Abstract
Rationale: The cellular and molecular landscape and translational value of commonly used models of pulmonary arterial hypertension (PAH) are poorly understood. Single-cell transcriptomics can enhance molecular understanding of preclinical models and facilitate their rational use and interpretation.
Objectives: To determine and prioritize dysregulated genes, pathways, and cell types in lungs of PAH rat models to assess relevance to human PAH and identify drug repositioning candidates.
Methods: Single-cell RNA sequencing was performed on the lungs of monocrotaline (MCT), Sugen-hypoxia (SuHx), and control rats to identify altered genes and cell types, followed by validation using flow-sorted cells, RNA in situ hybridization, and immunofluorescence. Relevance to human PAH was assessed by histology of lungs from patients and via integration with human PAH genetic loci and known disease genes. Candidate drugs were predicted using Connectivity Map.
Measurements and Main Results: Distinct changes in genes and pathways in numerous cell types were identified in SuHx and MCT lungs. Widespread upregulation of NF-κB signaling and downregulation of IFN signaling was observed across cell types. SuHx nonclassical monocytes and MCT conventional dendritic cells showed particularly strong NF-κB pathway activation. Genes altered in SuHx nonclassical monocytes were significantly enriched for PAH-associated genes and genetic variants, and candidate drugs predicted to reverse the changes were identified. An open-access online platform was developed to share single-cell data and drug candidates (http://mergeomics.research.idre.ucla.edu/PVDSingleCell/).
Conclusions: Our study revealed the distinct and shared dysregulation of genes and pathways in two commonly used PAH models for the first time at single-cell resolution and demonstrated their relevance to human PAH and utility for drug repositioning.
Keywords: pulmonary hypertension, single-cell RNA sequencing, drug repurposing, monocrotaline, Sugen-hypoxia
At a Glance Commentary
Scientific Knowledge on the Subject
The cellular and molecular landscape and translational value of commonly used models of pulmonary arterial hypertension (PAH) are poorly understood. Single-cell transcriptomics can enhance molecular understanding of preclinical models and facilitate their rational use and interpretation.
What This Study Adds to the Field
Our study revealed the distinct and shared dysregulation of genes and pathways in two commonly used PAH models for the first time at single-cell resolution and demonstrated their relevance to human PAH and utility for drug repositioning.
Despite advances in the management of pulmonary arterial hypertension (PAH), it remains an incurable and progressive disease characterized by severe pulmonary vascular remodeling, poor quality of life, and guarded long-term prognosis (1). Notably, current therapies focus on relieving symptoms and do not reverse vascular remodeling, the key pathological feature of PAH. The lack of therapies targeting underlying mechanisms in PAH may be in part because of our limited understanding of the pathogenic cell types and their specific molecular pathways. It has been increasingly recognized that in addition to pulmonary vascular cells, other cell types in the lung, including various immune-cell populations, may also play an important role in PAH and other pulmonary diseases (2–7). However, to our knowledge, a comprehensive evaluation to systematically compare these various cell types has not been undertaken in the most widely used preclinical models of PAH, namely the monocrotaline (MCT) and Sugen-hypoxia (SuHx) rat models. Given that numerous novel therapies tested in PAH animal models have not translated to the bedside (8), a more comprehensive understanding of the cellular and molecular landscape of these models is needed to unravel mechanistic insights and enhance the ability of preclinical work to predict drug efficacy in humans (9).
In this study, we performed lung single-cell RNA sequencing (scRNA-seq) in MCT and SuHx rats to investigate altered cell types, genes, and pathways and further integrated the findings with human genetics to assess human relevance. We then identified potential drug-repurposing candidates through computational screening of drug transcriptional profiles against the dysregulated transcriptional programs revealed by scRNA-seq. Lastly, to facilitate dissemination of the data and findings, we offer an open-access online platform for the wider research community (http://mergeomics.research.idre.ucla.edu/PVDSingleCell/). Some of the results of these studies have been previously reported in the form of an abstract (10).
Methods
The main methods are below with additional details provided in an online supplement.
Animals
Adult male Sprague-Dawley rats (250–350 g) were used for all animal experiments, which were approved by the University of California, Los Angeles, Animal Research Committee. For the SuHx model, rats were injected subcutaneously with Sugen 5416 (20 mg/kg) followed by being kept in hypoxia at 10% O2 for 21 days and then by being kept in normoxia for 14 days. For the MCT model, rats were injected subcutaneously with MCT (60 mg/kg) followed by being kept in normoxia for 28 days. Age-matched control rats were kept in normoxia for 28 days. Echocardiography and right heart catheterization were performed. Lungs were then harvested and enzymatically dissociated into single-cell suspensions, which was followed by scRNA-seq (11) (n = 6/group).
scRNA-seq Analysis
Expression data was normalized, filtered, and clustered using the Seurat R package (R Foundation for Statistical Computing) (12). Cell types were identified on the basis of known cell-type marker genes. Cell-type proportions were quantified and compared between PAH models and control animals, as previously described (13). Global transcriptomic shifts between groups were assessed using a Euclidian distance method (14). Differentially expressed genes (DEGs) were determined for each cell type between control and either SuHx or MCT rats using MAST (Model-based Analysis of Single-Cell Transcriptomics) (15). To annotate DEGs for biological pathways or PAH relevance, gene-set enrichment analysis was performed using hallmark pathways from the Molecular Signature Database (16) as well as using human PAH-associated gene sets obtained from DisGeNET (17) and the Comparative Toxicogenomics Database (18).
scRNA-seq Validation in Rat and Human Lung Tissues
The identities of select cell types were validated using bulk RNA-seq on cells purified by fluorescence-activated cell sorting (FACS) from the lungs of an additional set of rats (n = 4/group). Select scRNA-seq DEGs from SuHx and MCT rats were validated by RNA in situ hybridization (ISH) and immunofluorescence using rat lung sections (n = 5–6 rats/group). The same DEGs were further evaluated by RNA ISH on human lung sections from patients with PAH compared with control patients (n = 7–8 subjects/group).
Integration of Rat PAH Single-Cell DEGs with Human PAH Genome-Wide Association Study
To evaluate the relevance of the rat DEGs to human PAH, we assessed the human orthologs of rat DEGs for enrichment of genetic variants associated with PAH from a human genome-wide association study (GWAS) (19) using marker set enrichment analysis in the Mergeomics R package (20).
Identification of Drugs Predicted to Reverse Rat Disease Signatures Using Connectivity Map
Signatures of MCT and SuHx DEGs for each cell type were queried against the full Connectivity Map (CMap) (21) database of compound expression signatures induced in human cell lines to prioritize those with highly matching or opposing signatures. Pattern-matching algorithms scored each reference perturbagen profile for the direction and strength of enrichment with query scRNA-seq DEG signatures. Perturbagens with positive or negative connectivity scores have similar or opposite signatures to that of the query (i.e., genes that are increased in the scRNA-seq DEG query are decreased by the perturbagen or vice versa).
Data Availability
The scRNA-seq data set and lists of cell type–specific marker genes and disease DEGs are available online at http://mergeomics.research.idre.ucla.edu/PVDSingleCell/CellBrowser/. Connectivity scores of the entire panel of perturbagens from the CMap analysis are available at http://mergeomics.research.idre.ucla.edu/PVDSingleCell/CMap/.
Results
scRNA-seq Identifies Diverse Cell Populations in the Rat Lung
The PAH phenotype in MCT and SuHx rats was confirmed by echocardiography (see Figures E1A, E1B, and E2B–E2L in the online supplement), immunohistochemistry (Figure E1C), and right heart catheterization (Figure E2A). The scRNA-seq of 18 lungs (6/group) profiled 33,392 cells (Figure 1A) after quality control (Figures E3A–E3D, E4A, and E4B), with even representation of groups (Figures E4C and E4D). After clustering on the basis of transcriptomic similarity, we identified 28 distinct cell types expressing established markers for epithelial, stromal, lymphoid, and myeloid cell populations and rare populations, including conventional dendritic cells (cDCs) and regulatory T cells (Tregs) (Figures 1B, 1C, and E5). Batch correction did not further optimize clustering and cell-type identification (Figure E4E) (12). Each cluster included cells from each group (Figures 1D and 1E). Compared with control animals, we observed a significant increase in the normalized cell fractions of interstitial macrophages (iMΦs) in MCT rats and alveolar macrophages (aMΦs) in SuHx rats (Figure 1E).
Figure 1.
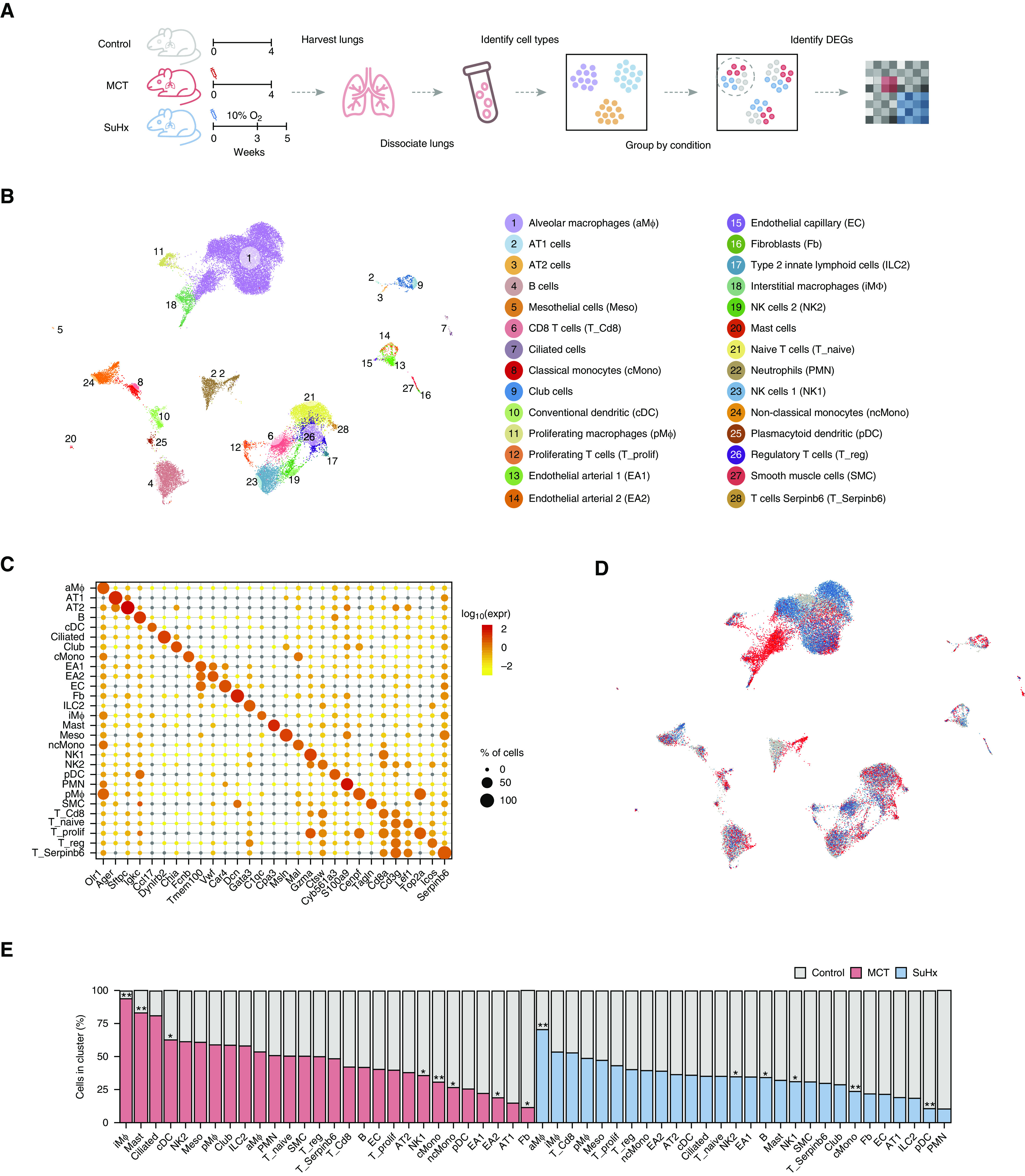
Single-cell RNA sequencing identifies diverse lung cell types in rat models of pulmonary arterial hypertension. (A) Schematic of study design for single-cell RNA sequencing analysis on the lungs of monocrotaline (MCT), Sugen-hypoxia (SuHx), and control rats (n = 6/group). (B) Uniform manifold approximation and projection plot showing lung cells from 18 rats with clusters labeled by cell type. (C) Dot plot highlighting log10 average expression of select marker genes used to identify cell clusters. The dot size corresponds to the percentage of cells expressing a gene in a given cluster. (D) Uniform manifold approximation and projection plot showing lung cells colored by disease condition: MCT in red, SuHx in blue, and control in gray (n = 6/group). (E) Bar table showing relative contributions of cells from disease models (MCT in red and SuHx in blue) versus the control model (gray) within each cell-type cluster. The cell-type cluster referred to on the y-axis is defined as the total number of cells of a cell type from the control model and either the MCT or SuHx model (but not both models). A significant increase in proportions of iMΦs in MCT and aMΦs in the SuHx model were noted relative to the control model. Wilcoxon rank-sum test: *P < 0.05 and **P < 0.01. DEG = differentially expressed gene; NK = natural killer cell.
FACS and Bulk RNA-seq Validate scRNA-seq Cell-Type Identities and Proportions
To validate the rare lung cell types identified from scRNA-seq, namely cDCs and Tregs, and their corresponding gene signatures, we performed bulk RNA-seq on FACS-purified cells and subsequent deconvolution using our scRNA-seq signatures as a reference (Figure 2A). We used canonical markers to isolate cDCs (CD64− CD11b/c+, RT1B+) and Tregs (CD4+, CD25+, CD278+) by FACS (Figures 2B and 2C). Deconvolution of FACS-purified transcriptomes showed strong enrichment for the correct cell types as identified by scRNA-seq, thus validating the accuracy of scRNA-seq cell signatures (Figure 2D). Furthermore, FACS-determined relative cell proportions between disease models and the control model showed a pattern similar to that from scRNA-seq (Figures 2E–2H). Specifically, both scRNA-seq and FACS showed significantly increased cDCs in MCT rats, but not in SuHx rats, and Tregs did not change in either model compared with the control model.
Figure 2.
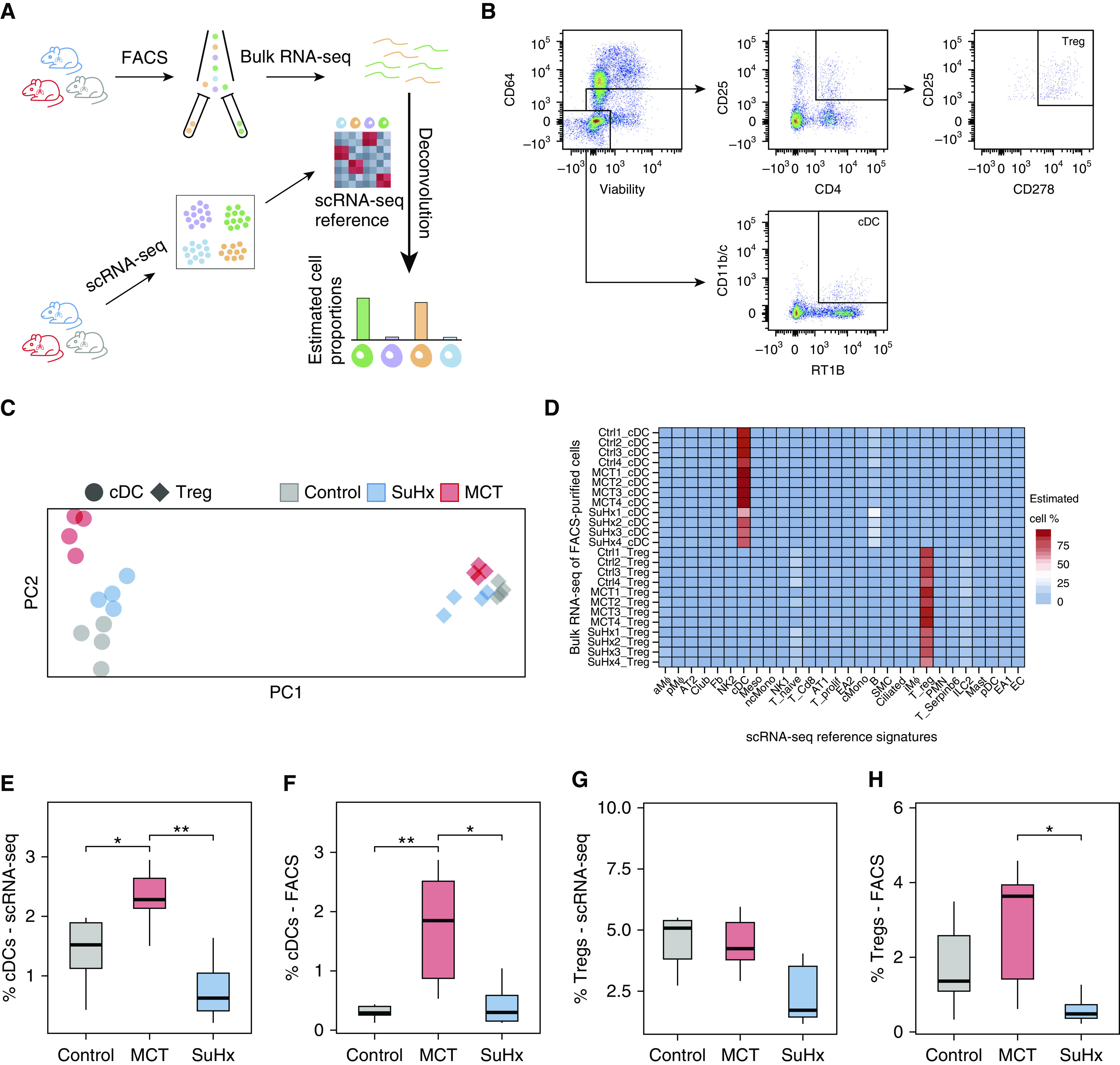
Fluorescence-activated cell sorting (FACS) and bulk RNA sequencing (RNA-seq) validate single-cell RNA-seq (scRNA-seq) cell-type identities and proportions. (A) Schematic showing the study design for deconvolution. The lungs from a separate set of Sugen-hypoxia (SuHx) (blue), monocrotaline (MCT) (red), and control (gray) rats (n = 4/group) underwent FACS using canonical cell-surface markers for two specific cell types, conventional dendritic cells (cDCs) and regulatory T cells (Tregs), after which bulk RNA-seq was performed on sorted cells to validate the cell-type identities in our scRNA-seq data using cell-type deconvolution of FACS-purified transcriptomes with CIBERSORTx (Stanford University). (B) Gating strategy for the isolation of CD64−, CD11b/c+, and RT1B+ cDCs and CD4+, CD25+, and CD278+ Tregs after gating for singlets and live cells. (C) Principal component (PC) analysis plot showing the clustering of bulk RNA-seq transcriptomes based on cell type (cDCs as circles and Tregs as diamonds) and disease condition. The first and second PCs (PC1 and PC2) explained 82% and 3% of the variance, respectively. (D) Heatmap showing deconvolution results using CIBERSORTx, in which relative proportions of cell types were estimated in each bulk RNA-seq sample of flow-sorted cells using a gene-expression signature for each cell type derived from scRNA-seq. High specificity for the correct cell types as identified by scRNA-seq was noted (83 ± 11% for cDCs; 77 ± 6% for Tregs). (E–H) FACS-determined relative cell-type proportions between disease models and the control model (F and H) showed a similar pattern to that of our scRNA-seq results (E and G). A significant increase in cDCs was noted in MCT compared with control rats. Furthermore, both methods consistently showed no significant changes in the number of cDCs in the SuHx model compared with the control model. The number of Tregs was also unchanged using either method in both disease models when compared with the control model. Wilcoxon rank-sum test: *P < 0.05 and **P < 0.01. FACS = fluorescence-activated cell sorter.
scRNA-seq Reveals DEGs with Cell-Type Specificity in PAH Models
A total of 4,724 and 2,324 DEGs were identified in MCT and SuHx rats (false discovery rate < 0.05), respectively, across 17 cell types (Figure 3A). There were 1,511 DEGs common in both models, of which 921 were regulated in the same direction. aMΦs, the largest cell cluster, had the most DEGs, likely due to high statistical power. We also assessed changes on a transcriptome scale within each cell type using a Euclidean distance-based approach that is less influenced by cluster size (14) (Figure 3B). Despite MCT rats having more DEGs, aMΦs and nonclassical monocytes (ncMonos) from the SuHx model demonstrated the strongest global transcriptomic shifts from the control model.
Figure 3.
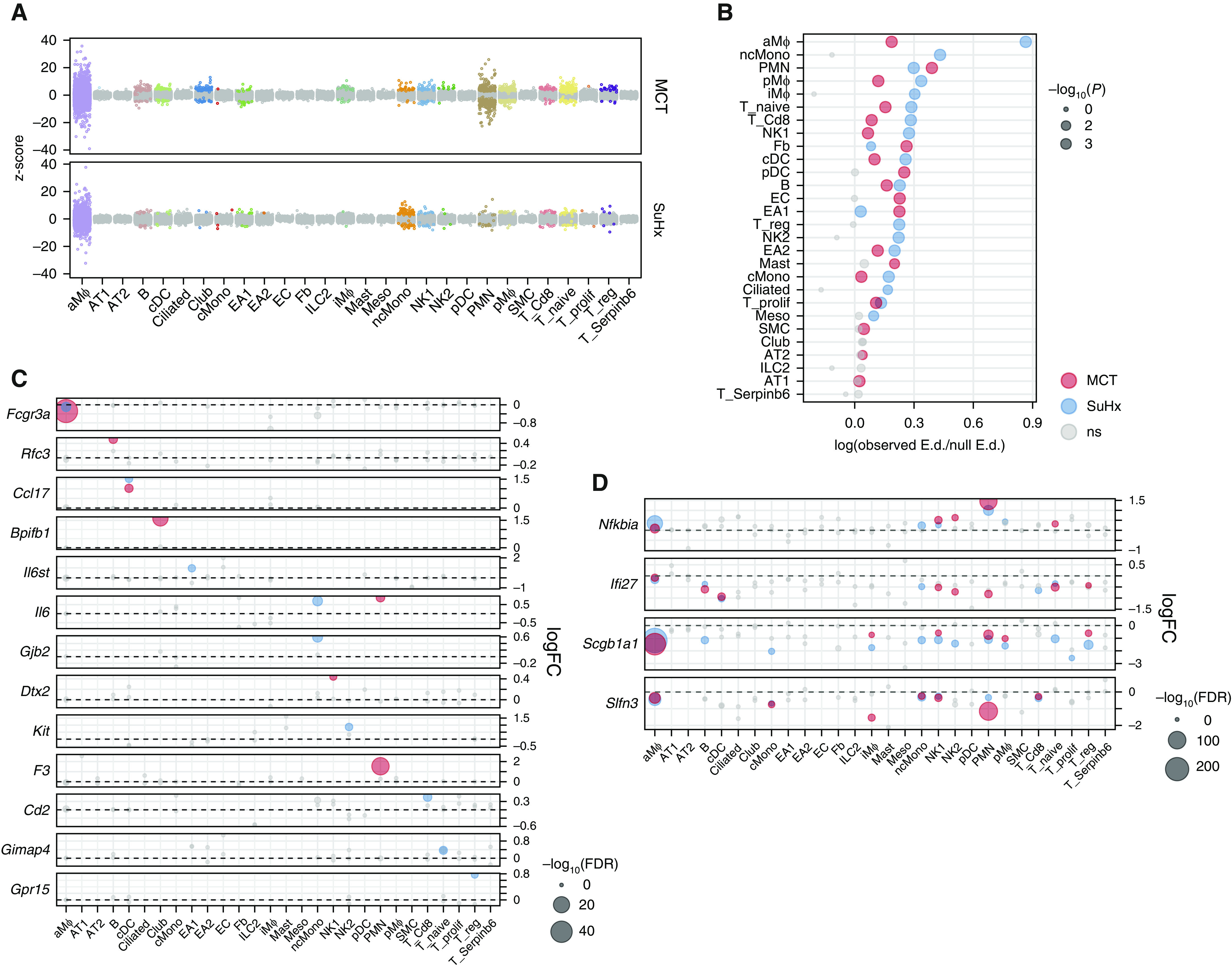
Single-cell RNA sequencing reveals differentially expressed genes (DEGs) in individual cell types of pulmonary arterial hypertension models. (A) Jitter plot showing changes in gene expression for each cell type due to monocrotaline (MCT) (top) or Sugen-hypoxia (SuHx) (bottom) conditions compared with the control condition. Each dot represents the differential expression MAST (Model-based Analysis of Single-Cell Transcriptomics) z-score of a gene. Dots indicating a false discovery rate (FDR) < 0.05 are in color. The gray dots indicate values that were not significant (ns). (B) Dot plot showing shifts in gene expression on a whole-transcriptome scale within each cell type for MCT (red) and SuHx (blue) models compared with the control model using a Euclidean distance (E.d.)-based statistical approach as previously described (14). The x-axis shows the log ratio of observed-to-null E.d. The alveolar macrophages and nonclassical monocytes from the SuHx model demonstrated the strongest global shifts in gene expression from the control model. (C) Dot plot comparing DEGs across cell types and disease models shows genes whose differential expression was specific to a disease model and a particular cell type. For example, Gpr15, which encodes an orphan G protein–linked receptor believed to be important in regulatory T cell (Treg) homing (22), was exclusively upregulated in Tregs from SuHx rats. (D) Dot plot showing DEGs consistent across immune-cell types. For instance, Ifi27, which encodes IFNα-inducible protein 27 and plays a role in apoptosis and vascular response to injury (23, 24), was downregulated across cell types in both models. (C and D) The horizontal dashed line for each gene represents zero logFC. (B–D) Gray dots indicate values that were ns, and the size of the dots corresponds to −log10(P) values (B) and −log10(FDR) values (C and D). logFC = log fold change.
A closer examination of DEGs revealed genes whose differential expression was model and cell-type specific (Figure 3C). In total, there were 2,088 and 574 DEGs specific to one cell type in MCT and SuHx rats, respectively. For example, Il6st, which encodes a signal transducer that mediates IL-6 signaling, was upregulated exclusively in a subpopulation of endothelial arterial type 1 (EA1) cells from SuHx; Il6 was specifically upregulated in SuHx ncMonos and MCT neutrophils, suggesting model-specific differences in IL-6 signaling. Gpr15 was exclusively upregulated in SuHx Tregs and encodes an orphan G protein–linked receptor implicated in Treg homing (22).
Furthermore, we identified 19 and 8 DEGs that were differentially expressed in the same direction in at least five cell types in either MCT or SuHx rats, among which 6 (Nfkbia, Scgb1a1, Ifi27, Slfn3, Mt-cox3, and AY172581.24) were altered across various immune cells in both models (Figure 3D). For example, Ifi27, which encodes IFNα–inducible protein 27 and plays a role in apoptosis and vascular response to injury (23, 24), was downregulated across cell types in both models and in human PAH lungs (Figure E6) (25).
scRNA-seq Reveals Pathways with Cell-Type Specificity in PAH Models
Pathway enrichment of DEGs revealed cell type–specific dysregulation of many pathways (Figure 4A). The most distinct difference between models was a strong downregulation of IFN signaling across multiple cell types in the MCT model that in the SuHx model was weaker or in the opposite direction (Figure 4B). The relevance of IFN downregulation to human PAH was demonstrated in EA1 cells as an example (Figures 4C and 4D). The most notable commonality between models was a widespread upregulation of TNFα/NF-κB signaling across cell types, most notably in SuHx ncMonos (Figure 4E and 4F).
Figure 4.
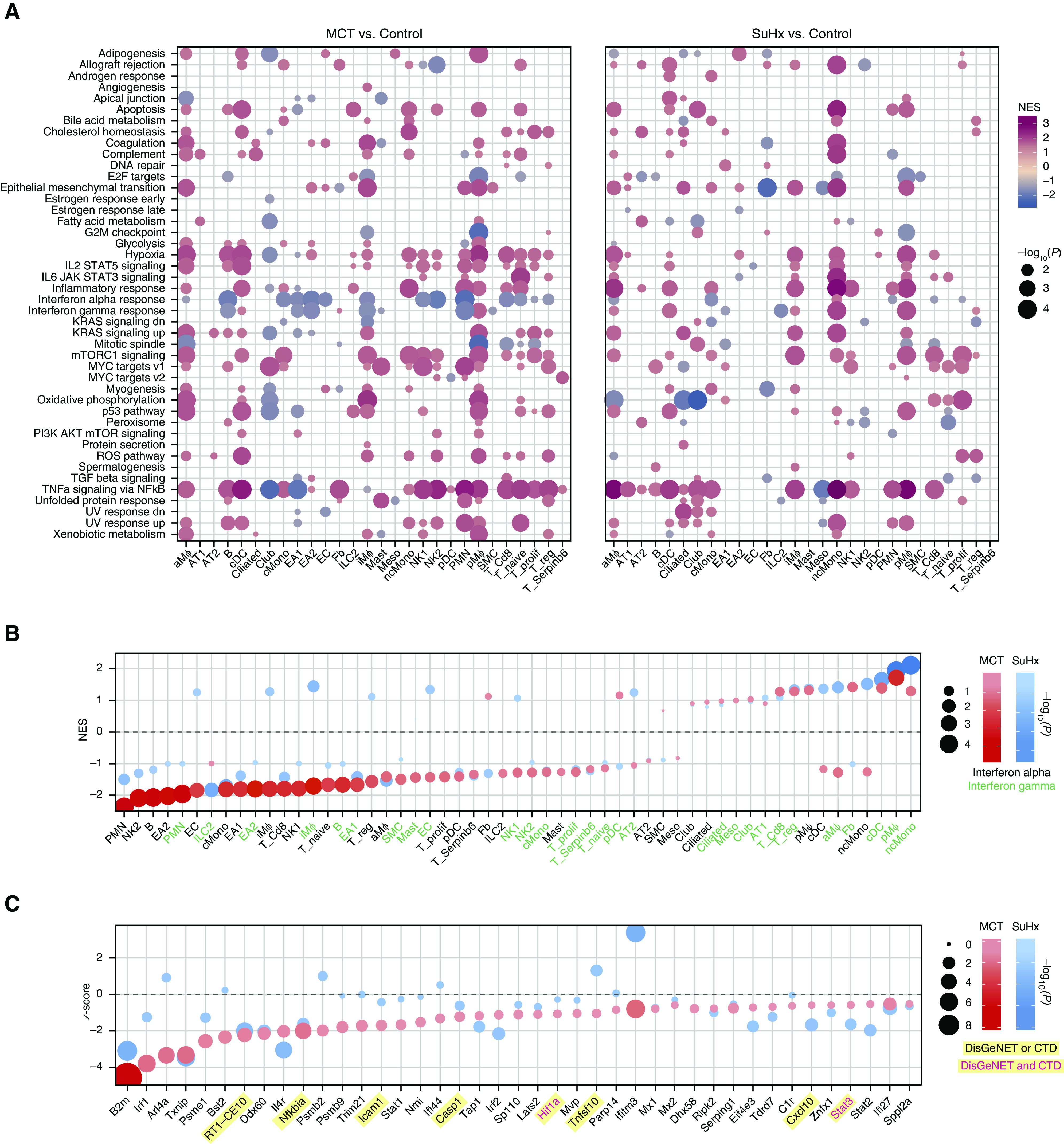
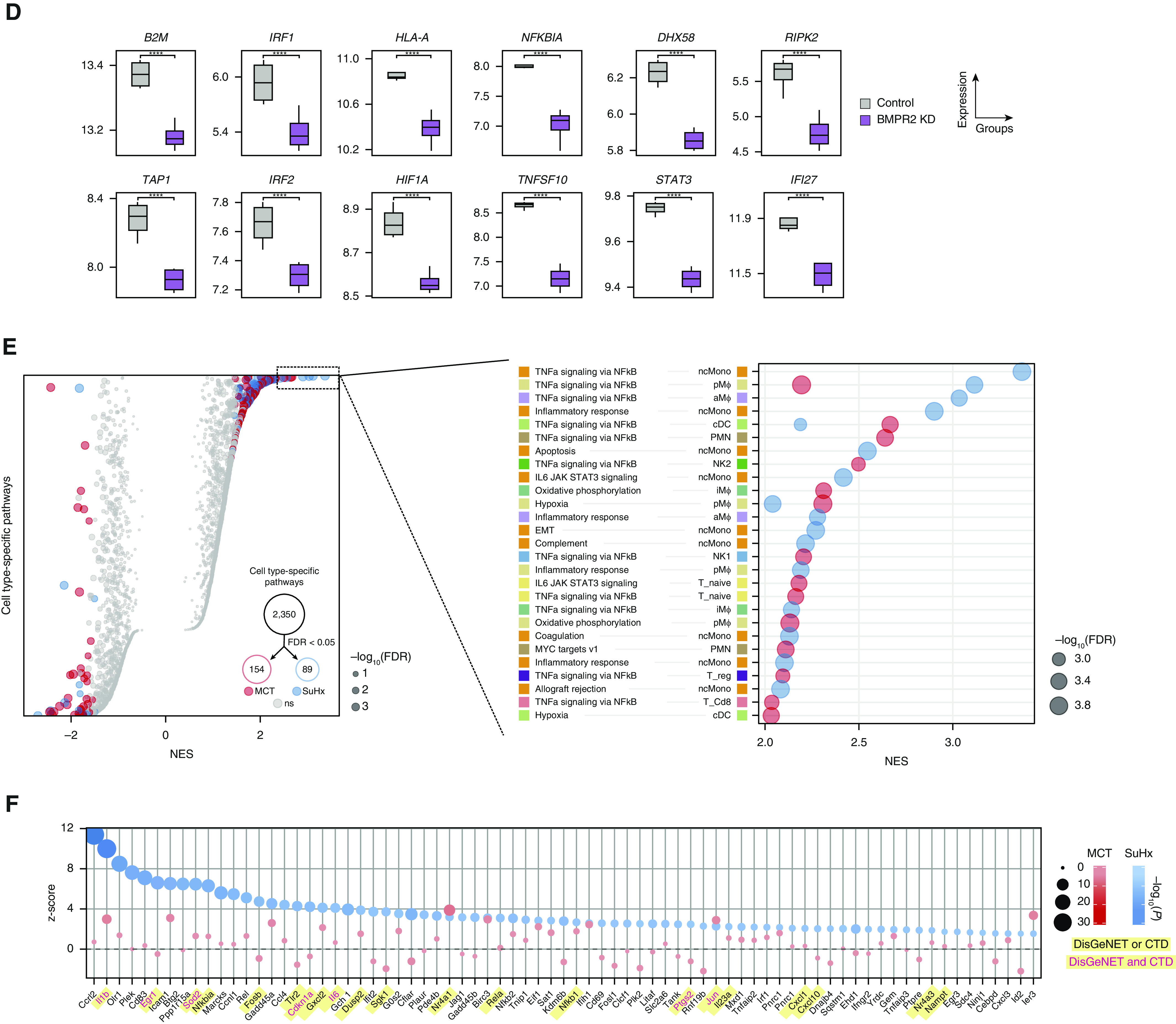
Single-cell RNA sequencing reveals pathways in individual cell types of pulmonary arterial hypertension models. (A) Heatmap showing cell type–specific pathway enrichment of gene signatures of monocrotaline (MCT) (left) and Sugen-hypoxia (SuHx) (right) models compared with the control model using gene-set enrichment analysis (GSEA) (P < 0.05) and hallmark pathways from the Molecular Signatures Database on the y-axis. The dot size corresponds to −log10(P), and color represents the normalized enrichment score (NES) from GSEA, indicating upregulation (red) or downregulation (blue). TNFα/NF-κB signaling was significantly upregulated across many cell types in both disease models. (B) Dot plot showing NES of IFNα (black text) and IFNγ (green text) response pathways across cell types in the MCT (red) and SuHx (blue) models, in which the size and color tint of dots represent strength of −log10(P) values. A strong downregulation of IFN pathways was seen across cell types in the MCT model. (C) Dot plot showing MAST (Model-based Analysis of Single-Cell Transcriptomics) z-scores of leading-edge genes accounting for the MCT EA1 downregulation of IFNγ response as determined by GSEA from the MCT (red) and SuHx (blue) models, in which the size and color tint of dots represent the strength of −log10(P) values. Gene labels highlighted in yellow represent human pulmonary arterial hypertension–associated genes from either (black text) or both (red text) of the Comparative Toxicogenomics Database and DisGeNET databases. (D) Boxplots showing RNA expression of human orthologs of select IFN leading-edge genes shown in C derived from a public microarray (Gene Expression Omnibus series 70456) in which primary human pulmonary arterial endothelial cells were transfected with control (gray) or BMPR2 (purple) siRNA (n = 4/group from 4 donors). P values were determined by using the limma R package: ****False discovery rate (FDR) < 0.05. (E) Dot plots showing all (left) and top 30 (right) cell type–specific pathways in descending order on the y-axis by NES in which positive scores indicate upregulation. Red (MCT) and blue (SuHx) dots met the FDR < 0.05 criterion, and gray dots were not significant (ns). The dot size indicates the strength of the FDR. The number of significant cell type–specific rat signatures by disease model is shown in the lower right (FDR < 0.05). In the left plot, dots on opposite sides of an NES of 0 for a given row represent opposite directionalities of cell type–specific enrichment of MCT and SuHx models. Many more cell type–specific pathways were significant in the MCT model compared with the SuHx model, but TNFα/NF-κB signaling in SuHx nonclassical monocytes (ncMonos) was the most prominently upregulated pathway overall (right). Cell-type colors correspond to those as labeled in Figure 1B. (F) Dot plot showing MAST z-scores of leading-edge genes accounting for the SuHx ncMono upregulation of TNFα/NF-κB signaling with figure legend as described in C. CTD = Comparative Toxicogenomics Database; KD = knockdown; NK = natural killer.
Validation of Select DEGs by RNA ISH and Immunofluorescence
Given the importance of ncMonos, suggested in our analyses above, we validated a DEG from ncMonos by RNA ISH on both rat and human lung sections (Table E1). We defined ncMonos as positive for both CD16 and Mal. Mal is the top marker for ncMonos in our scRNA-seq (Figure 1C) and is involved in the MyD88 pathway, important in human lung ncMonos (26). We validated the upregulation of Ccrl2, a top SuHx ncMono DEG, encoding a chemokine receptor–like protein whose function is unknown but is upregulated during monocyte-to-MΦ differentiation (Figure 5A) (27). We also validated the upregulation of Fabp4, a top MCT aMΦ DEG, encoding a fatty acid–binding protein involved in lipid metabolism and inflammation (Figure 5B) (28). We further demonstrate similar upregulation of both proteins by immunofluorescence in rat lungs (Figures 5C and 5D).
Figure 5.
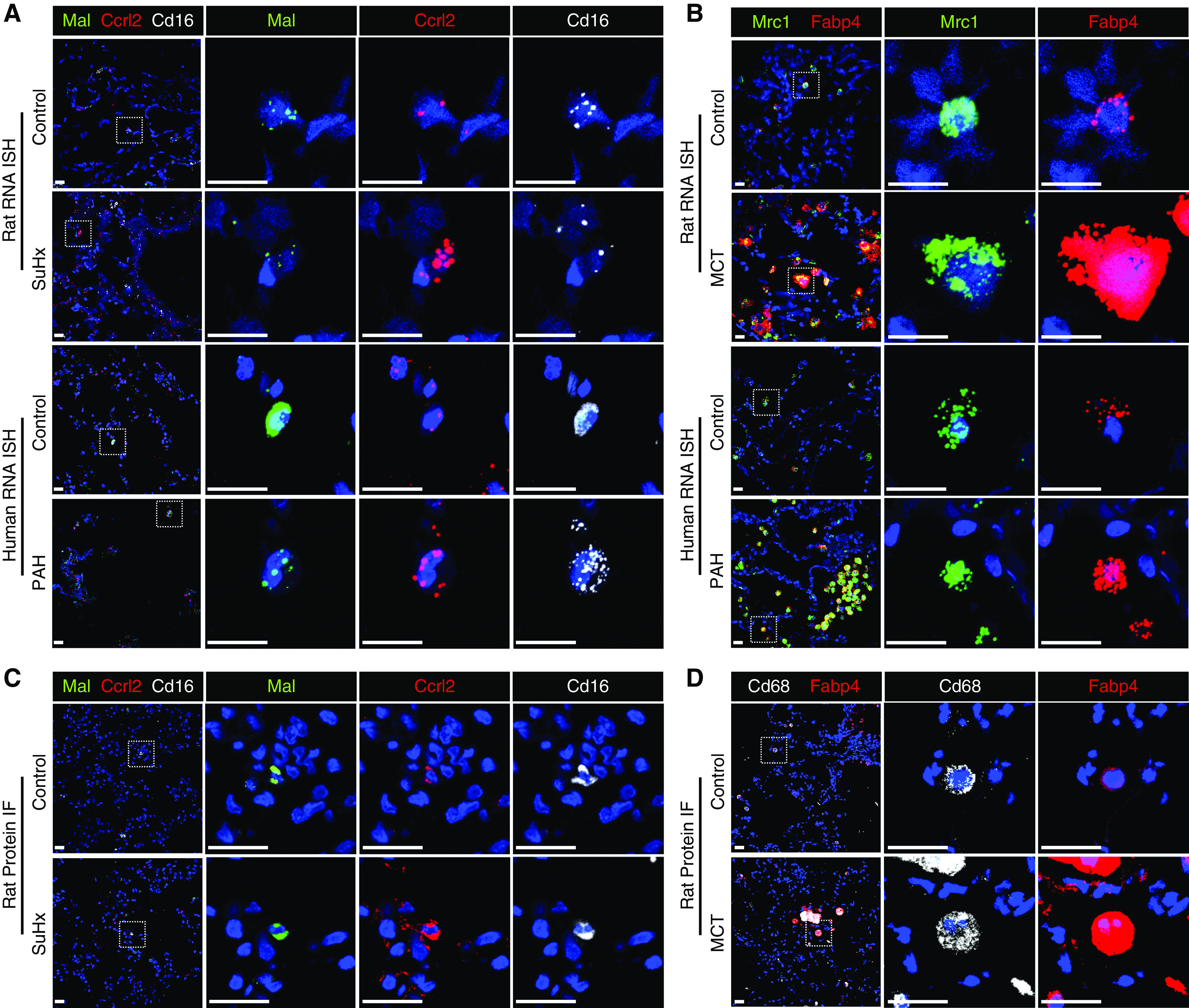
RNA ISH and immunofluorescence (IF) validate select differentially expressed genes. (A and B) The upregulation of Ccrl2 (red) in Sugen-hypoxia (SuHx) nonclassical monocytes (ncMonos) from single-cell RNA sequencing (scRNA-seq) was observed by RNAscope (Advanced Cell Diagnostics) in SuHx rats and patients with PAH (A) and by IF in SuHx rats (B). ncMonos were defined as cells positive for both Cd16 (white) and Mal (green). We chose Mal for double-labeling because it was the top marker gene specific for ncMonos in our data. (C and D) The upregulation of Fabp4 (red) in MCT alveolar macrophages (aMΦs) from scRNA-seq was demonstrated, in which aMΦs were defined as cells positive for Mrc1 (green) for rat and human RNAscope (C) or Cd68 (white) for rat IF (D). Both Mrc1 and Cd68 are canonical markers and were cell type–specific markers for aMΦs. The cell nuclei are labeled with DAPI (blue). Scale bars, 20 μm. ISH = in situ hybridization; MCT = monocrotaline; PAH = pulmonary arterial hypertension.
Integrative Analysis of Rat scRNA-seq DEGs with Human PAH Genes Supports the Relevance to Humans
We curated genes implicated in PAH from DisGeNET and the Comparative Toxicogenomics Database (Figure 6A) and demonstrated that the top pathways enriched in these public gene sets (Figure 6B) were also highly enriched in our rat scRNA-seq (Figures 4A and 4E). When directly testing rat DEGs for enrichment of these PAH genes, we noted a marked upregulation in SuHx ncMonos in particular (Figures 6C–6E).
Figure 6.
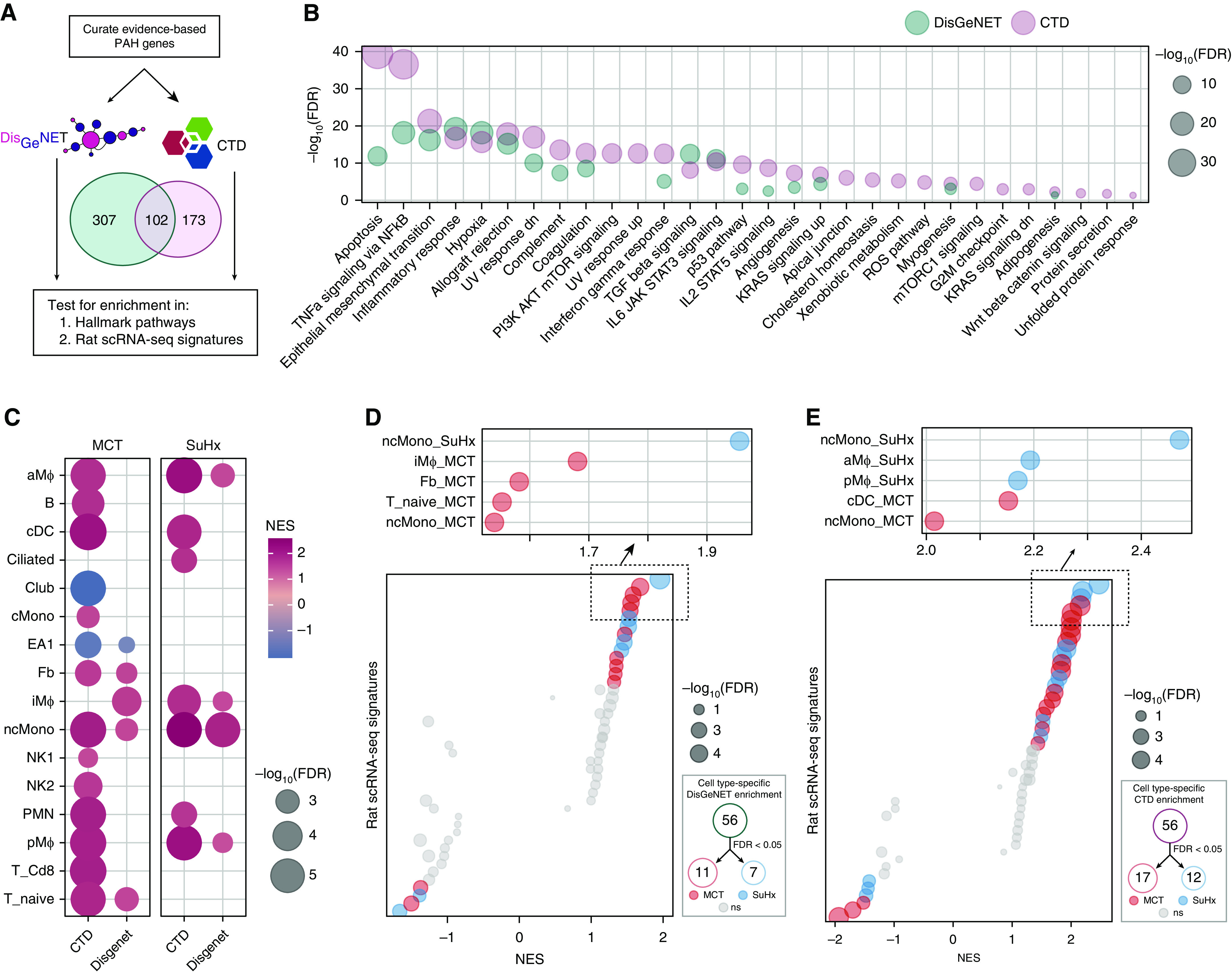
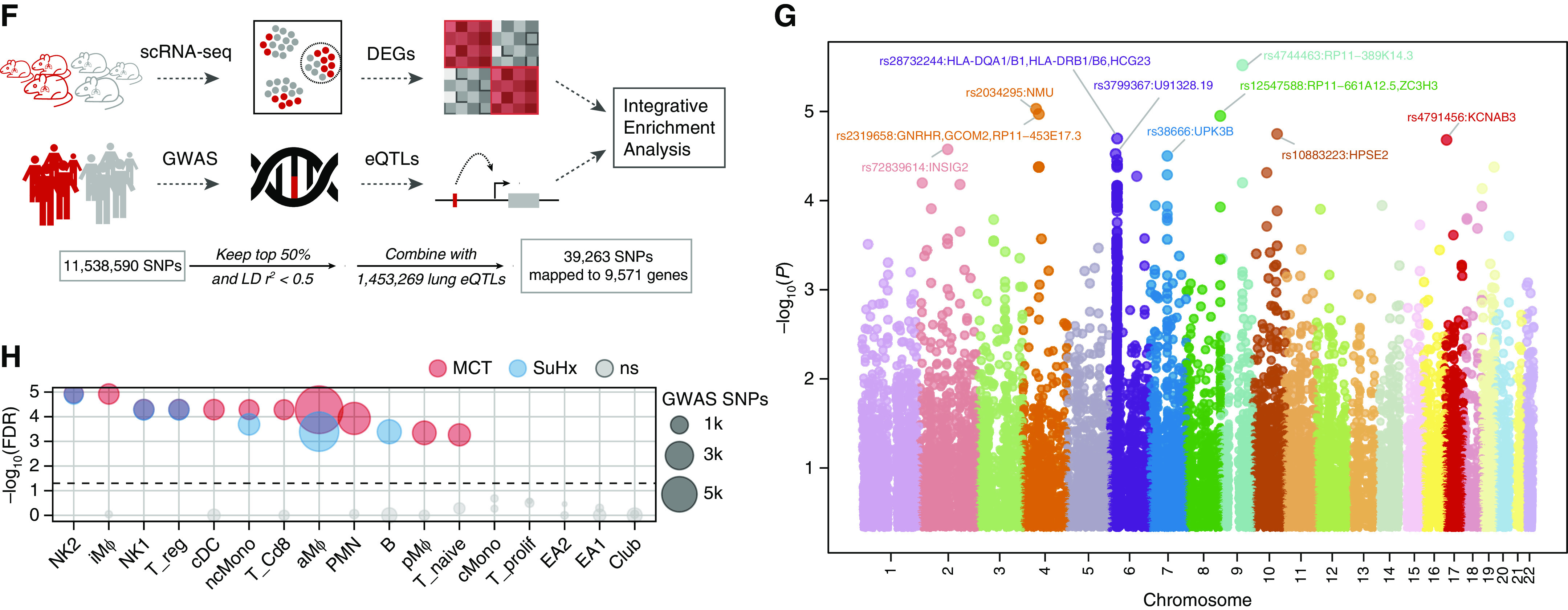
Integrative analysis of rat single-cell RNA sequencing (scRNA-seq) differentially expressed genes (DEGs) with human pulmonary arterial hypertension (PAH) genetics points to the relevance of the DEGs to human PAH. (A) Schematic of analytical approach whereby genes implicated in human PAH were curated from DisGeNET (409 genes) and Comparative Toxicogenomics Database (CTD) (275 genes), of which 102 genes were shared between the databases. These genes were then tested for enrichment in (B) Molecular Signatures Database hallmark pathways and (C–E) rat scRNA-seq signatures. (B) Dot plot showing pathways significantly enriched for PAH genes by hypergeometric test (false discovery rate [FDR] < 0.05), whether from the DisGeNET database (green) or the CTD database (purple). The dot size represents the −log10(FDR). These gene sets were highly enriched for known or implicated PAH pathways, such as apoptosis, NF-κB signaling, and endothelial-to-mesenchymal transition and were similar overall to those altered in Sugen-hypoxia (SuHx) and monocrotaline (MCT) rat lung scRNA-seq (Figure 4). (C) Heatmap showing most highly significant (FDR < 0.01) enrichment of PAH genes in MCT (left) and SuHx (right) cell type–specific signatures using gene-set enrichment analysis, in which red indicates upregulation and blue indicates downregulation. The dot size represents −log10(FDR). Significant upregulation of PAH genes was noted in myeloid cell types in both models, and in nonclassical monocytes (ncMonos) in particular. (D and E) Dot plots showing all (left) and top 5 (upper right) cell type–specific rat signature enrichment for PAH genes from the DisGeNET (D) and CTD (E) databases. The red (MCT) and blue (SuHx) dots indicate meeting the FDR < 0.05 criterion, and gray dots indicate values that were not significant (ns). The dot size represents −log10(FDR). A number of significant cell type–specific rat signatures by disease model are shown in the lower right (FDR < 0.05). In the left-sided plots, dots on opposite sides of an NES of 0 for a given row represent opposite directionalities of cell type–specific enrichment of MCT and SuHx models. The SuHx ncMonos DEGs were most highly enriched for PAH genes comparing both models. For the MCT model, DEGs from iMΦs and cDCs demonstrated the strongest enrichment for PAH genes. (F) Schematic of analytical approach for human PAH genome-wide association study (GWAS) integration. Human orthologs of rat scRNA-seq DEGs were assessed for enrichment of genetic variants associated with PAH in human GWAS to further assess human relevance of the rat signatures. GWAS SNPs were filtered by keeping the top 50% by P value strength and LD r2 < 0.5, after which SNPs were mapped to genes by integrating with lung-specific expression quantitative loci (eQTLs) curated from public databases. DEGs within each cell type (P < 0.01 to include DEGs from rare cell types with low statistical power) were then tested for enrichment of these GWAS-integrated expression SNPs (eSNPs). The GWAS P values of each eSNP set (by cell type and disease model) were then compared against that of eSNPs generated from random gene sets to assess the significance of enrichment for stronger GWAS association P values using a modified chi-square statistic. (G) Manhattan plot showing −log10(P) values of 39,263 eSNPs used for rat DEG enrichment analysis after GWAS SNP filtering and eQTL integration as described in F. SNPs are ordered along the x-axis according to their chromosomal location; the colors represent different chromosomes. Select eSNPs are labeled with their reference SNP identifiers and the corresponding genes they regulate. (H) Dot plot showing rat cell type–specific DEGs enriched for PAH-associated genetic variants from human GWAS. Red (MCT) and blue (SuHx) dots indicate an FDR < 0.05, and gray dots indicate values that were ns. The horizontal dashed line corresponds to an FDR = 0.05. The dot size is proportional to the number of enriched GWAS SNPs in the thousands. Significant enrichment for human PAH GWAS signals among DEGs in both rat disease models was noted from a number of immune-cell types of both myeloid and lymphoid origin, supporting that the rat disease signatures are relevant to PAH pathogenesis in humans. LD = linkage disequilibrium; NES = normalized enrichment score.
We further integrated rat scRNA-seq with a PAH GWAS using Mergeomics to assess human relevance (Figures 6F and 6G). We found significant enrichment for GWAS signals among DEGs in both models from a number of immune cells of both myeloid and lymphoid origins, supporting that DEGs from our rat models are relevant to PAH pathogenesis in humans (Figure 6H and Tables E2–E4).
scRNA-seq Uncovers Perturbations in Vascular Cell Types Relevant to Human PAH
Given the importance of pulmonary vascular cell types to PAH pathogenesis, we provide a closer examination of the endothelial arterial subpopulations of EA1 and EA2 cells, as well as of smooth muscle cells (SMCs) and fibroblasts (Figures 7A and 7B). We show model- and cell type–specific alterations of many established and unknown genes in PAH (Figure 7C) and a distinct pathway dysregulation (Figure 7D). We also highlight select DEGs that are similarly altered in public human cell type–specific data sets (Figure 7E). For example, Bmpr2, the most well-studied PAH gene, was downregulated in MCT EA1 cells but not in EA2 cells, whereas downregulation was not observed in SuHx vascular cells, which is consistent with a recent study (29). Cst3, encoding cystatin C, was upregulated in MCT and SuHx EA1 cells. Furthermore, Cst3 serum levels correlated with right ventricular indices and predicted mortality in patients with PAH, despite unknown mechanism in PAH (30, 31).
Figure 7.
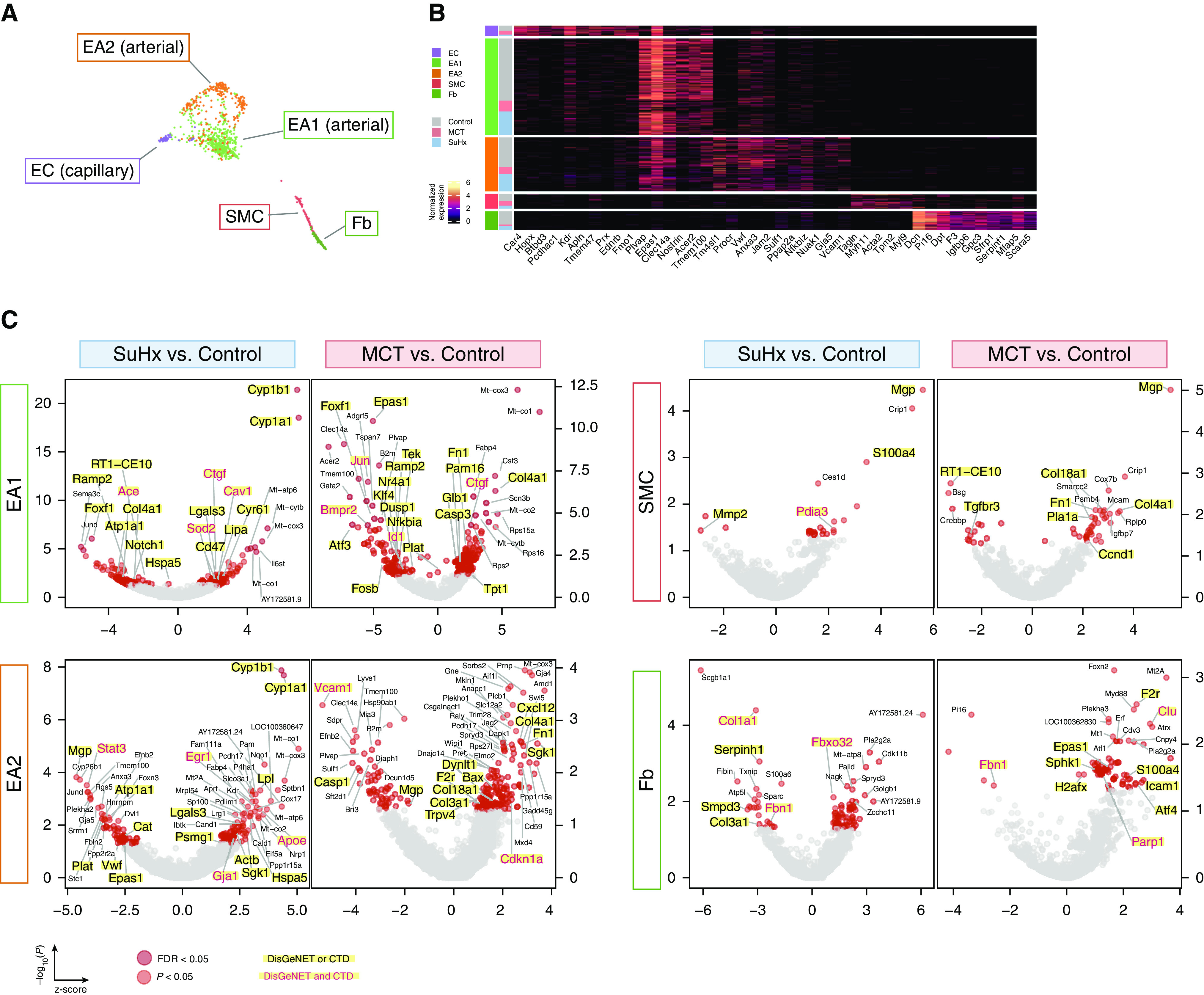
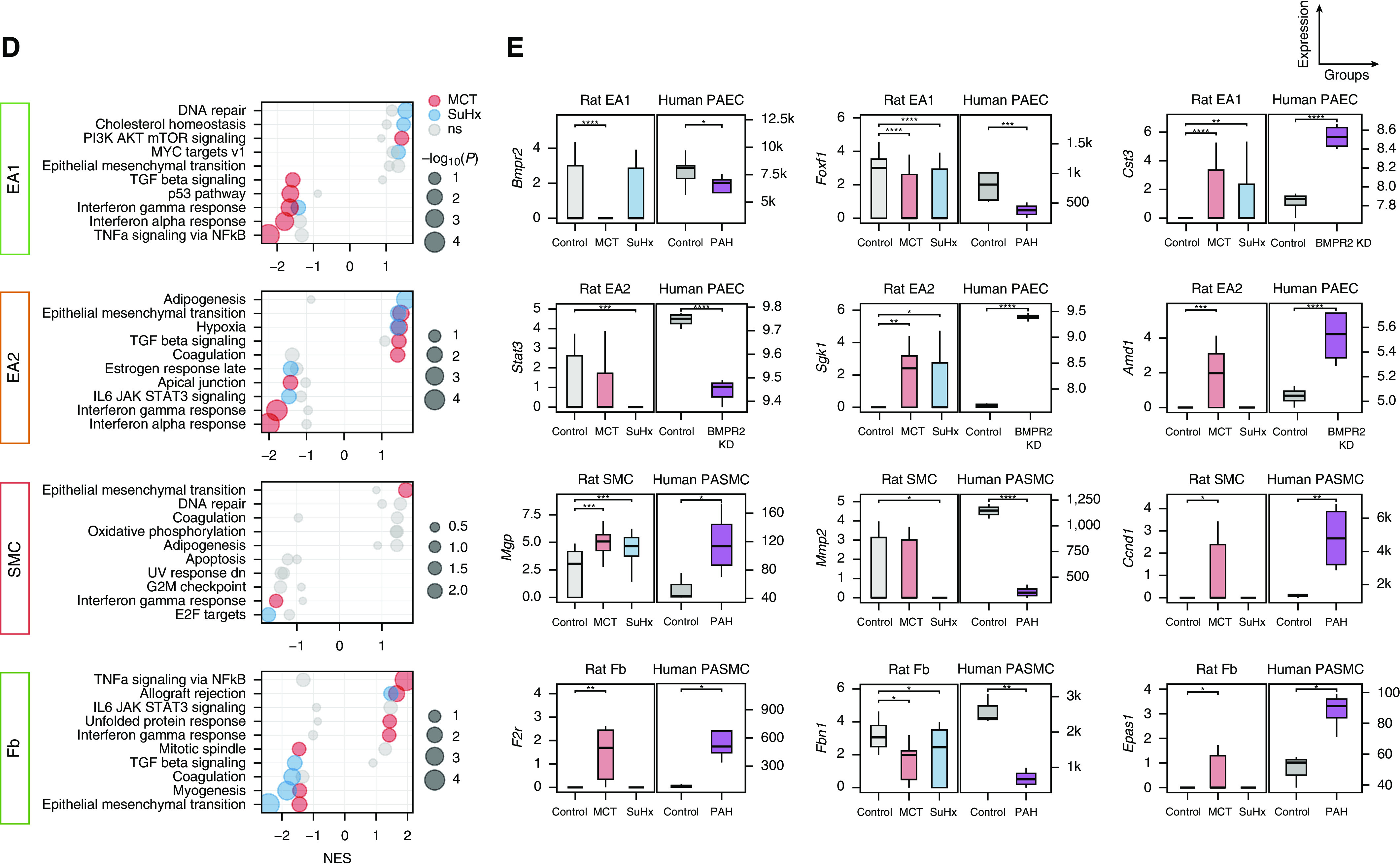
Single-cell RNA sequencing uncovers perturbations in lung vascular cell types relevant to human pulmonary arterial hypertension (PAH). (A) Uniform manifold approximation and projection plot showing vascular cells from 18 rat lungs with clusters labeled by cell type. (B) Heatmap showing normalized expression of top marker genes used to identify the vascular cell types, in which each row is an individual cell. Shown to the left are the condition and cell type to which each cell belongs. (C) Volcano plots showing differentially expressed genes (DEGs) within vascular cell types for the Sugen-hypoxia (SuHx) or monocrotaline (MCT) models versus the control model, in which the x-axis represents MAST (Model-based Analysis of Single-Cell Transcriptomics) z-scores and the y-axis indicates −log10(P). Significant upregulated (z > 0) or downregulated (z < 0) genes are shown as red (P < 0.05) or dark red (false discovery rate [FDR] < 0.05) dots. DEGs (P < 0.05) labeled and highlighted in yellow represent human PAH-associated genes from either (black text) or both (red text) of the CTD and DisGeNET databases. Otherwise, DEGs are labeled with their gene names if the FDR < 0.05 (endothelial arterial type 1 cell [EA1]) or P < 0.01 (EA2, SMC, Fb). (D) Dot plots showing the top five upregulated and top five downregulated pathways within vascular cell types as determined by gene-set enrichment analysis. Colored dots in red (MCT) or blue (SuHx) indicate significant values (P < 0.05), whereas gray dots represent values that were not significant (ns). The dot size corresponds to the −log10(P) value. (E) Box plots showing expression of select DEGs in rat lung vascular cell types with similar changes shown side by side in human orthologs from public cell type–specific expression data sets: BMPR2: Gene Expression Omnibus series (GSE) 126262, primary PAECs from two patients with PAH with BMPR2 mutations versus nine unused donor controls; FOXF1: GSE126262, primary PAECs from four male patients with PAH versus five male unused donor controls; CST3, STAT3, SGK1 and AMD1: GSE70456, four BMPR2 siRNA–transfected versus four control siRNA–transfected primary human PAECs from four donors; MGP, MMP2, CCND1, F2R, FBN1, and EPAS1: GSE2559, primary human PASMCs from two patients with PAH versus two normal subjects (n = 4 vs. 3, respectively, including BMP2-treated vs. untreated). P values from RNA sequencing (GSE126262) were determined by using DESeq2, whereas those from microarray (GSE70456 and GSE2559) were determined by using R limma: *P < 0.05, **P < 0.01, ***P < 0.001, and ****FDR < 0.05. CTD = Comparative Toxicogenomics Database; Fb = fibroblast; KD = knockdown; NES = normalized enrichment score; PAEC = pulmonary artery endothelial cell; PASMC = pulmonary arterial SMC; SMC = smooth muscle cell.
Integration of Rat DEGs with CMap Identifies Potential Candidate Drugs for Repositioning
To investigate the utility of scRNA-seq DEGs to identify the therapeutic potential of existing drugs for PAH, we screened all cell-specific transcriptional signatures against the molecular profiles of thousands of pharmacologic perturbagens tested in human cell lines from CMap, including 5 approved PAH drugs, 26 drugs tested in PAH clinical trials, and 15 drugs that have shown efficacy in PAH animal models (Figures 8A and 8B). Comparisons of drug connectivity profiles revealed interesting patterns. For example, bosentan, an endothelin receptor antagonist currently used in patients with PAH, and tacrolimus, a calcineurin inhibitor for which a phase 2 clinical trial for PAH was completed, had very similar connectivity profiles across cell types and disease models, suggesting converging pathways that are likely due to similar activation of BMP signaling (32, 33). In contrast, distinct connectivity profiles were observed among drugs from the same class, such as with treprostinil and iloprost (Figure 8B), both of which are synthetic analogs of prostacyclin but have opposing effects on a preclinical model of lung cancer (34), likely due to different off-target effects mediated by differential engagement of other eicosanoid receptors. Our analysis also predicted that sildenafil reverses MCT’s SMC signature most enriched for genes related to epithelial-to-mesenchymal transition (EMT), consistent with prior studies investigating sildenafil’s effect on pulmonary arterial SMCs (PASMCs) from humans (35–37) and MCT rats (38). Upregulated EMT genes in our MCT SMC signature were also found to be downregulated in a toxicogenomic microarray of sildenafil-exposed rat hepatocytes (Figures E7A–E7C) (39).
Figure 8.
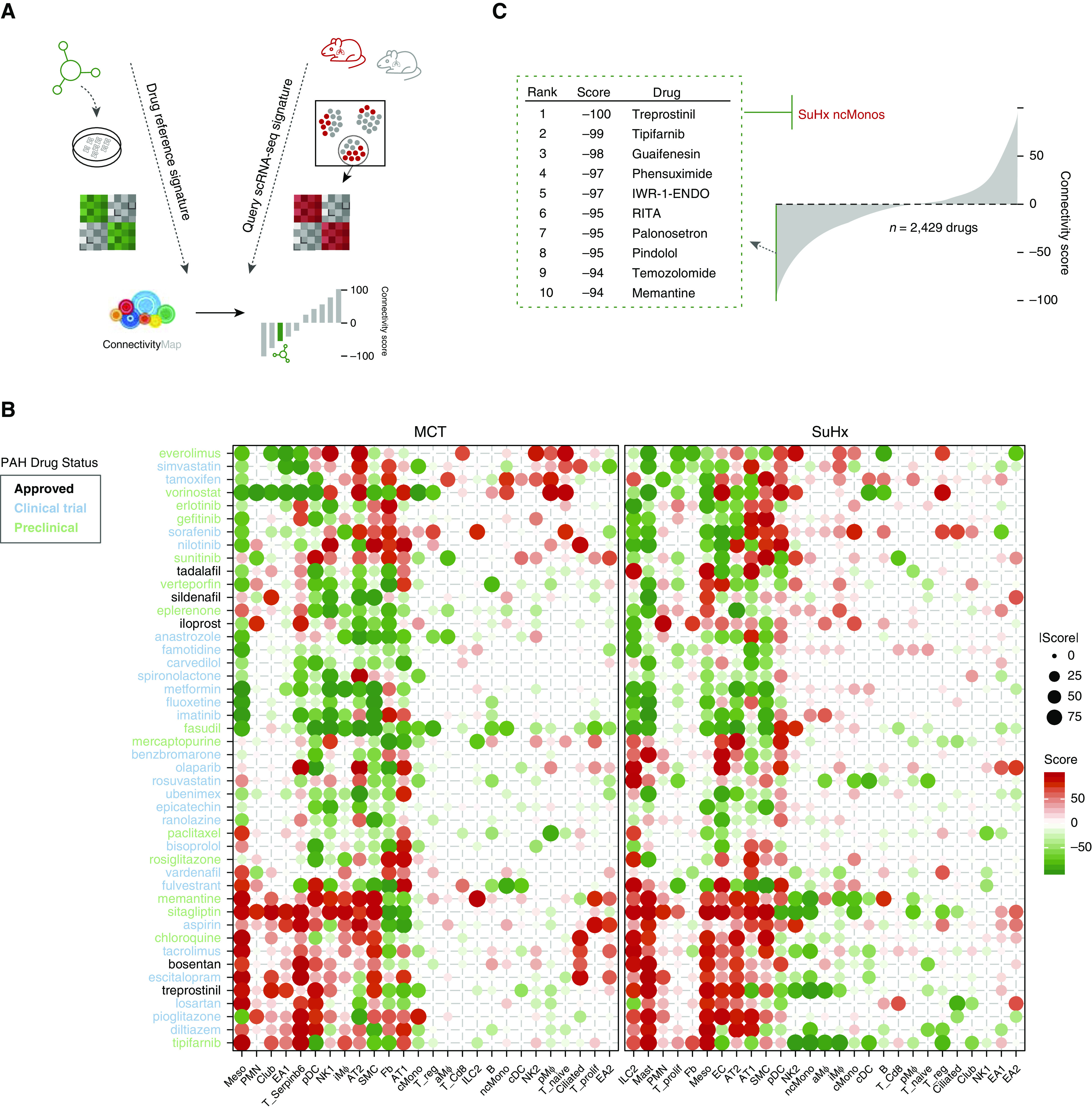
Integration of rat differentially expressed genes (DEGs) with Connectivity Map identifies potential candidate drugs for repositioning. (A) Schematic of analytical approach whereby signatures of rat DEGs (P < 0.01 to include DEGs from rare cell types with low statistical power) for each cell type for both Sugen-hypoxia (SuHx) and monocrotaline (MCT) models against the control model were queried against the full Connectivity Map database of compound and genetic perturbational expression signatures induced in human cell lines. The pattern-matching algorithms scored each reference perturbagen profile for the direction and strength of enrichment with the query single-cell RNA sequencing (scRNA-seq) DEG signature. Perturbagens with strongly positive connectivity scores have highly similar signatures to that of the query, whereas those perturbagens with strongly negative scores have signatures that strongly oppose that of the query (i.e., genes that are upregulated in the scRNA-seq DEG query are downregulated by treatment with the perturbagen or vice versa). (B) Heatmap showing connectivity scores of rat scRNA-seq DEGs to drugs approved for use in patients with pulmonary arterial hypertension (PAH) (black), drugs currently or previously in PAH clinical trials (blue), and preclinical drugs with demonstrated efficacy in PAH animal models (green). The size of dots corresponds to absolute values of the connectivity score. The PAH-related drugs showed distinct matching patterns to cell type–specific PAH rat signatures. For example, bosentan and tacrolimus had very similar connectivity profiles across cell types and disease models, although they come from different classes of drugs. (C) The top 10 drugs with the most negative connectivity scores, which are thus predicted to most strongly reverse the transcriptional signature of SuHx nonclassical monocytes (ncMonos), are shown (out of 2,429 compounds screened). The drugs predicted against SuHx ncMonos were of particular interest, given the strong upregulation of both NF-κB signaling and human PAH genes. The drug with the most negative connectivity score was treprostinil, one of the most effective PAH-targeted therapies currently in use in patients with advanced PAH.
We next examined drugs predicted to reverse the transcriptional signature of SuHx ncMonos, given that these cells were most highly enriched for genes and pathways relevant to PAH. Out of 2,429 compounds screened, the drug with the strongest reversal of SuHx DEGs in ncMonos was treprostinil, one of the most effective PAH-targeted therapies currently in use (Figure 8E). Two other top drugs screened against SuHx ncMonos were recently shown to attenuate or prevent PAH in animal models: tipifarnib, a farnesyltransferase inhibitor currently undergoing clinical trials for various cancers (40), and memantine, an NMDA receptor antagonist used to treat Alzheimer’s dementia (41). Our analysis also revealed novel candidates for repurposing, such as palonosetron, a serotonin-receptor antagonist used to treat chemotherapy-induced nausea, and guaifenesin, an expectorant used to loosen airway mucus via unknown mechanisms (Figure 8C).
Dissemination of scRNA-seq and Drug Repositioning Results
We implemented a web server to enable interactive browsing of the entire scRNA-seq data set, as well as searching and downloading of cell type–specific marker genes, disease signatures, and drug predictions (Figure 9).
Figure 9.
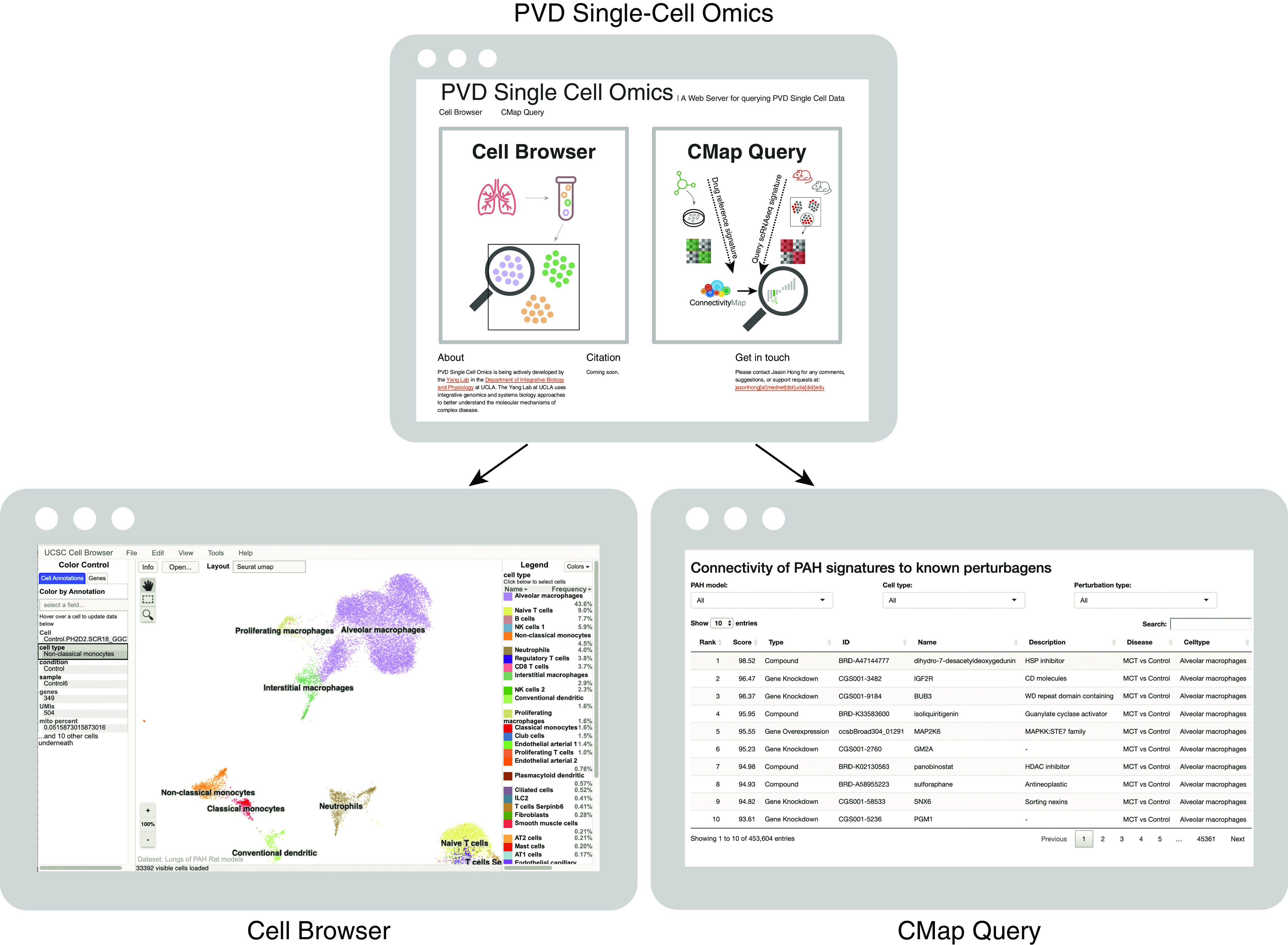
The PVD Single-Cell Omics website offers an open-access online platform. The entire rat single-cell RNA sequencing gene-expression data set and lists of cell type–specific marker genes and disease differentially expressed genes are available online in the form of an interactive cell browser at http://mergeomics.research.idre.ucla.edu/PVDSingleCell/CellBrowser/. Connectivity scores of the entire panel of perturbagens from the CMap analysis are available to query at http://mergeomics.research.idre.ucla.edu/PVDSingleCell/CMap/. CMap = Connectivity Map; PAH = pulmonary arterial hypertension; PVD = pulmonary vascular disease.
Discussion
In this study, we uncover comprehensive cellular landscapes of altered genes and pathways at single-cell resolution in the lungs of two widely used animal models of PAH. Comparison of landscapes between SuHx and MCT models reveals critical similarities and differences in key cell types. We integrated rat scRNA-seq with human data to determine human relevance and with pharmacotranscriptomic data to identify potential drug-repurposing candidates. Finally, we offer an open-access platform for the wider research community to access the data and findings.
The mechanisms leading to the PAH phenotype in MCT and SuHx rat models are believed to differ in that MCT is associated with endothelial toxicity and marked lung inflammation, whereas SuHx is characterized by angioproliferative pulmonary vascular disease (42). SuHx is overall believed to recapitulate PAH more closely than MCT, such as in the formation of neointimal plexiform lesions, the pathological hallmark of PAH. However, human relevance of these models is likely more nuanced at the cellular and molecular level. The lack of in-depth understanding of lung cell types and their respective pathway alterations in PAH limits our ability to rationally leverage these models in translational science. In this study, we found cell type–specific upregulation of known PAH pathways across multiple cell types in both MCT and SuHx models, most notably involving NF-κB signaling. Although prior studies have demonstrated NF-κB’s importance in PAH (43, 44), our study systemically resolves and implicates understudied cell types that most strongly mediate this critical pathway: SuHx ncMonos and MCT cDCs. Similar to cDCs in human PAH (45), cDCs in rat PAH were increased in MCT (but not SuHx) lungs compared with control lungs, as determined by scRNA-seq and FACS. In addition, iMΦs were also increased in MCT lungs but not in SuHx lungs, which is concordant with prior studies in MCT rats and human PAH (46). MCT iMΦ and cDC transcriptional signatures were significantly enriched for PAH GWAS single-nucleotide polymorphisms and known PAH genes, whereas those of SuHx signatures were not (Figures 6D, 6E, and 6H). Therefore, for the investigation of cell types such as cDCs or iMΦs, the MCT model may recapitulate human PAH better than the SuHx model does.
A rather unexpected finding from pathway analysis was the widespread downregulation of IFNα and IFNγ signaling across both immune and vascular cells, which was most notable in the MCT model. In endothelial cells, many of the genes accounting for the decrease in IFN signaling in our study were also downregulated in BMPR2-silenced human pulmonary arterial endothelial cells. A harmful downregulation of this pathway is supported by prior data showing that exogenous IFNα decreased proliferation in human pulmonary arterial endothelial cells and PASMCs in vitro and reversed PAH in animal models (47). However, other studies suggest that excess IFN signaling may contribute to PAH (48, 49). Further research is needed to dissect the nuanced role of this pathway in PAH.
In addition to resolving PAH-relevant cell types and pathways, scRNA-seq revealed many altered genes with cell-type and model specificity. Ccrl2, which has not been previously implicated in PAH, was the top upregulated gene contributing to the strong NF-κB pathway enrichment and transcriptional signature in SuHx ncMonos; we confirmed Ccrl2’s upregulation in human PAH ncMonos by lung histology. Fabp4, a fatty acid–binding protein that we found to be highly upregulated in MCT and human PAH aMΦs, is a transcriptional target of HIF-1α and has been implicated in regulating inflammatory cytokines and NF-κB signaling in aMΦs (28, 50). Further investigation of the role and mechanisms of altered genes, such as Ccrl2 in ncMonos and Fabp4 in aMΦs, is warranted.
Many of the DEGs identified in our study could be simply correlated with disease rather than pathogenic drivers of PAH. Given that genetic risk signals carry causal information, we integrated rat scRNA-seq with a human GWAS to infer causality of cell-type transcriptional programs in human PAH. Transcriptional signatures of ncMonos from SuHx and MCT models, along with numerous other myeloid and lymphoid cells, including iMΦs and cDCs from MCT rats and Tregs from MCT and SuHx rats, were significantly enriched for genetic variants from a human PAH GWAS, suggesting that these cell types and their corresponding pathways may play a causal role in PAH. The relevance of ncMonos is supported by increased ncMonos in patients with PAH (46), and its deficiency in HIF-1α leads to impaired maturation of iMΦs and disease attenuation in hypoxic mice (51). Blocking CX3CL1–CX3CR1 signaling, which is important for ncMono survival (52), decreased lung iMΦs and attenuated vascular remodeling in rodent models (46). Our unbiased comparative study further prioritizes ncMonos as a particularly important cell type in PAH pathogenesis.
To gain further translational insights, we leveraged a wealth of pharmaco transcriptomic data to query PAH signatures identified from all lung cell types in both models. Supporting this approach, our analysis predicted sildenafil’s action in reversing MCT’s SMC signature most enriched for EMT genes, consistent with prior studies investigating sildenafil’s effect on human PASMCs (35–37). Thus, sildenafil may have a therapeutic effect in PAH beyond pulmonary vasodilation. Indeed, targeted in vivo delivery of sildenafil to PASMCs was recently shown to inhibit vascular remodeling and improve survival in MCT rats (38). Further validating our approach, treprostinil, one of the most effective PAH-targeted therapies currently in use for patients with PAH, was the top drug predicted to reverse the SuHx ncMono disease signature that was most enriched for NF-κB signaling. Although treprostinil’s primary clinical effect in PAH is believed to be pulmonary vasodilation via the prostacyclin pathway, an additional therapeutic mechanism may be attenuation of NF-κB signaling, based on our results and previous human and murine studies (53–56).
Our analysis also predicted other top drugs that may reverse the SuHx ncMono signature for repositioning in PAH. Among these, palonosetron, a 5-HT3 receptor antagonist used for chemotherapy-induced nausea, may have therapeutic potential in PAH through inhibiting the upregulation in serotonin signaling, known to occur in PAH pathobiology, and/or through inhibiting NF-κB, the top pathway implicated in SuHx ncMonos. Indeed, another 5-HT3 receptor antagonist, tropisetron, modulates NF-κB in a rat model of type 2 diabetes (57). Another top prediction was guaifenesin, an expectorant which acts to loosen airway mucus through unknown mechanisms. Guaifenesin may act as a NMDA receptor antagonist targeting the dysregulation in glutamate–NMDA receptor signaling in PAH (41, 58) or may act through modulating pathways like TNFα/NF-κB signaling that are upregulated in SuHx ncMonos, as revealed in our analysis and suggested in a prior human study (59). Although guaifenesin is a common over-the-counter medicine used by patients with PAH on an as-needed basis for mucus relief, it may not be available at the doses and frequency needed to treat PAH. Such drug predictions warrant further investigation as potential repurposing candidates for PAH.
The overall strengths of our study include offering the first single-cell resolution landscape of two widely used rat models of PAH; comprehensive comparative and integrative omics analysis to prioritize cell types, genes, and pathways relevant to human PAH; high-throughput computational screening to identify potential drug repositioning candidates for future testing; and an open-access resource for the wider PAH research community. Select key findings were also experimentally validated using alternative methods, such as FACS with bulk RNA-seq, ISH, and immunofluorescence. There are also limitations to our study that underscore the need to further improve on current single-cell approaches, particularly the need to enzymatically dissociate the heterogenous cell populations that may be particularly fragile or tightly embedded in the extracellular matrix (3). Our lung dissociation yielded fewer vascular cells overall than immune cells, which limited statistical power within these cell clusters and potentially favored larger immune clusters in our comparative analyses. Alternative methods such as single-nucleus RNA-seq may mitigate such limitations. Despite this limitation, separate analysis of the captured vascular cells still provided valuable insights. Furthermore, our study unravels numerous genes, pathways, and drugs that warrant experimental and functional testing in future studies.
In conclusion, our scRNA-seq study of SuHx and MCT rat lungs dissects the distinct and shared dysregulation of gene-expression programs and pathway activation in individual cell populations, elucidates their relevance to human PAH pathobiology and drug repositioning, and will help guide the rational use of these preclinical models in future translational studies in PAH.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Dr. Ramin Salehirad and Dr. Min Zhou for their input on the flow cytometry experiments, Dr. Zeyneb Kurt for her guidance on Mergeomics, Dr. Min Li for her assistance with the lung dissociation protocol, and Dr. Gregory Fishbein for his help in identifying control human lung sections.
Footnotes
Supported by NIH/NHLBI grant T32 HL072752 (J.H.), an American Lung Association Catalyst Award (J.H.), NIH/NHLBI grant R01HL147883 (X.Y.), and NIH/NHLBI grants R01HL129051 and R01HL147586 (M.E.).
Author Contributions: J.H., D.A., M.E., and X.Y. contributed to the conception and design of the research and interpretation of the data. J.H. conducted or helped with all experiments, analyzed the data, made the figures, and wrote the manuscript. S.U. performed the animal experiments. G.R. and C.M.C. contributed to staining. G.R. contributed to the bulk RNA sequencing experiment. I.S.A. and G.D. performed the single-cell experiments. C.M.C. and M.B. contributed to lung dissociation experiments. C.H. analyzed drug data. C.H. and E.S. analyzed images. T.L. and E.S. prepared lung sections for staining. J.F.P. analyzed echo data. M.H., F.S., D.M., B.G., and D.-A.T. provided the genome-wide association study data. X.Y., M.E., L.M., R.C., and R.S. provided intellectual input.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202006-2169OC on October 6, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Benza RL, Miller DP, Barst RJ, Badesch DB, Frost AE, McGoon MD. An evaluation of long-term survival from time of diagnosis in pulmonary arterial hypertension from the REVEAL registry. Chest. 2012;142:448–456. doi: 10.1378/chest.11-1460. [DOI] [PubMed] [Google Scholar]
- 2.Thenappan T, Ormiston ML, Ryan JJ, Archer SL. Pulmonary arterial hypertension: pathogenesis and clinical management. BMJ. 2018;360:j5492. doi: 10.1136/bmj.j5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 5.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 6.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saygin D, Tabib T, Bittar HET, Valenzi E, Sembrat J, Chan SY, et al. Transcriptional profiling of lung cell populations in idiopathic pulmonary arterial hypertension. Pulm Circ. 2020;10:1–15. doi: 10.1177/2045894020908782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet S, Provencher S, Guignabert C, Perros F, Boucherat O, Schermuly RT, et al. Translating research into improved patient care in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2017;195:583–595. doi: 10.1164/rccm.201607-1515PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denayer T, Stöhr T, Van Roy M. Animal models in translational medicine: validation and prediction. New Horiz Transl Med. 2014;2:5–11. [Google Scholar]
- 10.Hong J, Arneson D, Ahn IS, Umar S, Diamente G, Bhetraratana M, et al. Lung single-cell transcriptomics of two animal models of pulmonary arterial hypertension reveals vulnerable cell types and pathways. Am J Respir Crit Care Med. 2020;201:A5867. [Google Scholar]
- 11.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 2019;573:75–82. doi: 10.1038/s41586-019-1404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arneson D, Zhang G, Ying Z, Zhuang Y, Byun HR, Ahn IS, et al. Single cell molecular alterations reveal target cells and pathways of concussive brain injury. Nat Commun. 2018;9:3894. doi: 10.1038/s41467-018-06222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, et al. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47:D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Germain M, Eyries M, Montani D, Poirier O, Girerd B, Dorfmüller P, et al. Genome-wide association analysis identifies a susceptibility locus for pulmonary arterial hypertension. Nat Genet. 2013;45:518–521. doi: 10.1038/ng.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 22.Kim SV, Xiang WV, Kwak C, Yang Y, Lin XW, Ota M, et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science. 2013;340:1456–1459. doi: 10.1126/science.1237013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosebeck S, Leaman DW. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008;13:562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- 24.Papac-Milicevic N, Breuss JM, Zaujec J, Ryban L, Plyushch T, Wagner GA, et al. The interferon stimulated gene 12 inactivates vasculoprotective functions of NR4A nuclear receptors. Circ Res. 2012;110:e50–e63. doi: 10.1161/CIRCRESAHA.111.258814. [DOI] [PubMed] [Google Scholar]
- 25.Mura M, Cecchini MJ, Joseph M, Granton JT. Osteopontin lung gene expression is a marker of disease severity in pulmonary arterial hypertension. Respirology. 2019;24:1104–1110. doi: 10.1111/resp.13557. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Z, Chiu S, Akbarpour M, Sun H, Reyfman PA, Anekalla KR, et al. Donor pulmonary intravascular nonclassical monocytes recruit recipient neutrophils and mediate primary lung allograft dysfunction. Sci Transl Med. 2017;9:eaal4508. doi: 10.1126/scitranslmed.aal4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan-Komito D, Valaris S, Kapellos TS, Recio C, Taylor L, Greaves DR, et al. Absence of the non-signalling chemerin receptor CCRL2 exacerbates acute inflammatory responses in vivo. Front Immunol. 2017;8:1621. doi: 10.3389/fimmu.2017.01621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makowski L, Brittingham KC, Reynolds JM, Suttles J, Hotamisligil GS. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor gamma and IkappaB kinase activities. J Biol Chem. 2005;280:12888–12895. doi: 10.1074/jbc.M413788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Happé C, Kurakula K, Sun X-Q, da Silva Goncalves Bos D, Rol N, Guignabert C, et al. The BMP receptor 2 in pulmonary arterial hypertension: when and where the animal model matches the patient. Cells. 2020;9:1422. doi: 10.3390/cells9061422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenster BE, Lasalvia L, Schroeder JD, Smyser J, Silveira LJ, Buckner JK, et al. Cystatin C: a potential biomarker for pulmonary arterial hypertension. Respirology. 2014;19:583–589. doi: 10.1111/resp.12259. [DOI] [PubMed] [Google Scholar]
- 31.Blok IM, van Riel ACMJ, Schuuring MJ, de Bruin-Bon RHACM, van Dijk APJ, Hoendermis ES, et al. The role of cystatin C as a biomarker for prognosis in pulmonary arterial hypertension due to congenital heart disease. Int J Cardiol. 2016;209:242–247. doi: 10.1016/j.ijcard.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Maruyama H, Dewachter C, Sakai S, Belhaj A, Rondelet B, Remmelink M, et al. Bosentan reverses the hypoxia-induced downregulation of the bone morphogenetic protein signaling in pulmonary artery smooth muscle cells. Life Sci. 2016;159:111–115. doi: 10.1016/j.lfs.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Spiekerkoetter E, Tian X, Cai J, Hopper RK, Sudheendra D, Li CG, et al. FK506 activates BMPR2, rescues endothelial dysfunction, and reverses pulmonary hypertension. J Clin Invest. 2013;123:3600–3613. doi: 10.1172/JCI65592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geraci MW. Targeting the prostacyclin/peroxisome proliferator-activated receptor gamma axis in lung cancer chemoprevention. Trans Am Clin Climatol Assoc. 2018;129:48–55. [PMC free article] [PubMed] [Google Scholar]
- 35.Tantini B, Manes A, Fiumana E, Pignatti C, Guarnieri C, Zannoli R, et al. Antiproliferative effect of sildenafil on human pulmonary artery smooth muscle cells. Basic Res Cardiol. 2005;100:131–138. doi: 10.1007/s00395-004-0504-5. [DOI] [PubMed] [Google Scholar]
- 36.Wharton J, Strange JW, Møller GMO, Growcott EJ, Ren X, Franklyn AP, et al. Antiproliferative effects of phosphodiesterase type 5 inhibition in human pulmonary artery cells. Am J Respir Crit Care Med. 2005;172:105–113. doi: 10.1164/rccm.200411-1587OC. [DOI] [PubMed] [Google Scholar]
- 37.Milara J, Escrivá J, Ortiz JL, Juan G, Artigues E, Morcillo E, et al. Vascular effects of sildenafil in patients with pulmonary fibrosis and pulmonary hypertension: an ex vivo/in vitro study. Eur Respir J. 2016;47:1737–1749. doi: 10.1183/13993003.01259-2015. [DOI] [PubMed] [Google Scholar]
- 38.Li B, He W, Ye L, Zhu Y, Tian Y, Chen L, et al. Targeted delivery of sildenafil for inhibiting pulmonary vascular remodeling. Hypertension. 2019;73:703–711. doi: 10.1161/HYPERTENSIONAHA.118.11932. [DOI] [PubMed] [Google Scholar]
- 39.Svoboda DL, Saddler T, Auerbach SS. An overview of national toxicology program’s toxicogenomic applications: drugMatrix and ToxFX. In: Hong H, editor. Advances in computational toxicology: methodologies and applications in regulatory science. Cham, Switzerland: Springer International Publishing; 2019. pp. 141–157. [Google Scholar]
- 40.Duluc L, Ahmetaj-Shala B, Mitchell J, Abdul-Salam VB, Mahomed AS, Aldabbous L, et al. Tipifarnib prevents development of hypoxia-induced pulmonary hypertension. Cardiovasc Res. 2017;113:276–287. doi: 10.1093/cvr/cvw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dumas SJ, Bru-Mercier G, Courboulin A, Quatredeniers M, Rücker-Martin C, Antigny F, et al. NMDA-type glutamate receptor activation promotes vascular remodeling and pulmonary arterial hypertension. Circulation. 2018;137:2371–2389. doi: 10.1161/CIRCULATIONAHA.117.029930. [DOI] [PubMed] [Google Scholar]
- 42.Stenmark KR, Meyrick B, Galie N, Mooi WJ, McMurtry IF. Animal models of pulmonary arterial hypertension: the hope for etiological discovery and pharmacological cure. Am J Physiol Lung Cell Mol Physiol. 2009;297:L1013–L1032. doi: 10.1152/ajplung.00217.2009. [DOI] [PubMed] [Google Scholar]
- 43.Farkas D, Alhussaini AA, Kraskauskas D, Kraskauskiene V, Cool CD, Nicolls MR, et al. Nuclear factor κB inhibition reduces lung vascular lumen obliteration in severe pulmonary hypertension in rats. Am J Respir Cell Mol Biol. 2014;51:413–425. doi: 10.1165/rcmb.2013-0355OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosokawa S, Haraguchi G, Sasaki A, Arai H, Muto S, Itai A, et al. Pathophysiological roles of nuclear factor kappaB (NF-kB) in pulmonary arterial hypertension: effects of synthetic selective NF-kB inhibitor IMD-0354. Cardiovasc Res. 2013;99:35–43. doi: 10.1093/cvr/cvt105. [DOI] [PubMed] [Google Scholar]
- 45.Marsh LM, Jandl K, Grünig G, Foris V, Bashir M, Ghanim B, et al. The inflammatory cell landscape in the lungs of patients with idiopathic pulmonary arterial hypertension. Eur Respir J. 2018;51:1701214. doi: 10.1183/13993003.01214-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Florentin J, Coppin E, Vasamsetti SB, Zhao J, Tai Y-Y, Tang Y, et al. Inflammatory macrophage expansion in pulmonary hypertension depends upon mobilization of blood-borne monocytes. J Immunol. 2018;200:3612–3625. doi: 10.4049/jimmunol.1701287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauer EM, Zheng H, Lotze MT, Bauer PM. Recombinant human interferon alpha 2b prevents and reverses experimental pulmonary hypertension. PLoS One. 2014;9:e96720. doi: 10.1371/journal.pone.0096720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.George PM, Oliver E, Dorfmuller P, Dubois OD, Reed DM, Kirkby NS, et al. Evidence for the involvement of type I interferon in pulmonary arterial hypertension. Circ Res. 2014;114:677–688. doi: 10.1161/CIRCRESAHA.114.302221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savale L, Sattler C, Günther S, Montani D, Chaumais M-C, Perrin S, et al. Pulmonary arterial hypertension in patients treated with interferon. Eur Respir J. 2014;44:1627–1634. doi: 10.1183/09031936.00057914. [DOI] [PubMed] [Google Scholar]
- 50.Hu B, Guo Y, Garbacz WG, Jiang M, Xu M, Huang H, et al. Fatty acid binding protein-4 (FABP4) is a hypoxia inducible gene that sensitizes mice to liver ischemia/reperfusion injury. J Hepatol. 2015;63:855–862. doi: 10.1016/j.jhep.2015.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu YA, Malakhau Y, Yu CA, Phelan SJ, Cumming RI, Kan MJ, et al. Nonclassical monocytes sense hypoxia, regulate pulmonary vascular remodeling, and promote pulmonary hypertension. J Immunol. 2020;204:1474–1485. doi: 10.4049/jimmunol.1900239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narasimhan PB, Marcovecchio P, Hamers AAJ, Hedrick CC. Nonclassical monocytes in health and disease. Annu Rev Immunol. 2019;37:439–456. doi: 10.1146/annurev-immunol-042617-053119. [DOI] [PubMed] [Google Scholar]
- 53.Raychaudhuri B, Malur A, Bonfield TL, Abraham S, Schilz RJ, Farver CF, et al. The prostacyclin analogue treprostinil blocks NFkappaB nuclear translocation in human alveolar macrophages. J Biol Chem. 2002;277:33344–33348. doi: 10.1074/jbc.M203567200. [DOI] [PubMed] [Google Scholar]
- 54.Watzinger K, Tancevski I, Sonnweber T, Löffler-Ragg J. Antiinflammatory properties of PAH drugs. Eur Respir J. 2016;48:PA1818. [Google Scholar]
- 55.Yeh C-H, Kuo C-H, Yang S-N, Huang M-Y, Wu H-C, Wang H-P, et al. Prostaglandin I2 analogs suppress tumor necrosis factor α production and the maturation of human monocyte-derived dendritic cells. J Investig Med. 2011;59:1109–1115. doi: 10.2310/JIM.0b013e3182281f62. [DOI] [PubMed] [Google Scholar]
- 56.Aronoff DM, Peres CM, Serezani CH, Ballinger MN, Carstens JK, Coleman N, et al. Synthetic prostacyclin analogs differentially regulate macrophage function via distinct analog-receptor binding specificities. J Immunol. 2007;178:1628–1634. doi: 10.4049/jimmunol.178.3.1628. [DOI] [PubMed] [Google Scholar]
- 57.Ali Mohamed R, Galal O, Refaat Mohammed A, Salah El-Abhar H. Tropisetron modulates peripheral and central serotonin/insulin levels via insulin and nuclear factor kappa B/receptor for advanced glycation end products signalling to regulate type-2 diabetes in rats. RSC Advances. 2018;8:11908–11920. doi: 10.1039/c7ra13105d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keshavarz M, Showraki A, Emamghoreishi M. Anticonvulsant effect of guaifenesin against pentylenetetrazol-induced seizure in mice. Iran J Med Sci. 2013;38:116–121. [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Cherryholmes G, Mao A, Marek C, Longmate J, Kalos M, et al. High plasma levels of MCP-1 and eotaxin provide evidence for an immunological basis of fibromyalgia. Exp Biol Med (Maywood) 2008;233:1171–1180. doi: 10.3181/0712-RM-328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data set and lists of cell type–specific marker genes and disease DEGs are available online at http://mergeomics.research.idre.ucla.edu/PVDSingleCell/CellBrowser/. Connectivity scores of the entire panel of perturbagens from the CMap analysis are available at http://mergeomics.research.idre.ucla.edu/PVDSingleCell/CMap/.


