Keywords: serrated polyposis syndrome, serrated polyps, RNF43, somatic mutation
Abstract
Aims
RNF43 is suggested to be involved in the serrated pathway towards colorectal cancer and encodes a transmembrane Ring‐type E3 ubiquitin ligase that negatively regulates the Wnt pathway. This study aimed to elucidate the role of RNF43 gene variants in serrated polyposis syndrome (SPS) and serrated polyps.
Methods and results
Three cohorts were tested. The first cohort included germline DNA of 26 SPS patients tested for pathogenic variants in RNF43 by Sanger sequencing all exons. In the second cohort we tested somatic DNA for RNF43 mutations from sporadic serrated lesions: 25 hyperplastic polyps, 35 sessile serrated lesions and 38 traditional serrated adenomas (TSA). In the third cohort we investigated RNF43 mutations in 49 serrated polyps and 60 conventional adenomas from 40 patients with Lynch syndrome. No germline RNF43 pathogenic variants were detected in our SPS cohort. In sporadic colorectal lesions we detected RNF43 deleterious frameshift mutations in three TSA and one SSL. The RNF43 mutations in previously described homopolymeric hot‐spots were detected in microsatellite‐instable (MSI) polyps and the other RNF43 mutations in microsatellite‐stable (MSS) serrated polyps. RNF43 hot‐spot mutations were discovered in seven serrated polyps and 12 conventional adenomas from Lynch patients.
Conclusion
Truncating germline RNF43 mutations are uncommon in SPS patients. Somatic mutations in RNF43 were found in sporadic TSA and SSL and both serrated polyps and adenomas from Lynch syndrome patients, suggesting that they do not develop early in the pathway to CRC and are not specific for serrated polyp subtypes.
Introduction
The Wnt signalling pathway plays a central role in colorectal carcinogenesis; but affected components differ between precursor lesions. APC mutations are present in the majority of conventional adenomas, while the precursors in the serrated pathway show aberrations in other genes of the Wnt signalling pathway. 1 RNF43 encodes a transmembrane Ring‐type E3 ubiquitin ligase that negatively regulates the Wnt pathway. 2 RNF43 was proposed as a candidate gene for serrated polyposis syndrome (SPS) in two whole‐exome sequencing studies. 3 , 4 A Spanish cohort discovered one SPS patient with a probably pathogenic RNF43 variant out of 96 screened SPS patients (1.0%) 5 and a large multinational cohort found five of 304 (1.6%) tested SPS patients with probably pathogenic RNF43 variants. 6 , 7 Worldwide, 16 carriers from 10 families have been reported to the present time. 3 , 4 , 5 , 6 , 8
SPS is a polyposis syndrome that is characterised by the presence of multiple serrated polyps. 9 Histological subtypes of serrated polyps are hyperplastic polyps (HP), sessile serrated lesions (SSL) and traditional serrated adenomas (TSA). 9 These lesions are morphologically marked by a saw‐tooth shape in the epithelium of the crypts, and can progress to CRC. 9 The increased risk of CRC in first‐degree relatives (FDR) of SPS patients and the occasional clustering of SPS cases within families suggests a partial hereditary aetiology. 10 , 11 , 12 Various modes of inheritance, such as autosomal dominant, recessive or co‐dominant, have been proposed. 10 , 11 , 12 Until now no definite inheritance has been proven. As a genetic test for SPS is therefore not available, the diagnosis is made on the basis of clinical criteria of the World Health Organisation (WHO) based on number, size and location of the serrated polyps. 9 , 13 Such a genetic test could aid in more accurate risk calculation and selection of patients for closer surveillance.
Somatic RNF43 mutations might be involved in the development of sporadic serrated polyps, similar to APC in familial adenomatous polyposis and sporadic conventional adenomas. Somatic RNF43 mutations can be found in approximately 18% of sporadic CRC, strongly associated with microsatellite instability (MSI) and mutually exclusive with APC mutations. 14 Sporadic MSI CRC can develop from serrated polyps, suggesting that RNF43 mutations might play a role in the serrated pathway to CRC. 1 RNF43 mutations in serrated polyp subtypes have been described mainly in TSA, less frequently in SSL and least often in HP. For SSL, most RNF43 mutations are described in MutL homologue 1 (MLH1)‐deficient SSL with dysplasia. 8 , 15 , 16 , 17
In this study we investigate RNF43 variants in germline DNA of SPS patients and somatic DNA from colorectal polyps. We used three cohorts: SPS patients, sporadic serrated polyps, and polyps from Lynch syndrome patients. Our first aim was to test whether germline RNF43 mutations are present in our cohort of SPS patients. Our secondary aim was to determine the frequency of somatic RNF43 mutations in both sporadic serrated polyps and lesions from Lynch syndrome patients and the relationship to MSI.
Methods
All patients were recruited at the Radboud University Medical Center, Nijmegen, the Netherlands; we assembled three independent cohorts to evaluate the role of RNF43, as shown in Figure 1.
Figure 1.
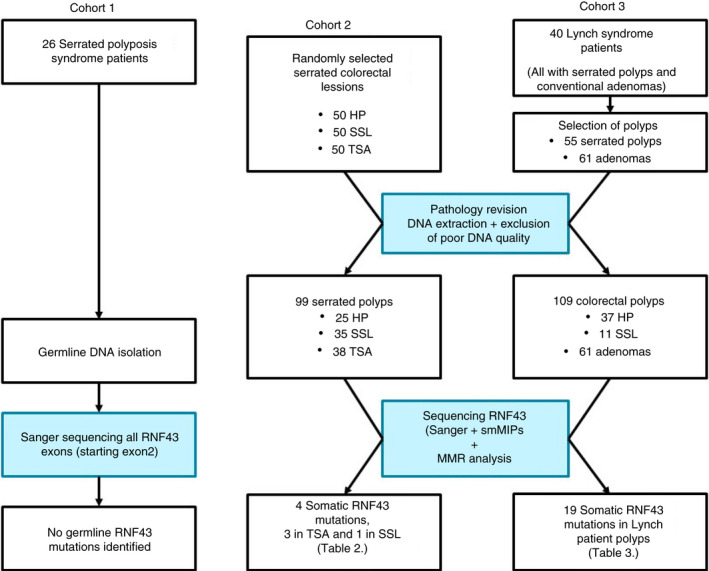
Flow‐chart of sample collection and analysis in the three cohorts.
SPS Patient (Germline) Cohort
In cohort 1 we used a candidate gene approach to detect germline mutations in SPS patients. Cohort 1 consisted of 26 adult patients without a known MUTYH, APC or other CRC predisposing germline mutation that fulfilled the WHO criteria for SPS. 3 Patients signed informed consent prior to isolation of DNA from peripheral blood using standard procedures. We collected patient clinical characteristics and family history during interviews and from the available medical files. Total polyp count, total serrated polyp count and total adenomatous polyp count were calculated from all endoscopies. Polymerase chain reaction (PCR) and Sanger sequencing were performed using standard procedures. 18 For germline DNA of SPS patients we performed Sanger sequencing of all exons, starting in exon 2 where the translation start site is located.
Polyp Cohorts
In order to search for somatic mutations in different polyp subtypes we used two cohorts. In cohort 2 we collected 150 sporadic serrated polyps from 98 patients, subdivided into 50 HP, 50 SSL and 50 TSA from our local archive. HP and SSL were consecutive lesions from 1 January 2013. Consecutive cases of TSA were included between 1 January 1995 and 1 July 2013. An expert pathologist (I.D.N.) revised all included colorectal lesions to verify the serrated subtypes; a second expert pathologist (G.A.M.) revised all TSA samples to confirm the diagnosis. In cohort 3 we collected conventional adenomas (n = 61) and serrated polyps (n = 55) from 40 patients with Lynch syndrome recruited from our local familial cancer registry. From each patient we included both serrated polyps and conventional adenomas.
DNA Sequencing
We isolated DNA from colorectal lesions after macrodissection from formalin‐fixed paraffin‐embedded slides, as described previously. 19 If necessary, we purified the DNA using the GenElute PCR Clean‐up kit (Sigma‐Aldrich, Saint Louis, MO, USA). Polyp samples with poor DNA concentration were excluded. We performed Sanger sequencing of codons 117 and 659, as these were previously described hot‐spot mutations. The sequences of primers used are summarised in Tables S1 and S2. For all available SSL and TSA additional sequencing was performed at a later stage, with single‐molecule molecular inversion probes (smMIPs). 20 A panel of smMIPs was used that included 56 smMIPs for RNF43 (covering exons 2–10 of ENST00000407977); two for codons 12 and 13 of KRAS; two for codon 61 of KRAS; and three for codon 600 of BRAF. The smMIP library preparations were performed manually, as described previously. 20 The smMIP library was sequenced at the Radboudumc Genomics Technology Center on a NextSeq500 (Illumina, San Diego, CA, USA), according to the manufacturer’s protocol (300 cycles high‐output sequencing kit) resulting in 2 × 150 base pairs (bp) paired‐end reads. Afterwards, BCL‐to‐FASTQ conversion and demultiplexing of barcoded reads was performed automatically (Illumina). Mapping of reads to the reference genome (GRCh37/hg19), consensus read building and variant calling was performed with SeqNext software from JSI medical systems (version 5.1.0, build 503; JSI Medical Systems, Ettenheim, Germany), with settings as previously described. 20 For variant calling, the following settings were used: required coverage/minimum absolute coverage: 20 combined; variants/minimum absolute coverage: five combined; minimum percentage coverage: 5% per dir. Subsequently, we removed synonymous variants, variants in untranslated regions (UTRs), variants in non‐coding regions and SNPs with AF > 0.01 (GnomAD database). Lastly, variants that were called based on <10% mutant reads and/or <10 total reads, but which could be cytosine deaminations, were also removed.
Microsatellite Stability Testing
All polyps were immunohistochemically stained with monoclonal antibodies against MLH1 (BD Pharmingen 551091, 1:40; BD Pharmingen, San Jose, CA, USA), PMS homologue 2 (PMS2) (BD Pharmingen 556415, 1:50), MutS homologue 2 (MSH2) (Calbiochem NA26, 1:40; Calbiochem, Darmstadt, Germany) and MSH6 (Abcam ab92471, 1:500; Abcam, Cambridge, UK) and developed with Powervision (LabVision, Cheshire, UK). We scored staining as positive, negative or unclear, as previously described. 21 When mismatch repair (MMR) protein staining was negative or unclear, we tested MSI using a panel of microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26 and BAT40). MSI status was characterised as MSI when more than one marker was unstable and microsatellite stable (MSS) with one or no unstable markers.
Statistics and Ethical Approval
Descriptive statistics were used to summarise patient and lesion characteristics. All analyses were performed with SPSS version 20. The rate of RNF43 mutations as a primary outcome for this study was determined as a percentage of the total number of polyps or patients. The local medical ethics committee approved the study protocol for germline DNA testing of SPS patients (NL44839.091.13) and the study protocol for testing of somatic polyp DNA (CMO 2015‐1882).
Results
RNF43 Germline Variants and SPS
We included 26 SPS patients whose clinical data are summarised in Table 1. This group shows heterogeneity on age, gender, total polyp counts and history of CRC. We detected no truncating germline variants in RNF43 in our study population. All detected single nucleotide polymorphisms (SNP) (rs3744093, rs2257205, rs76384648, rs2680701, rs34523089, rs2526374, rs9652855, rs61746279, rs2158460) showed allele frequencies similar to the frequencies in the HapMap‐CEU population or CSAgilent population (participants with European ancestry). 5 , 7
Table 1.
Characteristics of SPS patients
| Patient no. | Sex | Number of SP | Number of AD | WHO criteria 2010 diagnosis* | Age at first polyp | History of CRC | History of extracolonic cancer | Smoking status |
|---|---|---|---|---|---|---|---|---|
| SPS1 | Female | >31 | >13 | 3 | 47 | No | Breast cancer | Current smoker |
| SPS2 | Female | 9 | 0 | 2 | 24 | No | No | Current smoker |
| SPS3 | Female | 30 | 5 | 3 | 47 | No | No | No smoker |
| SPS4 | Male | 17 | 2 | 1 | 57 | No | No | No smoker |
| SPS6 | Female | 10 | 9 | 1 | 51 | No | No | No smoker |
| SPS7 | Female | 1 | 0 | 2 | 39 | No | No | No smoker |
| SPS9 | Female | Multiple | 0 | 1 + 3 | 59 | Yes, 3×** | No | No smoker |
| SPS10 | Male | 70 | 12 | 1 + 3 | 60 | No | No | Former smoker |
| SPS11 | Male | >63 | 2 | 1 + 3 | 45 | No | No | Current smoker |
| SPS12 | Female | 46 | 7 | 3 | 54 | No | Hodgkin lymphoma | Former smoker |
| SPS13 | Male | 22 | 5 | 3 | 56 | No | No | Former smoker |
| SPS19 | Female | 63 | 1 | 1 + 3 | 52 | Yes, 1× | No | Former smoker |
| SPS20 | Male | >46 | 11 | 1 + 3 | 64 | No | No | Former smoker |
| SPS21 | Female | >10/multiple | >4 | 3 | 45 | No | No | Current smoker |
| SPS22 | Female | 61 | 0 | 1 + 3 | 60 | No | No | Current smoker |
| SPS24 | Male | 25 | 6 | 1 + 3 | 71 | No | No | Former smoker |
| SPS25 | Male | 44 | 0 | 1 + 3 | 42 | No | No | No smoker |
| SPS26 | Male | 22 | 17 | 3 | 49 | No | No | Former smoker |
| SPS27 | Male | 21 | 1 | 3 | 57 | No | Pancreatic IPMN | Former smoker |
| SPS28 | Female | >25 | 7 | 1 + 3 | 67 | No | Adrenal incidentaloma | Current smoker |
| SPS29 | Male | >63 | 2 | 1 + 3 | 59 | No | No | Current smoker |
| SPS32 | Male | 30 | 9 | 3 | 50 | No | No | Former smoker |
| SPS33 | Female | 32 | 4 | 1 + 3 | 77 | No | No | Current smoker |
| SPS34 | Male | >25 | 2 | 1 + 3 | 75 | Yes, 1× | No | Former smoker |
| SPS36 | Male | 32 | 17 | 1 + 3 | 61 | No | No | Current smoker |
| SPS37 | Female | 21 | 4 | 3 | 73 | No | Breast cancer | Former smoker |
SP, Serrated polyp; SPS, Serrated polyposis syndrome; AD, Conventional adenoma; CRC, Colorectal carcinoma.
According to the WHO criteria: 1, ≥5 SP proximal to the sigmoid (≥2 larger than 10 mm); 2, ≥1 SP proximal to the sigmoid + first‐degree relative with SPS; 3, >20 SP of any size, distributed over the entire colon.
SPS09: one medullar carcinoma, one poorly differentiated adenocarcinoma and one mucinous carcinoma.
Somatic RNF43 Mutations and Sporadic Serrated Polyps
After histological review, samples with poor DNA quality were excluded and 98 serrated polyps were analysed. These were diagnosed as HP (n = 25), SSL (n = 35) and TSA (n = 38). Two SSL with low‐grade dysplasia were included. Of the TSA, two showed high‐grade dysplasia (HGD), one showed a transition to adenocarcinoma and 35 showed low‐grade dysplasia (LGD). The majority of HP (76%) and SSL (85.7%) were removed from the proximal colon, while TSA were most often removed from the distal colon (23.1% proximal). We detected loss of expression in at least one of the MMR genes in seven serrated polyps, while staining was unclear in eight serrated polyps. After MSI status testing, 10.2% were MSI (n = 10) (Table 2). Sanger sequencing showed somatic RNF43 mutations in two TSA (5.3%) and none in the other SP subtypes. A p.Arg117fs mutation constituting of a deletion of 1 bp in a homopolymeric tract of six C–G pairs was seen in one TSA. In one other TSA we detected a p.Gly659fs deletion of 1 bp in a MSI locus of seven C–G pairs. We detected loss of expression of MSH6 in both TSA with RNF43 mutations, and the TSA with p.Arg117fs mutations additionally showed loss of expression of MSH2 (Figures 2 and 3). Further investigation into the patients that presented with these TSA showed that they were both patients with known Lynch syndrome. Additional sequencing using smMIPs confirmed both these RNF43 mutations, and found an additional p.Glu318Ter variant in the previously mentioned TSA with the p.Gly659fs mutation. Furthermore, a p.Arg40Lysfs mutation in an MSS SSL and a p.Gly67Asp mutation in an MSS TSA were detected, both with a BRAF hot‐spot V600E variant. In total, two SSL (5.7%) and 10 TSA (26.3%) were BRAF V600E‐mutated and 12 TSA (31.6%) showed KRAS hot‐spot mutations (Table S3).
Table 2.
Characteristics of sporadic colorectal lesions
| HP (n = 25) | SSL (n = 35) | TSA (n = 38) | |
|---|---|---|---|
| Proximal location | 19 (76%) | 30 (85.7%) | 9 (23.7%) |
| Dysplasia | – | 2 (5.7%) | 38 (100%) |
| CRC in polyp | – | – | 1 (2.6%) |
| MSS* | 24 (96.0%) | 31 (86.6%) | 33 (81.6%) |
| RNF43 | – | 1 × p.Arg40Lysfs | 1 × p.Gly67Asp |
| MSI* | 1 (4.0%) | 4 (4.1%) | 5 (5.1%) |
| RNF43 | – | – |
1 × p.Arg117fs† 1 × p.Gly659fs + p.Glu318Ter† |
HP, Hyperplastic polyp; SSL, Sessile serrated lesion; TSA, Traditional serrated adenoma; CRC, Colorectal carcinoma; MSI, Microsatellite‐instable; MSS, Microsatellite‐stable; MMR, Mismatch repair.
Immunohistochemistry for MMR genes normal staining pattern and normal pentaplex.
Although cases were randomly selected based on histological diagnosis, these polyps turned out to be from known Lynch syndrome patients.
Figure 2.
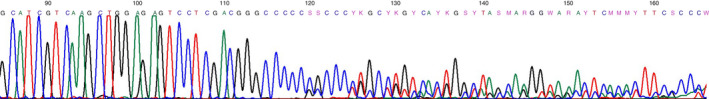
Deleterious frameshift mutation encoding p.Arg117fs.
Figure 3.
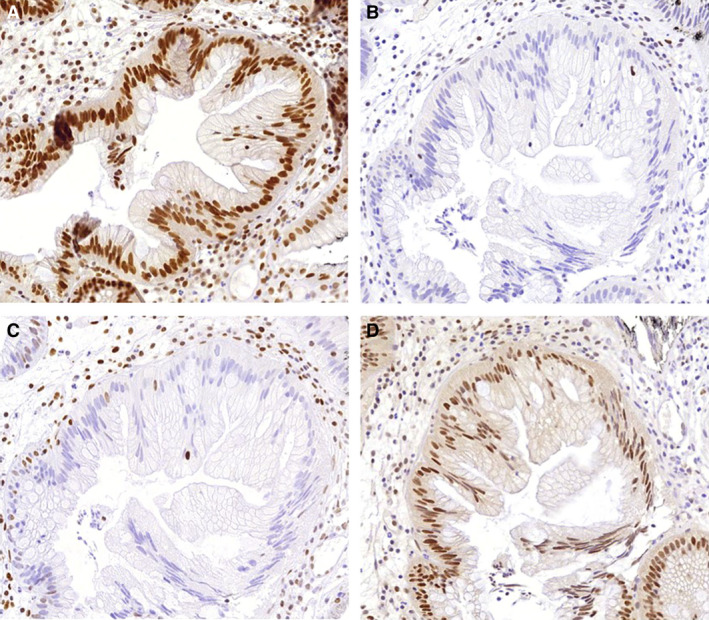
Immunohistochemistry of traditional serrated adenoma (TSA) with p.Arg117fs. A, MutL homologue 1 (MLH1) staining; normal expression of the MLH1 mismatch repair (MMR) protein. B, MutS homologue 2 (MSH2) staining; loss of expression of the MSH2 MMR protein. C, MSH6 staining; loss of expression of the MSH6 MMR protein. D, PMS homologue 2 (PMS2) staining; normal expression of the PMS2 MMR protein.
Somatic RNF43 Mutations in Lynch Syndrome‐Associated Polyps
Because two randomly selected TSAs came from Lynch syndrome patients, we established a third cohort. After histological revision and exclusion of samples with poor DNA quality, 48 serrated polyps and 61 conventional adenomas from Lynch syndrome patients were included. Of the serrated polyps, the majority (n = 37) were HP and 11 were SSL, one of which contained dysplasia. Within the subgroup of conventional adenomas the largest group were tubular adenoma (n = 57) and the rest (n = 3) were tubulovillous adenomas. One of the tubular adenomas contained HGD, the rest LGD. Using Sanger sequencing 15 p.Gly659 frameshift mutations were detected, which were found in 10 tubular adenomas, one SSL and four HP. p.Arg117 frameshifts were detected in six lesions, five tubular adenomas and one SSL. In three tubular adenomas both the p.Gly659 and p.Arg117 frameshift were present. Additional sequencing (only in SSL and TSA) using smMIPs confirmed the p.Arg117 frameshift mutation in an SSL and detected an additional p.Gly659Val frameshift mutation in a SSL with dysplasia previously not detected by Sanger sequencing (Table 3, Table S3). RNF43 mutations were discovered in both MSI and MSS polyps of all subtypes and the relationship between the RNF43 mutation and the MSI status is shown in Table 3 and Table S3. The three polyps with two mutations were all MSI. From the 16 polyps with one detected mutation eight were MSS (50%) and eight MSI (50%), as shown in Table S3.
Table 3.
RNF43 hot‐spot mutations in MSI and MSS polyps from Lynch patients
| RNF43 | MS staining | HP (n = 37) | SSL (n = 11) | TA (n = 58) | TVA (n = 3) | Total (n = 109) |
|---|---|---|---|---|---|---|
| No hot‐spot mutation | MSI | – | – | 7 | 2 | 9 (8.2%) |
| MSS | 28 | 7 | 36 | 1 | 72 (66.0%) | |
| Unclear | 5 | 1 | 3 | – | 9 (8.2%) | |
| p.Arg117fs | MSI | – | – | 2 | – | 2 (1.8%) |
| MSS | – | 1 | – | – | 1 (0.9%) | |
| p.Gly659fs | MSI | 1 | 1 | 4 | – | 6 (5.5%) |
| MSS | 3 | 1 | 3 | – | 7 (6.4%) | |
| Both hot‐spot mutations | MSI | – | – | 3 | – | 3 (2.7%) |
| MSS | – | – | – | – | 0 (0%) |
MSI, Microsatellite‐instable; MSS, Microsatellite‐stable; MS, Microsatellite; MMR, Mismatch repair; HP, Hyperplastic polyp; SSL, Sessile serrated lesion; TA, Tubular adenema; TVA, Tubulovillous adenoma.
Immunohistochemistry for MMR genes normal staining pattern.
Discussion
We set out to elucidate the relation of RNF43 mutations with the serrated pathway in both hereditary and sporadic serrated polyps. In our cohort of 26 SPS patients we detected no germline truncating RNF43 mutations. In unselected TSA and SSL, we observed sporadic RNF43 frameshift mutations in previously described hot‐spots (p.Arg117fs and G659fs) in TSA with MSI and mutations outside these hot‐spots in MSS serrated polyps. Somatic RNF43 mutations were more common in polyp DNA from Lynch patients, and were found in seven serrated polyps and 12 adenomas in our cohort of 109 lesions.
Somatic RNF43 mutations in approximately 18% of sporadic CRC are strongly associated with MSI and mutually exclusive with APC mutations. 14 It is not known which precursor lesions precede the RNF43‐mutated CRC because of the loss of typical histological morphology. In our cohort of sporadic serrated lesions, RNF43 mutations were found in a low percentage of both TSA and SSL (7.9 and 2.8%, respectively). This is in line with previous data that show that TSAs more commonly contain genetic alterations that lead to Wnt pathway activation, in contrast to other serrated polyps. 17 We detected a lower proportion of RNF43‐mutated TSA (7.9%) compared to previous studies that reported 24% and 38% RNF43 mutations. 16 , 17 We did not replicate the association of RNF43‐mutated TSA with the BRAF V600E mutation previously described, as in our cohort only one RNF43‐mutated TSA also showed a V600E BRAF mutation in addition to one RNF43‐mutated SSL with a BRAF V600E mutation. 22 We did not find histopathological differences in the cohorts to explain this. It is possible that inclusion bias due to the size of the cohorts or missed RNF43 lesions due to lower smMIPs sequencing coverage in our cohort contributed to these differences. Our result of 2.8% RNF43‐mutated SSL is similar to previously reported RNF43‐mutated non‐dysplastic SSL. 15 , 17 The low number of RNF43 mutations in SSL in our cohort can be explained because we mainly included SSL without dysplasia (33 of 35), with a low percentage of MLH1 hypermethylation, reflecting an early stage of these polyps. In contrast, in other cohorts that included dysplastic SSL with MLH1 hypermethylation, RNF43 mutations were reported to be as high as 86%. 15 This reflects that the RNF43 mutation is likely to be a later event in the serrated pathway.
One common pathway in the progression of precursor lesions towards CRC is activation of the Wnt signalling pathway. In sporadic adenomas, this is an early step in carcinogenesis mediated in the majority of cases by truncating mutations in APC. Mutations in RNF43 may lead to increased Wnt signalling due to increased presence of the Frizzled Wnt receptor on the cell surface. 2 , 23 Thus, RNF43 seem to be an alternative carcinogenesis pathway. However, recent studies show that the RNF43 G659fs mutation that is described as a RNF43 hot‐spot mutation probably has no effect on RNF43 activity and is likely to be a mutation secondary to MLH1 deficiency. 24 Our results in serrated polyps, with RNF43 mutations in previously described microsatellite hot‐spots and the association with MSI, suggest that these mutations indeed occur as a result of MMR deficiency. In contrast, we also describe RNF43 mutations outside these hot‐spots in MSS serrated polyps that cannot be explained by this pathophysiological mechanism.
The development of serrated lesions in Lynch syndrome patients is common. 25 RNF43 is less frequently mutated in hereditary MSI CRC compared to sporadic CRC, 26 arguing that sporadic microsatellite unstable CRC arise from serrated polyps, while Lynch syndrome cancers commonly derive from colorectal adenomas with APC or CTNNB1 mutations. 26 In our subset of serrated polyps and adenomas from Lynch syndrome patients we found RNF43 mutations, mainly in conventional adenomas, but also in four HP and three SSL. We did not find RNF43‐mutated HP in our sporadic cohort, but whether or not this is purely the influence of polyp selection is not clear. Due to the MSI in Lynch patients these lesions are at high risk for these mutations, but this does not seem to be a specific feature for serrated polyps. The presence of RNF43 mutations in different subtypes of CRC might become clinically relevant, because these tumour specifics are suggested be associated with the more aggressive CRC phenotype 27 , 28 and are suspected to raise susceptibility to porcupine inhibition. 29 As these polyps in Lynch syndrome patients are precursors to RNF43‐mutated CRC, it is relevant to know the difference between mutations spectra that might lead to RNF43‐mutated CRC in Lynch syndrome and their characteristics.
RNF43 is one of the candidate genes for SPS based on previous studies. 3 , 4 However, we and others could not confirm this in other SPS populations. 5 , 7 While the patients described in the initial discovery cohort 3 are comparable in age to our population, more recently described SPS patients with RNF43 pathogenic variants 4 seem to be more severely affected at a younger age. Although not completely clear in all publications, the patients previously described with RNF43 pathogenic variants present with mainly serrated polyps and sporadic adenomatous polyps, comparable to our patients. Overall, RNF43 variants are not a common germline defect predisposing for SPS. 3 , 4 , 5 , 7 , 8 Due to the clinical criteria used to define SPS, it is a heterogenous group and it is likely that only a minor subset will be attributable to high‐penetrance germline genetic defects. Previous evidence suggests that smoking might be one of the underlying causative factors for the development of SPS. 30 In our cohort, 77% of SPS patients were found to be current or former smokers, which is higher than the overall prevalence rate of 49% of ever smokers in Europe. 31 In previous studies it is unknown how many of the study participants were smokers, making it possible that smoking could have played a role in the development of serrated polyps in our cohort. 3 , 4 , 5 , 7 , 8 .
A limitation of the study is that we included only 26 SPS patients. As SPS is a rare disease and often goes unrecognised, this is a reasonable‐sized cohort for this analysis. 32 A strength of this study is that we had a large population of the rare serrated polyp subtype TSA. While the interobserver variability of the pathological diagnosis of serrated polyps is high, we tried to decrease the influence of the uncertainty by review by two expert pathologists. While the majority of HP are small, and found in the rectum and distal colon in clinical practice, our subset of HP mainly consisted of larger proximal lesions. The two reasons for this are that small rectal HP are often not removed by the clinician due to their benign nature and that the DNA quality of small HP was too poor to be included for sequencing. We started by only testing two locations (codons 117 and 659) of the RNF43 gene in the somatic polyp DNA using Sanger sequencing, but recognising the importance of RNF43 mutations outside the hot‐spots, we later expanded with smMIPs sequencing of the entire RNF43 gene. However, the coverage obtained with this sequencing method was below an average of 100 reads for the RNF43 gene in 32 of 63 sequenced polyps (Table S3). Thereby, we could have missed somatic RNF43 mutations in other coding parts of the gene.
We used the WHO 2010 criteria for SPS patients, as these were the leading criteria for research at the time of inclusion. Since then the new 2019 WHO criteria have been formulated, which differ from the 2010 criteria in all three criteria, 13 as can be seen in Table 4. The previous second criterion is removed, with the implication that family members of SPS patients are no longer seen as affected if they have at least one polyp. In our cohort, two patients were included based on this criterion. We decided to retain these patients in the study as they are related and both from a large SPS family, making them of interest for genetic research. All patients fulfilling criterion 3 in the WHO 2010 criteria also fulfilled the new 2019 criteria. Of the two patients fulfilling only criterion 1 in the 2010 criteria, one patient also fulfilled criterion 1 of the 2019 criteria; the other could not be definitely determined because the older endoscopy reports did not specify smaller lesions as smaller or larger than 5 mm. We do not think that the changed criteria would have influenced the results of the study if we had included them based on the 2019 criteria. We believe that the age of the patients is likely to be a more important limitation of the study.
Table 4.
Old and new WHO criteria for serrated polyposis
| 2010 criteria 9 | 2019 criteria 13 | ||
|---|---|---|---|
| 1 | At least five serrated polyps proximal to the sigmoid colon with two or more of these being >10 mm | 1 | ≥5 serrated lesions/polyps proximal to the rectum, all being ≥5 mm in size, with ≥2 being ≥10 mm in size |
| 2 | Any number of serrated polyps proximal to the sigmoid colon in an individual who has a first‐degree relative with serrated polyposis | NA | NA |
| 3 | >20 serrated polyps of any size, but distributed throughout the colon | 2 | >20 serrated lesions/polyps of any size distributed throughout the large bowel, with ≥5 being proximal to the rectum |
NA, Not applicable.
Conclusion
Truncating germline RNF43 variants are uncommon in SPS patients. Somatic deleterious frameshift mutations in RNF43 were found in both serrated polyps and adenomas from Lynch syndrome patients, suggesting that the correlation between RNF43 and MSI is stronger than the correlation with histological subtypes of precursor lesions.
Author contributions
YvH, LK, EVB and FS performed the research, YvH, PD, FN, TB, IN, NH, designed the research study, EVB, GM, IN, NH, FS contributed essential reagents or tools, YvH, LK, EVB, FS analysed the data, YvH, LK, TB, IN wrote the paper. All authors approved final version of the manuscript except for FN due to his passing.
Conflict of interest
The authors have no potential conflicts of interest to disclose.
Supporting information
Table S1. RNF43 primers germline DNA (SPS patients).
Table S2. RNF43 primers somatic DNA (colorectal lesions).
Table S3. Overview of all included polyps and analyses of cohort 2 and 3.
Acknowledgements
The authors are grateful to Annemarie Verschoor, Jody Salomon, Jeroen Dijkstra and Madeleine Berendsen for their help in laboratory work. Polat Dura is supported by a Mozaic Grant awarded by the Netherlands Organisation for Scientific Research (NWO, 017‐009‐074).
van Herwaarden Y J, Koggel L M, Simmer F, Vink‐Börger E M, Dura P, Meijer G A, Nagengast F M, Hoogerbrugge N, Bisseling T M & Nagtegaal I D (2021) Histopathology 78, 749–758. 10.1111/his.14286 RNF43 mutation analysis in serrated polyposis, sporadic serrated polyps and Lynch syndrome polyps
References
- 1. Crockett SD, Nagtegaal ID. Terminology, molecular features, epidemiology, and management of serrated colorectal neoplasia. Gastroenterology 2019; 157: 949–966. e4. [DOI] [PubMed] [Google Scholar]
- 2. Koo BK, Spit M, Jordens I et al. Tumour suppressor RNF43 is a stem‐cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012; 488; 665–669. [DOI] [PubMed] [Google Scholar]
- 3. Gala MK, Mizukami Y, Le LP et al. Germline mutations in oncogene‐induced senescence pathways are associated with multiple sessile serrated adenomas. Gastroenterology 2014; 146; 520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taupin D. A deleterious RNF43 germline mutation in a severely affected serrated polyposis kindred. Hum. Genome Var. 2015; 2; 15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quintana I, Mejias‐Luque R, Terradas M et al. Evidence suggests that germline RNF43 mutations are a rare cause of serrated polyposis. Gut 2018; 67; 2230–2232. [DOI] [PubMed] [Google Scholar]
- 6. International Society for Gastrointestinal Hereditary Tumours (InSiGHT). Fam. Cancer 2017; 16(Suppl. 1); 1–134. [DOI] [PubMed] [Google Scholar]
- 7. Buchanan DD, Clendenning M, Zhuoer L et al. Lack of evidence for germline RNF43 mutations in patients with serrated polyposis syndrome from a large multinational study. Gut 2017; 66; 1170–1172. [DOI] [PubMed] [Google Scholar]
- 8. Yan HHN, Lai JCW, Ho SL et al. RNF43 germline and somatic mutation in serrated neoplasia pathway and its association with BRAF mutation. Gut 2017; 66; 1645–1656. [DOI] [PubMed] [Google Scholar]
- 9. Snover DC. Serrated polyps of the colon and rectum and serrated polyposis. WHO classification of tumors of the digestive system. Lyon: International Agency for Research on Cancer (IARC), 2010. [Google Scholar]
- 10. Young J, Jenkins M, Parry S et al. Serrated pathway colorectal cancer in the population: genetic consideration. Gut 2007; 56; 1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Win AK, Walters RJ, Buchanan DD et al. Cancer risks for relatives of patients with serrated polyposis. Am. J. Gastroenterol. 2012; 107; 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boparai KS, Reitsma JB, Lemmens V et al. Increased colorectal cancer risk in first‐degree relatives of patients with hyperplastic polyposis syndrome. Gut 2010; 59; 1222–1225. [DOI] [PubMed] [Google Scholar]
- 13. Rosty N, Brosens LAA, Nagtegaal ID. Genetic tumour syndromes of the digestive system. In Arends MJ, Carneiro F, Lax SF eds. WHO Classification of Tumours Editorial Board, vol. 1, 5th edn. Lyon: IARC Publications, 2019. [Google Scholar]
- 14. Giannakis M, Hodis E, Jasmine MuX et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 2014; 46; 1264–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hashimoto T, Yamashita S, Yoshida H et al. WNT pathway gene mutations are associated with the presence of dysplasia in colorectal sessile serrated adenoma/polyps. Am. J. Surg. Pathol. 2017; 41; 1188–1197. [DOI] [PubMed] [Google Scholar]
- 16. Nakanishi H, Sawada T, Kaizaki Y et al. Significance of gene mutations in the Wnt signaling pathway in traditional serrated adenomas of the colon and rectum. PLoS One 2020; 15; e0229262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sekine S, Yamashita S, Tanabe T et al. Frequent PTPRK–RSPO3 fusions and RNF43 mutations in colorectal traditional serrated adenoma. J. Pathol. 2016; 239; 133–138. [DOI] [PubMed] [Google Scholar]
- 18. Lacko M, Roelofs HM, Te Morsche RH et al. Genetic polymorphism in the conjugating enzyme UGT1A1 and the risk of head and neck cancer. Int. J. Cancer 2010; 127; 2815–2821. [DOI] [PubMed] [Google Scholar]
- 19. Knijn N, Mekenkamp LJ, Klomp M et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br. J. Cancer 2011; 104; 1020–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eijkelenboom A, Kamping EJ, Kastner‐van Raaij AW et al. Reliable next‐generation sequencing of formalin‐fixed, paraffin‐embedded tissue using single molecule tags. J. Mol. Diagn. 2016; 18; 851–863. [DOI] [PubMed] [Google Scholar]
- 21. Overbeek LI, Ligtenberg MJ, Willems RW et al. Interpretation of immunohistochemistry for mismatch repair proteins is only reliable in a specialized setting. Am. J. Surg. Pathol. 2008; 32; 1246–1251. [DOI] [PubMed] [Google Scholar]
- 22. Sekine S, Yamashita S, Yamada M et al. Clinicopathological and molecular correlations in traditional serrated adenoma. J. Gastroenterol. 2020; 55(4): 418–427. [DOI] [PubMed] [Google Scholar]
- 23. Jiang X, Hao HX, Growney JD et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc. Natl Acad. Sci. USA 2013; 110; 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tu J, Park S, Yu W et al. The most common RNF43 mutant G659Vfs*41 is fully functional in inhibiting Wnt signaling and unlikely to play a role in tumorigenesis. Sci. Rep. 2019; 9; 18557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vleugels JLA, Sahin H, Hazewinkel Y et al. Endoscopic detection rate of sessile serrated lesions in Lynch syndrome patients is comparable with an age‐ and gender‐matched control population: case–control study with expert pathology review. Gastrointest. Endosc. 2018; 87; 1289–1296. [DOI] [PubMed] [Google Scholar]
- 26. Fennell LJ, Clendenning M, McKeone DM et al. RNF43 is mutated less frequently in Lynch syndrome compared with sporadic microsatellite unstable colorectal cancers. Fam. Cancer 2018; 17; 63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eto T, Miyake K, Nosho K et al. Impact of loss‐of‐function mutations at the RNF43 locus on colorectal cancer development and progression. J. Pathol. 2018; 245; 445–455. [DOI] [PubMed] [Google Scholar]
- 28. Matsumoto A, Shimada Y, Nakano M et al. RNF43 mutation is associated with aggressive tumor biology along with BRAF V600E mutation in right‐sided colorectal cancer. Oncol. Rep. 2020; 43; 1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koo BK, van Es JH, van den Born M et al. Porcupine inhibitor suppresses paracrine Wnt‐driven growth of Rnf43;Znrf3‐mutant neoplasia. Proc. Natl Acad. Sci. USA 2015; 112; 7548–7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ij JE, Rana SA, Atkinson NS et al. Clinical risk factors of colorectal cancer in patients with serrated polyposis syndrome: a multicentre cohort analysis. Gut 2017; 66; 278–284. [DOI] [PubMed] [Google Scholar]
- 31. Special Eurobarometer 385. Attitudes of Europeans Towards Tobacco. Report. European Commission, Directorate‐General Health and Consumers. Special Eurobarometer 2012. Brussels: EU Open Data Portal. [Google Scholar]
- 32. van Herwaarden YJ, Pape S, Vink‐Borger E et al. Reasons why the diagnosis of serrated polyposis syndrome is missed. Eur. J. Gastroenterol. Hepatol. 2019; 31; 340–344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. RNF43 primers germline DNA (SPS patients).
Table S2. RNF43 primers somatic DNA (colorectal lesions).
Table S3. Overview of all included polyps and analyses of cohort 2 and 3.


