Summary
Ferns appear in the fossil record some 200 Myr before angiosperms. However, as angiosperm‐dominated forest canopies emerged in the Cretaceous period there was an explosive diversification of modern (leptosporangiate) ferns, which thrived in low, blue‐enhanced light beneath angiosperm canopies. A mechanistic explanation for this transformative event in the diversification of ferns has remained elusive.
We used physiological assays, transcriptome analysis and evolutionary bioinformatics to investigate a potential connection between the evolution of enhanced stomatal sensitivity to blue light in modern ferns and the rise of angiosperm‐dominated forests in the geological record.
We demonstrate that members of the largest subclade of leptosporangiate ferns, Polypodiales, have significantly faster stomatal response to blue light than more ancient fern lineages and a representative angiosperm. We link this higher sensitivity to levels of differentially expressed genes in blue‐light signaling, particularly in the cryptochrome (CRY) signaling pathway. Moreover, CRYs of the Polypodiales examined show gene duplication events between 212.9–196.9 and 164.4–151.8 Ma, when angiosperms were emerging, which are lacking in other major clades of extant land plants.
These findings suggest that evolution of stomatal blue‐light sensitivity helped modern ferns exploit the shady habitat beneath angiosperm forest canopies, fueling their Cretaceous hyperdiversification.
Keywords: blue‐light signaling, cryptochrome, fern evolution, photosynthesis, stomata
Short abstract
See also the Commentary on this article by Chater, 230: 886–888.
Introduction
Molecular and fossil evidence places the origin of ferns at around 400 Ma, more than 200 Myr before the appearance of angiosperms (Kenrick & Crane, 1997; Soltis et al., 1999). The rise of angiosperms in the Cretaceous period led to darker forest understories and coincided with a decline in fern and gymnosperm species richness, but leptosporangiate ferns (Polypodiidae) (Christenhusz et al., 2011; The Pteridophyte Phylogeny Group, 2016) subsequently proliferated to become the second most species‐rich group of vascular plants, and now comprise the vast majority of extant fern species (Schneider et al., 2004; Smith et al., 2006; Schuettpelz & Pryer, 2009).
Many unique functional and physiological changes appear to have rapidly evolved with the explosive diversification and radiation of modern (leptosporangiate) ferns, including nutrient and water‐uptake mechanisms, alteration in sporophytic hydraulic and stomatal regulation, and modification of reproductive systems (Watkins et al., 2007a, 2007b,2007a, 2007b; Creese et al., 2014; Franks & Britton‐Harper, 2016; Cai et al., 2017a). However, despite suggestions that the success of leptosporangiate ferns was an opportunistic response to the dominance of angiosperms (Schneider et al., 2004), mechanistic evidence for the drivers of this formative event in plant diversification has remained elusive.
Plant photosynthesis is regulated by stomata, small pores on plant leaves and stems formed by guard cells (Willmer & Fricker, 1996). Evolution has shaped the morphology and regulatory mechanisms governing stomatal movement over the past 450 Myr. Understanding fern stomatal responses to environmental cues will provide crucial insights into their success as the second‐largest clade of land plants. It has been reported that, when subjected to water deficit, the stomata of ferns lack an active closing response (closure via metabolically driven reduction in guard cell osmotic pressure), and exhibit only hydropassive closure (closure via passive turgor loss to achieve equilibrium with the external water potential, without changing guard cell osmotic pressure) (Brodribb & McAdam, 2011), and other studies have reported that fern stomata are relatively or completely insensitive to blue light and CO2 (Doi et al., 2006; Doi & Shimazaki, 2008; Brodribb et al., 2009). However, it has since been shown that ferns exhibit significant variability in the sensitivity of stomatal responses to light, CO2 and vapor pressure deficit (Ruszala et al., 2011; Creese et al., 2014; Lind et al., 2015; Franks & Britton‐Harper, 2016; Chen et al., 2017; Horak et al., 2017; Cai et al., 2017a; Westbrook & McAdam, 2020). These studies were conducted on different life stages (sporophyte and gametophyte), but with relatively few fern species representing a small number of clades. Understanding this striking functional diversity amongst ferns requires novel, broad‐based investigations across diverse fern lineages.
The altered spectral quality of light beneath forest canopies exerts strong selective pressures on the evolution of plants and animals that occupy this habitat (Endler, 1993). Photosynthetic pigments in the upper canopy absorb relatively more red than blue light wavelengths and so shift the understory light conditions. Thus, understory plants have optimized the sensitivity of light‐harvesting and gas exchange control systems (Gommers et al., 2013). Red and blue light are key environmental cues for stomatal opening. For example, activation of the blue‐light receptor phototropins (PHOTs) in the guard cell plasma membrane drives K+, anion and water influx into guard cells, leading to an increase of turgor pressure and stomatal opening (Iino et al., 1985; Shimazaki et al., 1986; Kinoshita et al., 2001; Inoue & Kinoshita, 2017). In contrast to seed plants, the PHOT‐dependent stomatal response to blue light was found to be absent in a select group of leptosporangiate ferns, prompting researchers to propose that PHOT dependence appeared early in land plant evolution, but was lost in some leptosporangiate ferns (Doi et al., 2006, 2015; Doi & Shimazaki, 2008). To gain a comprehensive understanding of the evolution and behavior of the stomatal blue‐light response, including possible alternative blue‐light receptors, in the leptosporangiate fern clade (Fig. 1a), the evolution of these traits and characters must be reconstructed using representatives of all major lineages of ferns (Schuettpelz & Pryer, 2009).
Fig. 1.
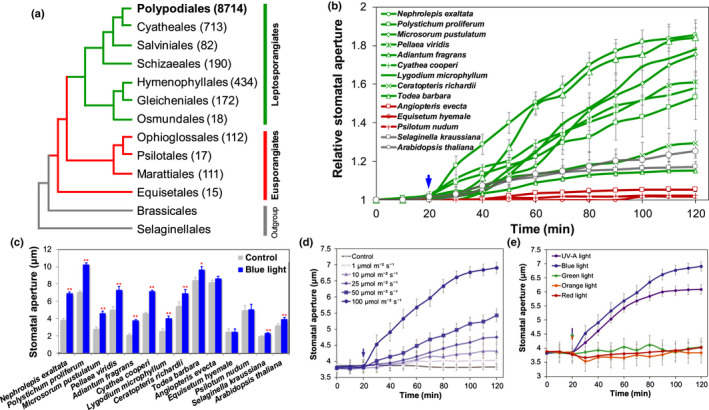
Light‐induced stomatal responses in different green plants. (a) The topology of fern orders. Thetree was constructed with Brassicales and Selaginellales as outgroups. The number in parentheses represents the number of fern species in each order. (b, c) Stomatal aperture response to blue light (100 μmol m−2 s−1) in epidermal peels of leptosporangiate ferns (Nephrolepis exaltata, Polystichum proliferum, Microsorum pustulatum, Pellaea viridis, Adiantum fragrans, Cyathea cooperi, Lygodium microphyllum, Todea barbara, Ceratopteris richardi) compared with eusporangiate fern lineages (Angiopteris evecta, Equisetum hyemale, Psilotum nudum), as well as a reference lycophyte (Selaginella kraussiana) and a model angiosperm (Arabidopsis thaliana). The control represents the average value of stomatal aperture in the first 20 min, whereas blue‐light treatment represents the stomatal aperture after 100 min blue‐light treatment (n = 4 biological replicates with 30–50 stomata for each plant species; data are means ± SE; *, P < 0.05; **, P < 0.01). (d) Dose response of stomata to blue light in N. exaltata. Stomatal aperture was measured in response to 1, 10, 25, 50 and 100 μmol m−2 s−1 blue light (n = 4 biological replicates with 30–50 stomata; data are means ± SE). (e) Response of N. exaltata stomata to different light wavelengths. The light treatments include red (675 ± 25 nm, 250 μmol m−2 s−1), orange (593 ± 40 nm, 150 μmol m−2 s−1), green (527 ± 20 nm, 100 μmol m−2 s−1) and blue light (475 ± 25 nm, 100 μmol m−2 s−1) and UV‐A (370 ± 36 nm, 75 μmol m−2 s−1). Four biological replicates with 30–50 stomata were used for each treatment; data are means ± SE.
Photoreceptors play key roles in plant evolution and diversification through the regulation of a range of plant functions, including growth and photosynthesis (Mathews, 2006; Christie, 2007). Plants use an array of photoreceptors, including PHOTs, phytochromes (PHYs), cryptochromes (CRYs), zeitlupes (ZTLs) and UV resistance locus8 (UVR8), to measure light quality and quantity and to regulate molecular, developmental and physiological processes (Moglich et al., 2010; Li et al., 2015). The biochemistry, structure and function of these photoreceptors have been characterized mainly in Arabidopsis thaliana (Lin, 2002; Chaves et al., 2011). The rapid radiation of ferns under the canopy of angiosperms is thought to be related to the diversification of PHOTs and PHYs (Li et al., 2018, 2015, 2014; Fiorucci & Fankhauser, 2017). In particular, the chimeric photoreceptor neochrome (NEO) fuses red‐sensing PHY at the N‐terminus and blue‐sensing PHOT at the C‐terminus to mediate phototropic responses (Kawai et al., 2003; Suetsugu et al., 2005). However, NEOs have so far been found only in the streptophyte alga Mougeotia scalaris (Zygnemetaceae, a possible sister lineage to land plants) (Wickett et al., 2014; Gitzendanner et al., 2018), hornworts and some leptosporangiate ferns belonging to the orders Cyatheales and Polypodiales. Possession of NEO represents a key innovation in the diversification of these ferns under low‐light conditions (Suetsugu et al., 2005; Kanegae et al., 2006; Li et al., 2014). The evolution of structural and functional diversity in PHYs may be an adaptation that enhances growth and survival of one subclade of leptosporangiate ferns, Polypodiales, in low light beneath the forest canopy (Possart & Hiltbrunner, 2013; Li et al., 2015). In green plants, CRYs are photoreceptors that mediate various blue light‐induced responses such as photomorphogenesis, flowering and circadian regulation (Lin, 2002; Chaves et al., 2011). Stomata of the Arabidopsis cry1cry2 double mutant showed reduced blue‐light response, and CRY1‐overexpressing plants showed hypersensitive stomatal opening to blue light (Mao et al., 2005). The evolution of CRYs has been dated to before the origin of land plants (Yang et al., 2017), but it is still not known whether evolution of CRYs played a significant role in fern diversification in the shadow of angiosperm‐dominated forests.
Here, we undertook a comprehensive evaluation of molecular and physiological evidence for stomatal blue‐light signaling and sensitivity across the full fern phylogeny. We hypothesized that rapid diversification of modern leptosporangiate ferns commencing around 200 Ma was linked to evolution of diversified blue‐light response machinery in these plants. Special consideration was given to evolution of the CRY family of blue‐light receptors across fern lineages and the potential advantage this gave to modern ferns as they diversified beneath angiosperm forest canopies.
Materials and Methods
Plant materials and growth
Sporophytes of Nephrolepis exaltata, Polystichum proliferum, Adiantum fragrans, Pellaea viridis, Ceratopteris richardii, Cyathea cooperi, Lygodium microphyllum, Todea barbara, Angiopteris evecta and Selaginella kraussiana were purchased from plant nurseries (Bunnings, Penrith, Australia; Rainforest Nursery, Upper Burringbar, Australia). Microsorum pustulatum, Equisetum hyemale and Psilotum nudum were collected from Western Sydney University (Richmond, NSW, Australia). Arabidopsis thaliana wild‐type Col‐0 seeds were sown in compost and grown for 4 wk before experiments. Plants were grown under 12 h : 12 h, day : night, 20 ± 1°C, 100 µmol m−2 s−1 photosynthetically active radiation and 60% relative humidity (RH). Plants were irrigated weekly and grown for at least 4 wk before commencing experiments.
Stomatal assay
Stomatal aperture assays were carried out on epidermal peels with viable stomata according to Cai et al. (2017a). Briefly, the epidermal peels were treated in low‐calcium measuring buffer (10 mM KCl, 50 μM Ca2+‐MES, pH 6.1) for 20 min in the dark as the control, followed by 100 min of light (blue, UV‐A, green, orange, red) treatments. Blue‐light treatments (0.1–100 μmol m−2 s−1) were applied using a light source (Abet Technologies, Milford, CT, USA) equipped with 475 ± 25 nm optical filter (Edmund Optics, Barrington, NJ, USA). The upper range of the blue‐light treatment is equivalent to the blue‐light photon flux in bright sunlight. Images of epidermal peels were recorded every 10 min using a microscope equipped with an NIS‐F1 charge coupled device camera and a DS‐U3 controller (Nikon, Tokyo, Japan). We tested whether the phenomenon of rapid stomatal opening in response to blue light is consistent across the extant fern lineages (Cai et al., 2017a), representing seven major orders (Polypodiales, Cyatheales, Schizaeales, Osmundales, Marattiales, Equisetales, Psilotales) of extant ferns that have radiated in diverse environmental conditions and temperature zones (Supporting Information Table S1), allowing a comprehensive assessment of responses to blue light in extant ferns.
Gas exchange
Measurement of stomatal conductance and photosynthetic rate was carried out using a gas exchange analyzer equipped with a 6400‐40 Leaf Chamber Fluorometer (Li‐Cor Inc., Lincoln, NE, USA), as in previous studies (Brodribb & McAdam, 2011; Cai et al., 2017a). The leaf chamber was clamped to healthy, newly matured fronds of fern plants maintained in well‐watered conditions. The leaf temperature was set at 24°C. The RH was maintained at c. 50%. The flow rate was set to 300 μmol s−1. CO2 concentration was maintained at 400 mol mol−1 using the CO2 mixer control. Data were recorded at 20 s intervals.
Measurement of reactive oxygen species (ROS)
The ROS production in guard cells was determined using fluorescent indicators 2′,7′‐dichlorodihydrofluorescein diacetate (H2DCFDA; Life Technologies) (Cai et al., 2017a). Epidermal strips of N. exaltata, A. evecta and A. thaliana were incubated in glass‐bottom Petri dishes containing low‐calcium measuring buffer (described earlier) with 20 µM H2DCFDA for 20 min in the dark. Confocal images were collected every 5 min with excitation at 488 nm and emission at 505–525 nm using an inverted confocal microscope (Leica Microsystems, Wetzlar, Germany). The ROS fluorescence of guard cells was estimated using Las‐ez software (Leica Microsystems) and Imagej software (NIH, Bethesda, MD, USA) by subtracting the florescence value of background from that of guard cells according to Chen et al. (2016).
RNA sequencing and quantitative RT‐PCR
RNA sequencing and analysis of leaf epidermis followed essentially the method of Chen et al. (2019). The lower epidermis of N. exaltata was peeled and floated on low‐calcium measuring buffer (see earlier) in a Petri dish, and irradiated with 100 µmol m−2 s−1 blue light for 20 min. After blue‐light treatment, the epidermis peels were collected and used for RNA extraction. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), and the cDNA libraries were constructed using NEBNext Ultra× RNA Library Prep Kit (Illumina, San Diego, CA, USA), following the manufacturer's procedure. RNA sequencing (RNA‐seq) was performed, and raw data obtained from the HiSeq 2500 platform were trimmed by removing empty reads, low‐quality bases (Q < 30 and length < 50 bp) and adaptor sequences. Reads mapping, assembly and abundance estimation of differentially expressed genes (DEGs) was performed by ‘Cuffdiff’ of Cufflinks v.2.1.1 with three biological replicates (a biological replicate in this case comprising material collected from one individual plant).
The fronds/leaves of N. exaltata, C. richardii, A. evecta and A. thaliana plants were irradiated with 100 µmol m−2 s−1 blue light for 20 min. The total RNA of the leaves was extracted using Trizol reagent (Life Technologies) following the manufacturer's procedure and the cDNA was synthesized using the SensiFAST Kit (Bioline, Eveleigh, NSW, Australia). The expression of the target genes was determined using a Rotor‐Gene Q6000 system (Qiagen). Beta‐tubulin was used as the reference gene for normalization of relative gene expression, as previously described (Cai et al., 2017a). Three independent biological replicates were used for each species and treatment.
Evolutionary bioinformatics, phylogeny and molecular dating
Comparative genetic analysis of gene families across the major green plant lineages and algae was described in Zhao et al. (2019). Genome sequence data of all species were obtained from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) and Ensembl Plants (http://plants.ensembl.org/index.html). The protein translations of CRYs were obtained from the OneKP database (www.onekp.com) and a fern transcriptome dataset (https://figshare.com/s/0f773861b6813f97ff63) (Table S2). Candidate protein sequences that satisfied the criteria of E‐value < 10−5 were selected by blastp searches.
Amino acid sequences translated from transcriptome data were used for construction of phylogenetic trees, and multiple phylogenetic trees were constructed and used in different figures for the data presentation and scientific interpretation. The amino acid sequence alignment was performed using Mafft. Conserved domains of aligned amino acid sequences were identified using Gblocks v.0.91b (http://phylogeny.lirmm.fr), and these were then used to construct phylogenetic trees. LG + I+G4 was the best‐fitting model of amino acid substitution under the Akaike information criterion using Iq‐tree v.1.6.8 (http://www.iqtree.org). The maximum likelihood (ML) tree was constructed using fasttree v.2.1 (http://www.microbesonline.org/fasttree/). Additional phylogenetic analysis was performed with full‐length sequences using Iq‐tree software with JTT + R6 model. The amino acids alignment of 2276 CRY sequences was performed using Pasta (https://github.com/smirarab/pasta). Phylogenic trees were displayed using iTol v.3.0 (https://itol.embl.de/).
The estimated time of gene duplication of CRY paralogs in each species was calculated according to the formula: T = K s/2λ × 10−6 Ma, where T is time of gene duplication, K s is the fraction of synonymous substitution per synonymous site, and λ is the rate of substitution (Lynch & Conery, 2000). The first CRYs duplication event (from CRY1/2/3/4/5 to CRY1/2/5, CRY3/4) occurred simultaneously with the divergence of the Euphyllophyta node (386–418 Ma, estimated from these species generated in TimeTree (http://www.timetree.org/)), λ for CRYs was estimated as 5.49 × 10−9–5.95 × 10−9. The alignment of paralogous pairs of CRYs was performed using Mafft (https://mafft.cbrc.jp/alignment/software/) and paraat 2.0 (http://bigd.big.ac.cn/tools/paraat), and K s of CRY paralogs was calculated using kaks_calculator 2.0 with the γ‐MYN model (https://sourceforge.net/projects/kakscalculator2/).
Results
Blue light induces distinct stomatal responses in diverse ferns
Most extant fern species are in the leptosporangiate order Polypodiales, accounting for 82.4% of fern species diversity (The Pteridophyte Phylogeny Group, 2016) (Fig. 1a). We first examined the general presence and characteristics of the stomatal blue‐light response in representative species of each major lineage of both eusporangiate and leptosporangiate ferns (Fig. 1a; Table S1), together with a lycophyte (S. kraussiana) and an angiosperm (A. thaliana). All of the leptosporangiate ferns examined here exhibited a significant (P < 0.01) stomatal opening in response to blue light, whereas the eusporangiate ferns did not show a significant response (Fig. 1b,c). The lycophyte (Selaginella) and angiosperm (Arabidopsis) also exhibited significant stomatal opening in response to blue light (Fig. 1b,c). Closer examination of N. exaltata, a leptosporangiate fern of Polypodiales, showed that its stomata opened in response to blue light and UV‐A light in a dose‐dependent manner (Figs 1d,e; S1). However, little stomatal opening was observed in response to green, orange or red light treatments in the epidermal peels of N. exaltata (Fig. 1e). Given the vital role of blue light in plant growth, development and stomatal regulation, we therefore concentrated on the effect of blue light in the following analysis.
The influence of blue‐light‐induced stomatal opening on gas exchange in N. exaltata was validated by measurements of net photosynthetic rate (A) and stomatal conductance (gs) on intact leaves (Figs 2a, S2a). Blue light at an intensity of 10 μmol m−2 s−1 (comparable to blue light received under 10% sunlight) was able to induce significant increase of A and gs, with both parameters saturating at 100 μmol m−2 s−1 (Fig. S2a). In contrast to epidermal peels (Fig. 1e), stomata of intact N. exaltata leaves responded strongly to red light (Fig. S2b), consistent with the known dependence of the stomatal red light response on mesophyll processes (Mott et al., 2008). Compared with the eusporangiate fern A. evecta (Marattiales) and the angiosperm A. thaliana, N. exaltata showed significantly faster responses of A and gs to blue light (Fig. 2a) and high‐intensity red light (600 μmol m−2 s−1) (Fig. S3). Exposing leaves to 100 μmol m−2 s−1 blue light while maintaining 600 μmol m−2 s−1 red light induced a substantial increase in g s for A. thaliana, but not in N. exaltata or other leptosporangiate ferns, including C. cooperi, L. microphyllum and T. barbara (Fig. S3).
Fig. 2.
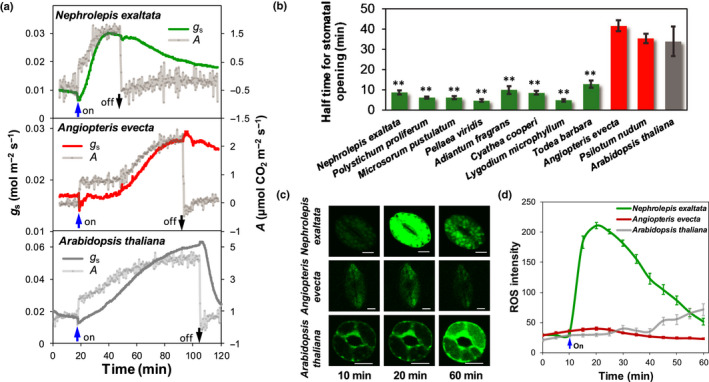
Gas exchange and guard cell reactive oxygen species production in response to blue light. (a) Stomatal conductance (g s) and net photosynthetic rate (A) in response to blue light in intact leaves of the leptosporangiate fern Nephrolepis exaltata, the eusporangiate fern Angiopteris evecta and the angiosperm Arabidopsis thaliana. Six biological replicates were performed and a typical result is shown. (b) Half‐time for stomatal opening under blue light, measured as g s in intact leaves, in the species from (b). For (a) and (b), measurement of g s and A commenced in the dark, as the control, followed by 100 μmol m−2 s−1 blue light. Data are means ± SE (n = 3–6 biological replicates). *, P < 0.05; **, P < 0.01. (c, d) Confocal images (c) and fluorescent probe intensity (d) of blue‐light‐induced reactive oxygen species (ROS) in guard cells of the leptosporangiate fern N. exaltata, the eusporangiate fern A. evecta and the angiosperm A. thaliana. Scale bar in (c) is 10 μm. For (d), n = 4 biological replicates with 30–50 stomata; data are means ± SE.
After establishing a general stomatal opening response to blue light in a diverse array of leptosporangiate ferns, we sought to compare more broadly the rate of the stomatal blue‐light response in leptosporangiate ferns with other lineages. Measurements on intact fern leaves revealed that stomata in leptosporangiate ferns open more rapidly in response to blue light compared with the eusporangiate ferns and the flowering plant A. thaliana (Fig. 2b). Importantly, while no responsiveness was observed in epidermal peels (Fig. 1c), stomata in intact leaves of the eusporangiate ferns (Angiopeteris evecta and P. nudum, representing Marattiales and Psilotales, respectively) opened in response to blue light, albeit substantially more slowly than in the leptosporangiate ferns (Fig. 2b).
Capacity for rapid blue‐light response in Polypodiales ferns
Blue‐light‐induced stomatal opening is usually associated with H+‐pumping (Kinoshita & Shimazaki, 1999; Shimazaki et al., 2007) and the transport of ions mediated by major K+, anion, and Ca2+ channels and co‐transporters (Chen et al., 2012; Hills et al., 2012; Wang et al., 2017). Guard cell apoplastic pH changes occurred within 30 s of blue‐light treatment, suggesting the rapid activation of H+ pumping in the Polypodiales fern N. exaltata under blue light (Fig. S4a). Blue‐light‐activated stomatal opening was significantly reduced by H+ pump inhibitors carbonyl cyanide m‐chlorophenylhydrazine (CCCP) and sodium orthovanadate (NaVO4), but it was less affected by the photosynthesis inhibitor 3‐(3,4‐dichlorophenyl)‐1,1‐dimethylurea (DCMU) (Fig. S4b). Blue‐light‐induced stomatal opening in N. exaltata also showed significant inhibition by K+ channel blocker tetraethylammonium (TEA), anion channel blocker niflumic acid (NFA) and Ca2+ channel blocker lanthanum chloride (LaCl3) (Fig. S4b). Thus, rapid change of guard cell apoplastic pH in response to blue light and inhibition of blue‐light‐induced stomatal opening by different inhibitors indicate the capacity for fast turnover of cellular signal transduction, supporting rapid blue‐light response in Polypodiales ferns.
Guard cell chloroplasts can provide ATP to fuel blue light‐dependent stomatal opening (Schwartz & Zeiger, 1984; Shimazaki & Zeiger, 1985; Lawson, 2009; Suetsugu et al., 2014). In this study, guard cells of the leptosporangiate fern P. proliferum contained around 100 chloroplasts, which is significantly higher than we observed in either a lycophyte or an angiosperm (Fig. S5a). The number of chloroplasts appeared to be a genetic feature of stomata not related to the blue‐light stomatal response (Fig. S5b,c). However, the generally high number of chloroplasts in fern guard cells (Figs 2, S5) may augment the rapid stomatal response to blue light via photosynthesis‐dependent stomatal opening.
Reactive oxygen species are essential secondary messengers and signaling molecules in various plant responses to environmental conditions (Moller & Sweetlove, 2010). We found a significant and rapid increase of ROS production which peaked at 20 min in guard cells of N. exaltata. By contrast, guard cells of the eusporangiate fern A. evecta showed only minimal ROS production (Fig. 2c,d). Arabidopsis thaliana showed distinct kinetics of blue‐light‐induced ROS production that increased gradually over 50 min (Fig. 2c,d). The speed and timing of ROS production in guard cells of ferns suggest that rapid production of ROS under blue light in Polypodiales may assist in rapid stomatal opening.
Conservation of the blue‐light signaling pathway in green plants
To elucidate the molecular mechanisms associated with the rapid stomatal response to blue light observed in Polypodiales, we conducted a genetic similarity analysis (Chen et al., 2017; Cai et al., 2017a, 2017b,2017a, 2017b) of key protein families related to the blue‐light signaling pathway (Inoue & Kinoshita, 2017). We analyzed protein sequences from 53 protein families in nine major categories (e.g. PHOT signaling pathway, CRY signaling pathway, starch breakdown‐mediated stomatal opening, HRB1/PP7‐regulated stomatal opening, red/UV‐B photoreceptors, other blue‐light‐related pathways, transporters and ion channels) from 36 green plant species (Fig. 3a; Tables S3–S6). Overall, the presence of 26 of 29 protein families is evolutionarily conserved across all land plant lineages examined, including liverworts, mosses, lycophytes, ferns, gymnosperms and angiosperms when using E value < 10−5 as a selection criterion (Tables S3–S4). The two aquatic ferns examined, Azolla filiculoides and Salvinia cucullata from the order Salviniales, contained all 29 protein families with high similarity to angiosperms (Fig. 3a; Table S4). We then selected key species representing major clades of plant evolution to compare the average size of each of the protein families involved in blue‐light signaling. We examined protein families of nine categories, including photoreceptors (CRYs, PHOTs, ZTLs, UVRs and PHYs; Fig. 2b; Table S3–S6), and found a similar number of PHOTs, ZTLs, UVR8 and PHYs across all clades but substantially more CRYs in Polypodiales (Figs 3b, 4, 5).
Fig. 3.
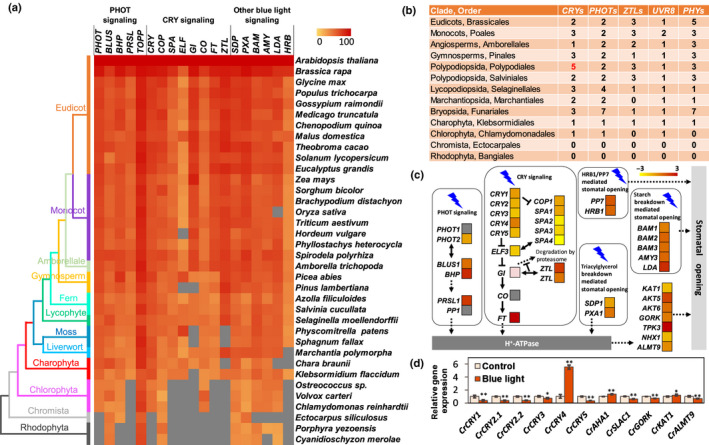
Comparative genomics and transcriptome analysis of the blue‐light signaling pathway. (a) Similarity heat map of blue‐light‐responsive protein families in different plant and algal species. Colored squares indicate protein sequence similarity from zero (yellow) to 100% (red); gray indicates no match of sequences in this species. (b) Number of photoreceptors in the major land plant and algal lineages. Arabidopsis thaliana, Oryza sativa, Amborella trichopoda, Tsuga heterophylla, Adiantum capillus‐veneris, Azolla filiculoides, Selaginella moellendorffii, Marchantia paleacea, Physcomitrella patens, Klebsormidium flaccidum, Chlamydomonas reinhardtii, Ectocarpus siliculosus and Porphyra yezoensis were representative species in each order. The number of cryptochromes (CRYs) in Adiantum capillus‐veneri is marked in red. (c) Transcriptome of differentially expressed genes (DEGs) of epidermal layers of the fern Nephrolepis exaltata in the key blue‐light signaling pathways (based on Arabidopsis) in response to 100 μmol m−2 s−1 blue light. (d) Gene expression of cryptochromes (CRYs) in response to blue light in the fern Ceratopteris richardii. Data are the average of three biological replicates and three technical replicates, and the error bars represent SD. *, P < 0.05; **, P < 0.01.
Fig. 4.
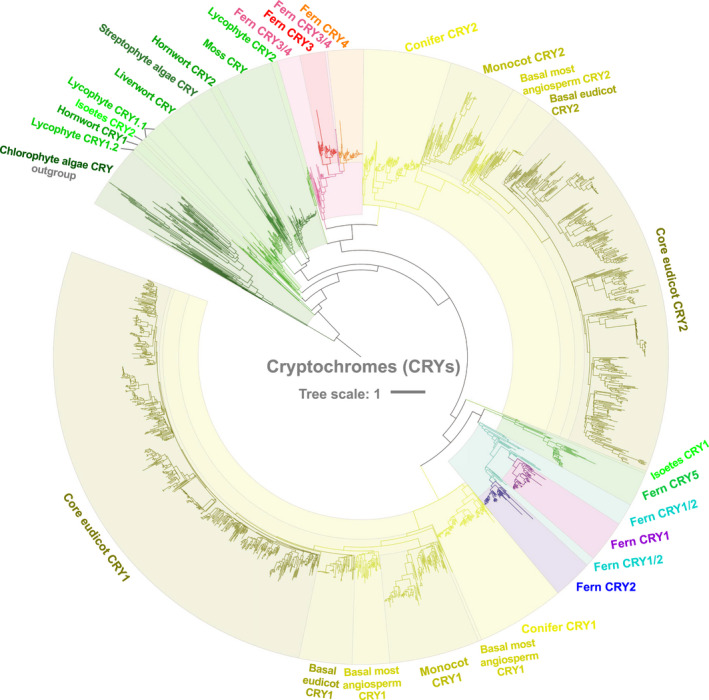
Phylogeny of cryptochromes (CRYs) reconstructed from 2276 protein sequences of green plants. These protein translations were obtained from the OneKP database (2167 sequences) and a fern transcriptome dataset (109 sequences). Detailed information on the phylogeny of CRYs in green plants is given in Supporting Information Fig. S7.
Fig. 5.
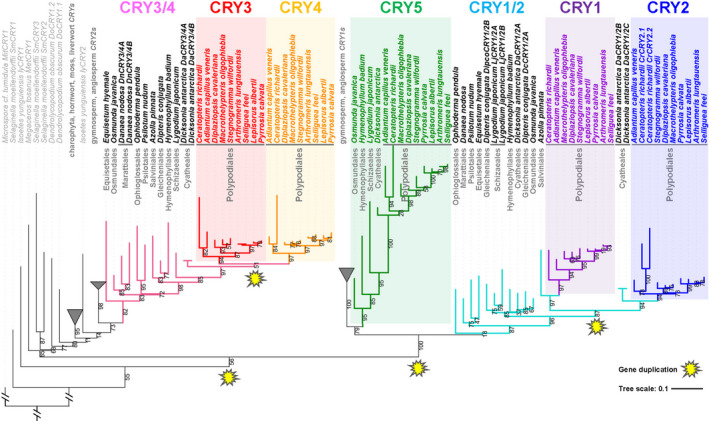
Duplication of cryptochrome (CRYs) in green plants. Four gene duplication events of CRYs are marked in yellow at the nodes of duplication. The bootstrap values (%) were displayed as branch labels. Microspora cf. tumidula (Chlorophyta) MitCRY1 was designated as the root of the phylogenetic tree.
To further explore the molecular mechanisms underlying the observed rapid stomatal response to blue light in ferns (Fig. 2a,b), we conducted RNA sequencing in N. exaltata and quantitative PCR (qPCR) experiments in N. exaltata, A. evecta, C. richardii and A. thaliana under 1 h of 100 μmol m−2 s−1 blue‐light treatment. RNA sequencing of frond epidermal tissue showed significant numbers of DEGs in response to blue light. Positive regulator genes (such as FT, GI and ZTL) and negative regulator genes (such as COP1, SPAs and ELF) of the CRY signaling pathway were significantly upregulated or downregulated, respectively (Fig. 3c; Table S7). In qPCR experiments, the fern species N. exaltata and A. evecta showed distinct blue light‐induced gene expression patterns with more upregulation in DEGs encoding blue‐light signaling components, photoreceptors and membrane transporters as compared with Arabidopsis (Fig. S6). Most interestingly, blue light‐induced expression of all six CrCRYs of C. richardii (Fig. 3d) was similar to those in the transcriptome of leaf epidermis of N. exaltata (Fig. 3c). Blue‐light upregulation of a unique CrCRY4 in C. richardii of Polypodiales was particularly strong, with 5.5‐fold higher upregulation (Fig. 3d). The blue‐light‐induced expression patterns of these genes in ferns were comparable to blue‐light‐induced gene expression in key green plant species such as A. thaliana, Oryza sativa, Physcomitrella patens, Chlamydomonas reinhardtii and Phaeodactylum tricornutum, indicating a conserved evolution of these genes in blue‐light signaling (Table S7).
Cryptochrome duplication is linked to adaptation of Polypodiales ferns in the understory
Cryptochromes emerged as the only blue‐light receptor family that showed significant difference in numbers between the Polypodiales and other green plant species (Fig. 3b–d). We then conducted phylogenetic and evolutionary analyses of CRYs to determine when this increase in gene copy numbers occurred (Figs 4, 5, S7–S10). A total of 2167 protein translations of CRYs from the OneKP transcriptome database and 109 protein translations from fern transcriptome datasets were used to reconstruct the phylogeny of CRYs in green plants (Figs 4, S7a–h). Importantly, CRYs in Chlorophyta and Charophyta form two independent clades (Figs 4, S7), which may indicate a potential evolutionary split between these two groups early in green plant evolution. The CRYs in liverworts and mosses were derived from within streptophytes, and those in ferns exhibited strong diversity and multiple duplications (Figs 4, 5, S7–S9).
There are two copies of CRYs in the three major lineages of ferns that are successively sister to the leptosporangiates (e.g. Ophioglossales, Psilotales and Equisetales), as well as in some lycophytes, mosses, liverworts, streptophyte algae and chlorophyte algae (Figs 3b, 5). The number of CRYs in the leptosporangiate fern lineages Osmundales, Schizaeales, Gleicheniales and Hymenophyllales increased to three following a gene‐duplication event (Figs 5, 6). Significantly, the number of CRYs increased from two to six in the leptosporangiate fern clades Cyatheales and Polypodiales (Figs 4, 5, 6), and these duplications are shared, reflecting a close relationship between Polypodiales and Cyatheales, the first and second largest orders of ferns, respectively (Fig. 1a). Our molecular dating estimates suggest that the first duplication of CRYs occurred around 386–418 Ma in the Devonian Period (Fig. 6; Tables S8–S9), based on evidence that CRY1/2/5 and CRY3/4 first originated in Isoetales (Figs 4, 5, 6, S8). From our molecular dating studies, the second duplication event of CRYs was estimated to have occurred between 373.9 and 404.9 Ma, leading to the rise of the unique CRY5 in the ancestor of Polypodiales, Osmundales and Hymenophyllales (Figs 4, 5, 6, S8). Most importantly, two duplication events for CRYs appear to have occurred at 196.9–212.9 Ma and 151.8–164.4 Ma, when CRY1, CRY2, CRY3 and CRY4 were duplicated mainly in the Polypodiales, possibly contributing to the successful adaptation and explosive radiation of ferns in the late Mesozoic and Cenozoic era. In comparison to the two duplication events of CRYs in Polypodiales, there was little change in the number of photoreceptors in other protein families (PHOTs, ZTLs, UVR8 and PHYs) during the evolution of green plants (Tables S3–S4).
Fig. 6.
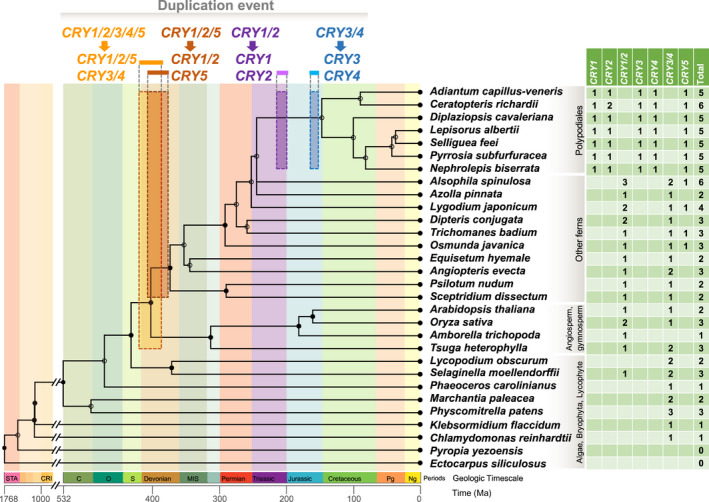
The calibrated time of duplication of cryptochromes (CRYs) for the evolution and diversification of ferns. The phylogenetic tree and the time of species diversification were generated by TimeTree (http://www.timetree.org/; for more detail see Supporting Information Tables S8, S9). Four duplication events are labeled with orange, brown, purple and cyan boxes. The width of the boxes represents the estimated time‐frame for gene duplication, and the length of the boxes represents the coverage of lineages. The estimated times of gene duplication were 386–418 Ma (orange), 373.9–404.9 Ma (brown), 196.9–212.9 Ma (purple) and 151.8–164.4 Ma (cyan). Geological timescale abbreviations: Ng, Neogene; Pg, Paleogene; MIS, Mississippian; S, Silurian; O, Ordovician; C, Cambrian; CRI, Cryogenian; STA, Statherian.
Discussion
Rapid stomatal blue‐light responses confer ecological advantages for leptosporangiates
We found widespread rapid stomatal sensitivity to blue light in leptosporangiate ferns, together with significantly slower stomatal sensitivity in eusporangiate ferns (Figs 1, 2). Until now, leptosporangiate fern stomata were considered generally unresponsive to blue light (Doi et al., 2015, 2006; Doi & Shimazaki, 2008) except Regnellidium diphyllum and Marsilea minuta (Marsileaceae, Salviniales) (Westbrook & McAdam, 2020). Marsileaceae have larger blue‐light stomatal responses and higher photosynthetic capacities than other leptosporangiate ferns, possibly as a result of their amphibious lifestyles and having many anatomical features similar to angiosperms (Westbrook & McAdam, 2020). However, the conclusions from previous studies (Doi et al., 2006, 2015; Doi & Shimazaki, 2008) were based on applying low (c. 5 μmol m−2 s−1) blue light with a background of high‐intensity red light. Following a similar protocol but using high‐intensity blue light (100 μmol m−2 s−1), our results showed no additive effect of blue light on stomatal apertures that were already open under red light (Fig. S3). The blue‐light response in leptosporangiate ferns therefore appears to be an adaptation to conditions where incident light falling on leaves is highly depleted in red wavelength intensities relative to blue, as in the forest understory (Endler, 1993).
The advantages of a rapid blue‐light stomatal response are two‐fold. First, gas‐exchange data for the leptosporangiate fern N. exaltata shows that, for the same intensity, blue light alone triggers a greater increase in stomatal conductance than red light alone (Figs 2, S2–S3). Second, rapid stomatal response is an advantage in conditions where plants rely on utilization of brief periods of localized leaf illumination (sun flecks), which typifies the light environment beneath dense forest canopies (Way & Pearcy, 2012). It is notable that the rate of stomatal opening observed in leptosporangiate ferns in response to blue light (Fig. 2b) is comparable to that seen in modern grasses, which have an exceptionally rapid stomatal response to red plus blue light (Franks & Farquhar, 2007). Our results are also consistent with a recent report comparing stomatal effects on light induction of photosynthesis in 15 species of ferns, gymnosperms and angiosperms. It was found that species with fast stomatal opening, such as ferns, forgo less photosynthesis during photosynthetic induction. Shade‐adapted species possess stomata that are fast‐opening but slow‐closing, indicating ecological adaptation to maximize light fleck use (Deans et al., 2019).
The association of stomatal responses of ferns with different stimuli suggests coordination of hydraulic and photosynthetic signaling networks modulating fern stomatal responses. In a global comparison (Wright et al., 2004), ferns presented significantly lower photosynthetic capacities and intrinsic water‐use efficiency than any functional group within seed plants for similar leaf mass per area and nitrogen contents. For instance, three fern species had values for Rubisco specificity factor (SC/O) similar to those typical of seed plants, but values of A, gs, mesophyll conductance (g m) and maximum velocity of carboxylation (V c,max) were within the lowest range of those observed in seed plants (Gago et al., 2013). However, fern species showed a wider range of stomatal responses than previously reported for angiosperms (Figs 1, 2). The striking variation in sensitivity of fern stomata to different environmental stimuli and in the magnitude of their responses across species and habitats demonstrates the capacity of ferns for diverse optimization of hydraulic and metabolic needs at both growth and evolutionary timescales.
Many plant species, and most ferns, cannot outcompete tall trees for access to light and have developed strategies for shade tolerance to cope with dim light and to optimize light capture (Gommers et al., 2013). The Cretaceous decline in diversity and range of many eusporangiate fern species followed by the diversification of leptosporangiate ferns points to a fundamental shift in functional traits that distinguishes leptosporangiate from other fern lineages. Fern species that dominated in the early Mesozoic may have lacked the adaptive capability and sensitivity to grow in habitats shaded by dense forest canopies (Augustynowicz & Gabrys, 1999; Watkins & Cardelús, 2012). Evolution of enhanced stomatal light response mechanisms, linked to the duplication of CRYs in ferns, may have been a key factor that triggered the diversification of leptosporangiate ferns, particularly Polypodiales, during the competitive displacement of older fern lineages.
Multiple duplications of CRYs contribute to fern radiation in the understory
We have discovered that Polypodiales acquired four more copies of blue‐light photoreceptor genes in the CRY family for a total of six, tripling the number of CRYs in this clade relative to other fern lineages. This increase in molecular machinery for driving the stomatal response to blue light is consistent with the heightened sensitivity of Polypodiales fern stomata to blue light (Figs 1, 2, 3, 4, 5). Therefore, it is likely that Polypodiales ferns have experienced at least one gene duplication while radiating in the subcanopy of angiosperm forests (Fig. 6). Gene duplication is generally viewed as the source of new material for the evolution of new gene functions (Lynch & Conery, 2000; Ohno, 2013). Analysis of P. patens cry1a and cry1b loss‐of‐function mutants demonstrated that PpCRY1a and PpCRY1b mediate blue‐light responses in a redundant manner (Imaizumi et al., 2002; Lin, 2002). The CRY family appears to have evolved via an initial duplication on the fern stem lineage, producing the ancestral CRY1/2 and CRY3/4 paralogs. CRY5 originated from duplication of the CRY1/2 paralog. Two additional duplications produced CRY1 and CRY2 on the stem branch of Cyatheales and Polypodiales, and CRY3 and CRY4 on the stem branch of Polypodiales, after the divergence from Cyatheales. Both Cyatheales and Polypodiales are within the leptosporangiates, comprising the majority of extant fern species (Imaizumi et al., 2002; Lin, 2002). The number of CRYs is fairly consistent across chlorophyte and streptophyte algae as well as land plants, with the exception of ferns (Figs 3, 4, 5). Polypodiales acquired as many as six copies of CRYs after these four gene duplications, including two unique duplications that we estimate occurred in the Mesozoic era (151.8–212.9 Ma) (Figs 5, 6). In Asplenium yunnanense (a member of Polypodiales), the sequence, structure and phylogenetic analysis of AyCRYs indicated that these proteins possess the typical photolyase homology region and C‐terminal region of CRY (CCT) domain characteristics and comprised two distinct groups that are separate from other plants (Imaizumi et al., 2000; Lin, 2002). Thus, the large number and remarkable differences in protein structure of CRYs in ferns may have promoted the evolution of novel functions to improve the utilization of light in shady habitats.
In some green plants, NEO1/PHY3 contains a PHY sensory module and a PHOT domain (Nozue et al., 1998; Christie, 2007; Yang et al., 2017), and NEOs have originated twice in green plant evolution (in Zygnematalean algae and hornworts). Hornwort NEOs were subsequently transferred horizontally to some ferns, conferring a significantly higher light sensitivity on that lineage and its descendants (Li et al., 2014). Molecular dating has placed the divergence time between fern and hornwort NEOs at around 178 Ma (Li et al., 2014), which fits between the two duplication events calculated here for CRYs during the evolution of Polypodiales ferns, at 196.9–212.9 Ma and 151.8–164.4 Ma (Fig. 6). Therefore, NEOs probably also played a critical role in facilitating the diversification of ferns under angiosperm‐dominated canopies (Schneider et al., 2004; Kanegae et al., 2006; Schuettpelz & Pryer, 2009). However, NEOs have so far only been found in a few species (Suetsugu et al., 2005; Yang et al., 2017), which may limit their potential role for adaptation of modern ferns to low light. By contrast, the multiple duplication of CRYs and the significant correlation between number of CRYs and number of species in each of the 11 fern orders (Fig. S11) may represent the key mechanistic innovation underlying the adaptation and radiation of Polypodiales in low, blue‐enhanced light.
Conclusion
The duplication of CRYs around 200 Ma may have been a critical event leading to evolution of enhanced stomatal responses to blue light in Polypodiales ferns, fueling their diversification as angiosperms came to dominate global forest canopies. Our new findings are consistent with an evolutionary model in which rapid stomatal response to blue light and the duplication of CRYs conferred an ecological advantage on Polypodiales ferns, assisting their exploitation of the forest understory habitat as they diversified through the Cretaceous and Cenozoic geological time periods.
Author contributions
Z‐HC, SC, FW and PJF designed and supervised this study. SC, YH, FC and CZ performed the experiments. SC, YH, XZ and GC performed the data analysis. JHL‐M contributed to the OneKP database. DBM, DES and PSS guided the analysis of phylogeny. MRB guided the stomatal physiology work. Z‐HC, SC, PJF and ES wrote the manuscript with inputs from GZ, SS, JMC, EN, FD and DX.
Supporting information
Fig. S1 UV‐A light induces stomatal opening in different fern species.
Fig. S2 Stomatal conductance and photosynthetic rate respond to increasing intensity of blue light and red light in Nephrolepis exaltata.
Fig. S3 Stomatal conductance and photosynthetic rate in response to red light followed by blue light in ferns and Arabidopsis thaliana.
Fig. S4 Blue‐light ‐induced stomatal opening, H+ pumping from guard cells, and the effect of inhibitors in the polypod fern Nephrolepis exaltata.
Fig. S5 Effects of number of chloroplasts on blue‐light‐induced stomatal opening in ferns.
Fig. S6 Heat map of differentially expressed genes (DEGs) in the blue‐light signaling pathways in the ferns Nephrolepis exaltata and Angiopteris evecta compared with the angiosperm Arabidopsis thaliana.
Fig. S7 The phylogeny of cryptochromes (CRYs) in green algae, bryophytes, lycophytes, ferns and gymnosperms and angiosperms.
Fig. S8 Phylogeny and duplication of cryptochromes (CRYs) in green algae, bryophytes, lycophytes, ferns and gymnosperms and angiosperms.
Fig. S9 Phylogenetic tree of cryptochromes (CRYs) using iq‐tree software.
Fig. S10 Alignment and functional domain analysis of cryptochromes (CRYs) in Ceratopteris richardii and Arabidopsis thaliana.
Fig. S11 Correlation analysis between number of CRYs and number of species in each of the orders of ferns.
Table S1 Information of the fern species and their habitats.
Table S2 Details of sequences used in the phylogenetic trees.
Table S3 Number of predicted proteins in 36 species with E‐value < 10−5.
Table S4 Similarity for the evolution of proteins involved in blue‐light‐induced stomatal opening in 36 species with E‐value < 10−5.
Table S5 Information on blue‐light‐related genes for evolutionary bioinformatic analysis.
Table S6 Information on blue‐light‐related gene families for evolutionary bioinformatic analysis.
Table S7 Comparative transcriptome analysis of different plant species exposed to blue light (data are from publicly available datasets).
Table S8 The estimated time of duplication of cryptochrome blue light receptors (CRYs) in diverse ferns, isoetales, conifer and angiosperms.
Table S9 The estimated time of node in TimeTree.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank Dr Anya Salih (WSU Confocal Bio‐imaging Facility), Linda Westmoreland, Renee Smith, Jennie Nelson, Elaine Chua and David Randall for their technical support. This work is funded by the Australian Research Council (DE1401011143, DP150104007, DP170100460) and National Natural Science Foundation of China (31571578, 31620103912, 31771687). The authors declare there are no competing interests.
See also the Commentary on this article by Chater, 230: 886–888.
Contributor Information
Peter J. Franks, Email: peter.franks@sydney.edu.au.
Feibo Wu, Email: wufeibo@zju.edu.cn.
Zhong‐Hua Chen, Email: z.chen@westernsydney.edu.au.
Data availability
The original data files of the transcriptome are available on NCBI BioProject (PRJNA563243). The GenBank IDs for the cryptochromes are CrCRY1 (MN403054), CrCRY2.1 (MN403055), CrCRY2.2 (MN403056), CrCRY3 (MN403057), CrCRY4 (MN403058) and CrCRY5 (MN403059).
References
- Augustynowicz J, Gabrys H. 1999. Chloroplast movements in fern leaves: correlation of movement dynamics and environmental flexibility of the species. Plant, Cell & Environment 22: 1239–1248. [Google Scholar]
- Brodribb T, McAdam SA. 2011. Passive origins of stomatal control in vascular plants. Science 331: 582–585. [DOI] [PubMed] [Google Scholar]
- Brodribb TJ, McAdam SAM, Jordan GJ, Feild TS. 2009. Evolution of stomatal responsiveness to CO2 and optimisation of water‐use efficiency among land plants. New Phytologist 183: 839–847. [DOI] [PubMed] [Google Scholar]
- Cai S, Chen G, Wang Y, Huang Y, Marchant DB, Wang Y, Yang Q, Dai F, Hills A, Franks PJ et al. 2017a. Evolutionary conservation of ABA signaling for stomatal closure. Plant Physiology 174: 732–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Papanatsiou M, Blatt MR, Chen ZH. 2017b. Speedy grass stomata: Emerging molecular and evolutionary features. Molecular Plant 10: 912–914. [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annual Review of Plant Biology 62: 335–364. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang Y, Wang X, Yang Q, Quan X, Zeng J, Dai F, Zeng F, Wu F, Zhang G et al. 2019. Leaf epidermis transcriptome reveals drought‐induced hormonal signaling for stomatal regulation in wild barley. Plant Growth Regulation 87: 39–54. [Google Scholar]
- Chen ZH, Chen G, Dai F, Wang Y, Hills A, Ruan YL, Zhang G, Franks PJ, Nevo E, Blatt MR. 2017. Molecular evolution of grass stomata. Trends in Plant Science 22: 124–139. [DOI] [PubMed] [Google Scholar]
- Chen ZH, Hills A, Bätz U, Amtmann A, Lew VL, Blatt MR. 2012. Systems dynamic modeling of the stomatal guard cell predicts emergent behaviors in transport, signaling, and volume control. Plant Physiology 159: 1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Wang Y, Wang JW, Babla M, Zhao C, García‐Mata C, Sani E, Differ C, Mak M, Hills A et al. 2016. Nitrate reductase mutation alters potassium nutrition as well as nitric oxide‐mediated control of guard cell ion channels in Arabidopsis. New Phytologist 209: 1456–1469. [DOI] [PubMed] [Google Scholar]
- Christenhusz MJM, Zhang XC, Schneider H. 2011. A linear sequence of extant families and genera of lycophytes and ferns. Phytotaxa 19: 5–22. [Google Scholar]
- Christie JM. 2007. Phototropin blue‐light receptors. Annual Review of Plant Biology 58: 21–45. [DOI] [PubMed] [Google Scholar]
- Creese C, Oberbauer S, Rundel P, Sack L. 2014. Are fern stomatal responses to different stimuli coordinated? Testing responses to light, vapor pressure deficit, and CO2 for diverse species grown under contrasting irradiances. New Phytologist 204: 92–104. [DOI] [PubMed] [Google Scholar]
- Deans RM, Brodribb TJ, Busch FA, Farquhar GD. 2019. Plant water‐use strategy mediates stomatal effects on the light induction of photosynthesis. New Phytologist 222: 382–395. [DOI] [PubMed] [Google Scholar]
- Doi M, Kitagawa Y, Shimazaki K. 2015. Stomatal blue light response is present in early vascular plants. Plant Physiology 169: 1205–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Shimazaki K. 2008. The stomata of the fern Adiantum capillus‐veneris do not respond to CO2 in the dark and open by photosynthesis in guard cells. Plant Physiology 147: 922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Wada M, Shimazaki K. 2006. The fern Adiantum capillus‐veneris lacks stomatal responses to blue light. Plant Cell and Physiology 47: 748–755. [DOI] [PubMed] [Google Scholar]
- Endler JA. 1993. The color of light in forests and its implications. Ecological Monogragphy 63: 1–27. [Google Scholar]
- Fiorucci AS, Fankhauser C. 2017. Plant strategies for enhancing access to sunlight. Current Biology 27: R931–R940. [DOI] [PubMed] [Google Scholar]
- Franks PJ, Britton‐Harper ZJ. 2016. No evidence of general CO2 insensitivity in ferns: one stomatal control mechanism for all land plants? New Phytologist 211: 819–827. [DOI] [PubMed] [Google Scholar]
- Franks PJ, Farquhar GD. 2007. The mechanical diversity of stomata and its significance in gas‐exchange control. Plant Physiology 143: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago J, Coopman RE, Cabrera HM, Hermida C, Molins A, Conesa MÀ, Galmés J, Ribas‐Carbó M, Flexas J. 2013. Photosynthesis limitations in three fern species. Physiologia Plantarum 149: 599–611. [DOI] [PubMed] [Google Scholar]
- Gitzendanner MA, Soltis PS, Wong GK, Ruhfel BR, Soltis DE. 2018. Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. American Journal of Botany 105: 291–301. [DOI] [PubMed] [Google Scholar]
- Gommers CM, Visser EJ, St Onge KR, Voesenek LA, Pierik R. 2013. Shade tolerance: when growing tall is not an option. Trends in Plant Science 18: 65–71. [DOI] [PubMed] [Google Scholar]
- Hills A, Chen ZH, Amtmann A, Blatt MR, Lew VL. 2012. OnGuard, a computational platform for quantitative kinetic modeling of guard cell physiology. Plant Physiology 159: 1026–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak H, Kollist H, Merilo E. 2017. Fern stomatal responses to ABA and CO2 depend on species and growth conditions. Plant Physiology 174: 672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino M, Ogawa T, Zeiger E. 1985. Kinetic‐properties of the blue‐light response of stomata. Proceedings of the National Academy of Sciences, USA 82: 8019–8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kadota A, Hasebe M, Wada M. 2002. Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens . Plant Cell 14: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kanegae T, Wada M. 2000. Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus‐veneris . Plant Cell 12: 81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue SI, Kinoshita T. 2017. Blue light regulation of stomatal opening and the plasma membrane H+‐ATPase. Plant Physiology 174: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanegae T, Hayashida E, Kuramoto C, Wada M. 2006. A single chromoprotein with triple chromophores acts as both a phytochrome and a phototropin. Proceedings of the National Academy of Sciences, USA 103: 17997–18001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Kanegae T, Christensen S, Kiyosue T, Sato Y, Imaizumi T, Kadota A, Wada M. 2003. Responses of ferns to red light are mediated by an unconventional photoreceptor. Nature 421: 287–290. [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. 1997. The origin and early evolution of plants on land. Nature 389: 33–39. [Google Scholar]
- Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K. 2001. phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki KI. 1999. Blue light activates the plasma membrane H+‐ATPase by phosphorylation of the C‐terminus in stomatal guard cells. EMBO Journal 18: 5548–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson T. 2009. Guard cell photosynthesis and stomatal function. New Phytologist 181: 13–34. [DOI] [PubMed] [Google Scholar]
- Li FW, Brouwer P, Carretero‐Paulet L, Cheng S, de Vries J, Delaux PM, Eily A, Koppers N, Kuo LY, Li Z et al. 2018. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nature Plants 4: 460–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, Graham SW, Wong GK, Pryer KM, Mathews S. 2015. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nature Communications 6: 7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FW, Villarreal JC, Kelly S, Rothfels CJ, Melkonian M, Frangedakis E, Ruhsam M, Sigel EM, Der JP, Pittermann J et al. 2014. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proceedings of the National Academy of Sciences USA 111: 6672–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CT. 2002. Blue light receptors and signal transduction. Plant Cell 14: S207–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind C, Dreyer I, López‐Sanjurjo EJ, von Meyer K, Ishizaki K, Kohchi T, Lang D, Zhao Y, Kreuzer I, Al‐Rasheid KA et al. 2015. Stomatal guard cells co‐opted an ancient ABA‐dependent desiccation survival system to regulate stomatal closure. Current Biology 25: 928–935. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ. 2005. From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proceedings of the National Academy of Sciences, USA 102: 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S. 2006. Phytochrome‐mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Molecular Ecology 15: 3483–3503. [DOI] [PubMed] [Google Scholar]
- Moglich A, Yang X, Ayers RA, Moffat K. 2010. Structure and function of plant photoreceptors. Annual Review of Plant Biology 61: 21–47. [DOI] [PubMed] [Google Scholar]
- Moller IM, Sweetlove LJ. 2010. ROS signalling–specificity is required. Trends in Plant Science 15: 370–374. [DOI] [PubMed] [Google Scholar]
- Mott KA, Sibbernsen ED, Shope JC. 2008. The role of the mesophyll in stomatal responses to light and CO2 . Plant, Cell & Environment 31: 1299–1306. [DOI] [PubMed] [Google Scholar]
- Nozue K, Kanegae T, Imaizumi T, Fukuda S, Okamoto H, Yeh KC, Lagarias JC, Wada M. 1998. A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proceedings of the National Academy of Sciences, USA 95: 15826–15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 2013. Evolution by gene duplication. Berlin, Germany: Springer Science & Business Media. [Google Scholar]
- Possart A, Hiltbrunner A. 2013. An evolutionarily conserved signaling mechanism mediates far‐red light responses in land plants. Plant Cell 25: 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszala EM, Beerling DJ, Franks PJ, Chater C, Casson SA, Gray JE, Hetherington AM. 2011. Land plants acquired active stomatal control early in their evolutionary history. Current Biology 21: 1030–1035. [DOI] [PubMed] [Google Scholar]
- Schneider H, Schuettpelz E, Pryer KM, Cranfill R, Magallón S, Lupia R. 2004. Ferns diversified in the shadow of angiosperms. Nature 428: 553–557. [DOI] [PubMed] [Google Scholar]
- Schuettpelz E, Pryer KM. 2009. Evidence for a Cenozoic radiation of ferns in an angiosperm‐dominated canopy. Proceedings of the National Academy of Sciences, USA 106: 11200–11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz A, Zeiger E. 1984. Metabolic energy for stomatal opening. Role of photophosphorylation and oxidative phosphorylation. Planta 161: 129–136. [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Doi M, Assmann SM, Kinoshita T. 2007. Light regulation of stomatal movement. Annual Review of Plant Biology 58: 219–247. [DOI] [PubMed] [Google Scholar]
- Shimazaki K, Iino M, Zeiger E. 1986. Blue light‐dependent proton extrusion by guard‐cell protoplasts of Vicia faba . Nature 319: 324–326. [Google Scholar]
- Shimazaki K‐I, Zeiger E. 1985. Cyclic and noncyclic phosphorylation in isolated guard cell protoplasts from Vicia faba L. Plant Physiology 78: 211–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG. 2006. A classification for extant ferns. Taxon 55: 705–731. [Google Scholar]
- Soltis PS, Soltis DE, Chase MW. 1999. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402: 402–404. [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Mittmann F, Wagner G, Hughes J, Wada M. 2005. A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proceedings of the National Academy of Sciences, USA 102: 13705–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N, Takami T, Ebisu Y, Watanabe H, Iiboshi C, Doi M, Shimazaki K. 2014. Guard cell chloroplasts are essential for blue light‐dependent stomatal opening in Arabidopsis. PLoS ONE 9: e108374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Pteridophyte Phylogeny Group . 2016. A community‐derived classification for extant lycophytes and ferns. Journal of Systematics and Evolution 54: 563–603. [Google Scholar]
- Wang Y, Hills A, Vialet‐Chabrand S, Papanatsiou M, Griffiths H, Rogers S, Lawson T, Lew VL, Blatt MR. 2017. Unexpected connections between humidity and ion transport discovered using a model to bridge guard cell‐to‐leaf scales. Plant Cell 29: 2921–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JE Jr, Cardelús CL. 2012. Ferns in an angiosperm world: Cretaceous radiation into the epiphytic niche and diversification on the forest floor. International Journal of Plant Sciences 173: 695–710. [Google Scholar]
- Watkins JE Jr, Mack MC, Sinclair TR, Mulkey SS. 2007a. Ecological and evolutionary consequences of desiccation tolerance in tropical fern gametophytes. New Phytologist 176: 708–717. [DOI] [PubMed] [Google Scholar]
- Watkins JE Jr, Rundel PW, Cardelús CL. 2007b. The influence of life form on carbon and nitrogen relationships in tropical rainforest ferns. Oecologia 153: 225–232. [DOI] [PubMed] [Google Scholar]
- Way DA, Pearcy RW. 2012. Sunflecks in trees and forests: from photosynthetic physiology to global change biology. Tree Physiology 32: 1066–1081. [DOI] [PubMed] [Google Scholar]
- Westbrook AS, McAdam SA. 2020. Atavistic stomatal responses to blue light in Marsileaceae. Plant Physiology 184: 1378–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, Mirarab S, Nguyen N, Warnow T, Carpenter E, Matasci N, Ayyampalayam S, Barker MS, Burleigh JG, Gitzendanner MA et al. 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proceedings of the National Academy of Sciences, USA 111: E4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmer CM, Fricker M. 1996. Stomata, 2nd edn. London, UK: Chapman & Hall. [Google Scholar]
- Wright IJ, Reich PB, Westoby M, Ackerly DD, Baruch Z, Bongers F, Cavender‐Bares J, Chapin T, Cornelissen JH, Diemer M et al. 2004. The world‐wide leaf economics spectrum. Nature 428: 821–827. [DOI] [PubMed] [Google Scholar]
- Yang Z, Liu B, Su J, Liao J, Lin C, Oka Y. 2017. Cryptochromes orchestrate transcription regulation of diverse blue light responses in plants. Photochemistry and Photobiology 93: 112–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Wang Y, Chan KX, Marchant DB, Franks PJ, Randall D, Tee EE, Chen G, Ramesh S, Phua SY et al. 2019. Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land. Proceedings of the National Academy of Sciences, USA 116: 5015–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 UV‐A light induces stomatal opening in different fern species.
Fig. S2 Stomatal conductance and photosynthetic rate respond to increasing intensity of blue light and red light in Nephrolepis exaltata.
Fig. S3 Stomatal conductance and photosynthetic rate in response to red light followed by blue light in ferns and Arabidopsis thaliana.
Fig. S4 Blue‐light ‐induced stomatal opening, H+ pumping from guard cells, and the effect of inhibitors in the polypod fern Nephrolepis exaltata.
Fig. S5 Effects of number of chloroplasts on blue‐light‐induced stomatal opening in ferns.
Fig. S6 Heat map of differentially expressed genes (DEGs) in the blue‐light signaling pathways in the ferns Nephrolepis exaltata and Angiopteris evecta compared with the angiosperm Arabidopsis thaliana.
Fig. S7 The phylogeny of cryptochromes (CRYs) in green algae, bryophytes, lycophytes, ferns and gymnosperms and angiosperms.
Fig. S8 Phylogeny and duplication of cryptochromes (CRYs) in green algae, bryophytes, lycophytes, ferns and gymnosperms and angiosperms.
Fig. S9 Phylogenetic tree of cryptochromes (CRYs) using iq‐tree software.
Fig. S10 Alignment and functional domain analysis of cryptochromes (CRYs) in Ceratopteris richardii and Arabidopsis thaliana.
Fig. S11 Correlation analysis between number of CRYs and number of species in each of the orders of ferns.
Table S1 Information of the fern species and their habitats.
Table S2 Details of sequences used in the phylogenetic trees.
Table S3 Number of predicted proteins in 36 species with E‐value < 10−5.
Table S4 Similarity for the evolution of proteins involved in blue‐light‐induced stomatal opening in 36 species with E‐value < 10−5.
Table S5 Information on blue‐light‐related genes for evolutionary bioinformatic analysis.
Table S6 Information on blue‐light‐related gene families for evolutionary bioinformatic analysis.
Table S7 Comparative transcriptome analysis of different plant species exposed to blue light (data are from publicly available datasets).
Table S8 The estimated time of duplication of cryptochrome blue light receptors (CRYs) in diverse ferns, isoetales, conifer and angiosperms.
Table S9 The estimated time of node in TimeTree.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The original data files of the transcriptome are available on NCBI BioProject (PRJNA563243). The GenBank IDs for the cryptochromes are CrCRY1 (MN403054), CrCRY2.1 (MN403055), CrCRY2.2 (MN403056), CrCRY3 (MN403057), CrCRY4 (MN403058) and CrCRY5 (MN403059).


