Abstract
Aims
A variety of tissue clearing techniques have been developed to render intact tissue transparent. For thicker samples, additional partial tissue delipidation is required before immersion into the final refractive index (RI)‐matching solution, which alone is often inadequate to achieve full tissue transparency. However, it is difficult to determine a sufficient degree of tissue delipidation, excess of which can result in tissue distortion and protein loss. Here, we aim to develop a clearing strategy that allows better monitoring and more precise determination of delipidation progress.
Methods
We combined the detergent sodium dodecyl sulphate (SDS) with OPTIClear, a RI‐matching solution, to form a strategy termed Accurate delipidation with Optimal Clearing (Accu‐OptiClearing). Accu‐OptiClearing allows for a better preview of the final tissue transparency achieved when immersed in OPTIClear alone just before imaging. We assessed for the changes in clearing rate, protein loss, degree of tissue distortion, and preservation of antigens.
Results
Partial delipidation using Accu‐OptiClearing accelerated tissue clearing and better preserved tissue structure and antigens than delipidation with SDS alone. Despite achieving similar transparency in the final OPTIClear solution, more lipids were retained in samples cleared with Accu‐OptiClearing compared to SDS.
Conclusions
Combining the RI‐matching solution OPTIClear with detergents, Accu‐OptiClearing, can avoid excessive delipidation, leading to accelerated tissue clearing, less tissue damage and better preserved antigens.
Keywords: antigenicity, delipidation, opacity, protein loss, three‐dimensional imaging, transparent brain
Our new strategy of using Accurate dilapidation with Optimal Clearing combines SDS, OPTIClear and refractive index‐matching solution. The new strategy provides fast, non‐swell/shrink, and no protein loss protocol for making the brain to be semi‐transparent. This opens a new avenue of fast pathological analysis of the brain without many thin tissue sectioning.
![]()
1. INTRODUCTION
Studying the structure and function of the brain in three dimensions (3D) has been a promising yet challenging task. Recently, a number of tissue clearing techniques have been developed to turn intact tissues optically transparent, including CLARITY, passive clarity technique (PACT), iDISCO, uDISCO and CUBIC‐X [1, 2, 3, 4, 5]. These techniques, combined with the recent development in fluorescent protein labelling and imaging techniques, make whole‐brain imaging much more accessible to standard laboratories [5, 6].
When visualising a thick tissue, the heterogeneity in refractive index (RI) between different regions and media induces heterogeneous light scattering in every direction, which fails to cancel out each other and generates optical opacity. Therefore, to render tissue transparent, reducing light scattering within tissues and between media is crucial. In theory, this could be solved by homogenising the RI within the tissue. The simplest way to achieve RI homogenisation is to immerse tissues into a solution to match the heterogeneous RI values to each other, that is, RI‐matching agents. However, clearing by simple immersion in RI‐matching agents have limited tissue clearing efficacies [7, 8, 9]. To clear larger samples, different approaches have been attempted. Organic solvent‐based clearing techniques such as iDISCO, uDISCO and FDISCO have adopted the use of sequential organic solvents that allow gradual dehydration and lipid removal, thus allow a robust efficacy in tissue clearing [3, 4, 10]. The downside being the toxic and corrosive nature of the solvents used, making them incompatible with microscope hardware and difficult to handle.
The strategy of lipid removal can also be employed in aqueous‐based clearing techniques such as CLARITY and CUBIC. However, the process is much slower and requires the use of elevated temperatures or electric fields to speed up lipid removal [1, 6, 11, 12]. In addition, it is well known that long duration of denaturing detergent treatment results in tissue damage, loss of antigenicity, quenched endogenous fluorescence and sample expansion in various stages of clearing. In some cases, the entire tissue collapses and dissolves [13]. To protect the tissue structure and composition, extra protein crosslinking steps have been added before delipidation, including the use of prolonged formaldehyde fixation [14], acrylamide hydrogel [1, 2], glutaraldehyde [11] and epoxy resin [6]. This partially solves the issue of tissue antigen preservation, but the tissue still swells during delipidation. Minimising changes in tissue size during the tissue clearing process is desirable for morphological analyses [15, 16]. It also adds significant complexity and time in tissue processing, and the use of toxic chemicals.
We therefore wondered if it is possible to simplify the delipidation step by combining delipidation and RI‐matching steps. To resolve this, we have developed a novel delipidation strategy termed Accurate delipidation with Optimal Clearing (Accu‐OptiClearing) by performing delipidation under a RI‐homogenising environment. By doing so, the degree of delipidation that is adequate for the final RI‐matching step and imaging can be visibly determined, providing better predictions on tissue transparency in the subsequent final RI‐matching step.
2. MATERIALS AND METHODS
2.1. Animals
All animals used in this study were approved and handled in accordance with the guidelines provided by the Committee on the Use of Live Animals in Teaching and Research (CULATR) in the University of Hong Kong (CULATR number 4014‐16), which is accredited by the AAALAC. We used the following animals in this study: Sprague‐Dawley (SD) rats (250 ~ 300 g, male), 3xTg mice (PS1M146 V, APPswe and tauP301L, 14‐month‐old, female), Tg(Thy1‐GFP) mice (3‐month‐old, male), wild‐type AB zebrafish (Danio rerio, 1‐year‐old), and Tg(fli1a:EGFP) y1 zebrafish (Danio rerio, 1 year old).
2.2. Perfusion and tissue preparation
Adult mice and rats were euthanised by intraperitoneal injection of sodium pentobarbital (120 mg/kg), perfused transcardially with 0.9% saline, followed by perfusion–fixation using 4% PB‐buffered paraformaldehyde (PFA). The brains were immediately harvested and post‐fixed with 4% PFA solution for at least 3 days at 4°C before proceeding to the clearing procedure. To section into a consistent thickness of tissues for subsequent clearing, whole rat or mouse brains after PFA post‐fixation were washed with phosphate‐buffered saline (PBS) (3 × 10 minutes) at room temperature before dissection guided by brain matrices with divisions at 1 mm intervals (Zivic Instruments).
For adult zebrafish (Danio rerio), fish were euthanised by the rapid cooling method in ice‐cold water for 30–40 minutes. The whole fish body used for clearing was prepared by removing the skin directly following the rapid cooling, while the whole fish brains were harvested under a stereomicroscope (Nikon SMZ 800 N) after euthanasia. Whole brain or whole fish were post‐fixed with 4% PFA for 1 or 3 days at 4°C, respectively, before proceeding to the clearing procedure.
2.3. Preparation of RI‐matching reagents
OPTIClear was prepared as described previously [14]. Briefly, a 20% (w/v) N‐methylglucamine (Sigma‐Aldrich #66930), 32% (w/v) Iohexol (Sigma‐Aldrich #D2158) and 25% (v/v) 2,2’‐thiodiethanol (TDE) (Sigma‐Aldrich #88559) solution using distilled water was made with pH adjusted to seven using concentrated hydrochloric acid.
RIMS [2], ScaleS4 [7], PROTOS [11], SeeDB2S [16], OPTIVIEW [17] and CUBICScale2 [18] were all prepared as described previously.
2.4. Accu‐OptiClearing procedures
The post‐fixed tissues were cleared by incubating in a mixture of different detergents and OPTIClear. The detergents used in the study included sodium dodecyl sulphate (SDS) (Sigma‐Aldrich #L5750), 1,2‐Hexandiol (HxD) (Sigma‐Aldrich #213691), Tween®20 (Sigma‐Aldrich #P1379) and CHAPS (Roth #1479.4).Unless specified otherwise, the detergent used in the study is referring to SDS. To prepare Accu‐OptiClearing agents, corresponding detergents were added during the preparation of OPTIClear (4% SDS‐OPTIClear: 4% SDS w/v OPTIClear; HxD‐OPTIClear: 10% HxD v/v OPTIClear; Tween‐20‐OPTIClear: 10% Tween®20 v/v OPTIClear; CHAPS‐OPTIClear: 10% CHAPS w/v OPTIClear). The pH was then adjusted 7–8 using concentrated hydrochloric acid before use.
For 4%SDS‐OPTIClear (Accu‐OptiClearing), the incubation time varied depending on myelin density, tissue sizes and thickness. Based on our observation, a 1 mm3 tissue block can be cleared by incubating overnight and a 4 mm thick coronal rat brain tissue section requires 3 days to clear. During incubation in OPTIClear‐based solution, all tissues were placed in glass containers (SciChem #BGL021020, 30 ml) covered with aluminium foil. We define partial delipidation to be adequate when the whole tissue turns visually transparent. At that point, tissues were washed with PBS (3 × 10 minutes, room temperature) to remove excess SDS, followed by immunostaining, and finally RI‐matching by incubating tissues in OPTIClear at 37°C for at least 6 hours before imaging. As a rule, the volume of OPTIClear used was roughly three times that of the tissue block to be cleared. See Table S1 for examples of clearing durations and volume of OPTIClear needed for different tissue sizes.
2.5. Immunofluorescence staining protocol
All immunofluorescence on rodent tissues was performed on 1 mm thick sections. The staining of pTauSer396 and amyloid‐β was performed on 3xTg mice (PS1M146 V, APPswe and tauP301L). Other immunostaining experiments were performed on SD rat tissue. For immunostaining on zebrafish brain tissues, wild‐type AB zebrafish at about 1 year old was sacrificed and the brains were harvested under stereomicroscope (Nikon).
The immunostaining step was performed after clearing with Accu‐OptiClearing strategy (4% SDS‐OPTIClear incubation at 37°C). After clearing, tissues were washed with PBS to remove excess clearing solution (3 × 10 minutes at room temperature), and then blocked with blocking solution (0.6 M glycine, 0.2% v/v Triton X‐100, 3% v/v goat/donkey serum, with 0.01% w/v sodium azide in 1x PBS) overnight at 37°C. The tissues were then stained with primary antibody in antibody diluent (PBS buffer, containing 0.2% v/v Tween 20, 3% donkey/goat serum (Vector Laboratories #S‐1000) and 0.01% w/v sodium azide (Sigma‐Aldrich #S2002) for at least 2 days, with daily supplementation to the dilution of 1:100. Tissue blocks (1 mm3) were submerged in 200 µl antibody diluent, at either 25ºC or 37ºC, depending on the antibodies used for staining. See Table S2 for list of primary and secondary antibodies used and their corresponding incubation temperature. Primary antibodies were removed by washing with 0.2% PBS‐Tween®20 (on shaker for 6 × 30 minutes at room temperature and then left overnight). The procedures for secondary antibodies follow that for primary antibodies. All tissues were counterstained with 4’,6‐diamidino‐2‐phenylindole (DAPI) by adding DAPI into antibody diluent (1 µg/ml) during secondary antibody staining. After washing off secondary antibodies in PBST on a shaker for 6 × 30 minutes, the tissues were incubated with OPTIClear for RI‐matching for at least 6 hours before visualisation under confocal microscopy.
2.6. DiI staining protocol
A 0.1 mg/ml DiI staining solution was prepared by dissolving 1,1’‐Dioctadecyl‐3,3,3’,3’‐tetramethylindocarbocyanine perchlorate (DiI) (ThermoFisher Scientific #D282) in a 1:1 v/v mixture of DMSO and distilled H2O. To perform DiI staining, 1 mm thick coronal sections were incubated in DiI staining solution for 1 day at 37°C, washed with destaining solution (1:1 mixture of DMSO and distilled H2O) and delipidated with the clearing solutions to be tested at 37°C until they become visually transparent. Finally, the tissues were washed with PBS (3 × 10 minutes at room temperature) and then incubated in OPTIClear for at least 6 hours at 37°C before imaging.
2.7. Neuronal tracing using virus
We used AAV‐DJ/8‐GFP (SignaGen Laboratories) in our study for neuronal tracing. For virus injection, SD rats (about 180 g, male) were anaesthetised with a mixture of ketamine (80 mg/kg) and xylazine (8 mg/kg). Viruses (titre: 1.21 × 1013 viral genome (VG)/ml) were injected bilaterally into the site of nucleus basalis of Meynert (nbM) (coordinates: 1.4 mm posterior, 2.6 mm lateral and 7 mm ventral to bregma) using a Hamilton syringe 1800 series (Hamilton Company) at a rate of 1 µl/min, 1 µl in total per injection site. One week after viral injection, the same site was re‐injected with saline (0.9% w/v NaCl). Three weeks later, the rats were euthanised and their brains harvested as described above and sectioned into 4 mm sagittal sections. The sections were cleared with Accu‐OptiClearing for 5 days before proceeding to mounting and confocal imaging.
2.8. Confocal microscopy
The cleared brain slices were mounted on 35 mm glass‐bottom dishes (MatTek #P35G‐1.5‐14‐C) filled with OPTIClear (about 100 µl is enough) and covered with 24 mm x 24 mm coverslips (Thermo Scientific #630‐2104). All imaging experiments were performed with an inverted confocal microscope (LSM 800; Carl Zeiss) equipped with the objectives Plan‐Apochromat 10× Ph1 M27 (NA 0.45; working distance, 2.0 mm), Plan‐Apochromat 20× Ph2 M27 (NA 0.8; working distance, 0.46 mm), Plan‐Apochromat 40× Oil DIC M27 (NA 1.4; working distance, 0.13 mm) and Plan‐Apochromat 63× Oil DIC M27 (NA 1.4; working distance, 0.19 mm). The laser excitation wavelengths used were 405 nm, 488 nm, 543 nm and 594 nm. The image was acquired using Zen 2.3 Blue Edition (Carl Zeiss).
2.9. Quantifications
2.9.1. Measurement of light absorbance by cleared tissues
SD rat brain coronal sections (4 mm thick) were cleared with different concentrations (8%, 4%, 2% or 1% w/v) of SDS and SDS‐OPTIClear (8%, 4%, 2%, or 1% w/v SDS, 20% (w/v) N‐methylglucamine, 32% (w/v) iohexol and 25% (v/v) 2,2’‐thiodiethanol (TDE)) (n replicates per group = 6) for 3 days at 37ºC. After clearing, tissue sections were trimmed to a round shape that can perfectly fit into a 96‐well plate well and incubated with the corresponding clearing solutions to be tested. It was ensured that the tissue filled up the well and no trapped bubbles. Absorbance at 750 nm was measured using a LabSystems Multiskan microplate reader (LabX). Uncleared tissues with the same thickness and region were used as controls. To confirm the absorbance reading was not due to the use of different detergents, blanks were included by filling up wells with the amount of clearing solutions only.
For zebrafish, wild‐type AB zebrafish (Danio rerio, 1‐year‐old) were cleared with 4% SDS alone and Accu‐OptiClearing, respectively (n replicates per group = 3) with different time points (0 day, 2 days, 4 days or 8 days) at 37ºC. The centre of the fish body was punched to obtain a round shape of tissues that fitted into the 96‐well plates and absorbance was measured as described above.
2.9.2. Measurement of tissue size change
4 mm thick SD rat brain coronal sections were cleared with different concentrations (8%, 4%, 2% or 1% w/v) of SDS and SDS‐OPTIClear (n replicates per group = 3) for 3 days at 37ºC. Bright‐field images of tissues were taken before and after clearing. The area of tissues was measured using the ‘polygon selections’ function in Fiji (Image J; NIH).
2.9.3. Measurement of protein loss
4 mm thick SD rat brain coronal sections were cleared with different concentrations (8%, 4%, 2% or 1% w/v) of SDS and SDS‐OPTIClear (n replicates per group = 6) for 3 days at 37ºC. The collected clearing solutions were then prepared with Laemmli buffer (6×) and the prepared samples (48 μl per well) loaded into 10% stain‐free acrylamide gels (Mini‐PROTEAN® TGX, BioRad #1610173). The gel was run at 120 V for about 1.5 hours and visualised using stain‐free‐enabled imager (GelDoc Go Imaging System; BioRad #12009077). Fresh clearing solutions were applied as controls for normalisation. For zebrafish, wild‐type AB zebrafish (Danio rerio, 1‐year‐old) was cleared with 4% SDS alone and 4%SDS with OPTIClear (about 2 ml per fish), respectively (n replicates per group = 3) with different time points (0 day, 2 days, 4 days, or 8 days) at 37ºC. The clearing solutions were collected and prepared with the same protocol as the rat tissues.
2.9.4. Tissue distortion assessment
To assess tissue distortion after clearing, 1 mm‐thick SD rat brain tissues were first stained with TO‐PRO®‐3 (Invitrogen #T3605) (1:100 in PBS) at 37°C overnight. The tissues were then cleared by incubating with the clearing solutions to be tested with triplicates at 37°C overnight, washed briefly with PBS (3 × 10 minutes, at room temperature), and incubated with OPTIClear at room temperature for 6 hours. The tissues were imaged with confocal microscopy at three different time points, namely after TO‐PRO‐3 staining, after tissue delipidation, after RI‐matching with OPTIClear.
To access any distortion of cell morphology after clearing, 1 mm thick tissue sections from Tg(Thy1‐GFP) mice harvested, fixed and washed with PBS as described above. The hippocampal region was imaged under confocal microscopy before delipidation. The imaged tissue was then delipidated with the clearing solutions to be tested until the whole tissue turned visually transparent and imaged for the second time. Finally, the tissues were washed briefly in PBS (3 × 10 minutes, at room temperature), RI‐homogenised with OPTIClear at room temperature for 6 hours and imaged for the final round. During the imaging experiment, we ensured that the imaging setup and orientation were identical for subsequent manual overlay of the neuron of interest.
2.9.5. Magnetic resonance imaging
Rat brain hemispheres were fixed and harvested as described above. The hemispheres were then immobilised in 1% agarose gel in a 50 ml Falcon tube before MRI imaging. After the first round of MRI imaging, the agarose gel was slowly melted at 37°C and the retrieved hemispheres were cleared with the clearing solutions to be tested for about 2 weeks at 37°C, washed with PBS and incubated in OPTIClear before they were immobilised with 1% agarose gel in Falcon tubes and the MRI imaging experiment repeated.
All MRI experiments were performed on a 7 T scanner with a maximum gradient of 360 mT/m (70/16 PharmaScan, Bruker Biospoin GmbH, Germany). DW images were acquired with a spin‐echo single‐shot EPI sequence. A diffusion encoding scheme that consisted of 30 diffusion gradient directions with b‐value of 1200 s/mm2, and five non‐diffusion‐weighted images was used. Other imaging parameters included: TR/TE = 3000/35.7 ms, slice thickness = 1 mm, FOV = 20 mm, data matrix = 128 × 128 and image resolution = 156 × 156 μm2 and number of averages = 3.
2.10. Statistical analysis
Image analysis was performed using Fiji (Image J; NIH), Zen Black (Carl Zeiss) and Zen Blue (Carl Zeiss) software. Maximum intensity projections and z‐depth colour coding were performed using the Zen Blue software. The 3D renderings were performed with Zen Blue or Imaris (Bitplane). Stitching was performed using Zen Blue or Zen Black. Statistical analyses were performed with GraphPad Prism version 7.0.0 (GraphPad Software). All statistics were performed in two‐way ANOVA, with Tukey's multiple comparison test.
3. RESULTS
3.1. The addition of OPTIClear in SDS‐based delipidation solution reduces clearing time and protein loss by partially delipidating tissues
In order to simplify and shorten the delipidation procedure of tissue clearing, we tried to combine the delipidation and RI‐matching procedure into one step, which is to clear tissues with a mixture of sodium dodecyl sulphate (SDS) and RI‐matching solutions. To do so, we screened different RI‐matching solutions by combining SDS with different RI‐matching solutions and observed changes in tissues after clearing. The RI‐matching solutions, RIMS, PROTOS, and SeeDB2S formed precipitates upon mixing with 8% w/v SDS, and thus are not suitable for our purpose. Combining SDS with ScaleS4, OPTIVIEW and CUBICScale2 caused tissue expansion similar to SDS alone Figure S1. Among the tested RI‐matching solutions, OPTIClear was found to work best when paired with SDS, showing good clearing without tissue swelling Figure 1A. Therefore, SDS‐OPTIClear are the ideal candidates for a new delipidation solution.
Figure 1.
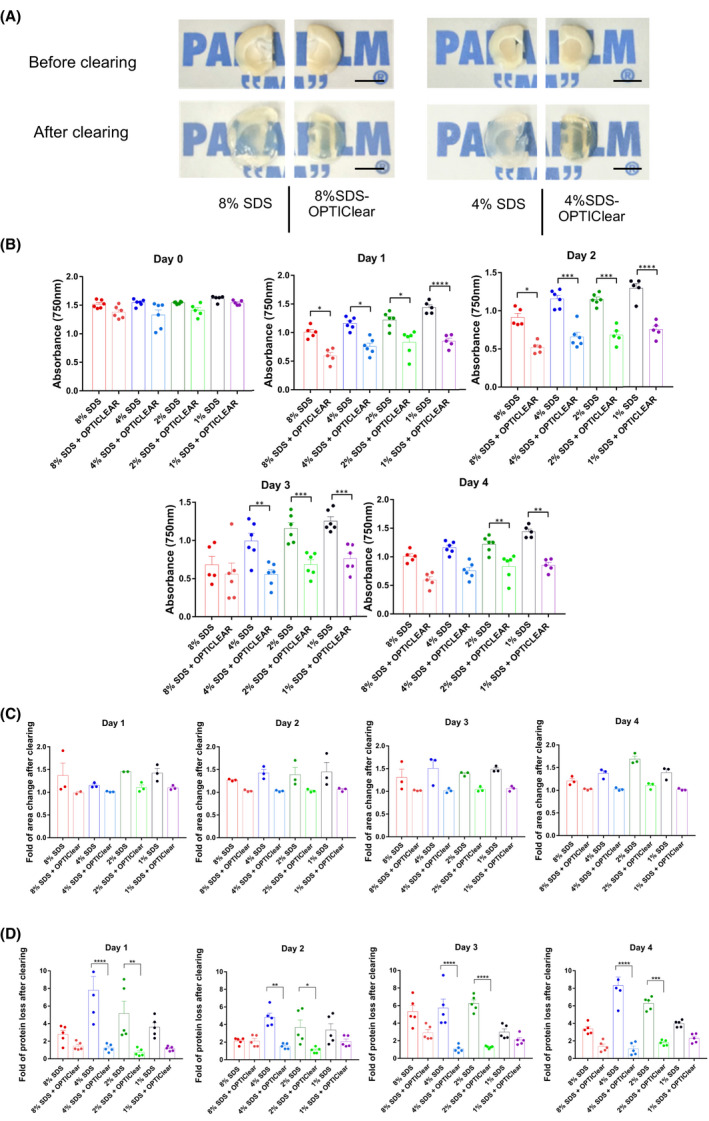
Tissue clearing with different concentrations of SDS vs SDS‐OPTIClear. (A) Representative images of 4 mm‐thick rat brain tissues before and after clearing with SDS and SDS‐OPTIClear respectively. Scale bar = 5 mm. (B) Absorbance at 750 nm for tissues cleared at varying concentrations of SDS vs SDS‐OPTIClear (n = 5–6). Lower absorbance level indicates higher transparency level. (C) Fold of area change after clearing with SDS compared to SDS‐OPTIClear (n = 3). Tissue sizes were quantified and normalised to the size of tissue before clearing. (D) Fold of protein loss after clearing for SDS only compared to SDS‐OPTIClear (n = 5–6). Solutions used for clearing were collected and then added to a stain‐free gel for electrophoresis, band intensities were quantified and normalised with the protein band intensity of sample solution collected at day 0 (before clearing). Mean ± SEM for all graphs. Two‐way ANOVA, Tukey's multiple comparison test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001
To test the clearing effect of adding OPTIClear into SDS‐based delipidation solution, 4 mm thick brain slices were divided and cleared with 1%, 2%, 4% or 8% w/v SDS with or without OPTIClear. The brains were perfused with 4% paraformaldehyde without hydrogel‐embedding treatment. All brain slices were cleared at 37°C. We then compared different parameters, including the degree of transparency Figure 1B, tissue size by area Figure 1C and protein loss after clearing Figure 1D. The transparency level was quantified by measuring the absorbance level at 750 nm, a wavelength along the infrared region to avoid interference from the natural yellow hue of cleared tissues. Low absorbance correlated with highly transparent tissue. We found that tissues cleared with SDS‐OPTIClear cleared faster than SDS alone, regardless of SDS concentration Figure 1A,B). By day 1, the absorbance level of tissues cleared with SDS‐OPTIClear has already reduced to almost half of the absorbance level before clearing. The lowest absorbance level was observed at day 2 in the 8% SDS‐OPTIClear samples and day 3 in the 4% SDS‐OPTIClear samples. In contrast, the 8% and 4% SDS‐only groups had significantly higher absorbance levels compared to their counterparts with OPTIClear even at day 3 and beyond, which would lead to longer delipidation.
Furthermore, we observed that there was no significant change in the tissue size throughout the clearing process in the SDS‐OPTIClear group Figure 1C. To assess the protein loss, we tried to perform quantitative protein assays such as the BCA assay but failed due to the reaction of OPTIClear with the BCA reagents. Therefore, gel electrophoresis of used clearing solutions was performed. In the SDS only groups, protein loss was positively correlated with the SDS concentration used, except in the 8% SDS group. However, with the use of SDS‐OPTIClear, protein loss was significantly less than that with SDS alone Figure 1D. By the end of day 4, the protein loss level was about eightfold higher in 4% SDS only groups, while in 4% SDS‐OPTIClear, the protein loss level remained constantly low throughout the clearing process.
The shortened duration of SDS treatment suggests that the tissue was partially rather than completely delipidated in the SDS‐OPTIClear group. Inspired by Leuze et al. who have previously demonstrated that lipid removal by CLARITY drastically reduced the diffusion coefficient and anisotropy on magnetic resonance imaging (MRI) [19], we performed MRI on pre‐ and post‐delipidated tissues by SDS‐OPTIClear vs SDS. We found more residual lipids in the SDS‐OPTIClear group Fig S2.To further validate the partial delipidation in OPTIClear‐mediated clearing, 1 mm thick tissues were stained with the lipophilic dye DiI for 1 day before clearing with either 4% SDS‐OPTIClear, 8% SDS‐OPTIClear, 4% SDS only or 8% SDS only Fig. S3. DiI fluorescence signals were well preserved in both PBS and OPTIClear controls,they were obviously diminished in tissues cleared with SDS only under identical imaging settings. In contrast, the DiI signal of tissues cleared using SDS‐OPTIClear was largely preserved, consistent with the MRI results that only partial delipidation was required to achieve full transparency during OPTIClear‐mediated clearing.
3.2. Optimised delipidation strategy better protects tissue morphology and antigenicity
Although tissues did not expand during clearing with SDS‐OPTIClear, further experiments were needed to assess the morphological changes during clearing. Treweek et al. observed transient morphological changes using SDS in which treated tissues swell before shrinking back to normal size after RI‐matching [15]. However, this kind of shrinking back to original size after RI‐matching may be incomplete and could further distort tissue structure. To assess the deformation of tissue architecture after clearing, tissues were first stained with TO‐PRO®‐3 [3, 4] and cleared with either 4% SDS‐OPTIClear, 8% SDS‐OPTIClear, 4% SDS only or 8% SDS only before RI‐matching. Non‐SDS treated tissues incubated with PBS or OPTIClear were used as controls Figure 2A.
Figure 2.
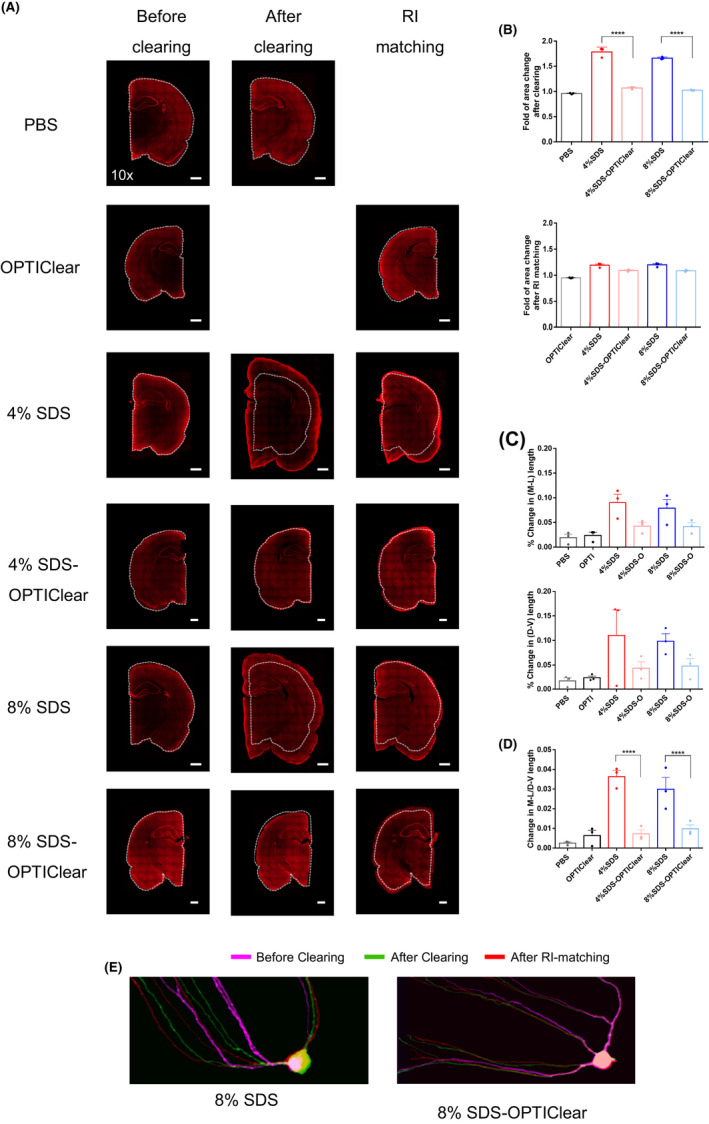
Tissue distortion level after clearing with different conditions. 1 mm‐thick rat tissues were stained with TO‐PRO®‐3 and then imaged before delipidation, after delipidation and after RI‐matching in OPTIClear. (A) Representative images of TO‐PRO®‐3 stained tissues under different stages of clearing. White dotted lines marked the original boundary of tissues. Scale bars = 1 mm (n = 3). (B) Fold tissue area changes after tissue clearing and RI‐matching, respectively. (C) Percentage changes in dorsal–ventral (D‐V) and mediolateral (M‐L) lengths after RI‐matching respectively (n = 3). (D) Percentage changes in the ratio of D‐V and M‐L length after RI‐matching (n = 3). (E) Merged images of GFP‐labelled hippocampal neurons under different stages of clearing. The same neuron was imaged before and after clearing, and after RI‐matching. Images were labelled with different colours (see Fig. S3 and merged together. Higher degree of overlapping indicates lower distortion level in morphology. Scale bars = 20 µm. Mean ±S.E.M. for all graphs. Two‐way ANOVA, Tukey's multiple comparison test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001
Consistent with others’ observations, while tissues were greatly expanded after clearing with SDS alone, they shrunk back to size after RI‐matching with OPTIClear Figure 2B. However, the ratio of medial–lateral (M‐L) and dorsoventral (D‐V) dimensions (see methods) was changed significantly. Such structural distortions can lead to errors during image analysis Figure 2C. In contrast to SDS alone, tissues cleared using SDS‐OPTIClear managed to maintain their tissue morphology throughout the process. The results suggest that adding OPTIClear to SDS not only avoids over‐delipidation and tissue damage, it also protects tissues from expansion and deformation.
To further assess the deformation of cell morphology during clearing, tissues from Tg(Thy1‐GFP) mice were cleared with either 8% SDS only or 8% SDS‐OPTIClear before RI‐matching. The images taken during different stages of clearing were imaged at the same orientation with identical imaging setups and manually overlaid to demonstrate the level of distortion in morphology Figure 2E, Fig. S4, with the more overlapping of the signal, the higher degree of preservation of cell morphology. Consistent with the observations in TO‐PRO®‐3 staining, while neurons were expanded after clearing with SDS alone, they could not shrink back to the original shape after RI‐matching with OPTIClear. On the contrary, neurons cleared with SDS‐OPTIClear maintained the cell morphology throughout the clearing and RI‐matching. This result further suggested that SDS‐OPTIClear prevented tissues from expansion and thus distortion in cell morphology.
Next, we tested whether immunostaining would be affected by such this clearing strategy. Previously, we reported the failure of immunofluorescence staining with the use of antibodies including anti‐Choline acetyltransferase (ChAT) and Iba‐1 after SDS treatment [14]. This could be due to antigen destruction or loss caused by excessive SDS treatment. In Iba‐1, ChAT and TOMM20‐stained rat hippocampus cleared with 4% SDS‐OPTIClear, immunofluorescence had higher signal‐to‐background ratios compared to that cleared with 4% SDS only Figure 3, Fig. S5, and microglia morphology was better demonstrated Figure 3A. Intriguingly, in these cases, the 4% SDS‐OPTIClear group achieved better antibody penetration depths than 4% SDS Figure 3 despite partial delipidation, even though tissues treated with OPTIClear expectedly resulted in superficial staining Fig. S6. For example, Iba‐1 staining was barely observed after clearing with 4% SDS alone. However, the fluorescent signal can be visualised up to 150 µm in 4% SDS‐OPTIClear. To further validate the improvement in immunostaining, staining with different antibodies including amyloid‐β (Aβ), glia fibrillary acidic protein (GFAP), Zonula occludens‐1 (ZO‐1) and synaptic markers synaptophysin and synapsin‐1 was performed Fig. S7. While Aβ and ZO‐1 staining failed in tissues treated with SDS only, all antibodies were compatible with the 4% SDS‐OPTIClear treatment, with similar signal intensities compared with the ones cleared with SDS alone.
Figure 3.
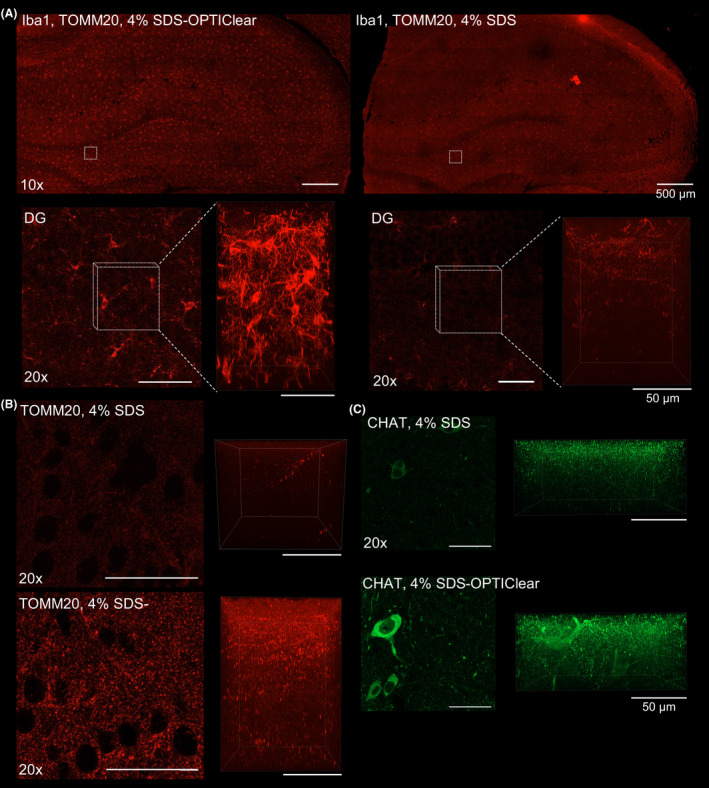
Comparison between 3D immunostaining of tissue cleared with SDS and SDS‐OPTIClear. (A) Iba‐1 staining of tissues cleared with 4% SDS‐OPTIClear (left panel) and 4% SDS only (right panel). Upper panel: images taken with a 10× objective. Lower panel: zoomed‐in image of the dentate gyrus (DG) taken with a 20× objective (left image), Z‐stack images taken with a 63× objective with z‐depth = 161.7 μm (right image). (B) TOMM‐20 staining of regions cleared with 4% SDS‐OPTIClear (left panel) and 4% SDS only (right panel). Left image: 20× images of DG regions. Right image: z‐stack of the images showed on the left (z‐depth = 125.84 μm). (C) ChAT staining taken with a 20× objective. Left panel: 20× images of the nbM cleared with 4% SDS and 4% SDS‐OPTIClear. Right panel: z‐stack of the images showed in the left panel (z‐depth = 72 μm). The same imaging and display settings were used for both 4% SDS and 4% SDS with OPTIClear conditions
3.3. The Accu‐OptiClearing strategy is compatible with other detergents and protocols
We next asked if this strategy of merging delipidation and RI‐matching step can be generalised to other detergents and protocols. While SDS has been widely used in various protocols [1, 2, 6, 11], other detergents have been tried, including 1,2‐Hexandiol (HxD) and Tween®20, which are used in CUBIC‐based clearing protocols [20, 21], and CHAPS, which is used in SHANEL [22]. Therefore, we combined OPTIClear with detergents including HxD, Tween‐20 and CHAPS and compared with conditions of the corresponding detergents alone. To do so, 2 mm thick coronal rat brain slices were divided into identical halves along the sagittal midline and cleared with 10% HxD‐OPTIClear, 10% Tween‐20‐OPTIClear, 10% CHAPS‐OPTIClear and the corresponding detergents only respectively (one brain slice per reagent tested). After 3 days of incubation with clearing solutions at 37ºC, an improvement in the clearing efficiency was evident compared to the corresponding detergents alone Figure 4A, suggesting that our strategy is generalisable to other clearing methods. This strategy is also generalisable to larger intact samples. For example, half rat brain hemispheres were cleared with SDS‐OPTIClear and HxD‐OPTIClear, respectively, at 37ºC, with a renewal of solutions twice per week. After 14 days, HxD‐OPTIClear cleared half brains better than SDS‐OPTIClear with little tissue swelling Figure 4B. Exploring detergents alternatives or combinations thereof may further improve clearing speed, efficacy and preservation of tissue structure and antigens.
Figure 4.
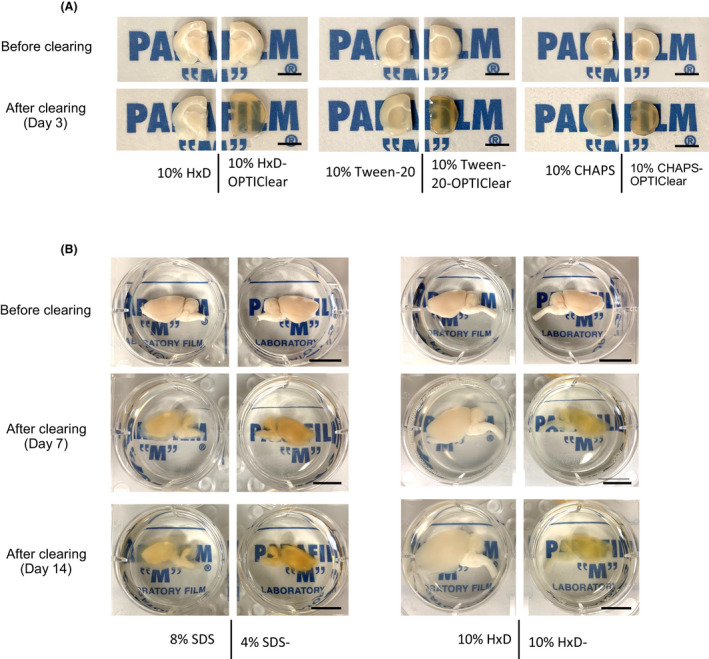
Comparison of clearing efficiency between combinations of OPTIClear with different detergents. (A) Representative images of tissues before and after clearing (scale bar = 5 mm) (B) Comparison of clearing efficiency between combinations of OPTIClear with SDS and 1,2‐Hexandiol (HxD). Representative images of rat brain hemispheres before and after clearing were shown. (scale bar = 10 mm)
3.4. Applications of Accu‐OptiClearing in whole‐mount adult zebrafish and rodent brain tissues
One of the advantages of tissue clearing is the ability to visualise targets in a comprehensive three‐dimensional manner without the need for serial sectioning. With Accu‐OptiClearing, whole adult zebrafish can be cleared with significantly shorter time and less protein loss Fig. S8 after physically removing the cuticle. Using the Tg(fli1a:EGFP) y1 fish line, it is possible to image whole zebrafish brain and spinal cord vessels labelled with EGFP up to 400 µm deep under confocal microscopy Figure 5A,B. By performing whole‐mount immunofluorescence using anti‐tyrosine hydroxylase (TH) antibodies, TH‐positive neurons can also be visualised in the harvested whole brain with a penetration depth of over 500 µm Figure 5C.
Figure 5.
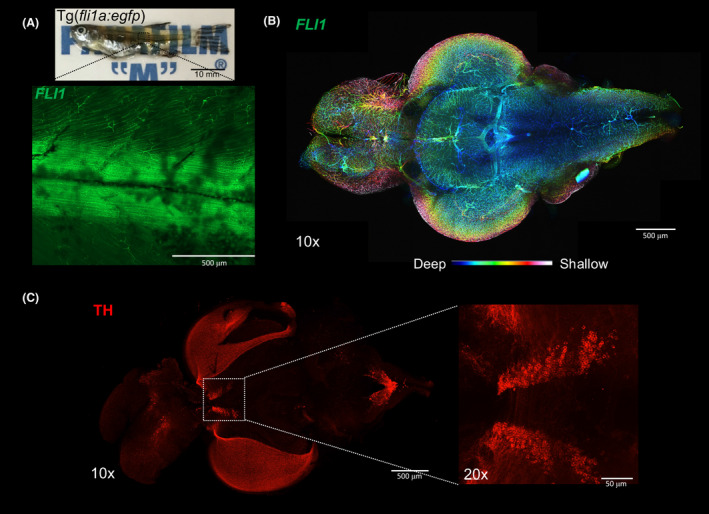
Zebrafish (Danio rerio) imaging after tissue clearing. (A) Whole Tg(fli1a:EGFP) y1 (1.5 years old) after clearing with 4% SDS‐OPTIClear, scale bar = 10 mm. Zoomed‐in 10× image showing EGFP fluorescence in blood vessels. (B)Colour‐coded image labelling the blood vessels in whole zebrafish brain, scale bar = 500 μm. (C) Whole brain immunostained with antibodies against tyrosine hydroxylase, scale bar = 500 μm, higher magnification (20×) showing posterior tuberculum, scale bar = 50 μm
Similarly, endogenous fluorescence and immunofluorescence are equally compatible with mouse tissues cleared with Accu‐OptiClearing. Phosphorylated tau can be readily visualised via 3D immunofluorescence in triple transgenic mice (3xTg), an AD model known for the presence of both Amyloid‐beta (Aβ) plaques and neurofibrillary tangles (NFT) Figure 6A–C. Using AAV‐GFP viral tracing, fine morphological details of GFP‐labelled magnocellular cholinergic neurons in the rat nucleus basalis of Meynert (nbM) can be visualised Figure 6D–F, with axonal projections up to 3,000 µm in length. Through this method, the cellular morphology and projections from nbM were clearly visible. Individual neurons can be isolated for further analysis of fibre lengths and morphology at high resolution and with great accuracy Fig. S9, Movie S1). Compared to traditional serial sectioning and staining, this approach provides a more comprehensive anatomical framework for understanding neuronal networks.
Figure 6.
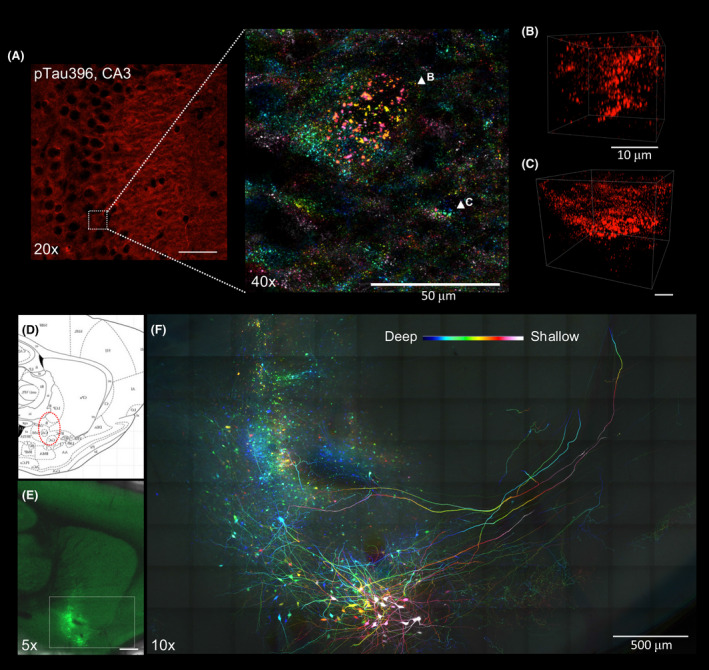
Applications of Accu‐OptiClearing in rodent brain tissues. Visualisation of phosphorylated tau aggregation in 3xTg mice. (A) Image of the hippocampal CA3 region stained with anti‐phosphorylated tau antibody. Inset showing colour‐coded projection of phosphorylated tau‐positive aggregations (indicated by arrowheads in B and C). Scale bar, 50 μm (for A), 10 μm (for B and C). Long‐range tracing of rat nucleus basalis of Meynert neurons with AAV‐GFP. (D) Sagittal rat brain atlas indicating the nbM region in red dotted circle. (E) Tiled Z‐stack image showing AAV‐GFP‐labelled nbM neurons three weeks after injection into the nbM region (scale bar = 1 mm). (F)Colour‐coded projection image with higher magnification of the region shown in E (scale bar = 500 μm, z‐depth = 205 μm)
4. DISCUSSION
In recent years, tissue clearing has emerged as a promising approach to image tissue structures and molecules in their native 3D context. However, the problem of tissue distortion and protein loss during clearing made the anatomical data generated by these new techniques questionable. Thus, turning tissues transparent with minimal deformation and protein loss has been a key consideration when optimising a clearing technique. Although numerous optimisations and massive chemical screening for other detergents have been reported [20], antigen loss and tissue swelling were still prominent. Attempts at tissue protection prior to delipidation introduced technical constraints with complex protocols and created diffusion barriers to molecular probes [1, 6, 11].
Here, we introduce a novel strategy that avoids excessive delipidation while achieving the same transparency level, by combining the delipidation and RI‐matching procedure into one step. With the combination of SDS‐OPTIClear, a RI‐matching solution that has been proven to render human brain tissue transparent with minimal structural disturbance [14], we demonstrated multiple benefits on increased clearing speed, reduced tissue destruction and distortion and enhanced immunostaining quality. Although tissue swelling/shrinkage can be favourable for the purpose of shortening the imaging time [4] or magnifying fine structures for higher resolution imaging [23], here we present a complementary clearing protocol that minimises the tissue swelling or shrinkage throughout the clearing process, such that the original tissue architecture can be preserved and investigated when desired.
We also demonstrated that complete tissue delipidation is not necessary to achieve full transparency. MRI studies and DiI staining suggest that lipids were not completely removed from the tissues, supporting our view that partial delipidation can be adequate for achieving the necessary tissue transparency for imaging and permeabilisation for probe penetration. Importantly, excessive delipidation should be achieved to avoid damages to tissue structure and antigens. Our strategy provides a convenient visual readout for the appropriate endpoint of partial delipidation. The Accu‐OptiClearing strategy can also be generalised to other detergents, including HxD, Tween®20 and CHAPS, and adaptable to a wide range of tissue types. We also showed an improvement in clearing efficacy when replacing SDS with HxD in clearing rat brain hemispheres, suggesting the Accu‐OptiClearing strategy can be further optimised with different detergents. Interestingly, HxD has been previously reported for its ability to preserve lipoproteins of the myelin sheath during clearing [21]. As OPTIClear is also compatible with lipophilic tracers, the combination of HxD and OPTIClear may facilitate myelin studies in 3D.
We demonstrated that our Accu‐OptiClearing strategy is directly applicable to scientifically relevant imaging studies of the brain in different species. In zebrafish (Danio rerio), we have efficient clearing with better preservation of proteins compared to conventional delipidation. In mice, we managed to visualise phosphorylated tau‐positive aggregations in AD disease mouse models in 3D. Accu‐OptiClearing can also be combined with viral tracers to visualise neural circuits and connectomes, as demonstrated in this study for tracing cholinergic neuron projections in the nbM.
Although clearing with Accu‐OptiClearing promotes clearing efficiency and preserves tissue morphology, limited antibody penetration remains a barrier to deep 3D tissue imaging. Also, we did not validate our protocol for its application to human brain tissue. Since OPTIClear is a RI‐matching agent optimised for brain tissues for compartment‐specific RI adjustment, its formulation may require adjustments when one applies Accu‐OptiClearing to other tissues with less lipid than brain tissue. Finally, in pigment‐rich tissues such as muscle and kidney, Accu‐OptiClearing may not provide the expected benefits as light is also blocked by absorption.
In summary, we have developed the Accu‐OptiClearing strategy as a general solution to balance the benefits of tissue delipidation and risks of damaging tissue antigens and structures. In coordination with advances in microscopy and tissue clearing chemistry, the Accu‐OptiClearing strategy is applicable in both research laboratories and clinical settings for post‐mortem 3D tissue analysis.
5. DESCRIPTION OF ETHICAL APPROVAL
All animals used in this study were approved and handled in accordance with the guidelines provided by the Committee on the Use of Live Animals in Teaching and Research (CULATR, 4014‐16) in our University, which is accredited by the AAALAC.
CONFLICT OF INTEREST
None of the authors have a conflict of interest.
AUTHORS’ CONTRIBUTIONS
HML conceived the Accu‐OptiClearing idea. HML, KL and RCCC conceived the experimental designs. KL and MHS performed tissue clearing experiments.MHS performed intracerebral injection. KL performed confocal imaging of cleared tissues. KL, VWSM and WCSC performed and supervised immunofluorescent staining. ESKH performed magnetic resonance imaging. KL, HML, KEW and YSH analysed the data. HML and RCCC supervised the experiments. RCCC provided financial support. All authors contributed significantly to the discussion on the experimental work and writing of the manuscript.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1111/nan.12673.
Supporting information
Fig S1‐S9
Table S1‐S2
Video S1
ACKNOWLEDGEMENTS
The study was supported by Innovative and Technology Fund (ITS/381/15, InP/220/16, InP/248/16, InP/172/17).
Lee K, Lai HM, Soerensen MH, et al. Optimised tissue clearing minimises distortion and destruction during tissue delipidation. Neuropathol Appl Neurobiol.2021;47:441–453. 10.1111/nan.12673
Krit Lee and Hei Ming Lai have equal contributions to the study.
Contributor Information
Hei Ming Lai, Email: hmlai@cuhk.edu.hk.
Raymond Chuen‐Chung Chang, Email: rccchang@hku.hk.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Chung K, Wallace J, Kim S‐Y, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013; 497(7449): 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang B, Treweek JB, Kulkarni RP, et al. Single‐cell phenotyping within transparent intact tissue through whole‐body clearing. Cell. 2014; 158(4): 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier‐Lavigne M. IDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014; 159(4): 896–910 [DOI] [PubMed] [Google Scholar]
- 4. Pan C, Cai R, Quacquarelli FP, et al. Shrinkage‐mediated imaging of entire organs and organisms using uDISCO. Nat Methods. 2016; 13(10): 859–867 [DOI] [PubMed] [Google Scholar]
- 5. Murakami TC, Mano T, Saikawa S, et al. A three‐dimensional single‐cell‐resolution whole‐brain atlas using CUBIC‐X expansion microscopy and tissue clearing. Nat Neurosci. 2018; 21(4): 625–637 [DOI] [PubMed] [Google Scholar]
- 6. Park Y‐G, Sohn CH, Chen R, et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat Biotechnol. 2019; 37(1): 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hama H, Hioki H, Namiki K, et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci. 2015; 18(10): 1518–1529 [DOI] [PubMed] [Google Scholar]
- 8. Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology‐preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013; 16(8): 1154–1161 [DOI] [PubMed] [Google Scholar]
- 9. Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: a detergent‐ and solvent‐free clearing method for neuronal and non‐neuronal tissue. Development. 2013; 140(6): 1364–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qi Y, Yu T, Xu J, et al. FDISCO: advanced solvent‐based clearing method for imaging whole organs. Sci Adv. 2019;5(1):eaau8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murray E, Cho JH, Goodwin D, et al. Simple, scalable proteomic imaging for high‐dimensional profiling of intact systems. Cell. 2015; 163(6): 1500–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee E, Choi J, Jo Y, et al. ACT‐PRESTO: rapid and consistent tissue clearing and labeling method for 3‐dimensional (3D) imaging. Sci Rep. 2016; 6(1): 18631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu AKL, Hurry MED, Ng OTW, et al. Bringing CLARITY to the human brain: visualization of Lewy pathology in three dimensions. NeuropatholApplNeurobiol. 2016; 42(6): 573–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lai HM, Liu AKL, Ng HHM, et al. Next generation histology methods for three‐dimensional imaging of fresh and archival human brain tissues. Nat Commun. 2018; 9(1): 1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Treweek JB, Chan KY, Flytzanis NC, et al. Whole‐body tissue stabilization and selective extractions via tissue‐hydrogel hybrids for high‐resolution intact circuit mapping and phenotyping. Nat Protoc. 2015; 10(11): 1860–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ke M‐T, Nakai Y, Fujimoto S, et al. Super‐resolution mapping of neuronal circuitry with an index‐optimized clearing agent. Cell Rep. 2016; 14(11): 2718–2732 [DOI] [PubMed] [Google Scholar]
- 17. Isogai Y, Richardson DS, Dulac C, Bergan J. Optimized protocol for imaging cleared neural tissues using light microscopy. Methods Mol Biol. 2017; 137–153 [DOI] [PubMed] [Google Scholar]
- 18. Susaki EA, Tainaka K, Perrin D, et al. Whole‐brain imaging with single‐cell resolution using chemical cocktails and computational analysis. Cell. 2014; 157(3): 726–739 [DOI] [PubMed] [Google Scholar]
- 19. Leuze C, Aswendt M, Ferenczi E, et al. The separate effects of lipids and proteins on brain MRI contrast revealed through tissue clearing. NeuroImage. 2017; 156:412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tainaka K, Murakami TC, Susaki EA, et al. Chemical Landscape For Tissue Clearing Based On Hydrophilic Reagents. Cell Rep. 2018; 24(8): 2196–2210.e9 [DOI] [PubMed] [Google Scholar]
- 21. Inoue M, Saito R, Kakita A, Tainaka K. Rapid chemical clearing of white matter in the post‐mortem human brain by 1,2‐hexanediol delipidation. Bioorganic Med Chem Lett. 2019; 29(15): 1886–1890 [DOI] [PubMed] [Google Scholar]
- 22. Zhao S, Todorov MI, Cai R, et al. Cellular and molecular probing of intact human organs. Cell. 2020; 180(4): 796–812.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ku T, Swaney J, Park J‐Y, et al. Multiplexed and scalable super‐resolution imaging of three‐dimensional protein localization in size‐adjustable tissues. Nat Biotechnol. 2016; 34(9): 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S9
Table S1‐S2
Video S1
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


