Abstract
Rationale: Macrophages are the most abundant immune cell in the alveoli and small airways and are traditionally viewed as a homogeneous population during health. Whether distinct subsets of airspace macrophages are present in healthy humans is unknown. Single-cell RNA sequencing allows for examination of transcriptional heterogeneity between cells and between individuals. Understanding the conserved repertoire of airspace macrophages during health is essential to understanding cellular programing during disease.
Objectives: We sought to determine the transcriptional heterogeneity of human cells obtained from BAL of healthy adults.
Methods: Ten subjects underwent bronchoscopy with BAL. Cells from lavage were subjected to single-cell RNA sequencing. Unique cell populations and putative functions were identified. Transcriptional profiles were compared across individuals.
Measurements and Main Results: We identify two novel subgroups of resident airspace macrophages—defined by proinflammatory and metallothionein gene expression profiles. We define subsets of monocyte-like cells and compare them with peripheral blood mononuclear cells. Finally, we compare global macrophage and monocyte programing between males and females.
Conclusions: Healthy human airspaces contain multiple populations of myeloid cells that are highly conserved between individuals and between sexes. Resident macrophages make up the largest population and include novel subsets defined by inflammatory and metal-binding profiles. Monocyte-like cells within the airspaces are transcriptionally aligned with circulating blood cells and include a rare population defined by expression of cell–matrix interaction genes. This study is the first to delineate the conserved heterogeneity of airspace immune cells during health and identifies two previously unrecognized macrophage subsets.
Keywords: alveolar macrophages, RNA sequencing, bronchoalveolar lavage
At a Glance Commentary
Scientific Knowledge on the Subject
Airspace macrophages are traditionally thought to exist as a homogeneous population during health. Whether macrophage subsets exist within and between healthy individuals is currently unknown.
What This Study Adds to the Field
This study is the first to examine immune cell heterogeneity within the healthy human lung airspaces. Using single-cell RNA sequencing, we define novel airspace macrophage and monocyte populations conserved across 10 healthy individuals. These data provide insight into the diversity of BAL cells in health and inform future translational studies of human lung disease.
Airspace macrophages (AMs) are essential regulators of lung function. During health, AMs engulf inhaled particles, recycle surfactant proteins, support tissue integrity, and survey the airways and alveoli for pathogens and danger signals. Upon exposure to inflammatory stimuli, they rapidly adapt their transcriptional programs, becoming innate effectors and master regulators of the host inflammatory and immunologic responses. Accordingly, they serve as sensitive indicators of lung health and are attractive targets for diagnostic and therapeutic studies of lung diseases. However, whether all AMs in the healthy airspaces are similarly programed to perform these myriad functions or whether specific functions may be ascribed to cellular subsets is unknown.
AMs are traditionally thought to exist as a homogeneous population during health (1–5). But emerging evidence suggests genetic variance and microenvironmental cues may influence macrophage plasticity (6, 7). The effects of varied environmental exposures, antecedent infections, and genetic differences on the programing states of human AMs is not well understood. We hypothesized that transcriptionally distinct subsets of AMs exist and vary in composition between healthy individuals. To test this, we undertook a comprehensive analysis of the unique and conserved features of AMs in a cross-section of healthy adults.
Tissue-resident AMs have been shown to self-renew with little input from circulating monocytes during homeostasis in mice (8–10). Although monocytes may survey the lung tissue (3), they have not been shown to enter the airspaces, mature, or contribute to the AM pool in the absence of inflammation. In humans, the presence of monocytes in the airspaces at the time of autopsy or after sham exposure has been attributed to low levels of inflammation inherent in mechanical ventilation or saline inhalation (4, 5, 11–14). Therefore, whether monocytes are consistently present in the healthy airspaces or emerge only after exposure to environmental irritants is unknown. Furthermore, whether specific monocyte subsets are recruited to the airspaces or whether this trafficking results in changes in monocyte programing has not been determined.
In this study, we used single-cell RNA sequencing (RNA-seq) to systematically assess the heterogeneity of BAL cells from 10 healthy adults. We confirmed that the vast majority of lavaged cells are mononuclear phagocytes and focused our analyses on these populations. We identified specific subsets of resident AMs, present in all subjects, with putative proinflammatory and metal-binding functions. Furthermore, we defined monocyte-like cells in the airspaces and directly compared them with peripheral blood. In doing so, we identified the unique and shared gene expression of monocyte subsets in the airspaces and peripheral blood and provide evidence for steady-state maturation of monocytes into macrophages within the healthy airspaces. Together, these data define the conserved transcriptional landscape of airspace macrophages and monocytes in healthy adults and provide a framework for the study of their programing during disease.
Methods
Subject Selection
Healthy subjects aged 18–50 years who were able to give informed consent were eligible for the study. Subjects underwent a comprehensive medical history and physical examination, spirometry, and ECG within 2 weeks of bronchoscopy. Eleven subjects were recruited to participate in the study. One subject was excluded based on abnormal spirometry and ECG results. Screening data were independently reviewed by investigators (K.J.M. and W.J.J.) and determination of eligibility was made by consensus. Exclusion criteria are listed in the online supplement.
Study Intervention
Ten participants underwent flexible fiberoptic bronchoscopy and lavage with serial installation and aspiration of normal saline in the apical medical subsegment of the lingua. All subjects tolerated the procedure without adverse events.
Data Analysis
Details of the bronchoscopy technique, single-cell isolation, RNA sequencing, and computational analyses are available in the online supplement. Briefly, single-cell data were normalized, integrated, and clustered using the Seurat (15, 16) pipeline. Cluster markers were identified by comparing normalized expression in each cluster with all others using a Wilcoxon rank sum test. Markers were identified as genes exhibiting significant upregulation when compared against all other clusters and defined by having a Bonferroni adjusted P value < 0.05 (adjusted for 18,004 genes), a log fold change >0.25, and detectable expression in >10% of cells. This analysis was then repeated separately for each subject to determine if the identified markers were consistent across subjects and to ensure that a single outlying subject did not drive marker results. Markers were considered “conserved” if they had adjusted P values < 0.05 for all 10 subjects.
Data Availability
RNA sequencing data have been deposited in the Gene Expression Omnibus, accession code GSE151928. The data set can be visualized at https://healthy-bal.cells.ucsc.edu.
Ethical Approval
The study was approved and monitored by the National Jewish Health Institutional Review Board (FWA00000778). Written informed consent was obtained from all participants.
Results
BAL Cell Populations Are Highly Conserved between Healthy Subjects
We performed single-cell RNA-seq on previously frozen BAL cells from four females and six males ranging from 26 to 50 years old (mean, 34; SD, 8 yr). Demographic data and bronchoscopic yield from study participants are shown in Table 1. We analyzed cell profiles that passed strict quality control thresholds, totaling 49,384 cells, and partitioned them into clusters using the 3,000 most informative genes (Figure 1A). The percentage of cells within each cluster was comparable between individuals (Figure 1B), and each cluster was composed of cells from all 10 subjects, with the exception of cluster 12, which contained cells from 9 of 10 subjects (Figures 1B and 1C).
Table 1.
Characteristics of Subjects and BAL Yield
| Subject | Race or Ethnicity | BMI (kg/m2) | BAL Return Volume (ml) | BAL Cell Count |
|---|---|---|---|---|
| 1 | White | 25 | 95 | 1.46 × 107 |
| 2 | White | 30 | 46 | 6.10 × 107 |
| 3 | Hispanic | 30 | 129 | 1.26 × 107 |
| 4 | Hispanic | 28 | 157 | 1.61 × 107 |
| 5 | White | 23 | 158 | 1.86 × 107 |
| 6 | Asian | 25 | 165 | 4.50 × 106 |
| 7 | Hispanic | 33 | 133 | 3.40 × 106 |
| 8 | White | 23 | 130 | 1.13 × 107 |
| 9* | Asian | 27 | 100 | 1.10 × 107 |
| 10* | White | 31 | 133 | 6.70 × 107 |
Definition of abbreviation: BMI = body mass index.
Cells from subjects 9 and 10 were processed in separate batches from the remaining subjects.
Figure 1.
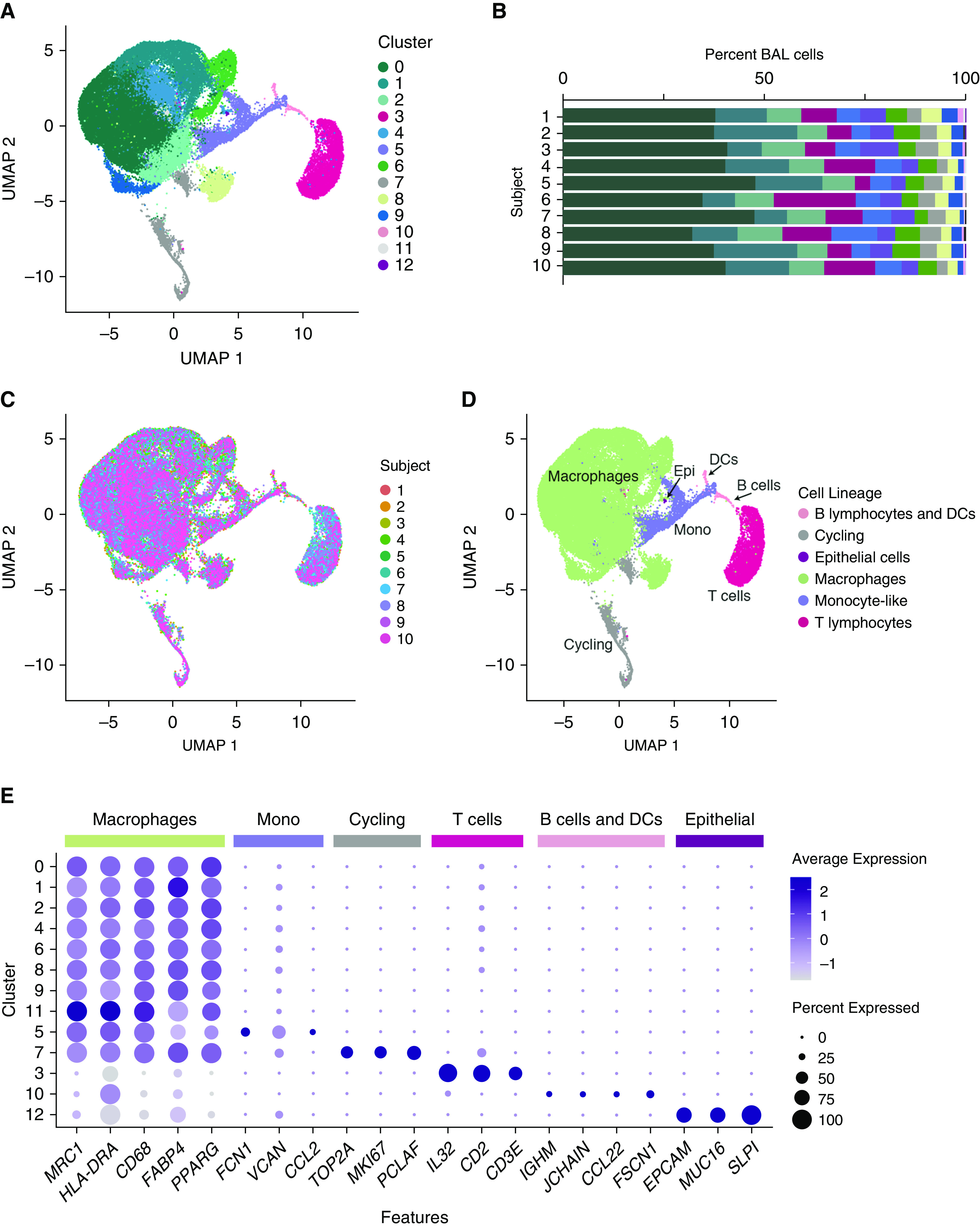
Single-cell transcriptional profiling of healthy human BAL cells. Ten healthy subjects underwent endoscopic BAL and single cells were isolated and assessed by single-cell RNA sequencing. (A) Uniform manifold approximation and projection (UMAP) plot shows clustering of 49,384 cells based on gene expression. Color specifies assignment of cells to one of 13 clusters inferred using shared nearest neighbor clustering. (B) Relative percentage of cells within each cluster by individual subject. (C) Subject information overlaid on UMAP plot. (D) Cell lineage inferred from expression of marker gene signatures. (E) Dot plot comparing expression of marker genes across cell clusters, where the x-axis denotes marker genes and the y-axis refers to clusters in A. Colored bars across the top of the figure denote inferred annotations of clusters based on the corresponding marker genes. Dot size is proportional to the percentage of cells expressing the gene in each cluster. Color intensity is proportional to averaged scaled log-normalized expression within a cluster. DCs = dendritic cells; Epi = epithelial cells; Mono = monocyte-like cells.
Clusters were numbered in order of size and annotated by comparing transcriptional signatures with known lineage markers and previously published data sets (17–19) (Figures 1D and 1E). Macrophages, monocyte-like cells (with expression of both monocyte and macrophage markers), lymphocytes, dendritic cells (DCs), cycling cells composed of multiple cell types, and a small population of epithelial cells were identified.
To verify that cell preservation techniques did not bias our results, we directly compared freshly isolated BAL cells with those from the same sample after freezing and subsequent thawing. Clustering of cells from each condition separately, or following integration of data sets, revealed similar populations between conditions (see Figures E1A–E1D in the online supplement). Furthermore, annotation of resultant clusters demonstrated similar composition of cell types between conditions, confirming preservation of cellular heterogeneity following freezing of BAL cells (Figures E1E–E1J).
Discrete AM Subsets Are Present during Health
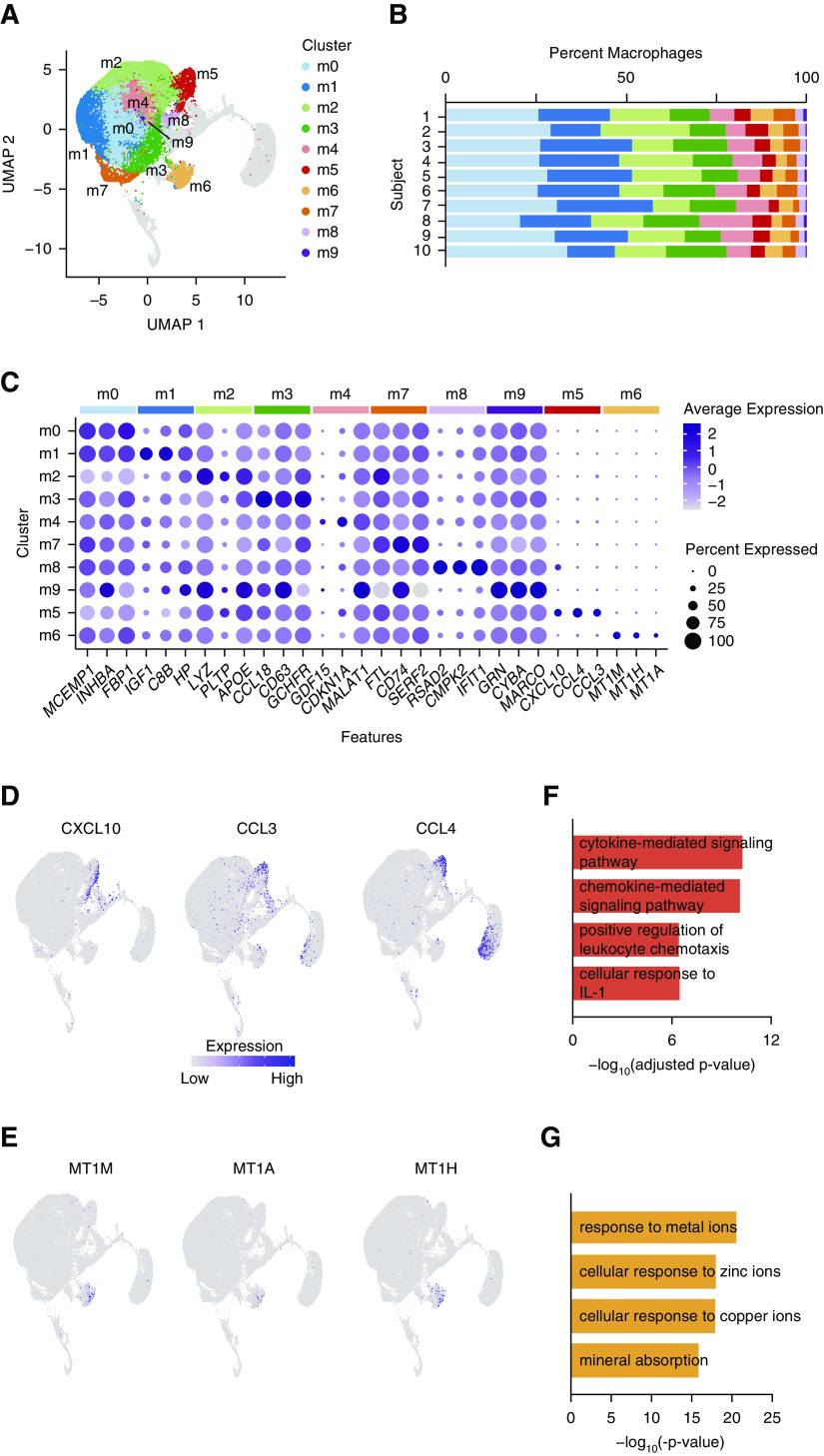
Because macrophage-specific markers were expressed at high levels in several clusters, we sought to determine whether heterogeneity exists among AMs during health. We reclustered populations with the highest expression of macrophage markers (clusters 0–2, 4, 6, 8, 9, and 11) and identified 10 macrophage subclusters (m0–9). Each subset contained cells from all 10 subjects (Figures 2A and 2B). We next defined the characteristics that distinguish each subcluster from other macrophages. We identified differentially expressed gene (DEG) profiles unique to each group and found that 8 of the 10 clusters (m0–4 and m7–9) exhibited overlapping gradients of expression of many DEGs and putative markers (Figure 2C and Table E1). Because these clusters made up the dominant population of cells within each sample and demonstrated relatively minor differences in expression of marker genes, we hypothesized that they represent the transcriptional spectrum of AMs during health. Indeed, functional enrichment analyses of DEGs from these eight clusters revealed interrelated pathways consistent with known AM functions, including cellular adhesion, phagocytosis, and lipid metabolism (Table E2). Together, these data suggest that these eight clusters represent a continuum of homeostatic resident AMs, without exclusive cellular markers or putative functions to indicate distinct cell types.
Figure 2.
Distinct populations of airspace macrophages exist during health. (A) Cells identified as macrophages (Figure 1 clusters 0–2, 4, 6, 8, 9, and 11) were reclustered. Color specifies assignment of cells to one of 10 clusters. (B) Relative proportion of cells within each cluster by individual subject. (C) Dot plot comparing expression of marker genes across cell clusters, where the x-axis denotes marker genes and the y-axis refers to clusters in A. Colored bars across the top of the figure denote the subcluster from which the marker genes were identified. Dot size is proportional to the percentage of cells expressing the gene in each cluster. Color intensity is proportional to averaged scaled log-normalized expression within a cluster. (D and E) Feature plots demonstrating scaled log-normalized expression of selected genes differentially expressed in clusters m5 (D) and m6 (E). (F and G) Functional enrichment analysis of cluster markers. Representative pathways are shown for m5 (F) and m6 (G). m0–9 = macrophage clusters 0–9; UMAP = uniform manifold approximation and projection.
We next examined the transcriptional profiles of the two macrophage subclusters with discrete DEG profiles (m5 and m6). We first explored m5 and compared cells in this cluster with the composite of all other AM clusters. We identified 175 genes that had increased expression in cluster m5, 48 of which were conserved across all subjects. These conserved DEGs include proinflammatory cytokines, chemokines, and inflammatory signaling molecules (Figures 2C and 2D and Table E1). Functional enrichment analysis of DEGs revealed pathways involved in inflammation, cytokine signaling, and leukocyte chemotaxis (Figure 2E and Table E2). Taken as a whole, these data suggest the presence of a small but conserved subset of proinflammatory AMs present in asymptomatic, healthy individuals.
Next, we identified the unique transcripts expressed by cells in cluster m6. Compared with all other AM clusters, 10 DEGs define cluster m6, 8 of which are conserved across all subjects (Table E1). Notably, all eight conserved DEGs are members of the metallothionein gene family and encode metal-binding proteins important in processing heavy metals such as zinc and cadmium. Expression of these genes is highly restricted to cluster m6 (Figures 2C and 2F), suggesting a unique role of these AMs in heavy metal processing. In support of this hypothesis, the cluster is defined by high expression of SLC30A1, a metal transport protein, and pathway enrichment analysis of DEGs identified cellular responses to metals and mineral absorption pathways (Figure 2G and Table E2). In sum, these data identify a novel subpopulation of AMs defined by expression of metallothionein and metal transport genes.
Unique Subsets of Monocyte-like Cells Are Present in Healthy Airspaces
The presence of rare monocyte-like cells within healthy human airspaces has been described (4, 11, 12, 20). However, little is known regarding their origin, fate, or function in the healthy lung. We started our analysis of monocyte-like cells by first determining whether heterogeneity was present within the population. Reclustering monocyte-like cells (Figure 1 cluster 5) revealed five subclusters (mo0–4), composed of cells from each subject (Figures 3A and 3B). Analysis of DEGs within each subcluster (Table E3) revealed two subsets defined by classical DC markers (mo3–4; Figure 3C), both of which have previously been described in BAL during health. Together with the CCL22-expressing or “activated” DC subset in Figure 1 (cluster 10), these three DC subclusters reflect the major conventional DC populations previously described in the healthy human airspaces (5, 12, 21–23). The remaining three clusters (mo0–2) had high expression of both monocyte (e.g., CSF1R) and macrophage (e.g., MARCO and VSIG4) genes.
Figure 3.
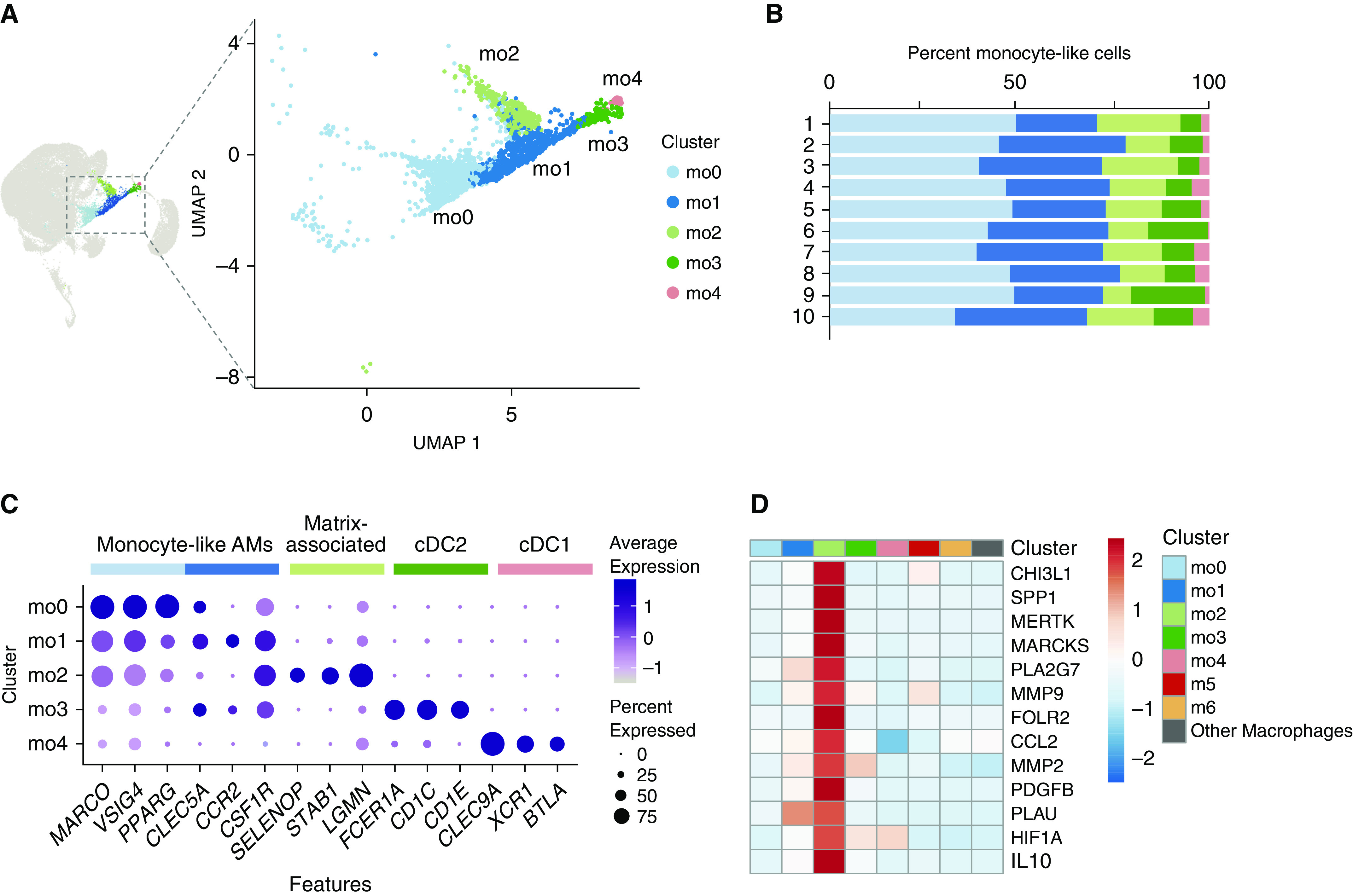
Monocyte-like cells are composed of dendritic cells and monocyte-like airspace macrophages. (A) Cells identified as monocyte-like (Figure 1 cluster 5) were reclustered. Color specifies assignment of cells to one of five clusters. (B) Relative proportion of cells within each cluster by individual subject. (C) Dot plot comparing expression of marker genes across cell clusters, where the x-axis denotes marker genes and the y-axis refers to clusters in A. Colored bars across the top of the figure denote inferred annotations of clusters based on the corresponding marker genes. Dot size is proportional to the percentage of cells expressing the gene in each cluster. Color intensity is proportional to averaged scaled log-normalized expression within a cluster. (D) Heat map demonstrating expression of select cell–matrix interaction genes (“other macrophages” are composed of clusters m0–4 and m7–9). The heat map is colored by the average scaled log-normalized expression in each cluster. AM = airspace macrophage; cDC = conventional dendritic cell; m0–9 = macrophage clusters 0–9; mo0–4 = monocyte-like clusters 0–4; UMAP = uniform manifold approximation and projection.
We next examined the defining characteristics of clusters mo0–2 and found that in addition to known monocyte genes such as STAB1 and LGMN, cluster mo2 was uniquely defined by DEGs involved in cell–matrix interactions, such as MMP14, SPP1, and ECM1 (Table E3). Cell–matrix interactions have been implicated in disease processes such as chronic lung inflammation and interstitial fibrosis (24, 25), and SPP1-expressing macrophages have been described as a rare population that exists during health and expands during pathologic conditions (20, 26, 27). To explore whether this small subset was also programed to interact with the cell matrix during health, we assembled a panel of genes associated with matrix deposition in the lung (20, 26, 28–36). We found that these genes were uniquely expressed in cluster mo2 compared with other monocyte-like subsets and all other resident AM populations (Figure 3D). In comparison, the transcriptional profiles that defined the remaining monocyte-like cell clusters (mo0–1) had substantial overlap, suggesting cells within these clusters may exist on a transcriptional continuum. Taken as a whole, our data show that monocyte-like cells are composed of four subgroups: two classical DC subsets (mo3 and mo4), and two monocyte subsets with overlapping expression of both monocyte and macrophage genes separated by the absence (mo0–1) or presence (mo2) of cell–matrix interaction genes.
To further assess the relationship between monocyte subsets, we performed trajectory analysis of monocyte and resident AM cell subclusters and visualized their transcriptional profiles along a pseudotemporal trajectory. We identified a trajectory branch primarily composed of monocytes. Cells furthest from AMs were primarily from mo2, followed by mo1 cells. Cells from mo0 were positioned between mo1 cells and AMs and appeared to overlap with AMs (Figures 4A and 4B and E2). Examination of DEGs along this trajectory in the direction of resident AMs demonstrates increasing expression of bona fide macrophage markers, such as complement genes, MARCO, and PPARG, over pseudotime (Figure 4C and Table E4). Furthermore, identification of inferred regulatory networks in each cluster revealed increasing activity of transcription factors associated with macrophage maturation, such as PU.1 (SPI1) and CEBPB moving from mo1 to mo0 to AMs (Figure 4D and Table E5). Together, these data imply that mo1 and mo0 represent monocytes along a continuum of maturation toward mature AMs.
Figure 4.
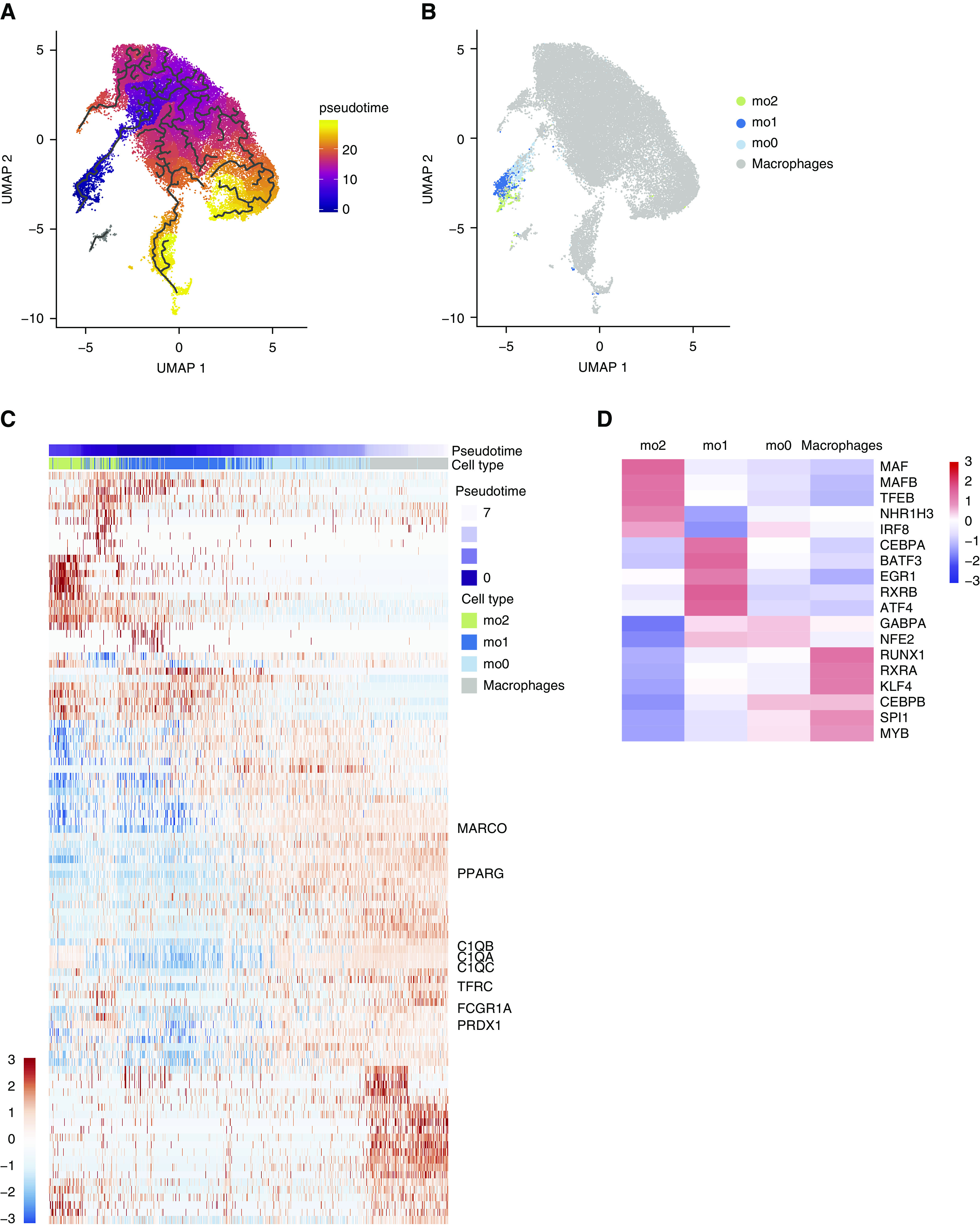
Airspace monocyte-to-macrophage maturation trajectory. (A) UMAP of 36,452 macrophages and monocytes colored by pseudotime. Black lines represent the principal graph trajectories. (B) UMAP colored by monocyte subclusters (from Figure 3 clusters mo0–2) or macrophages (Figure 2 clusters m0–9). (C) Heat map of the top 100 pseudotime differentially expressed genes across the monocyte-to-macrophage branch. The heat map is colored by scaled log-normalized expression for each cell. (D) Heat map of scaled regulon activity scores for select transcription factor motifs implicated in monocyte-to-macrophage maturation (“macrophages” are composed of clusters m0–4 and m6–9). m0–9 = macrophage clusters 0–9; mo0–2 =monocyte-like cell clusters 0–2; UMAP = uniform manifold approximation and projection.
Monocyte-like Cells Are Aligned but Distinct from Circulating Blood Cells
The finding of monocyte-like cells within the airspaces of healthy adults, and the suggestion of maturation into AMs, implies a steady-state trafficking of monocytes into the airspaces. To identify putative precursors of monocyte-like cells within the BAL, we integrated the transcriptional signatures of monocyte-like cells from BAL with publicly available single-cell RNA-seq data of peripheral blood mononuclear cells (PBMCs) from a healthy male and compared cell populations between the two compartments. BAL monocyte-like cells aligned exclusively with monocyte and DC populations in the PBMC data set (Figure E3). Extraction and reclustering of overlapping populations from both compartments resulted in 11 distinct subclusters, herein referred to as p0–10 (Figure 5A). Ten of the subclusters contained at least 1% of cells from both the BAL and PBMC cells (Figures 5B and 5C). In contrast, p6 was composed almost entirely of circulating blood cells.
Figure 5.
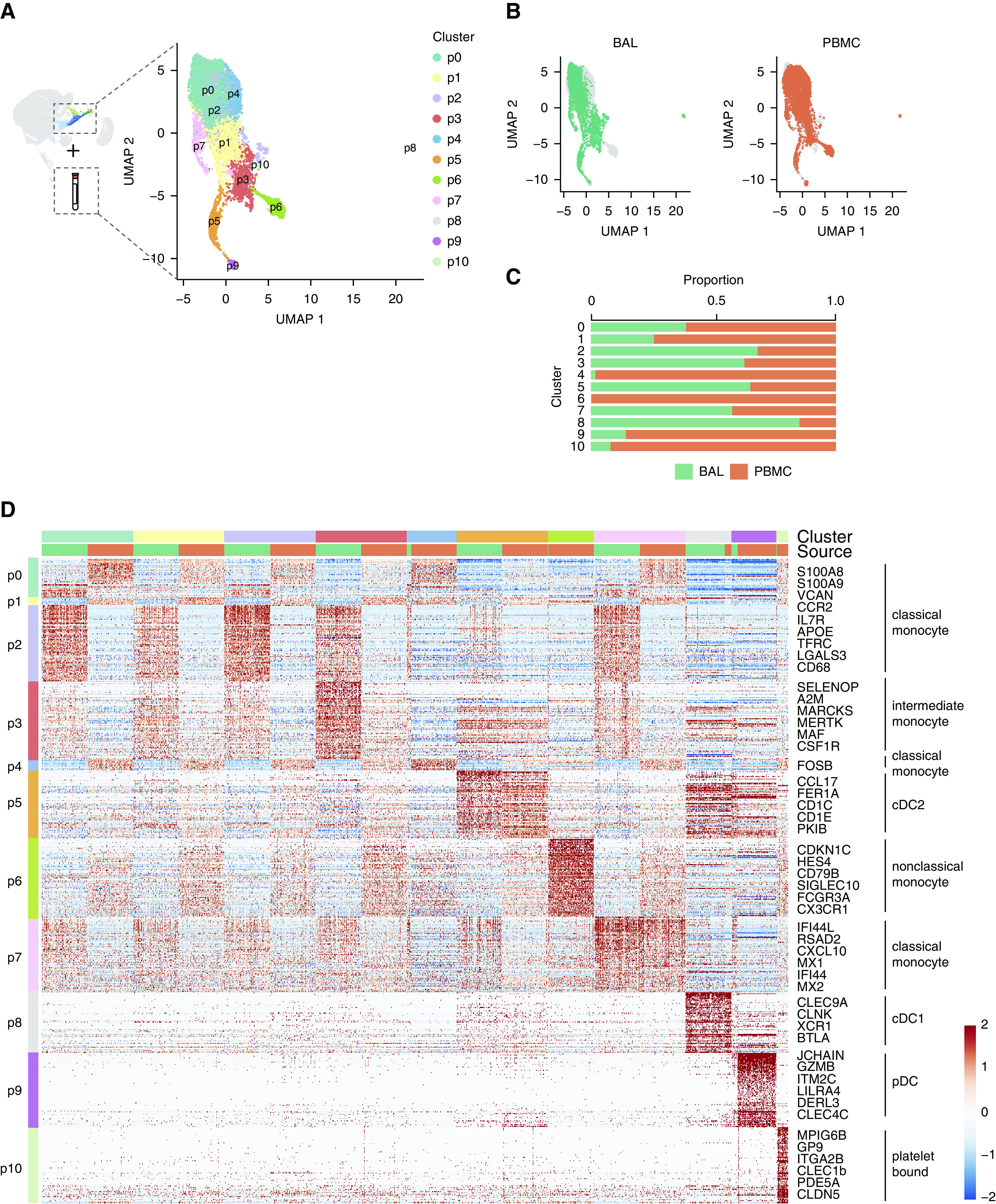
Monocyte-like cells are aligned but distinct from circulating blood cells. (A) Monocyte-like cells from BAL (Figure 1 cluster 5) were integrated with peripheral blood monocytes and reclustered. Color specifies assignment of cells to one of 11 clusters, labeled p0–11. (B) Contributions of cells from BAL or PBMCs to UMAP projection. (C) Relative proportion of cells within each cluster from BAL or PBMCs. (D) Heat map of cell clusters with select marker genes and annotations highlighted. Up to 100 cells were randomly selected from BAL and PBMC samples in each cluster for inclusion in the heat map. Up to the top 100 markers significant in at least two individual samples were included for each cluster. The heat map is colored by scaled log-normalized expression. cDC = conventional dendritic cell; PBMC = peripheral blood mononuclear cell; pDC = plasmacytoid dendritic cell; UMAP = uniform manifold approximation and projection.
To correlate the integrated clusters with well-defined monocyte and DC populations in the blood, we performed annotation using data sets of PBMC single-cell gene expression (18, 37) (Figure 5D and Table E6). Five subclusters (p0, 1, 2, 4, 7) were defined by markers of classical, CD14hi monocytes and included cells from mo0–2 in the BAL (Figure E4). One subcluster (p6) was identified as nonclassical, CD16hi or so-called “patrolling monocytes.” Importantly, this cluster was composed almost exclusively of PBMCs, supporting the notion of selective trafficking of monocytes to the airspaces, rather than the nondiscriminant spillage of blood into airspaces during bronchoscopy.
Subcluster p3 was defined by markers of intermediate, CD14+ CD16+ monocytes and contained mainly matrix-associated cells (mo2) from the BAL (Figure E4). This result suggests that matrix-associated cells are closely related to intermediate CD14+ CD16+ monocytes in the peripheral blood.
Three dendritic cell subclusters comprising both PBMC and BAL cells (p5, p8, and p9; Figure 5D) were also identified. One small subcluster, composed mostly of PBMCs, shared markers of both classical monocytes and platelets and was labeled as “platelet bound” (p10, Figure 5D and Table E6).
In sum, these data support the notion that monocyte-like cells within the BAL make up a heterogeneous group that are closely aligned with specific circulating monocyte subsets.
AM Programing Is Conserved between Sexes
Finally, we examined whether global differences in BAL cell abundance or transcriptional programing exist between sexes. We found no statistically significant differences in the proportion of cells within BAL subsets between males and females (Figure E5A). Because myeloid cells represent the vast majority of BAL cells in the data set, we compared overall DEGs between combined AM populations in males versus females. Of 18,004 genes examined, we detected 19 genes with significantly increased expression in females (0.11%) and 25 (0.14%) genes with increased expression in males (Figure E5B and Table E7). Examination of individual transcripts revealed more than half of DEGs in each group were encoded on sex chromosomes. Pathway enrichment did not identify putative functions related to these DEGs. Together, these data suggest that cellular composition and programing of AMs is highly conserved between sexes.
Discussion
In this study, we used single-cell RNA-seq to define the cellular landscape of BAL among healthy adults. By integrating data across 10 individuals, our study was powered to identify rare populations and define cellular variability between subjects. By isolating cells from the entire cellular fraction of BAL rather than sorting on traditional cellular markers, we avoided selection bias to produce a comprehensive data set. From our analyses, we describe for the first time two unique populations of resident AMs defined by proinflammatory and metal-binding signatures and further define a rare matrix-associated subset of monocyte-like cells whose transcriptional signature is notable for genes implicated in diseases with enhanced lung matrix deposition. By performing trajectory analyses and comparing monocyte-like cells with peripheral blood, our data give strong support to the notion of steady-state monocyte to AM maturation in the healthy human airspaces. Remarkably, our findings are highly conserved across all 10 subjects, despite varying sex, ages, and genetic backgrounds.
The finding of AMs with high expression of metallothionein genes suggests these cells may be exposed to particulates containing heavy metals such as zinc, cadmium, or iron. Although it has long been assumed that the presence of heavy metals in polluted air influences AM programing (38, 39), to our knowledge, the extent to which polluted air may induce metallothionein gene expression in vivo has not been examined. Our study does not determine whether metallothionein expression represents a transient response of AMs to metal exposure or whether cells within this subset have been permanently programed by a particular microenvironmental signal or anatomic location, such as the proximal airways or bronchoalveolar duct junctions, locations particularly prone to deposition of particulates (40). Notably, subpopulations of metallothionein-expressing AMs are present in other published single-cell RNA-seq data sets from human lung digests (26). Further investigation of the molecular signals that induce specific metallothionein isoforms and the effects of such signaling on AM function may inform our understanding of chronic airway disease and guide preventive health efforts.
In a similar vein, our finding of a unique subset of AMs with inflammatory cytokine and chemokine expression implies that a proportion of AMs continually respond to danger and pathogen-associated signals inevitably encountered by the lungs via inhalation of ambient air. In fact, the DEG profile that defines this cluster, including genes such as CXCL8, SOD2, and TNF, mimics the expression profile of human AMs treated with bacterial pathogens in vitro (41). However, an alternative explanation is that this gene expression signature denotes recent migration or differentiation from monocytes. Indeed, this AM subset is also defined by high expression of PTGS2, CD83, and TNFAIP3, which in mice mark a population of classical monocytes that survey the lung and transport antigen to lymph nodes during health (3). Whether this population represents a dynamic programing shift in response to stimuli or denotes a specialized function as an antigen-presenting cell in humans is yet to be determined.
Although our study is not sufficient to definitively ascribe the origin or differentiation program of cellular subsets, our results present compelling evidence of ongoing trafficking and maturation of circulating blood cells in the airspaces of asymptomatic, healthy adults. By comparing monocyte-like BAL cells directly with PBMCs, we have identified the shared and unique aspects of cells within each compartment. Moreover, using trajectory and transcriptional regulatory network analyses, our data suggest that circulating monocytes may differentiate into two distinct populations within the airspaces—matrix-associated monocytes or mature AMs. The suggestion of steady-state monocyte to AM maturation may be in contrast to studies in mice, where local replication of embryonic-derived, tissue-resident AMs provides self-renewal (8, 10). Although our data also demonstrate a population of proliferative AMs (Figure 1 clusters 8 and 9), the rate and source of AM replenishment in healthy humans remains unknown. Therefore, whether monocyte maturation within healthy human airspaces represents a response to imperceptible environmental irritants or a necessary contribution to maintain the AM niche over the lifespan is not clear. The implications for understanding the mechanisms of monocyte trafficking, differentiation, and programing within the airspaces are many, given our evolving appreciation of monocyte-derived cells as the main drivers of inflammation and repair within the lung (1, 27, 42–48).
The presence of a matrix-associated monocyte population has been described in several recent studies examining immune cell heterogeneity during fibrosis in the lung and other organs (20, 26, 28, 37, 49). Additional studies comparing PBMCs and BAL cells from the same individuals are needed to determine the relative expression of these “profibrotic” genes in airspace monocytes compared with putative precursors in the blood. These data may reveal whether a particular microenvironmental cue or cellular interaction drives the “profibrotic” signature upon arrival to the lung or whether these cells represent an immature precursor to other monocyte subsets. Identifying the signals that drive expression of matrix-associated genes and elucidating the temporal and causal relationship of these genes to monocyte programing will be important to understand the maintenance of normal lung architecture versus fibrosis.
Perhaps the most notable finding of this study is the high degree of conservation of cell types and programing states among healthy subjects. Our subjects represent adults in three decades of life, with varied genetic backgrounds. Our study was not designed to examine variations in gene expression along continuous variables such as age but did allow for comparisons between sexes. Our data demonstrate that the vast majority of AM genes are expressed at comparable levels between sexes. However, because we did not collect data on estrous cycles, we may not have detected dynamic regulation of gene expression by estrogens. Because all subjects were recruited from a single metropolitan area, were free of occupational exposures, and had no family history of respiratory illnesses, our study does not explore the role of such factors on AMs.
Our study examined cells contained in the lavage fluid from the distal airways. As such, it did not include cells from other compartments such as the interstitium. Likewise, it is possible that tightly adherent cell populations (such as sessile macrophages [50]) could exist in the airspaces but are not removed from the lungs by lavage. Here, we focused our analyses on myeloid cells, as they represent the majority of BAL cells. It is likely that additional heterogeneity is present within the lymphocyte populations and may reflect intersubject differences that were not evident in macrophages and monocytes.
Conclusions
In this study, application of single-cell RNA-seq to BAL of 10 healthy adults yielded the discovery of novel populations of proinflammatory and metallothionein-expressing resident AMs and further definition of monocyte-like cell subsets within the healthy airspaces. Our results establish that BAL cells exist in heterogeneous but conserved populations during health. Studying these subsets over the course of disease may further our basic understanding of lung pathology and provide specific immune targets for lung diseases.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Max Seibold for his expertise and advice in designing the study.
Footnotes
K.J.M. is supported by NIH grant K08 HL141648, the Boettcher Foundation’s Webb-Waring Biomedical Research Award, and a Parker B. Francis Research Opportunity Award. C.M.M. is supported by the Boettcher Foundation’s Webb-Waring Biomedical Research Award. A.L.M. is supported by NIH grant K99 HL141658. P.M.H. is supported by NIH grants AI141389 and HL149741. W.J.J. is supported by NIH grants HL140039, HL130938, and HL148335. Single-cell capture and next-generation sequencing were performed by the National Jewish Health Genomics Facility and the University of Colorado Cancer Center Genomics Shared Resource, which is supported by the NCI Cancer Center Support grant P30CA046934.
Author Contributions: K.J.M. designed the study, screened subjects, performed bronchoscopies, processed samples, analyzed data, prepared figures, and wrote the manuscript. C.M.M. performed biostatistical and computational analyses, prepared figures, and contributed to the manuscript. S.A.M. and J.D.M. processed samples and generated data. C.L.G. recruited, screened, and obtained informed consent from subjects and coordinated the study. A.L.M. and P.M.H. interpreted data and contributed to the manuscript. W.J.J. initiated, designed, and supervised the study, screened subjects, analyzed data, and wrote the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202005-1989OC on October 20, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Mould KJ, Jackson ND, Henson PM, Seibold M, Janssen WJ. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight. 2019;4:126556. doi: 10.1172/jci.insight.126556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;173:1307. doi: 10.1016/j.cell.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. doi: 10.1016/j.immuni.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu YR, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol. 2016;54:13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jardine L, Wiscombe S, Reynolds G, McDonald D, Fuller A, Green K, et al. Lipopolysaccharide inhalation recruits monocytes and dendritic cell subsets to the alveolar airspace. Nat Commun. 2019;10:1999. doi: 10.1038/s41467-019-09913-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sajti E, Link VM, Ouyang Z, Spann NJ, Westin E, Romanoski CE, et al. Transcriptomic and epigenetic mechanisms underlying myeloid diversity in the lung. Nat Immunol. 2020;21:221–231. doi: 10.1038/s41590-019-0582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarling JD, Lin HS, Hsu S. Self-renewal of pulmonary alveolar macrophages: evidence from radiation chimera studies. J Leukoc Biol. 1987;42:443–446. [PubMed] [Google Scholar]
- 10.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brittan M, Barr L, Conway Morris A, Duffin R, Rossi F, Johnston S, et al. A novel subpopulation of monocyte-like cells in the human lung after lipopolysaccharide inhalation. Eur Respir J. 2012;40:206–214. doi: 10.1183/09031936.00113811. [DOI] [PubMed] [Google Scholar]
- 12.Desch AN, Gibbings SL, Goyal R, Kolde R, Bednarek J, Bruno T, et al. Flow cytometric analysis of mononuclear phagocytes in nondiseased human lung and lung-draining lymph nodes. Am J Respir Crit Care Med. 2016;193:614–626. doi: 10.1164/rccm.201507-1376OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Grady NP, Preas HL, Pugin J, Fiuza C, Tropea M, Reda D, et al. Local inflammatory responses following bronchial endotoxin instillation in humans. Am J Respir Crit Care Med. 2001;163:1591–1598. doi: 10.1164/ajrccm.163.7.2009111. [DOI] [PubMed] [Google Scholar]
- 14.Bharat A, Bhorade SM, Morales-Nebreda L, McQuattie-Pimentel AC, Soberanes S, Ridge K, et al. Flow cytometry reveals similarities between lung macrophages in humans and mice. Am J Respir Cell Mol Biol. 2016;54:147–149. doi: 10.1165/rcmb.2015-0147LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 18.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J. 2019;54:1802441. doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naessens T, Morias Y, Hamrud E, Gehrmann U, Budida R, Mattsson J, et al. Human lung conventional dendritic cells orchestrate lymphoid neogenesis during chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2020;202:535–548. doi: 10.1164/rccm.201906-1123OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bigley V, McGovern N, Milne P, Dickinson R, Pagan S, Cookson S, et al. Langerin-expressing dendritic cells in human tissues are related to CD1c+ dendritic cells and distinct from Langerhans cells and CD141high XCR1+ dendritic cells. J Leukoc Biol. 2015;97:627–634. doi: 10.1189/jlb.1HI0714-351R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel VI, Booth JL, Duggan ES, Cate S, White VL, Hutchings D, et al. Transcriptional classification and functional characterization of human airway macrophage and dendritic cell subsets. J Immunol. 2017;198:1183–1201. doi: 10.4049/jimmunol.1600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgstaller G, Oehrle B, Gerckens M, White ES, Schiller HB, Eickelberg O. The instructive extracellular matrix of the lung: basic composition and alterations in chronic lung disease. Eur Respir J. 2017;50:1601805. doi: 10.1183/13993003.01805-2016. [DOI] [PubMed] [Google Scholar]
- 25.Heukels P, Moor CC, von der Thüsen JH, Wijsenbeek MS, Kool M. Inflammation and immunity in IPF pathogenesis and treatment. Respir Med. 2019;147:79–91. doi: 10.1016/j.rmed.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 28.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai T, Tanaka M, Hasui K, Shirahama H, Kitajima S, Yonezawa S, et al. Effect of an immunotoxin to folate receptor beta on bleomycin-induced experimental pulmonary fibrosis. Clin Exp Immunol. 2010;161:348–356. doi: 10.1111/j.1365-2249.2010.04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang J, Jung Y, Tighe RM, Xie T, Liu N, Leonard M, et al. A macrophage subpopulation recruited by CC chemokine ligand-2 clears apoptotic cells in noninfectious lung injury. Am J Physiol Lung Cell Mol Physiol. 2012;302:L933–L940. doi: 10.1152/ajplung.00256.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machacek C, Supper V, Leksa V, Mitulovic G, Spittler A, Drbal K, et al. Folate receptor β regulates integrin CD11b/CD18 adhesion of a macrophage subset to collagen. J Immunol. 2016;197:2229–2238. doi: 10.4049/jimmunol.1501878. [DOI] [PubMed] [Google Scholar]
- 32.Chang LC, Tseng JC, Hua CC, Liu YC, Shieh WB, Wu HP. Gene polymorphisms of fibrinolytic enzymes in coal workers’ pneumoconiosis. Arch Environ Occup Health. 2006;61:61–66. doi: 10.3200/AEOH.61.2.61-66. [DOI] [PubMed] [Google Scholar]
- 33.Li G, Jin F, Du J, He Q, Yang B, Luo P. Macrophage-secreted TSLP and MMP9 promote bleomycin-induced pulmonary fibrosis. Toxicol Appl Pharmacol. 2019;366:10–16. doi: 10.1016/j.taap.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Todd JL, Vinisko R, Liu Y, Neely ML, Overton R, Flaherty KR, et al. IPF-PRO Registry investigators. Circulating matrix metalloproteinases and tissue metalloproteinase inhibitors in patients with idiopathic pulmonary fibrosis in the multicenter IPF-PRO Registry cohort. BMC Pulm Med. 2020;20:64. doi: 10.1186/s12890-020-1103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago-Ruiz L, Buendía-Roldán I, Pérez-Rubio G, Ambrocio-Ortiz E, Mejía M, Montaño M, et al. MMP2 polymorphism affects plasma matrix metalloproteinase (MMP)-2 levels, and correlates with the decline in lung function in hypersensitivity pneumonitis positive to autoantibodies patients. Biomolecules. 2019;9:574. doi: 10.3390/biom9100574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sziksz E, Pap D, Lippai R, Béres NJ, Fekete A, Szabó AJ, et al. Fibrosis related inflammatory mediators: role of the IL-10 cytokine family. Mediators Inflamm. 2015;2015:764641. doi: 10.1155/2015/764641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang YC, Li Z, Carter JD, Soukup JM, Schwartz DA, Yang IV. Fine ambient particles induce oxidative stress and metal binding genes in human alveolar macrophages. Am J Respir Cell Mol Biol. 2009;41:544–552. doi: 10.1165/rcmb.2008-0064OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham JA, Gardner DE, Waters MD, Coffin DL. Effect of trace metals on phagocytosis by alveolar macrophages. Infect Immun. 1975;11:1278–1283. doi: 10.1128/iai.11.6.1278-1283.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adamson IYR, Vincent R, Bjarnason SG. Cell injury and interstitial inflammation in rat lung after inhalation of ozone and urban particulates. Am J Respir Cell Mol Biol. 1999;20:1067–1072. doi: 10.1165/ajrcmb.20.5.3468. [DOI] [PubMed] [Google Scholar]
- 41.Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci USA. 2002;99:1503–1508. doi: 10.1073/pnas.022649799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q, Luo AA, Qiu H, Han B, Ko BH, Slutsky AS, et al. Monocyte interaction accelerates HCl-induced lung epithelial remodeling. BMC Pulm Med. 2014;14:135. doi: 10.1186/1471-2466-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsiao HM, Fernandez R, Tanaka S, Li W, Spahn JH, Chiu S, et al. Spleen-derived classical monocytes mediate lung ischemia-reperfusion injury through IL-1β. J Clin Invest. 2018;128:2833–2847. doi: 10.1172/JCI98436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maus UA, Waelsch K, Kuziel WA, Delbeck T, Mack M, Blackwell TS, et al. Monocytes are potent facilitators of alveolar neutrophil emigration during lung inflammation: role of the CCL2-CCR2 axis. J Immunol. 2003;170:3273–3278. doi: 10.4049/jimmunol.170.6.3273. [DOI] [PubMed] [Google Scholar]
- 45.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mould KJ, Barthel L, Mohning MP, Thomas SM, McCubbrey AL, Danhorn T, et al. Cell origin dictates programming of resident versus recruited macrophages during acute lung injury. Am J Respir Cell Mol Biol. 2017;57:294–306. doi: 10.1165/rcmb.2017-0061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zasłona Z, Przybranowski S, Wilke C, van Rooijen N, Teitz-Tennenbaum S, Osterholzer JJ, et al. Resident alveolar macrophages suppress, whereas recruited monocytes promote, allergic lung inflammation in murine models of asthma. J Immunol. 2014;193:4245–4253. doi: 10.4049/jimmunol.1400580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCubbrey AL, Barthel L, Mohning MP, Redente EF, Mould KJ, Thomas SM, et al. Deletion of c-FLIP from CD11bhi macrophages prevents development of bleomycin-induced lung fibrosis. Am J Respir Cell Mol Biol. 2018;58:66–78. doi: 10.1165/rcmb.2017-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramann R, Machado F, Wu H, Kusaba T, Hoeft K, Schneider RK, et al. Parabiosis and single-cell RNA sequencing reveal a limited contribution of monocytes to myofibroblasts in kidney fibrosis. JCI Insight. 2018;3:e99561. doi: 10.1172/jci.insight.99561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westphalen K, Gusarova GA, Islam MN, Subramanian M, Cohen TS, Prince AS, et al. Sessile alveolar macrophages communicate with alveolar epithelium to modulate immunity. Nature. 2014;506:503–506. doi: 10.1038/nature12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in the Gene Expression Omnibus, accession code GSE151928. The data set can be visualized at https://healthy-bal.cells.ucsc.edu.