Summary
Quantitative information on the spatiotemporal distribution of polarised proteins is central for understanding cell‐fate determination, yet collecting sufficient data for statistical analysis is difficult to accomplish with manual measurements.
Here we present Polarity Measurement (Pome), a semi‐automated pipeline for the quantification of cell polarity and demonstrate its application to a variety of developmental contexts.
Pome analysis reveals that, during asymmetric cell divisions in the Arabidopsis thaliana stomatal lineage, polarity proteins BASL and BRXL2 are more asynchronous and less mutually dependent than previously thought. A similar analysis of the linearly arrayed stomatal lineage of Brachypodium distachyon revealed that the MAPKKK BdYDA1 is segregated and polarised following asymmetrical divisions.
Our results demonstrate that Pome is a versatile tool, which by itself or combined with tissue‐level studies and advanced microscopy techniques can help to uncover new mechanisms of cell polarity.
Keywords: BASL, BRXf, cell polarity, image quantification, stomatal lineage
Introduction
Cell polarity is the central mechanism responsible for organising a cell into subdomains with specialised functions. The subcellular enrichment of polarity proteins enables complex processes like cell migration, directional long‐range signal transduction, cell growth anisotropy and asymmetrical cell division (ACD) (Drubin & Nelson, 1996; Muroyama & Bergmann, 2019). Due to the sessile lifestyle of plants, cell polarity is particularly important for the proper execution of developmental and physiological programmes. Plants need to adjust their development based on extrinsic cues and the lack of cell migration puts greater emphasis on the orientation of ACDs and differential cell expansion for tissue patterning.
Although cell polarity can be manifested in the distribution of many components, including organelles, RNAs and metabolites, proteins polarly distributed at the cell cortex play especially prominent roles in plants. These ‘polarity proteins’ may be integral or peripheral membrane components and are often encoded only in plant genomes. Their distribution defines cellular and tissue‐level axes, and they direct the development of major body axes, tissue layers and the distribution of specialised cell types within a tissue (Muroyama & Bergmann, 2019). For example, in the stomatal lineage of the Arabidopsis leaf epidermis, polarly localised BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) and the BREVIS RADIX family (BRXf) are required to orient ACDs and ensure that the daughters of such divisions take on different fates (Dong et al., 2009; Rowe et al., 2019).
Many other proteins exhibiting polarised distributions in plants have been identified over the past decade, but our understanding of the mechanisms that control their distribution is still in its infancy (Dong et al., 2009; Scacchi et al., 2009; Zourelidou et al., 2009; Houbaert et al., 2018; Marhava et al., 2018; Denninger et al., 2019; Yoshida et al., 2019; Tan et al., 2020; van Dop et al., 2020; Zhang et al., 2020). Quantifying the distribution of polar proteins in time and space is a necessary first step in understanding how cell polarity is formed and coordinated with neighbouring or daughter cells. Comparing the polar domain size of different polarity proteins can help infer how protein complexes are assembled, while the temporal dynamics of the protein abundance along the membrane can reveal trafficking and post‐translational regulatory mechanisms.
In animal systems, quantification of polarity proteins has already challenged old paradigms and revealed unforeseen phenomena. For example, PAR proteins in animal cells have been known for nearly 30 yr, yet a recent 3D image segmentation and quantitative analysis of PAR proteins in the Caenorhabditis elegans embryo germline revealed that PAR polarity domain size does not scale with cell size and this intrinsic property regulates the timing of cell‐fate transition (Hubatsch et al., 2019).
Similar quantitative analyses in plants would help to identify critical players and novel mechanisms in the establishment of cell polarity. However, most studies so far have relied heavily on tedious visual inspection and remain qualitative (Wisniewska et al., 2006; Dong et al., 2009; Scacchi et al., 2009; Zhang et al., 2016; Marhava et al., 2018; Rowe et al., 2019; Yoshida et al., 2019; van Dop et al., 2020). Early efforts to define ‘polarity indices’ fall roughly into two categories: (i) those that compare protein abundance between the peak of the polarity ‘crescent’ and the opposite side of the cell; and (ii) those that measure the relative size of the crescent. Neither of these two types of polarity indices captures the full picture of protein distribution. Polarity indices that only compare protein abundance between poles may be insufficient to distinguish between the subtle contributions of the multitude of processes that shape the distribution (Marhavy et al., 2014; Zhang et al., 2015; Langowski et al., 2016; Houbaert et al., 2018; Denninger et al., 2019). Conversely, polarity indices that consider only the width of the crescent (Zhang et al., 2015) and do not compare protein abundance at the poles, are also insufficient to fully describe polarisation, as we will show in this work.
Here, we present Polarity Measurement (Pome), an image analysis pipeline designed to measure polarity in plant cells from confocal fluorescence images. Our tool converts the measurement of individual cells into data sets that can be interrogated with a variety of statistical tools. We demonstrate the use of Pome to systematically quantify the polarity of BASL and BRXf over time in the Arabidopsis stomatal lineage. Based on these detailed measurements, we modify previous models for how these proteins establish and maintain polarity in stomatal lineage cells. We also demonstrate how Pome can quantify aspects of asymmetric protein distributions in different tissues and organisms and again reveal that previous models for polarity establishment must be reconsidered. Finally, to facilitate community access and implementation of Pome, we provide our pipeline and documentation in an accessible format.
Materials and Methods
Plant material
All Arabidopsis lines used in this study were generated in the Col‐0 background. Construction of all polarity protein reporter lines (pBRXL2::BRXL2‐YFP pML1::mCherry‐RCI2A, pBASL::YFP‐BASL pML1::mCherry‐RCI2A, pBASL::Myr‐BRX‐YFP pML1::mCherry‐RCI2A brxq, pBRXL2::BRXL2‐YFP pML1::mCherry‐RCI2A basl‐2, pPIN2::PIN2‐GFP) have been reported elsewhere (Wisniewska et al., 2006; Rowe et al., 2019). The Brachypodium BdYDA1 reporter line pBdYDA1::BdYDA1‐YFP was generated in the Bd21‐3 ecotype as described elsewhere (Abrash et al., 2018).
Plant growth conditions
All Arabidopsis seeds were surface sterilised by bleach or 75% ethanol and stratified for 2 d. After stratification, seedlings were vertically grown on half½strength Murashige and Skoog (½MS) medium for 3–6 d under long‐day conditions (16 h : 8 h, light : dark, at 22°C).
Microscopy and image acquisition
All imaging experiments for Arabidopsis were performed on a Leica SP5 confocal microscope with HyD detectors using ×25 NA0.95 and ×40 NA1.1 water objectives with image size 1024 × 1024 and digital zoom from ×1 to ×2.5. For time‐lapse experiments, 3‐d post germination (dpg) seedlings were mounted in a custom imaging chamber filled with ½MS solution (Davies & Bergmann, 2014; Bringmann & Bergmann, 2017; Simmons et al., 2019). Laser settings for each reporter, except the membrane marker (pML1::mCherry‐RCI2A or PI), were adjusted to avoid over‐saturation. For the time‐lapse experiments reported in this study, 30 or 40 min intervals between each image stack capture were used.
Images of the BdYDA1 reporter in developing Brachypodium leaves were reanalysed from the same set of raw images reported in Fig. 3 of Abrash et al. (2018). Images of PAR dynamics in C. elegans embryos were reanalysed from the raw images of supplementary video 2 reported in Hubatsch et al. (2019).
Fig. 3.

Pome shows that BRXL2 can transiently polarise during asymmetrical cell divisions (ACDs) in the basl‐2 mutant. (a) BRXL2 localisation pattern during cell division in wild‐type (Col‐0) and the basl‐2 mutant. Cells marked with pML1::RCI2A‐mCherry (magenta) and pBRXL2::BRXL2‐YFP (green) are tracked before, during and after ACDs, where 00:00 (h:min) corresponds to the formation of the cell plate. (b) Pome measurements of BRXL2 polarity from cells in (a). The formation of a transient BRXL2 polar crescent in basl‐2 at time frame 00:00 and 00:40 is indicated by white arrows in (a) and (b). (c, d) BRXL2 SD and support vector machine (SVM) polarity index estimated for each time point in (a). The decision boundary of each classification model is labelled with a solid black line and the region is classified as polarised is marked in shaded orange. Bars in (a), 5 μm.
Pome analysis of fluorescence images
All raw fluorescence image Z‐stacks were projected with Sum Slices in Fiji unless noted otherwise. For all time‐lapse images, drift was corrected using the Correct 3D Drift plugin (Parslow et al., 2014). These time‐lapse images were then split into individual time frames and treated as still images for analysis afterwards. Each individual cell of interest was then examined for manual correction before quantification, when regions containing interfering vesicles (images with FM4‐64 staining) or nucleus (images of BASL reporter) were manually removed. When running Pome (Supporting Information Methods S1) on a selected cell, the settings were adjusted per experiment. Information on how to determine Pome settings and examples of Pome data analysis are described in detail in the Pome user guide files (Notes S1, S2) accompanying this manuscript. The results of Pome measurement were then imported, summarised and analysed in RStudio.
Fitting the polarity protein distribution along the cell membrane to a Gaussian model
For polarity proteins in Arabidopsis and Brachypodium stomatal lineage, the mean florescence intensity of the polarity protein reporter along the cell membrane was fitted to a Gaussian model (Eqn 1) by nonlinear regression with the nls function from the stats package (R Core Team, 2020) in rstudio. Key parameters of this regression model, including SD α, centre µ, amplitude α and baseline value β, were estimated using the formula:
| (Eqn 1) |
Generating polarity classification matrixes
To determine if the parameters (σ, α and β) estimated by Pome were sufficient to distinguish between polarised (BRXL2 in stomatal lineage ground cells and BASL) and depolarised (BRXL2 in guard mother cells and Myr‐BRX) markers, we used measurements from 80 cells (20 cells per marker) to train a binary logistic regression model using the glm function from the stats package in rstudio. To visualise each parameter’s ability to classify polarity, a receiver operating characteristic (ROC) plot was generated with the roc function from the proc package (Robin et al., 2011). The resulting plot (Fig. 1h) suggest that σ alone could successfully discriminate between polarised and depolarised markers. Next, a simpler binary logistic regression model was built using only σ (Fig. 1i) to determine a suitable cut‐off value. To calculate the error rate of the polarity ~ σ logistic regression model, a 10‐fold cross‐validation prediction error was calculated with the training dataset with the cv.glm function in the boot package (Davison & Hinkley, 1997), where an average reporting error of 9.2% was reported.
Fig. 1.
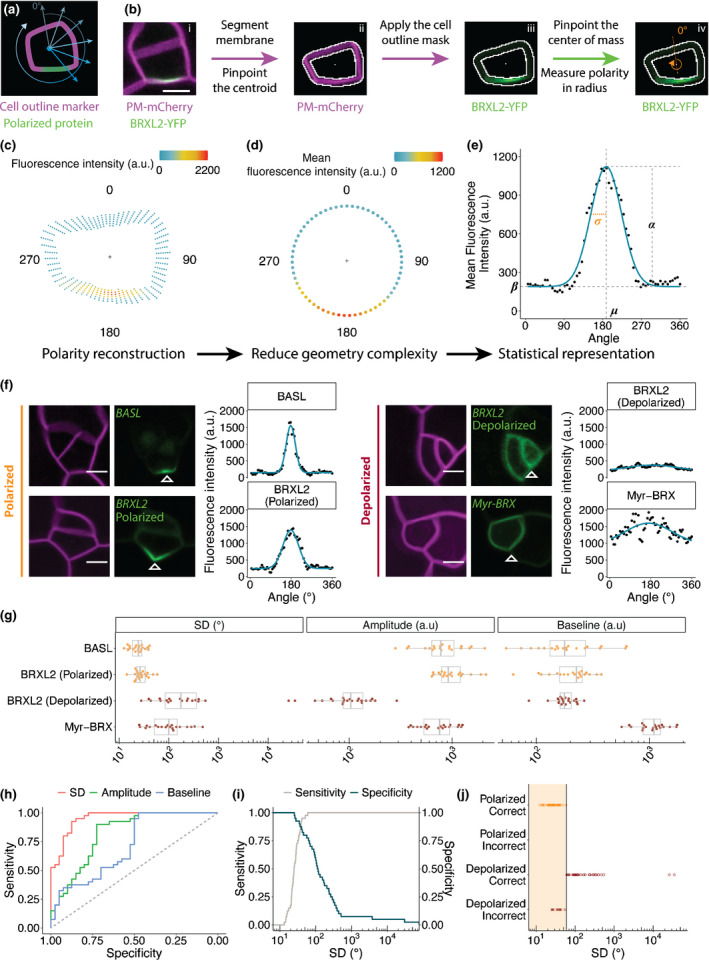
Pome measures polarity in cells of the Arabidopsis stomatal lineage. (a) Cartoon of cell polarity analysis in Pome. (b) Work flow. (i) Example of a two‐channel input image in which one channel is a plasma membrane marker (magenta) and the second is a protein whose polarity will be quantified, in this case BRXL2‐YFP (green). (ii) Result of the automatic segmentation and centroid detection. (iii) Application of the cell outline mask to the polarity channel. (iv) Quantification of fluorescence intensity along the membrane. A line is drawn from the cell centroid (white dots in (ii) and (iii)) to the BRXL2 centre of mass (green dot in (iv)) to define 0° and 180° angles. (c, d) Reconstruction and visualisation of cell polarity from Pome measurements. The cell centroid is marked with ‘+’. BRXL2 fluorescence intensity of each measured pixel (c) and the average BRXL2 fluorescence intensity of all the measured pixels at a given angle (d) are represented by dots coloured by their fluorescence intensity. (e) Curve fitting of the average pixel intensities to a Gaussian model (blue line). The four parameters from the regression model are labelled as: standard deviation (SD) σ, centre µ, amplitude α and baseline β. (f) Representative confocal images and Pome measurements of polarised and depolarised markers. Magenta: pML1::RCI2A‐mCherry; green: pBASL::YFP‐BASL (upper left), pBRXL2::BRXL2‐YFP (lower left and top right), pBASL::Myr‐BRX‐YFP (lower right). Open white triangles indicate the centres of the polarity domain suggested by Pome. (g) Comparison of the fitted parameter values for each marker (n = 20 cells/marker). (h) Receiver operating characteristic (ROC) curve generated from the binary logistic regression model built using the SD, amplitude and baseline. The closer the curve is to the upper left corner, the higher the predictive power of the parameter. (i) Sensitivity and specificity curves for a range of polarisation cut‐off values of SD. (j) Result of the classification of the training data set using a SD cut‐off value of 60°. The region classified as polarised is marked in shaded orange. Bars, in (b, f) are 5 μm.
For the support vector machine (SVM) model, the parameters from the same 80 cells in the binary logistic regression model were used as training data. To remove imaging setting and reporter basal expression bias, a normalised amplitude α′ was calculated by dividing the amplitude α by the mean fluorescence intensity of all angles. The SVM model was then built with the svm function in the e1071 package (Meyer et al., 2019). Next, linear coefficients were extracted from the model to calculate the slope (3.672) and intercept (0.069) of the decision boundary (Fig. S2b). We can then define an SVM polarity index as the distance to the SVM model decision boundary calculated with the formula:
| (Eqn 2) |
A positive SVM polarity index indicates a polarised cell, while a negative SVM polarity index indicates a depolarised cell. Higher SVM polarity index values indicate higher polarisation.
Statistical analysis
All statistical analyses were performed in rstudio. Unpaired Mann–Whitney U‐tests were conducted to compare two data samples with the compare_means function from the ggpubr package (Kassambara, 2020).
Results and Discussion
Pome: a semi‐automated tool to quantify cortical protein polarity
The Arabidopsis stomatal lineage is a powerful model for genetic analysis of asymmetrical divisions, but mechanistic dissection of cell polarity has been challenging. BASL and BRXf polarity (demonstrated with BRX and BRXL2) is required for ACDs (Dong et al., 2009; Rowe et al., 2019), but how these proteins initiate and maintain polarisation is not understood. The primary challenge hindering quantitative analysis in the stomatal lineage is the rare and transient nature of cells undergoing ACDs, and demands the imaging of a large number of samples to discern statistically significant differences between genotypes or conditions. Additionally, cell shapes in the stomatal lineage are irregular and variable, which can make drawing conclusions from visual inspection difficult and adds uncertainty to manual measurements.
To reduce the time required for quantification and to collect shape‐agnostic measurements, we developed Pome, a pipeline composed of an intuitive Fiji macro and an R script that automatically extracted cortical polarity information from user‐defined cells. Pome requires confocal images of a cell with two different channels. One of these channels must contain a uniform cell outline marker (e.g. integral plasma membrane reporter, FM 4‐64 or propidium iodide). The macro then quantifies the fluorescence of polar proteins in the second channel at different angles (Fig. 1a–d). This simplification provides comparable results that are independent of cell shape. Users can specify the number of measurements (i.e. angles) along the membrane.
The macro calculates an angle of polarisation with respect to the field of view after extracting the centroid and the centre of mass of the polar protein in the measured cell from the outline marker and the polar protein channels, respectively. A line from the centroid to the centre of mass defines the 0° and 180° angles used for reporting results (Fig. 1b). Further analysis of these angles of polarisation could reveal the existence of tissue‐level coordination, as has been shown by previous studies (Bringmann & Bergmann, 2017; Mansfield et al., 2018).
Initially, we used Pome to quantify the distribution of BRXL2 in stomatal lineage ground cells (SLGCs) and found that its protein abundance at the membrane is well represented by a Gaussian model (Fig. 1e). This allowed us to describe the BRXL2 localisation with three parameters: SD, σ, amplitude (α) and baseline intensity (β) (Fig. 1e).
To evaluate the capacity of Pome to distinguish among degrees of polarity, we applied it to three additional cases: BASL (polarised), BRXL2 in guard mother cells (GMCs, depolarised) and myristoylated BRX (Myr‐BRX, depolarised) (Figs 1f,S1). These cases present a variety of challenges to polarity quantification, including signal interference from the nucleus (BASL), weak expression causing a low signal‐to‐noise ratio (depolarised BRXL2 in GMCs) and signals from abutting membranes in two neighbouring cells interfering with each other signals (myr‐BRX). We selected 20 cells for each marker and measured their distribution along the cell periphery. The distributions of these markers were different, but they can all be described by a Gaussian model (Fig. 1f). The SD, amplitude and baseline intensity (Fig. 1g) for BASL and the polarised BRXL2 were statistically indistinguishable (Fig. S2a). The distribution of the depolarised (cytosolic) BRXL2 and Myr‐BRX were clearly different from each other and from the distribution of the polarised BASL and BRXL2. Note that the SD of depolarised BRXL2 and Myr‐BRX were statistically indistinguishable, but their amplitudes and baselines were not, reflecting differences in fluorescence levels and possibly protein abundance (Fig. 1g).
Next, we compared the SD, amplitude and baseline intensity between our polarised and depolarised datasets using binary logistic regression and generated a ROC curve for each parameter to test its ability to classify polarity (Fig. 1h), where the closer the curve is to the upper left corner on the ROC plot, the higher the predictive power of the parameter. These results suggested that SD alone is predictive of polarity. Furthermore, SD is nearly independent of the expression level of the marker and the imaging setting, making it a reliable polarity classifier. To determine the cut‐off value for polarity classification using SD as the sole classifier, we plotted the sensitive (true positive rate) and specificity (true negative rate) for each SD cut‐off value. We found that a cut‐off of 42° can correctly discriminate between a polarised and a depolarised marker with a 95% classification sensitivity and 83% specificity, while a cut‐off of 60° achieved 100% classification sensitivity and 78% specificity (Fig. 1i,j). Alternatively, an SVM algorithm can be used to generate a polarity classification model with 100% classification sensitivity and 100% specificity using the SD and normalised amplitude (α′ amplitude divided by mean fluorescence intensity) (Fig. S2b,c).
Characterising the dynamics of polarity proteins in the stomatal lineage
We implemented Pome to analyse the dynamics of BASL and BRXf distribution. Biochemical studies indicated that BASL and BRXf interact, while genetic analysis indicated that they require each other to remain polarised (Rowe et al., 2019). BASL and BRXf have been shown to occupy the same general region of the cell cortex, but the evidence is limited to single time point images and qualitative interpretation. As a result, it is unclear if their polarity domains always overlap or whether one protein polarises first and then recruits the other.
To address this question, we first measured the distribution of BRXL2 before and after an ACD, from the initial formation of its polar crescent to its complete dissociation from the membrane (Fig. 2a). We fitted the fluorescence measurements of each time frame to a Gaussian model and extracted the SD, amplitude and baseline (Fig. 2b). We found that BRXL2 became polarised 5 ± 2.5 h before the formation of the new cell wall and remained polarised for over 9 ± 2.5 h afterward, using the SD cut‐off of 60° from our binary logistic regression. The SD and baseline values of the BRXL2 distribution remained remarkably constant over this period, while the amplitude increased and then decreased. This result could be consistent with scenarios in which BRXL2 is locally delivered to the crescent and retained there, or uniformly delivered to the membrane but transported into the crescent more rapidly than it can diffuse away.
Fig. 2.
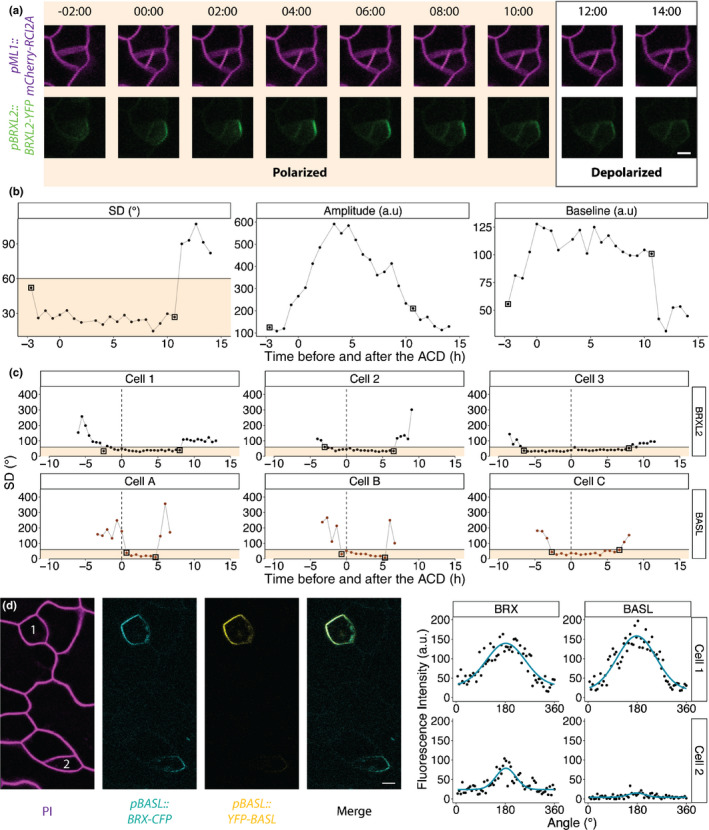
Pome reveals differences in BRXL2 and BASL polarity dynamics during asymmetrical cell divisions (ACDs). (a) Time course of BRXL2 localisation during an ACD. Cells are marked with pML1::RCI2A‐mCherry (magenta) and pBRXL2::BRXL2‐YFP (green). Time point 00:00 (h:min) marks the formation of the cell plate. (b) Changes in SD, amplitude and baseline of the BRXL2 distribution determined by Pome. (c) Plotted measurements of individual cells, each expressing either BRXL2 (top cells, 1–3) or BASL (bottom cells, A–C). Vertical dashed lines indicate cell plate formation. Solid horizontal lines indicate a SD cut‐off value of 60° and all values in shaded orange are classified as polarised. The 60° SD cut‐off value that achieved 100% classification sensitivity is chosen because of the lower image resolution and lower signal‐to‐noise ratio of time‐lapse images compared with still images. For each cell, the first and last time‐points showing polarisation are marked by black boxes. (d) BRX (cyan) and BASL (yellow) localisation pattern during the late G2/early mitotic phase in two meristemoids. Note that in cell 2, BRX is polarised while BASL is not. Bars in (a, d), 5 μm.
Subsequently, we performed a similar characterisation of BASL dynamics. Comparison of the dynamics of BRXL2 and BASL showed that BRXL2 polarised earlier than BASL during ACDs and stayed polarised longer than BASL afterward (Figs 2c,S3). A dual‐reporter line expressing BRX‐CFP (a redundant homologue of BRXL2 in the stomata lineage) and BASL‐YFP further supported the hypothesis that BRXf polarises ahead of BASL during ACDs. In still images of the dual‐reporter line, from which we could obtain higher resolution images than in time‐lapse images, we identified a few cells at late G2/early mitotic phase where BRX, but not BASL, was polarised (Fig. 2d).
We also examined the dynamics of BRXL2 in basl‐2 null mutants, where BRXL2 was believed to be depolarised (Rowe et al., 2019). Strikingly, we found that, in some basl‐2 cells, BRXL2 could still polarise transiently during ACDs (Fig. 3a) and Pome could accurately describe these polarity changes (Fig. 3b). In these basl‐2 cells, BRXL2 was observed to polarise right after the formation of the cell wall that separates the new sister cells. This transient polarity coincided with the time window in which BRXL2 polarity is often enhanced in wild‐type cells (Fig. 3a,c,d). Whether this transient BRXL2 polar crescent in basl‐2 is functional, however, is debatable, as it did not show up in all the dividing cells and the basl‐2 mutant had evident ACD defects (Dong et al., 2009). Nevertheless, these results suggested that BRXL2 accumulates at the polar crescent first and then recruits BASL and the formation of this complex is required to retain both proteins at the crescent.
Applications of Pome in other developmental contexts
Pome was initially designed to address the challenges of measuring polarity in dispersed, asynchronously dividing, leaf epidermal cells. An automatic tool to quantify polarity, however, is useful for other developmental contexts and the simple design of Pome should make it adaptable to other tissues and organisms.
In Arabidopsis, the MAPKKK YODA (AtYDA) is polarised and segregated preferentially to larger daughter cells in the stomatal lineage ACDs by BASL (Zhang et al., 2015). In Brachypodium, a YODA homologue, BdYDA1, is also required for stomatal lineage ACDs (Abrash et al., 2018). However, the Brachypodium genome does not encode a BASL homologue (Bowles et al., 2020). It was therefore unclear whether BdYDA1 would be polarised in the Brachypodium stomatal lineage. Grass stomata are arranged in cell files and exhibit a base‐to‐tip gradient of development. Meristemoids undergo directional ACDs such that the smaller daughter cell (stomatal precursor, GMC) is oriented towards the tip and the larger one (pavement cell precursor) towards the base (Fig. 4a,b). To determine if BdYDA1 is polarised, we used Pome to quantify its distribution in individual meristemoids before ACDs and in the newly formed larger daughter cells resulting from this division (Fig. 4a,b).
Fig. 4.
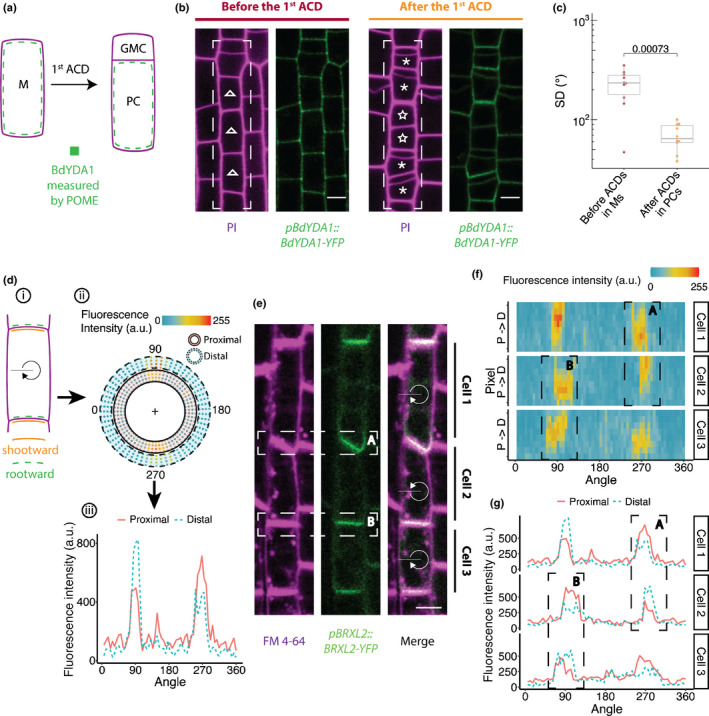
Application of Pome to quantifying polarity in diverse cell types. (a–c) Pome reveals BdYDA1 is polarised in Brachypodium stomatal lineage cells. (a) Schematic of the cell types in the Brachypodium stomatal lineage. Meristemoids (M) divide asymmetrically to generate one small guard mother cell precursor (GMC) and one large pavement cell precursor (PC). (b) pBdYDA1::BdYDA1‐YFP (green) localisation pattern in stomatal lineage cells (indicated by white‐dashed boxes) before (left) and after (right) the first asymmetric cell divisional (ACD). Cell boundaries are visualised by propidium iodide (PI, magenta). Ms are labelled with open triangles and PCs with asterisks. Note that not all the Ms have undergone ACDs in the right panel. The two Ms in the middle (labelled with stars) have not divided yet and exhibit no BdYDA1 polarity. (c) Plot of the SD indicates that BdYDA1 is not polarised in meristemoids (pre‐ACD), but is polarised in PC precursors (n = 10 cells/stage). (d–g) Application of Pome to determine shootward or rootward polarity in contiguous cells. (d) Scheme of how Pome can be used to determine which of two adjacent cells is expressing a polarised marker. (i) In contiguous cells, a polarised protein could be localised to the upper side (shootward, orange line) and/or the lower side (rootward, green dashed line). Pome can be applied to distinguish these different patterns. The black line indicates the 0° angle and the clockwise arrow indicates the direction of rotation for Pome measurement. Pome measurement of a rootward polarity protein containing cell is shown in ii) and iii). (ii) Reconstruction of marker localisation in proximal and distal membrane regions. Fluorescence in the proximal membrane region (within the solid rings) is contributed by the selected cell, while fluorescence in the distal membrane region (within the dashed rings) is contributed by the neighbouring cells. (iii) Comparison of the marker intensity in the proximal (solid red line) and distal regions (dashed blue line) at a particular angle indicates whether the signal at the membrane is mostly coming from the neighbouring cell (as shown at 90°) or from the target cell (as shown at 270°). (e) Rootward localisation of BRXL2 (green) in three adjacent lateral root cap cells in Arabidopsis. Plasma membrane visualised with FM4‐64 (magenta). Regions of interest (ROIs) analysed to determine on which side of the shared interphase a polarised protein resides are indicated by dashed boxes labelled as region A and region B in (e), (f) and (g). (f) Heat map of the fluorescence intensity of the BRXL2 localisation shown in (e). For each angle, the four innermost and outermost pixels are considered the proximal (P) and distal (D) membrane regions, respectively. (g) For the three cells shown in (e), the total fluorescence intensities of the proximal (solid red line) and distal (dashed blue line) membrane regions are plotted at each angle. Bars in (b, e), 5 μm.
BdYDA1 was not polarised in meristemoids but, after ACDs, polarised BdYDA1 was detected in the larger daughter cell (Fig. 4b,c). This segregation and polarisation of BdYDA1 is consistent with a role restricting stomatal fate and these larger daughters often wrongfully acquire stomatal identity in the loss‐of‐function mutant bdyda1 (Abrash et al., 2018). The position of polar BdYDA1, distal to the new division plane in the larger daughter cell, is equivalent to where AtYDA localises in a BASL‐dependent manner in Arabidopsis (Zhang et al., 2015). This leads to the interesting conclusion that MAPKKKs can and must polarise during stomatal ACDs to ensure differential fates in different plants, but BASL may be newly recruited into this role in dicots and alternative mechanisms must exist to polarise BdYDA1.
The expression of BdYDA1 in each of many contiguous stomatal cells represented a complication for automated polarity analysis. We had to rely on prior knowledge on oriented ACDs (Abrash et al., 2018) to infer in which cells and on what cell face BdYDA1 was localised. The problem of assigning polarity to individual contiguous cells is quite common. In Arabidopsis roots, for example, cells are also arranged in files and localisation of proteins to specific faces is required for the development and physiological functions of the root. Many proteins in the root and vascular systems, especially those involved in polar auxin transport, display polar localisation to ‘shootward’ and/or ‘rootward’ faces of the cell (Wisniewska et al., 2006; Scacchi et al., 2009; Zourelidou et al., 2009; Breda et al., 2017; Marhava et al., 2018) (Figs 4e, S4a). Determining whether a protein is present at both shootward and rootward faces in a cell within a contiguous file has been challenging.
To test whether Pome could be used to determine which side of the interface between two adjacent cells a protein resides, we quantified the localisation of BRXL2 and the transmembrane protein PIN2 in Arabidopsis roots. BRXL2 has been reported to localise rootward in roots (Marhava et al., 2020) and PIN2 to localise shootward in root epidermis cells (Wisniewska et al., 2006). We quantified the pixel intensity at multiple concentric ‘rings’ of the plasma membrane. Fluorescence in the proximal membrane region (within the solid rings in Fig. 4d) is contributed by the selected (target) cell, while fluorescence in the distal membrane region (within the dashed rings) is considered to be contributed by the neighbouring cell. With this modified Pome protocol, we validated BRXL2’s rootward localisation pattern in lateral root cap cells (Fig. 4e–g) and PIN2’s shootward localisation in root epidermal cells (Fig. S4). This modified version of Pome can also be applied to validate bi‐polar localisation, when polarity proteins are present at both shootward and rootward faces. In this case, no appreciable differences will be observable between proximal and distal measurements at the membrane sections shared between the continuous cell file and the signal intensity of the polarity reporter will be higher at such membrane sections. The same principle of capturing pixel intensity of a reporter across concentric ‘rings’ could also be used to determine the redistribution of proteins between membrane and cytoplasm, a common response to environmental perturbation in plants (Zourelidou et al., 2009; Marhava et al., 2018).
Conclusion
Plant proteins exhibit staggering variation in cortical polar domains. They can be found linked to dispersed ACDs, in contiguous cell files aligned with organ axes, or nestled in cell corners. Pome can quantify dynamic polarity in these different contexts and complements other recent advances in cell segmentation and quantitative image analysis (Wolny et al., 2020). Pome converts a once low‐throughput and qualitative procedure into a semi‐automated one that is amicable to statistical analysis. It could be even more powerful when combined with other microscopy techniques such as fluorescence resonance energy transfer (FRET) that directly measure protein interaction.
Ultimately, we expect fluorescence microscopy and tools like Pome to answer some and open new questions in cell biology. In our exploration of stomatal lineage proteins in two plant species, we were able to find that BASL, while a central hub in stomatal polarity, may be preceded by BRXL2 polarity in Arabidopsis and must be supplanted by other mechanisms for establishing polar localisation of the MAPKKK BdYDA1 in Brachypodium. Pome also allowed us to validate the rootward and shootward localisation of BRXL2 and PIN2, respectively, in Arabidopsis roots. Furthermore, the utility of Pome is not limited to plants, as we were also able to use it to quantify PAR‐2 polarity in C. elegans embryos (Hubatsch et al., 2019) (Fig. S5). The simplicity and versatility of our pipeline make it accessible to the broad research community regardless of previous programming skills.
Author contributions
YG, RV, EW, DCB and LSC designed the project, interpreted the data and wrote the manuscript; YG, RV and EW performed the experiments; YG, RA and LSC designed the image quantification pipeline.
Supporting information
Fig. S1 Localisation pattern of polarity proteins used to test Pome in the Arabidopsis stomatal lineage.
Fig. S2 Additional polarity parameters and classification models.
Fig. S3 Confocal images of BRXL2 and BASL localisation pattern during asymmetrical cell divisions (ACDs).
Fig. S4 Application of Pome in root epidermal cells to validate shootward localisation of PIN2.
Fig. S5 Application of Pome in quantifying PAR‐2 polarity in the Caenorhabditis elegans P cell lineage during embryo development.
Methods S1 Pome macro V1.
Notes S1 Brief user guide for Pome.
Notes S2 Pome data analysis in R.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Nathan Goehring for providing the raw images of PAR protein behaviours in Caenorhabditis elegans embryo and Ximena Anleu‐Gil for images of BdYDA1 in Brachypodium. We thank Katelyn McKown, Gabriel Amador, Andrea Mair and other members of the Bergmann Laboratory for valuable feedback on the manuscript. We thank Joao Romalho for providing suggestions on the user guide for Pome. EW is supported by the German Research Foundation grant no. 438457603. DCB is an investigator of the Howard Hughes Medical Institute. This material is based upon work supported by the National Science Foundation under grant no. 1942722 to LSC. The authors declare no competing interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Abrash E, Anleu Gil MX, Matos JL, Bergmann DC. 2018. Conservation and divergence of YODA MAPKKK function in regulation of grass epidermal patterning. Development 145: dev165860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles AMC, Bechtold U, Paps J. 2020. The origin of land plants is rooted in two bursts of genomic novelty. Current Biology 30: 530–536. [DOI] [PubMed] [Google Scholar]
- Breda AS, Hazak O, Hardtke CS. 2017. Phosphosite charge rather than shootward localization determines OCTOPUS activity in root protophloem. Proceedings of the National Academy of Sciences, USA 114: E5721–E5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M, Bergmann DC. 2017. Tissue‐wide mechanical forces influence the polarity of stomatal stem cells in Arabidopsis. Current Biology 27: 877–883. [DOI] [PubMed] [Google Scholar]
- Davies KA, Bergmann DC. 2014. Functional specialization of stomatal bHLHs through modification of DNA‐binding and phosphoregulation potential. Proceedings of the National Academy of Sciences, USA 111: 15585–15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AC, Hinkley DV. 1997. Bootstrap methods and their application. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Denninger P, Reichelt A, Schmidt VAF, Mehlhorn DG, Asseck LY, Stanley CE, Keinath NF, Evers JF, Grefen C, Grossmann G. 2019. Distinct RopGEFs successively drive polarization and outgrowth of root hairs. Current Biology 29: 1854–1865.e5. [DOI] [PubMed] [Google Scholar]
- Dong J, MacAlister CA, Bergmann DC. 2009. BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dop M, Fiedler M, Mutte S, de Keijzer J, Olijslager L, Albrecht C, Liao CY, Janson ME, Bienz M, Weijers D. 2020. DIX domain polymerization drives assembly of plant cell polarity complexes. Cell 180: 427–439.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. 1996. Origins of cell polarity. Cell 84: 335–344. [DOI] [PubMed] [Google Scholar]
- Houbaert A, Zhang C, Tiwari M, Wang K, de Marcos Serrano A, Savatin DV, Urs MJ, Zhiponova MK, Gudesblat GE, Vanhoutte I et al. 2018. POLAR‐guided signalling complex assembly and localization drive asymmetric cell division. Nature 563: 574–578. [DOI] [PubMed] [Google Scholar]
- Hubatsch L, Peglion F, Reich JD, Rodrigues NT, Hirani N, Illukkumbura R, Goehring NW. 2019. A cell size threshold limits cell polarity and asymmetric division potential. Nature Physics 15: 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. 2020. ggpubr: ‘ggplot2' based publication ready plots . [WWW document] URL https://CRAN.R‐project.org/package=ggpubr.
- Langowski L, Wabnik K, Li H, Vanneste S, Naramoto S, Tanaka H, Friml J. 2016. Cellular mechanisms for cargo delivery and polarity maintenance at different polar domains in plant cells. Cell Discovery 2: 16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield C, Newman JL, Olsson TSG, Hartley M, Chan J, Coen E. 2018. Ectopic BASL reveals tissue cell polarity throughout leaf development in Arabidopsis thaliana. Current Biology 28: 2638–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P, Aliaga Fandino AC, Koh SWH, Jelinkova A, Kolb M, Janacek DP, Breda AS, Cattaneo P, Hammes UZ, Petrasek J et al. 2020. Plasma membrane domain patterning and self‐reinforcing polarity in Arabidopsis. Developmental Cell 52: 223–235.e5. [DOI] [PubMed] [Google Scholar]
- Marhava P, Bassukas AEL, Zourelidou M, Kolb M, Moret B, Fastner A, Schulze WX, Cattaneo P, Hammes UZ, Schwechheimer C et al. 2018. A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558: 297–300. [DOI] [PubMed] [Google Scholar]
- Marhavy P, Duclercq J, Weller B, Feraru E, Bielach A, Offringa R, Friml J, Schwechheimer C, Murphy A, Benkova E. 2014. Cytokinin controls polarity of PIN1‐dependent auxin transport during lateral root organogenesis. Current Biology 24: 1031–1037. [DOI] [PubMed] [Google Scholar]
- Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F. 2019. e1071: Misc functions of the department of statistics, probability theory group (formerly: E1071) . [WWW document] URL https://CRAN.R‐project.org/package=e1071.
- Muroyama A, Bergmann D. 2019. Plant cell polarity: creating diversity from inside the box. Annual Review of Cell and Developmental Biology 35: 309–336. [DOI] [PubMed] [Google Scholar]
- Parslow A, Cardona A, Bryson‐Richardson RJ. 2014. Sample drift correction following 4D confocal time‐lapse imaging. Journal of Visualized Experiments 86. doi: 10.3791/51086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2020). R: A language and environment for statistical computing, v. 4.0.1. [WWW document] URL https://www.R‐project.org/.
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. 2011. pROC: an open‐source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe MH, Dong J, Weimer AK, Bergmann DC. 2019. A plant‐specific polarity module establishes cell fate asymmetry in the arabidopsis stomatal lineage. bioRxiv. doi: 10.1101/614636. [DOI]
- Scacchi E, Osmont KS, Beuchat J, Salinas P, Navarrete‐Gomez M, Trigueros M, Ferrandiz C, Hardtke CS. 2009. Dynamic, auxin‐responsive plasma membrane‐to‐nucleus movement of Arabidopsis BRX. Development 136: 2059–2067. [DOI] [PubMed] [Google Scholar]
- Simmons AR, Davies KA, Wang W, Liu Z, Bergmann DC. 2019. SOL1 and SOL2 regulate fate transition and cell divisions in the Arabidopsis stomatal lineage. Development 146: dev171066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Zhang X, Kong W, Yang XL, Molnar G, Vondrakova Z, Filepova R, Petrasek J, Friml J, Xue HW. 2020. The lipid code‐dependent phosphoswitch PDK1‐D6PK activates PIN‐mediated auxin efflux in Arabidopsis. Nat Plants 6: 556–569. [DOI] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J. 2006. Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Wolny A, Cerrone L, Vijayan A, Tofanelli R, Barro AV, Louveaux M, Wenzl C, Strauss S, Wilson‐Sanchez D, Lymbouridou R et al. 2020. Accurate and versatile 3D segmentation of plant tissues at cellular resolution. eLife 9: e57613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, van der Schuren A, van Dop M, van Galen L, Saiga S, Adibi M, Moller B, Ten Hove CA, Marhavy P, Smith R et al. 2019. A SOSEKI‐based coordinate system interprets global polarity cues in Arabidopsis. Nature Plants 5: 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Adamowski M, Marhava P, Tan S, Zhang Y, Rodriguez L, Zwiewka M, Pukysova V, Sanchez AS, Raxwal VK et al. 2020. Arabidopsis Flippases cooperate with ARF GTPase exchange factors to regulate the trafficking and polarity of PIN auxin transporters. The Plant Cell 32: 1644–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guo X, Dong J. 2016. Phosphorylation of the polarity protein BASL differentiates asymmetric cell fate through MAPKs and SPCH. Current Biology 26: 2957–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang P, Shao W, Zhu JK, Dong J. 2015. The BASL polarity protein controls a MAPK signaling feedback loop in asymmetric cell division. Developmental Cell 33: 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M, Muller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C. 2009. The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana . Development 136: 627–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Localisation pattern of polarity proteins used to test Pome in the Arabidopsis stomatal lineage.
Fig. S2 Additional polarity parameters and classification models.
Fig. S3 Confocal images of BRXL2 and BASL localisation pattern during asymmetrical cell divisions (ACDs).
Fig. S4 Application of Pome in root epidermal cells to validate shootward localisation of PIN2.
Fig. S5 Application of Pome in quantifying PAR‐2 polarity in the Caenorhabditis elegans P cell lineage during embryo development.
Methods S1 Pome macro V1.
Notes S1 Brief user guide for Pome.
Notes S2 Pome data analysis in R.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


