Abstract
The phenomenon that organisms can distinguish genetically related individuals from strangers (i.e., kin recognition) and exhibit more cooperative behaviours towards their relatives (i.e., positive kin discrimination) has been documented in a wide variety of organisms. However, its occurrence in plants has been considered only recently. Despite the concerns about some methodologies used to document kin recognition, there is sufficient evidence to state that it exists in plants. Effects of kin recognition go well beyond reducing resource competition between related plants and involve interactions with symbionts (e.g., mycorrhizal networks). Kin recognition thus likely has important implications for evolution of plant traits, diversity of plant populations, ecological networks and community structures. Moreover, as kin selection may result in less competitive traits and thus greater population performance, it holds potential promise for crop breeding. Exploration of these evo‐ecological and agricultural implications requires adequate control and measurements of relatedness, sufficient replication at genotypic level and comprehensive measurements of performance/fitness effects of kin discrimination. The primary questions that need to be answered are: when, where and by how much positive kin discrimination improves population performance.
Keywords: cooperative behaviour, crop breeding, group performance, inclusive fitness, kin discrimination, kin selection, plant communication, resource competition, root exudates, root interaction
Short abstract
Can plant distinguish genetically related individuals from strangers (kin recognition) and exhibit more cooperative behaviour towards their relatives (positive kin discrimination)? Having reviewed the 14 years of research on this issue, we conclude that the answer to this question is yes. Focusing on belowground interactions, we illustrate how this kin recognition involves reduced resource competition between related plants but may also involve interactions with symbionts (e.g., mycorrhizal networks). But the implications of kin recognition for evolution of plant traits, diversity of plant populations, ecological networks and community structures, as well as crop production, though widely suggested to be significant, still need to be explored. Addressing these outstanding questions requires approaches that quantify when, where and by how much positive kin discrimination improves population performance.
1. INTRODUCTION
Plants often grow in dense vegetation stands, such as grasslands, woodlands, forests or agricultural fields, where there are usually intensive interactions with neighbours. These interactions can occur in the form of resource competition, facilitative habitat modification or communication. The ubiquity and diversity of plant–plant interactions entail that the effects of a given set of traits for an individual cannot be viewed independently of the characteristics of its neighbours (e.g., Cabal, Martínez‐García, de Castro Aguilar, Valladares, & Pacala, 2020; Riechert & Hammerstein, 1983). Plant–plant interactions in turn also play a key role in driving ecosystem processes such as carbon uptake, water and nutrient balances, interactions with other non‐plant organisms as well as crop yields (see review Anten & During, 2011).
Much plant ecological research is built on the concept of individual selection, that is, plant traits of a given individual are (at least implicitly) viewed from the perspective of how these traits directly benefit the fitness of that individual. Cooperative behaviour (i.e., actions that benefit the group rather than the individual) and even altruistic behaviour (i.e., actions that benefit other individuals at a cost to the actor) are, however, quite common in nature. To explain the evolution of such ‘non‐selfish’ behaviour, Hamilton (1964) introduced the idea of ‘kin selection’. The basic concept of kin selection is that if one individual helps another with whom it shares a certain number of alleles, those alleles will be passed to the next generation not only through its own fitness but also through the enhanced fitness of that recipient. An important prerequisite for relatedness‐dependent behaviour (i.e., kin discrimination) to occur, in turn, is kin recognition (i.e., the ability to detect the difference in level of relatedness between oneself and another individual), though, alternatively, kin selection also can be favoured by population viscosity (Waldman, 1988).
Kin recognition has been demonstrated in a wide variety of organisms including animals (both vertebrates and invertebrates, Waldman, 1988), fungi (Malik & Vilgalys, 1999) and even bacteria (Smith & Dworkin, 1994). However, the idea that it may also occur in plants was long considered outlandish. This scepticism occurred despite the well‐known facts that plants can sense and respond to the presence of other plants, for example, through light signals (Pierik & de Wit, 2014) and physical contact (de Wit et al., 2012). Plants can even detect the status of neighbours, for example, whether a neighbour is attacked by a herbivore being conveyed through volatiles (Karban, Yang, & Edwards, 2014) or whether the neighbour is stressed by drought being conveyed through sounds (Jeong et al., 2014). The fact that plants can distinguish between self‐ and non‐self, and are thus capable of some level of identity recognition, has been evident from the observation that many species prevent self‐pollination (Fujii, Kubo, & Takayama, 2016). Research indicates that plants may also be able to distinguish between their own roots and those of a non‐self neighbour (see Chen, During, & Anten, 2012 for a review) and between their own herbivore‐induced volatiles and those produced by another plant (Karban & Shiojiri, 2009).
The first evidence of belowground kin recognition in plants came from a study of the annual Cakile edentula (Dudley & File, 2007), finding that plants reduced root allocation (i.e., produced less roots for a given aboveground mass) when interacting with half‐sibs from the same mother rather than with strangers from a different mother. A series of subsequent findings have strengthened the idea that kin recognition and associated kin discrimination occur in plants (Figure 1), though there is a heavy debate about methodology used (e.g., Ehlers & Bilde, 2019; Klemens, 2008; Simonsen, Chow, & Stinchcombe, 2014, and see details in Section 5). Kin discrimination has by now been studied for different traits and functions in a wide variety of wild as well as domesticated species.
FIGURE 1.
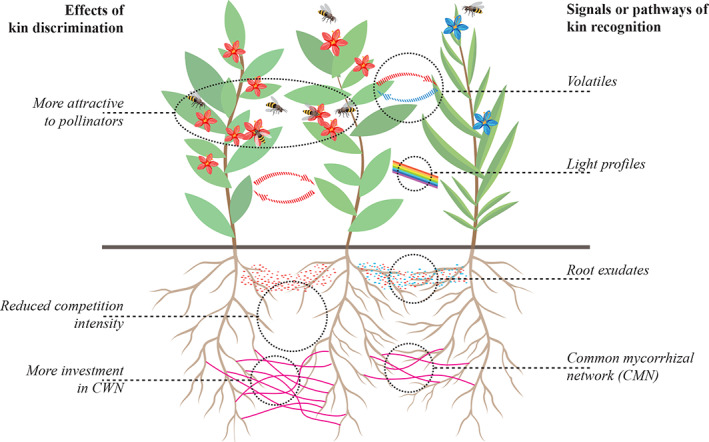
Summary of the physiological mechanisms for kin recognition and the associated kin discrimination effects in plants. Signals and pathways that are proven to mediate belowground kin recognition include root exudates and probably also common mycorrhizal networks. In addition, signals like volatiles and probably also profiles of reflected light can mediate aboveground kin recognition, but they are not the focus of this review. Effects of kin discrimination include not only a reduction of resource competition intensity but also cooperation to increase resource availability, such as more investments in common mycorrhizal network, and probably also in aboveground pollinator attraction (which is not the focus here) [Colour figure can be viewed at wileyonlinelibrary.com]
The existence of kin recognition in plants has important consequences for plant ecology and evolutionary biology, which go far beyond the interesting fact that plants can recognize their kin. First, kin selection may affect the genetic structure and diversity of a population, as it tends to favour lower genetic diversity at the group level (Platt & Bever, 2009). Second, cooperative traits tend to lead to greater performance at the population level (Anten & During, 2011) and thus may have important implications for ecosystem functioning. This, in turn, may also have implications for agriculture (Murphy, Swanton, Van Acker, & Dudley, 2017), since farmers aim for group performance (high yields or resource‐use efficiency at the crop‐stand level) rather than individual performance. It further raises the question, to what extent kin discrimination could contribute to better crop performance and could thus be a breeding target (Anten & Vermeulen, 2016; Denison, 2011).
The objectives of this paper are therefore to (a) critically appraise the evidence for kin discrimination and its underlying mechanisms in plants, (b) discuss potential ecological implications of kin discrimination and (c) discuss the extent to which kin discrimination could be a favourable trait in agricultural crops. While the degree of relatedness/kinship can range from the same genetically identical clone to different phylogenetic clades, we mainly focus on kin recognition at the intraspecific level. We first give a brief introduction to kin selection and related concepts and describe under what conditions it is most likely to occur. We then explore the evidence for kin recognition and discrimination in plants and critically appraise the methods that were used. In so doing, we explore different implications of kin recognition for resource acquisition. The first, and the one having received most attention in the literature, is plants exhibiting less competitive traits by producing smaller, shorter‐lived or less efficient resource harvesting structures when interacting with kin than when interacting with non‐kin. Secondly, we move beyond the direct plant–plant resource competition and explore how kin discrimination in plants may result in increasing the availability of resources to a group of related plants, and show how this can involve interactions with other organisms such as mycorrhizal symbionts. Thirdly, we briefly touch upon the environmental dependency and wider ecological implications of kin discrimination. We also raise concerns about limitations and drawbacks of the methodologies and interpretations in the current research field. Finally, we explore potential applications of kin selection in crops, discussing to what extent kin recognition would be a desirable crop trait.
2. SOME BASIC CONCEPTS
2.1. Cooperation versus cheating
An issue that is receiving increasing interest in the plant scientific literature is the conflict between individual selection and the performance of plant populations (Anten & During, 2011; McNickle & Dybzinski, 2013). Populations of plants with traits that are optimal in the sense of maximizing group fitness are often not resistant to invasion by a more competitive plant type which invests more in resource harvesting. Evolutionary game theory predicts that populations will thus evolve towards non‐optimal states. The result, that is, a population of plants overinvesting in resource harvesting, has been denoted as a tragedy of the commons (Hardin, 1968). The classic example is plant height: short stature and associated small investment in stem tissue would favour investment in fitness‐enhancing characteristics (e.g., seed production), but such communities can be invaded by taller plants (Falster & Westoby, 2003).
A suite of other traits have since been associated with such a tragedy of the commons: plants may produce more leaf area, larger specific leaf area and more roots, or larger flowers than what would maximize fitness of the population (see review by Anten & Vermeulen, 2016). As a result, vegetation stands may not be optimized in the sense of maximal fitness under a given set of conditions and this may have implications for ecosystem functioning (Anten & During, 2011; Farrior, Rodriguez‐Iturbe, Dybzinski, Levin, & Pacala, 2015).
2.2. Kin selection when and where
Nature is full of examples of cooperative or even altruistic behaviour, which run contrary to the above‐mentioned tragedy of the commons. Hamilton (1964) proposed that selection for altruistic behaviour may occur in populations of genetically related individuals (so‐called kin). The basic concept is that if one helps another individual with whom one shares a certain number of alleles, these alleles can be indirectly passed to the next generation through the enhanced fitness of that individual. Hamilton (1964) formalized this as a rule stating that altruistic behaviour will be selected if the product of fractional genetic relationship (r) and the fitness benefit for the recipient (B) is larger than the fitness cost incurred by the actor (C, the direct fitness effect for the actor):
| (1) |
in which r × B is also known as the indirect fitness effect. Hamilton's rule generally predicts that individuals that are genetically more closely related are also more likely to cooperate. However, it is important to consider that if closely related individuals are phenotypically more similar than distantly related ones, they would also compete more intensely with each other. In the case of the more distantly related plants, differences would lead to stronger niche differentiation (Platt & Bever, 2009; Van Dyken, 2010). This raises the question as to when kin selection is likely to occur despite the existence of kin competition. It is clear from Equation (1) that this is the case if r and/or B are high, or C is low.
Equation (1) also dictates an important boundary condition. Because r ≤ 1 (r = 1 when plants are genetically identical), it holds that if competition is a zero‐sum game, that is, fitness gains of one are equal to losses of the other (i.e., thus C = B), r × B is always smaller than or occasionally equal to C, and thus kin selection is unlikely to occur. For instance, let there be two genetically different plants (r < 1) that compete for a fixed amount of soil resources. If one plant reduces investment in resource harvesting (e.g., roots) while the other does not, and fitness costs (C, that is, acquiring less nutrients) for that plant are equal to the fitness gains (B) of the other plant, this can never have a selective benefit since r × B < C. Of course, C and B need not be equal even if the size of resource pool is fixed. For instance, since the production of resource‐harvesting structures is costly, the relationship between plant size and seed production tends to be curvilinear (see Section 6.1). In this case, two related plants might do better by mutually refraining from competition. This is because, if r is sufficiently high, the direct fitness gains of unilaterally increasing the size of resource harvesting structures could be smaller than the indirect fitness losses caused by reducing fitness of related neighbours (Ehlers & Bilde, 2019).
This discussion makes it clear that kin selection critically depends on the cost and benefit of the interaction and the ecological setting. Together, at least two ecological factors can be considered to favour kin selection. (a) A high r value, that is, plants tending to interact with genetically related neighbours. This is more likely to occur if dispersal rates are small and would increase, for example, with the level of self‐pollination; or if plants are mainly clonally reproduced. (b) Competition being a negative non‐zero sum game which entails that altruistic behaviour not only changes the competitive balance but also increases the availability of resources.
In addition, competition does not only occur between individual plants but also between groups of plants, especially when resources are highly mobile (e.g., competition for pollinators between mass‐flowering crops and grassland plants; Holzschuh, Dormann, Tscharntke, & Steffan‐Dewenter, 2011). Thus, even if cooperative behaviour would be selected against within a group, it can be selectively favoured between groups if cooperative groups are more productive than less cooperative groups (Maynard Smith, 1964; Nowak, Tarnita, & Wilson, 2010; Wilson, 1975). This multi‐level group selection may occur in the absence of kin selection, but the two are largely interchangeable. Multi‐level selection may also favour kin selection (Nowak et al., 2010). In both cases, it is important that genetic variation within groups is small compared to genetic variation between groups (Platt & Bever, 2009).
In summary, evolution of cooperative traits through kin selection is likely to occur
when costs of cooperation are low;
when cooperative interactions result not only in reduced intensity of competition but also in greater availability of resources;
when resources are mobile, making competition between groups more likely; and
in viscous plant populations in which there is a clear negative relationship between genetic relatedness and physical distance.
2.3. Repeated interaction as an alternative hypothesis
Though Hamilton's rule with the key concept of inclusive fitness provides the theoretical basis for the evolution of kin selection, there are still alternative but not exclusive explanations, such as long‐term benefits from repeated interactions (Dudley, 2015; Trivers, 1971). Game theoretical models predict that players falling into an unrepeated prisoners' dilemma are prone to cheat to maximize their short‐term interests, while those in iterated prisoners' dilemma tend to cooperate for their long‐term interests, as long as this accumulated benefits can exceed the costs of cooperation (Killingback & Doebeli, 2002). Therefore, reciprocal altruism can simply emerge in a group of individuals with intensively repeated interactions along their life spans, regardless of their relatedness (St‐Pierre, Larose, & Dubois, 2009).
In many cases, the concept of reciprocal altruism is restricted to cooperation among non‐kin. However, owing to the nature of dispersal, (half‐)sibs from the same mother or individuals from the same family are more likely to have overlapping living spaces, creating greater than random chances of repeated interactions. Thus, reciprocal altruism is still more likely to occur among relatives, potentially leading to the evolution of kin selection in plant species, especially those with viscous population structures.
3. BELOWGROUND KIN RECOGNITION AND COMPETITION FOR AVAILABLE RESOURCES
This section deals with the situation where plants exhibit less competitive traits by producing smaller or less efficient resource‐harvesting structures when interacting with closely related neighbours, and its mediation through kin recognition. We focus on kin recognition at the root level, as this has been most extensively studied. We refer to, for example, Crepy and Casal (2015) and Karban, Shiojiri, Ishizaki, Wetzel, and Evans (2013) for intriguing examples of aboveground kin recognition through light and volatile chemical signals, respectively, and to Torices, Gómez, and Pannell (2018) for an above‐ground example of kin‐recognition‐based cooperation for the attraction of pollinators (Figure 1).
3.1. Evidence for belowground kin recognition
Evidence accumulated in the last two decades has shown that changes in root traits of a given plant may occur in response to the identity of neighbouring plants. Early work in this regard showed that plant roots can distinguish whether roots, with which they come in contact, are from the same plant or from a different plant (non‐self) (e.g., Chen, Vermeulen, During, & Anten, 2015; Gersani, Brown, O'Brien, Maina, & Abramsky, 2001; Gruntman & Novoplansky, 2004; Holzapfel & Alpert, 2003). While their methodology was criticized (e.g., Chen et al., 2015; Chen et al., 2020; Hess & de Kroon, 2007; McNickle, 2020; Semchenko, Hutchings, & John, 2007), it clearly spurred a wave of interest in identity recognition at the root level.
The first study to document root‐mediated kin recognition was by Dudley and File (2007). They grew C. edentula plants either solitarily or paired with half‐sibs or strangers in pots, and found that plants interacting with strangers increased root allocation as compared to both solitary ones and those interacting with half‐sibs. This was viewed as evidence for kin recognition in plants. The responses of plants in belowground kin recognition also involve changes in root morphological and physiological traits beyond biomass allocation. Compared to interactions with strangers, interaction with relatives can stimulate some species to produce root systems with lower lateral root density (Palmer et al., 2016), less root branching intensity and shorter specific root length (Semchenko, Saar, & Lepik, 2014), as well as lower nutrient uptake rate (Zhang, Liu, Tian, Xu, & Ouyang, 2016) and lower water uptake rate (Takigahira & Yamawo, 2019).
Interestingly, belowground kin recognition also appears to result in expression of less competitive aboveground traits. For example, plants of the annual forest understory species Impatiens pallida allocated a larger fraction of their mass to roots when grown in kin groups than when grown in non‐kin groups (Murphy & Dudley, 2009). However, this was due to plants in kin interaction having smaller shoots rather than larger root systems, and these smaller shoots could be associated with less intense aboveground competition. In addition, plants growing with kin neighbours also tended to produce more branches which they interpreted as a response that further reduces mutual shading. Similarly, a reduction in competitive ability for light was observed in Fagus crenata seedlings, which produced leaves with relatively reduced chlorophyll contents (Takigahira & Yamawo, 2019), and in Lychnis flos‐cuculi which produced leaves with relatively low specific leaf areas (Lepik, Abakumova, Zobel, & Semchenko, 2012), when these plants had root interactions with a relative rather than with a stranger. From an evolutionary point of view, the aforementioned findings suggest that the expression of kin discrimination may reflect an adaptation for the acquisition of key limiting resources of a species in its specific habitat.
Meanwhile, it should be noted that although evidence of belowground kin recognition to date has been identified in various types of plant groups ranging from gymnosperms (e.g., Cycas edentata, Marler, 2013) to angiosperms (see above‐mentioned examples), and from wild species (e.g., Deschampsia cespitosa, Semchenko et al., 2014) to crops (e.g., wheat, Zhu & Zhang, 2013; soybean, Murphy, Van Acker, Rajcan, & Swanton, 2017, see more in Section 7), there are also a number of studies finding no evidence or even negative effects of kin interaction. For instance, in a pot‐based experiment with eight co‐occurring temperate grassland species, only one species demonstrated less competitive traits and a higher fitness gain in kin than in non‐kin interactions (Lepik et al., 2012). In another pot‐based experiment, the relatedness of neighbours at the accession level affected neither growth nor seed production of Arabidopsis thaliana (Masclaux et al., 2010). Moreover, Lupinus angustifolius potted with sibs or genotypes from the same population produced even fewer seeds than those potted with genotypes from different populations (Milla, Escudero, & Iriondo, 2011). These discoveries imply that the ability of kin recognition is not a universal feature in plant kingdom.
3.2. Mechanisms of belowground kin recognition
How do plant roots detect the level of relatedness in other plants? Most studies so far point to the involvement of root exudates. To date, more than 200,000 plant secondary metabolites have been identified, and a great portion of them are secreted as root exudates into the rhizosphere (Kessler & Kalske, 2018). Plants typically secrete a wide variety of them, and the composition of exudates differs between species and between genotypes within a species (Mommer, Kirkegaard, & van Ruijven, 2016).
The first evidence for the role of exudates in kin recognition came from Biedrzycki, Jilany, Dudley, and Bais (2010), who found stronger lateral root growth of A. thaliana seedlings when exposed to exudates from stranger's roots than from sib's. This difference disappeared when exudate solutions were collected from donor plants that were treated with a root secretion inhibitor. More recently, Yang, Li, Xu, and Kong (2018) also found that rice seedlings growing in solutions with exudates from a different cultivar allocated relatively more mass to roots than plants growing in solutions with exudates from the same cultivar.
Semchenko et al. (2014) took a somewhat different approach, collecting leachates from pots with different D. cespitosa plants and supplying these leachates to sibs or strangers. Plants exposed to leachates from strangers not only increased root allocation but also produced roots with a higher branching density and a higher specific root length than those exposed to kin leachates. All these studies clearly indicate that exudates play a role in kin recognition at the root level (Figure 1).
The role of exudates in kin recognition seems to be evident in a general sense, but the exact compounds that are involved or the ways they operate (e.g., dose dependent) are still unknown. To our knowledge, so far only one study (Yang et al., 2018) has specifically considered this question. It found secretion of the compound allantoin, a N‐rich compound involved in various plant metabolic processes, to be higher in rice plants interacting with neighbours from a different cultivar than those interacting with neighbours from the same cultivar. It was also found that an exogenous application of allantoin can cause changes in auxin synthesis and gene expression in the focal rice plants. These findings clearly demonstrated that allantoin production depends on the level of relatedness in the interaction and that the level of allantoin production is linked to kin recognition. More specifically, compared to monocultures, mixed‐culture stimulated plants to secrete greater amounts of allantoin, leading to higher gene expression levels of auxin transporters, which were in turn associated with more root growth but less yield.
Some recent neighbour detection studies also showed that lactone (e.g., (−)‐loliolide) and phytohormones (e.g., jasmonic acid and salicylic acid) in the root exudates of various weed species were the key chemicals that are sensed by allelopathic wheat and rice and can elicit them to produce more phytotoxins (Kong et al., 2018; Li, Xia, & Kong, 2016; Li, Zhao, & Kong, 2020). However, to what extent these chemicals are also involved in the kin recognition of wheat and rice is still unknown. To sum up, evidence is clearly mounting that plant roots are capable of recognizing kin and that this may involve a wide variety of belowground as well as aboveground traits. Root exudates play a role in the process, though more work is needed to identify the (combinations of) compounds involved.
4. BELOWGROUND KIN RECOGNITION AND COOPERATION TO INCREASE RESOURCE AVAILABILITY: THE CASE OF INVESTMENT IN COMMON MYCORRHIZAL NETWORKS
While the previous section dealt with variation in the investment in a plant's own resource‐harvesting structures using roots an example, here we discuss the investment in a common network that involves another life‐form using the example of mycorrhizal networks.
Plants form symbiotic associations with mycorrhizal fungi whereby the mycorrhiza provide water, nutrients and/or defence from pathogens to the plants, and the plants provide carbon in return (Smith & Read, 2008). One emerging property in this symbiosis is the formation of common mycorrhizal networks that connect different plants, allowing them to transfer N, water and even carbon between them. Such networks thus entail a common good: carbon investment of each plant into the network allows the mycorrhiza to grow larger and exploit larger volumes of soil, giving plants access to more water and nutrients. If these soil resources are limiting, the investment in turn provides a common benefit for all plants (Rankin, Bargum, & Kokko, 2007).
In theory, this system is open to cheating, and an associated tragedy of the commons (File, Klironomos, Maherali, & Dudley, 2012; Wyatt, Kiers, Gardner, & West, 2014). That is, if a plant unilaterally provides less carbon to the mycorrhiza, it will reduce its own costs while the resulting reduction in soil resources provided by the common mycorrhizal network are shared by the connected population, thus giving the cheater a relative benefit. Such selection for cheating can lead to collapse of the network. One way that stable symbiosis in the networks can be enforced is through sanctioning. If fungi can detect differences in carbon supply by plants and make nutrient provisioning dependent on it, plants that invest more in the common network would get a greater reward than those that do not (Wyatt et al., 2014). Kin selection, however, would be an alternative, there being added selective advantage to invest in a network shared with kin. Investment in the common mycorrhizal network is a cooperative behaviour that increases the availability of resources, making the interaction a non‐zero‐sum game whereby costs incurred by investing in this common network are smaller than the accumulated benefit.
To explore whether kin recognition may be involved in the functioning of the common mycorrhizal network, File et al. (2012) conducted experiments where Ambrosia artemisiifolia plants were grown either with half‐sibs or strangers from different mothers, and with or without mycorrhizal fungi. The size of the mycorrhizal networks, expressed as soil hyphal length and levels of root colonization, were greater in half‐sib groups than in stranger groups. Half‐sib groups also benefitted more from the presence of a common mycorrhizal network, for example, in terms of P uptake and suppression of pathogens. These intriguing effects of kin recognition and possible kin selection operating via mycorrhiza raise questions as to the mechanism of kin recognition. File et al. (2012) proposed that it could involve direct kin recognition, whereby plants recognize the presence of kin through exudates released by neighbouring plants into the soil, or through secondary metabolites actually being transported through the mycorrhizal hyphae, which induce them to invest resources into the common mycorrhizal network. The mechanism, however, could also be less direct, for example, kin recognition at root level inducing changes in root architecture, or anatomy making mycorrhizal colonization easier. Or, if kin interacting plants are already doing better in terms of assimilation, they will have more resources to invest in the networks.
To further explore whether kin‐related signals might be directly communicated via mycorrhizal hyphae, Pickles et al. (2017) grew sib and stranger seedling pairs of interior Douglas fir (Pseudotsuga menziesii var. glauca) in pots and provided one plant of the pair with 13C labelled CO2 and measured how much carbon was allocated into the ectomycorrhizal fungi biomass or transferred to the paired seedlings. They found that levels of 13C in the mycorrhizal biomass as well as in recipient plants were higher in sib pairs than in stranger pairs. They concluded that these results could indicate the presence of signalling compounds being transferred. Research on ectomycorrhizal host–symbiont interface reveals that ectomycorrhizal fungi produce signalling compounds, which are translocated into plant cell nuclei to alter the host transcriptome, for example, to promote ectomycorrhizal symbiosis and reduce jasmonic acid production (Plett et al., 2014). The results of Pickles et al. (2017) thus suggest that the greater carbon exchange between sibs is associated with increased transfer of signalling compounds through the network (Babikova et al., 2013; Plett et al., 2014) (Figure 1).
These findings together provide evidence that kin recognition between plants may involve other organisms that not only deliver signals for recognition but also provide rewards in kin selection. The extent to which this form of relatedness‐dependent cooperative behaviour (i.e., investing resources in the network) can increase the carrying capacity for the plant population through enhancement of resource availability, however, still needs to be demonstrated. Furthermore, several important questions still need to be answered. For example, what is the role of transferred carbon compounds? Do they act as cues that initiate/mediate the process of kin recognition between mycorrhiza‐connected plants? Or, are they simply the consequences of kin recognition between plants via direct root interaction/communication? Moreover, does this relatedness‐dependent investment of plants in a common mycorrhizal network commonly occur in nature; and to what extent does this phenomenon affect the stability and efficiency of the network?
5. KIN RECOGNITION IN A BROADER ECOLOGICAL CONTEXT
The effects of kin recognition on the performance of plant individuals have consequences on the dynamics and structures of plant populations and communities. For example, the preferential transfer of photosynthates to sibs via common mycorrhizal networks in Douglas fir (Pickles et al., 2017) can provide sibs with competitive advantages over unrelated neighbours. Recent evidence suggests that kin recognition can also make groups of related plants more competitive against hetero‐specific neighbours. For instance, in competition with Trifolium repens, sib pairs of Plantago asiatica directed more leaves towards T. repens than stranger pairs, leading to a reduction of growth in T. repens (Yamawo & Mukai, 2020). Positive discrimination can also facilitate the invasion success of exotic species. Evidence for this comes from a study showing that individuals of the South American alligator weed (Alternanthera philoxeroides) from genotypes introduced to North America grew larger in intra‐genotypic interaction than in inter‐genotypic interaction; but the trend was opposite for those from native genotypes in South America (Zhang et al., 2019). The aforementioned findings suggest that these relatedness‐dependent facilitative effects including support (e.g., resource transfer) and cooperation (e.g., reduced competition intensity) can potentially affect the distribution of genetic diversity in plant populations by reducing local genetic heterogeneity (Tedersoo, Bahram, & Zobel, 2020).
Relatedness‐dependent cooperation is probably also environment (or microhabitat) dependent. For instance, a higher survival rate of Medicago rigidula in intra‐genotypic than in inter‐genotypic competition seems to occur only when plants were grown in soils collected underneath allelopathic thyme shrubs (Ehlers, David, Damgaard, & Lenormand, 2016). Conversely, such a cooperation in the con‐generic species M. minima occurred only when plants were grown in non‐thyme soil. There is further evidence that the extent of kin discrimination is correlated with the nutrient availability in the habitats. For example, reduced root activity and nutrient uptake in sib‐pairs of sorghum, compared to non‐sib ones, occurred only under low nutrient availability (Li, Xu, & Feng, 2018). Similarly, intra‐cultivar interacting peas produced more seeds than inter‐cultivar interacting ones only at low nutrient availability (Pezzola, Pandolfi, & Mancuso, 2020). Palmer et al. (2016) further demonstrated that N and P are the key elements determining the nutrient availability effects. They grew A. thaliana seedlings either solitarily or paired with a sib, a relative (from a different mother in the same accession) or a stranger (from a different accession) in agar with the Murashige‐Skoog nutrient medium. They found that stranger‐paired plants had more lateral roots than other plants only when nutrient concentration was lower than 0.75 strength of the standard solution. Relative‐ and sib‐paired plants produced more lateral roots than solitary plants only when the solution was further diluted. They could reproduce these results by individually removing N and P from the full‐strength solution but not by removing other elements, showing that kin recognition depends not only on the amount of nutrients but also their composition, the presence of N and P being critical in this example. A great body of literature has demonstrated that local soil N and P availabilities are the key signals that modulate root architecture of plants (Jia & von Wirén, 2020; Péret et al., 2014; Tian, De Smet, & Ding, 2014). Moreover, the effects of kin recognition on the performance of plants can be density‐dependent. For instance, Lepik et al. (2012) showed that the level of inflorescence mass of T. repens increased with neighbour density only in sib groups; meanwhile its petiole length (a key trait that related to competition for light) was positively correlated with neighbour density only in stranger groups.
Clearly, kin recognition can have implications for the structure, diversity and functioning of communities likely extending to the ecosystem level, which we are only beginning to unravel. There is thus a clear need for systematic ecological field experiments across different systems and species, where genetic relatedness in populations is manipulated or at least known and a broad scale of community interactions are explored. Moreover, kin discrimination seems to be more important in regulating plant growth and community structure under stressful, especially infertile, conditions. This could imply that species originating from stressful habitats might be more likely to possess the ability of kin recognition, but this needs to be tested.
6. CRITICAL CONSIDERATIONS ABOUT CURRENT RESEARCH
6.1. Considerations about measuring kin recognition
A methodological concern with pot‐based studies of root‐mediated kin recognition is that in most cases roots of individual plants could not be separated and root mass needed to be measured at the pair (or group) level. This implies that the larger individual(s) contribute more to the results than smaller ones. Since size inequality is likely to be larger in non‐kin than in kin interactions and larger plants typically investing proportionally more in structural support (stems and coarse roots) and less in other parts (leaves, fine roots and potentially reproduction), the averaged proportional investment in seed production observed in non‐kin groups will inevitably tilt to that of larger individuals, for example, a lower seed production level (Klemens, 2008). Even if plant individuals can be separately harvested and measured, Jensen's inequality effect (Figure 2) suggests that, when plant size and seed production follow a curvilinear saturating relationship and competition between plants with greater differences in relatedness yields larger size asymmetry, plants in non‐kin interactions will naturally generate lower group seed production than those in kin interactions independently of kin recognition (Ehlers & Bilde, 2019; Simonsen et al., 2014). Notably, a pair‐wise family design (Bhatt, Khandelwal, & Dudley, 2011), whereby all combinations of lineages (either plants of the same or different families) are tested and differences in competitiveness are corrected for, could be a solution for the problem of size inequality.
FIGURE 2.
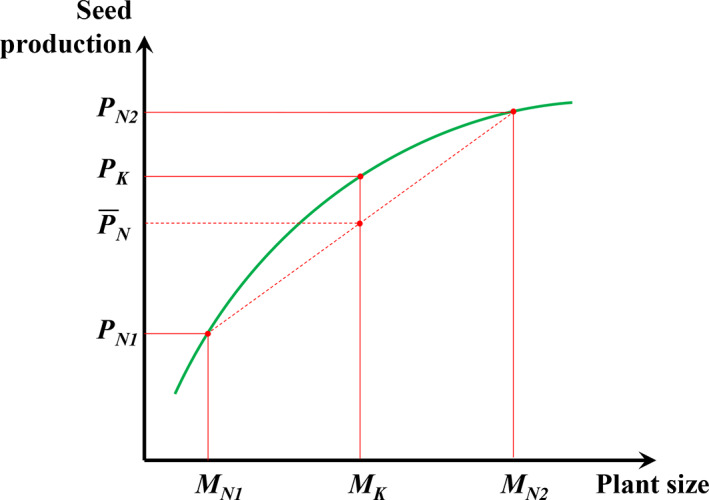
Jensen's inequality effect on the interpretation of difference in group seed production between kin pairs and non‐kin pairs. Plants in non‐kin pairs have lower degrees of genetic similarity and thus tend to have larger differences in competitive ability, leading to larger differences in plant sizes (e.g., MN1 for the weak competitor and MN2 for the stronger competitor) associated with larger differences in seed productions (e.g., PN1 and PN2; and their averaged seed production will be ). On the other hand, plants in kin pairs tend to have a similar plant size (MK) associated with a similar level of seed production (PK). When the group total mass (e.g., 2MK in a kin pair, and MN1 + MN2 in a stranger pair) is determined by the total amount of available resources in the group and the correlation between plant size and seed production follows a saturation curve, for a given amount of resources at the group level (i.e., MN1 + MN2 = 2MK), the group seed production of plants in non‐kin pairs will be inevitably lower than that in kin pairs (i.e., ) [Colour figure can be viewed at wileyonlinelibrary.com]
Some studies (Fang et al., 2013; Yang et al., 2018) used pots filled with transparent substrates, for example, agar or solutions, allowing them to visually observe kin recognition effects on root architecture and spatial arrangement. A problem, however, is that such solutions are very different from soil, which typically has lower oxygen levels or higher mechanical resistance (Chen, Hajiboland, Bahrami‐Rad, Moradtalab, & Anten, 2019). It is entirely unknown whether such differences may somehow interact with the kin versus non‐kin contrast.
Moreover, most of the work on kin recognition discussed in this paper concludes that genetically related plants exhibit more cooperative traits leading to higher group performance. However, such plants are also phenotypically more similar, and classic ecological theory dictates that these plants exhibit less niche differentiation and complementarity, leading to stronger competition (Chase & Leibold, 2003). Thus, kin interaction effects and complementarity effects are to some extent confounded, and this may cause complications in the interpretation of experimental results. For instance, increased root production of plants observed in non‐kin groups than in kin groups can also be a result of belowground niche complementarity between different genotypes, families or varieties in the former (Dudley, Murphy, & File, 2013). The outcome of kin interaction, as compared to non‐kin interaction, in most cases is likely determined by both kin recognition and niche effects (Ehlers & Bilde, 2019). Depending on the relative size of two opposite effects, the outcome of kin interaction can be positive, neutral or even negative as compared to non‐kin interaction. As one can partly mask the other, it could lead to false negatives, for example, wrongly concluding that kin recognition does not exist.
Concerns mentioned above can be largely attributed to the fact that it is impossible to exhaustively test all biotic and abiotic environmental determinants and separate kin recognition effect from them in the real plant–plant interactions. Mechanism‐driven approaches may thus provide some help. For instance, in the studies of belowground kin recognition, focal plants can be experimentally exposed to the key signals, for example, root exudates, manually collected from kin or non‐kin neighbours rather than expose focal plants to neighbours per se (e.g., Biedrzycki et al., 2010; Mercer & Eppley, 2014; Semchenko et al., 2014). This method has been successfully applied (see Section 3.2). However, it should be noted that the collected root exudates solutions (from, e.g., soil extracts, leachates or hydroponic medium) from different genotypes, families or varieties of plants may differ not only in the chemical composition of exudates which are supposedly involved in kin recognition but also in the composition and concentration of other compounds such as nutrients. Thus, we suggest that an adjustment of nutrient contents, at least the key elements N and P (Palmer et al., 2016), should be made for the collection of exudate solutions before treating focal plants with these solutions (Chen et al., 2020; Semchenko et al., 2014).
Finally, all studies mentioned so far were essentially pot‐based greenhouse studies, and translation from such studies to the field is notoriously difficult especially when the work involves roots. Yang et al. (2018) conducted a double experiment with rice wherein different combinations of kin (same variety) and non‐kin (different varieties) were grown in both pots and farm lands. Results were very consistent: plants in both conditions interacting with kin produced fewer roots and had greater seed production than those growing with non‐kin. While more work needs to be done, it does suggest that kin recognition operates under field conditions and can influence plant community performance.
6.2. Considerations about measuring fitness
While the previous section appraised the methods used to show kin recognition in plants, here we discuss the methods used to quantify the fitness benefits of kin discrimination. Most studies conducted experiments where they grew focal individuals, either in kin or non‐kin groups, and then measured the performance of either only focal individuals (e.g., Karban et al., 2013; Yang et al., 2018) or the whole group (e.g., Biernaskie, 2011; Dudley & File, 2007). Results were mixed, with some studies (e.g., File et al., 2012; Karban et al., 2013; Yang et al., 2018) finding plants in kin groups to produce more seeds or biomass than in non‐kin groups; while others (e.g., Biernaskie, 2011; Milla et al., 2011; Takigahira & Yamawo, 2019) did not find a difference.
Moreover, the basic set‐up of experiments comparing performance of plants in kin or non‐kin groups needs to be further considered. As noted, current studies on fitness effects of kin discrimination simply compare performance between kin‐ and non‐kin groups. This does not answer the question whether the genetic variation in kin discrimination for a given (set of) trait(s) is associated with a greater performance in kin groups. Addressing this question entails firstly quantifying genetic variation in kin discrimination within a population, and secondly, determining whether this variation correlates positively with performance in kin groups. We thus urge for a broader quantitative genetic approach to the research on the selection effects of kin recognition.
7. KIN SELECTION AND CROPS
Farmers generally aim to increase crop yields (or some other performance measure such as resource‐use efficiency) at the crop‐stand level. This entails that crops would need to have traits that enhance group rather than individual performance (e.g., Anten & Vermeulen, 2016; Denison, 2011; Donald, 1968; Weiner, Andersen, Wille, Griepentrog, & Olsen, 2010). Modern crop selection has to some extent unwittingly (i.e., without formal knowledge of how natural selection operates) already done this. The most notable example is the shortening of the stem of several major cereal crops (e.g., wheat and rice) during the green revolution (Khush, 1999). However, there may be ample scope for further crop improvement especially in traits other than stature, such as the size and architecture of root systems, and allocation to symbiotic relationships with soil biota (see review Anten & Vermeulen, 2016). As kin selection can lead to more cooperative traits in plants and greater group performance, its potential use in crop breeding could be considered (Murphy, Swanton, et al., 2017). Kin discrimination has been demonstrated in several crop species, for example, soybean (Murphy, Van Acker, et al., 2017), barley (Ninkovic, 2003), sorghum (Zhang et al., 2016), wheat (Zhu & Zhang, 2013, but see Fréville et al., 2019), rice (Yang et al., 2018) and Jerusalem artichoke (Helianthus tuberosus, Fukano, Guo, Noshita, Hashida, & Kamikawa, 2019), with findings that kin‐interacting plants tend to exhibit more cooperative root systems. A question then arises: to what would extent crop selection for kin recognition and discrimination be an effective means to increase crop yields?
To address this question, it is important to note that genetic diversity in crop stands tends to be much lower than in natural vegetation. Modern agriculture has increasingly shifted to mono‐species cropping; and for many crop species (e.g., maize, rice and wheat) inbred hybrid cultivars have been developed and are grown as populations of genetically almost identical plants. In such crop settings, plants rarely interact with non‐kin. On the other hand, genotype mixtures and even species mixtures are still used, especially by small‐holder farmers in poor countries (Joshi et al., 2020) as well as organic farmers (Bedoussac et al., 2015).
Secondly, when addressing the question whether kin recognition and associated discrimination is a useful crop trait, it is important to consider the alternatives to which this trait is compared. Conceptually, positively kin‐discriminating genotypes can be regarded as facultative cooperators, expressing more cooperative traits only when interacting with kin (i.e., a result of kin recognition). One can then consider two alternatives: obligate competitors and obligate cooperators, that is, plants exhibiting, respectively, competitive or cooperative traits irrespective of whether they interact with kin or non‐kin (though the level of cooperation could shift in response to some other factors). A crop stand of strongly related kin‐discriminating facultative cooperators would be expected to do better than a similar stand of obligate competitors. But the comparison between facultative and obligate cooperators is more complex. In a cultivar mixture, facultative cooperators would likely do worse than obligate cooperators, as the interaction with the less related other cultivars would induce expression of less cooperative traits in the facultative cooperator but not in the obligate one. In mono‐cultivar crops, the difference between the kin‐discriminating and obligate cooperators would at best be zero, but if there would be costs associated with kin recognition itself, facultative cooperators could still perform worse (Figure 3).
FIGURE 3.
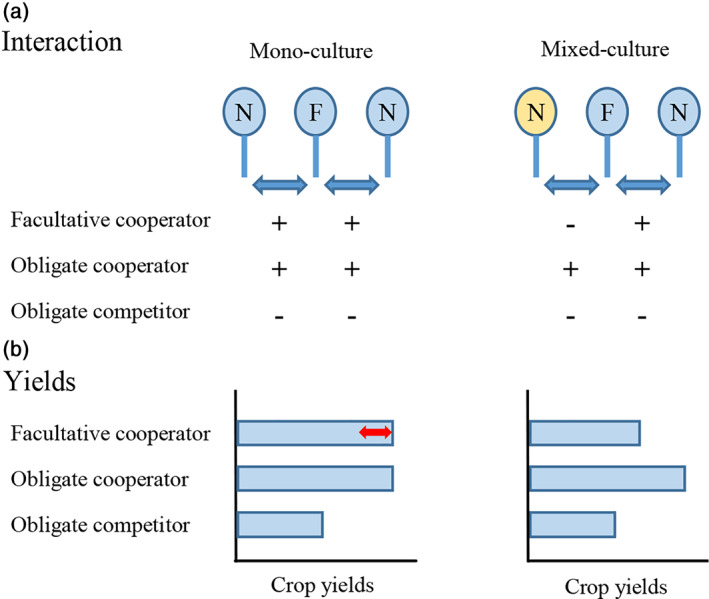
Illustration for the concept of (a) plant–plant interactions and (b) their consequences on crop yields of three hypothesized behaviour modes: facultative (i.e., kin‐dependent) cooperator, obligate cooperator and obligate competitor. The focal (F) plant is either grown in a mono‐culture where neighbours (N) are fully composed of relatives (kin, indicated by the blue colour), or grown in a mixed culture where neighbours (N) are composed of both relatives and strangers (non‐kin, indicated by the yellow colour). In (a), ‘+’ indicates a more cooperative trait expression and ‘−’ a more competitive one. In (b), the double‐headed arrow indicates potential variation in yields [Colour figure can be viewed at wileyonlinelibrary.com]
Unfortunately, to date there is little experimental evidence for whether kin discrimination increases crop yields. This is largely due to the methods that were used to determine the effects of kin recognition on performance. As noted in Section 6.2, most studies (e.g., Dudley & File, 2007; File et al., 2012; Yang et al., 2018) compared yields of a given genotype when interacting with relatives to its performance with strangers. However, answering the question about the usefulness of kin recognition in crops entails comparing yields of crop stands of a set of genotypes that differ in the ability to recognize kin. This entails a rather elaborate experiment (see also Section 6.2). Firstly, genetic variation in kin discrimination needs to be documented across a sufficiently large population of plants. For each target genotype, this ideally involves replication of kin and non‐kin neighbours. Subsequently, the extent to which genotypic variation in kin discrimination and crop yields in kin groups are correlated can be determined, which would give an indication whether kin recognition is positively associated with crop yields.
Even though, for crop yields, the constitutive expression of cooperative traits might be favourable compared to relatedness‐dependent expression of such traits, the latter might still be useful if it occurs much more commonly than the former. However, the extent to which the genetic basis for cooperative traits that occurs within the extant genetic diversity of a crop species is associated with kin selection is still unknown. Addressing this issue involves answering two questions. First, how closely are kin selection and evolution of cooperative traits in plants linked? A very tight link between the two would entail that at least in the wild ancestors of crop species, expression of cooperative traits would predominantly be linked to kin recognition. Second, to what extent has past crop selection unwittingly favoured or disfavoured kin recognition. This question could be addressed by comparing wild ancestors and domesticated cultivars of different release dates and testing whether kin recognition has increased over time. This requires experiments with ample replication at the genotypic level, which, to our knowledge, have not been published.
A final point of consideration is that the use of kin recognition in crops generally means low‐diversity crops. As noted, in so doing, one foregoes the potential benefits of diversity (e.g., niche differentiation in resource acquisition or variation in disease resistance). Interestingly, much work on making agriculture more sustainable through improvement of ecological relationships in crop systems has focused on enhancing diversity (Kiær, Skovgaard, & Østergård, 2009; Tooker & Frank, 2012), while the role of kin recognition has not been considered. Clearly, much more research is needed to indicate when, where and by how much kin recognition can contribute to increasing yields or other crop functions. This could include modelling studies that explore the extent to which selection for cooperative traits could contribute to yield increases.
8. CONCLUSIONS AND OUTLOOK
Since the pioneering work of Dudley and File (2007), kin discrimination has been documented to affect many traits in a wide variety of plant species among different taxonomic clades, life forms and origins (see citations in the preceding sections). Many cases have been documented of plants exhibiting more cooperative behaviour towards kin than towards strangers. These behaviours can reduce competition for a common resource pool or expand the availability of resources. While concerns still remain about some of the methods used, it is probably safe to say that belowground kin recognition in plants exists. However, our understanding of the mechanisms has so far been limited, except for the idea that, in general, root exudates play a role in kin recognition. More biochemical and molecular research is needed to identify the chemical cues that associate with or signify genetic relatedness, and how and where this is sensed in plants. Most of the work so far has been conducted with rather limited sets of genetic material and are being limited to (half‐)sibs versus strangers without quantifying genetic relatedness (but see, e.g., Karban et al., 2013). Expanding on this will not only help overcome some of the experimental biases that have been associated with research on kin discrimination (see examples of elegant designs from Bhatt et al., 2011; Ehlers & Bilde, 2019; Semchenko et al., 2014) but would also give a much better indication of the level at which plants can detect genetic relatedness. Kin recognition may also have far‐reaching implications for the structure and composition at the community level, as it can modify multi‐trophic interactions and potentially select populations with lower genetic diversity. Many more field experiments are needed to explore these effects. Finally, the existence of kin discrimination has been linked to breeding more cooperative crops that could form more productive stands with higher resource‐use efficiency. More research is needed to determine the extent to which the positive effects indeed exist and whether it would not be more effective to breed crops that express these traits constitutively rather than in a relatedness‐dependent way.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
This work was supported by the Wageningen University strategic funds and the National Natural Science Foundation of China (32071526). Bin J. W. Chen was supported by the Six Talent Peaks Program of Jiangsu Province (TD‐XYDXX‐006). We want to thank Johannes Ransijn and the members of the Centre for Crop Systems Analysis, Wageningen University for constructive discussion, and thank Beibei Tao for the artwork of Figure 1.
Anten NPR, Chen BJW. Detect thy family: Mechanisms, ecology and agricultural aspects of kin recognition in plants. Plant Cell Environ. 2021;44:1059–1071. 10.1111/pce.14011
Niels P. R. Anten and Bin J. W. Chen contributed equally to this study.
Funding information Six Talent Peaks Program of Jiangsu Province, Grant/Award Number: TD‐XYDXX‐006; National Natural Science Foundation of China, Grant/Award Number: 32071526; Wageningen University strategic funds
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- Anten, N. P. R. , & During, H. J. (2011). Is analysing the nitrogen use at the plant canopy level a matter of choosing the right optimization criterion? Oecologia, 167, 293–303. 10.1007/s00442-011-2011-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anten, N. P. R. , & Vermeulen, P. J. (2016). Tragedies and crops: Understanding natural selection to improve cropping systems. Trends in Ecology & Evolution, 31, 429–439. 10.1016/j.tree.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Babikova, Z. , Gilbert, L. , Bruce, T. J. A. , Birkett, M. , Caulfield, J. C. , Woodcock, C. , … Johnson, D. (2013). Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecology Letters, 16, 835–843. 10.1111/ele.12115 [DOI] [PubMed] [Google Scholar]
- Bedoussac, L. , Journet, E.‐P. , Hauggaard‐Nielsen, H. , Naudin, C. , Corre‐Hellou, G. , Jensen, E. S. , … Justes, E. (2015). Ecological principles underlying the increase of productivity achieved by cereal‐grain legume intercrops in organic farming. A review. Agronomy for Sustainable Development, 35, 911–935. 10.1007/s13593-014-0277-7 [DOI] [Google Scholar]
- Bhatt, M. V. , Khandelwal, A. , & Dudley, S. A. (2011). Kin recognition, not competitive interactions, predicts root allocation in young Cakile edentula seedling pairs. New Phytologist, 189, 1135–1142. 10.1111/j.1469-8137.2010.03548.x [DOI] [PubMed] [Google Scholar]
- Biedrzycki, M. L. , Jilany, T. A. , Dudley, S. A. , & Bais, H. P. (2010). Root exudates mediate kin recognition in plants. Communicative & Integrative Biology, 3, 28–35. 10.4161/cib.3.1.10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biernaskie, J. M. (2011). Evidence for competition and cooperation among climbing plants. Proceedings of the Royal Society B: Biological Sciences, 278, 1989–1996. 10.1098/rspb.2010.1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal, C. , Martínez‐García, R. , de Castro Aguilar, A. , Valladares, F. , & Pacala, S. W. (2020). The exploitative segregation of plant roots. Science, 370, 1197–1199. 10.1126/science.aba9877 [DOI] [PubMed] [Google Scholar]
- Chase, J. M. , & Leibold, M. A. (2003). Ecological niches: Linking classical and contemporary approaches. Chicago: University of Chicago Press. [Google Scholar]
- Chen, B. J. W. , During, H. J. , & Anten, N. P. R. (2012). Detect thy neighbor: Identity recognition at the root level in plants. Plant Science, 195, 157–167. 10.1016/j.plantsci.2012.07.006 [DOI] [PubMed] [Google Scholar]
- Chen, B. J. W. , During, H. J. , Vermeulen, P. J. , de Kroon, H. , Poorter, H. , & Anten, N. P. R. (2015). Corrections for rooting volume and plant size reveal negative effects of neighbour presence on root allocation in pea. Functional Ecology, 29, 1383–1391. 10.1111/1365-2435.12450 [DOI] [Google Scholar]
- Chen, B. J. W. , During, H. J. , Vermeulen, P. J. , de Kroon, H. , Poorter, H. , & Anten, N. P. R. (2020). The analysis of plant root responses to nutrient concentration, soil volume and neighbour presence: Different statistical approaches reflect different underlying basic questions. Functional Ecology, 34, 2210–2217. 10.1111/1365-2435.13664 [DOI] [Google Scholar]
- Chen, B. J. W. , Hajiboland, R. , Bahrami‐Rad, S. , Moradtalab, N. , & Anten, N. P. R. (2019). Presence of belowground neighbors activates defense pathways at the expense of growth in tobacco plants. Frontiers in Plant Science, 11, 751. 10.3389/fpls.2019.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B. J. W. , Vermeulen, P. J. , During, H. J. , & Anten, N. P. R. (2015). Testing for disconnection and distance effects on physiological self‐recognition within clonal fragments of Potentilla reptans . Frontiers in Plant Science, 6, 215. 10.3389/fpls.2015.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepy, M. A. , & Casal, J. J. (2015). Photoreceptor‐mediated kin recognition in plants. New Phytologist, 205, 329–338. 10.1111/nph.13040 [DOI] [PubMed] [Google Scholar]
- de Wit, M. , Kegge, W. , Evers, J. B. , Vergeer‐van Eijk, M. H. , Gankema, P. , Voesenek, L. A. C. J. , & Pierik, R. (2012). Plant neighbor detection through touching leaf tips precedes phytochrome signals. Proceedings of the National Academy of Sciences of the United States of America, 109, 14705–14710. 10.1073/pnas.1205437109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison, R. F. (2011). Past evolutionary tradeoffs represent opportunities for crop genetic improvement and increased human lifespan. Evolutionary Applications, 4, 216–224. 10.1111/j.1752-4571.2010.00158.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald, C. M. (1968). The breeding of crop ideotypes. Euphytica, 17, 385–403. [Google Scholar]
- Dudley, S. A. (2015). Plant cooperation. AoB PLANTS, 7, plv113. 10.1093/aobpla/plv113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, S. A. , & File, A. L. (2007). Kin recognition in an annual plant. Biology Letters, 3, 435–438. 10.1098/rsbl.2007.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, S. A. , Murphy, G. P. , & File, A. L. (2013). Kin recognition and competition in plants. Functional Ecology, 27, 898–906. 10.1111/1365-2435.12121 [DOI] [Google Scholar]
- Ehlers, B. K. , & Bilde, T. (2019). Inclusive fitness, asymmetric competition and kin selection in plants. Oikos, 128, 765–774. 10.1111/oik.06390 [DOI] [Google Scholar]
- Ehlers, B. K. , David, P. , Damgaard, C. F. , & Lenormand, T. (2016). Competitor relatedness, indirect soil effects and plant coexistence. Journal of Ecology, 104, 1126–1135. 10.1111/1365-2745.12568 [DOI] [Google Scholar]
- Falster, D. S. , & Westoby, M. (2003). Plant height and evolutionary games. Trends in Ecology & Evolution, 18, 337–343. 10.1016/S0169-5347(03)00061-2 [DOI] [Google Scholar]
- Fang, S. , Clark, R. T. , Zheng, Y. , Iyer‐Pascuzzi, A. S. , Weitz, J. S. , Kochian, L. V. , … Benfey, P. N. (2013). Genotypic recognition and spatial responses by rice roots. Proceedings of the National Academy of Sciences of the United States of America, 110, 2670–2675. 10.1073/pnas.1222821110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrior, C. E. , Rodriguez‐Iturbe, I. , Dybzinski, R. , Levin, S. A. , & Pacala, S. W. (2015). Decreased water limitation under elevated CO2 amplifies potential for forest carbon sinks. Proceedings of the National Academy of Sciences of the United States of America, 112, 7213–7218. 10.1073/pnas.1506262112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- File, A. L. , Klironomos, J. , Maherali, H. , & Dudley, S. A. (2012). Plant kin recognition enhances abundance of symbiotic microbial partner. PLoS One, 7, e45648. 10.1371/journal.pone.0045648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fréville, H. , Roumet, P. , Rode, N. O. , Rocher, A. , Latreille, M. , Muller, M.‐H. , & David, J. (2019). Preferential helping to relatives: A potential mechanism responsible for lower yield of crop variety mixtures? Evolutionary Applications, 12, 1837–1849. 10.1111/eva.12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, S. , Kubo, K.‐i. , & Takayama, S. (2016). Non‐self‐ and self‐recognition models in plant self‐incompatibility. Nature Plants, 2, 16130. 10.1038/nplants.2016.130 [DOI] [PubMed] [Google Scholar]
- Fukano, Y. , Guo, W. , Noshita, K. , Hashida, S. , & Kamikawa, S. (2019). Genotype‐aggregated planting improves yield in Jerusalem artichoke (Helianthus tuberosus) due to self/non‐self‐discrimination. Evolutionary Applications, 12, 508–518. 10.1111/eva.12735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersani, M. , Brown, J. s. , O'Brien, E. E. , Maina, G. M. , & Abramsky, Z. (2001). Tragedy of the commons as a result of root competition. Journal of Ecology, 89, 660–669. 10.1046/j.0022-0477.2001.00609.x [DOI] [Google Scholar]
- Gruntman, M. , & Novoplansky, A. (2004). Physiologically mediated self/non‐self discrimination in roots. Proceedings of the National Academy of Sciences of the United States of America, 101, 3863–3867. 10.1073/pnas.0306604101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, W. D. (1964). The genetical evolution of social behaviour. Journal of Theoretical Biology, 7, 1–52. 10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- Hardin, G. (1968). The tragedy of the commons. Science, 162, 1243–1248. 10.1126/science.162.3859.1243 [DOI] [PubMed] [Google Scholar]
- Hess, L. , & de Kroon, H. (2007). Effects of rooting volume and nutrient availability as an alternative explanation for root self/non‐self discrimination. Journal of Ecology, 95, 241–251. 10.1111/j.1365-2745.2006.01204.x [DOI] [Google Scholar]
- Holzapfel, C. , & Alpert, P. (2003). Root cooperation in a clonal plant: Connected strawberries segregate roots. Oecologia, 134, 72–77. 10.1007/s00442-002-1062-x [DOI] [PubMed] [Google Scholar]
- Holzschuh, A. , Dormann, C. F. , Tscharntke, T. , & Steffan‐Dewenter, I. (2011). Expansion of mass‐flowering crops leads to transient pollinator dilution and reduced wild plant pollination. Proceedings of the Royal Society B: Biological Sciences, 278, 3444–3451. 10.1098/rspb.2011.0268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, M.‐J. , Cho, J.‐I. , Park, S.‐H. , Kim, K.‐H. , Lee, S. K. , Kwon, T.‐R. , … Siddiqui, Z. S. (2014). Sound frequencies induce drought tolerance in rice plant. Pakistan Journal of Botany, 46, 2015–2020. [Google Scholar]
- Jia, Z. , & von Wirén, N. (2020). Signaling pathways underlying nitrogen‐dependent changes in root system architecture: From model to crop species. Journal of Experimental Botany, 71, 4393–4404. 10.1093/jxb/eraa033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, B. K. , Gurung, S. B. , Vista, S. P. , Paneru, P. B. , Gurung, R. , & Pant, S. (2020). Cultivar mixture. In Joshi B. K., Gauchan D., Bhandari B., & Jarvis D. (Eds.), Good practices for agrobiodiversity management (pp. 65–70). Kathmandu: NAGRC, LI‐BIRD and Alliance of Bioversity International and CIAT. [Google Scholar]
- Karban, R. , & Shiojiri, K. (2009). Self‐recognition affects plant communication and defense. Ecology Letters, 12, 502–506. 10.1111/j.1461-0248.2009.01313.x [DOI] [PubMed] [Google Scholar]
- Karban, R. , Shiojiri, K. , Ishizaki, S. , Wetzel, W. C. , & Evans, R. Y. (2013). Kin recognition affects plant communication and defence. Proceedings of the Royal Society B: Biological Sciences, 280, 20123062. 10.1098/rspb.2012.3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban, R. , Yang, L. H. , & Edwards, K. F. (2014). Volatile communication between plants that affects herbivory: A meta‐analysis. Ecology Letters, 17, 44–52. 10.1111/ele.12205 [DOI] [PubMed] [Google Scholar]
- Kessler, A. , & Kalske, A. (2018). Plant secondary metabolite diversity and species interactions. Annual Review of Ecology, Evolution, and Systematics, 49, 115–138. 10.1146/annurev-ecolsys-110617-062406 [DOI] [Google Scholar]
- Khush, G. S. (1999). Green revolution: Preparing for the 21st century. Genome, 42, 646–655. 10.1139/g99-044 [DOI] [PubMed] [Google Scholar]
- Kiær, L. P. , Skovgaard, I. M. , & Østergård, H. (2009). Grain yield increase in cereal variety mixtures: A meta‐analysis of field trials. Field Crops Research, 114, 361–373. 10.1016/j.fcr.2009.09.006 [DOI] [Google Scholar]
- Killingback, T. , & Doebeli, M. (2002). The continuous prisoner's dilemma and the evolution of cooperation through reciprocal altruism with variable investment. American Naturalist, 160, 421–438. 10.1086/342070 [DOI] [PubMed] [Google Scholar]
- Klemens, J. A. (2008). Kin recognition in plants? Biology Letters, 4, 67–68. 10.1098/rsbl.2007.0518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, C.‐H. , Zhang, S.‐Z. , Li, Y.‐H. , Xia, Z.‐C. , Yang, X.‐F. , Meiners, S. J. , & Wang, P. (2018). Plant neighbor detection and allelochemical response are driven by root‐secreted signaling chemicals. Nature Communications, 9, 3867. 10.1038/s41467-018-06429-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepik, A. , Abakumova, M. , Zobel, K. , & Semchenko, M. (2012). Kin recognition is density‐dependent and uncommon among temperate grassland plants. Functional Ecology, 26, 1214–1220. 10.1111/j.1365-2435.2012.02037.x [DOI] [Google Scholar]
- Li, J. , Xu, X. , & Feng, R. (2018). Soil fertility and heavy metal pollution (Pb and Cd) alter kin interaction of Sorghum vulgare . Environmental and Experimental Botany, 155, 368–377. 10.1016/j.envexpbot.2018.05.009 [DOI] [Google Scholar]
- Li, L.‐L. , Zhao, H.‐H. , & Kong, C.‐H. (2020). (−)‐Loliolide, the most ubiquitous lactone, is involved in barnyardgrass‐induced rice allelopathy. Journal of Experimental Botany, 71, 1540–1550. 10.1093/jxb/erz497 [DOI] [PubMed] [Google Scholar]
- Li, Y.‐H. , Xia, Z.‐C. , & Kong, C.‐H. (2016). Allelobiosis in the interference of allelopathic wheat with weeds. Pest Management Science, 72, 2146–2153. 10.1002/ps.4246 [DOI] [PubMed] [Google Scholar]
- Malik, M. , & Vilgalys, R. (1999). Somatic incompatibility in fungi. In Worrall J. J. (Ed.), Structure and dynamics of fungal populations (pp. 123–138). Dordrecht: Springer. [Google Scholar]
- Marler, T. E. (2013). Kin recognition alters root and whole plant growth of split‐root Cycas edentata seedlings. HortScience, 48, 1266–1269. 10.21273/hortsci.48.10.1266 [DOI] [Google Scholar]
- Masclaux, F. , Hammond, R. L. , Meunier, J. , Gouhier‐Darimont, C. , Keller, L. , & Reymond, P. (2010). Competitive ability not kinship affects growth of Arabidopsis thaliana accessions. New Phytologist, 185, 322–331. 10.1111/j.1469-8137.2009.03057.x [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J. (1964). Group selection and kin selection. Nature, 201, 1145–1147. 10.1038/2011145a0 [DOI] [Google Scholar]
- McNickle, G. G. (2020). Interpreting plant root responses to nutrients, neighbours and pot volume depends on researchers' assumptions. Functional Ecology, 34, 2199–2209. 10.1111/1365-2435.13517 [DOI] [Google Scholar]
- McNickle, G. G. , & Dybzinski, R. (2013). Game theory and plant ecology. Ecology Letters, 16, 545–555. 10.1111/ele.12071 [DOI] [PubMed] [Google Scholar]
- Mercer, C. A. , & Eppley, S. M. (2014). Kin and sex recognition in a dioecious grass. Plant Ecology, 215, 845–852. 10.1007/s11258-014-0336-9 [DOI] [Google Scholar]
- Milla, R. , Escudero, A. , & Iriondo, J. M. (2011). Congruence between geographic range distribution and local competitive ability of two Lupinus species. American Journal of Botany, 98, 1456–1464. 10.3732/ajb.1000519 [DOI] [PubMed] [Google Scholar]
- Mommer, L. , Kirkegaard, J. , & van Ruijven, J. (2016). Root–root interactions: Towards a rhizosphere framework. Trends in Plant Science, 21, 209–217. 10.1016/j.tplants.2016.01.009 [DOI] [PubMed] [Google Scholar]
- Murphy, G. P. , & Dudley, S. A. (2009). Kin recognition: Competition and cooperation in Impatiens (Balsaminaceae). American Journal of Botany, 96, 1990–1996. 10.3732/ajb.0900006 [DOI] [PubMed] [Google Scholar]
- Murphy, G. P. , Swanton, C. J. , Van Acker, R. C. , & Dudley, S. A. (2017). Kin recognition, multilevel selection and altruism in crop sustainability. Journal of Ecology, 105, 930–934. 10.1111/1365-2745.12787 [DOI] [Google Scholar]
- Murphy, G. P. , Van Acker, R. , Rajcan, I. , & Swanton, C. J. (2017). Identity recognition in response to different levels of genetic relatedness in commercial soya bean. Royal Society Open Science, 4, 160879. 10.1098/rsos.160879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninkovic, V. (2003). Volatile communication between barley plants affects biomass allocation. Journal of Experimental Botany, 54, 1931–1939. 10.1093/jxb/erg192 [DOI] [PubMed] [Google Scholar]
- Nowak, M. A. , Tarnita, C. E. , & Wilson, E. O. (2010). The evolution of eusociality. Nature, 466, 1057–1062. 10.1038/nature09205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, A. G. , Ali, M. , Yang, S. , Parchami, N. , Bento, T. , Mazzella, A. , … Massa, N. (2016). Kin recognition is a nutrient‐dependent inducible phenomenon. Plant Signaling & Behavior, 11, e1224045. 10.1080/15592324.2016.1224045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret, B. , Desnos, T. , Jost, R. , Kanno, S. , Berkowitz, O. , & Nussaume, L. (2014). Root architecture responses: In search of phosphate. Plant Physiology, 166, 1713–1723. 10.1104/pp.114.244541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzola, E. , Pandolfi, C. , & Mancuso, S. (2020). Resource availability affects kin selection in two cultivars of Pisum sativum . Plant Growth Regulation, 90, 321–329. 10.1007/s10725-019-00562-7 [DOI] [Google Scholar]
- Pickles, B. J. , Wilhelm, R. , Asay, A. K. , Hahn, A. S. , Simard, S. W. , & Mohn, W. W. (2017). Transfer of 13C between paired Douglas‐fir seedlings reveals plant kinship effects and uptake of exudates by ectomycorrhizas. New Phytologist, 214, 400–411. 10.1111/nph.14325 [DOI] [PubMed] [Google Scholar]
- Pierik, R. , & de Wit, M. (2014). Shade avoidance: Phytochrome signalling and other aboveground neighbour detection cues. Journal of Experimental Botany, 65, 2815–2824. 10.1093/jxb/ert389 [DOI] [PubMed] [Google Scholar]
- Platt, T. G. , & Bever, J. D. (2009). Kin competition and the evolution of cooperation. Trends in Ecology & Evolution, 24, 370–377. 10.1016/j.tree.2009.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett, J. M. , Daguerre, Y. , Wittulsky, S. , Vayssières, A. , Deveau, A. , Melton, S. J. , … Martin, F. (2014). Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proceedings of the National Academy of Sciences of the United States of America, 111, 8299–8304. 10.1073/pnas.1322671111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin, D. J. , Bargum, K. , & Kokko, H. (2007). The tragedy of the commons in evolutionary biology. Trends in Ecology & Evolution, 22, 643–651. 10.1016/j.tree.2007.07.009 [DOI] [PubMed] [Google Scholar]
- Riechert, S. E. , & Hammerstein, P. (1983). Game theory in the ecological context. Annual Review of Ecology and Systematics, 14, 377–409. 10.1146/annurev.es.14.110183.002113 [DOI] [Google Scholar]
- Semchenko, M. , Hutchings, M. J. , & John, E. A. (2007). Challenging the tragedy of the commons in root competition: Confounding effects of neighbour presence and substrate volume. Journal of Ecology, 95, 252–260. 10.1111/j.1365-2745.2007.01210.x [DOI] [Google Scholar]
- Semchenko, M. , Saar, S. , & Lepik, A. (2014). Plant root exudates mediate neighbour recognition and trigger complex behavioural changes. New Phytologist, 204, 631–637. 10.1111/nph.12930 [DOI] [PubMed] [Google Scholar]
- Simonsen, A. K. , Chow, T. , & Stinchcombe, J. R. (2014). Reduced plant competition among kin can be explained by Jensen's inequality. Ecology and Evolution, 4, 4454–4466. 10.1002/ece3.1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. R. , & Dworkin, M. (1994). Territorial interactions between two Myxococcus species. Journal of Bacteriology, 176, 1201–1205. 10.1128/jb.176.4.1201-1205.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. E. , & Read, D. J. (2008). Mycorrhizal symbiosis (3rd ed.). London, England: Academic Press. [Google Scholar]
- St‐Pierre, A. , Larose, K. , & Dubois, F. (2009). Long‐term social bonds promote cooperation in the iterated Prisoner's Dilemma. Proceedings of the Royal Society B: Biological Sciences, 276, 4223–4228. 10.1098/rspb.2009.1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takigahira, H. , & Yamawo, A. (2019). Competitive responses based on kin‐discrimination underlie variations in leaf functional traits in Japanese beech (Fagus crenata) seedlings. Evolutionary Ecology, 33, 521–531. 10.1007/s10682-019-09990-3 [DOI] [Google Scholar]
- Tedersoo, L. , Bahram, M. , & Zobel, M. (2020). How mycorrhizal associations drive plant population and community biology. Science, 367, eaba1223. 10.1126/science.aba1223 [DOI] [PubMed] [Google Scholar]
- Tian, H. , De Smet, I. , & Ding, Z. (2014). Shaping a root system: Regulating lateral versus primary root growth. Trends in Plant Science, 19, 426–431. 10.1016/j.tplants.2014.01.007 [DOI] [PubMed] [Google Scholar]
- Tooker, J. F. , & Frank, S. D. (2012). Genotypically diverse cultivar mixtures for insect pest management and increased crop yields. Journal of Applied Ecology, 49, 974–985. 10.1111/j.1365-2664.2012.02173.x [DOI] [Google Scholar]
- Torices, R. , Gómez, J. M. , & Pannell, J. R. (2018). Kin discrimination allows plants to modify investment towards pollinator attraction. Nature Communications, 9, 2018. 10.1038/s41467-018-04378-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers, R. L. (1971). The evolution of reciprocal altruism. The Quarterly Review of Biology, 46, 35–57. 10.1086/406755 [DOI] [Google Scholar]
- Van Dyken, J. D. (2010). The components of kin competition. Evolution, 64, 2840–2854. 10.1111/j.1558-5646.2010.01033.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman, B. (1988). The ecology of kin recognition. Annual Review of Ecology and Systematics, 19, 543–571. 10.1146/annurev.es.19.110188.002551 [DOI] [Google Scholar]
- Weiner, J. , Andersen, S. B. , Wille, W. K.‐M. , Griepentrog, H. W. , & Olsen, J. M. (2010). Evolutionary agroecology: The potential for cooperative, high density, weed‐suppressing cereals. Evolutionary Applications, 3, 473–479. 10.1111/j.1752-4571.2010.00144.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D. S. (1975). A theory of group selection. Proceedings of the National Academy of Sciences of the United States of America, 72, 143–146. 10.1073/pnas.72.1.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt, G. A. K. , Kiers, E. T. , Gardner, A. , & West, S. A. (2014). A biological market analysis of the plant‐mycorrhizal symbiosis. Evolution, 68, 2603–2618. 10.1111/evo.12466 [DOI] [PubMed] [Google Scholar]
- Yamawo, A. , & Mukai, H. (2020). Outcome of interspecific competition depends on genotype of conspecific neighbours. Oecologia, 193, 415–423. 10.1007/s00442-020-04694-w [DOI] [PubMed] [Google Scholar]
- Yang, X.‐F. , Li, L.‐L. , Xu, Y. , & Kong, C.‐H. (2018). Kin recognition in rice (Oryza sativa) lines. New Phytologist, 220, 567–578. 10.1111/nph.15296 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Liu, Q. , Tian, Y. , Xu, X. , & Ouyang, H. (2016). Kin selection or resource partitioning for growing with siblings: Implications from measurements of nitrogen uptake. Plant and Soil, 398, 79–86. 10.1007/s11104-015-2641-z [DOI] [Google Scholar]
- Zhang, Z. , Zhou, F. , Pan, X. , van Kleunen, M. , Liu, M. , & Li, B. (2019). Evolution of increased intraspecific competitive ability following introduction: The importance of relatedness among genotypes. Journal of Ecology, 107, 387–395. 10.1111/1365-2745.13016 [DOI] [Google Scholar]
- Zhu, L. , & Zhang, D.‐Y. (2013). Donald's ideotype and growth redundancy: A pot experimental test using an old and a modern spring wheat cultivar. PLoS One, 8, e70006. 10.1371/journal.pone.0070006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


