher order structures is tightly controlled in space and time by a plethora of actin binding proteins (ABPs). [4]
While actin may serve as a potential entry for drug design due to its involvement in many cellular processes, it is also a highly challenging target owing to its high abundance, its dynamics, and its remarkable conservation of sequence and 3D structure among species. Actin stabilizers such as phalloidin (1, Figure 1) are thus generally toxic. Recently, optojasps, cell‐permeable and photo‐switchable small molecules derived from the natural F‐actin inhibitor jasplakinolide (JASP, 2, Figure 1) [5] were shown to provide direct optical spatiotemporal control of the actin cytoskeleton. [6] The design of optojasps originated from in silico molecular modelling data of the JASP binding site and thorough analysis of structure–activity‐relationship data. [7] It features a modified cyclodepsipeptide macrocycle with a bistable azobenzene attached via a linker chain to the Ala residue position (3, Figure 1). The diazene bond can be isomerized from its trans‐configured ground state to a high‐energy cis form that thermally reverts back to trans with a specific half life, or by irradiation with green light. [6]
Figure 1.
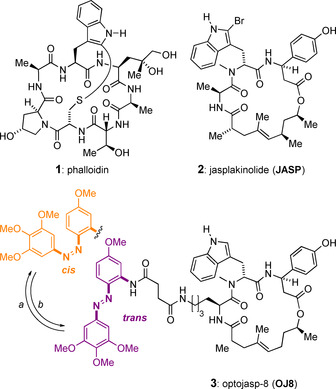
Chemical structures of F‐actin stabilizing natural product toxins and of optojasp‐8. Conditions: a) hν=380 nm; b) hν=520 nm or ΔT (t 1/2=15 min in PBS buffer at 37 °C). [6]
Recent high‐resolution electron cryo microscopy (cryo‐EM) structures of JASP‐stabilized F‐actin allowed a near‐atomic analysis of the JASP binding pocket and its interactions with F‐actin,[ 2 , 8 , 9 ] highlighting the potential of cryo‐EM for structure‐based drug design. [10] Surprisingly, these data suggested the photoswitch moiety of functional optojasps to be placed in the interior of the actin filament. [6]
Ligand switchability in photopharmacology has been conceptualized well, [11] but elucidiating the molecular recognition of photoswitches on the molecular level has rarely been achieved. For optojasps, such data should facilitate their optimization and allow examining the effects of photoswitching on the overall structure of F‐actin. We hence strived to structurally characterize optojasp‐stabilized F‐actin by cryo‐EM. We were able to solve structures of F‐actin in complex with optojasp‐8 (OJ8, 3) [6] in both its inactive (trans‐3, “dark” PDB 7AHQ and EMD‐11790) and its active (cis‐3, “bright”, PDB 7AHN and EMD‐11787) state when bound to F‐actin by cryo‐EM at 3.6 Å (trans) and 2.9 Å (cis) resolution, respectively (Figure S1 and Table S1). Interestingly, while the cis‐state is significantly more potent than the trans state of OJ8, both states could be enriched in an actin‐bound form, in line with the residual activity of the trans state of this optojasp. [6]
Cryo‐EM structures require averaging of many helical protein segments that cannot be individually assessed for presence or kind of a bound small molecule. Hence, sample preparation and imaging for the dark state was conducted under red light to prevent unwanted isomerization (trans‐3). For the bright state, OJ8 containing samples were repetitively activated by 380 nm light pulses. F‐actin was then plunge frozen, and directly imaged to prohibit thermal relaxation (cis‐3). Since modelling of the actin‐bound ligand is crucial for understanding of the molecular recognition and for subsequent design efforts, we used state‐of‐the‐art molecular dynamics‐based modelling [12] in combination with energetic geometry restraints to determine the conformation of OJ8 (see Supporting Information for details).
In its trans state, each OJ8 ligand binds in regular intervals to three actin subunits from both strands of the double‐stranded F‐actin helix (Figure 2 A, Figure S2A and Movie S1A,B). Its binding changes neither the overall architecture of F‐actin nor its helical symmetry (Table S1). The binding pocket and the conformation of the bound macrocycle closely resembles the one described previously for cLys‐JASP, the core of OJ8.[ 2 , 9 ] In analogy to cLys‐JASP, trans‐3 stabilizes a transient state of partial ATP‐hydrolysis which is characterized by incomplete release of phosphate (Figure S2A) and increased stability compared to ADP‐bound F‐actin.[ 2 , 13 ] F‐actin adopts the open DNase‐I‐binding loop (D‐loop) state when trans‐3 is bound (Figure 2 A, Figure S2A and Movie S1A,B) and thereby generally resembles the structure previously reported for cLys‐JASP‐stabilized F‐actin.[ 2 , 9 ] Taken together, the addition of an azobenzene photoswitch to JASP does not modify the mechanism by which JASP‐like ligands stabilize actin filaments, namely by introducing additional contacts within and between strands.
Figure 2.
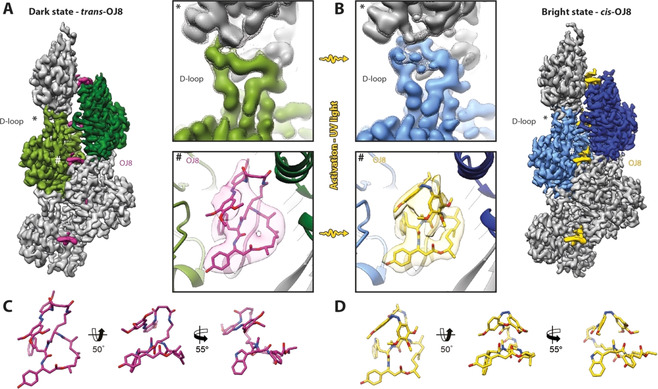
High‐resolution structures of OJ8‐stabilized F‐actin. A,B) Cryo‐EM structures of OJ8‐stabilized F‐actin in the (A) dark state, trans‐OJ8 (magenta), and (B) bright state, cis‐OJ8 (yellow). To highlight the double stranded helical arrangement of F‐actin, the central and subsequent actin subunit are colored in shades of green (dark state) and blue (bright state), respectively. The central D‐loop (*) and OJ8 binding site (#) are enlarged for direct comparison of the structures before and after activation of OJ8 with UV light (380 nm). (Top) While the D‐loop solely adopts the open conformation with trans‐OJ8 bound, it is mixed with a considerably higher population of the closed D‐loop state after irradiation. The map is additionally shown at a lower threshold (mesh) to highlight the second, less populated D‐loop conformation of the cis‐OJ8 structure. (Bottom) The azobenzene photoswitch stacks onto the macrocycle and changes its configuration from trans to cis upon UV‐irradiation. C,D) Atomic wire models of trans‐ and cis‐OJ8. For details also see Figure S2 and Movies S1,2.
Binding of cis‐3 occurs at the same binding site and also does not alter the overall arrangement of F‐actin (Figure 2 B, Figure S2B and Movie S1C,‐D) or its symmetry (Table S1). Interestingly, photoswitching leaves the conformation of the bound macrocycle unchanged and solely affects the azobenzene photoswitch (Figure 2), as hypothesized earlier. [6] Thus, the mode of ligand binding, which primarily features interactions between the macrocycle and F‐actin,[ 2 , 9 ] is conserved for both isomeric states of OJ8 (Figure 2, Figure S2 and Movie S1).
While cis‐3 stabilizes the transient state of partial ATP‐hydrolysis also seen for trans‐3 (Figure S2), it differs markedly from trans‐3 in modulating the conformation of the D‐loop of F‐actin (Figure 2). The D‐loop has been shown to be highly dynamic and polymorphic.[ 14 , 15 ] Different D‐loop conformations were observed in G‐actin, including an α‐helical and a β‐turn fold, but in most structures this region is not resolved. [14] The D‐loop is at a major interface of the F‐actin double helix. Yet, different D‐loop conformations have also been reported for F‐actin,[ 8 , 15 , 16 , 17 ] including the open and closed state.[ 2 , 9 , 18 ] The conformation of the D‐loop was inter alia proposed to be linked to the protein sequence,[ 8 , 18 ] to the nucleotide state,[ 2 , 14 , 16 , 17 ] and to binding of small molecules.[ 2 , 9 , 19 , 20 ]
Although both the trans‐ and the cis‐isomer of 3 share most of their interactions with F‐actin with respect to the macrocyclic ligand (Figure 2 A,B, Figure S2), photoswitching changes the conformation of the D‐loop of F‐actin (Figure 2). In contrast to the exclusively open D‐loop state found in F‐actin stabilized by trans‐3, we find a mixed conformation of actin with a higher population of the closed D‐loop state for cis‐3 (Figure 2, Figure S2, Movie S2), although F‐actin was prepared similarly. The closed conformation, typical for ADP‐bound F‐actin [2] and for phalloidin bound, aged F‐actin, [9] was not observed in any previous JASP‐F‐actin structure. This suggests that the interactions of cis‐3 with F‐actin do not stabilize the open D‐loop state to the same extent as trans‐3 or JASP itself,[ 2 , 9 ] eventually allowing a partial relaxation of the D‐loop toward the closed state. This indicates that F‐actin‐bound cis‐3, probably as a result of the more compact, globular azobenzene moiety, is considerably more “phalloidin‐like” than JASP, or than trans‐3 (Figure 2, Figure 3 A).
Figure 3.
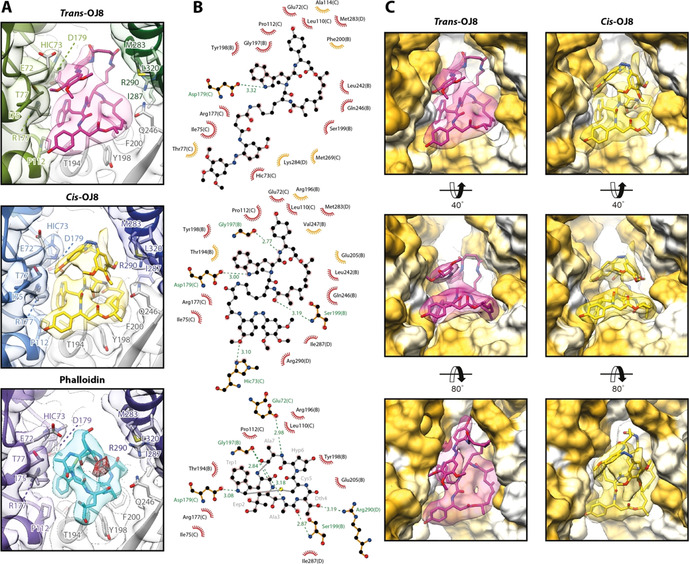
Interactions at the binding site of OJ8 in comparison to phalloidin. A) The binding site of trans‐3 (magenta) and cis‐3 (yellow) in comparison to the one of phalloidin (cyan, PDB: 6T1Y, EMDB: 10363, [9] ). Due to the scorpion tail‐like fold of the photoswitch, OJ8 resembles the three‐dimensional arrangement of phalloidin more closely than the flat conformation of JASP. The conformation of cis‐3 is remarkably similar to phalloidin (see text). B) 2D ligand‐protein interaction diagrams of the ligands shown in (A). Both hydrophobic contacts (red arcs with rays) and putative hydrogen bonds (dashed green lines) are depicted. Chain IDs are stated in brackets for protein residues. Residues that only participate in interactions with either trans‐3 (top) or cis‐3 (center) are highlighted in orange. Note that all residues involved in the binding of phalloidin (bottom), also interact with cis‐3. C) Surface representation of the OJ8 binding site from three different perspectives. While trans‐3 adopts an elongated conformation, partially filling the cavity between the actin strands (into the plane of projection), the conformation of cis‐3 is more compact, thereby increasing contacts to the subunit displayed in the upper right corner (SU D, pointed end direction), also see Movie S1.
The cyclodepsipeptide macrocycle of OJ8 is bound rigidly and approximately takes on a stoichiometric ratio in both isomeric states as its density appears at the same threshold as the protein backbone (Movie S1). By contrast, the connecting linker remains unresolved in both structures, indicating considerable flexibility as usually observed for loops that have little to no contact with surrounding protein residues (Figure 2, Figure S2 and Movie S1). The flexibility of the azobenzene extension depends on its isomeric state; while bound cis‐3 appears remarkably rigid with its density appearing at the same threshold as the macrocycle, the density for trans‐3 appears only at much lower density thresholds (Movie S1). This suggests that cis‐3 synergistically stabilizes F‐actin in a very regular, ordered manner, likely due to tighter interactions with the protein assembly. Intriguingly, we could not find residual density for the trans configuration in our bright state structure (cis‐3), although OJ8 can only be activated by light to a 94:6 cis/trans ratio at best (photostationary equilibrium, Table S2).
The trans configuration of the azobenzene moiety closely resembles available X‐ray structures[ 21 , 22 ] (Figure S3A). By contrast, the bound azobenzene of cis‐3 differs markedly from X‐ray structures[ 23 , 24 ] (Figure S3B, also see Methods) showing almost 90° torsion angles, which are appreciable even when considering the non‐atomic experimental resolution (2.9 Å). When conjugated to JASP and bound to F‐actin, the symmetry of the cis‐form is reduced due to shearing and twisting (Table S3). While the resulting overall conformation may be energetically unfavorable when isolated, the binding energy is apparently increased due to a complementary shape match of cis‐3 with F‐actin.
Our data clearly show that the azobenzene photoswitch folds back in a scorpion tail‐like manner. It stacks onto the cyclodepsipeptide macrocycle, thereby creating a compact, space‐filling structure that actively complements the more disk‐like appearance of JASP (Figure 2). While the azobenzene in its trans configuration non‐specifically protrudes into the cavity between the two strands of F‐actin, cis‐3 is more compact, extending toward the pointed end of the filament (Figure 3, Movie S1). Interestingly, phalloidin (1) that stabilizes F‐actin at the same binding pocket as JASP,[ 9 , 25 ] not only shares its interacting residues with cis‐3, but the hydroxyl group of the hydroxyproline is placed in the same region as the polar, cis‐configured N=N double bond (Figure 3 A,B). The close resemblance of the F‐actin bound cis‐3 and phalloidin, as well as the presence of the closed D‐loop state, suggests that both ligands do not stabilize the open D‐loop state to the same extent as JASP or trans‐3. Such ligand‐induced modulation may have relevance for the functional difference of F‐actin stabilizing compounds. [26] We have previously shown that the addition of JASP to F‐actin‐ADP results in strong binding of coronin‐1B, which is usually limited to F‐actin‐ADP‐Pi. [2] Based on this, we proposed that nucleotide‐sensitive ABPs such as coronin distinguish the nucleotide state of F‐actin based on the conformation of the D‐loop, [2] which serves as a hub for ABP binding. Unfortunately, total internal reflection fluorescence (TIRF) microscopy, which is considered the state‐of‐the‐art method to measure the binding of ABPs to F‐actin, [2] was found incompatible with the photoswitchable ligand when measuring the binding of coronin‐1B to F‐actin (Figure S4, also see Methods), clearly indicating the limits of current biochemistry methods in photopharmacology.
The large cavity between the two strands of F‐actin, which is incompletely filled by OJ8 in either configuration (Figure 3 C), suggested that also the bound ligand should be responsive to light. Indeed, ion‐pair reversed‐phase chromatography showed reversible photoswitching of OJ8 inside actin filaments (Figure S5). This may suggest that optojasps can be incorporated in the trans state during F‐actin polymerization and may then be switched to the more strongly stabilizing cis form. Furthermore, the azobenzene “extension” does clearly not only provide a passive element of steric blockade (trans), but can also contribute actively to binding (cis). As a result, activity of both the cis‐ and the trans‐state of the optojasp depend on switch topology and linker length, cogently explaining the complex structure–activity‐correlation observed for optojasps. [6]
In summary, for the first time both isomeric states of a protein‐bound bioactive photoswitch were resolved by cryo‐EM. Notably, the metastable cis‐state of the optojasp photoswitch was characterized, which in this case actively contributes to binding by providing a fitting shape for a cavity in F‐actin. This does not only result in a better fit and higher activity for the cis‐isomer, but also in switching the F‐actin into a state resembling the phalloidin‐bound form. This conformational modulation of the whole F‐actin offers interesting explanations for different in cellulo activities observed previously for F‐actin ligands.[ 6 , 26 ] Such effects may now be further explored by structure‐based optimization of the optojasps. In combination, our data showcase the impact that cryo‐EM can have on understanding drug functioning on complex protein targets as well as on structure determination of ligands not accessible by Xray crystallography, opening up new prospects for structure‐based drug design.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary
Supplementary
Acknowledgements
We thank O. Hofnagel, D. Prumbaum and N. Bleimling for assistance with data collection, W. Linke and A. Unger (Ruhr‐Universität Bochum, Germany) for providing muscle acetone powder, T. Croll (University of Cambridge, UK) for providing custom restraints for ISOLDE and H. Görls (FSU Jena) for the structure query of cis‐azobenzene in the CSD. This work was supported by the Max Planck Society (to S.R.), the state of Thuringia (to H.‐D.A., grant 43‐5572‐321‐12040‐12) and the European Research Council under the European Union's Seventh Framework Programme (FP7/ 2007‐2013, grant 615984 to S.R.). S.P. was supported as a fellow of Studienstiftung des deutschen Volkes. V.N. received a DAAD predoctoral fellowship. A.B. was supported by an EMBO long‐term fellowship. Open access funding enabled and organized by Projekt DEAL.
S. Pospich, F. Küllmer, V. Nasufović, J. Funk, A. Belyy, P. Bieling, H.-D. Arndt, S. Raunser, Angew. Chem. Int. Ed. 2021, 60, 8678.
Dedicated to Professor Peter B. Dervan on the occasion of his 75th birthday
Contributor Information
Prof. Dr. Hans‐Dieter Arndt, Email: hd.arndt@uni-jena.de.
Prof. Dr. Stefan Raunser, Email: stefan.raunser@mpi-dortmund.mpg.de.
References
- 1. Svitkina T., Cold Spring Harbor Perspect. Biol. 2018, 10, a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Merino F., Pospich S., Funk J., Wagner T., Küllmer F., Arndt H.-D., Bieling P., Raunser S., Nat. Struct. Mol. Biol. 2018, 25, 528–537. [DOI] [PubMed] [Google Scholar]
- 3. Combeau C., Carlier M. F., J. Biol. Chem. 1988, 263, 17429–17436. [PubMed] [Google Scholar]
- 4. Merino F., Pospich S., Raunser S., Semin. Cell Dev. Biol. 2020, 102, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crews P., Manes L. V., Boehler M., Tetrahedron Lett. 1986, 27, 2797–2800. [Google Scholar]
- 6. Borowiak M., Küllmer F., Gegenfurtner F., Peil S., Nasufovic V., Zahler S., Thorn-Seshold O., Trauner D., Arndt H.-D., J. Am. Chem. Soc. 2020, 142, 9240–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tannert R., Milroy L.-G., Ellinger B., Hu T.-S., Arndt H.-D., Waldmann H., J. Am. Chem. Soc. 2010, 132, 3063–3077. [DOI] [PubMed] [Google Scholar]
- 8. Pospich S., Kumpula E.-P., von der Ecken J., Vahokoski J., Kursula I., Raunser S., Proc. Natl. Acad. Sci. USA 2017, 114, 10636–10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pospich S., Merino F., Raunser S., Structure 2020, 28, 437–449. [DOI] [PubMed] [Google Scholar]
- 10. Merino F., Raunser S., Angew. Chem. Int. Ed. 2017, 56, 2846–2860; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 2890–2905. [Google Scholar]
- 11. Hüll K., Morstein J., Trauner D., Chem. Rev. 2018, 118, 10710–10747. [DOI] [PubMed] [Google Scholar]
- 12. Croll T. I., Acta Crystallogr. Sect. D 2018, 74, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pollard T. D., J. Cell. Biol. 1986, 103, 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kudryashov D. S., Reisler E., Biopolymers 2013, 99, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das S., Ge P., Oztug Durer Z. A., Grintsevich E. E., Zhou Z. H., Reisler E., Structure 2020, 28, 586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. von der Ecken J., Müller M., Lehman W., Manstein D. J., Penczek P. A., Raunser S., Nature 2015, 519, 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou S. Z., Pollard T. D., Proc. Natl. Acad. Sci. USA 2019, 116, 4265–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ren Z., Zhang Y., Zhang Y., He Y., Du P., Wang Z., Sun F., Ren H., Plant Cell 2019, 31, 2855–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumari A., Kesarwani S., Javoor M. G., Vinothkumar K. R., Sirajuddin M., EMBO J. 2020, 39, e104006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Belyy A., Merino F., Sitsel O., Raunser S., PLoS Biol. 2020, 18, e3000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown C. J., Acta Crystallogr. 1966, 21, 146–152. [Google Scholar]
- 22. Bouwstra J. A., Schouten A., Kroon J., Acta Crystallogr. Sect. C 1983, 39, 1121–1123. [Google Scholar]
- 23. Mostad A., Rømming C., Acta Chem. Scand. 1971, 25, 3561–3568. [DOI] [PubMed] [Google Scholar]
- 24. Hampson G. C., Robertson J. M., J. Chem. Soc. 1941, 409–413. [Google Scholar]
- 25. Lynen F., Wieland U., Justus Liebigs Ann. Chem. 1938, 533, 93–117. [Google Scholar]
- 26. Wang S., Crevenna A. H., Ugur I., Marion A., Antes I., Kazmaier U., Hoyer M., Lamb D. C., Gegenfurtner F., Kliesmete Z., Ziegenhain C., Enard W., Vollmar A., Zahler S., Sci. Rep. 2019, 9, 9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary
Supplementary


