Abstract
The World Health Organization guidelines recommend that individuals living with HIV receive ≥ 6 months of isoniazid preventive therapy, including pregnant women. Yet, plasma isoniazid exposure during pregnancy, in the antiretroviral therapy era, has not been well‐described. We investigated pregnancy‐induced and pharmacogenetic‐associated pharmacokinetic changes and drug‐drug interactions between isoniazid and efavirenz in pregnant women. Eight hundred forty‐seven women received isoniazid for 28 weeks, either during pregnancy or at 12 weeks postpartum, and 786 women received efavirenz. After adjusting for NAT2 and CYP2B6 genotype and weight, pregnancy increased isoniazid and efavirenz clearance by 26% and 15%, respectively. Isoniazid decreased efavirenz clearance by 7% in CYP2B6 normal metabolizers and 13% in slow and intermediate metabolizers. Overall, both isoniazid and efavirenz exposures were reduced during pregnancy, but the main determinants of drug concentration were NAT2 and CYP2B6 genotypes, which resulted in a five‐fold difference for both drugs between rapid and slow metabolizers.
Study Highlight.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Genetic variability greatly impacts isoniazid and efavirenz exposure, and it may affect interactions between these two commonly co‐administered drugs. Very little knowledge is available on the effect of pregnancy on their pharmacokinetics (PKs).
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ We investigated the magnitude of pregnancy‐induced changes in isoniazid and efavirenz PKs, accounting for drug‐drug interactions and pharmacogenetics.
WHAT DOES THIS STUDY ADDS TO OUR KNOWLEDGE?
☑ Pregnancy increases clearance of isoniazid (by 26%) and efavirenz (by 15%), reducing both drug’s exposures. Isoniazid co‐administration reduces efavirenz clearance, and this reduction is larger in slow and intermediate CYP2B6 metabolizer.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ The modest pregnancy‐related decreases in both drugs exposures may be of clinical importance in rapid NAT2 acetylator or CYP2B6 metabolizer, who already experience low exposure and are, hence, at risk of treatment failure. Isoniazid‐induced increase in efavirenz exposure may raise the chance of toxicity among slow CYP2B6 metabolizers. Both findings suggest potential role for genetic testing and therapeutic drug monitoring.
HIV and tuberculosis are leading causes of morbidity and mortality among women of reproductive age, particularly among those living in low‐income and middle‐income countries. 1 , 2 Pregnant women living with HIV have an increased susceptibility to tuberculosis (TB) infection and progression from latent TB infection to active disease. 3 To reduce the risk of HIV‐associated TB, the World Health Organization (WHO) recommends 6 months of isoniazid preventive treatment (IPT) along with antiretroviral therapy (ART). 4 This recommendation was also made for pregnant women, based on limited data. Both antiretrovirals and anti‐TB regimens often include drugs that inhibit or induce liver enzymes, including cytochrome‐P450 (CYP) isoenzymes, causing potential drug‐drug interactions (DDIs). 5 The situation is further complicated in pregnancy, which is associated with physiological changes, altered body weight and composition, changes in plasma protein concentrations, and other processes that can affect drug pharmacokinetics (PKs). 6 , 7 , 8 , 9
The metabolism of both efavirenz (a commonly used antiretroviral to treat HIV) and isoniazid are affected by genetic polymorphisms in specific drug‐metabolizing genes, and plasma drug levels can be further influenced by pregnancy. Studies to date, although small in number of participants and limited in geographic diversity have shown, a predicted two‐fold increase in CYP2B6 activity during pregnancy. 7 Dooley et al. 10 observed a 19% increase in plasma efavirenz clearance during pregnancy compared with the postpartum period, and Olagunju et al. 11 reported similar findings. Furthermore, an ancillary metabolic pathway of efavirenz, CYP2A6, is inhibited by isoniazid, thus potentially causing DDIs. 12 There is currently little data regarding the effect of pharmacogenomics and pregnancy on isoniazid PKs and on the DDI between isoniazid and efavirenz in pregnancy and postpartum.
As part of the recently completely IMPAACT study, P1078 TB APPRISE trial, which investigated the safety and efficacy of administering IPT to pregnant women living with HIV, and reported a significantly higher rate of adverse pregnancy outcomes in women who received IPT during pregnancy, 13 we analyzed the key pharmacogenetic polymorphisms of efavirenz and isoniazid metabolism, the changes in the drug concentrations and PKs of these two drugs during pregnancy, and their DDIs.
METHODS
Participants and study design
This analysis used PK data collected in IMPAACT study P1078. The study design is reported in the primary manuscript 13 summarized here, and we report the PK procedure. The IMPAACT study P1078 was a prospective, double‐blind, placebo‐controlled, randomized, noninferiority trial conducted in 8 countries, and had a total of 13 sites. Local and collaborating institutional review boards approved the trial, and all women provided informed consent. Full details on ethics approval are provided in the primary manuscript. 13
HIV‐positive pregnant women ≥ 18 years old were recruited if gestational age was 14 to 34 weeks, on WHO‐recommended HIV treatment for prevention of mother‐to‐child transmission, weighing > 35 kg. At study entry, women were randomized either to arm A (immediate 28 weeks of 300‐mg isoniazid daily treatment, then placebo) or arm B (deferred isoniazid treatment, placebo until week 12 postpartum, then 28 weeks of 300 mg isoniazid). Adherence to isoniazid (or placebo) was monitored by self‐report and pill count defined as the percentage of expected doses/pills taken during the entire treatment period, whereas ART adherence was self‐reported.
Sample collection
Intensive samples were captured at predose, 1, 2, 4, 6, 8, and 12 hours postdosing, whereas sparse sampling occurred at least 2 hours postdosing. Both sparse and intensive samples were captured on two visits; during pregnancy (between 28 and 40 weeks’ gestation) and week 16 postpartum (+ 4 weeks). Blood samples were centrifuged at 800 G for 10 minutes, and plasma was aliquoted into cryovials and immediately frozen at −70°C to await further processing. Plasma concentrations were determined by liquid chromatography‐tandem mass spectrometry. Isoniazid calibration ranged from 0.105 to 25.0 mg/L, the interday accuracy and precision ranged from 92.2% to 104.5%, and 6.5% to 10.8%, respectively. Efavirenz calibration ranged from 0.0195 to 20.0 mg/L, the interday accuracy and precision ranged from 95.2% to 100.2%, and 3.2% to 11.3%, respectively.
Composite CYP2B6 genotype was defined based on combinations of four polymorphisms as follows: normal (1: 15582CC‐516GG‐983TT or 2: 15582CT‐516GG‐983TT); intermediate (3: 15582TT‐516GG‐983TT; 4: 15582CC‐516GT‐983TT; 5: 15582CC‐516GG‐983CT; 6: 15582CT‐516GT‐983TT; or 7: 15582CT‐516GG‐983CT); and slow metabolizer genotype (8: 15582CC‐516TT‐983TT; 9: 15582CC‐516GT‐983CT; 10: 15582CC‐516GG‐983CC, each with or without 11: −48GT; and 12: −48GG). 14 Individuals with 983CC were considered ultra‐slow metabolizers. (The term “normal” is used for consistency with standard nomenclature, not to suggest that others are abnormal.) For NAT2, genotypes were categorized based on combinations of rs1801279 (NAT2*14), rs1801280 (NAT2*5), rs1799930 (NAT2*6), and rs1799931 (NAT2*7), as slow, homozygous for the variant allele at any of the four loci (i.e., AA, CC, AA, and AA, respectively), or heterozygous at two or more loci; intermediate, heterozygous at a single locus; or rapid, no variant allele at any locus (i.e., GG, TT, GG, and GG, respectively). 15 , 16 Genotyping was done in Vanderbilt Technology for Advanced Genomics (VANTAGE) using MassARRAY iPLEX Gold (Agena Bioscience, San Diego, CA) and Taqman (ThermoFisher Scientific, Waltham, MA).
Data analysis
Isoniazid and efavirenz concentrations were interpreted using population PK modeling using NONMEM version 7.4.3. 17 Perl‐speaks‐NONMEM version 4.8.1, Pirana, and R with the package Xpose4 were used to facilitate the model development process, data manipulation, and generation of model diagnostics. 18 To describe the PK of both isoniazid and efavirenz, one‐compartment and two‐compartment models were tested with first‐order absorption (with or without lag time or a chain of transit compartments), and first‐order elimination. Because both drugs are mainly hepatically cleared, a well‐stirred liver model 19 was tested to capture the effect of first‐pass metabolism. The liver hepatic blood flow Q h 20 was assumed to be 90 L/h in a typical individual and adjusted for the effect of body size using allometric scaling. 21 The free fraction of efavirenz and isoniazid in plasma were fixed to 0.5% 22 and 95%, 23 respectively. The prehepatic bioavailability of a typical individual was fixed to a reference value of 1.
The intensive data was used to develop the base model because these women were monitored closely, dosing was observed, and richer sampling schedule allowed for the identification of the structural model. This model was then used to explore the sparse data and identify implausible values, which were flagged as outliers and removed from model development analysis. An extreme value of conditional weighted residuals 24 (i.e., absolute value ≥ 4) was used as a criterion to identify outliers. Because conditional weighted residuals values for data observations arising from the postulated model are assumed to be normally distributed, randomly obtaining an absolute value ≥ 4 has a probability of < 0.00633%, only a chance of 1 of 10,000. We assumed that PKs of both drugs were at steady‐state at the time of the PK visit unless the participant had been on efavirenz for < 16 days, 25 or missed dose in the previous 3 days had been reported.
Random effects at occasion‐level (each dose), visit‐level (each PK sampling visit), and/or subject‐level were included on the PK parameters if statistically significant using a lognormal distribution. Between‐occasion variability was tested on absorption parameters, whereas between‐visit variability and between‐subject variability (BSV) were tested on disposition parameters. A combination of proportional and additive error was used to model unexplained residual variability.
Model development was guided by evaluating the drops in objective function value (ΔOFV) of nested models. The OFV was assumed as χ2‐distributed, hence with a 3.84 drop in OFV being significant at P < 0.05 for one additional parameter (i.e., 1 degree of freedom). Besides statistical significance, a set of diagnostic plots, including visual prediction checks and physiological plausibility of the results were considered in model development decisions.
Because enzyme metabolic status has been widely reported to critically affect PKs of both drugs, NAT2 genotype effect on isoniazid clearance and CYP2B6 and CYP2A6 genotype on efavirenz clearance were tested early in the model development process. 12 , 26 Allometric scaling was applied on all clearance and volume of distribution parameters 27 to account for body size effect. Other covariate effects on the PK parameters, including pregnancy and DDI, were investigated using a stepwise approach with forward inclusion (P < 0.05) and backward elimination (P < 0.01).
Concentrations below limit of quantification were handled similarly to the M6 method by Beal, 28 to mitigate the effect of the M6 method imputation the additive error for the imputed values was inflated by lower limit of quantification/2. Only a subset of participants was included in the genotype study, those with missing genotypic information were assigned a phenotype using the mixture model. 29 Participants with missing weight and height were assigned the typical values depending on whether pregnant or postpartum. The original protocol focused on IPT, hence did not include capturing ART PK information (i.e., efavirenz dosing time) for sparse sampling. This was later amended, resulting in 32% of the women having a missing efavirenz dosing time in at least one PK visit during the early phase of the trial. Women with missing dose times during one visit were assumed to take medication at the same time throughout the trial consistently; therefore, dosing time of the recorded visit was imputed on the missing visit. For women with missing dosing time at both visits, a median dosing time (i.e., 20:30) from the data was imputed.
RESULTS
Study profile
PK samples were available from 847 women on 300 mg isoniazid (748 of whom were treated with efavirenz (600 mg dose), 80 with nevirapine, 17 with lopinavir, and 2 with atazanavir based ART. Of the 847 on isoniazid, 32 underwent intensive sampling (at predose, 1, 2, 4, 6, 8, and 12 hours postdosing), providing 300 observations, whereas 815 underwent sparse sampling (at least 2 hours postdosing), providing 1,015 observations, of which 88 were identified as implausible outliers (outliers were identified per the description in Methods) and were removed from the model‐building process. Two hundred ten women had isoniazid profiles available during both pregnancy and postpartum, because they had started isoniazid at a late gestational age such that their 28 weeks of isoniazid had not elapsed prior to their postpartum PK visit.
The number of participants on efavirenz in the different arms (isoniazid or placebo treatment) varied because of ART switching over the course of the study, but the majority were on efavirenz. Of 786 women on efavirenz, 21 underwent intensive sampling, providing 266 observations, and 765 underwent sparse sampling, providing 1,363 observations, of which 15 were identified as implausible outliers (outliers were identified per the description in Methods) and were removed from the model‐building process. Baseline characteristics of all women are summarized in Table 1 .
Table 1.
Demographic, clinical, and laboratory characteristics of women on isoniazid and efavirenz during pregnancy and at postpartum
| Characteristics (median and range, or n and %) | Participants in isoniazid PK analysis | Participants in efavirenz PK analysis | ||
|---|---|---|---|---|
| Pregnancy (n = 420) | Postpartum (n = 637) | Pregnancy (n = 712) | Postpartum (n = 670) | |
| Age, years | 29 (18–45) | 29 (19–42) | 29 (18–45) | 29 (18–45) |
| Weight, kg | 68 (39–167) | 62 (38–165) | 67 (42–164) | 61 (37–114) |
| Body mass index, kg/m2 | 27 (18–61) | 24 (16–45) | 27 (18–61) | 25 (16–49) |
| Gestation/postnatal age, weeks, at PK sampling time | 26 (14–34) | 16 (7–23) | 26 (14–34) | 16 (7–23) |
| Baseline viral load, copies/mL | < 40 (< 40–237,000) | < 40 (< 40–465,000) | < 40 (< 40–237,000) | < 40 (< 40–465,000) |
| Drug regimen | ||||
| On isoniazid | 420 (100%) | 637 (100%) | 352 (49%) | 540 (80%) |
| On efavirenz | 371 (88%) | 563 (88%) | 712 (100%) | 670 (100%) |
| On nevirapine | 11 (3%) | 47 (8%) | ‐ | ‐ |
| On lopinavir/ritonavir | 5 (1%) | 5 (1%) | ‐ | ‐ |
| On atazanavir/ritonavir | 2 (0%) | 2 (0%) | ‐ | ‐ |
| Days on EFV at PK sampling time | 125 (18–3,800) | 264 (1–4,228) | 125 (18–3,800) | 408 (1–4,228) |
| Drug metabolizing genotype | NAT 2 acetylation status | CYP2B6 metabolizer status | ||
|---|---|---|---|---|
| Rapid | 52 (12%) | 70 (11%) | 168 (24%) | 146 (22%) |
| Intermediate | 140 (33%) | 202 (32%) | 299 (42%) | 264 (40%) |
| Slow | 159 (39%) | 199 (31%) | 118 (16%) | 102 (15%) |
| Ultra‐slow | ‐ | ‐ | 1 (0%) | 2 (0%) |
| Missing | 69 (16%) | 166 (26%) | 126 (18%) | 156 (23%) |
| CYP2A6 metabolizer status | ||||
|---|---|---|---|---|
| Normal | 501 (70%) | 439 (65%) | ||
| Intermediate | 81 (11%) | 71 (11%) | ||
| Slow | 4 (1%) | 4 (1%) | ||
| Missing | 126 (18%) | 156 (23%) | ||
Two hundred ten women had isoniazid profiles during both pregnancy and postpartum.
Five hundred ninety‐six women had efavirenz profiles available during both pregnancy and postpartum.
EFV, efavirenz; PK, pharmacokinetic.
Distribution of drug metabolizer genotypes
The distribution of drug metabolizer genotypes for NAT2, CYP2B6, and CYP2A6 (classified into three phenotypes as described in Methods), stratified by country, is illustrated in Figure 1 . Rapid isoniazid acetylator NAT2 genotypes (had site range of 4–31%) were the least prevalent group in all eight countries, which varied considerably by sites. Intermediate metabolizer CYP2B6 (had site range of 43–63%), were frequent in all countries genotypes as were CYP2A6 normal metabolizer genotypes (had site range of 76–93%).
Figure 1.
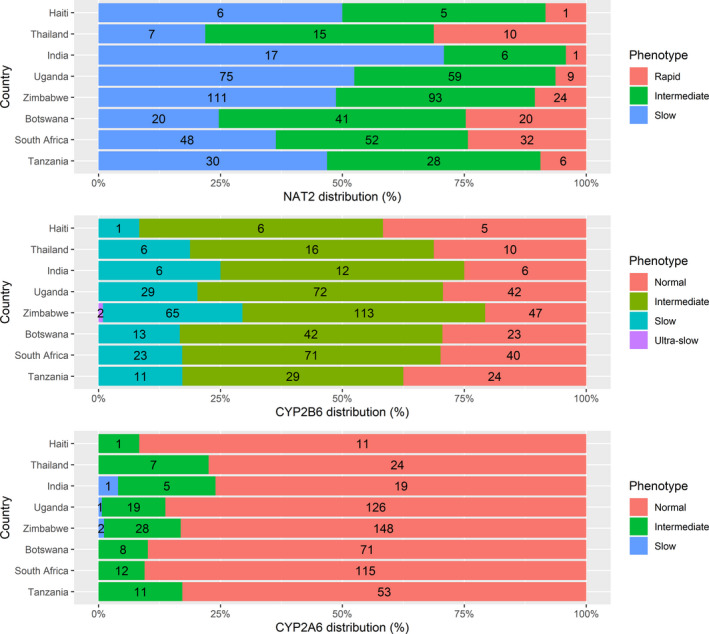
Distribution of the enzyme metabolizer genotypes for NAT2, CYP2B6, and CYP2A6 in the participants across the eight countries involved in the study. [Colour figure can be viewed at wileyonlinelibrary.com]
Structural model
A two‐compartment disposition model best described the PKs of both isoniazid and efavirenz with first‐order absorption through a chain of transit compartments. 30 Drug elimination implemented with a well‐stirred liver model, 19 was able to describe both hepatic clearance and first‐pass extraction with the parameter of hepatic intrinsic clearance, as shown in Figure 2 . Parameter estimates for isoniazid and efavirenz, and the precisions for each are presented in Table 2 and Table 3 , respectively.
Figure 2.
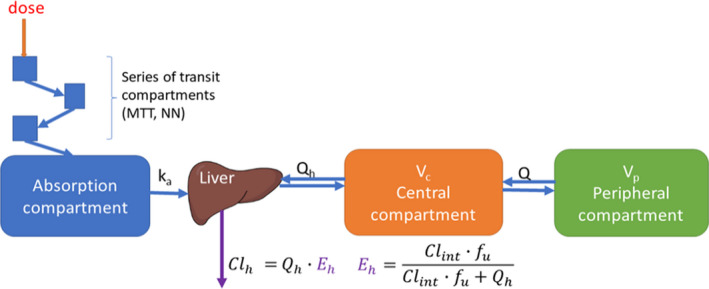
Schematic representation of the pharmacokinetic model of both efavirenz and isoniazid. The absorption is described with a series of transit‐compartment to capture the delay in absorption, and a rate constant K a. The hepatic extraction (Eh) is responsible for both first‐pass metabolism and the systemic elimination with first‐order kinetics. V c represents the volume of distribution in the central compartment. Drug transfer between the central and peripheral compartment is defined by intercompartmental clearance Q/F, were F represents the oral bioavailability. [Colour figure can be viewed at wileyonlinelibrary.com]
Table 2.
Final PK parameter estimates for isoniazid
| Parameter | Typical value (95% CI a ) | Variability b , %CV (95% CI a ) |
|---|---|---|
| CLint c , L/hour, NAT2 rapid | 72.3 (61.5–86.7) | 69.2 (64.2–74.2) e |
| CLint c , L/hour, NAT2 intermediate | 38.5 (34.6–43.2) | |
| CLint c , L/hour, NAT2 slow | 14.5 (13.1–16.0) | |
| V c d , L | 37.6 (33.9–40.7) | |
| V p d , L | 13.3 (10.5–16.9) | |
| Q/F c , L/hour | 3.32 (2.53–4.54) | |
| k a, 1/hour | 2.69 (1.91–3.51) | 145 (116–172) f |
| MTT, hours | 0.342 (0.209–0.459) | 116 (98.7–150) f |
| NN | 48.4 (22.2–83.8) | |
| QH c , L/hour | 90 FIXED | |
| fu, % | 95 FIXED | |
| Prehepatic relative bioavailability | 1 FIXED | 12.3 (8.20–15.7) f |
| Proportional error, % | 13.2 (11.3–15.3) | |
| Additive error, mg/L | 0.0378 (0.0335 −0.0449) | |
| Pregnancy effect on CL, % | +26.2 (19.8–33.2) |
%CV, percent coefficient of variation; CI, confidence interval; CL, clearance; CLint, clearance intrinsic; f u, unbound fraction of isoniazid in plasma 50; INH, isoniazid; k a, first‐order rate constant of INH absorption; MTT, absorption mean transit time; NN, number of absorption transit compartment; PK, pharmacokinetic; Q/F, apparent intercompartmental clearance for INH; QH, blood liver flow 40; Vc, apparent central volume of distribution for INH; Vp, apparent peripheral volume of distribution for INH.
The bold values represent significant covariates in the models.
The 95% CIs were obtained with the Standardized Infection Ratio procedure.
Variability was modeled with log‐normal distribution and is presented as an approximate percentage CV.
Clearance parameters are allometrically scaled based on fat‐free mass (typical value reported for 39 kg, which was the median fat‐free mass weight of the study population).
Volume of distribution parameters are scaled based on weight (typical value reported for 67 kg, which was the median weight of the study population).
Between subject variability.
Between occasion variability.
Table 3.
Final parameter estimates for efavirenz
| Parameter | Typical value (95% CI a ) | Variability b , %CV (95% CI) |
|---|---|---|
| CLint c , L/hour, CYP2B6 normal | 2,690 (2,300–3,030) | 53.8 (48.9–59.2) e |
| CLint c , L/hour, CYP2B6 intermediate | 1,940 (1,790–2,100) | |
| CLint c , L/hour, CYP2B6 slow | 545 (487–624) | |
| V c d , L | 135 (109–165) | |
| V p d , L | 512 (487–623) | |
| Q/F c , L/hour | 26.9 (19.8–36.5) | |
| K a, 1/hour | 1.75 Fixed | 180 (114.9–227) f |
| MTT, hour | 1.78 (1.20–2.39) | 131 (103–166) f |
| NN | 48.4 (11.3–64.7) | |
| QH c , L/hour | 90 Fixed | |
| f u, % | 0.5 Fixed | |
| Prehepatic relative bioavailability | 1 Fixed | 23.2 (20.7–26.1) f |
| Proportional error, % | 6.91 (4.72–9.45) | |
| Additive error, mg/L | 0.353 (0.303–0.408) | |
| Pregnancy effect on CL, % | +15.9 (9.75 to 21.9) | |
| INH effect on CL/F, L/hour, in CYP2B6 fast metabolizers, % | −6.87 (−12.1 to −1.13) | |
| INH effect on CL/F, L/hour in CYP2B6 intermediate and slow metabolizers, % | −13.4 (−17.3 to −9.06) |
%CV, percent coefficient of variation; CI, confidence interval; CL, clearance; CLint, clearance intrinsic; f u, unbound fraction of efavirenz in plasma; INH, isoniazid; K a, first‐order rate constant of INH absorption; MTT, absorption mean transit time; NN, number of absorption transit compartment; Q/F, apparent intercompartmental clearance for INH; Qh, blood liver flow; V c, apparent central volume of distribution for INH; V p, apparent peripheral volume of distribution for INH.
The bold values represent significant covariates in the models.
The 95% CIs were obtained with the Standardized Infection Ratio procedure.
Variability was modeled with log‐normal distribution and is presented as an approximate percentage CV.
Clearance parameters are allometrically scaled based on fat‐free mass (typical value reported for 39 kg, which was the median fat‐free mass weight of the study population).
Volume of distribution parameters are scaled based on weight (typical value reported for 67 kg, which was the median weight of the study population).
Between subject variability.
Between occasion variability.
Isoniazid pharmacokinetics: Covariate effects
Allometric scaling was applied to all clearance, volume, and liver parameters to account for body size effect, improving the fit, and explaining part of BSV (ΔOFV = −44). When fat‐free mass replaced bodyweight for allometric scaling of clearance (but not volume) parameters, it improved the fit of the model (ΔOFV = −6.27). NAT2 genotype significantly affected clearance of isoniazid (ΔOFV = −273, χ2 df = 2, P ≪ 0.001), as intrinsic clearance varied greatly among rapid, intermediate, and slow acetylators. After adjusting for body size and NAT2 genotype, pregnancy was found to increase clearance by 26% (ΔOFV = −49.6, χ2 df = 1, P ≪ 0.001). There was no significant difference in clearance among participants on the four different ART (efavirenz, nevirapine‐based, lopinavir/ritonavir‐based, and atazanavir/ritonavir‐based) regimens (ΔOFV = −6.34, χ2 df = 3, P = 0.0964). Inclusion of pregnancy on any other parameter, including the central compartment volume of distribution did not improve model fit.
Figure 3 displays the model‐predicted individual exposures of isoniazid stratified by both NAT2 genotype and pregnancy status. The 0–24‐hour area under the concentration‐time curve (AUC0–24) of isoniazid boxplot shows that isoniazid exposure differs among the three genotypes, and decreased exposures were observed during antepartum compared with postpartum in all three genotypes. Combining all genotypes, the overall median (interquartile range) of isoniazid AUC0–24 antepartum was 8.05 mg·h/L (4.43–16.7 mg·h/L), compared with 11.1 mg·h/L (6.26–23.9 mg·h/L) postpartum. Maximum concentrations (Cmax) during pregnancy and postpartum were 2.89 mg/L (1.97–4.13 mg/L) and 3.69 mg/L (2.64–5.13 mg/L), respectively. The postpartum AUC0–24 was 1.4‐fold greater compared with antepartum, whereas slow acetylator AUC0–24 was 5‐fold greater compared with rapid acetylators. An isoniazid AUC0–24 of 10.52 mg∙h/L has been associated with 90% of early bactericidal activity in patients with active TB. 31
Figure 3.
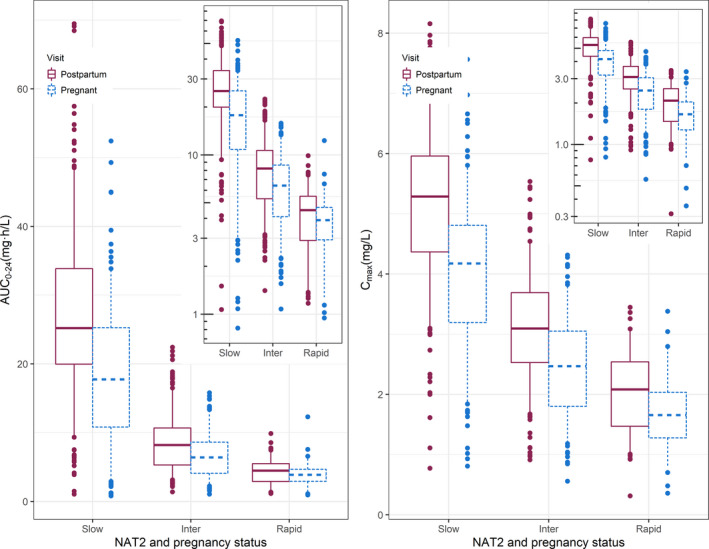
Isoniazid exposures stratified by NAT2 genotype and pregnancy. The box plot (with box representing median and interquartile range and whiskers the 5th‐95th interval) summarizes isoniazid maximum concentration (Cmax) on the right and 0–24‐hour area under the concentration‐time curve (AUC0–24) on the left for the three genotypes (slow, intermediate, and rapid acetylator) for both the antepartum (red solid line) and postpartum (blue dashed lines) visit. AUC0–24 was calculated by integrating between the 0 and 24 hours after dosing time points. The inset panel shows the same values on the log‐scale. [Colour figure can be viewed at wileyonlinelibrary.com]
Efavirenz pharmacokinetics: Covariate effects
Efavirenz had a structural model similar to isoniazid, except the first‐order absorption rate constant was unstable when sparse and intensive data were pooled together; so, the value observed in the intensive PK data analysis was used as a fixed constant. Allometric scaling with bodyweight on all clearance and volume parameters improved the model fit and explained part of the BSV (ΔOFV = −28.6). Using fat‐free mass instead of total bodyweight for clearance (but not volume) parameters further improved the model (ΔOFV = −12.5). CYP2B6 genotype significantly affected the clearance of efavirenz (ΔOFV = −399, χ2 df = 2, P ≪ 0.001). After adjusting for body size and CYP2B6 genotype, pregnancy was found to increase the clearance of efavirenz by 16% (ΔOFV = −75.5, χ2 df = 1, P ≪ 0.001). Clearance in women co‐administrated isoniazid and efavirenz was 7% lower in normal metabolizers and 13% lower in slow and intermediate metabolizers (ΔOFV = −39.2, χ2 df = 2, P ≪ 0.001) regardless of pregnancy status. Effect of CYP2A6 phenotype on the clearance and bioavailability of efavirenz was investigated among CYP2B6 slow metabolizers, in whom CYP2A6 may be more important for efavirenz clearance, but no significant effect was observed.
The model‐predicted individual exposures are shown in Figure 4 , where panels a and b stratify by CYP2B6 genotype and pregnancy status, whereas panels c and d stratify by CYP2B6 genotype and isoniazid co‐administration. Figure 4 shows that efavirenz varied among the three genotypes, and panels a and b show a decrease in exposures during pregnancy. The overall exposure of efavirenz was higher postpartum compared with pregnancy with median (interquartile range) postpartum AUC0–24 of 70.6 mg·h/L (47.9–118 mg·h/L), compared with 55.8 mg·h/L (38.6–92.7 mg·h/L) antepartum. Concentrations at 12 hours postdosing (C12) antepartum and postpartum were 2.18 mg·h/L (1.48–3.68 mg/L) and 2.69 mg·h/L (1.87–4.71 mg/L), respectively. Among normal CYP2B6 metabolizers 13 women (9%) had postpartum efavirenz C12 of < 1 mg/L compared with 25 women (15%) antepartum.
Figure 4.
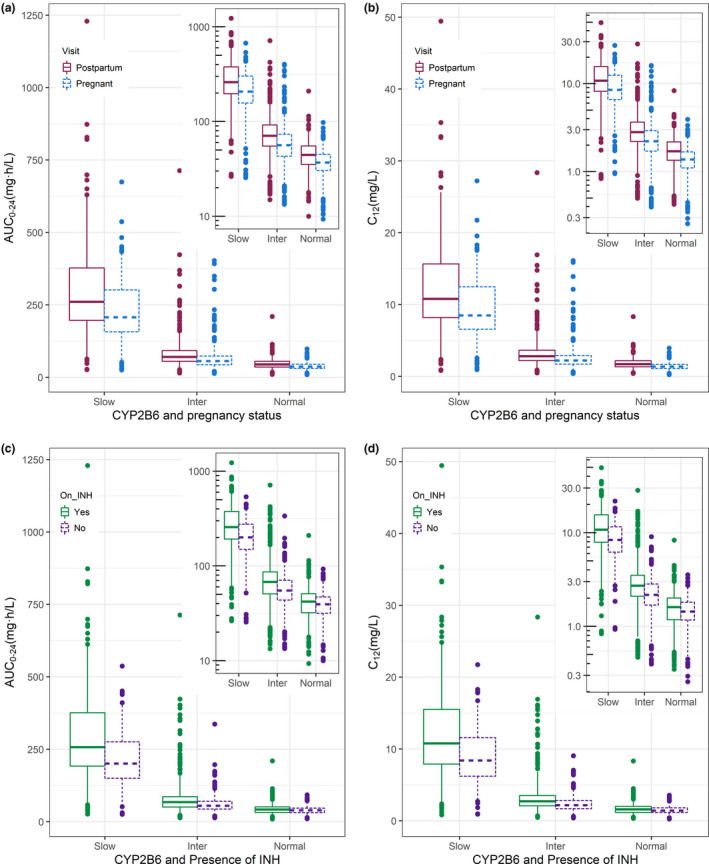
Efavirenz exposures stratified by CYP2B6 genotype, pregnancy and isoniazid co‐administration. (a, b) Displays box plot (with box representing median and interquartile range and whiskers the 5th‐95th interval) summarizing 0–24‐hour area under the concentration‐time curve (AUC0–24) and concentrations at 12 hours postdosing (C12), respectively, for the three genotypes (slow, intermediate, and normal metabolizer) stratified by pregnancy (solid line) and postpartum (dashed lines) visit. (c, d) Stratify AUC0‐24 and C12, respectively, using INH co‐administration. [Colour figure can be viewed at wileyonlinelibrary.com]
We observed a DDI consisting of increased exposure of efavirenz in participants on concomitant isoniazid displayed in Figure 4 c,d. The difference in exposure in the slow and intermediate metabolizers was more apparent than in the normal metabolizers resulting in high concentration in slow CYP2B6 metabolizers. For example, choosing a threshold of 15 mg/L (similar to the upper quartile of C12 in CYP2B6 slow metabolizers co‐administered isoniazid in Figure 4 d), among CYP2B6 slow metabolizers in the absence of isoniazid, efavirenz C12 was > 15 mg/L in 4 individuals (9%), compared with 45 individuals (35%) with the presence of isoniazid. As a point of reference, a therapeutic range for plasma efavirenz concentrations of 1–4 mg/L has been suggested, 32 although concentrations somewhat < 1 mg/L do not consistently predict treatment failure and concentrations > 4 mg/L do not consistently predict toxicity. The overall median (interquartile range) of AUC0–24 was 66.8 mg·h/L (45.2–114 mg·h/L) when isoniazid was present and 55.4 mg·h/L (39.7–98.1 mg·h/L) when absent, corresponding to a 1.2‐fold increase in efavirenz AUC0–24 due to the presence of isoniazid. All four panels in Figure 4 show that, whereas pregnancy and isoniazid affected efavirenz exposure, these effects were modest compared with CYP2B6 genotype, which resulted in a 5‐fold difference between slow to normal metabolizers.
DISCUSSION
We performed the largest study of its kind, assessing isoniazid and efavirenz PK, genetic polymorphisms, and DDIs in a geographic diverse population of pregnant and postpartum women living with HIV in high TB burden regions of the world. We show that pregnancy modestly increases the clearance of both isoniazid and efavirenz, therefore increasing the risk of low (and possibly subtherapeutic) drug exposure during pregnancy. Notably, although we also show that drug metabolizer genotypes profoundly affect plasma exposure of these two drugs, to an extent far greater than of pregnancy. For this reason, the risk of lower plasma drug exposure during pregnancy is likely greatest only in women who are CYP2B6 normal metabolizers or NAT2 rapid acetylator, because they clear the drug faster compared with the other genotypes and are already exposed to lower concentrations. Additionally, we report that efavirenz exposure is increased by isoniazid co‐administration, especially in CYP2B6 intermediate and slow metabolizers. Last, high concentrations observed in slow CYP2B6 and NAT2 genotype increases the risk of adverse effects.
The increase in clearance of isoniazid and efavirenz may be due to several physiological changes related to pregnancy (e.g., plasma albumin levels, 6 bodyweight and composition, plasma volume, 9 hepatic blood flow, 9 and induction or inhibition of drug metabolizing enzymes). The pregnancy‐induced increase in weight was accounted for in the model using allometric scaling, whereas no unbound concentration was captured, therefore, we could not confirm or disprove the effect of pregnancy on the protein‐binding of efavirenz. Antepartum increase in hepatic blood, 9 potentially increase the hepatic clearance of drugs with high hepatic extraction ratio, may be more apparent in low binding drugs like isoniazid compared with efavirenz. The other reason for changes in drug clearance during pregnancy is induction or inhibition of drug‐metabolizing enzymes. Pregnancy increases the activity of CYP2B6 7 and CYP3A4 6 (major and minor route of efavirenz metabolism 33 ). Tsutsumi et al. 8 observed a reduction in NAT2 activity during pregnancy, but the reduction was clinically not significant. The reported clearance increase is modest compared with the large BSV in drug exposure due to host genetics. However, these changes may be of relevance for rapid/normal metabolizers, who already experience lower exposure, as further increase in clearance might increase their risk for subtherapeutic concentrations.
Similar to previous reports, 10 , 12 , 34 we observed a DDI between isoniazid and efavirenz, as isoniazid decreases the clearance of efavirenz, particularly for intermediate and slow efavirenz metabolizers. This is thought to be due to isoniazid’s inhibition of CYP2A6. 35 However, for individuals who are CYP2B6 slow metabolizers, CYP2A6 assumes increased importance in clearing efavirenz, so that CYP2B6 slow metabolizers on isoniazid may have a higher risk of efavirenz toxicity. Our analysis did not detect any difference in isoniazid exposure among different ARTs (efavirenz‐based, nevirapine‐based, lopinavir/ritonavir‐based, and atazanavir/ritonavir‐based), which contrasts with previous reports. 36 , 37 However, it may be that our analysis was not powered to detect these differences because 88% of the participants were on efavirenz‐based ART.
Previously reported isoniazid exposures varied widely between studies. This might be due to effects of large BSV and between‐occasion variability , different proportions of NAT2 acetylator genotypes, different drug formulations, DDIs, and instability of isoniazid in plasma, 38 which makes PK studies of isoniazid somewhat challenging. This wide range is well summarized in a recent review by Daskapan et al. 39 Comparing our results with previous reports on nonpregnant populations, 39 , 40 , 41 we observed lower isoniazid exposures during pregnancy and postpartum visits, whereas the ratio in exposures between slow and rapid NAT2 genotype was similar (clearance for rapid acetylators was observed to be 4‐fold faster than those of slow acetylators). It is unclear if the relatively low concentrations we observed at both visits are due to efavirenz exposure, as most women were on efavirenz‐based regimen. It is also possible that the effect of pregnancy on isoniazid clearance takes time to reverse fully, and women in the postpartum visit may also have clearance levels higher than the general (nonpregnant, adult) population. It has been previously discussed that using the postpartum period as a control to study effects of pregnancy may be suboptimal because of factors that include lactation and delayed reversal of pregnancy‐related physiological changes. 42 Efavirenz exposures in our study were in line with previous reports in nonpregnant individuals. 33 , 43 , 44 Similarly, the ratio between normal and slow CYP2B6 metabolizers exposures was similar, with a C12 ratio of 5.
In our study, we report genotype frequencies from a large cohort of patients with wide geographic representation. A frequency of NAT2 slow acetylators above 50% worldwide has been reported. 45 Studies have shown greater than two‐fold difference in the prevalence of NAT2 slow acetylators in agriculturists (slow acetylators more prevalent) compared with pastoralists in central Asian population and sub‐Saharan African populations. 46 A pooled analysis by Sabbagh et al. 45 shows the highest level of within‐population diversity of NAT2 genotype in Africans. In our study population, NAT2 slow acetylators were most prevalent, followed by intermediate. Among CYP genes in humans, CYP2B6 is one of the most polymorphic gene. 47 A meta‐analysis by Zhou et al. 48 reported CYP2B6 normal metabolizer status in 61%, 38%, 76%, 45%, and 58% in Europeans, Africans, East Asians, South Asians, and admixed Americans, respectively. In comparison, 65%, 65%, 31%, 66%, and 72% distribution of CYP2A6 normal functioning gene were reported in Europeans, Africans, East Asians, South Asians, and admixed Americans, respectively. In the present study, intermediate followed by normal metabolizer CYP2B6 genotypes were frequent in all countries, as were CYP2A6 normal metabolizer genotypes.
Our study had some limitations, much of which we believe have been mitigated by use of a model‐based approach analysis. Genotype data was not available in a subset of patients, 179 (21%) for isoniazid, and 173 (22%) for efavirenz, but this was addressed by imputation using a mixture model to assign individuals with missing information to a phenotype group. Blood draws during sparse sampling visits were scheduled for 2 hours after isoniazid dose. Because efavirenz is typically taken at night, this resulted in concentrations drawn 10–14 hours after efavirenz dose. Exact time of dose was not always reliably recorded for sparse sampling visits. For a portion of the study, timing of ART dosing was not recorded, so that information was imputed. Although imputed data cannot be expected to be fully accurate, our approach was feasible to implement and expected to be robust because most women were taking efavirenz in the evening and were sampled the following morning, hence the slow terminal half‐life (40–55 hours 49 ) and accumulation of efavirenz minimized the impact of imprecise dosing time information. The uncertainty around self‐reported or imputed dosing times and sampling schedule chosen for the sparse data may be the reasons for high variability observed in the absorption parameters, but this had a minor effect in our analysis because the structural model was built using the intensive data which had rich and reliable information for the entire PK profile. Similarly, the method we used for the identification and exclusion of implausible outlier values prevented these few samples from affecting the development of the PK models.
In conclusion, our study showed modest reductions in isoniazid and efavirenz exposure during pregnancy compared with postpartum. Although the size of this effect is modest and unlikely to be of clinical significance for most patients, lower exposure may be important in normal CYP2B6 and rapid NAT2 acetylator, for whom the resulting concentrations may be subtherapeutic, thus possibly leading to ineffective treatment and development of drug resistance. We also confirm the previously reported increased efavirenz exposure in individuals on concomitant isoniazid. This effect was overall modest but was most pronounced in CYP2B6 intermediate and slow metabolizers, who already had higher efavirenz levels and may, therefore, be at higher risk of efavirenz‐related toxicities.
Funding
Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Network (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (NIAID) with co‐funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH), all components of the National Institutes of Health (NIH), under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC), and UM1AI106716 (IMPAACT LC), NICHD contract number HHSN275201800001I. Amita Gupta was also supported by NIH UM1AI069465. The University of Cape Town Clinical PK Laboratory is supported in part via the Adult Clinical Trial Group (ACTG), by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under award numbers UM1 AI068634, UM1 AI068636, and UM1 AI106701; as well as the Infant Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT), funding provided by National Institute of Allergy and Infectious Diseases (U01 AI068632), The Eunice Kennedy Shriver National Institute of Child Health and Human Development, and National Institute of Mental Health grant AI068632.
Conflict of Interest
All authors declared no competing interests for this work.
Author Contributions
K.G., P.D., A.G., D.H., C.L.W., and D.C. wrote the manuscript. A.G., D.H., R.B., G.M., L.A., K.M., S.B., A.W., T.R.S., G.H., P.J.P., N.C., and C.L.W. designed the research. L.W., J.N., C.L.W., C.O.M., T.C., D.H., R.B., N.C., G.M., L.A., K.M., S.B., T.V., L.S.C., G.R.M., A.V., B.T.M., L.A., R.B., N.N., V.R., E.K., M.M., V.C., M.N., T.M., F.T., A.H., K.S., B.Z., D.C., P.J.P., T.R.S., G.T., A.W., and A.G. performed the research. K.G., P.D., G.M., and L.A. analyzed the data.
Supporting information
Fig S1‐S2
Table S1
Table S2
Table S3
Supplementary Material
Acknowledgments
The study team is very grateful to the women who participated in this study. We appreciate the contributions of the following individuals: Gary Maartens and Marilyn Solomons from the University of Cape Town. The IMPAACT team working at the sites in South Africa, Tanzania, Botswana, Zimbabwe, Uganda, Thailand, India, and Haiti. Kamunkhwala Gausi acknowledges her PhD funders, the Virtual consortium whose aim is to investigate the challenges of TB treatment for individuals on second‐line ART while promoting African leadership and capacity building. Computations were performed using facilities provided by the University of Cape Town’s ICTS High Performance Computing team: hpc.uct.ac.za.
References
- 1. World Health Organization (WHO) . Tuberculosis Report 2017 Global (Geneva, 2017) <http://apps.who.int/bookorders>. [Google Scholar]
- 2. Selwyn, P.A. et al. A prospective study of the risk of tuberculosis among intravenous drug users with human immunodeficiency virus infection. N. Engl. J. Med. 320, 545–550 (1989). [DOI] [PubMed] [Google Scholar]
- 3. Singh, N. & Perfect, J.R. Immune reconstitution syndrome and exacerbation of infections after pregnancy. Clin. Infect. Dis. 45, 1192–1199 (2007). [DOI] [PubMed] [Google Scholar]
- 4. World Health Organization . Latent Tuberculosis Infection Updated and Consolidated Guidelines For Programmatic Management (Geneva, 2018) <https://www.who.int/tb/publications/2018/latent‐tuberculosis‐infection/en/>. [PubMed] [Google Scholar]
- 5. Khoo, S.H. , Gibbons, S. , Seden, K. & Back, D.J. Systematic review: drug‐drug interactions between antiretrovirals and medications used to treat TB, malaria, hepatitis B&C and opioid dependence <https://www.who.int/hiv/topics/treatment/drug_drug_interactions_review.pdf> (2014).
- 6. Pinheiro, E.A. & Stika, C.S. Drugs in pregnancy: pharmacologic and physiologic changes that affect clinical care. Semin. Perinatol. 44, 151221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dickmann, L.J. , Isoherranen, N. , Trager, W.F. , Levy, R.H. & Keirns, J.J. Quantitative prediction of CYP2B6 induction by estradiol during pregnancy: potential explanation for increased methadone clearance during pregnancy. Drug Metab. Dispos. 41, 270–274 (2013). [DOI] [PubMed] [Google Scholar]
- 8. Tsutsumi, K. et al. The effect of pregnancy on cytochrome P4501A2, xanthine oxidase, and N‐acetyltransferase activities in humans. Clin. Pharmacol. Ther. 70, 121–125 (2001). [DOI] [PubMed] [Google Scholar]
- 9. Jeong, H. Altered drug metabolism during pregnancy: hormonal regulation of drug‐metabolizing enzymes. Expert Opin. Drug Metab. Toxicol. 6, 689–699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dooley, K.E. et al. Pharmacokinetics of efavirenz and treatment of HIV‐1 among pregnant women with and without tuberculosis coinfection. J. Infect. Dis. 211, 197–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olagunju, A. et al. Pharmacogenetics of pregnancy‐induced changes in efavirenz pharmacokinetics. Clin. Pharmacol. Ther. 97, 298–306 (2015). [DOI] [PubMed] [Google Scholar]
- 12. Luetkemeyer, A.F. et al. Combined effect of CYP2B6 and NAT2 genotype on plasma efavirenz exposure during rifampin‐based antituberculosis therapy in the STRIDE study. Clin. Infect. Dis. 60, 1860–1863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta, A. et al. Isoniazid preventive therapy in HIV‐infected pregnant and postpartum women. N. Engl. J. Med. 381, 1333–1346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holzinger, E.R. et al. Genome‐wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet. Genomics 22, 858–867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boukouvala sotiria, E.M. et al. Database of arylamine N‐acetyltransferases (NATs). Democritus University Thrace (2016) <https://nat.mbg.duth.gr/>.
- 16. McDonagh, E.M. et al. PharmGKB summary: very important pharmacogene information for N‐acetyltransferase 2. Pharmacogenet. Genomics 24, 409–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boeckmann, A.J. , Beal, S.L. & Sheiner, L.B. NONMEM user’s guide, part V. Introductory Guide. NONMEM Project Group 48, (2011) 10.1017/CBO9781107415324.004. [DOI] [Google Scholar]
- 18. Keizer, R.J. , Karlsson, M.O. & Hooker, A. Modeling and simulation workbench for NONMEM: Tutorial on Pirana, PsN, and Xpose. CPT Pharmacometrics Syst. Pharmacol. 2, e50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gordi, T. et al. A semiphysiological pharmacokinetic model for artemisinin in healthy subjects incorporating autoinduction of metabolism and saturable first‐pass hepatic extraction. Br. J. Clin. Pharmacol. 59, 189–198 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang, J. , Jamei, M. , Yeo, K.R. , Rostami‐Hodjegan, A. & Tucker, G.T. Misuse of the well‐stirred model of hepatic drug clearance. Drug Metab. Dispos. 35, 501–502 (2007). [DOI] [PubMed] [Google Scholar]
- 21. Mehvar, R. Clearance concepts: fundamentals and application to pharmacokinetic behavior of drugs. J. Pharm. Pharm. Sci. 21, 88s–102s (2018). [DOI] [PubMed] [Google Scholar]
- 22. Alghamdi, W.A. , Al‐Shaer, M.H. & Peloquin, C.A. Protein binding of first‐line antituberculosis drugs. Antimicrob. Agents Chemother. 62, e00641‐18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sturkenboom, M.G.G. et al. Quantification of isoniazid, pyrazinamide and ethambutol in serum using liquid chromatography‐tandem mass spectrometry. J. Appl. Bioanal. 1, 89–98 (2015). [Google Scholar]
- 24. Hooker, A.C. , Staatz, C.E. & Karlsson, M.O. Conditional weighted residuals (CWRES): a model diagnostic for the FOCE method. Pharm. Res. 24, 2187–2197 (2007). [DOI] [PubMed] [Google Scholar]
- 25. Eckhardt, B.J. & Gulick, R.M. Drugs for HIV infection. Infect. Dis. (Auckl) 2, 1293–1308.e2 (2017). [Google Scholar]
- 26. Parkin, D.P. et al. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am. J. Respir. Crit. Care Med. 155, 1717–1722 (1997). [DOI] [PubMed] [Google Scholar]
- 27. Anderson, B.J. & Holford, N.H.G. Mechanism‐based concepts of size and maturity in pharmacokinetics. Annu. Rev. Pharmacol. Toxicol. 48, 303–332 (2008). [DOI] [PubMed] [Google Scholar]
- 28. Beal, S.L. Ways to fit a PK model with some data below the quantification limit. J. Pharmacokinet. Pharmacodyn. 28, 481–504 (2001). [DOI] [PubMed] [Google Scholar]
- 29. Keizer, R.J. , Zandvliet, A.S. , Beijnen, J.H. , Schellens, J.H.M. & Huitema, A.D.R . Performance of methods for handling missing categorical covariate data in population pharmacokinetic analyses. AAPS J. 14, 601–611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Savic, R.M. , Jonker, D.M. , Kerbusch, T. & Karlsson, M.O. Implementation of a transit compartment model for describing drug absorption in pharmacokinetic studies. J. Pharmacokinet. Pharmacodyn. 34, 711–726 (2007). [DOI] [PubMed] [Google Scholar]
- 31. Donald, P.R. et al. The influence of dose and N‐acetyltransferase‐2 (NAT2) genotype and phenotype on the pharmacokinetics and pharmacodynamics of isoniazid. Eur. J. Clin. Pharmacol. 63, 633–639 (2007). [DOI] [PubMed] [Google Scholar]
- 32. Marzolini, C. et al. Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV‐1‐infected patients. AIDS 15, 71–75 (2001). [DOI] [PubMed] [Google Scholar]
- 33. Desta, Z. et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2B6 and efavirenz‐containing antiretroviral therapy. Clin. Pharmacol. Ther. 106, 726–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bertrand, J. et al. Dependence of efavirenz‐ and rifampicin‐isoniazid‐based antituberculosis treatment drug‐drug interaction on CYP2B6 and NAT2 genetic polymorphisms: ANRS 12154 study in Cambodia. J. Infect. Dis. 209, 399–408 (2014). [DOI] [PubMed] [Google Scholar]
- 35. Wen, X. , Wang, J.‐S. , Neuvonen, P.J. & Backman, J.T. Isoniazid is a mechanism‐based inhibitor of cytochrome P450 1A2, 2A6, 2C19 and 3A4 isoforms in human liver microsomes. Eur. J. Clin. Pharmacol. 57, 799–804 (2002). [DOI] [PubMed] [Google Scholar]
- 36. Sekaggya‐Wiltshire, C. et al. Low anti‐tuberculosis drug concentrations in HIV‐Tuberculosis co‐infected adults with low body weight. (11th International Workshop On Clinical Pharmacology of Tuberculosis Drugs, The Hague, The Netherlands, October 23, 2018) <http://regist2.virology‐education.com/abstractbook/2018/abstractbook_11tbpk.pdf> Abstract 39. [Google Scholar]
- 37. Chirehwa, M.T. et al. Effect of efavirenz‐based antiretroviral therapy and high‐dose rifampicin on the pharmacokinetics of isoniazid and acetyl‐isoniazid. J. Antimicrob. Chemother. 74, 139–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poole, N.F. & Meyer, A.E. Stability of isoniazid in aqueous solutions and plasma. Exp. Biol. Med. 104, 560–562 (1960). [DOI] [PubMed] [Google Scholar]
- 39. Daskapan, A. et al. A systematic review on the effect of HIV infection on the pharmacokinetics of first‐line tuberculosis drugs. Clin. Pharmacokinet. 58, 747–766 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zabost, A. et al. Correlation of N‐acetyltransferase 2 genotype with isoniazid acetylation in polish tuberculosis patients. Biomed Res. Int. 2013, 1–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seng, K.‐Y. et al. Population pharmacokinetic analysis of isoniazid, acetylisoniazid, and isonicotinic acid in healthy volunteers. Antimicrob. Agents Chemother. 59, 6791–6799 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Kock, M. , Tarning, J. , Barnes, K.I. & Denti, P. Response to “lactation status and studies of pyrimethamine pharmacokinetics in pregnancy”. CPT Pharmacometrics Syst. Pharmacol. 6, 731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Orrell, C. et al. Effect of mid‐dose efavirenz concentrations and CYP2B6 genotype on viral suppression in patients on first‐line antiretroviral therapy. Int. J. Antimicrob. Agents 47, 466–472 (2016). [DOI] [PubMed] [Google Scholar]
- 44. Dickinson, L. et al. Pharmacokinetic and pharmacodynamic comparison of once‐daily efavirenz (400 mg vs. 600 mg) in treatment‐naive HIV‐infected patients: results of the ENCORE1 study. Clin. Pharmacol. Ther. 98, 406–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sabbagh, A. , Darlu, P. , Crouau‐Roy, B. & Poloni, E.S. Arylamine N‐acetyltransferase 2 (NAT2) genetic diversity and traditional subsistence: a worldwide population survey. PLoS One 6, e18507 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patin, E. et al. Sub‐Saharan African coding sequence variation and haplotype diversity at the NAT2 gene. Hum. Mutat. 27, 720 (2006). [DOI] [PubMed] [Google Scholar]
- 47. Zanger, U.M. & Klein, K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front. Genet. 4, 24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou, Y. , Ingelman‐Sundberg, M. & Lauschke, V.M. Worldwide distribution of cytochrome P450 alleles: a meta‐analysis of population‐scale sequencing projects. Clin. Pharmacol. Ther. 102, 688–700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Squibb, B.‐M. SUSTIVA ® (efavirenz) Capsules and Tablets. Full Prescribing Information (Princeton, NJ, 2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2
Table S1
Table S2
Table S3
Supplementary Material


